Summary
Liquid granules rich in intrinsically disordered proteins and RNA play key roles in critical cellular functions such as RNA processing and translation. Many details of the mechanism via which this occurs still remain to be elucidated. Motivated by the lacuna in the field, and by the prospects of developing de novo artificial granules that provide extrinsic control of translation, we report a bottom-up approach to engineer ribonucleoprotein granules comprised of a recombinant RNA-binding IDP that exhibits phase behavior in water. We developed a kinetic model to illustrate that these granules inhibit translation through reversible or irreversible sequestration of mRNA. Within monodisperse droplets capable of transcription and translation, we experimentally demonstrate temporal inhibition of translation, by using designer IDPs that exhibit tunable phase behavior. This work lays the foundation for developing artificial granules that promise to further our mechanistic understanding of their naturally occurring counterparts.
Graphical Abstract

eTOC
Simon et al have developed artificial protocells containing engineered IDP-RNA rich liquid granules that dynamically regulate translation. A kinetic model describing the mechanism for regulating translation is provided. This development provides a new platform for interrogating and better understanding the properties and functions of the myriad granules found in nature.
Introduction
The interior of living cells is a complex milieu containing thousands of proteins, nucleic acids, lipids, and small molecules (Theillet et al., 2014). These molecules are often organized spatially and temporally within the cytoplasm and nucleus through the use of distinct membraneless organelles (Luby-Phelps, 2013; Toretsky and Wright, 2014). Membraneless organelles, also referred to as granules, are localized protein-and-RNA-rich liquid compartments lacking an enclosing membrane that assemble and disassemble in response to environmental cues, fuse with one another upon contact, and rapidly exchange their contents with the surroundings to regulate various biochemical processes (Anderson and Kedersha, 2006; Brangwynne, 2011; Brangwynne et al., 2009). It is often the case that the underlying drivers comprising membraneless ribonucleoprotein (RNP) granules are intrinsically disordered proteins (IDPs) that phase separate in solution to form distinct liquid compartments (Brangwynne et al., 2015; Uversky et al., 2015). An increasing body of work exists, however, demonstrating that granule formation is influenced by macromolecules other than disordered proteins, including RNA-RNA interactions, folded RNA-binding domain interactions with RNA, and intrinsically disordered regions (IDRs) within folded RNA helicases or RNA-binding proteins (Polymenidou, 2018; Protter and Parker, 2016; Protter et al., 2018; Wang et al., 2018).
A number of studies have investigated both the molecular driving forces behind granule formation and their resultant biophysical properties. From these, a rich picture emerges: within the cell, IDRs interact with RNA molecules and together undergo phase separation that is driven by IDR-IDR interactions to form biomolecular condensates that have viscosity values similar to dense solutions like honey (Brangwynne et al., 2009; Elbaum-Garfinkle et al., 2015). These viscous networks with specified mesh sizes selectively partion macromolecules of interest, inhibiting transport into and out of the granules (Nott et al., 2016; Wei et al., 2017). Changing the sequence, and therefore the macromolecular interactions, of the IDRs and RNAs can modulate granule viscosity and mesh size, which endows them with an ability to dynamically modify the inclusion/exclusion of molecules as needed (Guo and Shorter, 2015; Wei et al., 2017; Zhang et al., 2015). Recent work by Protter and coworkers investigated RNP granule assembly in eukaryotic cells to understand the role of various macromolecules in their formation. Their findings indicate that RNP granule assembly is driven by highly specific interactions (protein-protein and protein-RNA), which can be enhanced by IDRs in proteins capable of promiscuous or weak interactions based on small structural elements that become effective at high local concentrations (Protter et al., 2018). The authors suggest this assembly mechanism may be shared by other macromolecular complexes rich in IDRs.
To provide consistency in nomenclature herein, we adopted a naming convention summarized in reviews by the Rosen and Brangwynne groups (Banani et al., 2017; Shin and Brangwynne, 2017). Here, we use the term ‘biomolecular condensates’ to describe all micron scale compartments lacking an enclosing membrane that serve to concentrate one or more proteins, nucleic acids, or both. Biomolecular condensates can refer to intracellular compartments, as well as extracellular biomaterials and bodies/puncta. This uniformity in naming convention applies to all such organelles comprised of proteins and RNAs involved in myriad functions, from regulating signal transduction to RNA metabolism (Banani et al., 2017). Therefore, throughout this manuscript when utilizing the term ‘biomolecular condensates’ we are broadly referring to membraneless organelles that are rich in proteins and nucleic acids.
While our understanding of the physicochemical drivers of macromolecular phase separation underlying organelle formation and the biophysical properties of biomolecular condensates is growing rapidly, our understanding of their function is relatively limited. Studies have shown that biomolecular condensates play a key role in various intracellular functions such as RNA storage, processing, and decay and the inhibition of translation (Anderson and Kedersha, 2009; Buchan, 2014; Sheu-Gruttadauria and MacRae, 2018; Su et al., 2016). These functions appear to be regulated by spatiotemporal granule assembly/disassembly and the biophysical properties of the granules. Recent work by the Drummond lab has further uncovered the role of biomolecular condensates inside cells; for example, Riback et al showed that poly (A)-binding (Pab1) proteins in yeast release mRNA under stress conditions and upon mRNA release, phase separate into liquid and hydrogel granules. The released chaperone mRNAs permit higher levels of translation that produce disperse Pab1 quinary assemblies, which then re-bind the mRNAs and complete an autoregulatory circuit to maintain cell fitness (Riback et al., 2017). Such studies uncovering the complex functions and regulatory dynamics of biomolecular condensates are continuously growing and providing us with a clearer understanding of their diverse roles in biology. However, the complexity and difficulties in probing biomolecular condensates in vivo makes it challenging to study the link between biomolecular condensate formation and properties with their function. Several reports have sought to develop model biomolecular condensates within protocells using macromolecular crowding agents (Deng and Huck, 2017; Long et al., 2005; Sokolova et al., 2013; Strulson et al., 2012); however, these studies have only explored the use of synthetic polymers and/or short polyelectrolytes to drive macromolecular crowding and, as such, are insufficient in providing an understanding of the relationship between material properties of condensates and their biochemical functions. Because these studies used abiotic components, they are incompatible with our twin goals of: (1) exploiting liquid-liquid phase separation of IDPs to control function within artifical protocells, and (2) accomplishing the first step in our longer term goal of reintroducing these components back into live cells to create genetically encoded artificial liquid granules that carry out new functions that are programmed into their phase behavior and thereby provide an additional level of extrinsic control of cellular behavior.
Motivated by this goal, we present a de novo engineered model system of genetically engineered biomolecular condensates that inhibit translation within droplet-based protocells. We show that artificial IDPs that exhibit lower critical solution temperature (LCST) phase behavior in a physiologically relevant regime can indeed interact with and reversibly enrich messenger RNA (mRNA) into messenger RNP (mRNP) granules within monodisperse droplet microenvironments. Guided by a simple kinetic model, we show experimentally in a cell-free translation system that these mRNP granules or biomecular condensates suppress the translation of superfolder green fluorescent protein (sfGFP) within microdroplet-based protocells by sequestering the sfGFP mRNA.
Results
IDP Design
We chose elastin-like polypeptides (ELPs) as our artificial IDPs to create engineered mRNP granules or biomolecular condensates. ELPs are composed of the repetitive sequence Val-Pro-Gly-Xaa-Gly, where Xaa is any amino acid except proline (Meyer and Chilkoti, 1999). Importantly, ELPs exhibit LCST phase behavior in water; below their cloud point temperature (TCP) they are soluble while above their TCP they undergo simple coacervation to form a protein-rich coacervate (Meyer and Chilkoti, 2004). We selected ELPs as our granule-forming IDPs for a number of reasons: (1) their LCST phase behavior has been extensively characterized and can therefore be easily tailored (McDaniel et al., 2013; Meyer and Chilkoti, 2004); (2) they can be produced in large quantities by recombinant expression in E. coli (MacEwan and Chilkoti, 2010); (3) they can be easily fused to other proteins and peptides, and these ELP fusion proteins exhibit temperature triggered coacervation (Christensen et al., 2009; Christensen et al., 2013; Hassouneh et al., 2010; Meyer and Chilkoti, 1999); (4) their chemical diversity and hence function can be expanded upon at the translational level by incorporation of unnatural amino acids (Amiram et al., 2015; Costa Simone et al., 2018) and by post-translational modifications (Luginbuhl Kelli et al., 2017; Mozhdehi et al., 2018); and (5) we have previously investigated their use as building blocks of model liquid granules within water-in-oil microdroplets (Simon et al., 2017). Here again, we chose to study engineered biomolecular condensates within water-inoil emulsion droplets, as they provide a discrete, well defined and highly uniform microenvironment that is ideal to observe the dynamics of biomolecular condensate formation under precisely controlled in vitro conditions that are challenging to control in vivo.
One hallmark of ELPs is their inert nature, meaning that they exhibit minimal interactions with other biomacromolecules (MacEwan and Chilkoti, 2010). To confer ELPs with the capacity to bind RNA, we designed an ELP (40 repeats of Val-Pro-Gly-Val-Gly) with a C-terminal RNA-binding domain (RBD) in the form of an RGG box from the C-terminus of PGL-1, a constituent protein of P granules in Caenorhabditis elegans (Kawasaki et al., 1998) (Figure 1A, Table 1, Supplementary Table 1). This RBD is enriched in Arg-Gly-Gly repeats, which are known to bind RNA through a variety of intramolecular interactions (Chong et al., 2018; Thandapani et al., 2013).
Figure 1. Engineered RNA-binding intrinsically disordered proteins.
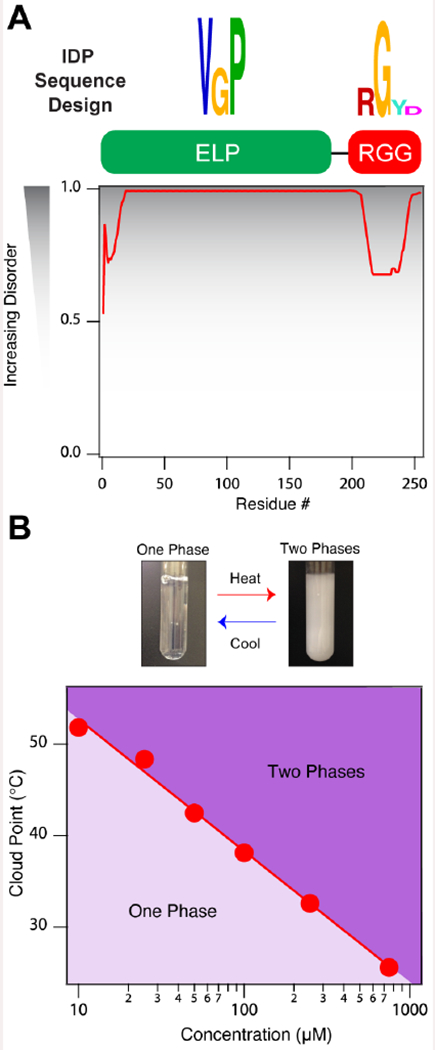
(A) Artificial RNA-binding IDP sequence design and PONDR results of ELP1-RBD indicate that the chains have a high propensity for disorder in solution. (B) Cloud point temperature (TCP) as a function of concentration for ELP-RBD illustrating ELP1-RBDs have LCST phase behavior in 1x PBS. Solid line represents the best logarithmic fit. Related to Supplementary Table 1 and Supplementary Figures 1 and 2.
Table 1.
Sequence of the IDPs and RNA molecules used in this study.
| Molecule | Sequence |
|---|---|
| ELP1 | (VPGVG)40 |
| ELP1-RBD | (VPGVG)40-GGGGGRGGYGGGDRGGRGGYGGDRGGRGGYGGGDRGGRGGYGGDRGRGGYGGRGGRGGGY |
| ELP2-RBD | (VPGSG)40-GGGGGRGGYGGGDRGGRGGYGGDRGGRGGYGGGDRGGRGGYGGDRGRGGYGGRGGRGGGY |
| ELP3-RBD | (VPGAG)80-GGGGGRGGYGGGDRGGRGGYGGDRGGRGGYGGGDRGGRGGYGGDRGRGGYGGRGGRGGGY |
| ssRNA1 | UAGCCGAAAAUGCAC |
This fusion protein, ELP1-RBD, was expressed in Escherichia coli cells in large quantities and purified (see Supplementary Figure 1A–B) in a facile manner using inverse transition cycling, a non-chromatographic method for purifying ELPs (Meyer and Chilkoti, 1999). In aqueous solution, ELP1-RBD is predicted to be disordered using the Predictor of Naturally Disordered Regions or PONDR algorithm (Li et al., 1999; Peng et al., 2006; Romero et al., 1997; Xue et al., 2010) (Figure 1A, Supplementary Figure 1C) and exhibits LCST phase behavior similar to that of canonical ELPs (Figure 1B, Supplementary Figure 2), which suggests that its phase behavior and thermally-triggered phase separation process is driven by the ELP segment.
IDP Binding and Phase Separation Behavior
After expressing and purifying ELP1-RBD, we evaluated its ability to bind single stranded RNA (ssRNA) and whether or not this interaction affects its phase behavior. In a gel retardation assay, ELP1-RBD completely retards the migration of enhanced green fluorescent protein (eGFP) mRNA (see Supplementary Table 2 for the mRNA sequence) on an agarose gel (Figure 2A), indicating a strong interaction between the ELP1-RBD and mRNA chains. In contrast, ELP without the C-termini RBD, termed ELP1, does not retard the migration of eGFP mRNA, illustrating that the RBD sequence is solely responsible for mediating the protein-mRNA interactions.
Figure 2. Recombinant ELP-RBDs efficiently bind and coacervate single stranded RNA.
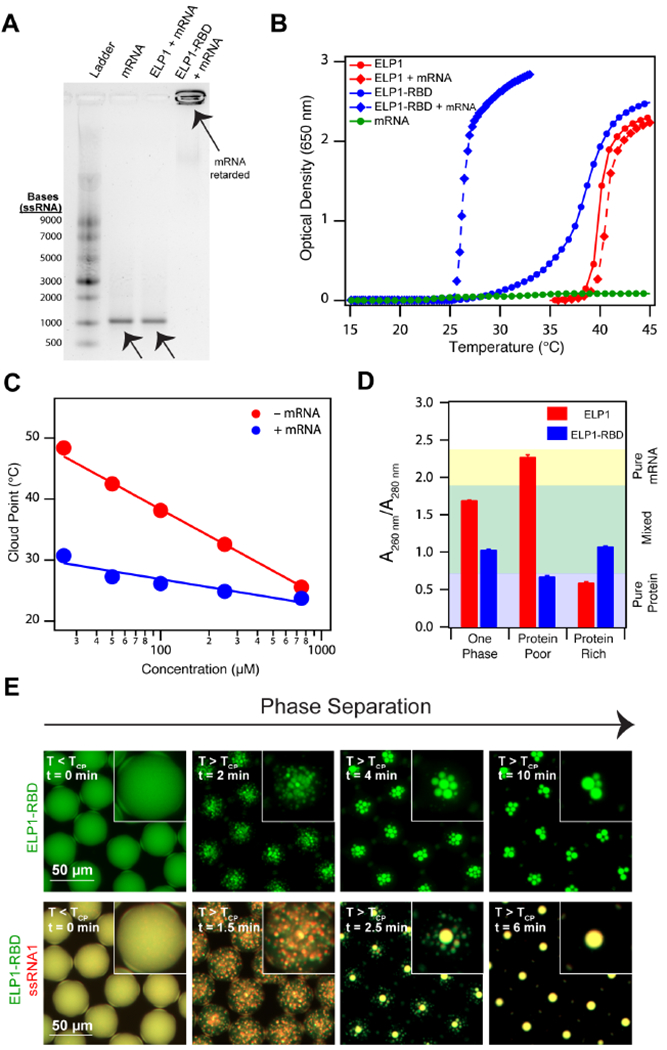
(A) 1% agarose gel stained with SYBR® Green II illustrating ELP1-RBD binds mRNA, whereas a control ELP lacking the RBD does not. ELP1 and ELP1-RBD concentration is 500 μM. eGFP mRNA concentration is 100 μg ml−1. (B) Optical density as a function of temperature of ELP1 with (red diamonds) and without (red circles) mRNA, ELP1-RBD with (blue diamonds) and without (blue circles) mRNA, and mRNA (green circles) in 1x PBS. Protein concentrations are 100 μM and eGFP concentration is 100 μg ml−1. (C) TCP as a function of concentration for ELP1-RBD (red points) and ELP1-RBD in the presence of mRNA (blue points) in 1x PBS. eGFP mRNA concentration is 100 μg ml−1. The solid lines represent the best logarithmic fit. (D) Nucleic acid quantitation of mixtures of ELP1 + mRNA (red) and ELP1-RBD + mRNA (blue) as a mixed, one phase solution before phase separation (left), a protein-poor phase after phase separation (middle), and a protein-rich phase after phase separation (right) in bulk. Starting ELP1 and ELP1-RBD concentrations are 500 μM and eGFP mRNA concentration is 100 μg ml−1. Error bars represent the SEM (n=3). (E) Fluorescence microscopy images of Alexa Fluor® 488-labeled ELP1-RBD (green)and Alexa Fluor® 594-labeled ssRNA1 (red) in microdroplets demonstrate that ELP1-RBD can co-phase separate with ssRNA upon heating above the TCP. ELP1-RBD concentration is 500 μM and ssRNA1 concentration is 2.5 μM. Related to Supplementary Tables 1 and 2, Supplementary Figures 1 and 2, and Supplementary Movies 1–3.
In temperature-controlled turbidity assays, we observed a marked depression in ELP1-RBD’s concentration-dependent cloud point behavior in the presence of mRNA (Figure 2B, Figure 2C, Supplementary Figure 2). Here, the TCP’s are an estimate of the slope of the lower concentration branch of the binodal curve – the thermodynamic boundary separating the one phase and two phases region on the temperature-concentration phase diagram (Rubinstein and Colby, 2003; Simon et al., 2017). Therefore, the slope of the lower concentration arm of the binodal of ELP1-RBD shifts to lower values in the presence of mRNA, indicating a lower temperature barrier to initiate phase separation. Consistent with our observations in the gel retardation assay, the presence of mRNA does not alter the TCP behavior of ELP1 (see Figure 2B, Figure 2C, and Supplementary Figure 2), further demonstrating the importance of the RBD in mediating ELP-mRNA interactions. It is important to note that the presence of mRNA flattens the slope of the TCP curve of ELP1-RBD (Figure 2C), an effect that is likely due to charge neutralization in the RBD sequence upon binding the mRNA that makes self-interactions between ELP1-RBD chains effectively more attractive (Rubinstein and Colby, 2003). This is further evidenced by static light scattering measurements that indicate ELP1-RBD + mRNA chains result in nanoscale brush-like structures composed of 1 mRNA chain binding on average 10 ELP1-RBD chains (see Table 2, Supplementary Figure 2A–D). Here, we determined the 10:1 ratio of ELP1-RBD:mRNA using a stoichiometric ratio of the MW of the condensate assembly to the MW of the ELP1-RBD and mRNA chains in solution (as determined by static light scattering measurements), assuming interactions only take place between ELP1-RBD and mRNA and not between ELP1-RBD chains. Our results pose an interesting contrast to a recent report mapping the binodal curves of LAF-1, a constituent IDR of Caenorhabditis elegans P granules, in the absence and presence of RNA. There, in the presence of various model ssRNAs, the authors observed a shift only in the higher concentration branch of the binodal curve (Wei et al., 2017).
Table 2. Static light scattering characterization of ELP1-RBD and ELP1-RBD + mRNA at 15 °C.
ELP1-RBD concentration is 500 μM and eGFP mRNA concentration is 100 μg ml−1. Related to Supplementary Figure 2.
| Rg (nm) | Rh (nm) | Rg/Rh | MW (g mol−1) | Nagg | |
|---|---|---|---|---|---|
| ELP1-RBD | 36.7 | 19.3 | 1.9 | 2.7 x 104 | 1 ELP1-RBD |
| ELP1-RBD + mRNA | 33.4 | 33.5 | 1.0 | 5.4 x 105 | 1 mRNA + 10 ELP1-RBD |
A critical step in suppressing translation with biomolecular condensates is the ability to sequester transcripts within IDP-rich organelles. We first investigated the ability of ELP1-RBD to enrich mRNA within its coacervates using a bulk phase separation assay. After heating to a temperature above the TCP of ELP1-RBD in the presence of mRNA, we centrifuged the sample and separated the protein-rich phase from the protein-poor phase. Using nucleic acid quantitation, we observed a mixture of protein and mRNA in the protein-rich phase, but only residual protein in the protein-poor phase (Figure 2D). In contrast, a solution of ELP1 + mRNA that is heated above the TCP of the ELP1 separate into purely protein in the protein-rich phase and purely mRNA in the protein-poor phase (Figure 2D). Together, this demonstrates the ability of ELP1-RBD — and conversely the inability of ELP1 — to enrich mRNA in its coacervates upon heating above the TCP of ELP1-RBD.
To visualize the heat-triggered phase separation and organelle formation of ELP1-RBD, we encapsulated fluorescently-labeled ELP1-RBD within water microdroplets, which we have previously shown do not impact phase behavior of ELPs (Simon et al., 2017). Upon rapidly heating above the TCP of ELP1-RBD, ELP1-RBD undergoes phase separation by spinodal decomposition (Simon et al., 2017) to form liquid organelles (Figure 2E, Supplementary Movie M1). In the presence of model fluorophore-labeled ssRNAs, termed ssRNA1 (see Table 1 for sequence), ELP1-RBD co-phase separates with ssRNA1 into biomolecular condensates (Figure 2E, Supplementary Movie M2, Supplementary Figure 2V). In both instances, cooling the solution below the TCP of the ELP1-RBD/ssRNA1 complex dissolves the granules (Supplementary Movies M1, M2) due to ELP1-RBD chain re-solubilization, demonstrating the reversible nature of the compartments. We replaced ELP1-RBD with ELP1 and observed phase separation of only the ELP1 component upon heating, while ssRNA1 remained soluble within the entire droplet (Supplementary Figure 2U, Supplementary Movie M3), further confirming the ability of ELP1-RBD, but not ELP1, to bind and co-phase separate with ssRNA.
Granule Formation and Inhibition of Translation
To study the effects of mRNA sequestration within granules on translation, we developed a droplet-based cell-free translation protocell system (Figure 3) that does not partition the cell-free expression components present inside the droplet into the organic phase (Supplementary Figure 4L). To guide the experimental analysis, we developed a kinetic model to examine the impact of granule formation on protein translation (see STAR Methods for a detailed description of the model). For the simplest case, we encapsulated in vitro transcription and translation (IVTT) components and plasmids encoding superfolder GFP (sfGFP) within microdroplets to create monodisperse protocells (see Figure 3 for a representative schematic and Figure 4A for images). In the absence of granules, our model predicts a sigmoidal increase in protein concentration over time (Figure 4B inset). The plateau in concentration at later time points is likely due to a depletion in substrate (e.g., nucleoside triphosphates or amino acids), which is consistent with previous reports of GFP production within crowded polymeric microdroplet reactors (Sokolova et al., 2013). Consistent with the model prediction, we observed a sigmoidal increase in fluorescence over time within droplets —that is proportional to sfGFP concentration— that were heated to the optimal temperature (T = 37 °C) for enzyme activity (see Figure 4A–B, Supplementary Movie M4, Supplementary Figure 1D–E).
Figure 3. Engineered ribonucleoprotein granules for inhibiting translation within droplet-based protocells.

(A) Schematic of microfluidic-generated droplets (left) and the inhibition of translation with messenger ribonucleoprotein granules inside the microdroplet protocells (right). Artificial RNA-binding IDPs coacervate mRNA transcripts upon heating and suppress translation, and coacervation is reversed by cooling, leading to resumption of tranlsation.
Figure 4. Engineered mRNP granules suppress sfGFP translation.
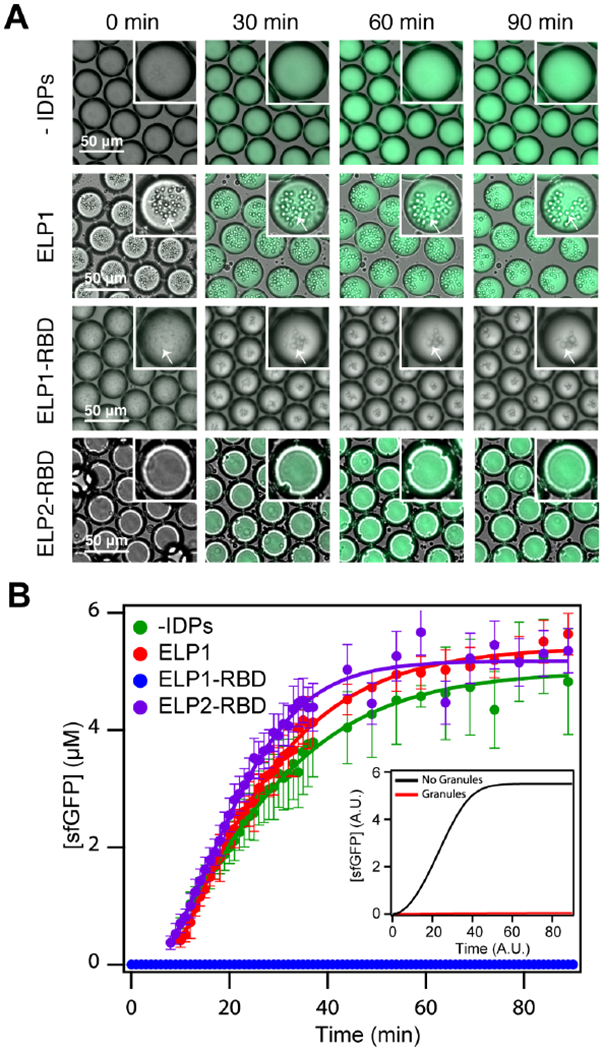
(A) Overlaid brightfield and fluorescence time-lapse images of droplets containing IVTT components and superfolder GFP plasmid (1 μg 50 μL−1) in the absence of an IDP (top row), with ELP1 (second row), with ELP1-RBD (third row), and ELP2-RBD (fourth row). White arrows indicate liquid coacervates. ELP1, ELP1-RBD, and ELP2-RBD concentration is 500 μM, and the temperature is 37°C. (B) Superfolder GFP concentration as a function of time in droplets without an IDP (green points), with ELP1 (red points), with ELP1-RBDs (blue points), and with ELP2-RBDs (purple points). Solid lines represent the best sigmoidal fit and the error bars represent the standard deviation (n ≥ 50 droplets). The inset shows simulated sfGFP concentration in the droplets (black) or the irreversible suppression of translation by granules (red). See STAR Methods for details on model formulation and analysis. Related to Supplementary Table 1, Supplementary Figures 2–3, and Supplementary Movies M4–7.
In the presence of ELP1-RBD granules that strongly sequester mRNA, our model predicts complete suppression in sfGFP fluorescence (see Figure 4B inset). To test this notion, we used ELP1-RBD, which we showed to be highly effective in sequestering mRNA and forming biomolecular condensates at body temperature in the IVTT mixture (Figure 4A, Supplementary Figure 2O–T). Here, the presence of ELP1-RBD granules completely silences sfGFP production within the protocells over the entire time scale of 0-90 min (Figure 4A–B, Supplementary Movie M5). Silencing translation in this fashion suggests efficient binding between the mRNA and ELP1-RBD chains and subsequent mRNA sequestration within the ELP1-RBD granules. It is likely that these protein-mRNA-rich granules are spatially separated from key translational machinery components as we observed little interaction between our disordered proteins and ribosomes or tRNA (Supplementary Figure 4F–I). Thus, our understanding is that spatial sequestration of the mRNA ‘turns off’ translation, though a complete understanding of the biophysical properties of these engineered granules requires further investigation.
The binding and co-phase separation of mRNA with ELP1-RBD into biomolecular condensates are critical and necessary steps in suppressing mRNA translation. Droplets containing IVTT components and plasmids supplemented with non-mRNA-binding ELP1s, which forms granules at T = 37 °C (Figure 4A, Supplementary Figure 2O–T), produce sfGFP over time similarly to droplets without IDPs (Figure 4B, Supplementary Movie M6). Additionally, protocell droplets supplemented with ELP2-RBDs (see Table 1 and Supplementary Table 1 for sequence) – mRNA-binding ELPs (see Supplementary Figure 3) that do not exhibit LCST phase behavior between 0-90 °C in aqueous solution (Supplementary Figure 3A–B) and hence do not form granules at T = 37 °C (Figure 4A, Supplementary Figure 3F–H) — similarly express sfGFP in a sigmoidal fashion over time when heated (Figure 4B, Supplementary Movie M7). Our model and these experiments together demonstrate that the presence of soluble mRNA-binding ELPs and ELP-only granules themselves do not inhibit mRNA translation; it is both the binding of the mRNA by ELP1-RBD followed by the LCST phase transition of the complex that leads to sequestration of the mRNA into biomolecular condensates that shuts off translation and suppresses sfGFP translation.
To demonstrate dynamic inhibition of sfGFP translation we engineered an mRNA-binding ELP, termed ELP3-RBD (see Table 1, Supplementary Table 1, Supplementary Figures 4–5), that has a TCP between 30 °C and 37 °C in the IVTT mixture (Supplementary Figure 5A–C) and thus a weaker, reversible sequestration mechanism than ELP1-RBD. In contrast to ELP1-RBD, ELP3-RBD does not exhibit a large difference in TCP in the presence of mRNA over a large concentration range and in the IVTT mixture (see Supplementary Figure 5G–K). ELP3-RBD was specifically designed at the sequence level so that its mRNA bound complex would have a transition temperature in the range of 30-40 °C at the typical operating concentration range of 250 to 750 μM so that raising the temperature above 37 °C would cause coacervation of the ELP3-RBD-mRNA complex, leading to formation of an mRNA rich granule. Conversely lowering the temperature below 35 °C would reverse coacervation and release the sequestered mRNA into the aqueous compartment of the protocell. Hence, we hypothesized that changing the solution temperature above and below the cloud point temperature of the ELP3-RBD-mRNA complex should allow us to reversibly toggle between soluble ELP3-RBD chains and ELP3-RBD granules while still keeping the transcription and translation machinery active (Figure 5A); at lower temperatures translation should be ON when the ELP3-RBD-mRNA complex is soluble and acessible to the translational machinery in the protocell, while at higher temperatures translation should be turned OFF when the ELP3-RBDs-mRNA complex forms granules which should sequester the mRNA away from the translational machinery (Figure 5A). To ensure the sequestration was limited to the mRNA transcripts and not tRNA, we performed gel retardation on the mRNA and tRNA in the presence of ELP3-RBD and found minimal to no interaction between the tRNA and ELP3-RBD (Supplementary Figure 4D–H), thus confirming the sequestration mechanism is due to coacervation of the transcripts and not other IVTT RNA components.
Figure 5. On-demand mRNP granule assembly and disassembly temporally inhibits translation.
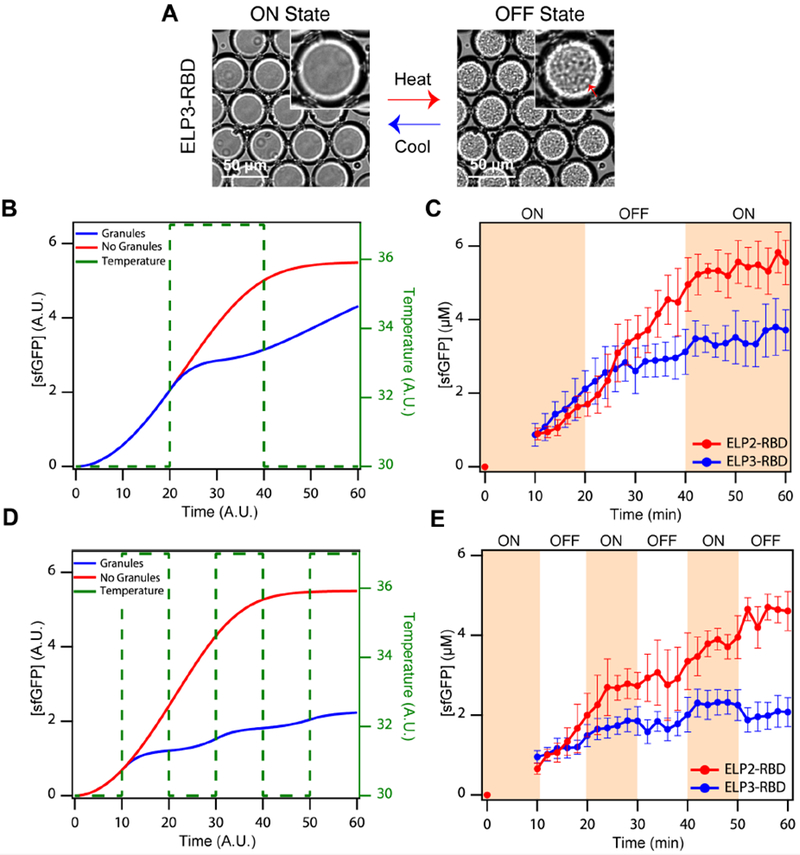
(A) Brightfield images of droplets containing IVTT components and ELP3-RBD at low and high temperatures. At low temperatures, sfGFP translation is ‘ON’ due to the lack of mRNP granules, while at high temperatures, ELP3-RBD forms mRNP granule condensates and translation is switched ‘OFF’ (red arrow indicates liquid granules). (B) Simulated sfGFP concentrations in the absence (blue) and presence (red) of periodic, temperature-modulated granule formation, following a 67% ON/33% OFF pattern, as indicated by the green dashed line. (C) Experimental demonstration corresponding to (B): red points and lines indicate droplets containing ELP2-RBD (no granule formation); blue points and lines indicate droplets containing ELP3-RBD (exhibiting temperature-responsive granule formation). (D) Simulated sfGFP concentrations in the absence (blue) and presence (red) of periodic, temperature-modulated granule formation, following a 50% ON/50% OFF pattern, as indicated by the green dashed line. (E) Experimental demonstration corresponding to (D). Data are presented in the same manner as in (C). Both simulations and experiments indicate tunable, temporal suppression of translation in droplets containing ELP3-RBD but not in droplets containing ELP2-RBD. The error bars represent the standard deviation (n ≥ 50 droplets). See Supplemental Information for further details on model formation and analysis. Related to Supplementary Table 1, Supplementary Figures 3–5, and Supplementary Movies M8–11.
We first modeled a 67% ON, 33% OFF pattern in the form of 20 minute ON → 20 minute OFF → 20 minute ON. Our model predicts that during the first ON stage at temperature T < TCP, translation is identical in systems with soluble RNA-binding IDPs and without IDPs (Figure 5B). However, during the OFF stage at temperature T > TCP, droplets without IDPs continue to express the gene in a sigmoidal fashion, whereas droplets with IDPs form biomolecular condensates and as a result, translation plateaus during this time window due to transcript sequestration within the granules (see Figure 5A, 5B). Our model captures a time lag in gene suppression response upon a step increase in temperature, which is due to the minutes long time scale of IDP-rich granule assembly (Simon et al., 2017). In the final ON stage, the model predicts a plateau in translation in droplets without granules due to substrate depletion and a slight increase in translation in droplets with soluble IDPs due to dissolution of the mRNA-IDP-rich granules (Figure 5B).
Experimentally, we validated these results by comparing protocell droplets containing either ELP2-RBDs or ELP3-RBDs subjected to the same temporal temperature profile (see Figure 5C, Supplementary Movie M8, M9). When compared, the overall sfGFP production is 27% less in protocells containing ELP3-RBD biomolecular condensates compared to protocells containing soluble ELP2-RBDs (Supplementary Figure 4J), which corresponds to a value slightly lower than the theoretical difference (33% less) in translation for 1/3 of the overall time. This difference between the observed and theoretical difference in sfGFP production may be due to the time scale of mRNP granule assembly upon heating in the OFF cycle mentioned above, where gene suppression does not begin immediately upon chaning the temperature to T > TCP due to the time required for mRNP granule formation and effective sequestration of the transcripts, which is captured in our model. We do not believe discrepancies between the model and experiments can be attributed to affects of mRNA on the phase behavior of ELP3-RBD inside the droplets. Quantification of the mRNA concentration throughout the experiment suggests that the experimental levels of mRNA transcript are far lower than those utilized during in vitro testing (Supplementary Figure 20). We would therefore expect mRNA to have a minimal effect on TCP observed throughout the experiment (Supplementary Figure 5J–K).
We then utilized our model to predict protein translation in a 50% ON, 50% OFF pattern in the form of a cycle consisting of 10 minute ON → 10 minute OFF → 10 minute ON →10 minute OFF → 10 minute ON → 10 minute OFF. Consistent with the model of the previous pattern, we predict an increase in protein concentration in droplets containing granule-forming IDPs during the ON stages at low temperatures where the IDPs remain soluble and a plateau in protein concentration during the OFF stages at high temperatures where the IDPs coalesce into granules with the transcripts (see Figure 3 and 5D). In contrast, our model predicts a sigmoidal sfGFP concentration curve at all temperatures in droplets without granules that is similar in profile to the isothermal translation curve (see Figure 4B inset and Figure 5D). Experimentally, we observe similar translation profiles as those predicted using our model in droplets containing ELP3-RBDs (Figure 5E, Supplementary Movie M10) and slightly lower, linear protein translation than predicted in protocell droplets containing soluble ELP2-RBDs (Supplementary Movie M11), which may be due to the relatively frequent temperature changes that adversely affect the machinery enzymes. However, when compared, overall sfGFP concentration was 67% greater in droplets containing ELP2-RBDs compared to droplets containing ELP3-RBDs (Supplementary Figure 4K). This observed difference in overall sfGFP production, which is similar to the experimental results using a 67% ON, 33% OFF pattern, in droplets with ELP2-RBDs compared to ELP3-RBDs differs from the theoretical value presumably due to the time lag in granule assembly upon heating. The ability to temporally inhibit translation in this fashion is an important advance in creating functional cytomimetic biomolecular condensates.
Discussion
We have developed a bottom-up approach for engineering ribonucleoprotein granules that function to temporally inhibit translation. These RNP organelles are composed of de novo RNA-binding intrinsically disordered proteins that display LCST phase behavior in aqueous solution. By binding and sequestering mRNA within protein-rich liquid organelles inside protocell microdroplets we are able to suppress translation by segregating the transcripts from the translational machinery, which can be further programmed to suppress translation in various temporal patterns with subtle changes in temperature that result in on-demand assembly and disassembly of the granules. A recent publication by the Forman-Kay group demonstrates a similar phase separation mechanism for mediating translation in vitro (Tsang et al., 2019). Here, the authors show that phosphoregulated Fragile X Mental Retardation Protein phase separates with RNA into granules that control activity-dependent translation.
This manuscript describes de novo engineered liquid granules that exploit aqueous phase behavior to control an essential cellular function —translation— within an artificial protocell. There is, however, work to be done to extend the applicability of the system more broadly. The macromolecular concentrations used in this study are likely higher than intracellular concentrations of naturally occurring IDPs and ordered proteins containing IDRs. To address this limitation, new designer IDPs with more physiologically relevant phase behavior, in terms of temperature range and concentration, can be designed and utilized. Additionally, such physiologically-relevant IDPs can be expressed within cells and extrisically controlled to finely regulate various processes to demonstrate applicability in vivo.
The results from our synthetic system, do however, provide direct experimental demonstration of tunable inhibition of translation by RNP-mediated mRNA sequestration, which we found to be a more effective strategy than other granule-based mechanisms, such as molecular crowding with synthetic polymers. Controlling key cellular processes such as translation in an engineered system in this manner is a key step towards creating more complex, realistic artificial cells that hold significant promise for studying and furthering our understanding of the functional role these RNP compartments play in nature.
STAR Methods Section
Contact for Reagent and Resource Sharing
Please contact Ashutosh Chilkoti (ashutosh.chilkoti@duke.edu) for additional information regarding the experiments described within this manuscript and for plasmid requests.
Experimental Model and Subject Details
Modeling inhibition of sfGFP translation by ELP-RBD.
We developed a simple kinetic model to describe the key reactions involved in the inhibition of sfGFP translation by reversible formation of ELP-RBD granules. These reactions include:
Generation of sfGFP mRNA (M) through transcription by consuming nucleotide triphosphates (Sm),
Generation of sfGFP protein (G) through translation by consuming amino acids (Sg),
Temperature-mediated reversible aggregation of ELP-RBD (ERs) into granules (ERg), and
Reversible binding of ERg with M, forming a complex ME.
The resulting model consists of several coupled ordinary differential equations (ODEs), each corresponding to one molecular species:
| S1 |
| S2 |
| S3 |
| S4 |
| S5 |
| S6 |
Here, t is time and T indicates the temperature state (1 = high temperature, 0 = low temperature) as a function of time. Our model makes several assumptions:
Both transcription and translation follow Michaelis-Menten kinetics, with respect to Sm and Sg.
Both granule formation and binding of granules to mRNA follow mass-action kinetics.
Molecules are not degraded.
At time 0, only Sm (=1.0), Sg (=1.0), and ERs (=2.0) are present; the levels of all other molecules are set to 0.
According to the experimental configuration, T is modeled as a series of pulses with different durations.
The definitions and base-values of all parameters are listed in Supplementary Table 1. Our model is intended to capture the qualitative trends of translation with or without inhibition mediated by ELP-RBD granules. Model construction and numerical simulations were done in Dynetica (Eidum et al., 2014; You et al., 2003).
Method Details
Gene Synthesis.
Genes encoding for ELP1, ELP1-RBD, ELP2-RBD, and ELP3-RBD were first synthesized using recursive directional ligation by plasmid reconstruction, a method described elsewhere(McDaniel et al., 2010). In brief, we modified a pET-24+ cloning vector (pET-24+) to contain endonuclease recognition sites for AcuI, BseRI, and BglI. We digested the modified vector with BseRI and ligated the desired ELP or RBD repeat sequence into the vector. The ELP1/ELP2/ELP3 and RBD sequences were created by annealing together complementary ssDNA strands that encode for the desired amino acid sequence (e.g., either the base ELP1/ELP2/ELP3 repeat sequence or the RBD sequence) along with “sticky end” overhangs. The base ELP1/ELP2/ELP3 sequence was used to create separate plasmids containing ELP1/ELP2/ELP3 with the desired number of repeat pentamers by digesting one plasmid with AcuI and BglI to create an “A” population and a separate plasmid with BseRI and BglI to create a “B” population. The two populations of cut plasmids are complementary to one another such that when ligated together the length of the ELP1 sequence doubles. This digestion-ligation process was continued until the desired number of ELP/ELP2/ELP3 pentamers was achieved.
After the desired ELP1, ELP2, ELP3 and RBD sequences were created, an “A” population of the ELP-containing plasmid and a “B” population of the RBD-containing plasmid were created using the method above. The populations were ligated together to generate a plasmid encoding for ELP1-RBD (e.g., an N-terminal ELP1 region adjacent to a C-terminal RBD region). This protocol was also used to create a plasmid encoding for ELP2-RBD (e.g., an N-terminal ELP2 region adjacent to a C-terminal RBD region) and ELP3-RBD (e.g., an N-terminal ELP3 region adjacent to a C-terminal RBD region). The genes encoding the final sequences of ELP1, ELP2, ELP3, RBD, ELP-RBD, ELP2-RBD, and ELP3-RBD (see Supplementary Table 1) were confirmed by DNA sequencing.
Protein Expression, Purification and Characterization.
Separate liquid cell cultures (100 ml) of BL21 Escherichia coli strains each harboring ELP1, ELP1-RBD, ELP2-RBD, ELP3-RBD, or sfGFP plasmids were inoculated from frozen glycerol stocks and grown to confluence overnight. Cultures were then inoculated at a 1:20 dilution in 1L 2xYT media supplemented with 45 μg ml−1 kanamycin. Cells were grown at 37°C in a shaking incubator (220 r.p.m.) for 6 h, at which time protein expression was then induced by the addition of 1mM IPTG (final concentration). Cells were then incubated at 37 °C (shaking at 220 r.p.m.) for an additional 18 h. Cell pellets were harvested by centrifuging cultures at 3500 g’s and resuspending in 15 ml 1x PBS. Cells were then lysed by sonicating the cell solutions for 3 minutes (Misonix; Farmingdale, NY) and the nucleic acids were precipitated by adding 10% polyethylenimine (MP Biomedicals, Santa Ana, CA). Precipitated nucleic acids and cellular debris were removed by centrifuging solutions at 14,000 r.p.m. at 4°C.
ELP1, ELP1-RBD, ELP2-RBD, and ELP3-RBD proteins were then purified using inverse transition cycling, as described elsewhere(MacEwan et al., 2014; Meyer and Chilkoti, 2002). Briefly, solutions of proteins were heated and ammonium sulfate was added to induce protein phase separation, centrifuged to collect all insoluble material at 40°C, 20,000 x g (“hot spin”), and re-suspended in cold 1x PBS. Upon cooling, the ELP1/ELP1-RBD/ELP2-RBD/ELP3-RBD resolubilizes, while contaminants remain insoluble and can be removed by centrifugation at 4°C, 20,000xg (“cold spin”). In contrast, sfGFP proteins were purified using immobilized metal affinity chromatography (Bio-Rad Profinity resins) per the manufacturer’s instructions. Protein purity was characterized by 4–20% gradient tris-HCl (Biorad, Hercules, CA) sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE) and staining with either 0.5 M copper chloride or 0.1% w/v Coommassie Blue (Fisher Scientific, Hampton, NH). Protein yield was determined gravimetrically after dialysis into Nanopure water and lyophilization.
Droplet Formation.
To create water-in-oil emulsion droplets, two liquid phases – a dispersed, aqueous phase containing ELP1/ELP1-RBD/ELP2-RBD/ELP3-RBD/sfGFP/ssRNA1 (or combinations thereof) in 1x PBS or the S30 cell-free expression kit and an organic, continuous phase comprised of 75%/5%/20% vol/vol TEGOSOFT® DEC/ABIL® EM 90/mineral oil – were injected into the microfluidic droplet generators at constant flow rates using precision syringe pumps. The flow rates of the dispersed and continuous fluids were tuned to ensure droplet formation in the dripping regime; in these experiments, the dripping regime was achieved using a constant flow rate of 500 μL hr−1 for the organic continuous phase and 50-75 μL hr−1 for the aqueous, dispersed phase. The production of droplets within the microfluidic device was monitored using a 5× objective on an inverted microscope (Leica) equipped with a digital microscopy camera (Lumenera Infinity 3-1 CCD).
Circular Dichroism Spectroscopy.
Circular Dichroism (CD) spectroscopy was performed using an Aviv Model 202 instrument and a 1 mm quartz sample cell (Hellma). ELP1-RBD was prepared by dissolving the purified lyophilized product in ultrapure water to a final concentration of 10 μM. The CD spectra were obtained at 20 °C from 260 nm to 180 nm in 1 nm steps at a 0.5 second average time. The CD spectra were corrected for the ultrapure water buffer signal at 20 °C. This data collection was repeated in triplicate, and the average of the three measurements was represented as a mean residue molar ellipticity ([Θ]).
Light Scattering.
Dynamic light scattering (DLS) measurements were performed over a temperature range of 25–50 °C using a Wyatt DynaPro temperature-controlled microsampler (Wyatt Technology, Santa Barbara, CA). Samples for the DLS system were prepared in 1x PBS and filtered through 0.02 μm Whatman Anotop sterile syringe filters (GE Healthcare Life Sciences, Pittsburgh, PA) into a 12 μL quartz crystal cuvette (Wyatt Technology, Santa Barbara, CA). 5 acquisitions were taken at each temperature, and the results presented represent the mean Rh of the sample at each temperature. The error bars represent the polydispersity percentage determined from the 5 data points taken at each temperature.
Static light scattering (SLS) measurements were performed using an ALV/CGS-3 goniometer system (Langen, Germany). Samples for the ALV/CGS-3 goniometer system were prepared in 1x PBS and filtered through 0.2 μm Whatman Anotop sterile syringe filters into a 10 mm disposable borosilicate glass tube (Fischer Scientific, Pittsburgh, PA). Simultaneous SLS measurements were obtained at angles between 30°−150° at 5° increments, with each angle consisting of 3 runs for 15 s, at 15°C. The differential refractive index (dn/dc) was determined by measuring refractive index at five different concentrations at 15°C using an Abbemat 500 refractometer (Anton Paar, Graz, Austria). Static light scattering data were analyzed by partial Zimm plots using the ALVSTAT software to determine the radius of gyration (Rg) and molecular weight. Nagg was determined by subtracting the molecular weight of the mRNA (as determined from the sequence) from that of the biomolecular condensate molecular weight and then dividing the result by molecular weight of an individual ELP1-RBD chain, as calculated from the amino acid sequence. This approach assumes no interaction between ELP1-RBD chains and confines interactions to those between ELP1-RBD and mRNA.
Temperature-controlled Spectrophotometry.
Cloud point transition temperatures (Tt) were determined via temperature-controlled spectrophotometry using a Cary 300 (Agilent Technologies). Samples containing various concentrations of protein in 1x PBS or the S30 cell-free protein expression kit buffer were heated at 1°C min−1 and the absorbance at λ =650 nm was recorded every 0.1 °C. The cloud point was determined as the maximum in the first derivative of the absorbance as a function of temperature.
Gel Retardation Assay.
eGFP mRNA, ELP1 + eGFP mRNA, and ELP1-RBD/ELP2-RBD/ELP3-RBD + eGFP mRNA were visualized via 1% agarose gel electrophoresis. 100 μg ml−1 eGFP mRNA and 500 μM ELP1/ELP1-RBD/ELP2-RBD/ELP3-RBD in 1x PBS were loaded into a standard 1% agarose gel supplemented with 1x SYBR® Green II RNA stain. eGFP mRNA fragments were separated in a 1x TAE buffer at 130V for 40 minutes, and the lanes were compared to a ssRNA ladder for sizing.
Cell-free Expression.
sfGFP was expressed within droplets utilizing a commercially available cell-free expression kit (see materials). Solutions were prepared following the manufacturer’s protocol using a constant sfGFP plasmid concentration of 20 μg ml−1 and ELP1/ELP1-RBD/ELP2-RBD/ELP3-RBD proteins were added to their desired final concentration as indicated. Monodisperse droplets containing these mixtures were formed using the microfluidic device and imaged on a precision temperature-controlled microscope stage.
Heating and Imaging.
Emulsion samples were collected on a glass microscope slide and heated using a precision Peltier heating and cooling stage (Linkam LTS120) equipped with a Linkam PE95 digital temperature control unit. The spatial distribution of Alexa Fluor 488-labeled (25% molar fraction N-terminal labeled) ELP/ELP-RBD and Alexa Fluor 594-labeled (5’ conjugated) ssRNA1 was characterized via fluorescence microscopy using an upright Zeiss Axio Imager D2 microscope with a 20× objective and the appropriate filter set (ex 470/40, em 525/50). Similarly, fluorescence intensity of superfolder GFP over time was characterized via fluorescence microscopy using an upright Zeiss Axio Imager D2 microscope with a 20× objective and the appropriate filter set (ex 470/40, em 525/50). Coacervate formation was monitored via bright-field microscopy using an upright Zeiss Axio Imager D2 microscope with a 20× objective. Fluorescence intensity within droplets was characterized using MATLAB.
Amplification of sfGFP DNA.
25 μL of GoTaq® Green Master Mix, 2X, 2 μL of T7 promoter forward and reverse primers, 1 μL of sfGFP plasmid and 20 ul of Nuclease-free water was mixed in PCR tube. Conditions for amplification was as follows: 10 sec at 98 °C, annealing for 10 sec at 62 °C; polymerization for 15 sec at 72 °C. The cycle was repeated 35 times and later gel purified.
RNA synthesis.
HiScribe™ T7 High Yield RNA Synthesis Kit was used to synthesize sfGFP RNA from PCR product gene. 4 μL of nuclease-free water, 2 μl of 10X reaction buffer, 2 μl of each ATP, GTP, UTP and CTP (100 mM), 4 μL of template sfGFP DNA and 2 μL of T7 RNA polymerase Mix was mixed thoroughly and incubated for 2 hours at 37 °C. Reaction mixture was later purified using Oligo Clean & Concentrator kit (Zymo research). The mixture of synthesized RNA (100 μg ml−1) and ELP3-RBD (500 μM) showed similar binding behavior in standard 1% agarose gel.
tRNA Pull Down.
To track the tRNA during the phase separation, 300 μl solution containing ELP3-RBD (500 μM), mRNA (100 μg ml−1) and tRNA (100 μg ml−1) was prepared. NaCl was added to the mixture to increase the salt concentration to 1 M, that helped reducing the Tt below 40 °C which the was the temperature for hot centrifuge. 300 μl solution of mRNA (100 μg ml−1) in PBS 1X (control 1), tRNA (100 μg ml−1) in PBS 1X (control 2) and ELP3-RBD (500 μM)/tRNA (100 μg ml−1) (control 3) was made as control experiments. The 260/280 ration for control 1 and 2 samples were measured using Nanodrop. The 260/280 ratios of ELP3-RBD/tRNA/mRNA mixture as well as control 3 were measured at 4 °C. Later both samples were heated to 65 °C for 5 mins and later centrifuged at 40 °C and 13000 rcf. The supernatant (protein poor phase) was separated and 260/280 was measured. The palette was immersed in PBS 1x and left on ice to dissolve well. Later the 260/280 ration was measured. Same samples were loaded on 1% agarose gel (Fig A) to visualize the interaction of tRNA to ELP3-RBD.
Ribosome Pull Down.
1 ml solution containing fluorophore-labeled Ribosome (μg ml−1) and ELP3-RBD (500 μM) was made. NaCl was added to the mixture to increase the salt concertation to 1 M in order to reduce the Tt of the system. Mixture was left on ice for 10 mins before fluorescent signal was measured. The mixture was later heated to 65 °C for 5 mins and centrifuged for 10 mins at 40 °C and 13000 rcf. The supernatant was separated from the palette immediately and the fluorescent signal was measured. The palette redissolved in 1 ml PBS 1x, and left on ice for 10 mins. The fluorescent signal was later measured. Labeled Ribosome (100 μg ml−1) in PBS 1x was used as a control.
Quantification and Statistical Methods
sfGFP fluorescence intensity within droplets was quantified using ImageJ and MATLAB scripts to maintain uniformity and consistency across samples. For each data point shown, a minimum of 50 discrete droplets was used across multiple experiments to ensure representative populations. Errors bars shown throughout the manuscript represent either standard deviation or standard error of the mean, as indicated in the figure legend. Statistical significance, where shown, was determined using student’s t-test between different samples and statistical significant was determined from p-values less than 0.05 (5%), which is indicated in the figure legends.
Supplementary Material
Supplementary Movie 1, Related to Figure 2
Video M1: Heat-triggered phase separation and cooling-triggered solubilization of Alexa Fluor 488-labeled ELP1-RBD (CELP-RBD = 500 μM, T = 10 °C ➔ 40 °C ➔ 10 °C at 20 °C min−1).
Supplementary Movie 7, Related to Figure 4.
Video M7: sfGFP production within droplets at a constant temperature with ELP2-RBDs (CELP2-RBD = 500 μM T = 37 °C).
Supplementary Movie 8, Related to Figure 5.
Video M8: sfGFP production within droplets with ELP3-RBDs at a 67% ON/33% OFF pattern (CELP3-RBD = 500 μM, TON = 30 °C, TOFF = 38 °C). At T = 30 °C ELP3-RBDs are soluble and do not form granules. At T = 38 °C ELP3-RBDs phase separate and form granules.
Supplementary Movie 9, Related to Figure 5.
Video M9: sfGFP production within droplets with ELP2-RBDs at a 67% ON/33% OFF pattern (CELP3-RBD = 500 μM, TON = 30 °C, TON = 38 °C). ELP3-RBDs are soluble at T = 30 °C and T = 38 °C and therefore do not have an OFF state.
Supplementary Movie 10, Related to Figure 5.
Video M10: sfGFP production within droplets with ELP3-RBDs at a 50% ON/50% OFF pattern (CELP3-RBD = 500 μM, TON = 30 °C, TOFF = 38 °C). At T = 30 °C ELP3-RBDs are soluble and do not form granules. At T = 38 °C ELP3-RBDs phase separate and form granules.
Supplementary Movie 11, Related to Figure 5.
Video M11: sfGFP production within droplets with ELP2-RBDs at a 50% ON/50% OFF pattern (CELP3-RBD = 500 μM, TON = 30 °C, TON = 38 °C). ELP3-RBDs are soluble at T = 30 °C and T = 38 °C and therefore do not have an OFF state.
Supplementary Movie 2, Related to Figure 2
Video M2: Heat-triggered co-phase separation and cooling-triggered solubilization of Alexa Fluor 488-labeled ELP1-RBD and Alexa Fluor 594-labeled ssRNA1 (CELP-RBD = 500 μM, CssRNA1 = 2.5 μM, T = 10 °C ➔ 40 °C ➔ 10 °C at 20 °C min−1).
Supplementary Movie 3, Related to Figure 2
Video M3: Heat-triggered phase separation and cooling-triggered solubilization of Alexa Fluor 488-labeled ELP1 and Alexa Fluor 594-labeled ssRNA1 (CELP = 500 μM, CssRNA1 = 2.5 μM, T = 20 °C ➔ 45 °C ➔ 10 °C at 20 °C min−1).
Supplementary Movie 4, Related to Figure 4.
Video M4: sfGFP production within droplets at a constant temperature without IDPs (T = 37 °C).
Supplementary Movie 5, Related to Figure 4.
Video M5: sfGFP production within droplets at a constant temperature with ELP1-RBDs (CELP-RBD = 500 μM T = 37 °C).
Supplementary Movie 6, Related to Figure 4.
Video M6: sfGFP production within droplets at a constant temperature with ELP1s (CELP = 500 μM T = 37 °C).
Highlights.
New engineered phase separating IDPs capable of binding RNA
IDPs co-phase separate with RNAs into liquid granules in response to heat
IDP-RNA rich granules regulate translation through mRNA sequestration
Droplet-based protocells temporally inhibit translation in a programmable manner
Acknowledgements
A.C. acknowledges support from the National Institutes of Health (NIH) (R01 GM61232 and NIGMS MIRA GM35- ) and from the National Science Foundation (NSF) Research Triangle MRSEC (DMR-1121107). J.R.S acknowledges the support of the NSF Graduate Research Fellowship Program (DGF1106401).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Declaration of Interests
The authors declare no competing interests.
References
- Amiram M, Haimovich AD, Fan C, Wang Y-S, Aerni H-R, Ntai I, Moonan DW, Ma NJ, Rovner AJ, Hong SH, et al. (2015). Evolution of translation machinery in recoded bacteria enables multi-site incorporation of nonstandard amino acids. Nature Biotechnology 33, 1272–1279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson P, and Kedersha N (2006). RNA granules. The Journal of Cell Biology 172, 803–808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson P, and Kedersha N (2009). RNA granules: post-transcriptional and epigenetic modulators of gene expression. Nature Reviews Molecular Cellular Biology 10, 430–436. [DOI] [PubMed] [Google Scholar]
- Banani SF, Lee HO, Hyman AA, and Rosen MK (2017). Biomolecular condensates: organizers of cellular biochemistry. Nature Reviews Molecular Cell Biology 18, 285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brangwynne CP. (2011). Soft active aggregates: mechanics, dynamics and self-assembly of liquid-like intracellular protein bodies. Soft Matter 7, 3052–3059. [Google Scholar]
- Brangwynne CP, Eckmann CR, Courson DS, Rybarska A, Hoege C, Gharakhani J, Julicher F, and Hyman AA (2009). Germline P granules are liquid droplets that localize by controlled dissolution/condensation. Science 324, 1729–1732. [DOI] [PubMed] [Google Scholar]
- Brangwynne CP, Tompa P, and Pappu RV (2015). Polymer physics of intracellular phase transitions. Nature Physics 11, 899–904. [Google Scholar]
- Buchan JR (2014). mRNP granules: assembly, function, and connections with disease. RNA Biology 11, 1019–1030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chong PA, Vernon RM, and Forman-Kay JD (2018). RGG/RG Motif regions in RNA binding and phase separation. Journal of Molecular Biology 430, 4650–4665. [DOI] [PubMed] [Google Scholar]
- Christensen T, Amiram M, Dagher S, Trabbic-Carlson K, Shamji Mohammed F, Setton Lori A, and Chilkoti A (2009). Fusion order controls expression level and activity of elastin-like polypeptide fusion proteins. Protein Science 18, 1377–1387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christensen T, Hassouneh W, Trabbic-Carlson K, and Chilkoti A (2013). Predicting Transition Temperatures of Elastin-Like Polypeptide Fusion Proteins. Biomacromolecules 14, 1514–1519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costa SA, Simon JR, Amiram M, Tang L, Zauscher S, Brustad EM, Isaacs FJ, and Chilkoti A (2018). Photo-crosslinkable unnatural amino acids enable facile synthesis of thermoresponsive nano- to microgels of intrinsically disordered polypeptides. Advanced Materials 30, 1704878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng N, Huck WTS Microfluidic formation of monodisperse coacervate organelles in liposomes Angew. Chem. Int. Ed. Engl, 56 (2017), pp. 9736–9740 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eidum D, Asthana K, Unni S, Deng M, and You L (2014). Construction, visualization, and analysis of biological network models in Dynetica. Quantitative Biology 2, 142–150. [Google Scholar]
- Elbaum-Garfinkle S, Kim Y, Szczepaniak K, Chen CC-H, Eckmann CR, Myong S, and Brangwynne CP (2015). The disordered P granule protein LAF-1 drives phase separation into droplets with tunable viscosity and dynamics. Proceedings of the National Academy of Sciences 112, 7189–7194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo L, and Shorter J (2015). It’s raining liquids: RNA tunes viscoelasticity and dynamics of membraneless organelles. Molecular Cell 60, 189–192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hassouneh W, Christensen T, and Chilkoti A (2010). Elastin-like polypeptides as a purification tag for recombinant proteins. Current Protocols Protein Science Chapter 6, Unit 6.11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawasaki I, Shim Y-H, Kirchner J, Kaminker J, Wood WB, and Strome S (1998). PGL-1, a predicted RNA-binding component of germ granules, is essential for fertility in C. elegans. Cell 94, 635–645. [DOI] [PubMed] [Google Scholar]
- Li X, Romero P, Rani M, Dunker AK, and Obradovic Z (1999). Predicting protein disorder for N-, C-, and internal regions. Genome Informatics Workshop on Genome Informatics 10, 30–40. [PubMed] [Google Scholar]
- Long MS, Jones CD, Helfrich MR, Mangeney-Slavin LK, and Keating CD (2005). Dynamic microcompartmentation in synthetic cells. Proceedings of the National Academy of Sciences 102, 5920–5925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luby-Phelps K (2013). The physical chemistry of cytoplasm and its influence on cell function: an update. Molecular Biology of the Cell 24, 2593–2596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luginbuhl KM, Mozhdehi D, Dzuricky M, Yousefpour P, Huang FC, Mayne NR, Buehne KL, and Chilkoti A (2017). Recombinant synthesis of hybrid lipid-peptide polymer fusions that self-assemble and encapsulate hydrophobic drugs. Angewandte Chemie International Edition 56, 13979–13984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacEwan SR, and Chilkoti A (2010). Elastin-like polypeptides: biomedical applications of tunable biopolymers. Biopolymers 94, 60–77. [DOI] [PubMed] [Google Scholar]
- MacEwan SR, Hassouneh W, and Chilkoti A (2014). Non-chromatographic purification of recombinant elastin-like polypeptides and their fusions with peptides and proteins from Escherichia coli. Journal of Visual Experiments. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McDaniel JR, MacKay JA, Quiroz FG, and Chilkoti A (2010). Recursive directional ligation by plasmid reconstruction allows rapid and seamless cloning of oligomeric genes. Biomacromolecules 11, 944–952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McDaniel JR, Radford DC, and Chilkoti A (2013). A unified model for de novo design of elastin-like polypeptides with tunable inverse transition temperatures. Biomacromolecules 14, 2866–2872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer DE, and Chilkoti A (1999). Purification of recombinant proteins by fusion with thermally-responsive polypeptides. Nature Biotechnology 17, 1112–1115. [DOI] [PubMed] [Google Scholar]
- Meyer DE, and Chilkoti A (2002). Protein purification by inverse transition cycling In Protein-Protein Interactions: A Molecular Cloning Manual (Cold Spring Harbor Laboratory Press; ), pp. 329–343. [Google Scholar]
- Meyer DE, and Chilkoti A (2004). Quantification of the effects of chain length and concentration on the thermal behavior of elastin-like polypeptides. Biomacromolecules 5, 846–851. [DOI] [PubMed] [Google Scholar]
- Mozhdehi D, Luginbuhl KM, Simon JR, Dzuricky M, Berger R, Varol HS, Huang FC, Buehne KL, Mayne NR, Weitzhandler I, et al. (2018). Genetically encoded lipid-polypeptide hybrid biomaterials that exhibit temperature-triggered hierarchical self-assembly. Nature Chemistry 10, 496–505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nott TJ, Craggs TD, and Baldwin AJ (2016). Membraneless organelles can melt nucleic acid duplexes and act as biomolecular filters. Nature Chemistry 8, 569–575. [DOI] [PubMed] [Google Scholar]
- Peng K, Radivojac P, Vucetic S, Dunker AK, and Obradovic Z (2006). Length-dependent prediction of protein intrinsic disorder. BMC Bioinformatics 7, 208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polymenidou M (2018). The RNA face of phase separation. Science 360, 859–860. [DOI] [PubMed] [Google Scholar]
- Protter DSW, and Parker R (2016). Principles and properties of stress granules. Trends in Cell Biology 26, 668–679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Protter DSW, Rao BS, Van Treeck B, Lin Y, Mizoue L, Rosen MK, and Parker R (2018). Intrinsically disordered regions can contribute promiscuous interactions to RNP granule assembly. Cell Reports 22, 1401–1412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riback JA, Katanski CD, Kear-Scott JL, Pilipenko EV, Rojek AE, Sosnick TR, and Drummond DA (2017). Stress-triggered phase separation is an adaptive, evolutionarily tuned response. Cell 168, 1028–1040. e1019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romero P, Obradovic Z, Kissinger C, Villafranca JE, and Dunker AK (1997). Identifying disordered regions in proteins from amino acid sequence. Paper presented at: Proceedings of International Conference on Neural Networks (ICNN’97). [Google Scholar]
- Rubinstein M, and Colby R (2003). Polymers physics (Oxford: ). [Google Scholar]
- Sheu-Gruttadauria J, and MacRae IJ (2018). Phase rransitions in the assembly and function of human miRISC. Cell 173, 946–957. e916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shin Y, and Brangwynne CP (2017). Liquid phase condensation in cell physiology and disease. Science 357. [DOI] [PubMed] [Google Scholar]
- Simon JR, Carroll NJ, Rubinstein M, Chilkoti A, and Lopez GP (2017). Programming molecular self-assembly of intrinsically disordered proteins containing sequences of low complexity. Nature Chemistry 9, 509–515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sokolova E, Spruijt E, Hansen MMK, Dubuc E, Groen J, Chokkalingam V, Piruska A, Heus HA, and Huck WTS (2013). Enhanced transcription rates in membrane-free protocells formed by coacervation of cell lysate. Proceedings of the National Academy of Sciences 110, 11692–11697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strulson CA, Molden RC, Keating CD, and Bevilacqua PC (2012). RNA catalysis through compartmentalization. Nature Chemistry 4, 941–946. [DOI] [PubMed] [Google Scholar]
- Su X, Ditlev JA, Hui E, Xing W, Banjade S, Okrut J, King DS, Taunton J, Rosen MK, and Vale RD (2016). Phase separation of signaling molecules promotes T cell receptor signal transduction. Science 352, 595–599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thandapani P, O’Connor TR, Bailey TL, and Richard S (2013). Defining the RGG/RG motif. Molecular Cell 50, 613–623. [DOI] [PubMed] [Google Scholar]
- Theillet F-X, Binolfi A, Frembgen-Kesner T, Hingorani K, Sarkar M, Kyne C, Li C, Crowley PB, Gierasch L, Pielak GJ, et al. (2014). Physicochemical properties of cells and their effects on intrinsically disordered proteins (IDPs). Chemical Reviews 114, 6661–6714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toretsky JA, and Wright PE (2014). Assemblages: functional units formed by cellular phase separation. Journal of Cellular Biology 206, 579–588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsang B, Arsenault J, Vernon RM, Lin H, and Sonenberg N (2019). Phosphoregulated FMRP phase separation models activity-dependent translation through bidirectional control of mRNA granule formation. Proceedings of the National Academy of Sciences 116, 4218–4227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uversky VN, Kuznetsova IM, Turoverov KK, and Zaslavsky B (2015). Intrinsically disordered proteins as crucial constituents of cellular aqueous two phase systems and coacervates. FEBS letters 589, 15–22. [DOI] [PubMed] [Google Scholar]
- Wang J, Choi J-M, Holehouse AS, Lee HO, Zhang X, Jahnel M, Maharana S, Lemaitre R, Pozniakovsky A, Drechsel D, et al. (2018). A molecular grammar governing the driving forces for phase separation of prion-like RNA binding proteins. Cell 174, 688–699.e616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei M-T, Elbaum-Garfinkle S, Holehouse AS, Chen CC-H, Feric M, Arnold CB, Priestley RD, Pappu RV, and Brangwynne CP (2017). Phase behaviour of disordered proteins underlying low density and high permeability of liquid organelles. Nature Chemistry 9, 1118–1125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xue B, Dunbrack RL, Williams RW, Dunker AK, and Uversky VN (2010). PONDR-FIT: A meta-predictor of intrinsically disordered amino acids. Biochimica et Biophysica Acta (BBA) - Proteins and Proteomics 1804, 996–1010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- You L, Hoonlor A, and Yin J (2003). Modeling biological systems using Dynetica—a simulator of dynamic networks. Bioinformatics 19, 435–436. [DOI] [PubMed] [Google Scholar]
- Zhang H, Elbaum-Garfinkle S, Langdon EM, Taylor N, Occhipinti P, Bridges AA, Brangwynne CP, and Gladfelter AS (2015). RNA controls polyQ protein phase transitions. Molecular Cell 60, 220–230. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Movie 1, Related to Figure 2
Video M1: Heat-triggered phase separation and cooling-triggered solubilization of Alexa Fluor 488-labeled ELP1-RBD (CELP-RBD = 500 μM, T = 10 °C ➔ 40 °C ➔ 10 °C at 20 °C min−1).
Supplementary Movie 7, Related to Figure 4.
Video M7: sfGFP production within droplets at a constant temperature with ELP2-RBDs (CELP2-RBD = 500 μM T = 37 °C).
Supplementary Movie 8, Related to Figure 5.
Video M8: sfGFP production within droplets with ELP3-RBDs at a 67% ON/33% OFF pattern (CELP3-RBD = 500 μM, TON = 30 °C, TOFF = 38 °C). At T = 30 °C ELP3-RBDs are soluble and do not form granules. At T = 38 °C ELP3-RBDs phase separate and form granules.
Supplementary Movie 9, Related to Figure 5.
Video M9: sfGFP production within droplets with ELP2-RBDs at a 67% ON/33% OFF pattern (CELP3-RBD = 500 μM, TON = 30 °C, TON = 38 °C). ELP3-RBDs are soluble at T = 30 °C and T = 38 °C and therefore do not have an OFF state.
Supplementary Movie 10, Related to Figure 5.
Video M10: sfGFP production within droplets with ELP3-RBDs at a 50% ON/50% OFF pattern (CELP3-RBD = 500 μM, TON = 30 °C, TOFF = 38 °C). At T = 30 °C ELP3-RBDs are soluble and do not form granules. At T = 38 °C ELP3-RBDs phase separate and form granules.
Supplementary Movie 11, Related to Figure 5.
Video M11: sfGFP production within droplets with ELP2-RBDs at a 50% ON/50% OFF pattern (CELP3-RBD = 500 μM, TON = 30 °C, TON = 38 °C). ELP3-RBDs are soluble at T = 30 °C and T = 38 °C and therefore do not have an OFF state.
Supplementary Movie 2, Related to Figure 2
Video M2: Heat-triggered co-phase separation and cooling-triggered solubilization of Alexa Fluor 488-labeled ELP1-RBD and Alexa Fluor 594-labeled ssRNA1 (CELP-RBD = 500 μM, CssRNA1 = 2.5 μM, T = 10 °C ➔ 40 °C ➔ 10 °C at 20 °C min−1).
Supplementary Movie 3, Related to Figure 2
Video M3: Heat-triggered phase separation and cooling-triggered solubilization of Alexa Fluor 488-labeled ELP1 and Alexa Fluor 594-labeled ssRNA1 (CELP = 500 μM, CssRNA1 = 2.5 μM, T = 20 °C ➔ 45 °C ➔ 10 °C at 20 °C min−1).
Supplementary Movie 4, Related to Figure 4.
Video M4: sfGFP production within droplets at a constant temperature without IDPs (T = 37 °C).
Supplementary Movie 5, Related to Figure 4.
Video M5: sfGFP production within droplets at a constant temperature with ELP1-RBDs (CELP-RBD = 500 μM T = 37 °C).
Supplementary Movie 6, Related to Figure 4.
Video M6: sfGFP production within droplets at a constant temperature with ELP1s (CELP = 500 μM T = 37 °C).


