Abstract
RNA structure is intimately connected to each step of gene expression. Recent advances have enabled transcriptome-wide maps of RNA secondary structure, termed RNA structuromes. However, previous whole-cell analyses lacked the resolution to unravel the landscape and also the regulatory mechanisms of RNA structural changes across subcellular compartments. Here we reveal the RNA structuromes in three compartments — chromatin, nucleoplasm and cytoplasm in human and mouse cells. The cytotopic structuromes substantially expand RNA structural information, and enable detailed investigation of the central role of RNA structure in linking transcription, translation, and RNA decay. We develop a resource to visualize the interplay of RNA-protein interactions, RNA modifications, and RNA structure, and predict both direct and indirect reader proteins of RNA modifications. We also validate a novel role of the RNA binding protein LIN28A as an N6-methyladenosine modification “anti-reader”. Our results highlight the dynamic nature of RNA structures and its functional significance in gene regulation.
RNAs fold into complex structures that are crucial for their functions and regulations including transcription, processing, localization, translation and decay1-6. Over the last few decades RNA structure has been studied extensively in vitro and in silico, and crystallography and cryo-EM structures of molecular machines such as the spliceosome and ribosome, containing RNAs at their core, have become available7,8. In recent years technologies have been developed to map RNA secondary structures for the whole transcriptome, i.e., RNA structuromes, by combining biochemical probing with deep sequencing9-16. These systems biology studies have revealed many novel insights on the RNA structure basis of gene regulation17-20. However, so far existing genome-wide structure probing studies have focused on whole-cell data, which only represents an ensemble average of RNA molecules in different subcellular compartments.
In fact, RNA undergoes a complex life cycle in eukaryotic cells, mirrored by its movement into distinct cytotopic locales4,21. RNA structure is thought to form co-transcriptionally on the chromatin template, undergo conformational changes resulting from RNA chemical modification and processing in the nucleus, and experience further changes in the cytoplasm during translation and RNA decay. Averaging the RNA structure signal in the entire cell may obscure these critical features. More importantly, detailed mapping of RNA structures in vivo will help to elucidate how they are regulated, which is essential to understanding the RNA structure basis for gene expression regulation.
An important driving force that regulates the landscape of RNA structural changes in post-transcription regulation are the RNA-binding proteins (RBPs). A study in Arabidopsis revealed that RNA secondary structure is anti-correlated with protein-binding density22. We recently used icSHAPE to probe RNA structuromes in mouse ES cells and examined the in vivo and in vitro structure profiles of RBFOX2, a splicing factor of the “feminizing locus on X” (Fox) family proteins; and HuR, an RBP that regulates transcript stability12. We implemented a machine learning algorithm and found that using structure signals significantly improved the prediction of RNA-binding sites of both RBPs, suggesting that RNA structure signature analysis is a powerful tool to investigate RNA–RBP interactions. However, in spite of these recent advances in our understanding of the association between RNA structure and RBP-binding, a compendium of the RNA structural basis of RBP binding is not available.
In addition to RBP binding, the modification and editing of RNAs are also an important mechanism for RNA structure regulation. RNA modification can regulate almost all RNA processes including RNA maturation, nuclear retention and exportation, translation, decay, and cell differentiation and reprogramming as well23,24. As one of the most abundant and important types of mRNA modification, N6-methyladenosine (m6A) has been shown to favor the unwinding of duplex RNAs by conformational switching12,25,26. The impact of structure destabilizing effect of m6A is exemplified by a study that investigated HNRNPC, a splicing factor that preferentially binds to single-stranded polyU tracts27. Biochemical studies showed that m6A modification can disrupt the local RNA structures and promote HNRNPC binding in nearby regions28. The study defined these m6A sites as “m6A-switches”, and identified the enrichment of tens of thousands of m6A-switches in the vicinity of HNRNPC binding sites, thereby altering HNRNPC-binding and splicing of the target mRNAs. However, whether RNA structural context is a general mechanism for the recognition of other “reader” proteins of m6A and other RNA modifications, is still unclear29.
Here we use in vivo click selective 2-hydroxyl acylation and profiling experiments (icSHAPE)12, a technique we developed to map RNA structure in vivo, in three compartments – chromatin, nucleoplasm and cytoplasm – in both mouse and human cells. Consequently, we were able to determine the precise relationship of RNA structure with cellular processes including transcription, translation and RNA decay in the compartment where they occur. Separately, we could quantify how RNA adopts different conformations across different cellular compartments, which we termed “structural change”, and investigate the sophisticated interplay of RNA structural changes, RNA modification and RBP binding.
Results
Cytotopic RNA structure maps substantially expand the scope and comprehensiveness of RNA structures.
To investigate the regulation of RNA structural changes in the cell, we performed icSHAPE to measure RNA secondary structure for transcripts isolated from three subcellular compartments and in two species (Figure 1a). After performing the icSHAPE reaction of living cells (hereafter “in vivo”), RNA fractionation30,31 enabled the study of RNA structural changes in distinct subcellular locations. Separately, we fractionated the three subcellular compartments, isolated and refolded naked RNA from each, and performed icSHAPE in vitro. This in vitro dataset served as a control for the RNA contents in each compartment. The use of both v6.5 mouse embryonic stem (mES) cells and human embryonic kidney (HEK293) cells allowed us to examine whether the structural patterns we observed are conserved across the two species and cell types.
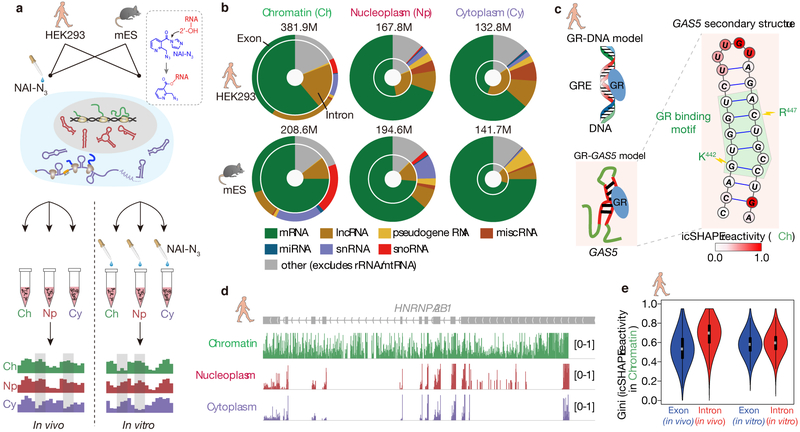
Fig 1 ∣. Chromatin fractions are enriched for pre-mRNA and lncRNA structures.
a, Experimental overview of the icSHAPE protocol. The dashed box highlights the chemical structure of NAI-N3 and its covalent bond with the 2'-OH group of RNA, which allows probing of RNA structures inside living cells. b, Donut charts showing read distributions of different RNA types in the three cellular compartments. The outer circles represent exon coverage while the inner circles represent intron coverage. c, GAS5 RNA secondary structure with icSHAPE reactivity scores shown in color. The nucleotides outlined in red interact with GR amino acids, shown in blue. d, UCSC tracks showing icSHAPE reactivity scores (y-axis), along the RNA sequence. 1 denotes unstructured (single-stranded) regions, and 0 denotes fully-structured regions. e, Violin plot of Gini index of icSHAPE data in exon versus in intron. The thick black bar in the center of the Violin plot represents the interquartile range, the thin black line extended from it represents the 95% confidence intervals, and the white dot is the median. The numbers of sliding windows (width = 20nt) on the respective regions from the left to the right are n=18930, n=5926, n=51409, n=82648.
We determined RNA structure, as previously described12,32, after enriching for messenger RNAs (mRNAs) and long noncoding RNAs (lncRNAs) by ribosome depletion, and sequencing the resulting icSHAPE libraries at high depth (~200 million reads per replicate, Supplementary Table S1). We first confirmed the quality of fractionation using RT-qPCR for landmark RNAs, and Western blots for specific proteins (Supplementary Figure 1). We used the icSHAPE pipeline12 (read depth = 100 as threshold) to calculate a score to represent the structural flexibility (indicative of unpaired RNA bases) of every nucleotide, and found good correlation across replicates (Pearson correlation coefficient r > 0.75 for the top 60% most-abundant transcripts in all replicates, Supplementary Figure 2). As expected, the correlations between replicates are higher than those across fractions (Supplementary Figure 3a-b), and they are also higher for more-abundant RNAs (Supplementary Figure 3c). We also noticed an even distribution of mapped across RNA populations (Supplementary Figure 3d, Supplementary Table S2). To further validate our structural data, we examined its agreement with known structures – two such RNAs are Ribonuclease P RNA and Signal recognition particle (SRP) RNA (Supplementary Figure 4a-b). Both RNAs are enriched in nucleoplasm, and indeed our nucleoplasm icSHAPE data closely match the existing structural models.
The chromatin RNA structurome is enriched for lncRNAs (Figure 1b, Supplementary Figure 4c). As an example, we examined the structure of the human growth arrest-specific 5 (GAS5) noncoding RNA, which acts as a decoy glucocorticoid response element (GRE) by binding to the DNA-binding domain of the glucocorticoid receptor (GR)33. Indeed, the expected GAS5 RNA structure is accurately recovered in the chromatin fraction, showing low icSHAPE scores for the double-stranded GR binding motif of the GAS5 RNA, and high reactivity score for the loop region (Figure 1c). Similar to lncRNAs, small nucleolar RNAs (snoRNAs) and small nuclear RNAs (snRNAs) are also enriched in the chromatin fraction, and to a smaller extent in the nucleoplasm fraction (both relative to the cytoplasm). Furthermore, intronic reads constitute the majority of the sequencing data in the chromatin fraction, but only ~15–20% of reads in the cytoplasmic fraction (Figure 1b, Supplementary Figure 4d). For example, we obtained intron structures for the transcript heterogeneous nuclear ribonucleoprotein A2/B1 (HNRNPA2B1) in the chromatin fraction, but these sequences were largely absent in the nucleoplasmic and cytoplasmic fractions (Figure 1d). Interestingly, we found that RNAs in vivo are much more folded in intron regions than in exon regions (average Gini index of 0.7 versus 0.5. A higher Gini index indicates a more structured region11), in contrast to in vitro conditions (both with average Gini index 0.6); this result holds true for both human (Figure 1e) and mouse (Supplementary Figure 4e). The finding that intronic regions are more folded in vivo is not likely due to differential RNA-binding-protein (RBP) binding in introns versus exons, as similar trends were observed when all known RBP-binding sites were excluded in the structural comparison (Supplementary Figure 4f-g), and RBP binding sites were found to have no bias for intron or exon regions (7.9% of exon regions and 7.2% of intron regions are bound by RBPs). Instead, these results may suggest distinct interplays between RNA structures, and transcriptional or splicing regulation in introns and exons. In summary, the RNA-structural profiles of the chromatin fraction provide a rich resource to interrogate structures of lncRNAs, pre-mRNAs including introns, and other chromatin-associated RNAs, expanding the scope of the RNA structurome.
RNA structure plays a central role in connecting many cellular events.
The cytotopic RNA structuromes allowed us to assess the roles of RNA structure (or lack thereof) in association with each step of the gene-expression life cycle, which takes place in distinct subcellular compartments. We obtained data on transcriptional rate, translational efficiency, and RNA half-life from previous studies in human and mouse34-36, and correlated data with the Gini index of icSHAPE reactivity. RNA structure in nascent RNA has been suggested to propel or impede RNA polymerase pausing at individual genes37. We therefore analyzed the relationship between transcription and 5'UTR (untranslated region) RNA structures of the chromatin-associated fraction, and found that lower transcriptional rate correlates modestly with more structure (r = –0.19, p = 1.5 × 10−6, Figure 2a, Supplementary Figure 5a-b). Next, many studies have found RNA secondary structure upstream of or at ribosome binding sites may affect translation differently15,17-19. Indeed we did observe that more 5'UTR RNA structure correlates with decreased translational efficiency in the cytoplasmic fraction (r = –0.31, p = 1.7 × 10−48, Figure 2b, Supplementary Figure 5c-d). Finally, as RNA degradation occurs in both nucleoplasm and cytoplasm via different pathways, we analyzed the dependence of RNA half-life on RNA structure in both fractions. We found that more-structured RNAs tended to have shorter half-lives in both the nucleus and cytoplasm (r = –0.23, p = 4.6 × 10−91 in nucleoplasm and r = –0.18, p = 1.1 × 10−36 in cytoplasm. Figure 2c-d, Supplementary Figure 5e-h). To further confirm our conclusion, we repeated the above analysis with a higher read depth cutoff (read depth = 200, Supplementary Figure 5i) and three other datasets (Supplementary Figure 5j-l). We also observed the same trends in mRNA 5'UTR, CDS and 3'UTR, suggesting that the degradation is not RNA-region specific and could possibly be targeted by dsRNase38 (Supplementary Figure 6a). However, more direct evidence is needed to establish a widespread role of dsRNase-dependent cleavage in transcript turnover.
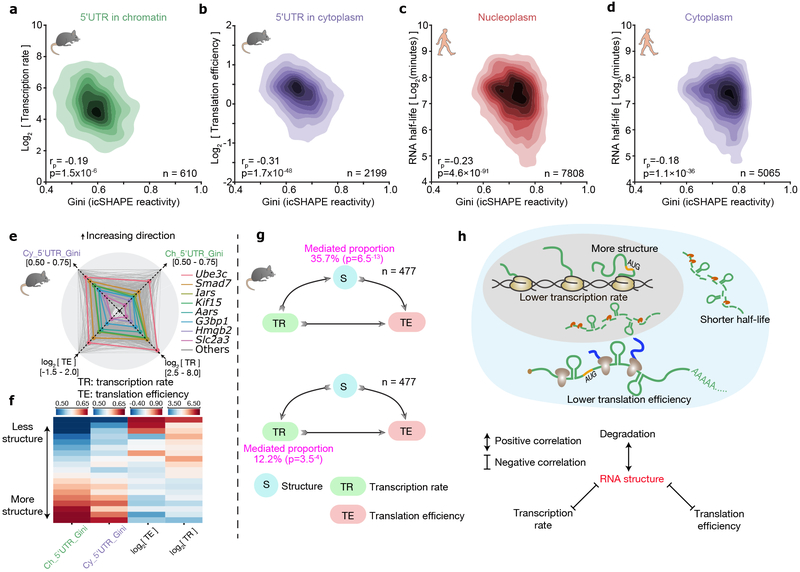
Fig 2 ∣. RNA structure plays a central role in connecting transcription, translation and RNA degradation.
a-d, Scatter plots of (a) transcription rate versus 5'UTR RNA structure in chromatin, (b) translational efficiency versus 5'UTR RNA structure in cytoplasm, (c) RNA half-life versus full-length-transcript RNA structure in nucleoplasm, and (d) RNA half-life versus RNA structure in cytoplasm. The 2-tailed p-value was calculated by python package function scipy.stats.pearsonr. rp is the Pearson correlation efficient. e, Radar diagram showing 5'UTR RNA structure in chromatin, 5'UTR RNA structure in the cytoplasm, transcription rate, and translational efficiency. Grey lines show all genes, and the colored lines highlight representative transcripts. f, Heatmap of 5'UTR RNA structure in chromatin, 5'UTR RNA structure in cytoplasm, transcription rate, and translational efficiency. Each strip represents an average of a bin comprising 5% data, ranked by RNA-structure reactivity in the chromatin fraction, 477 common transcripts are shown. g, Mediator model (above) and cofounding model (bottom) of RNA structure in connecting transcription rate and translation efficiency. P-values were calculated by two-sided t-test. h, Schematic showing RNA structure connects transcription, translation and RNA degradation.
Quantitative correlation analysis showed that the relationships among RNA structure, transcription and translation are not binary, as there is a general trend that an RNA with lower transcriptional rate tends to simultaneously be more structured and translated less efficiently (Figure 2e-f). The positive link between transcription and translation, two major events in gene expression, has been previously appreciated39(Supplementary Figure 6b). Recent studies have suggested different mechanisms, including m6A modification, that could account for this linkage by imprinting an mRNA transcript during its synthesis and later regulating its translation39-41. Our data suggest that genome-wide RNA structures formed at chromatin during transcription remain largely unchanged in the nucleoplasm and cytoplasm fractions, and might thus serve as a link between transcription and translation efficiencies. We therefore considered two models to explain our observations – in the first model RNA structure is a mediation factor that is affected by transcription, and it in turn affects translation; and in the second model RNA structure is a cofounding factor that has an effect on both transcription and translation (Figure 2g). Statistical analysis suggests that while both models could be true, the first (mediation) model can account for a larger fraction of the positive correlation between transcription and translation, and is statistically more significant. In summary, RNA structure plays a general role that connects many cellular events including transcription, translation and RNA degradation (Figure 2h).
Pervasive RNA structural changes across different cellular compartments.
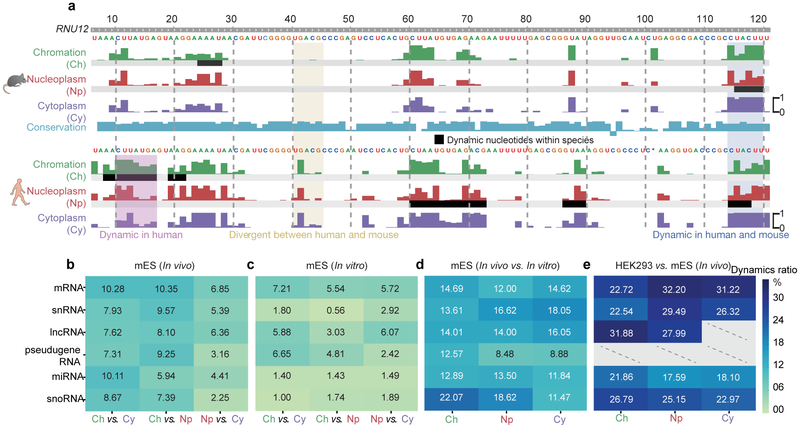
More importantly, cytotopic RNA structuromes also enabled us to examine how RNA adopts different conformations across different cellular compartments, which we term “structural change”. Overall, RNA structures seemed slightly more unfolded in the chromatin fraction (Supplementary Figure 7). As specific regions of an individual RNA can be regulated differently and display different patterns of structural changes, we implemented a statistical method to discover regions of structural variation (Method, Supplementary Table S3). As an example, we show that U12 snRNA displayed structural-change regions between compartments (Figure 3a, black bars). In addition, despite high evolutionary conservation of U12, the RNA structures showed shared and unique conformational changes in human and mouse. These findings suggest that both species-specific and conserved mechanisms may regulate RNA structures and structural change.
Fig 3 ∣. RNA structure differences in cellular context.
a, U12 small nuclear RNA (snRNA) structural change across cellular compartments, and the structural divergence in two species. Tracks show the icSHAPE score plotted along the RNA sequence. The black bars highlight RNA structural change regions. b-e, Heatmaps showing fractions of structurally-different regions across cellular compartments (b) in vivo, (c) in vitro, (d) between in vivo and in vitro, and (e), between human and mouse. Dashed lines represent insufficient data.
On a genome-wide scale, we found that different RNA categories showed different levels of structural change in vivo (Figure 3b). To begin to dissect the factors that regulate RNA structural change in cells, we used the same analysis pipeline to evaluate data obtained from fractionated, purified RNA that was refolded in vitro (Figure 3c), and compared RNA conformational changes observed between compartments in vivo and in vitro. In general, as expected, RNA structures vary less between the compartments in vitro relative to in vivo (comparing Figure 3b to Figure 3c), suggesting that fewer factors influence RNA folding in vitro versus in vivo. This finding is particularly true for highly-conserved small RNAs such as snoRNAs, micro RNAs (miRNAs) and snRNAs, suggesting that these functional RNAs adopt stable structures in vitro but are subjected to extensive regulation in vivo. The structural differences are magnified when directly comparing in vivo to in vitro icSHAPE data for each compartment (Figure 3d), and different RNA categories displayed varying levels of structural differences in vivo and in vitro, consistent with previous findings from whole-cell data12. Finally, we compared the levels of structural divergence between mouse and human for sequence-conserved regions. We used the same pipeline used above to call for regions of structural changes, and found even larger fractions of structural differences, suggesting substantial species-specific regulation of RNA structure (Figure 3e). Taken together, our analyses suggest that structural changes are pervasive, reflecting that many different factors may contribute to their regulation in different circumstances.
RNA modification and RBP binding underlie RNA structural changes.
RNA modification and RBP-binding are important factors that are known to influence RNA structure. To disambiguate their contributions to RNA structural change, we overlaid compartment-specific RNA structuromes with RNA modifications and RBP-binding sites. Figure 4a-c shows examples of focal conformational changes around known locations of m6A modification, pseudouridylation (Ψ) and heterogeneous nuclear ribonucleoprotein C (HNRNPC) binding.
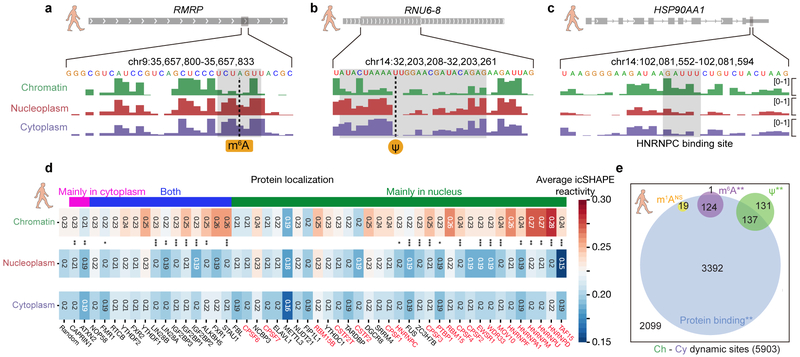
Fig 4 ∣. RNA modification and RBP binding underlie RNA structural changes.
a-c, RNA structural change at (a) an m6A-modified site, (b) a Ψ-modified site, and (c) an HNRNPC-binding site. Tracks show the icSHAPE score plotted along the RNA sequence. d, Heatmap of average icSHAPE scores in RBP binding regions in different cellular compartments, ranked by increasing structural change (from left to right) between the chromatin and the nucleoplasmic fractions. Proteins are annotated by their known localizations, with chromatin-associated RBPs shown in red. P-values were calculated by single-sided Mann-Whitney U test and corrected by the Bonferroni method. Source data for panel d are available online. e, The number and overlap of different types of RNA modification sites and RBP binding sites in regions with RNA structural change. P-values were calculated by a permutation test for 1,000 times. * p-value < 0.05; ** p-value < 1e-3; *** p-value < 1e-5.
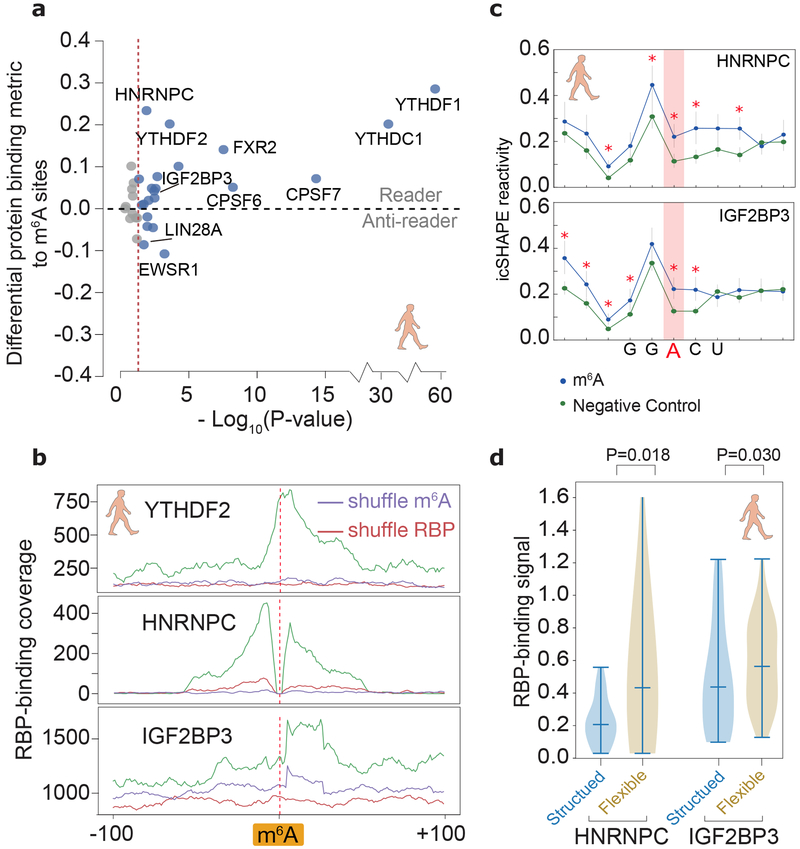
As m6A is well known as an RNA structure switch favoring unpairing of dsRNA,12,28 we compared the genome-wide structures for m6A methylated versus non-methylated sites with the same underlying sequence motif, and confirmed similar patterns of structure destabilization in all three fractions (Supplementary Figure 8a). Furthermore, the structural differences are largest in the nucleoplasm fraction, consistent with the finding that METTL3-METTL14 complex deposits m6A on nuclear RNA42. Following the structural changes of the same set of m6A sites from nucleoplasm and cytoplasm, we observed that RNA structure appears more open upon RNA migrating from the chromatin to the nucleoplasm, and thereafter remains the same (Figure 4a, Supplementary Figure 8a). This analysis agrees with that fact that the vast majority of m6A is deposited within the nucleus42. We repeated the analysis for pseudouridylation, another abundant RNA modification generated by the isomerization of uridine, which permits hydrogen bonding to the adjacent phosphate backbone. The extra hydrogen bond can rigidify RNA structure of Ψ-modified regions24. We found that in general these regions have higher icSHAPE reactivity (i.e. less structured), suggesting that modification hinders RNA structure folding freely, which again occurs predominantly in nucleus (Figure 4b, Supplementary Figure 8b). We note that our analysis is most powered to detect RNA modification effects where the modification events occur in a more homogenous fashion (i.e. the majority of the transcript copy have modifications at the site that occur in a specific cellular location). We may not be able to identify the structure-changing modifications that occur in a highly-variable manner for each transcript copy.
All RNAs associate extensively with proteins in cells, and RNA binding protein (RBP) interactions are both sensitive to and profoundly impacts RNA structure. Taking HNRNPC as an example, we first confirmed that it bound to a stem-loop structure, inferred from more single-stranded nucleotides with flanking dsRNA (Figure 4c). We also followed the structural transition of the binding sites from chromatin to nucleoplasm and cytoplasm. We found that HNRNPC binding sites are more open in chromatin, also consistent with its major localization in chromatin-associated pre-RNA (Supplementary Figure 8c). Our findings also suggest that HNRNPC binding could be a factor that accounts for the structural change around the binding sites. Indeed, there is a significant overlap between HNRNPC binding and structural variation sites (Supplementary Figure 9).
We extended the analysis to all RBPs with binding site information available from published RBP CLIP-seq experiments43. As shown in Figure 4d, occupancy of many RBPs are linked with RNA structural changes, while others preferentially bind to structurally-stable regions of RNA. For example, many chromatin-associated proteins (e.g. HNRNPD and others shown in red in Figure 4d) bind to more open RNA regions; these regions become more structured after dissociating from the chromatin and the proteins. In contrast, the double-stranded-binding RBP Staufen homolog 1 (STAU1), a protein that shuttles between the nucleus and the cytoplasm, appears to stabilize RNA structures upon its binding after RNA leaves chromatin. Thus, by determining the structuromes of multiple cytotopic localizations, our study provides an estimate of the relative contributions of known modification mechanisms and protein binding to RNA structural rearrangement (Figure 4e). Protein binding using existing CLIP-seq data can explain most of the RNA structural change sites (3392 of 5903), and many RNA modification sites with RNA structure changes overlap with protein binding sites. Our results thus suggest a complex interplay among RNA modification, protein binding and RNA structural change.
Structural analysis dissects different types of m6A readers.
Identifying RBPs that can read RNA modifications is of fundamental significance in the study of epitranscriptomics42 Using our cytotopic RNA structurome data to filter published CLIP-seq data, we computed the effect that m6A modification has on protein binding (Methods). Our analysis identified most of the known m6A readers, including the canonical YTH domain proteins, and the newly identified HNRNPC28 and the IGF2BP proteins44. All these readers bind to a region that contains one or more m6A sites stronger than a control (unmodified) site with the same m6A sequence motif (Figure 5a). Interestingly, the analysis also revealed several proteins with decreased bindings on modified m6A sites (termed “anti-readers”)42,45, including LIN-28 homolog A (LIN28A) and EW RNA binding protein 1 (EWSR1).
Fig 5 ∣. Structural analysis dissects different types of m6A readers.
a, Differential RBP binding to m6A sites and control sites containing an m6A motif. P-values are calculated to show the statistical significance of the binding differences by single-sided Mann-Whitney U test and corrected by the Benjamini/Hochberg method. Source data for panel a are available online. b, Metagene profiles of protein binding in m6A-flanking regions. c, Metagene profiles showing that RNA structures are different between known m6A-modified sites and unmodified sites (negative control), at m6A motifs overlapping a binding site of IGF2BP3 and HNRNPC. P-values were calculated by single-sided Mann-Whitney U test, red asterisks, p-values less than 0.01. The error bars represent the standard error of the mean. The numbers of HNRNPC and IGF2BP3 binding regions are 86 and 56, respectively. d, Violin plots of RBP-binding strengths of HNRNPC and IGF2BP3 in structured and flexible regions containing a m6A motif. Structured and flexible regions are defined as the RBP-binding regions at the bottom 30% or top 30% of average icSHAPE scores, respectively. P-values were calculated by single-sided Mann-Whitney U test. The numbers of HNRNPC and IGF2BP3 binding regions in comparison are 137 and 320, respectively.
The precise pattern of RBP binding peaks and RNA structure at m6A sites can further reveal the biochemical mechanism of the m6A readers (Figure 5b). While the canonical readers bind most strongly directly at the m6A sites, the binding of HNRNPC and IGF2BP readers peaks at a distance. Our icSHAPE data supports a previous study that suggested that HNRNPC acts as m6A reader not by recognizing the N6-methyl group, but rather by binding a purine-rich motif that becomes unpaired and accessible upon nearby m6A modification28 (Figure 5c). Similarly, our RNA-structural data suggest that IGF2BP proteins (here IGF2BP3) may also be able to read the structural changes induced by the so-called m6A-switch28 (Figure 5c). Furthermore, both HNRNPC and IGF2BP3 bind more tightly to flexible regions (Figure 5d).
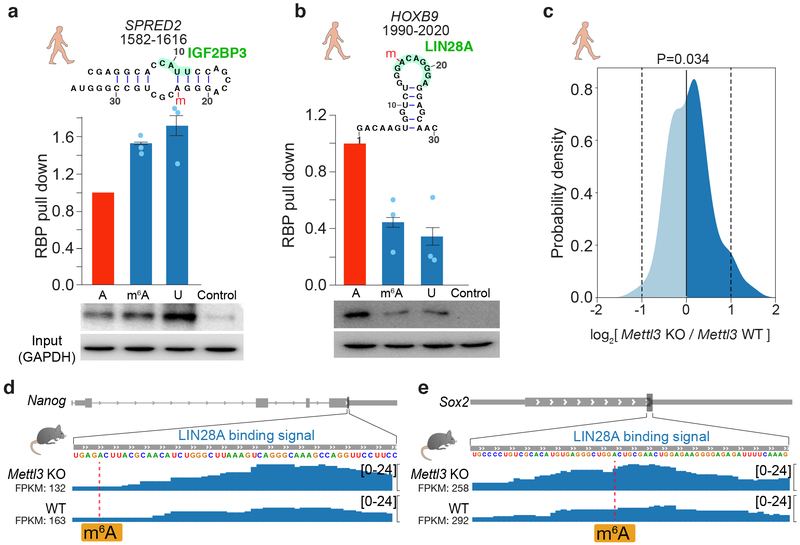
To validate the role of IGF2BP3 as a possible “indirect reader” and LIN28A as an “anti-reader” of m6A modification, we selected four endogenous m6A sites as targets. Each of the four targets contained three variants for the m6A site — an unmodified nucleotide, an m6A modification, and an adenosine-to-uracil mutation that mimics the disruption of base pairing (for IGF2BP3) or RBP binding (for LIN28A) (Figure 6a-b, Supplementary Data Set 1, Supplementary Figure 10a-b, Supplementary Note). We synthesized RNA oligonucleotides and used these RNA probes to retrieve RBPs from cell lysates. RNA pulldown analyses revealed that IGF2BP3 displays enhanced binding to the m6A-modified RNAs and uracil mutations relative to the unmethylated controls, confirming IGF2BP3 to be a m6A-switch reader to the hairpin probes (Figure 6a, Supplementary Figure 10a). IGF2BPs contain different RNA binding domains including two RNA recognition motifs (RRM) and four K-homology (KH) domains. A recent study suggested that the third and fourth KH domains of IGF2BP3 can recognize m6A directly via a GGAC motif44. Our data suggest that IGF2BP3 may also bind different RNA targets in a manner dependent on the m6A-structural switch, akin to the indirect m6A reader hnRNPC. Further experiments28 are necessary to validate the observations in vivo and to dissect the different domains of IGF2BP3 in reading m6A modifications.
Fig 6 ∣. Validation of IGF2BP3 as an indirect m6A reader and LIN28A as an anti-reader.
a-b, RNA pull-down assays and western blots for (a) IGF2BP3 and (b) LIN28A, using RNA probes that contain unmodified A, m6A, and U, respectively, derived from the indicated positions in the transcripts. m6A sites are marked with a red “m”. Histograms show mean of RNA pull-down from three independent replicates. The error bars represent standard error of mean (s.e.m.).Uncropped blots are shown in Supplementary Data Set 1. c. Density plot of LIN28A binding strength (log ratio) at m6A sites in Mettl3 knockout (KO) versus wild-type mES cells. P-value is calculated by two-sided t-test. The number of transcripts is 145. d-e, Signal tracks of Nanog and Sox2 showing LIN28A binding at specific loci in Mettl3 KO and wildtype mES cells.
Conversely LIN28A displayed reduced binding to the m6A-modified and uracil-mutant target RNAs, supporting the hypothesis that LIN28A is an anti-reader that requires an unmethylated adenosine for binding (Figure 6b, Supplementary Figure 10b). To confirm the anti-reader role of LIN28A, we performed LIN28A CLIP-seq experiments in the wild type and the m6A-methyltransferase Mettl3-knockout mES cells46. Many mRNAs containing one or more known m6A site showed increased binding to LIN28A when m6A deposition is abrogated, relative to the negative controls (p = 0.034, t test, Figure 6c-e, Supplementary Figure 10c-d). Increased LIN28A binding is not due to increased mRNA accumulation in Mettl3 KO ES cells (Figure 6c-e, Supplementary Figure 10c-d). LIN28A is an RBP known to enforce ES cell pluripotency and suppress ES cell differentiation47, while m6A is required for stem cell differentiation46. The negative regulation of m6A on LIN28A binding is consistent with the protein’s functional roles. For example, LIN28A is a well-studied inhibitor of primary microRNA processing48, and m6A was recently shown to promote pri-miRNA processing29. Thus, the discovery of LIN28A as an m6A anti-reader potentially unifies their functional and molecular mechanisms in pluripotency, microRNA biogenesis, and post-transcriptional gene regulation.
Discussion
Our analysis of RNA structuromes in different subcellular locations illuminated distinct RNA structural states in chromatin, nucleoplasm and cytosol. Fractionation enriched specific pools of RNAs, such as nuclear-enriched lncRNAs and pre-mRNAs including introns, thus substantially expand the scope and comprehensiveness of the RNA structuromes. Cytotopic RNA structuromes revealed the intimate connection between RNA structure and RNA processes such as transcription, translation, RNA degradation, RBP interaction and RNA modification. Through comparative analysis, we were able to dissect the role of RNA modifications and RNA-binding proteins in influencing structure, and resolved the different sets of direct and indirect RNA modification readers. We further found and validated a novel role of the pluripotency regulator LIN28A as an anti-reader for m6A modification.
How RNA structure is regulated in vivo had remained elusive, although this information is essential to revealing hidden roles of RNA structures in gene expression regulation. Our study presents the first landscape and regulation of RNA structuromes and their changes in mammalian cells. By comparative analysis we showed that the majority of the RNA structures are stable across three locations, suggesting that they have been largely determined since their biogenesis (Figure 3a-b). This structure stability could partially explain the correlations between different RNA events including transcription, translation and RNA decay (Figure 2h). Future studies involving structure perturbations that uncouple those functional correlations are required to test this hypothesis.
Nevertheless, our analysis has also revealed a large number of sites with RNA structure changes, which undergo conformational changes as RNAs transit from their sites of transcription on chromatin, are processed in the nucleus, and ultimately decoded in the cytoplasm. A recent study examined mRNA structure changes during zebrafish early embryogenesis and found translation to be a major driving force that shapes the landscape of mRNA structural changes49. Our cytotopic data offers an opportunity to validate the finding in mammalian cells. We found that the structural change between mRNAs in the chromatin fraction and the nucleoplasmic fraction is approximately the same as that between mRNAs in the nucleoplasmic fraction and the cytoplasmic fraction. Furthermore, the structural changes for mRNAs are similar to those for lncRNAs (Figure 3b). As translation remains a possible important biological process that helps to shape RNA structuromes, our observations suggest that other factors may play crucial roles that regulate RNA structure for both mRNAs and lncRNAs, in a similar fashion in mammalian cells.
Among many factors known to influence RNA structure, RNA modification and RBP-binding are important cis- and trans- regulators. Our comparative analysis illuminates their relative contributions to the observed RNA structure differences in different aspects. In vivo (Figure 3b) both RNA modification and RBP-binding are likely different in different compartments, whereas in vitro (Figure 3c) there are no RBP-binding to contribute to the structure changes. This difference in regulators may explain why RNA structures are more diverse in vivo. When comparing in vivo to in vitro structure (Figure 3d) RNA modification should remain unchanged, but there are no RBP-binding to contributes to the structural differences in vitro, thus suggesting that RBP-binding as a whole is an important regulator of RNA structure changes. And finally, both of RBP-binding and RNA modification are likely very different in mouse and human, which may account for the big structural divergences in the two species (Figure 3e).
Finally, the specific RNA regions that undergo structural transition at each subcellular location provide direct readouts of the molecular mechanisms that shape the gene expression program. The finding of LIN28A as a m6A anti-reader may have implications for human disease, as both LIN28A and m6A have been implicated in cancer progression, germ cell development, and metabolism50. In the future, studying RNA structural transitions together with RNA modifications and RBP binding in physiological states, and in the context of biological and structural perturbations, will help to elucidate the complex regulatory role of RNA structures in biology and medicine.
Methods
Cell culture and NAI-N3 modification in vivo
Human HEK293 cells were bought from Cell Bank, Shanghai Institutes for Biological Sciences, Chinese Academy of Sciences. The cells were cultured in DMEM supplemented with 10% FBS at 37°C with 5% CO2 in 15-cm plates. V6.5 mouse ES cells are from the Howard Chang laboratory. The cells were cultured at 37°C with 5% CO2 in 15-cm plates coated with 0.2% gelatin, in knockout DMEM (Gibco) media supplemented with 15% FBS, 1% PenStrep, 1% MEM NEAA, 1% Glutamax, 0.2% beta-mercaptoethanol, and 0.01% LIF. All cell lines are tested negative for mycoplasma contamination. Cells were cultured to 80~90% confluency, then rinsed, collected and treated with NAI-N3 as previously described by Flynn et. al. (2016)32.
Subcellular fractionation
Subcellular fractionation for HEK293 cells was performed as previously described31 with the following modifications. Cell pellets were first resuspended in 200 µL cold cytoplasmic lysis buffer using wide orifice tips and incubated on ice for 6 minutes. The subsequent steps were as described previously31, with the exception that for each collected subcellular fraction, 5% was used for immunoblot analysis, and then 1 mL Trizol LS (Life technologies) was added to the remaining aliquot for RNA purification using the QIAGEN RNA cleanup protocol.
Fractionation for mES cells was carried out as previously described30. We confirmed by imaging the efficacy of fractionation using DAPI to stain for intact nuclei, and used ER-tracker red (BODIPY TR Glibenclamide, Thermo Fischer Scientific) to confirm removal of ER contaminants in the nuclear fractionations. Ribolock RNase inhibitor (Thermo Fischer Scientific) was used to prevent RNA degradation.
Western blot and RT-qPCR
Western blot and quantitative RT-qPCR (RT-qPCR) of marker proteins/transcripts for HEK293-subcellular compartments were used to verify the subcellular fractionation results. Western blots were performed with antibodies for three proteins – GAPDH (Abcam), SNRP70 (Abcam) and histone H3 (Abcam). Samples of subcellular fractions were boiled at 95°C for 10 minutes, then spun at 14000g for 3 minutes at room temperature to minimize the influence of sticky DNA (especially in the chromatin samples) on Western blots. For every Western blot, 1% of the sample volume was used. For RT-qPCR, the same percentage (1%) of RNA samples was used. Then the ratio of each marker gene (GAPDH, U1, ACTIN [intron]) was calculated in the respective chromatin, nucleoplasmic and cytoplasmic fractions.
For mES cells, similar western-blot experiments were carried out using antibodies against actin (Abcam), histone H3 (Abcam) and SNRP70 (Abcam). We confirmed NAI-N3 treatment didn’t affect fractionation. Protein was quantified using the Pierce BCA protein assay kit (Thermo Fischer Scientific). We loaded the protein from each compartment in proportion to the amount obtained (roughly 1:1:2 for chromatin:nucleoplasm:cytoplasm).
NAI-N3 modification in vitro
For refolding, purified RNA was first denatured at 95°C for 1 min, quickly cooled to 4°C, and then incubated for 5 min in the folding buffer with 100 mM HEPES, 6 mM MgCl2, and 100 mM NaCl. Samples were then respectively modified for 5 minutes at 37°C with the NAI-N3 reagent and DMSO as control. The modified RNA was cleaned up and eluted in RNase-free water by using RNeasy mini columns (Qiagen).
IcSHAPE library construction of subcellular fractions
10 µg of RNA from each subcellular fraction for HEK293 cells was depleted of ribosomal RNA (rRNA) using the mouse/human ribominus kit (Invitrogen). About 500 ng RNA was recovered in the nucleoplasmic and cytoplasmic samples and about 2 µg RNA in the chromatin fractions. RNA from mES-cell fractions were depleted using the ribominus eukaryotic system v2 kit (Thermo Fischer Scientific). IcSHAPE sequencing libraries were then constructed from these RNA samples as previously described32. Libraries of mES-cells were sequenced on the Hiseq 4000 and libraries of HEK293 cells were sequenced on the Hiseq 2500 to ~200 million reads per replicate.
LIN28A plasmid transfection and RNA pull-down
30 µg pCMV-Flag human LIN28A vector was transfected into 15-cm plates with 90 µL polyethylenimine (PEI) dissolved in 1 mL opti-MEM, following the standard transfection protocol. Fresh medium was added after 6 hours, and cells were harvested after 48 hours. RT-qPCR (Takara) was performed to test the transfection efficiency.
The in vitro RNA pull-down assay was performed as described28. In summary 100 pmol RNA oligonucleotides were refolded by heating at 90°C for 1 minutes, and then incubated at 30°C for 5 minutes. 2 × 107 HEK293 cells were lysed in lysis buffer (150 mM NaCl, 1 mM EDTA, 1% Triton X-100, 0.5 mM DTT, 50 mM Tris-HCl, pH 7.5, 0.5% sodium deoxycholate) with 10 µL PMSF (Amresco), 10 µL phosphatase inhibitor cocktail (Promega), 2.5 µL SUPERase In inhibitor (Life Technologies) and 2.5 µL RNasin ribonuclease inhibitor (Promega) added. 10 µL refolded RNA and 1 mL cell lysate were incubated at 4°C for 2 hours. Then 100 µL pre-washed MyOneC1 streptavidin beads were added into the buffer and incubated at 4°C for 45 minutes. The beads were washed first with high salt buffer (50 mM Tris-HCl, pH 7.5, 1 M NaCl, 1% TRITON X-100) at room temperature for 4 minutes, and then 2 more times with low salt buffer, also at room temperature and for 4 minutes. Proteins were eluted in 15 µL elution buffer (1% SDS, 50 mM Tris-HCl pH 8.0, 1 M NaCl) at 95°C for 10 minutes. The eluted protein samples were quantified by western blot with IGF2BP3 antibody (Abcam) and LIN28A antibody (Abcam). Blotting membranes were stained by ECL-prime (RPN2232, GE Healthcare) and visualized by a digital imaging system. Control samples were prepared identically to the lysate samples, with the exception that no RNA oligonucleotides were added. The sequences of IGF2BP3-binding probes and LIN28A-binding probes are in the Supplementary Note.
CLIP-seq experiments
To carry out CLIP-seq experiments, mettl3 WT and KO mESCs46 were grown on gelatinized plates in Knockout DMEM media (Gibco) supplemented with 15% FBS, 1% penicillin-streptomycin (Gibco), glutamate, non-essential amino acids (NEAA), basal medium eagle (BME, Thermo Fischer Scientific), and ESGRO LIF (EMD Millipore). To harvest cells for irCLIP51, cells were initially washed with ice-cold PBS, then crosslinked on ice with UV-C (254 nm) at 0.3 J/cm2 in a Stratalinker, washed again with ice-cold PBS (+10 mM EDTA) for 5 minutes, and removed from the plate.
Immunoprecipitation of crosslinked protein-RNA complexes was performed for 3.5 hours at 4°C with anti-LIN28 antibody (Abcam ab46020). Washing of LIN28-RNA complexes, RNA trimming, on-bead biochemistry, and library generation was performed in triplicate as previously described51. Mouse-anti-GFP antibody (Abcam) was used to generate a negative-control library. Reverse transcription of LIN28 bound RNA was performed with SuperScript IV (Thermo Fischer Scientific) at 53°C for 30 minutes. Library amplification was done for a total of 14 cycles using Phusion HF polymerase master mix (NEB). Libraries were gel purified and submitted for sequencing. Libraries were sequenced on NextSeq 500 with custom sequencing primer P6_seq51.
Reads were mapped to mm10 assembly and PCR duplicates were removed using UMI-tools52). Reproducible RT stops were identified using the FAST-iCLIP pipeline53. Lin28a binding sites were called by Piranha54 with the parameter –s –b 50.
Reads mapping and filtering of icSHAPE data
Raw sequence reads were split by library barcodes, then collapsed to remove PCR duplicates and trimmed to remove 3' adaptors following the icSHAPE computational pipeline32. The genome and annotation files for human (hg38) and mouse (mm10) were downloaded from the GENCODE website and parsed to obtain transcriptome sequences by using cufflinks55 with the following parameter:
gffread -g genome.fa -s genome.size -W -M -F -G -A -O -E -w transcriptome.fa -d transcriptome.collapsed.info genome.gtf
Processed reads were mapped to the transcriptomes by using bowtie256 with icSHAPE suggested parameters (--non-deterministic --norc). IcSHAPE scores were then calculated as previously described, with enough coverage (read depth>100)32. To study intron structures, the full-gene sequences with introns were generated from the genome and annotation files. Processed reads were mapped to the full-gene sequences to calculate icSHAPE scores for both exons and introns.
Transcriptional rate, translational efficiency and half-life analysis
Publicly-available Ribo-seq (SRR315623) and RNA-seq (SRR315594) datasets were used to calculate translational efficiency (TE) for each transcript in mouse ES cells36,57, and a GRO-seq (SRR935117, SRR935118, SRR942449, SRR942450, SRR942451, SRR5655667,SRR5655668, SRR5655669, SRR5655670,SRR5655671) dataset was used to calculated transcriptional rate (TR), also in mES cells35,58,59. All sequenced reads were aligned to mouse protein-coding transcripts (including 5 mouse rRNA transcripts) by using bowtie2 with default parameters. For genes with multiple isoforms, the isoform with the longest coding sequence (CDS) was chosen as the reference. TE for each transcript was calculated as the reads per kilobase of mRNA per million reads (RPKM) of the CDS region in the Ribo-seq library, divided by the RPKM of the whole transcript in the RNA-seq library41. TR for each transcript was calculated as its number of transcripts (mapping reads decided by the transcripts length) in the GRO-seq library, the data was normalized by sequencing depth to compare between different samples. Pre-calculated half-life data for each transcript in HEK293 cells was collected from a previous study34.
All correlations and p-values were calculated with Python package Seaborn (https://seaborn.pydata.org).
Mediation and cofounding-factor analysis
The mediation and cofounding-factor analysis for the role of RNA structures in connecting transcription and translation were performed using methods described in the literature60,61. Data of translational efficiency (TE), transcriptional rate (TR) and RNA structure (in terms of Gini index of icSHAPE scores of the cytoplasmic 5'UTR) of 477 mouse transcripts were used for the analysis.
To test whether RNA structure is a mediation factor that is affected by transcription and then affects translation, two linear regressions were performed:
Here c is the regression coefficient relating TR to TE, and c' is the regression coefficient relating TR to TE adjusted for the mediator, i.e., RNA structure (S).
The value of the mediated effect was estimated by taking the difference in the coefficients, c – c’. The mediated proportion (1 – c’/c) is used to measure the effect size of mediation. The mediated effect (c – c’) divided by the standard error (, here σc is the standard error of c, σc′ is the standard error of c’, is the correlation of TR and S) was compared to the tN-2 distribution to determine whether the mediated effect is significant.
To test whether RNA structure is a cofounding factor that have an effect on both transcription and translation, another two linear regressions were performed:
The mediated proportion (1 – c’/c) was estimated, and significance test was conducted the same as described above.
Structural change site analysis
For each compartment, 2 DMSO replicates and 2 NAI-N3 replicates were used to calculate four icSHAPE scores combinatorically (by using 1 DMSO and 1 NAI-N3 sample every time). To compare the structural difference between two compartments, two sets of the four icSHAPE scores of each nucleotide were compared to calculate a p-value by singled-end t-test. All p-values are adjusted by the Bonferroni method. Those nucleotides with adjusted p-values < 0.05 and averaged icSHAPE score difference > 0.2 were defined as structural change sites. This calculation framework was used to define those sites with both large differences between the two compartments and small difference between replicates, as changed sites.
To obtain structural divergence between human and mouse, each transcript was pairwise-aligned by parsing .maf file from the UCSC genome browser website28. The structurally-divergent sites were called using the same approach.
Overlap of RNA modifications, RBP bindings and structural change sites
RBP binding data by CLIP-seq experiments were collected from CLIPdb43, and m1A, m6A and pseudouridylation sites were collected from literature62,63. Protein localization information was from the website GeneCards (genecards.org)64, which hosts the UniProt and COMPARTMENTS localization information. Some literatures were also referred to, especially for the protein localization in the chromatin fraction. To study if structural change sites are enriched in RBP binding region, m6A sites, m1A sites or pseudouridylation sites, change sites were random shuffled within the transcript for 1,000 times. Thereafter the number of change sites within an RBP-binding region or within the flanking region (10 nt) of a modification site was counted. The real overlapping number was compared with the overlapping numbers in all permutations to calculate the p-value. [* p-value < 0.05; ** p-value < 0.001; *** p-value< 0.00001]
M6A preference of RNA-binding proteins (RBPs)
True m6A modification sites with GGACU motifs were obtained from a published dataset62. To generate a control set of m6A modification sites, each transcript with true GGACU m6A sites was scanned by the GGACU motif to produce the same number of pseudo m6A sites with the same sequence motif, avoiding 20 nt flanking regions of a true m6A site.
For each RBP, the binding strength to each binding site was normalized to 0–1. And the binding sites were intersected with the true and control set of m6A modification sites. The binding strengths intersecting with true m6A were compared with the binding strengths intersecting with control (pseudo) m6A sites to calculate a p-value (single-end Mann-Whitney U test) and the mean difference.
Lin28a-binding peak calling and analysis
mES m6A modification sites were collected from a published dataset46. Those m6A sites with the ratio of binding strengths in Mettle3 KO versus WT less than 0.8 were filtered out.
Lin28a CLIP sequencing reads were mapped to the mouse genome (mm10) by using bowtie2 with default parameters. CLIP binding peaks were called with Piranha54 with parameters “-b 50 -s”. To study the correlation between m6A and Lin28a, their genome coordinates were mapped onto the transcriptome. M6A sites were shuffled within the same transcripts, keeping the same m6A sequence context (keeping the one base upstream and one base downstream of the m6A modification site unchanged). RBP-binding sites were also shuffled within the same transcripts. A Lin28a binding site was defined as overlapped with a m6A site if their distance was less than 50 nt.
Statistics and reproducibility
Figure 3b,c,d,e: Statistical comparisons of the compartment structural changes were carried out with the single-sided t-test adjusted by the Bonferroni method. Statistical significance was set to adjusted P value ≤ 0.05.
Figure 4d: Statistical comparisons between the average icSHAPE reactivities in RBP binding regions in different cellular compartments were assessed with the single-sided Mann-Whitney U test adjusted by the Bonferroni method. The exact P-values are represented in Source Data Figure 4d.
Figure 4e: Statistical significances of the overlap among RNA modifications, RBP bindings and structurally-change sites were assessed by a permutation test for 1,000 times. Exact p-values were:
protein binding: 0.0 for 1,000 permutation
m1A: 0.909
m6A: 0.0 for 1,000 permutation
ψ: 0.0 for 1,000 permutation
Figure 5a: Statistical significance of the binding differences was assessed by the single-sided Mann-Whitney U test adjusted by the Benjamini/Hochberg method. The exact p-values are presented in Source Data Fig5a.
Figure 5c: Statistical significance of structural difference was assessed by the single-sided Mann-Whitney U test. All difference with p-values lower than 0.01 are marked with red stars.
Figure 5d: Statistical significance of RNA-binding signal was assessed by the single-sided Mann-Whitney U test. The exact p-values were:
HNRNPC: 0.018
IGF2BP3: 0.030
Figure 6c: Statistical significance of LIN28A binding strength (log ratio) at the m6A sites in Mettl3 knockout (KO) versus wild-type mES cells was assessed by the two-sided t-test for the mean of one group of scores. The exact p-values were 0.034.
All tests are carried out with the scipy Python package (Scipy.org) with functions: scipy.stats.ttest_1samp, scipy.stats.mannwhitneyu, and scipy.stats.ttest_ind.
Code availability
All scripts can be found on GitHub at:
Data availability
All sequencing data are available through the Gene Expression Omnibus (GEO) under accession GSE117840 (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE117840). icSHAPE reactivity scores and Lin28A CLIP peaks can be found on the UCSC Genome Browser at:
Source data for Fig. 4d and Fig. 5a are available with the paper online.
Supplementary Material
Acknowledgments
We thank members of the Chang and Zhang labs for discussion. We thank Ryan Flynn for experimental advice. We thank Chao Dai, Yang Li, and Yang Yang for computational advice, and we acknowledge sequencing support from the genomics and synthesis biology core facility. This work is supported by NIH grants R01-HG004361 (H.Y.C), R35-CA209919 (to H.Y.C.) and R01GM127295 (to E.T.K.), and by the National Natural Science Foundation of China (Grants No. 31671355, 91740204, and 31761163007), and the National Thousand Young Talents Program of China (to Q.C.Z.). F.M.F. was supported by a NIH T32 Stanford Genome Training Program (SGTP) Fellowship and the Arnold O. Beckman Postdoctoral Fellowship. H.Y.C. is co-founder and serves on the SAB of Epinomics and Accent Therapeutics. Some sequencing data was generated on an Illumina Hiseq 4000 that was purchased with funds from NIH (award number S10OD018220). H.Y.C. is an Investigator of the Howard Hughes Medical Institute.
Footnotes
Competing interests
H.Y.C. is co-founder and serves on the SAB of Epinomics and Accent Therapeutics.
References
- 1.Sharp PA The centrality of RNA. Cell 136, 577–580, doi: 10.1016/j.cell.2009.02.007 (2009). [DOI] [PubMed] [Google Scholar]
- 2.Pan T & Sosnick T RNA folding during transcription. Annu Rev Biophys Biomol Struct 35, 161–175, doi: 10.1146/annurev.biophys.35.040405.102053 (2006). [DOI] [PubMed] [Google Scholar]
- 3.Warf MB & Berglund JA Role of RNA structure in regulating pre-mRNA splicing. Trends in biochemical sciences 35, 169–178, doi: 10.1016/j.tibs.2009.10.004 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Martin KC & Ephrussi A mRNA localization: gene expression in the spatial dimension. Cell 136, 719–730, doi: 10.1016/j.cell.2009.01.044 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kozak M Regulation of translation via mRNA structure in prokaryotes and eukaryotes. Gene 361, 13–37, doi: 10.1016/j.gene.2005.06.037 (2005). [DOI] [PubMed] [Google Scholar]
- 6.Garneau NL, Wilusz J & Wilusz CJ The highways and byways of mRNA decay. Nat Rev Mol Cell Biol 8, 113–126, doi: 10.1038/nrm2104 (2007). [DOI] [PubMed] [Google Scholar]
- 7.Ramakrishnan V Ribosome structure and the mechanism of translation. Cell 108, 557–572 (2002). [DOI] [PubMed] [Google Scholar]
- 8.Yan C et al. Structure of a yeast spliceosome at 3.6-angstrom resolution. Science 349, 1182–1191, doi: 10.1126/science.aac7629 (2015). [DOI] [PubMed] [Google Scholar]
- 9.Wan Y et al. Landscape and variation of RNA secondary structure across the human transcriptome. Nature 505, 706–709, doi: 10.1038/nature12946 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ding Y et al. In vivo genome-wide profiling of RNA secondary structure reveals novel regulatory features. Nature 505, 696–700, doi: 10.1038/nature12756 (2014). [DOI] [PubMed] [Google Scholar]
- 11.Rouskin S, Zubradt M, Washietl S, Kellis M & Weissman JS Genome-wide probing of RNA structure reveals active unfolding of mRNA structures in vivo. Nature 505, 701–705, doi: 10.1038/nature12894 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Spitale RC et al. Structural imprints in vivo decode RNA regulatory mechanisms. Nature 519, 486–490, doi: 10.1038/nature14263 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lu Z et al. RNA Duplex Map in Living Cells Reveals Higher-Order Transcriptome Structure. Cell 165, 1267–1279, doi: 10.1016/j.cell.2016.04.028 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zubradt M et al. DMS-MaPseq for genome-wide or targeted RNA structure probing in vivo. Nature methods 14, 75–82, doi: 10.1038/nmeth.4057 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mustoe AM et al. Pervasive Regulatory Functions of mRNA Structure Revealed by High-Resolution SHAPE Probing. Cell 173, 181–195 e118, doi: 10.1016/j.cell.2018.02.034 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Strobel EJ, Yu AM & Lucks JB High-throughput determination of RNA structures. Nature reviews. Genetics, doi: 10.1038/s41576-018-0034-x (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mortimer SA, Kidwell MA & Doudna JA Insights into RNA structure and function from genome-wide studies. Nature reviews. Genetics 15, 469–479, doi: 10.1038/nrg3681 (2014). [DOI] [PubMed] [Google Scholar]
- 18.Bevilacqua PC, Ritchey LE, Su Z & Assmann SM Genome-Wide Analysis of RNA Secondary Structure. Annual review of genetics 50, 235–266, doi: 10.1146/annurev-genet-120215-035034 (2016). [DOI] [PubMed] [Google Scholar]
- 19.Piao M, Sun L & Zhang QC RNA Regulations and Functions Decoded by Transcriptome-wide RNA Structure Probing. Genomics Proteomics Bioinformatics 15, 267–278, doi: 10.1016/j.gpb.2017.05.002 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wan Y, Kertesz M, Spitale RC, Segal E & Chang HY Understanding the transcriptome through RNA structure. Nature reviews. Genetics 12, 641–655, doi: 10.1038/nrg3049 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Buxbaum AR, Haimovich G & Singer RH In the right place at the right time: visualizing and understanding mRNA localization. Nat Rev Mol Cell Biol 16, 95–109, doi: 10.1038/nrm3918 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gosai SJ et al. Global analysis of the RNA-protein interaction and RNA secondary structure landscapes of the Arabidopsis nucleus. Molecular cell 57, 376–388, doi: 10.1016/j.molcel.2014.12.004 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Roundtree IA, Evans ME, Pan T & He C Dynamic RNA Modifications in Gene Expression Regulation. Cell 169, 1187–1200, doi: 10.1016/j.cell.2017.05.045 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhao BS, Roundtree IA & He C Post-transcriptional gene regulation by mRNA modifications. Nat Rev Mol Cell Biol 18, 31–42, doi: 10.1038/nrm.2016.132 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kierzek E & Kierzek R The thermodynamic stability of RNA duplexes and hairpins containing N6-alkyladenosines and 2-methylthio-N6-alkyladenosines. Nucleic Acids Res 31, 4472–4480 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Roost C et al. Structure and thermodynamics of N6-methyladenosine in RNA: a spring-loaded base modification. J Am Chem Soc 137, 2107–2115, doi: 10.1021/ja513080v (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Konig J et al. iCLIP reveals the function of hnRNP particles in splicing at individual nucleotide resolution. Nature structural & molecular biology 17, 909–915, doi: 10.1038/nsmb.1838 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Liu N et al. N(6)-methyladenosine-dependent RNA structural switches regulate RNA-protein interactions. Nature 518, 560–564, doi: 10.1038/nature14234 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Alarcon CR, Lee H, Goodarzi H, Halberg N & Tavazoie SF N6-methyladenosine marks primary microRNAs for processing. Nature 519, 482–485, doi: 10.1038/nature14281 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gagnon KT, Li L, Janowski BA & Corey DR Analysis of nuclear RNA interference in human cells by subcellular fractionation and Argonaute loading. Nat. Protoc. 9, 2045–2060, doi: 10.1038/nprot.2014.135 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bhatt DM et al. Transcript dynamics of proinflammatory genes revealed by sequence analysis of subcellular RNA fractions. Cell 150, 279–290, doi: 10.1016/j.cell.2012.05.043 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Flynn RA et al. Transcriptome-wide interrogation of RNA secondary structure in living cells with icSHAPE. Nature protocols 11, 273–290, doi: 10.1038/nprot.2016.011 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kino T, Hurt DE, Ichijo T, Nader N & Chrousos GP Noncoding RNA gas5 is a growth arrest- and starvation-associated repressor of the glucocorticoid receptor. Sci. Signal 3, ra8, doi: 10.1126/scisignal.2000568 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Schueler M et al. Differential protein occupancy profiling of the mRNA transcriptome. Genome Biol 15, R15, doi: 10.1186/gb-2014-15-1-r15 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Jonkers I, Kwak H & Lis JT Genome-wide dynamics of Pol II elongation and its interplay with promoter proximal pausing, chromatin, and exons. Elife 3, e02407, doi: 10.7554/eLife.02407 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ingolia NT, Lareau LF & Weissman JS Ribosome profiling of mouse embryonic stem cells reveals the complexity and dynamics of mammalian proteomes. Cell 147, 789–802, doi: 10.1016/j.cell.2011.10.002 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zhang JW & Landick R A two-way street: regulatory interplay between RNA polymerase and nascent RNA structure. Trends Biochem. Sci. 41, 293–310, doi: 10.1016/j.tibs.2015.12.009 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Houseley J & Tollervey D The many pathways of RNA degradation. Cell 136, 763–776, doi: 10.1016/j.cell.2009.01.019 (2009). [DOI] [PubMed] [Google Scholar]
- 39.Harel-Sharvit L et al. RNA polymerase II subunits link transcription and mRNA decay to translation. Cell 143, 552–563, doi: 10.1016/j.cell.2010.10.033 (2010). [DOI] [PubMed] [Google Scholar]
- 40.Zid BM & O'Shea EK Promoter sequences direct cytoplasmic localization and translation of mRNAs during starvation in yeast. Nature 514, 117–121, doi: 10.1038/nature13578 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Slobodin B et al. Transcription Impacts the Efficiency of mRNA Translation via Co-transcriptional N6-adenosine Methylation. Cell 169, 326–337 e312, doi: 10.1016/j.cell.2017.03.031 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Roundtree IA & He C RNA epigenetics--chemical messages for posttranscriptional gene regulation. Current opinion in chemical biology 30, 46–51, doi: 10.1016/j.cbpa.2015.10.024 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Yang YC et al. CLIPdb: a CLIP-seq database for protein-RNA interactions. BMC Genomics 16, 51, doi: 10.1186/s12864-015-1273-2 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Huang H et al. Recognition of RNA N(6)-methyladenosine by IGF2BP proteins enhances mRNA stability and translation. Nat Cell Biol 20, 285–295, doi: 10.1038/s41556-018-0045-z (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Edupuganti RR et al. N(6)-methyladenosine (m(6)A) recruits and repels proteins to regulate mRNA homeostasis. Nature structural & molecular biology 24, 870–878, doi: 10.1038/nsmb.3462 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Batista PJ et al. m(6)A RNA modification controls cell fate transition in mammalian embryonic stem cells. Cell stem cell 15, 707–719, doi: 10.1016/j.stem.2014.09.019 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Yu J et al. Induced pluripotent stem cell lines derived from human somatic cells. Science 318, 1917–1920, doi: 10.1126/science.1151526 (2007). [DOI] [PubMed] [Google Scholar]
- 48.Viswanathan SR & Daley GQ Lin28: A microRNA regulator with a macro role. Cell 140, 445–449, doi: 10.1016/j.cell.2010.02.007 (2010). [DOI] [PubMed] [Google Scholar]
- 49.Beaudoin JD et al. Analyses of mRNA structure dynamics identify embryonic gene regulatory programs. Nature structural & molecular biology 25, 677–686, doi: 10.1038/s41594-018-0091-z (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Shyh-Chang N & Daley GQ Lin28: primal regulator of growth and metabolism in stem cells. Cell stem cell 12, 395–406, doi: 10.1016/j.stem.2013.03.005 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Zarnegar BJ et al. irCLIP platform for efficient characterization of protein-RNA interactions. Nat. Methods 13, 489–492, doi: 10.1038/nmeth.3840 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Smith T, Heger A & Sudbery I UMI-tools: modeling sequencing errors in Unique Molecular Identifiers to improve quantification accuracy. Genome Res. 27, 491–499, doi: 10.1101/gr.209601.116 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Flynn RA et al. Dissecting noncoding and pathogen RNA-protein interactomes. RNA 21, 135–143, doi: 10.1261/rna.047803.114 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Murakami Y, Spriggs RV, Nakamura H & Jones S PiRaNhA: a server for the computational prediction of RNA-binding residues in protein sequences. Nucleic Acids Res 38, W412–416, doi: 10.1093/nar/gkq474 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Trapnell C et al. Differential gene and transcript expression analysis of RNA-seq experiments with TopHat and Cufflinks. Nat. Protoc. 7, 562–578, doi: 10.1038/nprot.2012.016 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Langmead B & Salzberg SL Fast gapped-read alignment with Bowtie 2. Nat. Methods 9, 357–359, doi: 10.1038/nmeth.1923 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Yoshikawa H et al. Efficient analysis of mammalian polysomes in cells and tissues using Ribo Mega-SEC. Elife 7, doi: 10.7554/eLife.36530 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Min IM et al. Regulating RNA polymerase pausing and transcription elongation in embryonic stem cells. Genes & development 25, 742–754, doi: 10.1101/gad.2005511 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Tastemel M et al. Transcription pausing regulates mouse embryonic stem cell differentiation. Stem Cell Res 25, 250–255, doi: 10.1016/j.scr.2017.11.012 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.MacKinnon DP, Fairchild AJ & Fritz MS Mediation analysis. Annu Rev Psychol 58, 593–614, doi: 10.1146/annurev.psych.58.110405.085542 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.MacKinnon DP, Lockwood CM, Hoffman JM, West SG & Sheets V A comparison of methods to test mediation and other intervening variable effects. Psychol Methods 7, 83–104 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Linder B et al. Single-nucleotide-resolution mapping of m6A and m6Am throughout the transcriptome. Nature methods 12, 767–772, doi: 10.1038/nmeth.3453 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Dominissini D et al. The dynamic N(1)-methyladenosine methylome in eukaryotic messenger RNA. Nature 530, 441–446, doi: 10.1038/nature16998 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Stelzer G et al. The GeneCards suite: from gene data mining to disease genome sequence analyses. Curr. Protoc. Bioinformatics 54, 1 30 31–31 30 33, doi: 10.1002/cpbi.5 (2016). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All sequencing data are available through the Gene Expression Omnibus (GEO) under accession GSE117840 (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE117840). icSHAPE reactivity scores and Lin28A CLIP peaks can be found on the UCSC Genome Browser at:
Source data for Fig. 4d and Fig. 5a are available with the paper online.