 Prenylation of aromatic compounds is a key tailoring reaction in biosynthesis of bioactive indole-diterpenes.
Prenylation of aromatic compounds is a key tailoring reaction in biosynthesis of bioactive indole-diterpenes.
Abstract
Prenylation of aromatic compounds is a key tailoring reaction in biosynthesis of bioactive indole-diterpenes. Here, we identify NodD1 as the enzyme responsible for the bisprenylation of nodulisporic acid F. This prenyltransferase showed a preference for its natural indole-diterpene substrate whereas other related enzymes were not able to catalyse this conversion.
Introduction
Indole-diterpenes (IDTs) are a structurally diverse group of fungal-derived natural products with a range of potentially useful bioactive properties including anti-MRSA,1 anti-H1N1,2 and insecticidal3 activities. Several different indole diterpene scaffolds have inhibitory effects on glioblastoma,4 prostate cancer5 or breast cancer6,7 cell lines. Mechanisms of action include Eg5 inhibition8,9 and modulation of the canonical Wnt (β-catenin dependent) signalling pathway. Paspaline and emindole SB are the simplest stable indole diterpenes so far identified and have the structural requirements for anti-cancer bioactivity and both have been identified as interesting compounds for further study. Unlike paspaline which is a precursor of many known indole diterpenes, few emindole SB derived compounds have been identified and there is a need to understand the potential for biosynthetic modification of this compound in order that greater chemical diversity can be generated for further bioactivity studies.
The biosynthetic pathway for IDTs, shown in Fig. S1,† begins with the dedicated biosynthesis of geranylgeranyl pyrophosphate, which is condensed with an indole moiety to produce 3-geranylgeranyl indole. Following epoxidation of the geranylgeranyl tail, specific cyclisation reactions deliver a variety of IDT skeletons. Different patterns of prenylation, hydroxylation, reduction, elimination and halogenation of these scaffolds lead to the diverse repertoire of compounds belonging to this group (Fig. 1).
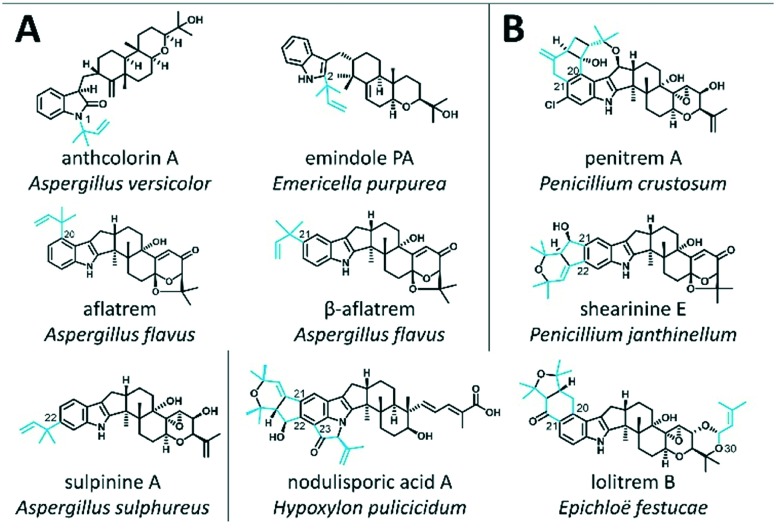
Fig. 1. Examples of known IDTs for which the biosynthesis involves the action of PTs. Compounds are categorised by whether the PTs act as the ultimate biosynthetic step (A) or in conjunction with oxidase enzymes to generate cyclic structures (B). Prenyl groups are highlighted in blue.
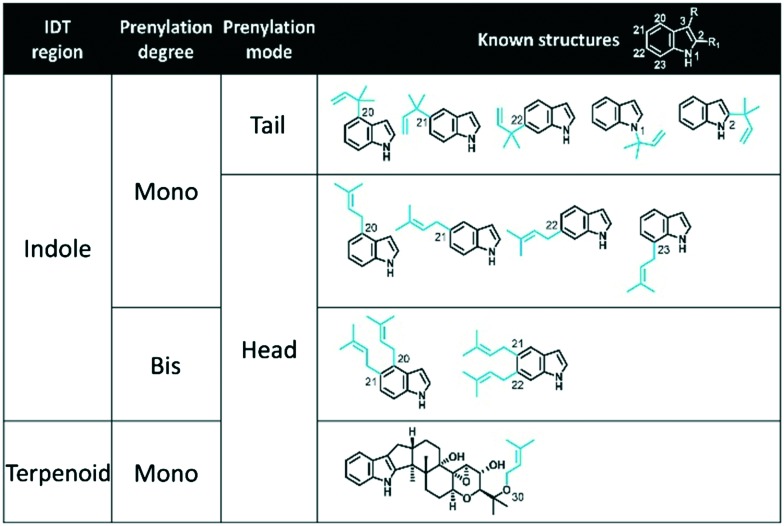
Indole-diterpene prenyl transferases (IDT PTs) encompass one of the most important groups of enzymes involved in IDT biosynthesis, acting as key tailoring enzymes for the generation of chemical diversity that is associated with specific bioactive properties.10 IDT PTs catalyse the prenylation of IDTs either in ultimate biosynthetic steps (e.g. emindole Ps, aflatrems, anthcolorins, sulpinines) as shown in Fig. 1A, or in conjunction with oxidase enzymes to generate cyclic structures (e.g. penitrems, shearinines, nodulisporic acids, and lolitrems) as shown in Fig. 1B. Using DMAPP as the prenyl donor, these ‘decoration’ IDT PTs prenylate the indole component of the IDT core compound in any of the six possible locations using either ‘head’ or ‘tail’ isoprenoid condensations (respectively described as ‘regular’ or ‘reverse’ prenylations) to generate mono-prenylated, bisprenylated or a mixture of mono- and bisprenylated products (Table 1). Although the majority of IDT PTs prenylate at various positions on the indole core, one IDT PT, LtmF,11 prenylates lolitrems and terpendole-type IDTs on the terpenoid component (shown at the bottom of Table 1). Prenylation of non-cyclic carbons by aromatic PTs is unusual but not unprecedented.12
Table 1. Known prenylations involved in IDT biosynthetic pathways. Prenyl groups are highlighted in blue. R and R1 on the top white structure indicate locations where the ‘terpenoid’ component of IDTs attaches to the indole component. Prenylations have been grouped by the region of the IDT they prenylate (i.e. ‘indole’ or ‘terpenoid’ component), degree of prenylation (i.e. mono- or bisprenylation), and prenylation mode (i.e. ‘head’ or ‘tail’ isoprenoid condensation). Carbon numbering is based on the conventional IDT numbering scheme. When comparing IDTs to non-IDTs decorated by aromatic PTs, C1,2,3 = C1,2,3; C20 = C4; C21 = C5; C22 = C6 and C23 = C7.
|
To date, the products of seven genes encoding these tailoring IDT PTs have been functionally analysed in vivo (PaxD from Penicillium paxilli,13 AtmD required for aflatrem biosynthesis in Aspergillus flavus,14 JanD for shearinine biosynthesis in P. janthinellum,15 PtmD and PtmE for penitrem biosynthesis in P. simplicissimum,16 LtmE and LtmF for lolitrem biosysnthesis in Epichloe festucae11), and five have been characterised in vitro (PaxD,15,17,18 AtmD,17 JanD,15 PtmE16 and AmyD18). Notably, all the IDT PTs analysed to date decorate paspaline-derived IDT skeletons (Fig. S1†) when performing their natural biosynthetic functions. Of these, AtmD has shown the highest degree of substrate promiscuity, where, as shown in Fig. S2,† not only can it prenylate the indole ring of uncyclized indole terpenes, including farnesyl indole and the IDT precursor geranylgeranylindole, but it is also observed to alter its mode of prenylation from tail to head depending on the substrate it is presented with (e.g. paspalinine vs. paspaline respectively).17 This remarkable substrate promiscuity may make AtmD a useful template for creating IDT PTs with novel substrate specificities. However, there is still very little known about IDT PTs and more information is required to rationally engineer mutant enzymes of altered specificity to increase IDT diversity.
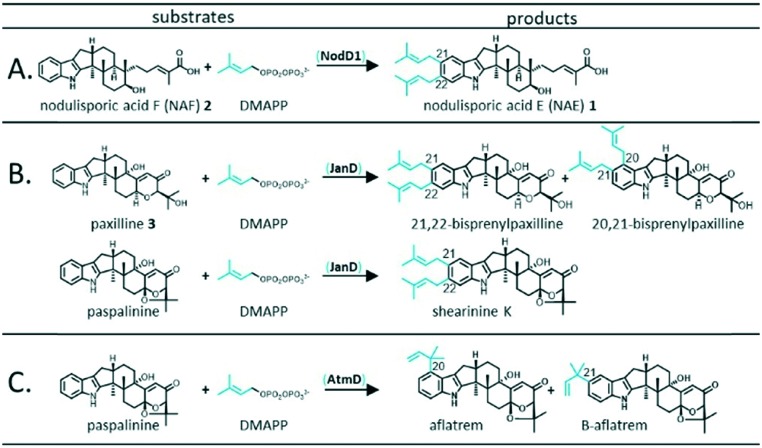
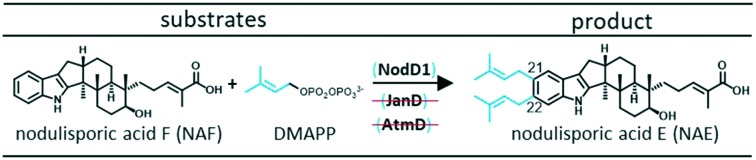
The primary goal of this study was to explore substrate specificity of an IDT PT, NodD1 from Hypoxylon pulicicidum, involved in the biosynthesis of the non-paspaline-derived IDT, nodulisporic acid E (NAE) 1 (Fig. 2A). In line with our overarching goal to better understand the structural features of IDT PTs that determine substrate specificities, we also analysed the abilities of JanD and AtmD, which standardly prenylate paspaline-derived scaffolds (Fig. 2B and C respectively) to accept an alternative non-paspaline-derived IDT, nodulisporic acid F (NAF) 2.
Fig. 2. Standard in vivo prenylation mechanisms observed for NodD1 (A), JanD (B) and AtmD (C). As shown in Fig. S2,† alternative in vitro prenyl transferase activity has been observed for AtmD.17.
Results
Previously, we proposed that one of the two putative PT-encoding genes (nodD1 or nodD2) identified in the nodulisporic acid biosynthetic gene cluster (nod) was likely involved in the bisprenylation of NAF 2 to form NAE 1.19 The slightly higher level of sequence homology seen for NodD1 compared with PaxD and JanD (Table S1†), enzymes that mediate bisprenylation of structurally similar IDTs with similar regioselectivity to NAE 1 biosynthesis (i.e. bisprenylation at C21 and C22 of the indole moiety), suggested that NodD1 was the most likely candidate for this enzymatic step.
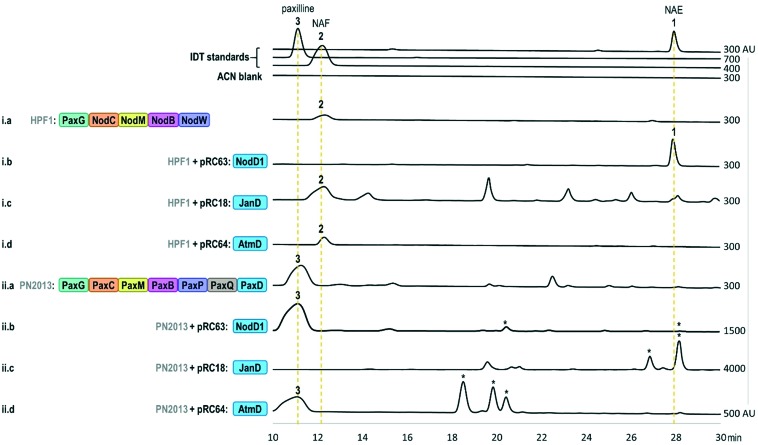
In turn, using our modular cloning system,20 we heterologously expressed the gene encoding NodD1 (nodD1) in a strain of Penicillium paxilli (HPF1) that we previously engineered to produce NAF 2.19 As shown in trace i.b of Fig. 3, LC-MS results revealed a new compound which matched the expected mass of the bisprenylated-NAF derivative, NAE 1 (Fig. S6,† HRMS [M + H]+ 572.4101 m/z), corresponding to a molecular formula of C38H53NO3. Subsequent NMR analysis confirmed the new compound was NAE 1 (Table S2 and Fig. S7–S11†). Notably, the prenylation efficiency of NAF 2 by NodD1 was high, with nearly full conversion of NAF 2 to NAE 1 observed (see absence of NAF 2 in trace i.b of Fig. 3).
Fig. 3. HPLC analysis (230 nm) of fungal extracts. Prenylated products, as confirmed by MS fragmentation patterns, for which we do not have standards, are denoted by an asterisk (*). Note: Some traces contain peaks that have similar retention times to other prenylated IDTs, but MS results shown in Fig. S3–S5,† indicate that they are not prenylated IDT compounds. Strain names (HPF1 or PN2013) are shown in grey and plasmid names (e.g. pRC63) are shown in black with the corresponding names of IDT enzymes encoded by the genes of each strain and plasmid shown within the coloured boxes. The x-axis of each trace has a minimum value of –100 arbitrary units (AU) and the maximum x-axis value is listed on the right side of the trace. Additional information regarding the strains and plasmids can be found in Tables S4, S5 and S11.† .
Next, based on previously observed substrate promiscuity associated with IDT PTs,17 we were curious whether NodD1 would be able to prenylate paxilline 3. To this end, we expressed nodD1 in wildtype P. paxilli strain PN2013, for which paxilline 3 is the major IDT product. LC-MS analysis revealed that at least four novel prenylated IDT compounds (based on MS fragmentation patterns) with [M + H]+ ions of 504.3 m/z, 504.3 m/z, 502.3 m/z and 570.4 m/z were present in the transformant extracts and not in the extracts from the parental P. paxilli strain PN2013 (Fig. S4 and S5† traces ii.b vs. ii.a). In contrast to the high prenylation efficiency observed for NodD1 to prenylate NAF 2, very low titres were observed for the prenylated paxilline compounds hindering further compound characterisation by NMR analysis.
To further understand the substrate specificity/promiscuity of IDT PTs, we explored whether JanD and AtmD could prenylate NAF 2 to produce NAE 1 or any novel compounds. We specifically chose JanD because it has the highest amino acid sequence identity (53.2%, Table S1†) to NodD1 of all known IDT PTs and catalyses the bisprenylation of some paspaline-derived IDTs with the same regioselectivity as NodD1 (Fig. 2B; prenylation at C21 and C22 of the indole moiety).15 In contrast, AtmD mediates monoprenylation at one of three sites (Fig. 2C, C20, C21, and a third unidentified position) in the biosynthesis of the aflatrems, but was chosen because it is the most promiscuous IDT PT characterised to date (Fig. S2†).17 Accordingly, the genes encoding JanD and AtmD (janD and atmD) were individually heterologously expressed in the NAF 2-producing P. paxilli strain (HPF1) and extracts were analysed. As shown in traces i.c and i.d of Fig. 3, no new compounds with masses corresponding to those expected for prenylated-NAF compounds were detectable in the extracts of the janD- or atmD-transformants, indicating that their respective PTs are unable to prenylate NAF 2in vivo. As positive controls, janD and atmD were individually expressed in wildtype P. paxilli strain PN2013 and, in accordance with previous in vitro results from Liu et al. 2013 and 2016,15,17 prenylated-paxilline products were observed in the transformant extracts (shown in Fig. 3, traces ii.c and ii.d). Whereas, nearly full conversion of paxilline 3 to two bisprenylated-paxilline compounds (20–21 and 21–22 bisprenylated-paxillines) was observed in the janD transformants, the atmD transformants only partially converted paxilline 3 to three monoprenylated-paxilline compounds (20-, 21-, 22-monoprenylated-paxillines). In other words, these results imply that JanD may prenylate paxilline 3 more efficiently than AtmD in vivo, but further kinetic experiments are required to confirm this in vitro.
Discussion
By heterologous expression in a fungal host we have functionally analysed NodD1, one of two PTs that are predicted to act on an emindole SB derived structural core in the biosynthesis of the fully functionalised nodulisporic acids, such as nodulisporic acid A.19 Emindole SB has been identified for its anti-cancer properties6,7 and the nodulisporic acids are known for their highly potent toxicity towards invertebrates and absence of tremorgenic or other toxic properties against mammalian species that are associated with many of the paspaline-derived IDTs.3,21 Functional characterisation of NodD1 may provide valuable insights into the relationships between IDT PT sequences, reaction chemistries and substrate selectivities.
IDT PTs appear to be part of a broader group of microbial indole PTs known as the αββα (“ABBA”) PTs that prenylate an aromatic ring via Friedel–Crafts electrophilic alkylation.22 Despite low amino acid sequence homology, members of this group of soluble enzymes share a common structural architecture.23 The αββα PTs are found only in bacteria and fungi where they are involved exclusively in the biosynthesis of secondary metabolites. To date, the crystal structures of five fungal aromatic PTs have been reported. This group includes FgaPT2,23,24 FtmPT1 (ref. 25–27) and CdpNPT27,28 from Aspergillus fumigatus, AnaPT29 from Neosartoria fischeri, AtaPT30 from Aspergillus terreus. All these enzymes have been identified through genome mining and characterised in vitro making it difficult, in some cases, to identify their in vivo substrates and metabolic products with certainty; however, all can mediate the transfer of DMAPP onto the indole core of at least one indole containing substrate.30,31 Whereas FgaPT1 and FgaPT2 appear to demonstrate high substrate specificity and regioselectivity, CdpNPT, AnaPT and AtaPT are able to utilise a variety of indole containing substrates and even appear to catalyse both head and tail prenylations.23–28,32 AtaPT also has been shown to mediate C-and/or O-prenylation on non-indole containing substrates.30,31
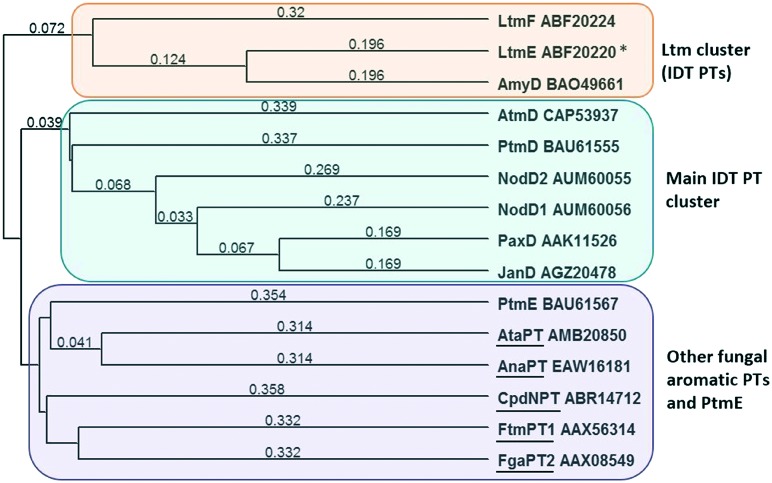
The low level of sequence identity seen between the IDT PTs and the other fungal aromatic PTs for which structures are known is consistent with the low level of sequence similarity that exists in the αββα PTs, despite their apparent common architecture and shared patterns of catalytic function. Phylogenetic analysis, shown in Fig. 4, demonstrated that, with the exception of PtmE, the known IDT PTs form two distinct clades with LtmE, LtmF (involved in the biosynthesis of lolitrems)11 and AmyD forming one clade (33–56% ID) and the remaining enzymes (PaxD, JanD, PenD, NodD1, NodD2 and AtmD) forming the second clade (29–66% ID), both of which are distinct from the five fungal PTs for which structures are known.
Fig. 4. Phylogenetic tree (uncorrected UPGMA) based for IDT PTs compared with fungal aromatic PTs with known structures (underlined). Numbers indicate difference in amino acid identity values over the entire sequence (Table S1†).
Several fungal ABBA PTs have been engineered to alter their substrate specificity and/or regioselectivity, which is highly desirable for creating novel chemical analogues and could be applied to the IDT PTs. However, with such high levels of sequence dissimilarity it is difficult to make predictions based on amino acid sequence data alone and it will be necessary to have a greater understanding of relationships between sequence and function and to determine the structural features that drive catalytic diversity in these enzymes.
Of the IDT enzymes, the aromatic PTs are the most well studied as they are not membrane associated and can therefore be characterised in vitro, making them a potential bioengineering starting point to generate diverse IDTs. Our results demonstrate clear differences in substrate specificity between NodD1, JanD and AtmD, with the latter two IDT PTs unable to prenylate a non-paspaline derived cyclic IDT core of NAF. Notably, NodD1 and JanD, while responsible for the substitution at the same position on the indole ring (i.e. C21 and C22), do not readily interchange IDT substrates. While NodD1 showed some ability to prenylate paxilline 3, JanD was unable to transform the less cyclised precursor, NAF 2, that possesses a partially uncyclised prenyl tail, and notably, a carboxylic acid functionality. Similarly, AtmD which displayed the ability to accept a variety of substrates during an in vitro study,17 was unable to accept NAF 2 as a substrate.
Conclusions
The IDT PTs offer great potential for the ability to create novel compounds via combinatorial biosynthesis using enzymes from different pathways and through creation of enzymes with novel catalytic functions. Prenylation of the emindole SB derived compound nodulisporic acid F provides the first report of an IDT PT that acts on a non-paspaline IDT core providing insight into the ability to modify compounds with anti-cancer therapeutic potential. This also provides evidence to further delineate the pathway for the biosynthesis of the nodulisporic acids, compounds with pesticidal properties which have so far been untapped. The strong preference of NodD1 for its natural substrate, NAF 2, and the inability of the related enzymes JanD and AtmD to prenylate this compound may help contribute to a greater understanding of how primary structure relates to the substrate specificities and regioselectivities displayed by these enzymes. There is much to explore and apply with this important group of enzymes as we begin to access the huge chemical and bioactive diversity that exists in the IDTs.
Conflicts of interest
There are no conflicts to declare.
Supplementary Material
Acknowledgments
This work was supported by the New Zealand Ministry of Business, Innovation, and Employment (RTVU1809).
Footnotes
†Electronic supplementary information (ESI) available: Including NMR spectra for NAE, sequence information for prenyltransferases, LCMS data, experimental and molecular biology details. See DOI: 10.1039/c9md00143c
References
- Ogata M., Ueda J.-y., Hoshi M., Hashimoto J., Nakashima T., Anzai K., Takagi M., Shin-ya K. J. Antibiot. 2007;60:645–648. doi: 10.1038/ja.2007.83. [DOI] [PubMed] [Google Scholar]
- Fan Y., Wang Y., Liu P., Fu P., Zhu T., Wang W., Zhu W. J. Nat. Prod. 2013;76:1328–1336. doi: 10.1021/np400304q. [DOI] [PubMed] [Google Scholar]
- Meinke P. T., Smith M. M., Shoop W. L. Curr. Top. Med. Chem. 2002;2:655–674. doi: 10.2174/1568026023393714. [DOI] [PubMed] [Google Scholar]
- Abdullaev I. F., Rudkouskaya A., Mongin A. A., Kuo Y. H. PLoS One. 2010;5:e12304. doi: 10.1371/journal.pone.0012304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivanets E. V., Yurchenko A. N., Smetanina O. F., Rasin A. B., Zhuravleva O. I., Pivkin M. V., Popov R. S., von Amsberg G., Afiyatullov S. S., Dyshlovoy S. A. Mar. Drugs. 2018;16:232. doi: 10.3390/md16070232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sallam A. A., Houssen W. E., Gissendanner C. R., Orabi K. Y., Foudah A. I., El Sayed K. A. MedChemComm. 2013;4:1360–1369. doi: 10.1039/C3MD00198A. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sallam A. A., Ayoub N. M., Foudah A. I., Gissendanner C. R., Meyer S. A., El Sayed K. A. Eur. J. Med. Chem. 2013;70:594–606. doi: 10.1016/j.ejmech.2013.09.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tarui Y., Chinen T., Nagumo Y., Motoyama T., Hayashi T., Hirota H., Muroi M., Ishii Y., Kondo H., Osada H., Usui T. ChemBioChem. 2014;15:934–938. doi: 10.1002/cbic.201300808. [DOI] [PubMed] [Google Scholar]
- Nakazawa J., Yajima J., Usui T., Ueki M., Takatsuki A., Imoto M., Toyoshima Y. Y., Osada H. Chem. Biol. 2003;10:131–137. doi: 10.1016/s1074-5521(03)00020-6. [DOI] [PubMed] [Google Scholar]
- Singh S. B., Ondeyka J. G., Jayasuriya H., Zink D. L., Ha S. N., Dahl-Roshak A., Greene J., Kim J. A., Smith M. M., Shoop W., Tkacz J. S. J. Nat. Prod. 2004;67:1496–1506. doi: 10.1021/np0498455. [DOI] [PubMed] [Google Scholar]
- Saikia S., Takemoto D., Tapper B. A., Lane G. A., Fraser K., Scott B. FEBS Lett. 2012;586:2563–2569. doi: 10.1016/j.febslet.2012.06.035. [DOI] [PubMed] [Google Scholar]
- Chen J., Morita H., Wakimoto T., Mori T., Noguchi H., Abe I. Org. Lett. 2012;14:3080–3083. doi: 10.1021/ol301129x. [DOI] [PubMed] [Google Scholar]
- Scott B., Young C. A., Saikia S., McMillan L. K., Monahan B. J., Koulman A., Astin J., Eaton C. J., Bryant A., Wrenn R. E., Finch S. C., Tapper B. A., Parker E. J., Jameson G. B. Toxins. 2013;5:1422–1446. doi: 10.3390/toxins5081422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tagami K., Minami A., Fujii R., Liu C., Tanaka M., Gomi K., Dairi T., Oikawa H. ChemBioChem. 2014;15:2076–2080. doi: 10.1002/cbic.201402195. [DOI] [PubMed] [Google Scholar]
- Liu C., Minami A., Dairi T., Gomi K., Scott B., Oikawa H. Org. Lett. 2016;18:5026–5029. doi: 10.1021/acs.orglett.6b02482. [DOI] [PubMed] [Google Scholar]
- Liu C., Tagami K., Minami A., Matsumoto T., Frisvad J. C., Suzuki H., Ishikawa J., Gomi K., Oikawa H. Angew. Chem., Int. Ed. 2015;54:5748–5752. doi: 10.1002/anie.201501072. [DOI] [PubMed] [Google Scholar]
- Liu C., Minami A., Noike M., Toshima H., Oikawa H., Dairi T. Appl. Environ. Microbiol. 2013;79:7298–7304. doi: 10.1128/AEM.02496-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu C., Noike M., Minami A., Oikawa H., Dairi T. Biosci., Biotechnol., Biochem. 2014;78:448–454. doi: 10.1080/09168451.2014.882759. [DOI] [PubMed] [Google Scholar]
- Van de Bittner K. C., Nicholson M. J., Bustamante L. Y., Kessans S. A., Ram A., van Dolleweerd C. J., Scott B., Parker E. J. J. Am. Chem. Soc. 2018;140:582–585. doi: 10.1021/jacs.7b10909. [DOI] [PubMed] [Google Scholar]
- van Dolleweerd C. J., Kessans S. A., Van de Bittner K. C., Bustamante L. Y., Bundela R., Scott B., Nicholson M. J., Parker E. J. ACS Synth. Biol. 2018;7:1018–1029. doi: 10.1021/acssynbio.7b00363. [DOI] [PubMed] [Google Scholar]
- Shoop W. L., Gregory L. M., Zakson-Aiken M., Michael B. F., Haines H. W., Ondeyka J. G., Meinke P. T., Schmatz D. M. J. Parasitol. 2001;87:419–423. doi: 10.1645/0022-3395(2001)087[0419:SEONAA]2.0.CO;2. [DOI] [PubMed] [Google Scholar]
- Tello M., Kuzuyama T., Heide L., Noel J. P., Richard S. B. Cell. Mol. Life Sci. 2008;65:1459–1463. doi: 10.1007/s00018-008-7579-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Metzger U., Schall C., Zocher G., Unsöld I., Stec E., Li S.-M., Heide L., Stehle T. Proc. Natl. Acad. Sci. U. S. A. 2009;106:14309–14314. doi: 10.1073/pnas.0904897106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Unsold I. A. Microbiology. 2005;151:1499–1505. doi: 10.1099/mic.0.27759-0. [DOI] [PubMed] [Google Scholar]
- Jost M., Zocher G., Tarcz S., Matuschek M., Xie X., Li S.-M., Stehle T. J. Am. Chem. Soc. 2010;132:17849–17858. doi: 10.1021/ja106817c. [DOI] [PubMed] [Google Scholar]
- Pan L. L., Song L. F., Miao Y., Yang Y., Merz, Jr. K. M. Biochemistry. 2017;56:2995–3007. doi: 10.1021/acs.biochem.7b00248. [DOI] [PubMed] [Google Scholar]
- Yin W. B., Ruan H. L., Westrich L., Grundmann A., Li S. M. ChemBioChem. 2007;8:1154–1161. doi: 10.1002/cbic.200700079. [DOI] [PubMed] [Google Scholar]
- Schuller J. M., Zocher G., Liebhold M., Xie X., Stahl M., Li S. M., Stehle T. J. Mol. Biol. 2012;422:87–99. doi: 10.1016/j.jmb.2012.05.033. [DOI] [PubMed] [Google Scholar]
- Yin W.-B., Xie X.-L., Matuschek M., Li S.-M. Org. Biomol. Chem. 2010;8:1133–1141. doi: 10.1039/b922440h. [DOI] [PubMed] [Google Scholar]
- Chen R., Gao B., Liu X., Ruan F., Zhang Y., Lou J., Feng K., Wunsch C., Li S. M., Dai J., Sun F. Nat. Chem. Biol. 2017;13:226–234. doi: 10.1038/nchembio.2263. [DOI] [PubMed] [Google Scholar]
- Bonitz T., Alva V., Saleh O., Lupas A. N., Heide L. PLoS One. 2011;6:e27336. doi: 10.1371/journal.pone.0027336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grundmann A., Kuznetsova T., Afiyatullov S., Li S. M. ChemBioChem. 2008;9:2059–2063. doi: 10.1002/cbic.200800240. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.