Abstract
ALIS are large, transient, cytosolic aggregates that serve as storage compartments for ubiquitin-tagged defective ribosomal products. We determined the importance of the protein p62 in the formation of ALIS and demonstrated that two domains of p62—PB1 and UBA—are essential for ALIS assembly. Those two major binding domains of p62, also known as sequestosome 1, were shown to play a critical role in the formation of autophagosomes or cytoplasmic aggregates. Specifically, the PB1 domain is essential for self-oligomerization, and the UBA domain allows p62 to bind to polyubiquitin chains or ubiquitinated proteins. After stimulation of RAW 264.7 macrophages with lipopolysaccharide, we observed a significant decrease in the number of cells with ALIS. Importantly, cells overexpressing either a PB1 mutant or UBA-deleted p62 construct also exhibited a substantially diminished number of cells containing ALIS. Since both p62 and ubiquitin are found in ALIS, we evaluated the dynamics of YFP-tagged p62 in ALIS. In contrast to the findings of a previous study that evaluated GFP-tagged ubiquitin motility in ALIS, we determined that YFP-tagged p62 has very limited mobility. Lastly, we determined that GST–tagged full-length p62 binds to Lys-63-linked polyubiquitin chains but not to Lys-48-linked chains. Overall, our findings provide insight on the essential role that p62, particularly its PB1 and UBA domains, has in the formation of ALIS.
Keywords: ALIS, PB1, UBA, p62, FRAP, ubiquitin
INTRODUCTION
Living cells are equipped with numerous mechanisms that help ensure proper protein synthesis and folding. If the quality checkpoints of those processes are perturbed, eventual protein misfolding can be toxic to the cell [1]. One mechanism by which the cell can eradicate misfolded proteins, otherwise known as DRiPs [2], is by ubiquitination and shuttling of DRiPs into cytosolic aggregates [1]. The cytosolic aggregates store ubiquitinated proteins that are targeted for degradation through autophagy or the proteasome [1]. One type of cytosolic aggregate that forms after maturation of dendritic cells, macrophage cell lines, or upon stimulation of toll-like receptor 4 is called ALIS [3,5]. ALIS can also form in nonimmune cells [6]. Unlike aggresomes, ALIS do not localize to the pericentriolar area, are not encased in vimentin, and are unaffected by actin or microtubule cytoskeletal disruptors [2]. Moreover, ALIS are transient; they can form within 4 hours of stimulation and begin to dissipate 20–44 hours later [5,6].
P62 (also known as sequestosome 1) is a 440-amino-acid protein that contains several binding motifs that allow the protein to serve as a scaffold for signaling and ubiquitin binding [7]. Endogenous p62 colocalizes with the ubiquitin found in ALIS when macrophages are stimulated with LPS [8]. Fujita et al. identified the role of p62 in ALIS occurrence after RAW cells depleted of p62 failed to show ALIS formation after LPS stimulation [8]. Subsequently, others showed that the N-terminal PB1 and C-terminal UBA domains of p62 are key components in the formation of cytosolic aggregates or autophagic machinery [9,10,11]. The PB1 domain of p62 mediates self-oligomerization, serves as an interaction surface for other PB1-containing proteins [12], associates with various kinases, such as atypical protein kinase C, and is an important factor in a number of signaling cascades [7,9]. Mutations in the PB1 domain of p62 prevent oligomerization of p62 [12] and ultimately inhibit recruitment to the site where autophagic machinery form following serum starvation [11].
The UBA domain allows p62 to associate with ubiquitin and ubiquitin-tagged proteins, resulting in the formation of cytosolic aggregates [9,10]. Such formation of p62 cytosolic aggregates implies that they have a role in cell survival, as p62 interacts with ubiquitin-linked cargos and shuttles them to the proteasome or lysosome [10,13]. In NIH3T3 cells, expression of either p62 in which the UBA domain was deleted or p62 with an I431A UBA mutant, which disrupts folding of the UBA domain, altered the localization pattern; rather than the typical cytoplasmic aggregates seen in cells with wildtype p62, the localization pattern of UBA-mutant or UBA-null p62 was diffuse [9]. Currently, the roles of the PB1 and UBA domains of p62 in ALIS formation are not known.
Because ALIS are motile and can interact with each other, fusion/interaction events among neighboring ALIS regulate their aggregation process [3]. Previous studies examined the mobility of GFP-tagged ubiquitin in ALIS by using FRAP. Those studies revealed that the fluorescence intensity of GFP-tagged ubiquitin found in ALIS recovered almost completely, which emphasizes the dynamic nature of ubiquitin in ALIS assembly and/or the recruitment of ubiquitin-tagged proteins to ALIS [3]. Both p62 and ubiquitin are present in ALIS, however, the dynamics of p62 in ALIS have not been investigated.
In the present study, we used confocal immunofluorescence microscopy to investigate the role of p62 and its PB1 and UBA domains in ALIS assembly. We also utilized FRAP to examine the dynamics of p62 in ALIS. Finally, we assessed the capacity of full-length p62 to bind to either Lys-63-linked or Lys-48-linked polyubiquitin chains.
METHODS
Antibodies and Reagents
The following primary antibodies were obtained commercially: guinea pig anti-p62 (GP62-C; Progen, Heidelberg, Germany); mouse anti-monoubiquitin and polyubiquitin conjugates (BML-PW8810; Enzo Life Sciences, Farmingdale, NY); rabbit anti-p62 (Cell Signaling Technologies, Danvers, MA); mouse anti-β-tubulin (E7; Developmental Studies Hybridoma Bank, University of Iowa, Iowa City, IA); and anti-mouse ubiquitin (P4D1; Covance, Princeton, NJ). The following secondary antibodies used for immunoblotting were obtained from Thermo Fisher Scientific (Carlsbad, CA): goat anti-rabbit IgG horseradish peroxidase (HRP) and goat anti-mouse IgG HRP. Secondary goat anti-guinea pig IgG HRP was purchased from Santa Cruz Biotechnology (Santa Cruz, CA). The following secondary antibodies used for immunofluorescence were obtained from Thermo Fisher Scientific: Alexa-Fluor 568 (goat anti-rabbit IgG, goat anti-guinea pig IgG, and goat anti-mouse IgG), Alexa-Fluor 488 (goat anti-rabbit IgG and goat anti-mouse IgG), and Alexa-Fluor 633 (goat anti-mouse IgG).
Reagents were procured from the following vendors: nocodazole, LPS, 2-mercaptoethanol, and puromycin from Sigma (St. Louis, MO); pGEX-6P-1 vector and PreScission Protease from GE Healthcare Life Sciences (Waukesha, WI); PEI (linear, molecular weight 25,000) from Polysciences, and ProLong® Gold Antifade Reagent with DAPI from Thermo Fisher Scientific; GST-Spr-19S/55a from Enzo Life Sciences; DMEM/High Glucose from HyClone (Lafayette, CO); FBS from Gemini Bio-Products (West Sacramento, CA); cell culture plates from Sarstedt (Numbrecht, Germany); polybrene, Immobilon-P membrane, and Luminata™ Forte Enhanced Chemiluminescence (ECL) reagent from Millipore (Billerica, MA); protease inhibitor cocktail from Roche (Indianapolis, IN); and nitrocellulose membrane (0.45 μm) from Thermo Fisher Scientific.
Ubiquitin Binding Assays
Pull-down experiments were performed with 4 μg of GST-fusion proteins immobilized on glutathione beads incubated with 0.7 μg of polyubiquitin chains, incubated for 2 h in THG buffer (0.5% Triton X-100, 50 mM HEPES, pH 7.5, 150 mM NaCl, and 10% glycerol), and washed three times. Bound proteins were processed as described previously [14].
Cell Culture
RAW 264.7 cells were used from passages 7–15 and maintained in DMEM/High Glucose medium supplemented with 10% FBS. To induce ALIS formation, cells were treated with 10 ng/ml of LPS for the indicated times.
Generation of p62 Knockdown Cells Using CRISPR-Cas9 Genome Editing
We knocked down p62 in RAW 264.7 cells by using LentiCRISPRv2 (Addgene plasmid 52961), a gift from Feng Zhang [15]. Guide RNA sequences were created using Massachusetts Institute of Technology’s CRISPR design tool (see http://www.crispr.mit.edu). An oligonucleotide that targeted p62 (5′-ATGGTGGGCGATGTTCCCGC-3′) was annealed and cloned into LentiCRISPRv2. Lentivirus was prepared by transfecting into HEK293T cells equal amounts of vesicular stomatitis virus G, pCMV delta R8.2 (Addgene plasmid 12263), a gift from Didier Trono, and LentiCRISPRv2 backbone containing the p62 guide RNA. Lentivirus production was performed as previously described [15].
Mammalian and Bacterial Expression Vectors
We used PCR to generate a pGW1-Myc p62 mammalian expression vector that amplifies human p62 cDNA, and then we cloned it in-frame into the EcoRI site of pGW1-Myc vector. To generate the p62-D69A mutant, we used QuikChange site-directed mutagenesis to change aspartic acid residue 69 to alanine in pGW1-Myc-p62, according to the manufacturer’s protocol (Agilent Technologies, Santa Clara, CA). To generate the pGW1-Myc-p62ΔUBA construct, a fragment of human p62 cDNA (residues 1–386) was amplified by PCR and cloned in-frame into the EcoRI site of the pGW1-Myc vector. The full length of p62 cDNA was cloned into the EcoRI site of the pGEX-6P-1 vector, and this plasmid was named pGEX-p62.
Transfection
Control and p62-knockdown RAW 264.7 cells were seeded at 80% confluence onto 35-cm dishes in complete medium without puromycin. Cells were transfected with 1.5 μg of either empty Myc, wildtype Myc-p62, Myc-p62 D69A, Myc-p62 K7A, or Myc-p62 UBA-deletion (ΔUBA) vector by using PEI.
Western Blot
Cells were lysed and scraped into 1X Laemmli sample buffer supplemented with 1 mM NaVO4 and 5% 2-mercaptoethanol. Lysates were processed as described previously [14].
Immunofluorescence and Confocal Microscopy
RAW 264.7 cells were grown on untreated glass coverslips, fixed with 4% paraformaldehyde for 25 minutes, and permeabilized using 0.15% Triton X-100 for 15 minutes. Cells were then stained, mounted and imaged as described previously [14].
Photobleaching
The fluorescent recovery after photobleaching (FRAP) experiments were based on our published methods [16,17] with minor modifications. Briefly, transfected RAW 264.7 cells were cultured in 35-mm glass-bottom microwell dishes (MatTek; Ashland, MA) and treated with 10 ng/ml LPS for 3–5 hours. The culture medium was removed, and Tyrode’s buffer containing 10 μg/ml nocodazole was added. The cells were incubated at 4°C for 15 min to minimize ALIS movement prior to imaging. YFP-p62 was excited at λ = 514 nm and detected at λ = 527 nm. The digital zoom was set at 3.0×, and the image area acquired before and after photobleaching was set at 150 × 150 pixels to minimize the time needed to acquire images. After obtaining 5 prebleach images, a circular area with a diameter of 10 pixels was photobleached with 300 iterations at 100% laser power (9.0 A) and 100% transmission. The recovery of fluorescence intensity in the bleached area was recorded at the same laser power but using 5% transmission, and images were obtained every 30 sec for 1,050 sec (17.5 min). Fluorescence intensities of the prebleached, bleached, control (i.e., a region of fluorescence outside of the bleached region), and background regions of interest were measured with the Fiji software plugin [18] for ImageJ, and results were analyzed with Microsoft Excel software. Small amounts of drift were corrected by using the Fiji registration plug-in; experiments in which drift could not be corrected were rejected.
RESULTS
Knockdown of p62 Leads to Diminished ALIS Formation
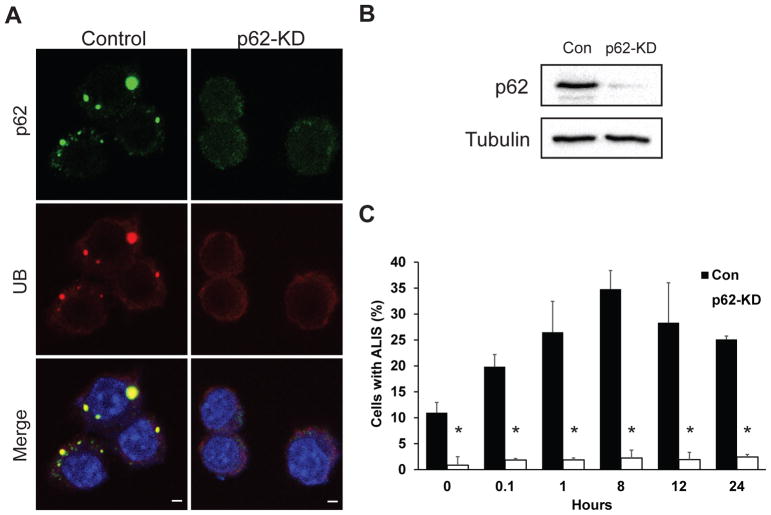
We examined the distribution of endogenous p62 and its role in ALIS formation after LPS stimulation of the murine macrophage cell line RAW 264.7. In RAW cells expressing endogenous p62 (control) that were stimulated with LPS, p62 colocalized with ubiquitin in ALIS (Fig. 1A; top row). We also examined ALIS formation in cells depleted of p62 (p62-knockdown). Utilizing CRISPR/Cas9 genome editing, we knocked down p62 gene expression in RAW 264.7 macrophages and observed a significant reduction of p62 protein (Fig. 1A; bottom row and Fig. 1B). Densitometric analysis of the western blot revealed 94% knock down of p62 compared to control cells. P62-knockdown cells were treated with LPS and immunofluorescently stained for endogenous p62 and ubiquitin to observe the effect that depleted p62 has on ALIS occurrence. At all time points, we observed significantly lower numbers of cells with ALIS compared to control cells. For example, puncta indicative of ALIS appeared in less than 3% of p62-knockdown cells compared to the 35% in control cells at the 8 hour time point (Fig. 1A; bottom row), indicating that the knockdown of p62 significantly diminished ALIS occurrence.
Fig. 1. p62 colocalizes with ALIS and is important for its formation.
A. Parental RAW 264.7 macrophages and macrophages depleted of p62 were treated with 10 ng/mL of LPS for 1 h, fixed, and immunostained for p62 (green) and conjugated ubiquitin (red). Images are representative of at least three independent experiments. Scale bars: 5 μm. B. RAW 264.7 cells were depleted of p62 via CRISPR/Cas9 genome editing, and knockdown was verified through immunoblotting using anti-p62 (top panel) or anti-tubulin (lower panel) antibodies. C. Control (Con) or p62-knockdown (KD) cells were treated with 10 ng/mL LPS for the indicated times. Cells were then scored for the presence of ALIS as described in Materials and Methods. ALIS were defined as aggregates containing conjugated ubiquitin. Error bars represent mean ± SEM from at least three separate experiments with more than 100 cells scored per experiment. *p < 0.05.
We also determined the presence of ALIS in cells expressing p62 and in p62-knockdown cells over 24 hours. Our study showed the presence of ALIS in 20% of control RAW cells as early as 5 minutes after LPS stimulation (Fig. 1C). Furthermore, ALIS occurrence peaked at 8 hours after LPS stimulation (~35% cells with ALIS) and started to dissipate shortly thereafter (Fig. 1C). Depletion of p62 resulted in significantly lower occurrence of ALIS during the 24-hour period (~3%; Fig. 1C). Overall, these findings suggest that endogenous p62 plays a critical role in the formation of ALIS in RAW cells.
p62 Dynamics in ALIS
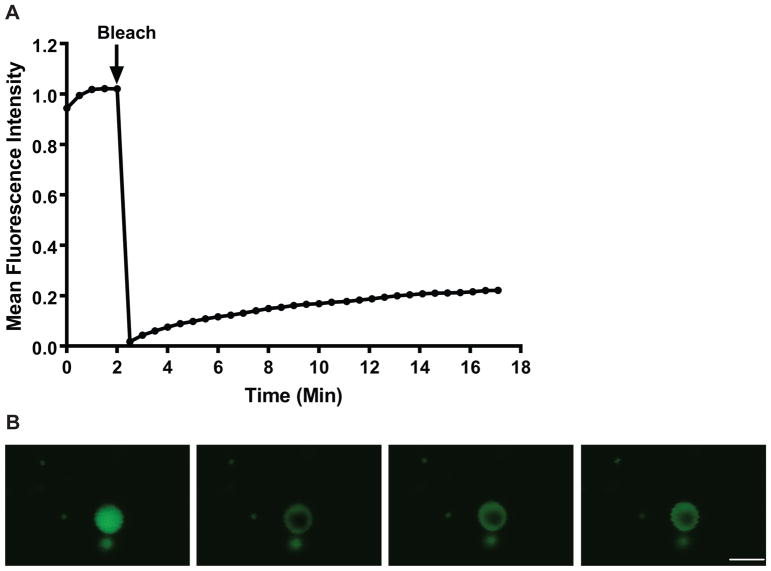
FRAP was used to examine the dynamics of p62 in ALIS after 3–5 hours of treatment with LPS. Images were obtained in a control region (i.e., a region of fluorescence outside the bleached region) before and after photobleaching to measure the change in fluorescence (if any) due to repeated imaging and the effects of photobleaching the ALIS on non-aggresome-like structures. We did not observe a change in fluorescence in the control region in any of the cells examined (data not shown). In addition, images of a background region (i.e., a region of little to no fluorescence outside the bleached region) were obtained before and after photobleaching to measure the change in fluorescence (if any) due to repeated imaging. We did not observe a change in the fluorescence in the background region (data not shown). To determine the recovery of fluorescence in the bleached area over time, images were obtained before and after photobleaching (Fig. 2, Supplemental Video 1). FRAP analysis of YFP-p62 revealed that the fluorescence within bleached ALIS recovered slowly, with an average half-time of recovery (t1/2) = 361.69 ± 20.69 sec (6.03 ± 0.35 min). Moreover, YFP-p62 was not a very mobile protein, as the average mobile fraction (Mf) = 26.08 ± 1.45%. No difference was detected in the t1/2 and Mf values in experiments in which cells were imaged for 17.5 min compared to experiments in which the cells were imaged for 35 min (data not shown).
Fig. 2. The use of FRAP to study the dynamics of YFP-p62 in ALIS.
A. FRAP data from 10 independent experiments resulted in approximately 26% fluorescence recovery 17 minutes after bleaching. B. Representative images of ALIS at prebleach (1.5 min) and postbleach (0, 6, and 14min). Scale bar: 5 μm and is valid for all panels.
The PB1 and UBA Domains of p62 Are Necessary for ALIS Assembly
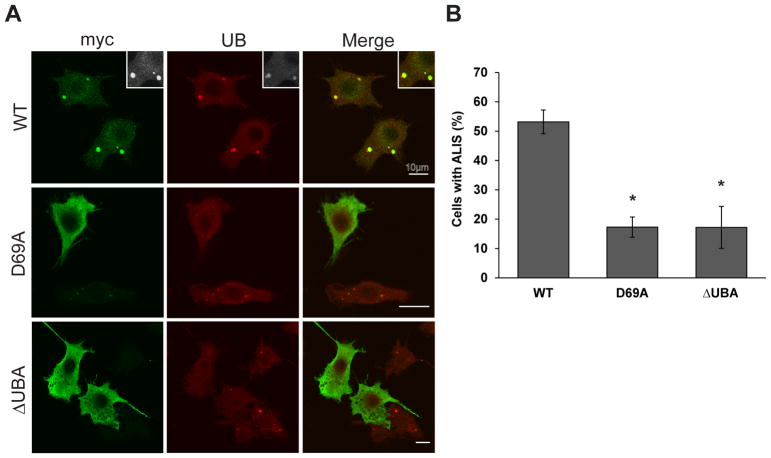
It is known that p62 mediates the formation of cytoplasmic bodies and autophagosomes through its PB1 and UBA domains [10,12]. However, it is currently not known whether those domains are also necessary for the formation of ALIS. To determine the importance of p62’s PB1 and UBA domains in ALIS assembly, parental RAW 264.7 cells were transfected with one of the following Myc-tagged constructs: wildtype (WT) p62, p62 PB1 mutant (K7A or D69A), or ΔUBA-p62. Confocal microscopic analyses of transfected RAW cells stained with anti-Myc and anti-ubiquitin antibodies revealed that overexpressed WT p62 localized with endogenous ubiquitin found in ALIS after 12 hours of LPS treatment (Fig. 2A; top row). However, none of the mutant p62 proteins (K7A, D69A, and ΔUBA) colocalized with endogenous ubiquitin in puncta (Fig. 2A), which indicated an attenuated capacity for cells transfected with mutant p62 constructs to form ALIS after LPS stimulation. We quantified the percentage of cells containing ALIS and found that in RAW macrophages overexpressing the p62 mutant constructs (K7A, D69A, or ΔUBA), there were significantly fewer (70–80% reduction; p value < 0.05) cells with ALIS than in overexpressed WT p62-transfected cells (Fig. 2B). Together, these results indicate that ALIS occurrence requires the PB1 and UBA domains of p62.
P62 Preferentially Binds to K63-linked Ubiquitin Chains
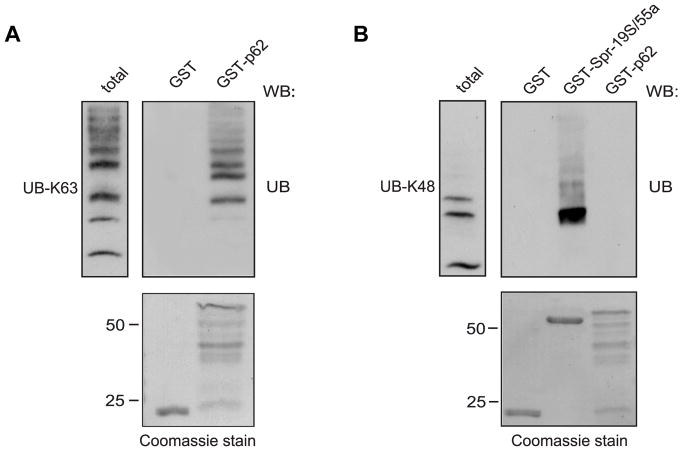
Many studies have reported that p62 can bind to either Lys-48-linked or Lys-63-linked polyubiquitin chains [19,20]. Additionally, many mono- and polyubiquitinated proteins are found in dendritic ALIS formations [3]. Therefore, we tested whether GST-tagged full-length p62 can bind directly to either Lys-63-linked or Lys-48-linked ubiquitin chains. As shown in Fig. 4A, GST-tagged full-length p62 bound directly to Lys-63-linked ubiquitin chains but not Lys-48-linked polyubiquitin. However, the positive control, 19S proteasomal subunit, Rpn 10/S5a, showed robust binding to Lys-48-linked ubiquitin chains (Fig. 4B) indicating that, overall, p62 binds preferentially to Lys-63-linked ubiquitin chains.
Fig. 4. p62 preferentially binds to Lys-63-linked polyubiquitin.
A. GST-p62 and GST alone were incubated with Lys-63-linked ubiquitin chains (UB-K63) After incubation and washes, the bound proteins were eluted with Laemmli buffer, resolved by SDS-PAGE, and western blotted (WB) with anti-ubiquitin antibody. Coomassie blue-stained gel (bottom) showed equivalent amounts of fusion proteins used for pull-down experiments. B. GST-p62, GST-Spr-19S/55a, and GST alone were incubated with Lys-48-linked ubiquitin chains (UB-K48). After incubation and washes, the bound proteins were processed and evaluated as described above.
DISCUSSION
The findings of this study showed the importance of p62 expression in ALIS formation. RAW macrophages stimulated with LPS exhibited a significant increase in the number of cells with ALIS, whereas in cells depleted of p62, considerably fewer cells contained ALIS. This finding agrees with a previous study which determined that ALIS formation is a p62-dependent event through toll-like receptor 4 pathway activation [8]. An important aspect of our study revealed the importance of the PB1 and UBA domains of p62 in ALIS formation. Among the cells overexpressing myc-tagged p62 PB1 mutant, D69A or K7A (data not shown), or myc-tagged p62 ΔUBA, there were significantly fewer cells containing ALIS compared with cells overexpressing WT p62. Previous studies emphasized the importance of the PB1 and UBA domains of p62 in cytoplasmic body and autophagosome formation [9,11]. Itakura and Mizushima examined the effect of p62 PB1 mutants (K7A or D69A) on the formation of autophagosomes [11]. Mutations in the PB1 domain prevented p62 localization at the autophagosome formation site in mitochondria [11]. Similar observations revealed that PB1 mutations and a deleted UBA domain in p62 reduced the number of cytoplasmic bodies in NIH3T3 fibroblasts [9]. Overall, expression of p62 with intact PB1 and UBA domains is critical for autophagosomes, cytoplasmic bodies, and ALIS formation.
In the present study, we also evaluated the dynamics of p62 in ALIS by utilizing FRAP. RAW 264.7 cells transfected with YFP-tagged p62 recovered approximately 26% of their fluorescence after photobleaching, which suggests that p62 is not a very mobile protein. The limited mobility of p62 can be attributed to its overall structural organization. Ciuffa et al. showed that p62 assembled into long, flexible, helical filaments through its PB1 domain [21]. However, interactions with polyubiquitin chains disrupted the p62 scaffolds and truncated the filament length [21]. Therefore, it is possible that the assembly and local disruption of p62 filaments account for the limited mobility of p62 found in ALIS. Others observed the dynamic nature of ubiquitin found in ALIS and found 90% fluorescence recovery of GFP-tagged ubiquitin after photobleaching [3]. Overall, in ALIS, ubiquitin is substantially more mobile [3] than p62 (this study), which is anticipated, as ubiquitin is much smaller (approximately 7 kDa) than p62 (approximately 62 kDa). Ubiquitin can function as a monomer or in conjugated forms, such as polyubiquitin chains, or as a tag on other proteins [22]. Therefore, ubiquitin’s small size indicates that it has the propensity to move more freely than the p62 polymers. Thus, it is possible that the dynamics of p62 are limited due to its large size and ability to form long, flexible polymers.
Our studies also demonstrated an interaction between GST-tagged full-length p62 and Lys-63-linked polyubiquitin chains that is consistent with p62’s roles in autophagy and receptor trafficking. In contrast, GST-tagged full-length p62 did not interact with Lys-48-linked polyubiquitin chains. These results indicate preferential binding of full-length p62 to Lys-63-linked polyubiquitin chains. Although previous studies demonstrated that p62’s UBA domain bound to either Lys-63-linked or Lys-48-linked polyubiquitin chains [19,20], those studies utilized constructs that contained only the UBA domain of p62 rather than full-length p62. Therefore, it is possible that the UBA domain might have different binding propensities than UBA in the context of full-length p62 to Lys-63-linked or Lys-48-linked polyubiquitin chains. Alternatively, the binding preference of full-length p62 to Lys-63-linked chains over Lys-48-linked chains might be due to the folding conformations of each type of polyubiquitin. Lys-63-linked ubiquitin chains have an extended and open conformation [22]. In contrast, Lys-48-linked ubiquitin chains form a closed conformational shape, rendering the structure largely unable to interact with other protein partners [22]. The closed folding conformation of Lys-48-linked chains impedes binding with full-length p62. Therefore, full-length p62 can easily interact with Lys-63-linked polyubiquitin due to its extended and open conformational structure.
In summary, we demonstrated the essential role of p62 in the formation of ALIS. Mutations in p62’s PB1 domain and deletion of the UBA domain significantly decreased the number of cells containing ALIS. Hence, the PB1 and UBA domains of p62 are critical for ALIS assembly. We also investigated the dynamics of p62 in ALIS and found that p62 is not a mobile protein. Finally, we observed preferential binding of p62 to Lys-63-linked polyubiquitin, likely due to its open and extended conformational shape compared with the closed conformational shape of Lys-48-linked ubiquitin chains.
Supplementary Material
The video is a recording of a typical ALIS prior to and after photobleaching of YFP-tagged p62. YFP-p62 fluorescence is slow to recover after the photobleaching, and p62 is not a very mobile protein. For this experiment, Mf = 25.62%. Scale bar: 5 μm.
Fig. 3. The PB1 and UBA domains of p62 are critical for ALIS assembly.
A. Parental RAW 264.7 cells were transfected with myc-tagged constructs (p62 WT, p62 D69A, or p62 ΔUBA) and treated with 10 ng/mL of LPS for 12 h. Transfected cells were fixed and immunostained for myc (green) and conjugated ubiquitin (red) Images are representative of at least three experiments. Scale bar: 10 μm. B. Quantification of ALIS in RAW 264.7 macrophages transfected with indicated vectors. Cells were scored for the presence of ALIS as described in Materials and Methods. ALIS were defined as aggregates containing conjugated ubiquitin. Error bars represent mean ± SEM from at least three separate experiments with more than 100 cells scored per experiment. *p < 0.05.
HIGHLIGHTS.
P62’s PB1 and UBA domains are required for the induction of ALIS.
YFP-tagged p62 is not a very mobile protein in ALIS.
P62 binds preferentially to K63-linked ubiquitin chains.
Acknowledgments
We thank Sabrina Imam and Dr. Edward M. Campbell (Department of Microbiology and Immunology, Loyola University Chicago) for their assistance in generating our p62 knockdown strain of RAW 264.7 cells via CRISPR/Cas9 genome editing. We also thank Dr. William Simmons (Department of Molecular Pharmacology and Therapeutics, Loyola University Chicago) for advice and guidance in preparation of the manuscript. This work was supported by the NIH (1R01NS073967-01A1).
Abbreviations
- ALIS
aggresome-like induced structures
- DALIS
dendritic cell aggresome-like induced structures
- DRiPs
defective ribosomal products
- GFP
green fluorescent protein
- GST
glutathione S-transferase
- FRAP
fluorescence recovery after photobleaching
- LPS
lipopolysaccharide
- PB1
Phox and Bem1
- PEI
polyethylenimine
- UBA
Ubiquitin association
- YFP
yellow fluorescent protein
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Garcia-Mata R, Gao YS, Sztul E. Hassles with taking out the garbage: aggravating aggresomes. Traffic. 2002;3:388–396. doi: 10.1034/j.1600-0854.2002.30602.x. [DOI] [PubMed] [Google Scholar]
- 2.Schubert U, Anton LC, Gibbs J, Norbury CC, Yewdell JW, Bennink JR. Rapid degradation of a large fraction of newly synthesized proteins by proteasomes. Nature. 2000;404:770–774. doi: 10.1038/35008096. [DOI] [PubMed] [Google Scholar]
- 3.Lelouard H, Ferrand V, Marguet D, Bania J, Camosseto V, David A, Gatti E, Pierre P. Dendritic cell aggresome-like induced structures are dedicated areas for ubiquitination and storage of newly synthesized defective proteins. J Cell Biol. 2004;164:667–675. doi: 10.1083/jcb.200312073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Canadien V, Tan T, Zilber R, Szeto J, Perrin AJ, Brumell JH. Cutting edge: microbial products elicit formation of dendritic cell aggresome-like induced structures in macrophages. J Immunol. 2005;174:2471–2475. doi: 10.4049/jimmunol.174.5.2471. [DOI] [PubMed] [Google Scholar]
- 5.Szeto J, Kaniuk NA, Canadien V, Nisman R, Mizushima N, Yoshimori T, Bazett-Jones DP, Brumell JH. ALIS are stress-induced protein storage compartments for substrates of the proteasome and autophagy. Autophagy. 2006;2:189–199. doi: 10.4161/auto.2731. [DOI] [PubMed] [Google Scholar]
- 6.Lelouard H, Gatti E, Cappello F, Gresser O, Camosseto V, Pierre P. Transient aggregation of ubiquitinated proteins during dendritic cell maturation. Nature. 2002;417:177–182. doi: 10.1038/417177a. [DOI] [PubMed] [Google Scholar]
- 7.Bitto A, Lerner CA, Nacarelli T, Crowe E, Torres C, Sell C. p62/SQSTM1 at the interface of aging, autophagy, and disease. Age (Dordr) 2014 doi: 10.1007/s11357-014-9626-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fujita K, Maeda D, Xiao Q, Srinivasula SM. Nrf2-mediated induction of p62 controls Toll-like receptor-4-driven aggresome-like induced structure formation and autophagic degradation. Proc Natl Acad Sci U S A. 2011;108:1427–1432. doi: 10.1073/pnas.1014156108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bjorkoy G, Lamark T, Brech A, Outzen H, Perander M, Overvatn A, Stenmark H, Johansen T. p62/SQSTM1 forms protein aggregates degraded by autophagy and has a protective effect on huntingtin-induced cell death. J Cell Biol. 2005;171:603–614. doi: 10.1083/jcb.200507002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Paine MG, Babu JR, Seibenhener ML, Wooten MW. Evidence for p62 aggregate formation: role in cell survival. FEBS Lett. 2005;579:5029–5034. doi: 10.1016/j.febslet.2005.08.010. [DOI] [PubMed] [Google Scholar]
- 11.Itakura E, Mizushima N. p62 Targeting to the autophagosome formation site requires self-oligomerization but not LC3 binding. J Cell Biol. 2011;192:17–27. doi: 10.1083/jcb.201009067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lamark T, Perander M, Outzen H, Kristiansen K, Overvatn A, Michaelsen E, Bjorkoy G, Johansen T. Interaction codes within the family of mammalian Phox and Bem1p domain-containing proteins. J Biol Chem. 2003;278:34568–34581. doi: 10.1074/jbc.M303221200. [DOI] [PubMed] [Google Scholar]
- 13.Seibenhener ML, Babu JR, Geetha T, Wong HC, Krishna NR, Wooten MW. Sequestosome 1/p62 is a polyubiquitin chain binding protein involved in ubiquitin proteasome degradation. Mol Cell Biol. 2004;24:8055–8068. doi: 10.1128/MCB.24.18.8055-8068.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]; 15 Long J, Gallagher TR, Cavey JR, Sheppard PW, Ralston SH, Layfield R, Searle MS. Ubiquitin recognition by the ubiquitin-associated domain of p62 involves a novel conformational switch. J Biol Chem. 2008;283:5427–5440. doi: 10.1074/jbc.M704973200. [DOI] [PubMed] [Google Scholar]
- 14.Karlsson AB, Washington J, Dimitrova V, Hooper C, Shekhtman A, Bakowska JC. The role of spartin and its novel ubiquitin binding region in DALIS occurrence. Mol Biol Cell. 2014;25:1355–1365. doi: 10.1091/mbc.E13-11-0705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sanjana NE, Shalem O, Zhang F. Improved vectors and genome-wide libraries for CRISPR screening. Nat Methods. 2014;11:783–784. doi: 10.1038/nmeth.3047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bakowska JC, Jupille H, Fatheddin P, Puertollano R, Blackstone C. Troyer syndrome protein spartin is mono-ubiquitinated and functions in EGF receptor trafficking. Mol Biol Cell. 2007;18:1683–1692. doi: 10.1091/mbc.E06-09-0833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.O’Connor C, Pertel T, Gray S, Robia SL, Bakowska JC, Luban J, Campbell EM. p62/sequestosome-1 associates with and sustains the expression of retroviral restriction factor TRIM5alpha. J Virol. 2010;84:5997–6006. doi: 10.1128/JVI.02412-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Schindelin J, Arganda-Carreras I, Frise E, Kaynig V, Longair M, Pietzsch T, Preibisch S, Rueden C, Saalfeld S, Schmid B, Tinevez JY, White DJ, Hartenstein V, Eliceiri K, Tomancak P, Cardona A. Fiji: an open-source platform for biological-image analysis. Nat Methods. 2012;9:676–682. doi: 10.1038/nmeth.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Isogai S, Morimoto D, Arita K, Unzai S, Tenno T, Hasegawa J, Sou YS, Komatsu M, Tanaka K, Shirakawa M, Tochio H. Crystal structure of the ubiquitin-associated (UBA) domain of p62 and its interaction with ubiquitin. J Biol Chem. 2011;286:31864–31874. doi: 10.1074/jbc.M111.259630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ciani B, Layfield R, Cavey JR, Sheppard PW, Searle MS. Structure of the ubiquitin-associated domain of p62 (SQSTM1) and implications for mutations that cause Paget’s disease of bone. J Biol Chem. 2003;278:37409–37412. doi: 10.1074/jbc.M307416200. [DOI] [PubMed] [Google Scholar]
- 21.Ciuffa R, Lamark T, Tarafder AK, Guesdon A, Rybina S, Hagen WJ, Johansen T, Sachse C. The selective autophagy receptor p62 forms a flexible filamentous helical scaffold. Cell Rep. 2015;11:748–758. doi: 10.1016/j.celrep.2015.03.062. [DOI] [PubMed] [Google Scholar]
- 22.Alfano C, Faggiano S, Pastore A. The Ball and Chain of Polyubiquitin Structures. Trends Biochem Sci. 2016;41:371–385. doi: 10.1016/j.tibs.2016.01.006. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
The video is a recording of a typical ALIS prior to and after photobleaching of YFP-tagged p62. YFP-p62 fluorescence is slow to recover after the photobleaching, and p62 is not a very mobile protein. For this experiment, Mf = 25.62%. Scale bar: 5 μm.