Abstract
Under denaturing conditions such as low pH and elevated temperatures, proteins in vitro can misfold and aggregate to form long rigid rods called amyloid fibrils; further self-assembly can lead to larger structures termed spherulites. Both of these aggregates resemble amyloid tangles and plaques associated with Alzheimer’s disease in vivo. The ability to form such aggregates in a multitude of different proteins suggests that it is a generic ability in their mechanism to form. Little is known about the structure of these large spherulites ranging from 5 to 100 microns and whether they can reproducibly form in amyloid β (1-40) (Aβ40), a 40-amino acid residue peptide, which is one of the major components of Alzheimer’s amyloid deposits. Here, we show that spherulites can readily form in Aβ40 under certain monomerization and denaturing conditions. Using polarized and nonpolarized Raman spectroscopy, we analyzed the secondary structure of spherulites formed from three different proteins: insulin, β-lactoglobulin (BLG), and Aβ40. Visually, these spherulites have a characteristic “Maltese Cross” structure under crossed polarizers through an optical microscope. However, our results indicate that insulin and Aβ40 spherulites have similar core structures consisting mostly of random coils with radiating fibrils, whereas BLG mostly contains β-sheets and fibrils that are likely to be spiraling from the core to the edge.
1. Introduction
A number of degenerative diseases such as Alzheimer’s disease (AD), type II diabetes, and Parkinson’s disease are associated with aggregates of excess protein deposited in the tissue or organs, making protein aggregation an active and very important field of research. These aggregates contain β-strands that lie perpendicular to the fibril axis; they are usually long (∼1 μm) and with an unbranched morphology, with fibril diameters ranging from 10 to 20 nm.1 Amyloid fibrils are thought to form via a polymerization mechanism, in which the protein first (partially) misfolds and forms stable nuclei, followed by the attachment of monomers to the growing ends.2
However, larger-scale structures are seen in the post-mortem brains of Alzheimer’s patients.3−5 Sections of brain tissue viewed under crossed polarizers show that a structure is present resembling the “spherulites” seen in many other in vitro systems including insulin,6 β-lactoglobulin (BLG),7 hen egg white lysozyme,8 human proislet amyloid polypeptide (ProIAPP1-48),9 and amyloid β(1-42) (Aβ42).5 These proteins have all been shown to demonstrate that the amyloid fibrils further self-assemble to form highly ordered spherical aggregates, microns or tens of microns in size, which have been named spherulites after similar structures that commonly form from viscous or impure polymer melts.10
Spherulitic structures have been observed in a wide range of alloys,11 volcanic rocks,12 synthetic polymers,13 liquid crystals,8,14 and gels,15 with the literature on these structures dating back as far as the 19th century. Starch granules have also been described as spherulites, along with certain carbohydrates such as chitin, which forms such structures through lateral molecular association.16 Between crossed polarizers, spherulites exhibit characteristic “Maltese cross” patterns, which rotate as the polarizers are rotated.6
The characteristic Maltese cross pattern has been observed in a rat model of AD,17 in brains of human patients suffering from Creutzfeldt–Jakob disease,18 and in neuritic plaques, a hallmark of AD,3,4 as well as in other protein systems in vivo,19,20 suggesting that the mechanism to form both spherulites in vitro and plaques found in many amyloid diseases may be similar.21
House et al. were the first to observe spherulites formed from in vitro Aβ42 and compare them to spherulites in the human brain tissue.5 During this study, House et al. showed that copper abolished the secondary structure β-sheet of preformed Aβ42 amyloid fibrils, whereby copper binds to fibrillar Aβ42. These findings are significant to the field and could potentially be beneficial for AD therapy.5 In collaboration with House et al., we have formed in vitro spherulitic aggregates in Aβ42 and confirmed the characteristic Maltese cross pattern.4 In this study, the Aβ42 spherulites were formed under near physiological conditions, after preformed aggregates were first abolished following an addition of an excess of copper in Aβ42. Interestingly, the in vitro Aβ42 spherulites were near-identical structures to that observed in 30 μm-thick sections of diseased brain tissue from patients with AD.4,21 More recently, Exley et al. also have shown that ProIAPP1-48 could form spherulites and that copper was a potent in vitro inhibitor.9,22 This has major implications for type 2 diabetes because IAPP and ProIAPP1-48 are associated with the death of β cells in the pancreas.22 Large spherically symmetric structures have been reported to form in amyloid β(1-40) (Aβ40),23,24 but their structures have not been shown to exhibit the Maltese cross patterns found in vivo.3
One challenge, readily discernible in the literature, is the irreproducibility of the starting material when working with Aβ.25 Studying Aβ in vitro requires a well-defined material, but the intrinsic propensity of Aβ to self-associate creates substantial experimental problems when attempting to reproducibly form aggregates.26 This issue with Aβ studies has led to the adoption of peptide preparation procedures that vary from group to group. The overall aim in each case is to remove all preformed aggregates to give a reproducible homogeneous initial state of the peptide that will give reliable and robust kinetics, but the variability in subsequent outcomes suggests that this may not have been achieved. The preparation method for forming spherulites in Aβ42 has been well documented by Exley et al.4 Here, we focus on the in vitro preparation method for forming spherulites in Aβ40.
The observed Maltese cross pattern was mathematically shown by Morse et al. to be caused by the uniaxial behavior of the building blocks (amyloid fibrils in the case of protein aggregates) that are oriented radially within the spherulites.27 Light paths passing through a spherulite encounter many fibrils orientated at different angles. The overall retardation along the light path is the sum of the individual retardations from every contributing fibril. At the center of the spherulite, the total retardation is zero due to the cancellation of effects in different directions; it reaches a maximum at approximately two-thirds from the center of the spherulite and then decreases to zero at the periphery, as the decrease in the thickness counteracts the birefringence of the fibers.27−30
The dark centers of the spherulites formed from different proteins have various core sizes that do not correlate with the diameter of the aggregates.6,31 These non-birefringent cores still form in insulin even when filtration is used before denaturing, suggesting that the core is due to some structure of the protein and not from any external effect. The non-birefringent core in insulin is thought to be due to collapsed fibrils that have lost their orientation because of stress as the spherulite continues to grow;7 however, this hypothesis has not been proven. We have previously shown that although the optical birefringence patterns of insulin and β-lactoglobulin spherulites are different, the cores of the spherulites are too small to directly compare the birefringence signals.32
Many techniques, including nuclear magnetic resonance (NMR), Fourier transform infrared (FTIR), and Raman spectroscopy, have been used to compare the secondary structure of native proteins with that of denatured proteins that form amyloid fibrils.33−44 In particular, both native insulin and BLG show a remarkable spectral change in the amide I region because of carbonyl stretching (C=O) after forming amyloid fibrils.34,41,45,46 The peaks of interest are those assigned to the β-sheet, α-helix, and random coil structures.
Spatially localized Raman measurements allow for a secondary structure or crystallinity to be investigated at different points within a given aggregate. Galiotis et al. successfully used this technique to evaluate the crystallinity in isotactic polypropylene (iPP) along the diameter of a spherical aggregate.47 By analyzing the intensity signal of the 809 cm–1 band (corresponding to crystallinity) normalized by the 830, 841, and 854 cm–1 bands (corresponding to noncrystallinity) along the diameter, they showed that crystallinity was symmetric and peaked at the nucleus. Using a setup similar to that used by Galiotis et al., we use Raman spectroscopy to analyze the secondary structure of the amyloid fibrils that are thought to be radially orientated within a spherulite. The utility of Raman spectroscopy as a beneficial technique to analyze anisotropic biological specimens was also recently reported by Lednev et al. who showed that the structural organization of insulin fibrils could be revealed using this methodology.46
Previous preliminary work on insulin alone showed a dramatic increase in the random coils and a decrease in the β-sheet content in the central region.32 Here, we show the secondary structure as a function of position across the diameter of the spherulites in insulin, BLG, and Aβ40, and how these structures differ from one another. The orientations of the fibrils within a spherulite are also compared using polarized Raman spectroscopy.
2. Results and Discussion
2.1. Secondary Structure of Insulin Spherulites
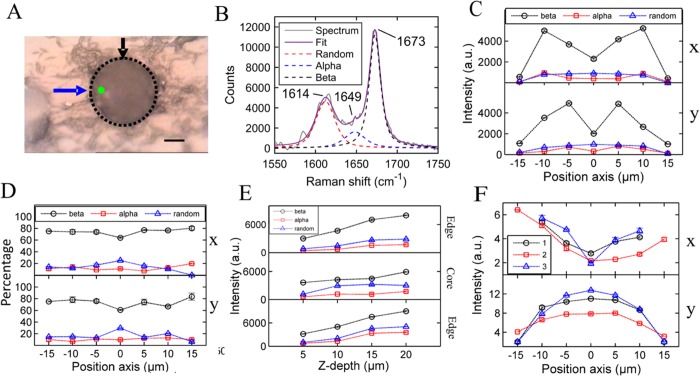
An insulin spherulite (formed under acidic conditions and elevated temperatures, see Experimental Section) visualized using the microscope attached to a Raman spectrometer is shown in Figure 1A, where the dotted line highlights the boundary of the spherulite. To compare the secondary structures across the spherulite, spectra were recorded, analyzed, and fitted every 5 μm along the x- and y-axes as indicated by the blue and black arrows, respectively. A typical Raman spectrum was recorded from the insulin spherulite (indicated by the green dot in Figure 1A), and fitted peaks are shown in Figure 1B, where the main peaks of interest are in the amide I region (1600–1700 cm–1).
Figure 1.
(A) Image of a typical insulin spherulite from the optical microscope attached to the Raman spectroscope. The blue and black arrows indicate the x- and y-directions, respectively. The scale bar is 10 μm. (B) Amide spectra region from the location indicated by the green dot in (A). Exposure time of 100 s. Assigned peaks, individual fits, and an overall fit are shown in the figure. (C) Raman peak intensities for the β-sheet, α-helix, and random coil across the insulin spherulite in the x-direction (top) and y-direction (bottom), where 0 μm marks the center. The β-sheet intensity drops significantly at the center of the spherulite, whereas the random coil intensity increases slightly. (D) Percentage contributions of the β-sheet, α-helix, and random coil in the x- and y-directions. The largest contribution is from the β-sheet, which decreases at the center. (E) Raman peak intensities for the β-sheet, α-helix, and random coil in the insulin spherulite as the material below the focus point increases (increasing z depth), where 0 μm is the coverslip plane. (F) Polarized β-sheet intensity across the spherulite in the x and y paths as indicated in Figure S1F. The β-sheet intensity is a minimum at the medians of the x-paths, suggesting that the fibrils are perpendicular to the direction of polarization at this point.
A dominant peak is clearly seen at approximately 1673 cm–1, corresponding to the β-sheet structure.48 Because native insulin is mainly α-helical (peak at 1649 cm–1) and does not contain any structures incorporating such a large quantity of β-sheets,49 the spherulite must contain a substantial quantity of denatured proteins in the form of amyloid fibrils. Raman peaks assigned to the β-sheet, α-helix, and random coils in the amide I region are given in Table 1.
Table 1. Raman Band Assignments48.
| wavenumber/cm–1 | structure assignment |
|---|---|
| 1673 | β-sheet |
| 1649 | α-helix |
| 1614 | random coils |
| 1603 | phenylalanine |
| 1585 | aromatics |
The absolute intensity (which is affected by the density and the thickness of the material) of the peaks can be plotted at each location across the spherulite in both the x- and y-directions, as shown in Figure 1C. Although there is a drop in intensity for both β-sheet and α-helix at the center, the signal does not drop to zero, suggesting that amyloid is present in the section through the core region. However, the random coil intensity is a maximum at the core, suggesting that more amorphous material is located at the center of the spherulite where the birefringence is lower, along with a small proportion of amyloid.
On analyzing the absolute signal across the spherulites, we can gain further structural information following a normalization procedure. Figure S1A shows a schematic representation of the laser penetration through the spherulite. The absolute intensity of the secondary structures is higher in the thickest region of the spherulite (region two in Figure S1A). To account for this thickness change across the spherulite whereby the edges have a lower mass thickness, it is necessary to normalize with respect to the sum of all three secondary structures so that the difference in sample illumination will be negligible. For the β-sheet intensity, the normalized intensity will be
| 1 |
where Iβ is the intensity of the β-sheet structure, Iα is the intensity of the α helix structure, and IR is the intensity of the random coil structure. At positions 1 and 2 in Figure S1A, the normalized intensities are
| 2 |
and
| 3 |
where λ and γ are factors that take into account the volume fraction of the material, where γ > λ. Both factors cancel out, and therefore the volume of penetration is not an issue when analyzing the normalized contribution of the secondary structures at positions along the spherulite.
Figure 1D shows the normalized intensities representing the percentage of each type of secondary structure in the x- and y-directions, where 100% is the total contribution of the three combined secondary structures. The largest contribution at any location in the spherulite is from the β-sheets, which makes up approximately 75% of the structure near the spherulite edge, dropping to approximately 60% at the core. The random coil contribution is a maximum at this central point, increasing from approximately 15% to 25% near the edge, indicating the presence of an amorphous material at the core. The same trend was found in the y-direction. The increase in the amorphous material at the core would explain the dark center in the Maltese cross patterns readily observed in the spherulites, because one would expect its birefringence to be low.
The secondary structure distribution as a function of position was similar in two more insulin spherulites (see Figure S1B,C). The same trend in all spherulites suggests that the non-birefringent core contains a larger amount of random coils and less β-sheets, and hence there is no uniform structure across the whole spherulite. Thus, the non-birefringent core is likely to be made up of a disordered material and a small quantity of amyloid fibrils. A higher proportion of random coils at the core suggests that the spherulite formation may begin with the amorphous protein acting as a nucleus with the fibrils then growing out from this center50 or the collapse and disorganization of the fibrils at the center.
The intensities of the β-sheet, α-helix, and random coil signals were compared with increasing z-depth within a spherulite at three independent locations, two avoiding the center (black arrows) and one through the center (red arrow), as shown in the schematic spherulite in Figure S1D.
Figure 1E (top and bottom panels) shows that the intensity increases for all assigned secondary structures as the thickness and therefore the scattering volume increases at the left hand and right hand sides of the spherulite (black arrows on Figure S1D). The intensities were not normalized because the volume illumination is crucial for the depth analysis. The volume of material for which light is scattered will increase by 20 μm3 as the depth is increased by 5 μm at each position and the beam spot size is approximately 4 μm2 in cross section regardless of depth. A different trend is observed in Figure 1E (middle panel) when increasing the depth through the core (red arrow on Figure S1D) compared with the case when the core is avoided. The gradient of intensity of the β-sheet (and the α-helix) decreases when the laser penetrates through the approximate location of the core and then increases as it leaves the core. This difference in trend at the core, compared with the edge of the spherulite, can be explained by the decrease in the β-sheets and increase in the random coils within the core. The β-sheet signal does not increase when going through the core (z-depth ≈ 5–10 μm), consistent with the drop in the β-sheet intensity observed in the x- and y-directions. This analysis is consistent with a spherical core of amorphous/random coil material surrounded above and below by the radiating β-sheet-rich fibrils.
2.2. Amyloid Fibril Orientations in Insulin Spherulites
The Maltese cross extinction pattern suggests that the spherulite is composed of radially orientated amyloid fibrils.6 To confirm this using polarized Raman spectroscopy, the intensity signal of the β-sheet content can be compared across the spherulite, indicating the orientation of the fibrils. This relationship is given by the formula
| 4 |
where a is a constant and θ is the angle between the molecular bonding of the protein backbone and the direction of the polarized laser (and therefore analyzer because both are parallel to each other).51 A maximum intensity will arise when the C=O carbonyl groups and hence the fibrils are parallel to the direction of polarization (Figure S1E).52
Figure S1F shows an insulin spherulite visualized using confocal microscopy in 2D; the direction of the laser polarization is given by the black double-headed arrow, which is parallel to the defined x-axis of the spherulite. The intensity across the spherulite is recorded along six paths; three in the x-direction, X1, X2, and X3, and three in the y-direction, Y1, Y2, and Y3, where X2, and Y2 go through the core as shown in Figure S1F.
The intensities were normalized with respect to the random coil intensity only at the corresponding positions because the α-helix signals are very weak when using polarized laser Raman spectroscopy. Opposing trends are apparent when comparing the β-sheet intensity in the x- and y-directions. The minimum and maximum β-sheet intensities occur at the middle of all x and y paths, respectively. In the x-direction (paths that are parallel to the direction of polarization), the overall signal is higher than in the y-direction. This suggests that the C=O groups within the fibrils are mostly aligned with the x-direction. The β-sheet intensity increases at either side of the minima in the x-direction, suggesting that the fibrils are more aligned with the direction of polarization away from the median of the X paths. At the core, more fibrils may be disordered and from nonpolarized Raman, we can see that there is also more amorphous material giving the lowest intensity signal.
In the y-direction, the scattering from the spherulite increases at the center, whereas the nonpolarized data across the diameter decreases at the center. Because the form of the polarized and nonpolarized signals is not the same across the identical path (Y2 path), the polarized data must be a result of the orientation of fibrils from both the x- and y-directions combined. The β-sheet intensity is a maximum at the middle of the three paths along the Y-direction, implying that the fibrils are most aligned with the direction of polarization at this point. The β-sheet intensity is a minimum at the edges. The results from the polarizing data suggest that the fibrils radiate out from the center of the spherulite (see Figure S2A for schematic interpretations), consistent with the earlier observations.50
2.3. Secondary Structure and Fibril Orientations in BLG
Like insulin, BLG also forms spherulites readily under acidic conditions and elevated temperatures in vitro. However, unlike insulin spherulites, Figure 2A shows that the absolute intensity of all secondary structures is maximum at the core of the BLG spherulite. This suggests that BLG spherulites may have more β-sheet material in general at their centers in contrast to insulin spherulites that have greater core diameters.
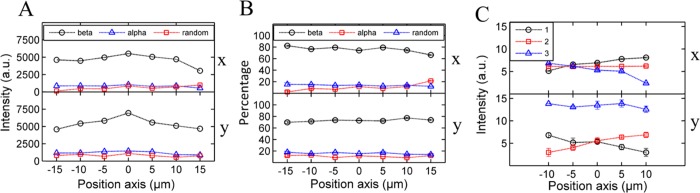
Figure 2.
(A) Raman peak intensities for the β-sheet, α-helix, and random coil across a BLG spherulite in the x-direction (top) and y-direction (bottom), where 0 μm marks the center. (B) Percentage of the β-sheet, α-helix, and random coil across a BLG spherulite in the x-direction (top) and y-direction (bottom), where 0 μm marks the center. (C) Polarized β-sheet intensity across a BLG spherulite to indicate the orientation of the fibrils in the x- and y-axes.
Both insulin and BLG spherulites consist of approximately 75% of β-sheet overall. However, the BLG levels of all three secondary structures seem to be constant across the spherulite (Figure 2B; results reproduced in two more BLG spherulites, Figure S2B,C) in contrast to insulin, which decreases in the β-sheet and increases in the random coil content toward the center.
Polarized Raman spectroscopy showed that the insulin spherulites possessed radial symmetry, as discussed in the previous section (Figure 1F). BLG differs in its internal structure (Figure 2C) because it has fibrils aligned more at one side of the spherulite in both the x and y-directions. It has been well documented in the lab of Mezzenga et al. that at the single fibril level, the structure and stiffness of the BLG fibrils can be altered depending on the environmental denaturing conditions (incubation temperature, pH, ionic strength, and surface interface).53−57 For example, BLG fibrils can form a twisted ribbon or helical ribbon structure when grown under mild or severe denaturing conditions, respectively.54,55 Our observations suggest that the fibril may spiral out from the core (not necessarily twisting along the fibril axis), as shown schematically in Figure S2D; this structure would then give β-sheet a maximum when the fibrils at the end of the spherulite were curved so that the fibrils were parallel with the orientation of polarization. The structure of the bending fibrils and no core is consistent with the results of the work by Poulin et al.: when fibrils radiate out from a seed, the splay energy is lower than the bending energy giving radial spherulites, whereas bending fibrils arise when there is no core and the bending energy is lower.58 The implied BLG structure is also similar to the bending extinction pattern observed from the phase transitions of radial hedgehogs to hyperbolic structures in nematic liquid crystals because of the reorientation of molecules.15
BLG spherulites shown here differ from insulin in terms of the structure of the core and orientation of the fibrils. These differences in the structure may reflect differences in the properties of the fibrils themselves or the way that they aggregate and pack. Although these structures appear similar through an optical microscope, detailed secondary structures show differences that cannot be detected by visual methods alone.
2.4. Secondary Structure and Fibril Orientations in Aβ40
The main issue when forming amyloid aggregates in vitro from Aβ is their irreproducibility apparently due to rapid self- and random aggregation.25 Here, we formed Aβ40 spherulites by employing a new reproducible monomerization and denaturing protocol. First, we tested two preparation methods aimed at eliminating pre-existing aggregates, creating a monomeric Aβ stock solution from peptide lyophilizates. The first method involves the use of trifluoroacetic acid (TFA) and HFIP separately to disrupt hydrophobic interactions in aggregated amyloid preparations, as shown in Table 2. This method has been previously shown to yield monomeric peptide solutions with α-helical and random coil secondary structures.25 The second method uses HFIP alone, as shown in Table 2, which avoids crossing the isoelectric point (pI) where the peptides are most insoluble, a condition which could lead to rapid aggregation and may prevent subsequent formation of long-range ordered structures.59
Table 2. Monomerize and Disaggregate Aβ Before Spherulite Formation25,60.
| method 1 | method 2 | process |
|---|---|---|
| dissolve 1 mg of peptide in 1 mL of TFA (takes approximately 10 min), sonicate for 30 s on ice, freeze peptide with liquid nitrogen | N/A | solubilizes Aβ effectively and disaggregates any pre-existing aggregates |
| lyophilize overnight | N/A | removes TFA |
| dissolve in 1 mL of HFIP and leave on ice for 5–10 min | dissolve in 1 mL of HFIP and leave on ice for 5–10 min | further disrupts any aggregates |
| divide into aliquots and dry using a rotatory evaporator at room temperature. | divide into aliquots and dry using a rotatory evaporator at room temperature. | removes HFIP |
Following both preparation methods, a 50 mM sodium phosphate buffer (pH 7.4) was prepared with the addition of 0.1 M NaCl and was added to the powdered Aβ40 peptide in 1.5 mL Eppendorfs to give concentrations of 30, 50, 150, and 300 μM. The lowest peptide concentration has been widely used for amyloid fibril experiments, without spherulites being identified; therefore, the higher concentrations (150 and 300 μM) may be necessary to form larger structures such as spherulites. The peptide concentrations were verified within a 5% error using a Nanodrop ND1000 spectrophotometer (see Experimental Section). The peptide solutions were incubated at 37 °C for 14 days, and samples produced using all incubation conditions were repeated three times. Using both TFA and HFIP separately before incubation resulted in all samples forming amyloid fibrils. However, only one repeat from two different conditions formed spherulites: 150 μM at 37 °C and 300 μM, at 37 °C. However, using HFIP alone to monomerize the protein resulted in all repeat samples incubated at 37 °C with 0.1 M NaCl at peptide concentrations of either 150 or 300 μM forming spherulites. It therefore seems that indeed avoiding the pI of the protein is vitally important for reproducible spherulite formation.
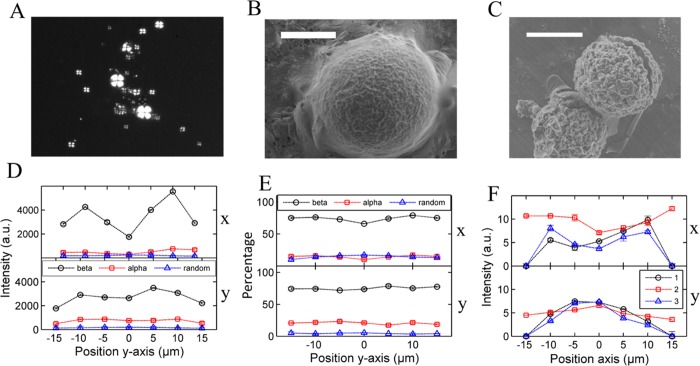
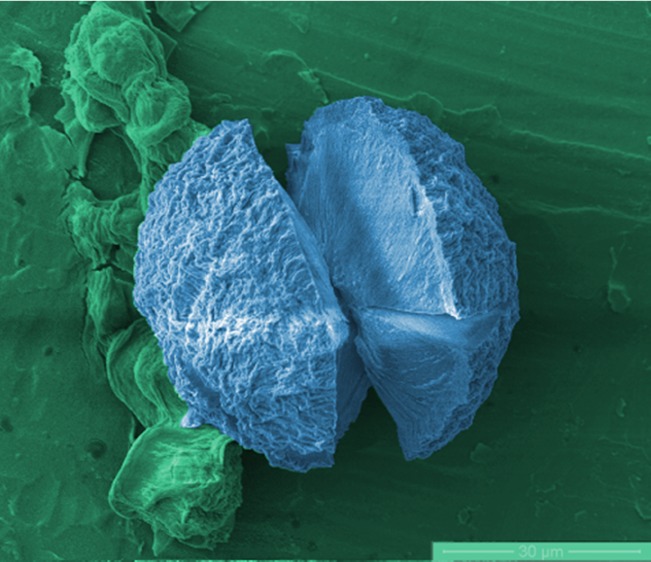
The characteristic Maltese cross pattern of Aβ40 spherulites (Figure 3A) resembled those readily observed in insulin and BLG;6,31 both have four quadrants of birefringence and a non-birefringent core. This second protocol prevents the peptide from crossing its pI (where the peptide is least soluble), and hence random aggregation may be slower, giving spherulites the time to form. This result indicates the importance of the preparation method and the environmental conditions when forming spherulitic amyloid aggregates. To gain further structural information, ESEM was used to image the spherulites formed in Aβ40 at a concentration of 300 μM, as shown in Figure 3B. For comparison, the spherulites formed in insulin are shown in Figure 3C. It is quite clear from the images that spherulites from both insulin and Aβ40 have similar morphologies from an external perspective.
Figure 3.
(A) Optical image of Aβ40 spherulites formed using hexafluoroisopropanol (HFIP) followed by 14 days of incubation at 37 °C with 0.1 M NaCl at a peptide concentration of 300 μM. (B) Environmental scanning electron microscopy (ESEM) images of a spherulite formed in the Aβ40 peptide at 300 μM, incubated for 14 days at 37 °C. The scale bar is 20 μm (C) Insulin spherulite formed at 1 mM, incubated for 24 h at pH 2. The scale bar is 20 μm (D) Raman peak intensities for the β-sheet, α-helix, and random coil across an Aβ40 spherulite in the x-direction (top) and y-direction (bottom), where 0 μm marks the center. (E) Percentage of the β-sheet, α-helix, and random coil across an Aβ40 spherulite in the x-direction (top) and y-direction (bottom), where 0 μm marks the center. (F) Polarized β-sheet intensity across an amyloid β (Aβ) spherulite to indicate the orientation of the fibrils in the x and y paths.
As with that of the insulin spherulites, Raman spectroscopy of the Aβ40 spherulite reveals a decrease in the absolute β-sheet intensity at the core (Figure 3D) and a slight increase in the random coil structure with the spherulite containing approximately 80% of a β-sheet structure, which drops to 70% at the core (Figure 3E). The random coil structure increases from approximately 5 to 10%. However, in contrast to insulin, the results suggest that there is more β-sheet content in an Aβ spherulite than in an insulin spherulite; the β-sheet percentage also decreases less across the Aβ spherulite, a result reproduced in two other spherulites (Figure S2E,F).
The polarized Raman secondary structure signals (Figure 3F) show the same trend as insulin spherulites. Hence, the proposed structure for Aβ40 spherulites is the same. Aβ40 spherulites are structurally more similar to those of insulin than BLG, having a core region with more random coils and less β-sheets, and radially orientated fibrils outside the core.
3. Experimental Section
3.1. Sample Preparation
BLG (product no. L0130) and bovine insulin (product no. I5500) were obtained from Sigma-Aldrich. These and all other chemicals were of analytical grade or better and used without further purification. Solutions were prepared by dissolving the required amount of protein powder in distilled and deionized water to give concentrations of 5.6 and 40 mg/mL for insulin and BLG, respectively. The pH was adjusted with HCl for insulin (pH 2) and BLG (pH 1.6). The samples were incubated for 24 h at 70 °C to form spherulites.
Aβ was purchased from Bachem (Bubendorf, Switzerland) as lyophilized trifluoroacetate salts (Batch H1194). Two methods were used to form spherulites, the first method eliminated pre-existing aggregates using TFA, followed by HFIP to disrupt hydrophobic interactions. The second method used HFIP alone, creating a monomeric Aβ stock solution, after which a 50 mM sodium phosphate buffer (pH 7.4) with the addition of 0.1 M NaCl was prepared and added to the Aβ40 powder peptide in 1.5 mL Eppendorfs to give final concentrations of 30, 50, 150, and 300 μM. All aliquot concentrations were confirmed by analyzing 1 μL of aliquots using a Nanodrop ND1000 spectrophotometer that is specifically designed for accurately measuring the concentrations of very small volumes. The Aβ40 peptide samples were then incubated at 37 °C for 14 days.
3.2. Raman Spectroscopy
A Renishaw System 2000 Raman spectrometer coupled to an Olympus microscope was used to monitor the secondary structure within the spherulites. A HeNe laser (λ = 633 nm), focused to a spot size of approximately 2 μm on the surface of the sample using a 50× objective lens, was used to excite Raman scattering from the spherulites. To obtain a clear Raman spectra, an exposure time of 100 s (five accumulations of 20 s) was used. This procedure was repeated three times at each location to give an average spectrum and the corresponding standard error of the mean (SEM). In sections where the orientation of fibrils was examined, the monochromatic light from the laser was polarized using a polarizing filter, and the analyzer was in place to detect only the polarized light from the scattered radiation.
3.3. Analysis of Data
Raman spectra were collected in the range of 1550–1750 cm–1, three times at each location. These spectra were fitted using mixed Gaussian and Lorentzian functions (see Figure 1B for an example of fitting) using the Wire 3.1 software (Renishaw, UK). For the secondary structure analysis, the absolute intensities were normalized by the sum of all secondary structures to account for the variation in density and to give a percentage contribution of each secondary structure, where 100% is the total contribution of all three secondary structures (see eqs 1 and 2 for the normalization procedure). Only in the case of analyzing the orientation of fibrils, using polarized Raman, were the β-sheet intensities normalized with the random coil peaks because the α-helix signal was weak. Spherulites were analyzed individually because they vary in size as do their cores.
3.4. ESEM
All spherulite imaging was carried out on an Electroscan ESEM 2010 microscope (FEI UK). Aliquots of the Aβ40, BLG, and insulin solutions were pipetted onto copper stubs, which were then placed inside of the chamber of the microscope. Using a Peltier-chip-controlled device located under the sample holder, samples were allowed to equilibrate to 2 °C and were kept sufficiently cold so that that liquid did not fully evaporate when the chamber pressure was pumped down. Drops of water were placed around the sample to maintain hydration while pumping down. The chamber was flooded with water vapor repeatedly until a pressure of less than 5 Torr was achieved.5
4. Conclusions
Insulin and Aβ40 spherulites are dominated at their edges by β-sheet structures, whose contribution decreases at the center but is not negligible. The random coil intensity increases at the center, suggesting that a combination of amorphous material and unaligned amyloid fibrils make up the core. The fibrils were shown to radiate out from the center in insulin and Aβ proteins. The increased disordered material and reduced amyloid fibril content at the core may explain the absence of birefringence in this region using cross-polarized light.
The insulin and Aβ40 spherulite formation may require amorphous protein to act as a nucleus from which subsequent fibril growth can occur. These fibrils at the center may collapse as the spherulite continues to grow, which accounts for the β-sheet presence at the core albeit reduced in amount. The collapsing fibrils lose their orientation showing no birefringence between crossed polarizers. The orientation of the fibrils was shown to be radial from the center because of the increasing and decreasing β-sheet intensities in different orientations depending on the alignment with the direction of polarization.
BLG showed a dramatic increase in the β-sheet intensity at the center, which may be due to this region of the spherulite being denser as when normalized, the β-sheet percentage remained almost constant. The amyloid fibrils in BLG did not have the same orientation as in insulin or Aβ40. Instead of radial symmetry, the observed β-sheet intensities imply a spiral of fibrils from the core to the edge.
Forming spherulites in Aβ40 is possible and reproducible under various conditions including those simulating physiological conditions in vitro. However, to accomplish this, we found that using HFIP alone before incubation is necessary for the formation of spherulites consistently in Aβ40, when incubated at high concentrations (150 or 300 μM) for 14 days at 37 °C. Passing through the pI seems to hinder reproducibility.
Although the proteins reported to form spherulites look macroscopically similar under polarized light, their internal structures may differ, and hence, their formation and packing cannot be assumed to be the same.
Acknowledgments
This work was supported by EPSRC. We thank Dr. Adam M Corrigan for helpful discussions.
Glossary
Abbreviations
- BLG
β-lactoglobulin
- Aβ40
amyloid β (1-40)
- (ProIAPP1-48)
human proislet amyloid polypeptide
- ESEM
environmental scanning electron microscopy
Supporting Information Available
The Supporting Information is available free of charge on the ACS Publications website at DOI: 10.1021/acsomega.6b00208.
Schematic representation of the laser penetration through the insulin spherulite; further spectra from insulin, BLG, and Aβ40 spherulites; proposed internal structure of the spherulites; and the direction of polarization relative to the amyloid fibrils (PDF)
Author Present Address
§ Medical Research Council Laboratory for Molecular Cell Biology and Department of Cell and Developmental Biology, University College London, Gower Street, WC1E 6BT, London, United Kingdom (D.C.).
Author Present Address
∥ College of Engineering, Mathematics and Physical Sciences, Harrison Building, University of Exeter, North Park Road, EX4 4QF, Exeter, United Kingdom (S.J.E.).
Author Contributions
All authors have given approval to the final version of the manuscript.
The authors declare no competing financial interest.
Supplementary Material
References
- Sunde M.; Serpell L. C.; Bartlam M.; Fraser P. E.; Pepys M. B.; Blake C. C. F. J. Mol. Biol. 1997, 273, 729–739. 10.1006/jmbi.1997.1348. [DOI] [PubMed] [Google Scholar]
- Collins S. R.; Douglass A.; Vale R. D.; Weissman J. S. PLoS Biol. 2004, 2, e321 10.1371/journal.pbio.0020321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin L.-W.; Claborn K. A.; Kurimoto M.; Geday M. A.; Maezawa I.; Sohraby F.; Estrada M.; Kaminsky W.; Kahr B. Proc. Natl. Acad. Sci. U.S.A. 2003, 100, 15294–15298. 10.1073/pnas.2534647100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Exley C.; House E.; Collingwood J. F.; Davidson M. R.; Cannon D.; Donald A. M. J. Alzheimer’s Dis. 2010, 20, 1159–1165. 10.3233/JAD-2010-091630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- House E.; Mold M.; Collingwood J.; Baldwin A.; Goodwin S.; Exley C. J. Alzheimer’s Dis. 2009, 18, 811–817. 10.3233/JAD-2009-1235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krebs M. R. H.; MacPhee C. E.; Miller A. F.; Dunlop I. E.; Dobson C. M.; Donald A. M. Proc. Natl. Acad. Sci. U.S.A. 2004, 101, 14420–14424. 10.1073/pnas.0405933101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Domike K. R.; Donald A. M. Biomacromolecules 2007, 8, 3930–3937. 10.1021/bm7009224. [DOI] [PubMed] [Google Scholar]
- Cannon D.; Donald A. M. Soft Matter 2013, 9, 2852–2857. 10.1039/c2sm26946e. [DOI] [Google Scholar]
- Exley C.; House E.; Patel T.; Wu L.; Fraser P. E. J. Inorg. Biochem. 2010, 104, 1125–1129. 10.1016/j.jinorgbio.2010.05.012. [DOI] [PubMed] [Google Scholar]
- Bassett D. C. J. Macromol. Sci., Part B: Phys. 2003, 42, 227–256. 10.1081/MB-120017116. [DOI] [Google Scholar]
- Bisault J.; Ryschenkow G.; Faivre G. J. Cryst. Growth 1991, 110, 889. 10.1016/0022-0248(91)90647-n. [DOI] [Google Scholar]
- Morse H. W.; Warren C. H.; Donnay J. D. H. Am. J. Sci. 1932, 23, 421. 10.2475/ajs.s5-23.137.421. [DOI] [Google Scholar]
- Sperling L. H.Introduction to Physical Polymer Science, 4th ed.; Spherulitic Morphology; Wiley: New York, 2006, chapter 6.5.1. [Google Scholar]
- Hutter J. L.; Bechhoefer J. Phys. Rev. E: Stat. Phys., Plasmas, Fluids, Relat. Interdiscip. Top. 1999, 59, 4342. 10.1103/PhysRevE.59.4342. [DOI] [Google Scholar]
- Lavrentovich O. D.; Terentjev E. M. Sov. Phys. JETP 1986, 64, 1237–1244. [Google Scholar]
- Marchant J. L.; Blanshard J. M. V. Starch/Staerke 1980, 32, 223–226. 10.1002/star.19800320703. [DOI] [Google Scholar]
- Snow A. D.; Sekiguchi R.; Nochlin D.; Fraser P.; Kimata K.; Mizutani A.; Arai M.; Schreier W. A.; Morgan D. G. Neuron 1994, 12, 219–234. 10.1016/0896-6273(94)90165-1. [DOI] [PubMed] [Google Scholar]
- Manuelidis L.; Fritch W.; Xi Y.-G. Science 1997, 277, 94–98. 10.1126/science.277.5322.94. [DOI] [PubMed] [Google Scholar]
- Acebo E.; Mayorga M.; Val-Bernal J. F. Pathology 1999, 31, 8–11. 10.1080/003130299105449. [DOI] [PubMed] [Google Scholar]
- Taniyama H.; Kitamura A.; Kagawa Y.; Hirayama K.; Yoshino T.; Kamiya S. Vet. Pathol. 2000, 37, 104–107. 10.1354/vp.37-1-104. [DOI] [PubMed] [Google Scholar]
- House E.; Jones K.; Exley C. J. Alzeihmers Dis. 2011, 25, 43–46. 10.3233/JAD-2011-110071. [DOI] [PubMed] [Google Scholar]
- Exley C.; Mold M.; Shardlow E.; Shuker B.; Ikpe B.; Wu L.; Fraser P. E. J. Diabetes Res. Clin. Metab. 2012, 1, 3. 10.7243/2050-0866-1-3. [DOI] [Google Scholar]
- Westlind-Danielsson A.; Arnerup G. Biochemistry 2001, 40, 14736–14743. 10.1021/bi010375c. [DOI] [PubMed] [Google Scholar]
- Yagi H.; Ban T.; Morigaki K.; Naiki H.; Goto Y. Biochemistry 2007, 46, 15009–15017. 10.1021/bi701842n. [DOI] [PubMed] [Google Scholar]
- Teplow D. B. Methods Enzymol. 2006, 413, 20–33. 10.1016/S0076-6879(06)13002-5. [DOI] [PubMed] [Google Scholar]
- Murphy R. M.; Pallitto M. M. J. Struct. Biol. 2000, 130, 109–122. 10.1006/jsbi.2000.4253. [DOI] [PubMed] [Google Scholar]
- Morse H. W.; Donnay J. D. H. Am. J. Sci. 1932, 23, 440. 10.2475/ajs.s5-23.137.440. [DOI] [Google Scholar]
- Gránásy L.; Pusztai T.; Tegze G.; Warren J. A.; Douglas J. F. Phys. Rev. E: Stat., Nonlinear, Soft Matter Phys. 2005, 72, 011605. 10.1103/physreve.72.011605. [DOI] [PubMed] [Google Scholar]
- Smith M. I.; Foderà V.; Sharp J. S.; Roberts C. J.; Donald A. M. Colloids Surf., B 2012, 89, 216–222. 10.1016/j.colsurfb.2011.09.018. [DOI] [PubMed] [Google Scholar]
- Domike K. R.; Hardin E.; Armstead D. N.; Donald A. M. Eur. Phys. J. E: Soft Matter Biol. Phys. 2009, 29, 173–182. 10.1140/epje/i2009-10465-y. [DOI] [PubMed] [Google Scholar]
- Domike K. R.; Donald A. M. Int. J. Biol. Macromol. 2009, 44, 301–310. 10.1016/j.ijbiomac.2008.12.014. [DOI] [PubMed] [Google Scholar]
- Krebs M. R. H.; Domike K. R.; Cannon D.; Donald A. M. Faraday Discuss. 2008, 139, 265–274. 10.1039/b715879c. [DOI] [PubMed] [Google Scholar]
- Hou L.; Shao H.; Zhang Y.; Li H.; Menon N. K.; Neuhaus E. B.; Brewer J. M.; Byeon I.-J. L.; Ray D. G.; Vitek M. P.; Iwashita T.; Makula R. A.; Przybyla A. B.; Zagorski M. G. J. Am. Chem. Soc. 2004, 126, 1992–2005. 10.1021/ja036813f. [DOI] [PubMed] [Google Scholar]
- Ortiz C.; Zhang D.; Ribbe A. E.; Xie Y.; Ben-Amotz D. Biophys. Chem. 2007, 128, 150–155. 10.1016/j.bpc.2007.03.012. [DOI] [PubMed] [Google Scholar]
- Zagorski M. G.; Barrow C. J. Biochemistry 1992, 31, 5621–5631. 10.1021/bi00139a028. [DOI] [PubMed] [Google Scholar]
- Zandomeneghi G.; Krebs M. R. H.; McCammon M. G.; Fändrich M. Protein Sci. 2004, 13, 3314–3321. 10.1110/ps.041024904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin S.-Y.; Chu H.-L. Int. J. Biol. Macromol. 2003, 32, 173–177. 10.1016/S0141-8130(03)00051-5. [DOI] [PubMed] [Google Scholar]
- Maiti N. C.; Apetri M. M.; Zagorski M. G.; Carey P. R.; Anderson V. E. J. Am. Chem. Soc. 2004, 126, 2399–2408. 10.1021/ja0356176. [DOI] [PubMed] [Google Scholar]
- Eker F.; Griebenow K.; Schweitzer-Stenner R. Biochemistry 2004, 43, 6893–6898. 10.1021/bi049542+. [DOI] [PubMed] [Google Scholar]
- Chen P.; Shen A.; Zhao W.; Baek S.-J.; Yuan H.; Hu J. Appl. Opt. 2009, 48, 4743–4748. 10.1364/AO.48.004743. [DOI] [PubMed] [Google Scholar]
- Ikeda S.; Li-Chan E. C. Y. Food Hydrocolloids 2004, 18, 489–498. 10.1016/j.foodhyd.2003.07.003. [DOI] [Google Scholar]
- Nonaka M.; Li-Chan E.; Naiki S. J. Agric. Food Chem. 1993, 41, 1178–1181. 10.1021/jf00032a002. [DOI] [Google Scholar]
- Buchanan L. E.; Carr J. K.; Fluitt A. M.; Hoganson A. J.; Moran S. D.; de Pablo J. J.; Skinner J. L.; Zanni M. T. Proc. Natl. Acad. Sci. U.S.A. 2014, 16, 5796–5801. 10.1073/pnas.1401587111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stroud J. C.; Liu C.; Teng P. K.; Eisenberg D. Proc. Natl. Acad. Sci. U.S.A. 2012, 20, 7717–7722. 10.1073/pnas.1203193109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lippert J. L.; Tyminski D.; Desmeules P. J. J. Am. Chem. Soc. 1976, 98, 7075–7080. 10.1021/ja00438a057. [DOI] [PubMed] [Google Scholar]
- Sereda V.; Sawaya M. R.; Lednev I. K. J. Am. Chem. Soc. 2015, 137, 11312–11320. 10.1021/jacs.5b07535. [DOI] [PubMed] [Google Scholar]
- Gatos K. G.; Minogianni C.; Galiotis C. Macromolecules 2007, 40, 786–789. 10.1021/ma0623284. [DOI] [Google Scholar]
- Dong J.; Wan Z.; Popov M.; Carey P. R.; Weiss M. A. J. Mol. Biol. 2003, 330, 431–442. 10.1016/S0022-2836(03)00536-9. [DOI] [PubMed] [Google Scholar]
- Blundell T. L.; Cutfield J. F.; Cutfield S. M.; Dodson E. J.; Dodson G. G.; Hodgkin D. C.; Mercola D. A.; Vijayan M. Nature 1971, 231, 506–511. 10.1038/231506a0. [DOI] [PubMed] [Google Scholar]
- Rogers S. S.; Krebs M. R. H.; Bromley E. H. C.; van der Linden E.; Donald A. M. Biophys. J. 2006, 90, 1043–1054. 10.1529/biophysj.105.072660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gommans H. H.; Alldredge J. W.; Tashiro H.; Park J.; Magnuson J.; Rinzler A. G. J. Appl. Phys. 2000, 88, 2509. 10.1063/1.1287128. [DOI] [Google Scholar]
- Lefèvre T.; Rousseau M.-E.; Pézolet M. Appl. Spectrosc. 2006, 60, 841–846. 10.1366/000370206778062039. [DOI] [PubMed] [Google Scholar]
- Adamcik J.; Mezzenga R. Soft Matter 2011, 7, 5437–5443. 10.1039/c1sm05382e. [DOI] [Google Scholar]
- Adamcik J.; Jung J.-M.; Flakowski J.; Rios P. D. L.; Dietler G.; Mezzenga R. Nat. Nanotechnol. 2010, 5, 423–428. 10.1038/nnano.2010.59. [DOI] [PubMed] [Google Scholar]
- Lara C.; Adamcik J.; Jordens S.; Mezzenga R. Biomacromolecules 2011, 12, 1868–1875. 10.1021/bm200216u. [DOI] [PubMed] [Google Scholar]
- Jordens S.; Riley E. E.; Usov I.; Olmsted P. D.; Mezzenga R. ACS Nano 2014, 8, 11071–11079. 10.1021/nn504249x. [DOI] [PubMed] [Google Scholar]
- Bolisetty S.; Adamcik J.; Mezzenga R. Soft Matter 2011, 7, 493–499. 10.1039/C0SM00502A. [DOI] [Google Scholar]
- Poulin P.; Stark H.; Lubensky T. C.; Weitz D. A. Science 1997, 275, 1770–1773. 10.1126/science.275.5307.1770. [DOI] [PubMed] [Google Scholar]
- Hortschansky P.; Schroeckh V.; Christopeit T.; Zandomeneghi G.; Fändrich M. Protein Sci. 2005, 14, 1753–1759. 10.1110/ps.041266605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brorsson A.C.; Kumita J. R.; MacLeod I.; Bolognesi B.; Speretta E.; Luheshi L. M.; Knowles T. P.; Dobson C. M.; Crowther D. C. Front. Biosci. 2010, 15, 373–396. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.