Abstract
Background
In present study, we explored the function of the metastasis-associated lung adenocarcinoma transcript 1 (MALAT1) gene in the development of non-small cell lung cancer (NSCLC).
Material/Methods
qRT-PCR was used to detect the MALAT1 mRNA expression level in cancer tissues and adjacent normal tissues of 115 NSCLC patients and in cell lines. MALAT1-mimic, MALAT1-inhibitor, and corresponding negative controls (NC) were utilized to transfect the H460 cells. Proliferation, migration, and invasion of H460 cells were evaluated by MTT method and Transwell assay. Expression levels of proteins in the ERK/MAPK signaling pathway were assessed by Western blot analysis.
Results
MALAT1 mRNA was upregulated in NSCLC tissues and cell lines compared to that in adjacent tissues and normal human bronchial cell line (BEAS-2B), respectively. Overexpression of MALAT1 significantly strengthened the proliferation, migration, and invasion ability of H460 cells. In comparison with the NC group, expression levels of CXCL5 and p-JNK proteins were elevated, while p-MAPK and p-ERK proteins were decreased in the MALAT1-mimic group. MALAT1 targets the 3′-untranslated region (UTR) fragment of the CXCL5 gene and inhibits its translation. Disturbance of the CXCL5 gene can reduce the protein expression of MAPK, p-MEK1/2, p-ERK1/2, and p-JNK, and inhibit the proliferation, migration, and invasion of MALAT1-mimic cells.
Conclusions
High MALAT1 expression promotes the proliferation, migration, and invasion of non-small cell lung cancer via the ERK/MAPK signaling pathway.
MeSH Keywords: Adenocarcinoma, Lung Neoplasms, MAP Kinase Signaling System, Small Cell Lung Carcinoma
Background
Non-small cell lung cancer (NSCLC) is a malignant tumor with high incidence and mortality rates [1]. Various factors have been found act as risk factors for the development of NSCLC [2,3]. DNA damage is the primary cause for cancer development [4]. Deficiency in DNA repair also leads to abnormal physiological processes of NSCLC [5]. Abnormal inflammation, formation of blood vessels, and hypoxia were found to play important roles in the development of cancer [6–8]. Therefore, any factors, especially genetic factors, involved in these processes may contribute to the development of NSCLC.
Metastasis-associated lung adenocarcinoma transcript 1 (MALAT1) is a long noncoding RNA (lncRNA) that is highly conserved in mammals [9]. MALAT1 is involved in numerous physiological processes, including transcription, splicing, translation, and epigenetic modulating of gene expression [10,11]. Numerous studies suggest that abnormal expression of MALAT1 is significantly correlated with different pathological processes [12,13]. Deficiency of MALAT1 can reduce the formation of blood vessels and blood flow perfusion in mice [14]. A recent study found that MALAT1 participates in inflammation pathways, cell cycle, cell death, and other pathways in brain injury [15]. Sallé-Lefort et al. found that MALAT1 expression level was obviously increased in hypoxia condition [16]. Many researches indicated that MALAT1 is increased in various cancers and predicts poor overall survival [17,18]. It acts as an oncogene in several cancers [19]. While, Kwok and colleagues suggested that MALAT1 acts as a tumor suppressor in colon and breast cancers [20]. Therefore, MALAT1 might play a crucial role in the development of NSCLC [21]. However, the exact mechanism of MALAT1 still not clear.
In the present study, we analyzed the mechanism of MALAT1 in NSCLC development and assessed the expression level and function of MALAT1 in NSCLC. The target gene and signaling pathway of MALAT1 were also assessed in this study.
Material and Methods
Sample collection
We recruited 115 NSCLC patients from the PLA General Hospital between January 2017 and December 2017. These patients did not receive any radiotherapy or chemotherapy before the operation. All the patients received radical lobectomy, and the cancer tissues and adjacent normal tissues (≥5 cm away from tumor tissues) were collected as samples. All tissue specimens were diagnosed by pathological examinations, and the results were confirmed by 2 pathologists. This study was approved by Ethics Committee of the PLA General Hospital. All patients signed informed consent before being enrolled in this study.
Cell culture
NSCLC cell lines (A549, H661, andH460) and a normal human bronchial cell line (BEAS-2B) were purchased from the Institute of Biochemistry and Cell Biology, Chinese Academy of Sciences. These cell lines were cultured in RPMI-1640 or DMEM medium with 10% fetal bovine serum (FBS). Cultural conditions were: saturated humidity with 5% CO2 at 37°C, replacing medium every 2 days until the cells fused into a single layer. Cells were digested by 0.25% trypsin, then subcultured at a 1: 3 ratio.
MALAT1 mRNA expression analysis
qRT-PCR (quantitative real-time reverse transcriptional polymerase chain reaction) was used to detect MALAT1 mRNA expression in tissues and cells. NSCLC cells were collected in exponential growth phase. Total RNA was extracted from these cells and tissues by Trizol. The PrimerScript RT reagent kit (Takara, Dalian, China) was used in the reverse transcription of MALAT1 cDNA, then the ABI 7900HTSequence Detection System was utilized for the detection of MALAT1 expression.
Transfection of MALAT1 into H460 NSCLC cells
Logarithmic cells with good growth state were digested for transfection. Overexpression vector (MALAT1-mimic), silent vector (MALAT1-inhibitor), and corresponding negative controls (NC) were diluted and mixed with Lipofectamine™ 2000, then cultured at room temperature for 20 min. This mixture (50 μmol/ml) was placed into 6-well plates with H460 cells (concentration 1×105/ml), fully mixed, and cultured for 6 h, then we replaced the medium and continued cultivated for 48 h. Transfection results were verified by qRT-PCR.
Cell proliferation analysis
(4,5-dimethylthiazol-2-yl)-2,5-diphenyltet-razolium bromide (MTT) was used for the cell proliferation analysis. We added 200 μl transfected H460 cells (1×104/ml) added into 96-well plates, cultured in a 37°C incubator with 5% CO2, and we replaced the medium every day. The medium was discarded at 24 h, 48 h, and 72 h. After washing in PBS, 20 μl MTT solution was added to every well and incubated at 37°C for 4h. Then, we discarded the solution, added 150 μl DMSO, oscillated in the dark for 10 min. The OD value of every well was assessed by enzyme-linked immunoassay at 490 nm.
Assay of invasion and migration
Invasion analysis was performed in Transwell chambers with Matrigel, and migration analysis was performed in Transwell chambers without Matrigel.
In 24-well plates, we placed 50 μl DMEM medium with 10% fetal bovine serum (FBS) and 200 μl cell suspension into the upper Transwell chamber, and we placed 500 μl DMEM medium (including 10% FBS) into the lower chamber, followed by incubation at 37°C for 48 h. The upper chambers were stained by crystal violet. We randomly selected 10 views, and a 200× inverted microscope was used to count the cells. Every experiment was performed in in triplicate.
Western blot analysis
Total proteins were extracted from the logarithmic growth phase cells. We used 12% SDS-PAGE electrophoresis to separate the proteins. After being transferred to polyvinylidene difluoride (PVDF) membranes, the proteins were blocked by 10% fat milk, and incubated by primary and secondary antibodies associated with the ERK/MAPK signaling pathway: CXCL5 antibody, MAPK antibody, p-MEK1/2 antibody, p-ERK1/2 antibody, and p-JNK antibody. Finally, the results were observed by use of an imager.
Dual-luciferase reporter analysis
To verify whether CXCL5 is the target gene of the MALAT1 gene, wide- and mutant-3′ untranslated regions (UTR) of the CXCL5 gene were transferred into dual-luciferase reporter vector (pmirGLO) and examined by direct sequencing. Transfection of CXCL5-pmirGLO into H460 cells were performed with Lipofectamine™ 2000 (Invitrogen). After being cultured at 37°C for 48 h, the cells were detected by a Dual-Luciferase Reporter Assay System.
Statistical analysis
All figures were plotted using GraphPad Prism. SPSS 18.0 was used to perform the data analysis. Continuous variables between different groups are presented by mean±SD and assessed by t test. Results had statistical significance when P<0.05.
Results
MALAT1 was upregulated in NSCLC tissues and cell lines
Expression levels of MALAT1 were significantly higher in NSCLC tissues than in adjacent normal tissues (Figure 1A, P<0.001), and it was also significantly higher in A549, H661, and H460 cell lines than in the normal bronchial cell line (BEAS-2B) (Figure 1B, P<0.001). In the 3 NSCLC cell lines, MALAT1 expression level was significantly higher in H460 cells and lower in A549 cells. Therefore, H460 cells were selected for further analysis.
Figure 1.
MALAT1 expression level in NSCLC tissues and cell lines. (A) Expression level of MALAT1 was significantly higher in NSCLC tissues than in adjacent tissues. (B) Expression level of MALAT1 was significantly higher in A549, H661, and H460 cell lines than that in normal bronchial cell line (BEAS-2B). * P<0.001.
Transfection of MALAT1 in H460 cells
qRT-PCR was used to assess the transfection results of H460 cells. We found that MALAT1 was overexpressed in MALAT1 mimic cells and reduced in MALAT1 inhibitor cells (Figure 2, P<0.05), suggesting that the transfection of MALAT1 was successful.
Figure 2.

MALAT1 transfection in H460 cells. * P<0.05.
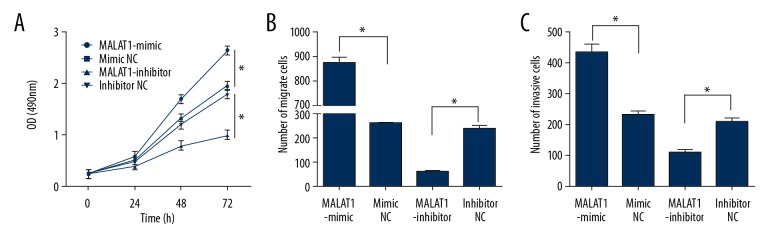
Overexpressed MALAT1 promoted the proliferation, invasion, and migration of H460 cells
MALAT1-mimic, MALAT1-inhibitor, and relative NC were transferred into H460 cells. We assessed the effects of MALAT1 expression levels on biological behaviors of H460 cells. We found that overexpressed MALAT1 promoted the proliferation, invasion, and migration of H460 cells; but low MALAT1 expression levels inhibited the proliferation, invasion, and migration of H460 cells (Figure 3, P<0.001).
Figure 3.
Effects of MALAT1 for biological behaviors of H460 cells. (A) Effects of MALAT1 for H460 proliferation. (B) Effects of MALAT1 for H460 migration. (C) Effects of MALAT1 for H460 invasion. * P<0.001.
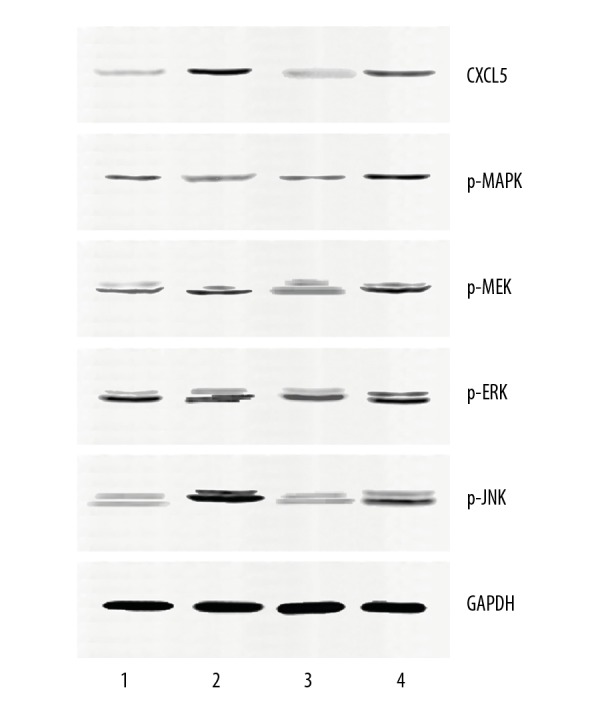
Effects of MALAT1 for proteins associated with ERK/MAPK signaling pathway
Western blot analysis showed that CXCL5 and p-JNK expressions were significantly increased, and p-MAPK and p-ERK were distinctly decreased in the MALAT1-mimic group. In the MALAT1-inhibitor group, CXCL5, p-JNK, and p-MEK1/2 expression levels were significantly lower than in the NC group (Figure 4, P<0.001).
Figure 4.

Effects of MALAT1 for proteins associated with the ERK/MAPK signaling pathway. 1: MALAT1-mimic; 2: Mimic NC; 3: MALAT1-inhibitor; 4: Inhibitor NC.
CXCL5 acts as the target gene of MALAT1
Dual-luciferase reporter analysis result indicated that low expression of MALAT1 could suppress the expression of luciferin, but when mutant 3′-UTR was combined with luciferin, the suppression was lost, suggesting that the 3′-UTR of CXCL5 acts as the target of MALAT1 and inhibits its expression (Figure 5).
Figure 5.
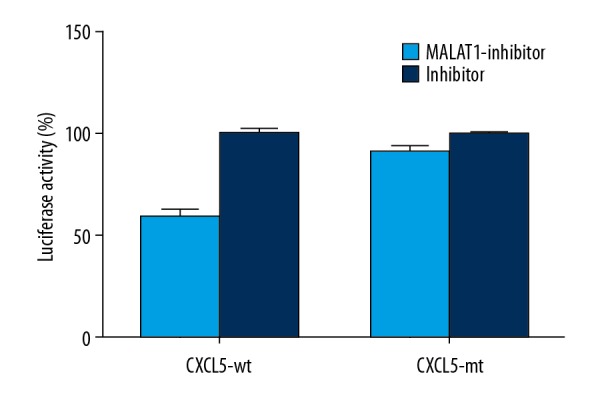
Dual-luciferase reporter analysis result.
Effects of CXCL5 on proteins associated with ERK/MAPK signaling pathway
CXCL5 was silenced by CXCL5 siRNA (siCXCL5) in MALAT1-mimic H460 cells, then we examined the CXCL5, MAPK, p-MEK1/2, p-ERK1/2, and p-JNK protein levels by Western blot analysis. The results showed that p-MEK1/2, p-ERK1/2, and p-JNK proteins were significantly lower in the MALAT1-mimic +siCXCL5 group compared to the than in MALAT1-mimic group (Figure 6, P<0.05).
Figure 6.
Effects of CXCL5 siRNA for proteins associated with the ERK/MAPK signaling pathway. 1: MALAT1-mimic +siCXCL5; 2: MALAT1-mimic; 3: Mimic NC+siCXCL5; 4: Mimic NC.
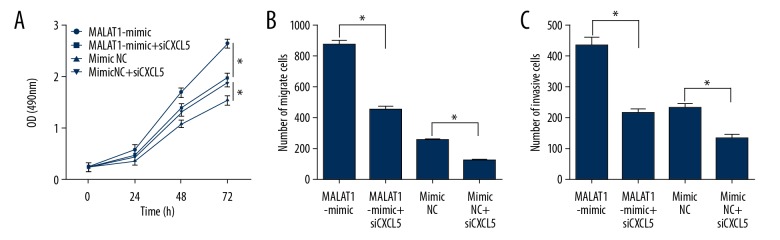
Silenced CXCL5 inhibited the proliferation, migration, and invasion of H460 cells
We also detected the alteration of H460 biological behaviors in MALAT1-mimic+siCXCL5, finding that knockout of CXCL5 inhibited the proliferation, migration, and invasion of H460 cells, which was caused by the MALAT1 overexpression (Figure 7).
Figure 7.
Knockout of CXCL5 in MALAT1-mimic cells inhibits the proliferation, migration, and invasion of H460 cells. (A) Effects of CXCL5 for H460 proliferation. (B) Effects of CXCL5 for H460 migration. (C) Effects of CXCL5 for H460 invasion. * P<0.001.
Discussion
lncRNA MALAT1 is significantly associated with several diseases. MALAT1 is upregulated in numerous tumors, and its abnormal expression causes many pathological alterations of tumors [22].
In the present study, we explored the expression of MALAT1 in tissues and different cell lines, showing that the expression level of MALAT1 was significantly increased in NSCLC tissues and cell lines, and the H460 cell line had the highest MALAT1 level. This agrees with a previous study performed by Guo et al., who suggested that MALAT1 was upregulated in NSCLC cells and tissues [23]. Elevated MALAT1 has also been discovered in esophageal squamous cell carcinoma (ESCC) tissues and influences tumor development [24]. We explored the effects of MALAT1 on the biological behaviors of H460 cells, finding that upregulated MALAT1 in H460 cells accelerates the biological behaviors of cells, including proliferation, invasion, and migration activity. Guo and colleagues indicated that knockout of MALAT1 inhibits the invasion and migration of NSCLC cells [23]. Proliferation and migration of prostate cancer (PC) cells were shown to be promoted by increased MALAT1 levels [25].
We found that the 3′-UTR of CXCL5 is the target gene of MALAT1. In the MALAT1-mimic group, CXCL5 and p-JNK were upregulated, while p-MAPK and p-ERK were downregulated, which is in accordance with results in different tumors. Guo and coworkers found that knockout of MALAT1 downregulates CXCL5 expression [23]. Leti et al. suggested that knockout of MALAT1 suppresses the CXCL5 expression in hepatic stellate cells [26]. Chen and colleagues found that knockdown of MALAT1 inhibits the activity of JNK [27]. However, Lei et al. indicated that low MALAT1 inhibits p-ERK expression and had no effects for p-JNK expression [28]. These results suggested that CXCL5 is the target of MALAT1, and that it plays an important role in tumor development via the ERK/MAPK signaling pathway.
When we knocked out the CXCL5 gene, p-MEK1/2, p-ERK1/2, and p-JNK proteins were distinctly lower in the MALAT1-mimic +siCXCL5 group than in the MALAT1-mimic group. We also assessed the biological behaviors of H460 cells in different groups. In the MALAT1-mimic+siCXCL5 cells, we found that the proliferation, migration, and invasion activities of H460 cells were reduced by silencing CXCL5. Guo and colleagues showed that knockout of CXCL5 in A549 cells inhibits migration and invasion [23]. Wang and colleagues suggested that CXCL5 facilitates the proliferation of H460 cells via activation of MAPK/ERK1/2 and PI3K/AKT pathways [29]. These results suggested that the function of MALAT1 for NSCLC biological behaviors was performed by the ERK/MAPK signaling pathway via the CXCL5 gene. A similar function has been found in other cancers. Du et al. suggested that MALAT1 acts as a suppressor for glioma through the ERK/MAPK signaling pathway [30,31].
Certain limitations of the present study must be considered. First, the function of MALAT1 for NSCLC development was only verified in the H460 cell line, which might have influenced the accuracy of our results. However, published articles have reported the oncogenic function of MALAT1 in other lung cancer cell lines, such as A549 [32] and H1299 [33], which was in accordance with our results. Secondly, effects of MALAT1 for other signaling pathways associated with tumor occurrence were did not assessed in the present study. Finally, the mechanism of MALAT1 in NSCLC development was not assessed in animal models.
Conclusions
We found that MALAT1 was upregulated in NSCLC tissues and cell lines, and it promotes the proliferation, invasion, and migration of H460 cells. CXCL5 acts as the target gene of MALAT1 and mediates the function of MALAT1 for the biological behaviors of H460 cells. Further studies focused on the association of MALAT1 with other signaling pathways should be performed in the future, so as to verify the mechanism of MALAT1 in NSCLC development.
Footnotes
Source of support: Departmental sources
References
- 1.Siegel RL, Miller KD, Jemal A. Cancer statistics, 2015. Cancer J Clin. 2015;65:5–29. doi: 10.3322/caac.21254. [DOI] [PubMed] [Google Scholar]
- 2.Kim JH, Ha M. The disease burden of lung cancer attributable to residential radon exposure in Korean homes. J Korean Med Sci. 2018;33:e223. doi: 10.3346/jkms.2018.33.e223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Liu X, Conner H, Kobayashi T, et al. Cigarette smoke extract induces DNA damage but not apoptosis in human bronchial epithelial cells. Am J Respir Cell Mol Biol. 2005;33:121–29. doi: 10.1165/rcmb.2003-0341OC. [DOI] [PubMed] [Google Scholar]
- 4.Kastan MB. DNA damage responses: Mechanisms and roles in human disease: 2007 G.H.A. Clowes Memorial Award Lecture. Mol Cancer Res. 2008;6:517–24. doi: 10.1158/1541-7786.MCR-08-0020. [DOI] [PubMed] [Google Scholar]
- 5.Weng MW, Lee HW, Park SH, et al. Aldehydes are the predominant forces inducing DNA damage and inhibiting DNA repair in tobacco smoke carcinogenesis. Proc Natl Acad Sci USA. 2018;115:E6152–61. doi: 10.1073/pnas.1804869115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gomes M, Teixeira AL, Coelho A, et al. The role of inflammation in lung cancer. Adv Exp Med Biol. 2014;816:1–23. doi: 10.1007/978-3-0348-0837-8_1. [DOI] [PubMed] [Google Scholar]
- 7.Piperdi B, Merla A, Perez-Soler R. Targeting angiogenesis in squamous non-small cell lung cancer. Drugs. 2014;74:403–13. doi: 10.1007/s40265-014-0182-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Guo Y, Wu M, Zhao J, Li Y. [Advances in hypoxia microenvironment and chemotherapy-resistant of lung cancer]. Zhongguo Fei Ai Za Zhi. 2014;17:265–68. doi: 10.3779/j.issn.1009-3419.2014.03.14. [in Chinese] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hutchinson JN, Ensminger AW, Clemson CM, et al. A screen for nuclear transcripts identifies two linked noncoding RNAs associated with SC35 splicing domains. BMC Genomics. 2007;8:39. doi: 10.1186/1471-2164-8-39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tripathi V, Shen Z, Chakraborty A, et al. Long noncoding RNA MALAT1 controls cell cycle progression by regulating the expression of oncogenic transcription factor B-MYB. PLoS Genet. 2013;9:e1003368. doi: 10.1371/journal.pgen.1003368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Biswas S, Thomas AA, Chen S, et al. MALAT1: An epigenetic regulator of inflammation in diabetic retinopathy. Sci Rep. 2018;8:6526. doi: 10.1038/s41598-018-24907-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wu Y, Huang C, Meng X, Li J. Long noncoding RNA MALAT1: Insights into its Biogenesis and Implications in Human Disease. Curr Pharm Des. 2015;21:5017–28. doi: 10.2174/1381612821666150724115625. [DOI] [PubMed] [Google Scholar]
- 13.Yoshimoto R, Mayeda A, Yoshida M, Nakagawa S. MALAT1 long non-coding RNA in cancer. Biochim Biophys Acta. 2016;1859:192–99. doi: 10.1016/j.bbagrm.2015.09.012. [DOI] [PubMed] [Google Scholar]
- 14.Zhang X, Tang X, Hamblin MH, Yin KJ. Long non-coding RNA Malat1 regulates angiogenesis in hindlimb ischemia. Int J Mol Sci. 2018;19 doi: 10.3390/ijms19061723. pii: E1723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Patel NA, Moss LD, Lee JY, et al. Long noncoding RNA MALAT1 in exosomes drives regenerative function and modulates inflammation-linked networks following traumatic brain injury. J Neuroinflammation. 2018;15:204. doi: 10.1186/s12974-018-1240-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Salle-Lefort S, Miard S, Nolin MA, et al. Hypoxia upregulates Malat1 expression through a CaMKK/AMPK/HIF-1alpha axis. Int J Oncol. 2016;49:1731–36. doi: 10.3892/ijo.2016.3630. [DOI] [PubMed] [Google Scholar]
- 17.Tian X, Xu G. Clinical value of lncRNA MALAT1 as a prognostic marker in human cancer: Systematic review and meta-analysis. BMJ Open. 2015;5:e008653. doi: 10.1136/bmjopen-2015-008653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wei Y, Niu B. Role of MALAT1 as a prognostic factor for survival in various cancers: A systematic review of the literature with meta-analysis. Dis Markers. 2015;2015 doi: 10.1155/2015/164635. 164635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Xiong Y, Wang J, Zhu H, et al. Chronic oxymatrine treatment induces resistance and epithelialmesenchymal transition through targeting the long non-coding RNA MALAT1 in colorectal cancer cells. Oncol Rep. 2018;39:967–76. doi: 10.3892/or.2018.6204. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 20.Kwok ZH, Roche V, Chew XH, et al. A non-canonical tumor suppressive role for the long non-coding RNA MALAT1 in colon and breast cancers. Int J Cancer. 2018;143:668–78. doi: 10.1002/ijc.31386. [DOI] [PubMed] [Google Scholar]
- 21.Tang Y, Xiao G, Chen Y, Deng Y. LncRNA MALAT1 promotes migration and invasion of non-small-cell lung cancer by targeting miR-206 and activating Akt/mTOR signaling. Anticancer Drugs. 2018;29:725–35. doi: 10.1097/CAD.0000000000000650. [DOI] [PubMed] [Google Scholar]
- 22.Zhao M, Wang S, Li Q, et al. MALAT1: A long non-coding RNA highly associated with human cancers. Oncol Lett. 2018;16:19–26. doi: 10.3892/ol.2018.8613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Guo F, Guo L, Li Y, et al. MALAT1 is an oncogenic long non-coding RNA associated with tumor invasion in non-small cell lung cancer regulated by DNA methylation. Int J Clin Exp Pathol. 2015;8:15903–10. [PMC free article] [PubMed] [Google Scholar]
- 24.Kang K, Huang YH, Li HP, Guo SM. Expression of UCA1 and MALAT1 long-chain non-coding RNAs in esophageal squamous cell carcinoma tissues is predictive of patient prognosis. Arch Med Sci. 2018;14:752–59. doi: 10.5114/aoms.2018.73713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chang J, Xu W, Du X, Hou J. MALAT1 silencing suppresses prostate cancer progression by upregulating miR-1 and downregulating KRAS. Onco Targets Ther. 2018;11:3461–73. doi: 10.2147/OTT.S164131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Leti F, Legendre C, Still CD, et al. Altered expression of MALAT1 lncRNA in nonalcoholic steatohepatitis fibrosis regulates CXCL5 in hepatic stellate cells. Transl Res. 2017;190:25–39e21. doi: 10.1016/j.trsl.2017.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chen J, Ke S, Zhong L, et al. Long noncoding RNA MALAT1 regulates generation of reactive oxygen species and the insulin responses in male mice. Biochem Pharmacol. 2018;152:94–103. doi: 10.1016/j.bcp.2018.03.019. [DOI] [PubMed] [Google Scholar]
- 28.Lei L, Zeng Q, Lu J, et al. MALAT1 participates in ultraviolet B-induced photo-aging via regulation of the ERK/MAPK signaling pathway. Mol Med Rep. 2017;15:3977–82. doi: 10.3892/mmr.2017.6532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wang L, Shi L, Gu J, et al. CXCL5 regulation of proliferation and migration in human non-small cell lung cancer cells. J Physiol Biochem. 2018;74:313–24. doi: 10.1007/s13105-018-0619-z. [DOI] [PubMed] [Google Scholar]
- 30.Han Y, Wu Z, Wu T, et al. Tumor-suppressive function of long noncoding RNA MALAT1 in glioma cells by downregulation of MMP2 and inactivation of ERK/MAPK signaling. Cell Death Dis. 2016;7:e2123. doi: 10.1038/cddis.2015.407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Du M, Chen W, Zhang W, et al. TGF-β regulates the ERK/MAPK pathway independent of the SMAD pathway by repressing miRNA-124 to increase MALAT1 expression in nasopharyngeal carcinoma. Biomed Pharmacother. 2018;99:688–96. doi: 10.1016/j.biopha.2018.01.120. [DOI] [PubMed] [Google Scholar]
- 32.Jen J, Tang YA, Lu YH, et al. Oct4 transcriptionally regulates the expression of long non-coding RNAs NEAT1 and MALAT1 to promote lung cancer progression. Mol Cancer. 2017;16:104. doi: 10.1186/s12943-017-0674-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chen W, Zhao W, Zhang L, et al. MALAT1-miR-101-SOX9 feedback loop modulates the chemo-resistance of lung cancer cell to DDP via Wnt signaling pathway. Oncotarget. 2017;8:94317–29. doi: 10.18632/oncotarget.21693. [DOI] [PMC free article] [PubMed] [Google Scholar]