Abstract
lα-Aspartic acid (Asp) residues in proteins are nonenzymatically isomerized to abnormal lβ-, dα-, and dβ-Asp isomers under physiological conditions. Such an isomerization of Asp residues is considered to be a trigger of protein denaturation because it either elongates the main chain or induces a different orientation of the side chain within the protein structure or both. However, previous studies have found no direct evidence of the effects of Asp isomers on protein function. Therefore, the production of Asp-isomer-containing proteins is required to verify the effects of Asp isomerization. Here, we describe the production of an Asp-isomer-containing protein using the expressed protein ligation. As a model protein, bovine pancreatic ribonuclease A (RNase A, EC 3.1.27.5), which catalyzes the cleavage of phosphodiester bonds in RNA, was used. In this study, lα-Asp at position 121 in RNase A was replaced by lβ-, dα-, and dβ-Asp. The objective aspartic acid at position 121 is located near the active site and related to RNA cleavage. The RNase A with lα-Asp at position 121 showed a normal activity. By contrast, the catalytic activity of lβ-, dα-, and dβ-Asp-containing RNase A was markedly decreased. This study represents the first synthesis and analysis of a protein containing four different Asp isomers.
Introduction
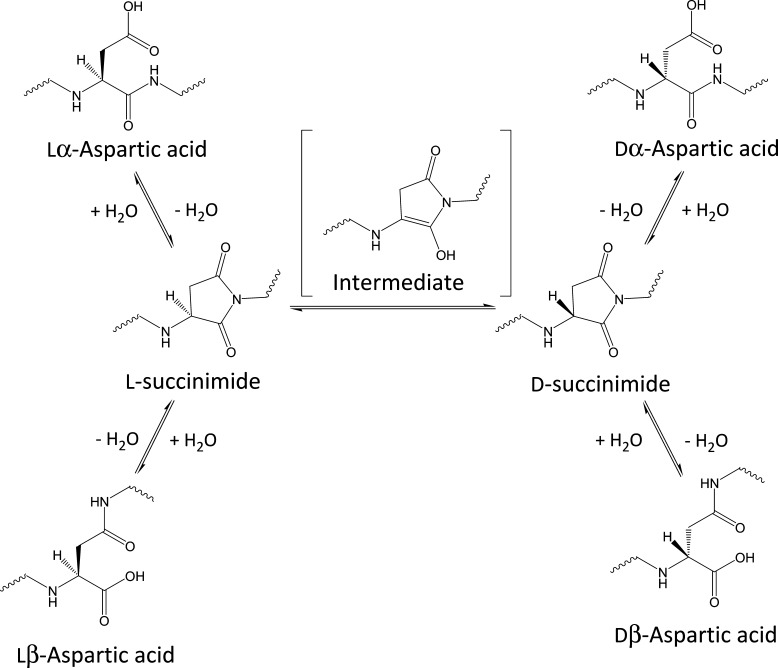
All amino acids except glycine have two enantiomers: l-amino acid and d-amino acid. l-Amino acid has been assumed to be an exclusive constituent of proteins in nature. However, recent studies have demonstrated the presence of many d-amino acid-containing proteins in various aged tissues, such as eye lens, brain, and skin.1−5 Above all, the d-aspartic acid (Asp) residue is considered to be associated with age-related disorders because many d-Asp residues have been found to accumulate in aged proteins as compared with other amino acid residues. Commonly, Asp residues in proteins are of the lα form; however, lα-Asp residues in proteins can be nonenzymatically isomerized to abnormal lβ-, dα-, and dβ-Asp isomers under physiological conditions via the formation of a succinimidyl intermediate over time, as shown in Figure 1(6) Isomerization of Asp is considered to be a trigger of protein denaturation as it induces elongation of the main chain and a different orientation of the side chain within the protein structure.
Figure 1.
Isomerization pathways of aspartic acid via succinimidyl intermediates.
Aki et al. studied the kinetics of isomerization of Asp residues in peptides, predicting that >60% of lα-Asp will isomerize to the lβ-Asp form within a few years in metabolically inert tissues.7 Therefore, isomerization of Asp residues is considered to be related to not only age-related disorders but also a wide range of protein-degenerative diseases, such as prion diseases. An analysis of changes in protein function due to isomerization of Asp is thus required.
Protein expression using recombinant DNA technology is the principal method of producing a target protein. Although it is the most frequently used approach, it is unable to produce d-Asp-containing proteins because this Asp isomer is a noncoded amino acid. In addition, although chemical synthesis is able to introduce Asp isomers into a peptide chain, the synthesis of a large polypeptide with more than 50 amino acid residues is technically difficult. It is also very difficult to extract the d-Asp-containing protein from aged tissue because of the very small amounts of these proteins. Given the above background, methods for the preparation of Asp isomer-containing protein are limited. Here, to overcome these limitations, we have focused on a combination of native chemical ligation (NCL) and chemical peptide synthesis to create an Asp-isomer-containing protein and evaluated the effect of Asp isomerization on this protein.
NCL, a coupling reaction that produces an amide bond between the thiol of an N-terminal cysteine (Cys) residue in one peptide and a C-terminal thioester in another peptide, was developed by Dawson et al.,8 which can be combined with expressed protein ligation (EPL), where the C-terminal thioester is produced by recombinant DNA technology. On the basis of these ligation reactions, several proteins have been synthesized.9−13 However, EPL is restricted to the synthesis of proteins containing Asp isomers because the cysteine residue must be located near the N- or C-terminus and the isomerization site of the Asp residue needs to be located outside this cysteine. Therefore, as an initial step to clarify the effect of Asp isomerization on protein function, we carried out a study on bovine pancreatic ribonuclease A (RNase A, EC 3.1.27.5), which has a native Asp residue that is amenable to EPL.
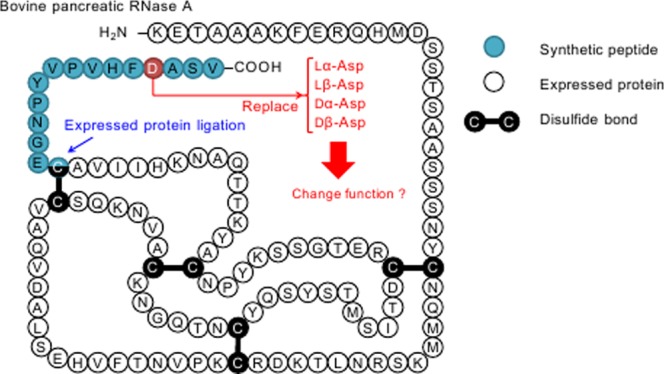
RNase A is a small monomeric protein composed of 124 amino acids with a molecular mass of 13 690. The three-dimensional structure of RNase A is fully defined by its amino acid sequence.14,15 Therefore, RNase A has been used extensively as a model protein to clarify structure–activity relationships, as summarized by Raines.16 The active site of RNase A has been clearly defined: two histidine (His12 and His119) and one lysine (Lys41) are the most important amino acids for the activity of RNase A. Structural analyses show that Asp121 forms a hydrogen bond with His119 during RNA cleavage by RNase A.17,18 In a site-directed mutagenesis study, where Asp121 was replaced by a glutamate residue (D121E), the catalytic activity of D121E decreased to 17% compared to that of wild-type RNase A. Furthermore, Asp121 is located near the C-terminus, and Cys110 is located on the inside of Asp12119 (Figure 2). Hence, Asp121 of RNase A is well positioned for isomer replacement via EPL.
Figure 2.
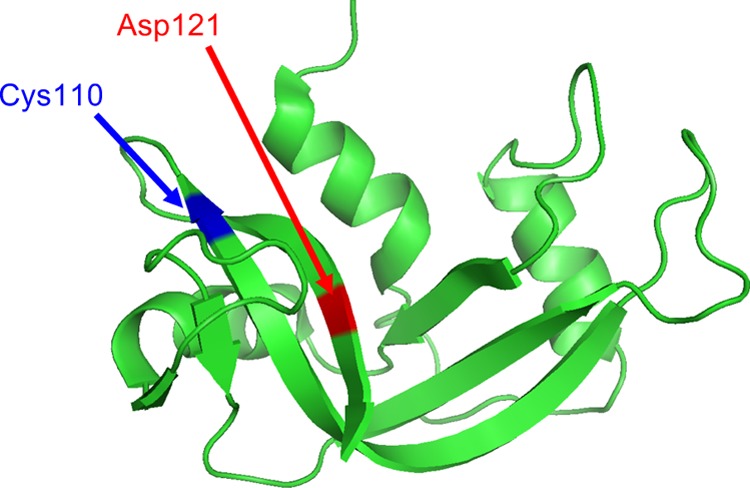
Structure of bovine pancreas ribonuclease A (EC3.1.27.5). The positions of Asp121 and Cys110 are shown in red and blue, respectively.
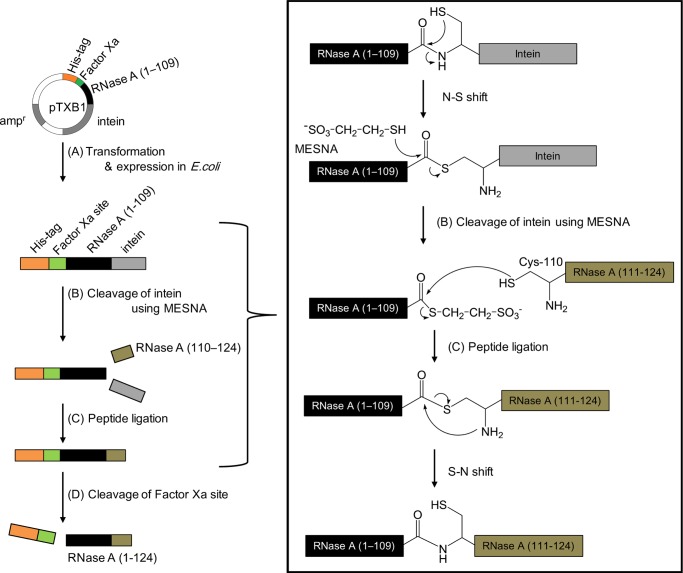
In this study, Asp121 of RNase A was replaced by three abnormal Asp isomers using EPL. The large segment of RNase A (residues 1–109) with a C-terminal thioester was expressed by the recombinant DNA technology. The short segment of RNase A corresponding to residues 110–124 with an N-terminal Cys residue was synthesized by the solid-phase peptide synthesis method, in which Asp121 was replaced by each of the Asp isomers. After their preparation, the large segment of RNase A was ligated to the short segment containing lα-Asp, lβ-Asp, dα-Asp, or dβ-Asp to form lα-RNase A, lβ-RNase A, dα-RNase A, and dβ-RNase A, respectively (Figure 3). The catalytic activities of different Asp-isomer-containing RNase A enzymes were then compared.
Figure 3.
Overview of the synthesis of RNase A by EPL.
In the present study, we report the novel preparation of a protein with each Asp isomer incorporated at a specific site coupled with an investigation of the function of the resultant variant proteins. The results clearly show that Asp isomerization affects the protein function.
Results
Synthesis of Asp-Isomer-Containing RNase A by EPL
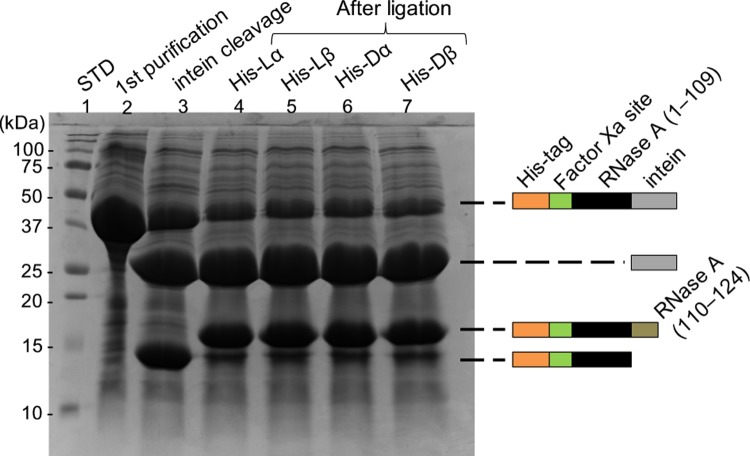
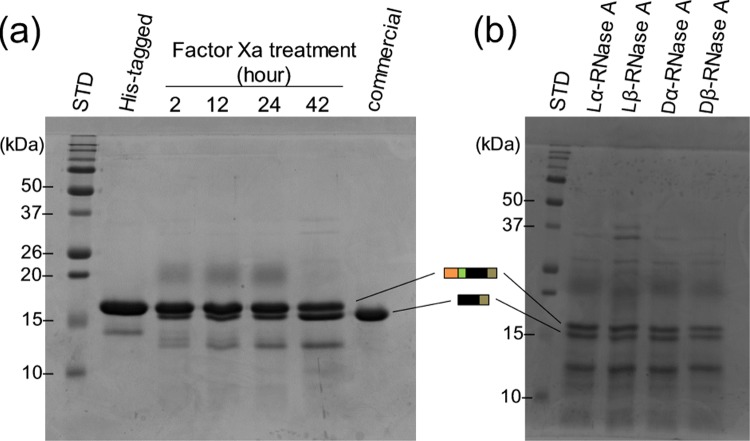
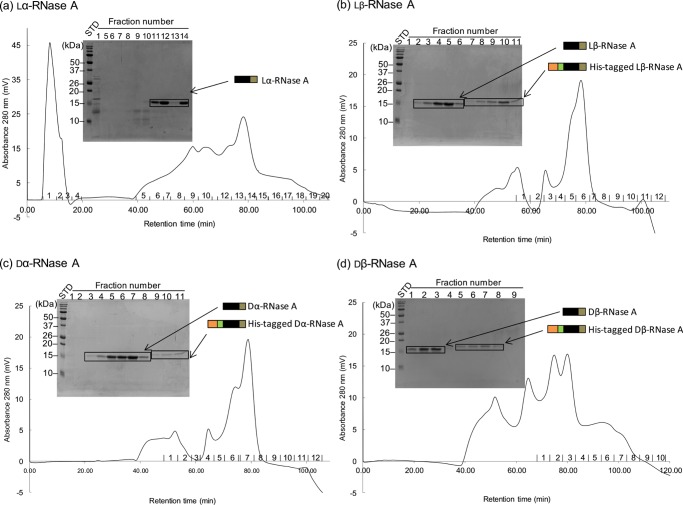
In this study, we synthesized an Asp-isomer-containing RNase A using EPL. The progression of the ligation reaction between the large segment and the short segment was monitored by sodium dodecyl sulfate-polyacrylamide gel electrophoresis using 15% polyacrylamide gels. Figure 4 shows the electrophoretogram of the large segment of RNase A (1–109) (lane 2), after the cleavage of intein (lane 3), and after the ligation of the short segment of RNase A (110–124) containing lα-, lβ-, dα-, and dβ-Asp at position 121 in lanes 4–7, respectively. Lane 3 indicates that intein was cleaved by 2-mercaptoethane sulfonate (MESNA) from RNase A (1–109), whereas lanes 4–7 indicate that the short segment of RNase A (110–124) was connected to the large segment of RNase A (1–109). The ligation efficiency did not differ among the different types of Asp isomer. After ligation, the His-tag was removed from RNase A (1–124) using factor Xa. About 50% of the His-tag was removed from lα-RNase A over a reaction time of 20 h (Figure 5a). Figure 5b shows the four isomeric RNases after removal of the His-tag by factor Xa. The four RNases, which still contained His-tag, were purified by ion-exchange chromatography using a cation-exchange column (TOYOPERL CM-650M; Tosoh Bioscience, Japan) (Figure 6). Figure 6 clearly shows the purity of the four RNases after ion-exchange chromatography.
Figure 4.
Electrophoretogram of the expressed intein-fused RNase A before and after peptide ligation using 15% polyacrylamide gel.
Figure 5.
Electrophoretogram of RNase A (1–124) before and after removal of the His-tag by factor Xa using 15% polyacrylamide gels. (a) Time course of the His-tag removal reaction from lα-RNase A by factor Xa. (b) Comparison of the molecular weights of four isomeric RNases after removal of the His-tag using factor Xa for 20 h.
Figure 6.
Elution profiles of ion-exchange chromatography using a cation-exchange column (CM-650M) with a linear gradient of 0–1 M NaCl in the presence of 20 mM Tris–HCl (pH 7.4) at a flow rate of 1 mL/min and electrophoretograms of purified Asp-isomer-containing RNase A (1–124) using 15% polyacrylamide gel; (a) lα-RNase, (b) lβ-RNase A, (c) dα-RNase A, and (d) dβ-RNase A.
Differential RNase Activity Depending on Asp Isomerization
The catalytic activity of RNase A toward 2′,3′-cCMP was measured by monitoring the increase in absorbance at 284 nm. Figure 7 shows the time course of the activity of the RNase A variants with different Asp isomers. The catalytic activity of lα-RNase A was at the same level as that of commercially available RNase A (Sigma-Aldrich, St. Louis, MO). On the other hand, replacement of the lα-Asp isomer at position 121 with lβ-, dα-, and dβ-Asp decreased the RNase activity.
Figure 7.
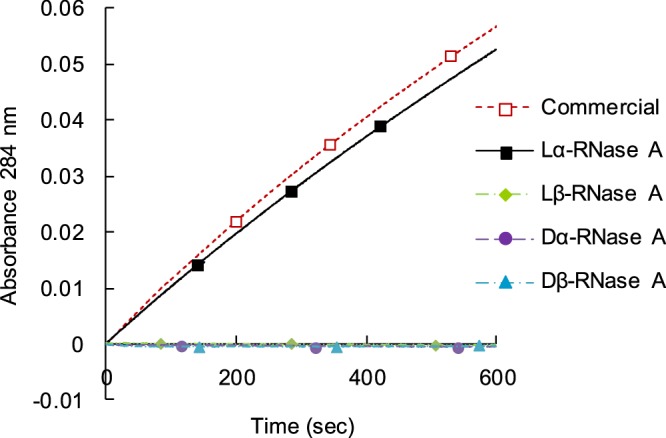
Comparison of the catalytic activities of four RNase A proteins containing different Asp isomers.
Differential RNase Solubility Depending on Asp Isomerization
The solubility of RNase A differed depending on the type of replacement of the Asp isomer. An aliquot of the RNase A (1–124) enzymes containing Asp isomers was stored in a lyophilized state after dialysis against water. Lyophilized RNase A containing lα-Asp and dα-Asp was soluble in water at a final concentration of 1 mg/mL. By contrast, RNase A containing lβ-Asp and dβ-Asp showed a very low solubility in water.
Discussion
In this study, a protein containing four different Asp isomers was initially synthesized, and it was revealed for the first time that isomerization of an Asp residue in the protein induced a marked change in protein function and solubility. RNase A was selected as an initial model protein to introduce four different Asp isomers because (1) it is amenable to EPL and (2) it is easy to evaluate the changes in RNase A function caused by Asp isomerization.
Our results clearly indicated that the catalytic activity of RNase A was completely lost by introducing abnormal Asp isomers at position 121. The loss of activity of the isomeric RNase A enzymes might be due to the structural changes caused by introducing isomerization at Asp121. We tried to analyze the structures of these proteins using X-ray crystallography, nuclear magnetic resonance analysis, circular dichroism spectroscopy, and infrared spectroscopy. However, structural information could not be obtained because of the low solubility of the isomeric RNase A proteins. In a previous study, the RNase activity of a D121E variant was found to decrease by approximately 83%.20 In common with gene mutation, the introduction of Asp isomers at position 121 clearly had a large effect on enzyme activity.
His12, His119, and Lys41 are the most important amino acids for the activity of RNase A. Asp121 forms a hydrogen bond with His119 during the RNA cleavage by RNase A. It is likely that the isomerization of Asp121 made it impossible for His119 to form the hydrogen bond that is needed for RNase activity. In addition to this functional change, the Asp isomerization affected the solubility of RNase A. Although Asp121 of RNase A is not isomerized in vivo, this study provides valuable information on the functional changes that are likely to occur in a protein due to the replacement of a single Asp isomer.
Asp isomers are often found in various metabolically inert tissues, such as eye lens, brain, skin, bone, artery wall, and tooth.1−5 In particular, they are found in metabolically inert proteins, such as crystallin from eye lens and β-amyloid protein from brain.21−27
In 2006, Fujii et al. demonstrated that the isomerization of one Asp residue in a partial peptide of αB-crystallin induced changes in the secondary structure, hydrophobicity, and chaperone activity.28 Molecular dynamics simulation also revealed that the inversion of l-leucine to d-leucine at position 2 in the tetrapeptide Leu-Leu-Gly-Asp induced a change in its three-dimensional structure.29 These structural changes may be a trigger of protein dysfunction and insolubilization. Indeed, our previous study supported the hypothesis that αB-crystallin and βA3-crystallin, in which an Asp residue is inverted from the l-form to the d-form, cannot assemble the crystallin complex in aged human lens.30 In addition, a large amount of Asp isomers has been found in the insoluble fraction of human cataract lens compared to that of the soluble fraction.31
In addition, it has been reported that, apart from isomerization, various amino acid modifications, such as deamidation, truncation, oxidation, and methylation, occur in human crystallin. These modifications are also considered to be a factor in the insolubilization of human lens.32−38
Asp isomers tend to accumulate in a metabolically inert tissue, such as crystallin, because the isomerization of Asp undergoes an equilibrium reaction. Therefore, the accumulation of proteins, in which function and solubility are decreased by the isomerization of Asp, may be a trigger of age-related diseases. It is considered that the dysfunction of RNase A observed in this study is also related to the denaturation of this enzyme. In terms of RNase A, isomerization from the α-form to the β-form (epimerization) may induce larger changes in the protein structure compared to those by the isomerization from the l-form to the d-form (inversion) because lyophilized RNase A containing β-Asp (lβ- and dβ-Asp) was insoluble, whereas RNase A containing dα-Asp remained soluble. Asp121 of RNase A is located near the C-terminus and forms a β-sheet structure through hydrogen bonds between NH and CO groups in the main chain of the protein. Because epimerization of an Asp residue extends the main chain of the protein, the β-sheet structure would be broken in RNase A containing lβ- and dβ-Asp. On the other hand, the overall structure might not be affected in RNase A containing dα-Asp. However, side-chain orientation is also important for RNase activity because the carboxylic group of Asp121 is related to the stabilization of the active center of RNase A. Thus, the inversion of Asp121 is considered to affect the stabilization of the active-site structure in RNase A. In this study, the Asp-isomer-containing RNase A proteins showed decreased solubility, suggesting that their structures might differ from those of lα-RNase A. lα-RNase A was undoubtedly folded to the proper structure because its enzymatic activity was the same as that of commercially available RNase A. This result suggested that protein disulfide isomerase (PDI) properly folded the chemically synthesized RNase A enzymes. On the one hand, PDI has the ability to refold not only RNase A but also other proteins by replacing non-native disulfides with the correct native ones.39−41 On the other hand, the enzymatic activity of the isomeric RNases was decreased despite using PDI. Therefore, isomerization of Asp121 in RNase A is considered to affect the RNase activity. Even if the isomeric RNases were misfolded, PDI might not refold them properly because it might not recognize a protein containing an Asp isomer. Nevertheless, although we could not determine the structure of the isomeric RNases, Asp isomerization undoubtedly changed the behavior of RNase. The selection of a suitable protein for analyzing the structural changes induced by the isomerization may help us to solve this problem. However, the isomerization of Asp is considered to be strongly related to protein aggregation, as observed for crystallin and β-amyloid protein. Therefore, it might be difficult to analyze the structural changes caused by the isomerization of Asp by avoiding aggregation of the protein.
This study demonstrated that EPL is a useful method to construct Asp-isomer-containing proteins. Synthesis of various proteins containing Asp isomer using EPL would be an effective way to verify the relationship between protein aggregation and isomerization of Asp.
Isomerization of Asp is one of the many post-translational modifications of proteins, including acetylation, phosphorylation, glycation, and deamidation. Asp isomers in aged proteins are produced spontaneously and slowly over time during the life span of an animal. Most cases of cataract and Alzheimer’s disease develop during the aging process regardless of the presence or absence of a genetic defect. Indeed, a large amount of Asp isomers are found in patients with these diseases. Therefore, it is strongly suspected that Asp isomerization is related to the aggregation and malfunction of aged proteins. Asp isomerization will be an important factor in age-related diseases even if an individual is born healthy. To clarify the effect of different Asp isomers on a protein, it is necessary to synthesize proteins containing each of the four Asp isomers. To date, however, limitations in the methods of protein synthesis have prevented the widespread use of this approach. For example, recombinant DNA technology cannot introduce Asp isomers into a protein, as the standard solid-phase peptide synthesis cannot prepare polypeptides longer than 100 amino acids. Therefore, EPL was used to overcome these problems in the present study. However, other approaches will be necessary to find further information about functional changes caused by Asp isomerization. For example, the isomerization site of Asp in crystallin in vivo is located on the inner side of crystallin. Thus, the synthesis of Asp-isomer-containing crystallin protein using EPL would be difficult because the site of the Asp isomer must be located near the N- or C-terminus and the cysteine residue required for EPL needs to be located inside the site of the Asp isomer. Thus, a method other than EPL is needed to synthesize crystallins containing Asp isomers. Human secretory phospholipase A2 has been synthesized by NCL,42 which might be applicable to crystallins and other proteins containing Asp isomers. The synthesis of crystallins and other proteins containing Asp isomers is a subject for our future studies.
In summary, it has been clarified that the introduction of Asp isomers into a protein decreases the protein function and leads to insolubilization. Exploring the role of d-amino acids in proteins in an l-amino acid world opens up a new field of protein science and will be useful to understand the mechanism underlying the onset of age-related diseases.
Experimental Procedures
Peptide Synthesis
The short segment of RNase A (residues 110–124; Cys-Gln-Gly-Asn-Pro-Tyr-Val-Pro-Val-His-Phe-Asp121-Ala-Ser-Val), containing four different Asp isomers at Asp121, was synthesized by 9-fluorenylmethyloxycarbonyl group (Fmoc)-based solid-phase peptide synthesis using an automated solid-phase peptide synthesizer (PSSM-8; Shimadzu, Japan). Fmoc amino acids and other coupling reagents were purchased from Watanabe Chemical Industries, Ltd. Fmoc-l-Asp(OtBu)-OH for use as lα-Asp was converted to Fmoc-l-Asp-OtBu-OH, Fmoc-d-Asp(OtBu)-OH, and Fmoc-d-Asp-OtBu-OH for use, respectively, as lβ-, dα-, and dβ-Asp. The coupling reaction was carried out by mixing each Fmoc amino acid (10 equiv), (benzotriazol-1-yloxy)tripyrrolidinophosphonium hexafluorophosphate (10 equiv), 1-hydroxybenzotriazole hydrate (10 equiv), and N-methylmorpholine (7.5 equiv) in N,N-dimethylformamide (DMF). The N-terminal Fmoc group was deblocked with 30% piperidine in DMF. A spontaneous cleavage of the peptide from the resin and the removal of the protective groups were achieved by the treatment of a mixture containing 90% trifluoroacetic acid (TFA), 5% 1,2-ethanedithiol, and 5% thioanisole for 2 h. The crude peptides were purified by reversed-phase high-performance liquid chromatography using a C18 column (Capcell Pak C18 ACR, 10 × 250 mm2; Shiseido, Japan) with a linear gradient of 0–50% acetonitrile in the presence of 0.1% TFA at a flow rate of 3.0 mL/min and detection at 230 nm.
Construction of Expression Plasmid Containing the Intein-Fused RNase A Gene
A complementary DNA (cDNA) of RNase A was purchased from Open Biosystems, Inc. Site-directed mutagenesis was carried out to delete the SapI restriction site in the RNase A gene. After mutagenesis, the cDNA of RNase A encoding amino acids 1–109 was amplified by polymerase chain reaction (PCR). During the PCR, an NdeI restriction site, His-tag, and factor Xa site were incorporated at the 5′ end, and an SapI restriction site was added at the 3′ end of the RNase A gene (1–109) (Figure S1). The amplified gene was cloned into a pTXB1 vector using NdeI and SapI. The primers used in the mutagenesis and PCR are given in Supporting Information.
Expression of the Intein Fusion Protein
The constructed plasmid was transformed into T7 express lysY/Iq Competent Escherichia coli (New England BioLabs, Ipswich, MA), and a large segment of RNase A (1–109) was expressed. The crude extract was purified by affinity chromatography using a Ni-sepharose column (HisPrep FF 16/10 Column; GE Healthcare UK Ltd) with a linear gradient of 0–250 mM imidazole in the presence of 20 mM Tris–HCl (pH 7.4), 500 mM NaCl, and 2 M urea at a flow rate of 1 mL/min using an AKTAprime system (GE Healthcare Life Sciences) (Figure S2).
EPL
The purified large segment of RNase A (1–109) dissolved in 20 mM Tris–HCl (pH 7.4), 500 mM NaCl, and 2 M urea was treated with MESNA for 20 h to cleave intein (Figure 3B). After cleavage, the short segment of RNase A (110–124) was added to the RNase A (1–109) solution (Figure 3C). The resulting RNase A (1–124) fused to His-tag was purified by affinity chromatography using a Ni-sepharose column (HisPrep FF 16/10 Column; GE Healthcare UK Ltd). After purification, the RNase A was treated by factor Xa (Merck Millipore, Darmstadt, Germany) in 50 mM Tris–HCl (pH 8.0), 100 mM NaCl, and 5 mM CaCl2 to cleave the His-tag (Figure 3D). The full-length RNase A was purified by ion-exchange chromatography using a cation-exchange column (TOYOPERL CM-650M; Tosoh Bioscience, Japan) with a linear gradient of 0–1 M NaCl in the presence of 20 mM Tris–HCl (pH 7.4) at a flow rate of 1 mL/min using an AKTAprime system (Figure 6).
Assay of RNA Cleavage Activity
The activity of RNase A was measured as described by Crook et al.43 In this study, 12 μg of RNase A was refolded using PDI (Takara Bio, Japan) in the presence of 20 mM reduced l-glutathione and 2 mM oxidized l-glutathione for 1 min at 25 °C. After refolding, the RNase A was added to 0.1 M cytidine 2′:3′-cyclic monophosphate (2′,3′-cCMP, Sigma-Aldrich, St. Louis, MO) dissolved in 0.1 M MOPS buffer (pH 7.0). The rate of increase in the absorbance at 284 nm was monitored by absorption spectrophotometry (UV-2700; Shimadzu, Japan).
Acknowledgments
This work was supported by a grant from the Ministry of Education, Culture, Sports, Science and Technology of Japan.
Glossary
Abbreviations
- 2′,3′-cCMP
cytidine 2′:3′-cyclic monophosphate
- cDNA
complementary DNA
- DMF
N,N-dimethylformamide
- dNTPs
deoxyribonucleotides
- EPL
expressed protein ligation
- Fmoc
9-fluorenylmethyloxycarbonyl group
- IPTG
isopropyl-β-d-thiogalactopyranoside
- LB
Luria-Bertani
- MESNA
2-mercaptoethane sulfonate
- NCL
native chemical ligation
- TE
Tris–EDTA
Supporting Information Available
The Supporting Information is available free of charge on the ACS Publications website at DOI: 10.1021/acsomega.6b00346.
Detailed description of the general experimental procedures, including the purification data of Asp isomers containing RNase A, is provided (PDF)
Author Present Address
∥ Department of Pharmaceutical Sciences, School of Pharmacy, International University of Health and Welfare, Kitakanemaru, Ohtawara, Tochigi 324-8501, Japan.
The authors declare no competing financial interest.
Supplementary Material
References
- Masters P. M. Stereochemically altered noncollagenous protein from human dentin. Calcif. Tissue Int. 1983, 35, 43–47. 10.1007/bf02405005. [DOI] [PubMed] [Google Scholar]
- Ritz S.; Turzynski A.; Schutz H. W.; Hollmann A.; Rochholz G. Identification of osteocalcin as a permanent aging constituent of the bone matrix: basis for an accurate age at death determination. Forensic Sci. Int. 1996, 77, 13–26. 10.1016/0379-0738(95)01834-4. [DOI] [PubMed] [Google Scholar]
- Matzenauer C.; Reckert A.; Ritz-Timme S. Estimation of age at death based on aspartic acid racemization in elastic cartilage of the epiglottis. Int. J. Legal Med. 2014, 128, 995–1000. 10.1007/s00414-013-0940-6. [DOI] [PubMed] [Google Scholar]
- Shapira R.; Chou C. H. Differential racemization of aspartate and serine in human myelin basic protein. Biochem. Biophys. Res. Commun. 1987, 146, 1342–1349. 10.1016/0006-291X(87)90797-2. [DOI] [PubMed] [Google Scholar]
- Spinelli P.; Brown E. R.; Ferrandino G.; Branno M.; Montarolo P. G.; D’Aniello E.; Rastogi R. K.; D’Aniello B.; Baccari G. C.; Fisher G.; D’Aniello A. D-aspartic acid in the nervous system of Aplysia limacina: possible role in neurotransmission. J. Cell. Physiol. 2006, 206, 672–681. 10.1002/jcp.20513. [DOI] [PubMed] [Google Scholar]
- Geiger T.; Clarke S. Deamidation, isomerization, and racemization at asparaginyl and aspartyl residues in peptides. Succinimide-linked reactions that contribute to protein degradation. J. Biol. Chem. 1987, 262, 785–794. [PubMed] [Google Scholar]
- Aki K.; Fujii N.; Fujii N. Kinetics of isomerization and inversion of aspartate 58 of alphaA-crystallin peptide mimics under physiological conditions. PLoS One 2013, 8, e58515 10.1371/journal.pone.0058515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dawson P. E.; Muir T. W.; Clark-Lewis I.; Kent S. B. Synthesis of proteins by native chemical ligation. Science 1994, 266, 776–779. 10.1126/science.7973629. [DOI] [PubMed] [Google Scholar]
- Torbeev V. Y.; Kent S. B. Convergent chemical synthesis and crystal structure of a 203 amino acid “covalent dimer” HIV-1 protease enzyme molecule. Angew. Chem., Int. Ed. Engl. 2007, 46, 1667–1670. 10.1002/anie.200604087. [DOI] [PubMed] [Google Scholar]
- Hondal R. J.; Nilsson B. L.; Raines R. T. Selenocysteine in native chemical ligation and expressed protein ligation. J. Am. Chem. Soc. 2001, 123, 5140–5141. 10.1021/ja005885t. [DOI] [PubMed] [Google Scholar]
- Schmohl L.; Wagner F. R.; Schumann M.; Krause E.; Schwarzer D. Semisynthesis and initial characterization of sortase A mutants containing selenocysteine and homocysteine. Bioorg. Med. Chem. 2015, 23, 2883–2889. 10.1016/j.bmc.2015.03.057. [DOI] [PubMed] [Google Scholar]
- Schwarzer D.; Zhang Z.; Zheng W.; Cole P. A. Negative regulation of a protein tyrosine phosphatase by tyrosine phosphorylation. J. Am. Chem. Soc. 2006, 128, 4192–4193. 10.1021/ja0585174. [DOI] [PubMed] [Google Scholar]
- Evans T. C. Jr.; Benner J.; Xu M. Q. Semisynthesis of cytotoxic proteins using a modified protein splicing element. Protein Sci. 1998, 7, 2256–64. 10.1002/pro.5560071103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smyth D. G.; Stein W. H.; Moore S. The sequence of amino acid residues in bovine pancreatic ribonuclease: revisions and confirmations. J. Biol. Chem. 1963, 238, 227–234. [PubMed] [Google Scholar]
- Kartha G.; Bello J.; Harker D. Tertiary structure of ribonuclease. Nature 1967, 213, 862–5. 10.1038/213862a0. [DOI] [PubMed] [Google Scholar]
- Raines R. T. Ribonuclease A. Chem. Rev. 1998, 98, 1045–1066. 10.1021/cr960427h. [DOI] [PubMed] [Google Scholar]
- Wlodawer A.; Sjolin L. Structure of ribonuclease A: results of joint neutron and X-ray refinement at 2.0-A resolution. Biochemistry 1983, 22, 2720–8. 10.1021/bi00280a021. [DOI] [PubMed] [Google Scholar]
- Quirk D. J.; Raines R. T. His ... Asp catalytic dyad of ribonuclease A: histidine pKa values in the wild-type, D121N, and D121A enzymes. Biophys. J. 1999, 76, 1571–1579. 10.1016/S0006-3495(99)77316-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurpiewska K.; Font J.; Ribo M.; Vilanova M.; Lewinski K. X-ray crystallographic studies of RNase A variants engineered at the most destabilizing positions of the main hydrophobic core: further insight into protein stability. Proteins 2009, 77, 658–669. 10.1002/prot.22480. [DOI] [PubMed] [Google Scholar]
- Wlodawer A.; Miller M.; Sjolin L. Active site of RNase: neutron diffraction study of a complex with uridine vanadate, a transition-state analog. Proc. Natl. Acad. Sci. U.S.A. 1983, 80, 3628–3631. 10.1073/pnas.80.12.3628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujii N.; Ishibashi Y.; Satoh K.; Fujino M.; Harada K. Simultaneous racemization and isomerization at specific aspartic acid residues in alpha B-crystallin from the aged human lens. Biochim. Biophys. Acta 1994, 1204, 157–163. 10.1016/0167-4838(94)90003-5. [DOI] [PubMed] [Google Scholar]
- Fujii N.; Satoh K.; Harada K.; Ishibashi Y. Simultaneous stereoinversion and isomerization at specific aspartic acid residues in alpha A-crystallin from human lens. J. Biochem. 1994, 116, 663–669. 10.1093/oxfordjournals.jbchem.a124577. [DOI] [PubMed] [Google Scholar]
- Fujii N.; Kawaguchi T.; Sasaki H.; Fujii N. Simultaneous stereoinversion and isomerization at the Asp-4 residue in betaB2-crystallin from the aged human eye lenses. Biochemistry 2011, 50, 8628–8635. 10.1021/bi200983g. [DOI] [PubMed] [Google Scholar]
- Mitkevich V. A.; Petrushanko I. Y.; Yegorov Y. E.; Simonenko O. V.; Vishnyakova K. S.; Kulikova A. A.; Tsvetkov P. O.; Makarov A. A.; Kozin S. A. Isomerization of Asp7 leads to increased toxic effect of amyloid-beta42 on human neuronal cells. Cell Death Dis. 2013, 4, e939 10.1038/cddis.2013.492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugiki T.; Utsunomiya-Tate N. Site-specific aspartic acid isomerization regulates self-assembly and neurotoxicity of amyloid-beta. Biochem. Biophys. Res. Commun. 2013, 441, 493–498. 10.1016/j.bbrc.2013.10.084. [DOI] [PubMed] [Google Scholar]
- Inoue K.; Hosaka D.; Mochizuki N.; Akatsu H.; Tsutsumiuchi K.; Hashizume Y.; Matsukawa N.; Yamamoto T.; Toyo’oka T. Simultaneous determination of post-translational racemization and isomerization of N-terminal amyloid-beta in Alzheimer’s brain tissues by covalent chiral derivatized ultraperformance liquid chromatography tandem mass spectrometry. Anal. Chem. 2014, 86, 797–804. 10.1021/ac403315h. [DOI] [PubMed] [Google Scholar]
- Masters P. M.; Bada J. L.; Zigler J. S. Jr. Aspartic acid racemisation in the human lens during ageing and in cataract formation. Nature 1977, 268, 71–73. 10.1038/268071a0. [DOI] [PubMed] [Google Scholar]
- Fujii N.; Fujii N.; Kida M.; Kinouchi T. Influence of Lbeta-, Dalpha- and Dbeta-Asp isomers of the Asp-76 residue on the properties of alphaA-crystallin 70–88 peptide. Amino Acids 2010, 39, 1393–1399. 10.1007/s00726-010-0597-0. [DOI] [PubMed] [Google Scholar]
- Gößler-Schöfberger R.; Hesser G.; Reif M. M.; Friedmann J.; Duscher B.; Toca-Herrera J. L.; Oostenbrink C.; Jilek A. A stereochemical switch in the aDrs model system, a candidate for a functional amyloid. Arch. Biochem. Biophys. 2012, 522, 100–106. 10.1016/j.abb.2012.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakaue H.; Takata T.; Fujii N.; Sasaki H.; Fujii N. Alpha B- and betaA3-crystallins containing d-aspartic acids exist in a monomeric state. Biochim. Biophys. Acta 2015, 1854, 1–9. 10.1016/j.bbapap.2014.10.006. [DOI] [PubMed] [Google Scholar]
- Fujii N.; Sakaue H.; Sasaki H.; Fujii N. A rapid, comprehensive liquid chromatography-mass spectrometry (LC-MS)-based survey of the Asp isomers in crystallins from human cataract lenses. J. Biol. Chem. 2012, 287, 39992–40002. 10.1074/jbc.M112.399972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilmarth P. A.; Tanner S.; Dasari S.; Nagalla S. R.; Riviere M. A.; Bafna V.; Pevzner P. A.; David L. L. Age-related changes in human crystallins determined from comparative analysis of post-translational modifications in young and aged lens: does deamidation contribute to crystallin insolubility?. J. Proteome Res. 2006, 5, 2554–2566. 10.1021/pr050473a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanson S. R.; Hasan A.; Smith D. L.; Smith J. B. The major in vivo modifications of the human water-insoluble lens crystallins are disulfide bonds, deamidation, methionine oxidation and backbone cleavage. Exp. Eye Res. 2000, 71, 195–207. 10.1006/exer.2000.0868. [DOI] [PubMed] [Google Scholar]
- Sharma K. K.; Santhoshkumar P. Lens aging: effects of crystallins. Biochim. Biophys. Acta 2009, 1790, 1095–1108. 10.1016/j.bbagen.2009.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lampi K. J.; Amyx K. K.; Ahmann P.; Steel E. A. Deamidation in human lens betaB2-crystallin destabilizes the dimer. Biochemistry 2006, 45, 3146–53. 10.1021/bi052051k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takata T.; Oxford J. T.; Demeler B.; Lampi K. J. Deamidation destabilizes and triggers aggregation of a lens protein, betaA3-crystallin. Protein Sci. 2008, 17, 1565–1575. 10.1110/ps.035410.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z.; Han J.; David L. L.; Schey K. L. Proteomics and phosphoproteomics analysis of human lens fiber cell membranes. Invest. Ophthalmol. Visual Sci. 2013, 54, 1135–1143. 10.1167/iovs.12-11168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Truscott R. J.; Mizdrak J.; Friedrich M. G.; Hooi M. Y.; Lyons B.; Jamie J. F.; Davies M. J.; Wilmarth P. A.; David L. L. Is protein methylation in the human lens a result of non-enzymatic methylation by S-adenosylmethionine?. Exp. Eye Res. 2012, 99, 48–54. 10.1016/j.exer.2012.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldberger R. F.; Epstein C. J.; Anfinsen C. B. Purification and Properties of a Microsomal Enzyme System Catalyzing the Reactivation of Reduced Ribonuclease and Lysozyme. J. Biol. Chem. 1964, 239, 1406–1410. [PubMed] [Google Scholar]
- Givol D.; Delorenzo F.; Goldberger R. F.; Anfinsen C. B. Disulfide Interchange and the Three-Dimensional Structure of Proteins. Proc. Natl. Acad. Sci. U.S.A. 1965, 53, 676–684. 10.1073/pnas.53.3.676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang J. G.; Wang C. C.; Tsou C. L. Formation of native insulin from the scrambled molecule by protein disulphide-isomerase. Biochem. J. 1988, 255, 451–455. 10.1042/bj2550451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hackeng T. M.; Griffin J. H.; Dawson P. E. Protein synthesis by native chemical ligation: expanded scope by using straightforward methodology. Proc. Natl. Acad. Sci. U.S.A. 1999, 96, 10068–10073. 10.1073/pnas.96.18.10068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crook E. M.; Mathias A. P.; Rabin B. R. Spectrophotometric assay of bovine pancreatic ribonuclease by the use of cytidine 2′:3′-phosphate. Biochem. J. 1960, 74, 234–238. 10.1042/bj0740234. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.