Abstract
Mitochondria are an iconic distinguishing feature of eukaryotic cells. Mitochondria encompass an active organellar network that fuses, divides, and directs a myriad of vital biological functions, including energy metabolism, cell death regulation, and innate immune signaling in different tissues. Another crucial and often underappreciated function of these dynamic organelles is their central role in the metabolism of the most abundant and biologically versatile transition metals in mammalian cells, iron. In recent years, cellular and animal models of mitochondrial iron dysfunction have provided vital information in identifying new proteins that have elucidated the pathways involved in mitochondrial homeostasis and iron metabolism. Specific signatures of mitochondrial iron dysregulation that are associated with disease pathogenesis and/or progression are becoming increasingly important. Understanding the molecular mechanisms regulating mitochondrial iron pathways will help better define the role of this important metal in mitochondrial function and in human health and disease.
Keywords: iron, mitochondria, metabolism
INTRODUCTION
Maternally inherited and thought to be of bacterial descent, mitochondria are double-membrane structures that are present in nearly all cells and possess their own genome, transcriptome, and proteome. Best known for their critical function in energy production via oxidative phosphorylation, mitochondria are essential for nutrient and oxygen sensing and for the regulation of critical cellular processes, including cell death and inflammation. Mitochondria were long considered the powerhouses of the cell; however, over the last decade it has become increasingly clear that mitochondria have a plethora of other biological functions inside the cell. Importantly, mitochondria are the sole site of heme synthesis and the primary generator of iron-sulfur (Fe-S) clusters, both of which are present in mitochondria and in the cytosol and required for a number of vital cellular processes (1).
The unique redox properties of iron allow for efficient electron transfer, which is beneficial to a number of diverse biological reactions. However, such reactive properties of iron may also promote the generation of reactive oxygen species (ROS), which in large doses can be damaging to intracellular systems. As a result, the acquisition, usage, and detoxification of iron must be tightly regulated to ensure that the metabolic needs of the cell are met and the risk of iron toxicity is minimized (2). Mitochondria are positioned at the center of such cellular iron homeostasis, where they provide the compartmentalization that is key to strictly regulating cellular iron levels. They also have the ability to catalyze electron transport via heme-Fe-S–containing proteins, using these processes in energy transduction.
Depending on the cell type, mitochondria contain up to 20–50% of total cellular iron (3, 4). The prevalence of iron-containing proteins in the mitochondria further supports the importance of iron in normal mitochondrial function (2). Within the mitochondrion, metal ions contribute to the metalation, folding, and stability of many intrinsic mitochondrial proteins, with mitochondrial iron levels intricately regulated by metallochaperones and metal transporters [e.g., mitoferrin-1 (MFRN1) and mitoferrin-2 (MFRN2)]. Important mitochondrial iron–containing proteins include the Fe-S cluster–containing proteins NADH ubiquinone oxidoreductase, Rieske iron–sulfur protein, and subunits of succinate dehydrogenase, all of which are involved in respiratory chain function. They also include enzymes involved in other processes, such as aconitase, lipoic acid synthase, and biotin synthase; iron-ion cofactor–containing proteins including iron monooxy-genases and dioxygenases; and heme-containing proteins including cytochrome bc1, cytochrome c, cytochrome c oxidase, and succinate dehydrogenase, all of which are involved in the electron-transport chain (2). As such, changes in mitochondrial iron concentrations can lead to alterations in Fe-S homeostasis, impaired Fe-S biogenesis, impaired heme synthesis, mitochondrial dysfunction, and increased oxidative stress.
Despite the clarity that mitochondria are the main site for dynamically active electron transport and redox activity requiring iron, knowledge about mitochondrial iron metabolism pathways in mammalian cells is minimal and remains largely confined to the heme synthesis pathway, with little known about the trafficking and storage of iron within this organelle. In this review, we discuss the role of mitochondrial iron pathways in human health and disease. We focus on the role of mitochondrial iron pathways in maintaining normal mitochondrial homeostasis and discuss what happens when these pathways are disrupted. Although much of the characterization of the mechanisms involved in maintaining mitochondrial iron homeostasis has been performed in yeast, the focus of this review is on mitochondrial iron in human health and disease. As such, where available, research done on higher eukaryotes is cited and, unless stated otherwise, mammalian gene/protein nomenclature is used to avoid confusion.
CELLULAR IRON METABOLISM
Iron is an essential metal for almost all organisms. It is one of the most abundant transition metals, but it is generally biologically unavailable. Therefore, organisms utilize large amounts of energy to acquire iron and make it bioavailable. The essential nature of iron is its involvement in many oxidation/reduction reactions. The ability of iron to participate in oxidation/reduction reactions is the feature that makes iron so useful in enzymatic reactions. That same feature of facile electron transfer makes iron potentially dangerous, as iron can donate electrons to O2 and H2O2 through Fenton chemistry to generate the hydroxyl radical or the hydroperoxyl radical and the superoxide anion (5). These molecules can oxidize nucleic acids, proteins, and lipids, resulting in damage, dysfunction, and cell death. Thus, iron acquisition, utilization, and storage are tightly regulated to prevent the damaging properties of iron.
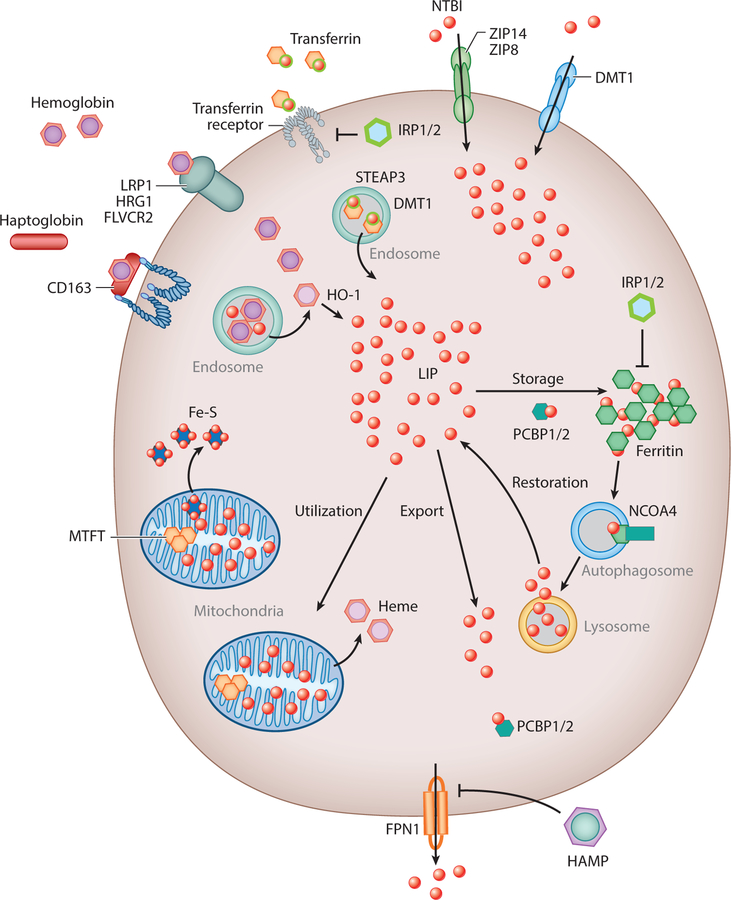
In eukaryotes, iron acquisition is mediated through metal or metal-siderophore transporters that transport iron across the plasma membrane at the cell surface or across membranes of intracellular organelles. Most of the original iron transporters were discovered in the model organism Saccharomyces cerevisiae (6), and homologous proteins have been identified in other eukaryotes, using bioinformatics/sequence homology, cloning, and complementation analysis. Most transporters identified predominantly transport-divalent iron, thus requiring enzymes to reduce the abundant ferric iron (Fe3+) to the more soluble ferrous iron (Fe2+). Reductases have been identified in all eukaryotes and can be found localized at the plasma membrane assisting in increased iron acquisition and on intracellular organelles where Fe2+ is either stored (vacuole/lysosome) or utilized for essential biologic processes, including but not limited to DNA replication, lipid synthesis, protein synthesis, heme synthesis, and Fe-S cluster synthesis. Some eukaryotes also have Fe3+ transporters for iron acquisition during times of limited iron access. Fe3+ acquisition can also be mediated by secretion of iron-binding molecules such as siderophores (7) or by the carrier protein transferrin (8). Once iron is acquired, it must be transported to sites of utilization such as the mitochondrion (heme and Fe-S cluster synthesis), to secretory pathways in the endoplasmic reticulum (in yeast) (9), or into the iron storage protein ferritin or into the vacuole/lysosome to protect cells against iron toxicity (Figure 1). In eukaryotes, iron acquisition can be regulated, but once acquired, there is no regulated mechanism for iron loss. In vertebrates, iron is only lost due to epithelial cell loss or through blood loss. There is no regulated mechanism of iron loss from an organism, but multicellular organisms must transfer iron from one tissue to another by cellular iron export into biological fluids (plasma). This process is mediated by the only identified plasma membrane iron exporter ferroportin1 (FPN1) (10). Iron in excess can replace other transition metals in their binding to proteins, resulting in defective protein function and inactive enzymatic activity. Furthermore, excess iron can accumulate in tissues resulting in tissue damage. Thus, in the absence of an export mechanism or in the absence of storage, malregulation of iron transport can lead to excess unsequestered iron. Excess iron in mammals is associated with many diseases, including hemochromatosis, β-thalassemia, diabetes, cardiomyopathy, and metabolic syndrome, as well as with many neurologic disorders (11).
Figure 1.
In eukaryotes, iron acquisition in the form of transferrin-bound or non-transferrin-bound (NTBI) iron is mediated through the metal or metal-siderophore transporter transferrin receptor, solute carrier family 39 member 8 (SLC39A8 or ZIP8), solute carrier family 39 member 14 (SLC39A14 or ZIP14), or solute carrier family 11 member 2 (SLC11A2 or DMT1). Heme-bound iron is acquired by the heme receptors/ transporters LDL receptor-related protein 1 (LRP1), solute carrier family 48 member 1 (SLC48A1 or HRG1), the hemoglobin scavenger receptor (CD163), and feline leukemia virus subgroup C cellular receptor family (FLVCR2). Heme is degraded by heme-oxygenase 1 to produce ferrous iron (Fe2+). Once inside the cell, transferrin-bound iron is degraded in the endosome by the metalloreductase six-transmembrane epithelial antigen of prostate 3 (STEAP3), capable of converting iron from an insoluble ferric (Fe3+) to a soluble ferrous (Fe2+) form. The labile iron pool (LIP) represents a pool of chelatable, redox-active iron, which is transitory. Iron is transported to sites of utilization such as the mitochondria (heme and Fe-S cluster synthesis) or into the iron storage proteins ferritin and mitochondrial ferritin (MTFT). Iron is exported from the cell by FPN1 (ferroportin1), which is inhibited by the iron hormone hepcidin (HAMP). Poly C-binding proteins 1/2 (PCBP1/2) act as iron chaperones that assist in the mineralization of ferritin whereas iron responsive element binding proteins 1 and 2 (IRP1/2) limit the transcription of ferritin in iron-depleted conditions. Similarly, in low-iron conditions, nuclear receptor coactivator 4 (NCOA4) protein assists in the release of iron from ferritin through an autophagy-mediated mechanism involving the lysosome.
Important to the regulation of cellular and mitochondrial iron homeostasis are the iron regulatory proteins (IRP1 and IRP2), which were first described in the late 1980s. These proteins bind to highly conserved hairpin structures termed iron responsive elements in the untranslated regions of target mRNAs involved in cellular iron metabolism, which allow for the tight regulation of mRNA translation through either stabilization or increased degradation of target mRNA. They are key regulators of iron sensing (hypoxia-inducible factor 2a), uptake [transferrin receptor 1, divalent metal transporter (DMT1), FPN1], heme synthesis (5-aminolevulinic acid synthase 2), and iron storage (ferritin) in all cells. Important for this discussion is that IRP1 is a 4Fe-4S cluster protein that acts as an aconitase under iron replete conditions where Fe-S clusters are efficiently synthesized. Under low-iron conditions, IRP1 loses the 4Fe-4S cluster, thus becoming an IRP protein that can now regulate mRNA translation. IRP2, on the other hand, accumulates in iron-limited conditions, thus regulating the expression of several iron homeostasis proteins that allow for increased iron acquisition (12).
The metallochaperone proteins, poly C-binding proteins (PCBPs), are also key regulatory proteins for cellular iron storage and turnover. PCBPs deliver iron to target proteins regulating iron storage, iron incorporation into proteins, and overall iron homeostasis. PCBP1, an iron chaper-one for ferritin that is only found in mammals, acts in a complex for efficient iron delivery to ferritin (13). Another key regulatory mechanism for ferritin-bound iron turnover is the activation of nuclear receptor coactivator 4 (NCOA4). NCOA4 protein assists in the release of iron from ferritin through an autophagy-mediated mechanism, termed ferritinophagy (14). Consequently, both PCBP1 and NCOA4 allow for the tight regulation of cellular iron homeostasis by regulating the delivery of iron for storage and breakdown of ferritin during iron limitation (Figure 1).
MITOCHONDRIAL IRON IMPORT
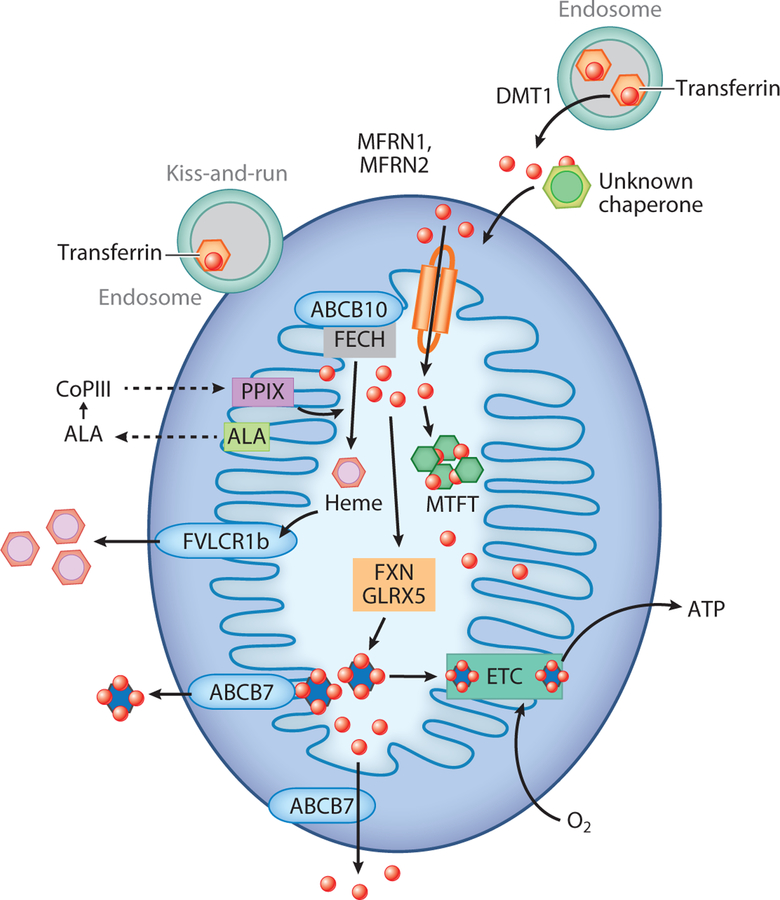
Iron can be internalized into animal cells through diferric transferrin-transferrin receptor–mediated endocytosis, heme import, and breakdown by heme oxygenase 1, or by direct plasma membrane transport of Fe2+ through divalent metal transporters such as DMT1, ZIP8, or ZIP14 (15, 16) (Figure 1). Homologous low-affinity cation transporters are present in all eukaryotes, whereas heme transporters are found only in higher eukaryotes, including Caenorhabditis elegans (17). Once iron is internalized into cells, it must be transported to sites of utilization. One essential site of iron utilization is the mitochondria, where heme and Fe-S cluster synthesis occur. Mitochondria are recognized by their iconic double-membraned structure consisting of an inner mitochondrial membrane (IMM) and outer mitochondrial membrane (OMM) (Figure 2). The OMM is highly permeable to ions and small uncharged metabolites due to the presence of voltage-dependent anion channels (VDACs/porins), which permit the transport of a variety of metabolites, including ATP, cations, pyruvate, and others. Conversely, the IMM forms a tight diffusion barrier to ions and, thus, any transport across the IMM is facilitated by hydrophobic mitochondrial carrier proteins (18).
Figure 2.
In eukaryotes, iron acquisition is mediated through metal or metal-siderophore transport. Cytosolic iron must first cross the semipermeable outer mitochondrial membrane and then be imported into the mitochondrial matrix for heme and Fe-S cluster synthesis through transferrin-mediated endosome-to-mitochondrial contact, termed kiss-and-run, and/or via divalent metal transporter (DMT1). Iron is transported across the tight diffusion barrier of the inner mitochondrial membrane by hydrophobic mitochondrial carrier proteins called mitoferrins (MFRN1, MFRN2), which may form a complex with ABCB10. In mitochondria, iron is inserted into protoporphyrin IX (PPIX) by ferrochelatase (FECH) to produce heme. The first step in the heme synthesis pathway involves the production of δ-aminolevulinic acid (ALA). ALA is then transported to the cytosol where the next four steps take place to form coproporphyrinogen III (CoPIII), which is transported back into mitochondria to form PPIX. Heme is transported outside of mitochondria via the 1b isoform of FLVCR (feline leukemia virus subgroup C cellular receptor) for incorporation into hemoproteins. Mitochondrial iron is also used for Fe-S cluster synthesis, which among other factors involves frataxin (FXN) and GLRX5 (glutaredoxin-related protein 5), which are exported into the cytosol by ABCB7. Fe-S clusters are essential for the efficient functional of the electron transport chain (ETC) to generate ATP in a process called oxidative phosphorylation. Excess iron can be stored in mitochondrial ferritin (MTFT).
Cytosolic iron must first cross the OMM and then be imported into the mitochondrial matrix for heme and Fe-S cluster synthesis. Two predominant hypotheses for iron delivery to the mitochondrial OMM exist. The first is the transient docking of endosomes containing transferrin-delivered iron to the OMM, a term referred to as kiss-and-run (19–22). The second hypothesis is that iron as Fe3+ is reduced in the endosome by STEAP3 (23). Then Fe2+ is transported across the endosomal membrane into the cytosol by DMT1, where Fe2+ is delivered to mitochondria by some unknown mechanism, although a chaperone-mediated mechanism has long been suggested (Figure 2). In addition, a form of DMT1 has been shown to localize to the mitochondria where it may mediate the transport of iron into the mitochondria (24). It is still unclear whether there is a single mechanism or multiple pathways to deliver iron across the OMM.
Iron import across the IMM was originally described in S. cerevisiae, where two homologous, high-affinity iron transporters, Mrs3 and Mrs4, were characterized (25, 26). Mrs3/Mrs4 orthologs, the mitoferrins (MFRNs), have been identified in all eukaryotes (1). MFRN1 or SLC25A37 has been shown to be important in erythroid cell development, as its loss results in profound anemia (27, 28). MFRN1 associates with a mitochondrial protein complex consisting of several proteins that assist in efficient heme synthesis (29–31). Recently purified recombinant MFRN1 has been shown to transport free iron and not a chelated iron complex, and it is selective for alkali divalent ions (32). There is some evidence in C. elegans that reductions in MFRNs and subsequent mitochondrial iron reductions result in abnormal development and extended life span (33). The role of the homolog MFRN2 or SLC25A28 remains to be determined, although studies suggest it may be important in other cell types (34–37). It is important to note that a phenotype for the loss of Mrs3 and Mrs4 proteins in yeast was only observed when cells were placed on low-iron growth medium. This suggests that other low-affinity mitochondrial iron importers may exist. In fact, the pyrimidine transporter Rim2 was found to transport iron in the absence of Mrs3 and Mrs4 (38). Similar to yeast, other mitochondrial iron importers may play a role in importing iron. In fact, recent studies suggest that the mitochondrial calcium uniporter may function as an Fe2+ importer (39).
MITOCHONDRIAL IRON UTILIZATION
Following import, mitochondrial iron is utilized in three metabolic pathways: heme synthesis, Fe-S cluster biogenesis, and mitochondrial iron storage into mitochondrial ferritin (MTFT). In Jurkat cells, respiratory complexes account for ∼17% of mitochondrial iron. Approximately 40% of mitochondrial iron is in the form of Fe3+ oxyhydroxide nanoparticles, with another 14% present as [Fe4S4]2+ clusters and heme b centers and about 15% as MTFT (4). Mitochondria are also thought to contain a free (labile or chelatable) iron pool that is extremely redox active, increases with aging, and drives the chains of oxidative events associated with disease (40, 41). This labile iron pool in mitochondria is thought to be low due to the tight coordination of the rate of influx with the rate of incorporation into heme and Fe-S clusters. However, chelatable iron is detectable in the mitochondria of cultured hepatocytes (42) and cardiomyocytes (3). Incorporation of iron into mitochondria may occur preferentially over incorporation of iron into the cytosolic labile iron pool (43).
Heme Synthesis
Heme is a complex of ferrous iron and tetrapyrrole ring of heme (protoporphyrin IX). It is an important prosthetic group for numerous vital proteins such as hemoglobin, myoglobin, cytochrome c, cytochrome p450, catalase, peroxidase, and others. Prosthetic heme acts in a number of catalytic and regulatory processes, including oxygen transport and storage, electron transfer for enzymatic redox reactions, signal transduction, ligand binding, and control of gene expression (2). Heme biosynthesis is a highly conserved process that occurs in all cells but is particularly active in erythroid cells and hepatocytes (44). Although the enzymatic reactions involved in heme biosynthesis are well understood, much remains to be learned about the transport of heme intermediates into and out of the mitochondrion. For a more detailed review of heme synthesis, the reader is referred elsewhere (44). Briefly, biosynthesis of protoporphyrin IX involves the sequential catalytic ability of eight enzymes and is strictly dependent on the mitochondrion, although four of the intermediate enzymatic steps occur within the cytosol (Figure 2). The first step in the heme synthesis pathway occurs in the mitochondrion and involves the production of δ-aminolevulinic acid (ALA) through the condensation of glycine and succinyl CoA by the enzyme 5-aminolevulinate synthase (ALAS). There are two genes for ALAS, the ubiquitously expressed ALAS1 and the erythroid-specific ALAS2 (44). ALA is then transported to the cytosol where the next four steps take place. First, ALA is converted to the monopyrrole porphobilinogen by ALA dehydratase. Next, porphobilinogen is converted into the cyclic tetrapyrrole uroporphyrinogen III by two subsequent enzymatic steps involving porphobilinogen deaminase and uroporphyrinogen III synthase. Uroporphyrinogen III is then decarboxylated to form coproporphyrinogen III (CoPIII), which is transported to mitochondria by either peripheral-type benzodiazepine receptors (45) or ABCB6 (46). However, human patients lacking ABCB6 have no evidence of anemia or other clinical abnormalities (47), making this function unlikely. The enzyme CoPIII oxidase, localized in the IMM space of mitochondria, catalyzes oxidative decarboxylation of CoPIII to protoporphyrinogen IX. Protoporphyrinogen III oxidase, an integral protein of the IMM, catalyzes the penultimate step in the heme pathway that generates protoporphyrin IX. The final step in this pathway involves insertion of one atom of Fe2+ into protoporphyrin IX by the IMM-associated enzyme ferrochelatase in the mitochondrial matrix (44). In nonerythroid cells, the rate of heme synthesis is dependent on the rate of 5-aminolevulinate synthesis via ALAS. In contrast, in erythroid cells, the rate-limiting step may be the delivery of transferrin iron (44).
Iron-Sulfur Cluster Biogenesis and Assembly
Fe-S cluster proteins are found in all kingdoms and are involved in cellular processes, including energy production, DNA maintenance, protein translation, lipid synthesis, and more. These proteins can be found in many subcellular compartments, so the synthesis and transport of Fe-S clusters to sites of incorporation into active proteins are very important and highly conserved processes. The biogenesis of Fe-S clusters begins in the mitochondrion and most of the proteins [18 known Fe-S cluster assembly proteins (ISC proteins)]. In addition, 11 known cytosolic Fe-S protein assembly factors (CIA proteins) involved in cluster synthesis are conserved in eukaryotes (reviewed in 48). Much of the original work on Fe-S cluster synthesis was performed in yeast, with contributions from many other organisms such as plants and humans. Fe-S biosynthesis is a complex, multistep process that involves a set of dedicated multimeric protein complexes. Mammalian Fe-S cluster synthesis involves two basic steps: nascent cluster assembly followed by its transfer into the apoproteins. Nascent assembly initiates on the main scaffold protein, Fe-S cluster assembly enzyme (ISCU). A cysteine desulfurase NFS1 forms a dimer to which monomers of the primary scaffold ISCU bind, and the structural component LYR motif-containing protein 4 (LYRM4) is required for the activity of NFS1 to provide sulfur, which is removed from cysteine for the nascent cluster formation. Frataxin is part of the core complex, and the 2Fe-2S cluster assembles upon ISCU when iron is provided together with the reducing equivalents. 2Fe-2S clusters are then transferred to recipient apoproteins. The release of the Fe-S cluster from the ISCU complex and loading onto recipient proteins are facilitated by chaperones HSPA9/HSC20, where ATP-bound HSPA9 hydrolyzes ATP to effect this transfer to the recipient. Other Fe-S clusters such as 4Fe-4S clusters that are needed in other proteins are formed after the 2Fe-2S clusters are released/transferred (reviewed in 49).
Loading of Fe-S clusters onto cytosolic or other organelle-localized proteins requires the function of the mitochondria Fe-S assembly machinery, but the molecular details are still to be defined, and some of the mitochondria cluster machinery is also found in the cytosol where it may facilitate Fe-S cluster formation. It is also hypothesized that Fe-S clusters are exported from the mitochondria. The exact protein(s) involved in this process are still being identified. The yeast protein Atm1 and its mammalian homolog ABCB7 have been suggested to be involved in mitochondrial Fe-S cluster export (50, 51); however, more investigation is needed into the exact substrate transported. Importantly, the mitochondrial Fe-S cluster synthesis machinery is necessary for cytosolic Fe-S protein assembly/function. Whether synthesized in the mitochondria and then exported or synthesized in the cytosol, Fe-S clusters are delivered to the CIA complex to load the cluster into recipient apoproteins. The CIA machinery is highly conserved and is composed of four main proteins: CIA1, nuclear prelamin A recognition factor–like (NARFL or NAR1), nucleotide-binding proteins 1 and 2 (NUBP1/2), and the electron transfer proteins NADPH-dependent diflavin oxidoreductase 1 (NDOR1) and cytokine-induced apoptosis inhibitor 1 (CIAPIN1). NDOR1 and CIAPIN1 are proposed to hand off Fe-S clusters to the main CIA complex, which acts as a scaffold to load Fe-S clusters onto cytosolic proteins. Similar to CIA proteins, nuclear Fe-S cluster proteins also require the mitochondrial Fe-S cluster machinery, which emphasizes the importance of mitochondrial iron homeostasis in these essential processes. For a more detailed review of Fe-S cluster synthesis in eukaryotes, see 48.
Mitochondrial Iron Storage
Iron within the mitochondria is either utilized for the biosynthesis of heme and Fe-S clusters, or it can be stored. The mitochondrion has not historically been hypothesized to be a site of storage, but in fact, as long as the Fe-S cluster synthesis machinery is working, increased mitochondrial iron imported through MFRN homologs does not seem to be detrimental (36, 52). Storage of iron in mitochondria of higher eukaryotes is mediated by MTFT, which is structurally similar to cytosolic H-chain ferritin (53). MTFT was first identified as being expressed in mitochondria of testes and erythroid cells (54), with more recent evidence for a role in brain (55). MTFT can act to store iron, and it has ferroxidase- and iron-binding activities similar to cytosolic ferritin (54, 56). Furthermore, overexpression of MTFT results in mitochondrial iron sequestration-limiting heme and Fe-S cluster synthesis that is associated with X-linked sideroblastic anemia (57). MTFT also appears to be important in neuroprotection, where it may play a role in protection against ROS within the mitochondria (58, 59). It is interesting to note that mitochondria may also have iron export machinery.
MITOCHONDRIAL IRON EXPORT
How iron is exported out of the mitochondria remains an evolving research field. The best iron export mechanism out of mitochondria involves the export of heme into the cytosol. Newly synthesized heme is delivered to numerous polypeptides in the cytosol, mitochondria, endoplasmic reticulum, and peroxisomes. Up until recently, the mechanisms by which heme is transported out of the mitochondrion were uncertain. However, recent studies have demonstrated that heme is transported out of the mitochondria via isoform b of FLVCR1 (feline leukemia virus subgroup cellular receptor 1) (60) (Figure 2). Suppression of Flvcr1b expression in vitro results in mitochondrial heme accumulation and termination of erythroid differentiation, whereas overexpression of Flvcr1b leads to intracellular heme accumulation and erythroid differentiation (60). As previously mentioned, the yeast protein Atm1 and its mammalian homolog ABCB7 may be involved in mitochondrial Fe-S cluster export (50, 51). Finally, the mitochondrial-localized ATP-binding cassette protein ABCB8 has been suggested to play a role in iron export, although these observations have only been reported in conditions of cardiotoxicity (61, 62), and so the normal role of ABCB8 remains to be elucidated. It is interesting to speculate regarding when iron export from mitochondria might happen.
MITOCHONDRIAL IRON AND MITOCHONDRIAL FUNCTION
Mitochondria are at the center of cellular metabolism, regulating the continuous aerobic oxidation of fatty acids and consuming the end products of glucose, glutamine, and amino acid degradation to aerobically produce ATP from oxygen and H2O. Given that iron is the most prevalent metal inside the mitochondrial matrix and serves to facilitate the complex redox chemistry of the electron transport chain, it is not surprising that mitochondrial iron levels and mitochondrial energy metabolism are tightly linked. Iron-protoporphyrin (heme) and Fe-S clusters are essential components of the IMM complexes of the electron transport chain. Specifically, complex I (NADH dehydrogenase) contains eight Fe-S clusters; complex II (succinate dehydrogenase) contains one heme b prosthetic group and three Fe-S clusters ([2Fe-2S], [4Fe-4S], [3Fe-4S]); complex III (ubiquinol: cytochrome c oxidoreductase) contains a cytochrome b subunit with two heme moieties, a cytochrome c1 subunit with one heme, and a Rieske protein subunit (UQCRFS1) with a [2Fe-2S] cluster; and complex IV (cytochrome c oxidase) contains two heme a moieties (63). Numerous studies demonstrate that alterations in mitochondrial iron content modify mitochondrial ATP production, via the tricarboxylic acid (TCA) cycle and/or glycolysis (64, 65). Most of these studies have centered on the role of frataxin. Frataxin appears to be a key activator of mitochondrial energy conversion and oxidative phosphorylation (66), and patients with frataxin deficiency have diminished skeletal muscle mitochondrial ATP production (67). Acute loss of Fe-S clusters leads to increased de novo fatty acid synthesis and lipid droplet accumulation, demonstrating the importance of mitochondrial iron homeostasis for many metabolic processes involving the mitochondrion (68). The reciprocal regulation of mitochondrial iron by mitochondrial energy metabolism pathways has also been observed. Examples include the loss of the TCA cycle enzyme fumarate hydratase, leading to defects in the biogenesis of Fe-S clusters that affect complex I function (69). In yeast, the mitochondrial acyl carrier protein, which has a well-known role in mitochondrial fatty acid synthesis, plays an evolutionarily conserved role in Fe-S biogenesis (70).
Mitochondrial Iron and Mitochondrial Dynamics, Biogenesis, and Turnover
Mitochondria undergo regular cycles of fission and fusion to alter their size and shape in response to the metabolic needs of the cell (71). Increased fusion or reduced fission results in the formation of elongated mitochondria, whereas increased fission or reduced fusion results in mitochondrial fragmentation. At the OMM, mitochondrial fusion is mediated by the dynamin-related GTPases mitofusin 1 and 2 (MFN1/2). At the IMM, mitochondrial fusion is regulated by the dynamin-related protein optic atrophy 1 (OPA1). Mitochondrial fission utilizes dynamin-related protein 1 (DRP1) from the cytosol to bind to the receptors mitochondrial fission protein 1 (FIS1), mitochondrial fission factor, and mitochondrial elongation factors 1 and 2 (MID51 and MID49) (71). While there is a clear association between mitochondrial fission and fusion and the metabolic state of the cell, little is known about mitochondrial iron content or usage and mitochondrial dynamics. There is some suggestion that mitochondrial iron levels may be linked to changes in mitochondrial dynamics, with the loss of MFN2 associated with mitochondrial iron overload (72), OPA1 cleavage associated with cellular iron overload (73), loss of FIS1 associated with iron chelation–induced mitochondrial elongation (74), and cellular iron overload associated with DRP1 (Ser637) dephosphorylation (75).
To allow the cell to maintain a healthy repertoire of mitochondria, mitochondrial fission and fusion processes are also in tandem with the formation of new mitochondria and the removal of damaged mitochondria. Mitochondrial biogenesis resulting from the growth and division of preexisting mitochondria is regulated by nuclear-encoded and mitochondrial-encoded proteins, namely the mitochondrial transcription factor TFAM, PPAR coactivator-1α (PGC-1α), AMPK, and nuclear respiratory factors 1 and 2 (NRF-1/2) (76). Impaired regulation of iron in mitochondria is closely linked with mitochondrial ΔΨ m and integrity. Increased uptake of iron into mitochondria depolarizes mitochondrial membranes and generates ROS through the Fenton reaction (77). An increase in this iron pool directly associates with an increase in oxidative damage. Such damaged or defective mitochondria are generally removed by selective encapsulation into double-membraned autophagosomes that are delivered to the lysosome for degradation in a process called mitophagy (78). When the ΔΨ m is low (i.e., under conditions of stress), damaged and depolarized mitochondria stabilize PTEN-induced kinase 1 (PINK1) or the BH-3-only BCL2 protein BNIP3 and the E3 ubiquitin ligase Parkin, which in turn recruit Parkin and the autophagy protein LC3B, respectively. Parkin ubiquitinates various OMM proteins, including MFN1/2, and recruits autophagosomes (79).
Interestingly, mitochondrial iron pathways may be closely linked with biogenesis and mitophagy (80, 81). Loss of frataxin in murine cerebellums results in severe deficits in mitochondrial biogenesis (loss of PGC-1α, TFAM, and NRF-1/2) with activation of mitochondrial fusion (81). Muscle tissue from patients deficient in the Fe-S cluster scaffold protein ISCU show enhanced expression of transcriptional coactivator PGC-1α and increased expression of mitochondrial fatty acid oxidation genes (82). Similarly, loss of frataxin in C. elegans results in mitophagy induction (83), and aconitase (with [4Fe-4S] cluster inactivation) is a dominant suppressor of Pink1 in Drosophila models (84). High doses of iron chelators, including deferiprone and deferoxamine, induce PINK1/Parkin-independent mitophagy involving the HIF-1–dependent gene BNIP3 (85). Loss of the cellular labile pool by the anticancer drug, KP46, also activated BNIP3L-mediated mitophagy (86). Conversely, increased MTFT in human ARPE-19 cells activates HIF-1α–dependent mitophagy (87). Collectively, these processes most likely represent an adaptive iron starvation response induced as a protective mechanism against mitochondrial stress that may contribute to the longevity of cells.
Mitochondria have developed a number of mechanisms to recognize and resolve dysfunction. Together, these mechanisms culminate with a mitochondrial stress response system that recovers organelles that are salvageable and degrades organelles that are beyond repair, ultimately yielding a healthier mitochondrial network (88). Mammalian mitochondrial stress responses include the ATF5-mediated mitochondrial unfolded protein response (UPRMT) and the ATF4-mediated stress pathway (88). Inside the mitochondria, the interaction between protein and iron homeostasis is strongly coupled (89, 90). Activation of the UPRMT proteins has been shown to stimulate heme synthesis by directly activating ALA synthase (mtCLPX in S. cerevisiae), a phenomenon that is conserved in mammalian homologs (91). In addition, frataxin deficiency causes upregulation of mitochondrial LON and CLPP proteases and severe loss of mitochondrial Fe-S proteins in murine hearts (92). While there is limited evidence for regulation of mitochondrial iron metabolism and mitochondrial behavior, function, and bioenergetics, the aforementioned findings suggest that there may be a range of cross talk and coregulatory systems linking iron to vital dynamic functions of the mitochondrion.
A Word of Caution: Methodology to Measure Mitochondrial Iron
To understand the genesis of mitochondrial iron overload, it is necessary to employ various techniques to quantify iron in mitochondria. A range of these techniques is elegantly summarized by Holmes-Hampton et al. (93) and includes colorimetric determination, atomic absorption spectroscopy, inductively coupled plasma-optical and plasma-mass emission spectroscopy, electron microscopy (in combination with iron histochemical staining such as Perls’s and Turnbull’s, X-ray microscopy, and electron energy loss spectroscopy), synchrotron X-ray fluorescence microscopy imaging, confocal Raman microscopy, ultraviolet (UV)-vis spectroscopy, electron paramagnetic resonance, X-ray absorption spectroscopy, and Mössbauer spectroscopy. Several of these methods can be used to measure iron in isolated mitochondria from cells and tissues with varying degrees of sensitivity. Caution must be exercised in the preparation of clean mitochondrial fractions for such measurements. Commercially available kits and subcellular fractionation protocols generally result in the isolation of crude mitochondrial preparations consisting of enriched mitochondrial fractions contaminated by other subcellular organelles, including peroxisomes and lysosomes (which also contain iron). Furthermore, downstream protocols are required that employ the use of Percoll gradients or digitonin treatment to generate purer fractions or mitoplasts (4). However, large amounts of starting material are required to accurately separate and measure nonheme or heme iron levels colorimetrically in isolated mitochondria or mitoplasts. In general, absolute iron concentrations in the mitochondria of mammalian cells are reported as mitochondrial iron to protein concentration ratios; however, normalization of mitochondrial iron to mitochondria to cell volume ratios have also been proposed (4). For lower levels of starting material, mitochondrial iron can be measured in mitoplasts by methods like atomic absorption spectroscopy (however, this method does not distinguish between heme and nonheme pools) or alternative approaches using fluorescence. For an extensive review on fluorescent ways to measure mitochondrial iron in live cells, the reader is referred elsewhere (94). Briefly, the most specific targeted fluorescent probes used to measure mitochondrial iron in live cells encompass mitochondria-targeted peptides incorporating dual fluorescent and selective iron chelators (40, 95). Other fluorescence-based systems have also been used including rhodamine B 4-[(1,10-phenanthrolin-5-yl) aminocarbonyl] benzyl ester derivatives (3, 96). However, limitations to using these probes include low sensitivity, difficulty in reversing the quenched signal under physiological conditions (difficulty of unequivocally assigning the quenched signal to labile iron), and interference by other metals such as copper (94).
MITOCHONDRIA AND CELLULAR IRON SENSING
Mitochondria perform a global regulatory role in iron homeostasis, as exemplified by disruptions in mitochondrial proteins that lead to perturbations in whole-cell iron processing (97). By producing Fe-S clusters and synthesizing heme, mitochondria play a central role in coordinating cellular iron homeostasis (iron import, utilization, storage, and export). Iron concentrations can rise to toxic levels in mitochondria of excitable cells, often leaving cytosolic iron depleted. In such circumstances, such as if Fe-S clusters or heme production cease to occur as a result of lack of iron, then the cell responds appropriately, through IRP1-mediated posttranscriptional regulation, by importing more iron (98). However, if Fe-S levels are depleted due to either mitochondrial dys-function or disturbances in Fe-S cluster homeostasis by ROS, iron uptake is enhanced, but the cellular response of importing more iron may become maladaptive and lead to excessive generation of ROS or other mechanisms. A number of mitochondrial proteins have been shown to play a role in the regulation of cellular iron homeostasis, which in turn has a global effect on systemic iron regulation. Mitochondrial dysfunction (such as mitochondrial DNA mutations) associates with systemic iron metabolism changes, including age-dependent macrocytic anemia, abnormal erythroid maturation, and defects in lymphopoiesis (99, 100). Loss of the mitochondrial complex III subunit Rieske Fe-S protein in fetal mouse hepatic stellate cells allows them to proliferate but impairs their differentiation, resulting in anemia and prenatal death (101). The IMM un-coupling protein UCP2 is a regulator of erythropoiesis, where inhibition of UCP2 function may contribute to the development of anemia (102). MFN2 is required for erythropoiesis (103) and is downregulated in progenitors from patients with refractory anemia with ring sideroblasts (72), suggesting feedback regulation by mitochondrial iron levels.
MITOCHONDRIAL IRON AND DISEASE
Mitochondria are found in all mammalian cells, but they are particularly abundant in the heart, muscles, and nervous system, where they support the energy demands of muscle contraction and neuronal function. Mitochondria also play a critical role in hemoglobin production in erythro-blasts. Consequently, muscle cells, cardiomyocytes, erythroblasts, and neurons are vulnerable to diseases associated with failure of mitochondrial iron homeostasis and consequent mitochondrial dysfunction. Failure to control mitochondrial iron leads to mitochondrial iron loss or excessive mitochondrial iron import (loading). Inborn errors that affect mitochondrial iron metabolism frequently result in a pathology of one or more of the above target tissues (63). Such mitochondrial diseases have provided important insights into the processes of mitochondrial iron metabolism and are discussed in detail in the following sections (Table 1).
Table 1.
Mitochondrial iron dysfunction and disease
| Pathobiology | Disease | Associated mitochondrial iron–related gene mutation | Evidence for mitochondrial iron dysfunction from human studies | Evidence for mitochondrial iron dysfunction from experimental approaches | Reference | ||||
|---|---|---|---|---|---|---|---|---|---|
| Red blood cell disorders | |||||||||
| Defective FECH activity with increased PPIX | Erythropoietic protoporphyria | FECH but may also be other contributory factors | Gives rise to inefficient heme synthesis | Deletion of Abcb10 in hematopoietic cells results in accumulation of PPIX and an increase of iron deposits inside mitochondria | 28, 91, 105, 107, 163, 164 | ||||
| Evidence for finely dispersed electron-dense deposits localized in mitochondria and increased MFRN1 expression in erythroblasts of patients | |||||||||
| ALAS2 gain-of-function mutation | |||||||||
| ClpX missense mutation | |||||||||
| In the presence of increased porphyrin synthesis, deletion of MFRN1 in hepatocytes results in a decreased ability to convert PPIX into heme, leading to protoporphyria | |||||||||
| Increased ALA activity and thus increased PPIX | X-linked protoporphyria | ALAS2 gain-of-function mutations | Increased erythrocyte protoporphyrin concentrations | NA | 106 | ||||
| Defects in adequately incorporating iron into heme | Sideroblastic anemia | DMT1, ALAS2, ABCB7, MFRN1, FXN, GLRX5, SLC25A38, TRNT1, SLC19A2, PUS1, YARS2 | Excessive deposits of nonheme iron in the mitochondria of red blood cell precursors in the bone marrow, resulting in a ring sideroblast | In an ALAS2-deficient cell line there are increased high-density deposits in the mitochondria as well as increased expression of MTFT | 106, 109–112, 165, 166; reviewed in 108 | ||||
| The pathogenesis of mitochondrial iron loading is not identical in all types of sideroblastic anemias | |||||||||
| In an ABCB7-deficient cell line there is a 6-fold increase of iron accumulation in the mitochondria which is poorly available to MTFT | |||||||||
| Tissue-speciftc mitochondrial iron disorders | |||||||||
| Reduced frataxin expression | Friedreich’s ataxia | FRDA GAA triplet repeat expansion | Mitochondrial iron accumulation, especially in tissues of high mitochondrial content, such as nerve and cardiac tissue | Lower levels of frataxin result in Fe-S cluster deficiency and intramitochondrial iron accumulation possibly involving MFRN2 | 116–118, 120 | ||||
| Inactivation of Frda in mice leads to early embryonic lethality without iron accumulation | |||||||||
| Cardiovascular and pulmonary diseases | |||||||||
| Alterations in mitochondrial iron pathways | Cardiomyopathy | TTP | Hearts from patients with doxorubicin-induced cardiomyopathy have higher mitochondrial iron levels than hearts from patients with other types of cardiomyopathies or normal cardiac function | Acute exhaustive exercise significantly increases MTFT expression in the murine heart and loss of ABCB8 in mouse hearts is associated with mitochondrial iron accumulation and cardiomyopathy | 61, 62, 121, 122, 123, 167 | ||||
| Mice with deletion of TTP display cardiac dysfunction with iron deficiency | |||||||||
| Increased mitochondrial iron in human hearts with ischemic cardiomyopathy | |||||||||
| Overexpression of ABCB8 decreases mitochondrial iron and protects against doxorubicin-induced cardiomyopathy in murine models | |||||||||
| Increased mitochondrial iron in mice after ischemia/ reperfusion | |||||||||
| miR-210- ISCU1/ 2 axis | Pulmonary hypertension | NFU1 | Cardiopulmonary exercise testing of a woman with homozygous ISCU mutations revealed exercise-induced pulmonary vascular dysfunction | Mice deficient in miR-210, via genetic/pharmacologic means or via an endothelial-specific manner, display increased ISCU1/2 and are resistant to Fe-S-dependent pathophenotypes and pulmonary hypertension | 124, 168, 169 | ||||
| Ten individuals with an NFU1 homozygous mutation develop fatal infantile encephalopathy and/or pulmonary hypertension | Heme biosynthesis modulation via δ-aminolevulinic acid administration attenuates chronic hypoxia-induced pulmonary hypertension | ||||||||
| IRP2 | Chronic obstructive pulmonary disease (COPD) | IRP2 | Increased expression of IRP2 in lung tissue of patients with COPD | Cigarette smoking increases mitochondrial iron levels, and mice deficient in IRP2 are protected from smoke-induced injury | 125, 170 | ||||
| Neurological diseases | |||||||||
| Alterations in mitochondrial iron pathways | Alzheimer’s disease (AD) | NA | AD brain has been found to contain increased levels of iron | MTFT overexpression attenuates Aβ-induced neurotoxicity in a murine AD model | 59, 129–131 | ||||
| Increased expression of MTFT has been observed in the brains of patients | |||||||||
| Alterations in mitochondrial iron pathways | Parkinson’s disease (PD) | NA | Brain tissues and peripheral cells reduced mitochondrial complex I activity | Activation of IRP1, increased DMT1 and TFR1 expression, increased iron uptake, and increased ROS generation | 58, 133, 134, 137–139 | ||||
| In a rodent rotenone model of PD, transferrin accumulates in dopamine neurons, with much of it accumulating in the mitochondria | |||||||||
| In murine models of PD (MPTP and 6- hydroxydopamine–induced PD), MTFT is increased | |||||||||
| TFR1-conditional knockout mouse model displays PD symptoms | |||||||||
| Alterations in mitochondrial iron pathways | Huntington’s disease (HD) | NA | Brain tissues show evidence of disrupted mitochondrial energy metabolism, decreased activities of mitochondrial respiratory complexes II–IV and aconitase, and increased transcript expression of MFRN2 | Mitochondrial iron accumulates in mouse and human HD brains with increased expression of MFRN2 and loss of frataxin | 143, 144 | ||||
| Deficiency in Fe-S cluster synthesis | Type 2 Wolfram syndrome | CISD2 | No reported evidence | Mitochondrial labile iron accumulation and oxidative stress | 145, 171 | ||||
| Iron chelation may preserve neuronal cell and β-cell function | |||||||||
| CISD2-null mice display adaptive enlargement of mitochondria with extensive crista structures | |||||||||
| Deficiency in Fe-S cluster synthesis | Hereditary myopathy with lactic acidosis | ISCU | Patients display decreased levels and activity of complexes I, II, and III of the ETC, as well as mitochondrial aconitase | Compromised Fe-S cluster synthesis and reduction in ETC redox properties, leading to loss of cellular respiration | 146, 147, 172–175 | ||||
| Adolescent onset autosomal recessive mitochondrial myopathy | FDX1L | Complete loss of ISCU in mice results in early embryonic death | |||||||
Abbreviations: ETC, electron transport chain; FECH, ferrochelatase; NA, not applicable; PPIX, protoporphyrin IX.
Red Blood Cell Disorders
Most iron in mammals is found in hemoglobin in red cells (1.5–2.5 g), and the daily amount of iron needed for erythropoiesis/heme synthesis is approximately 20–30 mg. Therefore, maintaining the ability to absorb iron in the gut and recycle iron from old red cells is critically important for red cell formation. Furthermore, the ability to import and incorporate the iron into mitochondrial synthesized heme and Fe-S clusters is essential. Defects associated with the loss of heme and Fe-S clusters can give rise to red cell disorders.
Protoporphyria.
Porphyrias are rare inherited disorders caused by excessive levels of free erythrocyte protoporphyrin IX (104). The excess protoporphyrin IX accumulates in skin and liver, resulting in light sensitivity and liver damage. Some genetic causes for protoporphyria are due to defects in enzymes in the heme biosynthesis pathway. Erythropoietic protoporphyria is due to a deficiency in the last enzyme of the heme biosynthesis pathway ferrochelatase, which is required for incorporating iron into the protoporphyrin IX ring (105). Importantly, ferrochelatase is an Fe-S cluster– containing enzyme, so defects in Fe-S cluster formation may give rise to defective ferrochelatase activity. Another genetic variant of protoporphyria is X-linked protoporphyria, which is due to increased aminolevulinic acid synthase 2 (ALAS2) enzyme activity (106). The consequence of increased ALAS2 activity is increased ALA levels and thus increased protoporphyrin IX with limited iron incorporation to make heme. Increased expression of Mfrn1 is associated with erythropoietic porphyria (107), but erythropoietic porphyria has also been reported in hepatocyte-specific Mfrn1 knockout mice fed ALA in the drinking water (28). These results suggest that iron delivery to mitochondria and heme and Fe-S synthesis are tightly regulated processes where increases or reductions in intermediate heme synthesis production can be detrimental to red cell biology.
Sideroblastic anemia.
Sideroblastic anemia is characterized by an impaired ability to produce normal red blood cells. The cells obtain normal amounts of iron, but they have a defect in adequately incorporating iron into heme (108). Causes for this disease can be genetic or acquired. Mutations in genes encoding the mitochondrial proteins involved in iron acquisition (DMT1), heme or Fe-S cluster synthesis/transport, such as ALAS2, ABCB7, MFRN1, frataxin, and GLRX5, have been found to be associated with sideroblastic anemia (106, 109–112; reviewed in 108). Acquired causes of sideroblastic anemia are found to be associated with nutritional metal deficiencies or overloads, alcohol consumption, myelodysplastic syndrome, and a side effect of certain medications, such as antibiotics and chelators (reviewed in 113).
TISSUE-SPECIFIC MITOCHONDRIAL IRON DISORDERS
Systemic iron overload commonly results from increased iron absorption secondary to other iron related syndromes such as hemochromatosis, anemias characterized by ineffective erythropoiesis or from hypertransfusion of red blood cells (63). In each of these conditions, excess Fe collects inside the cell accumulating both in the cytosol and in other intracellular compartments. Iron has an enormous damaging potential and even a small transit pool of cellular chelatable iron is believed to catalyze the generation of ROS within mammalian cells (42). Several laboratories have characterized an accumulation of cellular and mitochondrial iron with age in different organisms and tissues in parallel with an increase in oxidative damage (114). Mitochondrial iron accumulation is a hallmark of diseases associated with impaired Fe-S biogenesis, such as Friedreich’s ataxia and skeletal muscle myopathies, as discussed in detail below. However, the pathophysiological relevance of the mitochondrial iron loading and the underlying mechanisms and regulatory aspects of this process remain unknown.
Friedreich’s Ataxia
Friedreich’s ataxia is a rare disease leading to severe neuro- and cardio-degeneration and is the most common inherited spino-cerebellar ataxia resulting in confinement to a wheelchair and death due to cardiomyopathy (115). Friedreich’s ataxia is an autosomal recessive disease caused by an intronic GAA triplet repeat expansion within the first intron of the Friedreich’s ataxia gene FRDA. This hinders the transcription of the FRDA gene, causing a marked reduction in expression of frataxin (116). The end result of this expansion is insufficient frataxin that gives rise to mitochondrial iron accumulation, especially in tissues of high mitochondrial content, such as nerve and cardiac tissue (117), with FRDA deletion leading to lethality (118). While neurological impairment may be the most prominent finding, hypertrophic cardiomyopathy is the most common cause of death (115). Frataxin is an IMM and mitochondrial matrix protein (119). The exact biochemical role of frataxin remains to be defined, but its importance in Fe-S cluster synthesis is clear. Yeast knockout models, conditional frataxin deletion in murine models, and histological and biochemical data from heart biopsies or autopsies of Friedreich’s ataxia patients have shown that frataxin defects cause a specific Fe-S cluster protein deficiency and intramitochondrial iron accumulation (117), possibly involving MFRN2 (120).
Cardiovascular and Pulmonary Diseases
Many defects in mitochondrial iron metabolism are associated with cardiovascular and pulmonary diseases. Mitochondrial iron overload is linked to doxorubicin-induced cardiomyopathy, myocardial infarction, and cardiac ischemia/reperfusion injury in murine models; it is also associated with some mouse genetic models of spontaneous cardiomyopathy (62, 121, 122). Acute exhaustive exercise significantly increases MTFT expression in the murine heart (123), and loss of ABCB8 in mouse hearts is associated with mitochondrial iron accumulation and cardiomyopathy (61). Fe-S deficiencies with mitochondrial iron overload promote pulmonary hypertension, an increasingly prevalent vascular disease defined by increased pulmonary arterial pressure and lung vasculopathy that is triggered by varied and often disparate stimuli (124). Mitochondrial iron overload has also been associated with the pathogenesis of chronic obstructive pulmonary disease (COPD), a chronic inflammatory lung disease associated with cigarette smoking (125).
Neurological Disease
The accumulation of iron within the central nervous system is associated with neurodegenerative diseases, including Alzheimer’s disease, Parkinson’s disease, Huntington disease, amyotrophic lateral sclerosis, and neurodegeneration with brain iron accumulation. The brain is especially susceptible to iron accumulation with increased oxidative stress and oxidative damage because of its high rate of oxygen consumption and relatively poor ability to deal with ROS. While some neurodegenerative diseases are due to a primary defect in iron-related genes, e.g., neuroferritinopathy (Ft-L), aceruloplasminemia (ceruloplasmin), or pantothenate kinase-associated degeneration (PANK2), most have iron dysregulation as a secondary feature. It is difficult to attribute iron dysregulation to a single pathogenic mechanism. The precise role of iron in these disorders is therefore a subject of great debate, with the consensus being that iron accumulation is downstream of other primary causes, but that the consequences of iron accumulation are connected to neuronal death. Efforts to understand and ameliorate the effects of iron and mitochondrial iron dysregulation are therefore of broad interest, especially as there are no treatments that can slow the progression of these diseases (126). Here, we review the relationship between iron accumulation in the mitochondria and mitochondrial dysfunction in the development of the most prevalent neurodegenerative diseases associated with iron accumulation in the brain.
Alzheimer’s disease.
Alzheimer’s disease is characterized by neuronal loss and neuroinflammation and involves the progressive dysfunction and degeneration of neurons in various cortical regions, which leads to progressive memory loss and dementia (127). Although several genes have been linked to the hereditary Alzheimer’s disease, the etiology of the sporadic form remains a mystery. Both forms share similar neuropathological and molecular features, including extracellular deposition of amyloid beta (Aβ); intracellular accumulation of hyperphosphorylated tau protein; disturbances in mitochondrial structure, function, and metabolism; oxidative stress; impairment of N-methyl-d-aspartate receptor-related signaling pathways; abnormalities of lipid signaling and metabolism; and aberrant cell cycle control (128). In addition to these characteristic features, the Alzheimer’s disease brain has been found to contain increased levels of various metals including iron (129). Pathological levels of iron are found in association with Aβ in the extracellular space as well as within neurons containing neurofibrillary tangles (130). Although there is little to no concrete evidence for the increased accumulation of iron inside the mitochondria of Alzheimer’s disease patients and model systems, increased expression of MTFT has been observed in the brains of patients and is associated with the antioxidant role of this protein (131). Furthermore, MTFT overexpression attenuates Aβ-induced neurotoxicity (59) in a murine Alzheimer’s disease model, suggesting that cells respond to excess mitochondrial iron.
Parkinson’s disease.
Parkinson’s disease (PD) is the most common neurodegenerative movement disorder worldwide. PD is characterized by degeneration of dopaminergic neurons of the substantia nigra pars compacta (SNpc), resulting in lower dopamine levels in the corpus striatum. PD is also characterized by the presence of proteinaceous cytoplasmic inclusions, called Lewy bodies, and by the deregulation of basal ganglia circuitries that leads to the appearance of motor symptoms, including resting tremor, rigidity, bradykinesia, and postural instability (132, 133). While sporadic cases represent more than 90% of total PD patients, there are several inherited forms caused by mutations in single genes, including α-syn, PARKIN, PINK1, DJ-1, LRRK2, and ATP13A2. These genes have all been shown to disrupt mitochondrial function (133), and mitochondria isolated from human brain tissues and peripheral cells of sporadic PD patients exhibit reduced mitochondrial complex I activity (134). Such mitochondrial dysfunction via inhibition of complex I may lead to the subsequent activation of IRP1, increased DMT1 and TFR1 expression, increased iron uptake, and increased ROS generation (133). However, despite concrete evidence for cellular iron accumulation in PD, a number of studies demonstrate mitochondrial iron–specific changes in human and rodent PD models. Specifically, in vitro treatment of SH-SY5Y dopaminergic neuroblastoma cells with mitochondrial complex I inhibitors such as rotenone or MPP+ results in ROS production, increased mitochondrial iron uptake, and increased expression of MFRN2 (135, 136). In a rodent rotenone model of PD, transferrin accumulates in dopamine neurons, with much of it accumulating in the mitochondria (137). In murine models of PD (MPTP and 6-hydroxydopamine–induced PD), MTFT is increased as a protective strategy to limit injury possibly via the modulation of α-synuclein expression (58, 138). However, in a TFR1-conditional knockout mouse model, which could alleviate the iron uptake into neurons, mitochondria exhibited the disrupted morphologies followed by neuronal death, deterioration of motor activity, and the early death of mice, similar to the symptoms observed in PD (139).
Huntington’s disease.
Huntington’s disease (HD) is a progressive neurodegenerative disease caused by a trinucleotide CAG repeat expansion in exon 1 of the HTT gene (140). HD is characterized by progressive motor, psychiatric, and cognitive deterioration, as well as weight loss. Neurodegeneration, characterized by neuronal death and glial activation, primarily occurs in the striatum and cerebral cortex (141). Mutant huntingtin protein is expressed in neurons and glia (142), and there is considerable evidence for bioenergetic dysfunction in HD. Brains from patients with advanced HD obtained postmortem demonstrate the disruption of mitochondrial energy metabolism, which includes decreased activities of mitochondrial respiratory complexes II–IV and aconitase (143). Interestingly, transcriptomic analyses indicate increased transcript expression of MFRN2 in human HD (144). In addition, mitochondrial iron accumulates in mouse and human HD brains with increased expression of MFRN2 and loss of frataxin (144).
Type 2 Wolfram syndrome.
Type 2 Wolfram syndrome, also known as DIDMOAD (diabetes insipidus, diabetes mellitus, optic atrophy, and deafness), is a multisystem, neuronal, and β-cell–degenerative disorder of autosomal recessive inheritance. The β-cell degeneration resulting from cellular stress and apoptosis leads to severe insulin deficiency mimicking type 1 diabetes. Type 2 Wolfram syndrome is associated with a single missense CISD2 (CDGSH iron-sulfur domain-containing protein 2) gene mutation that truncates 75% of the protein in affected patients, leading to mitochondrial labile iron accumulation and oxidative stress (145).
Skeletal Myopathy
Abnormalities in mitochondrial iron homeostasis can also cause disorders of skeletal muscle with or without abnormalities in other tissues. Deficiency of the scaffold protein ISCU, due to a splicing error, causes autosomal recessive myopathy with mitochondrial iron overload (146). Other examples include hereditary myopathy with lactic acidosis (HML), a rare and recessively inherited disease characterized by early onset muscle weakness, exercise intolerance, and lactic acidosis (147). HML is associated with mutations in intron 5 of the ISCU gene, resulting in a decrease in ICSU expression in striated muscle and, as a consequence, impaired maturation of mitochondrial and extramitochondrial Fe-S enzymes and mitochondrial iron accumulation (147).
These findings demonstrate that mitochondrial iron accumulation is a common consistent pathogenic event that accompanies mitochondrial dysfunction in a range of diseases and tissue types. Such excess mitochondrial iron can cause oxidative stress and cellular damage, ultimately leading to increased morbidity and mortality. Many questions remain as to how or why this iron accumulates inside mitochondria and how these processes are regulated; these issues require more extensive investigation.
TARGETING MITOCHONDRIAL IRON FOR THERAPY
Removing surplus iron by way of chelation may reduce oxidative stress and has been a longtime therapeutic target for the treatment of many of the diseases described above. Early trials of the iron chelator deferoxamine have generally failed, as this chelating agent does not cross cell membranes well (148). Only chelators capable of penetrating the mitochondria are likely to be useful in the treatment of mitochondrial iron over load disorders. Deferiprone (e.g., Ferriprox) is an oral iron chelator that is capable of crossing the plasma membrane, slowing the targeting of mitochondrial nonheme iron deposits to prevent mitochondrial iron loading (149). Deferiprone is presently approved in the United States and Europe for the treatment of thalassemia major. Its ability to cross cellular barriers and the blood-brain barrier to redistribute intramitochondrial iron has rendered it useful in the treatment of numerous disorders associated with mitochondrial iron overload (150).
Several studies have evaluated the effect of deferiprone in a range of neurological diseases. Administration of deferiprone to Friedreich’s ataxia patients for six months resulted in a selective and significant reduction in foci of brain iron accumulation and initial functional improvements (151). Again, in Friedreich’s ataxia patients, combination therapy of deferiprone and idebenone demonstrated neurological stabilization, reduction in iron on magnetic resonance imaging (MRI) of the dentate nucleus, and decreased intraventricular septum thickness and left ventricular mass index (152). However, studies using triple therapy with deferiprone, idebenone, and riboflavin have demonstrated no clear neurologic or cardiac benefits (153). Deferiprone is also currently being investigated in patients with prodromal Alzheimer’s disease and mild Alzheimer’s disease in a phase 2 study (). In animal models, deferiprone showed promise by reducing Aβ and tau phosphorylation levels in the hippocampus of rabbits with Alzheimer’s disease (fed a cholesterol-enriched diet) (154). Deferiprone is also being evaluated for efficacy in the treatment of early onset PD (study ), with early studies from a randomized double-blind, placebo-controlled trial indicating seemingly positive results (155). Deferiprone also rescues mitochondrial markers of HD ex vivo and in vivo (144), justifying its use in the treatment of this disease. Although deferiprone appears to be relatively safe at lower doses, and some evidence supports its therapeutic benefit, there have been no systematic, large-scale, placebo-controlled studies to further evaluate its utility to treat Friedreich’s ataxia and other neurological disorders (150).
Recently, a number of novel mitochondrial iron chelators have been developed that may hold promise for the treatment of mitochondrial iron-related diseases. Deferiprone-resveratrol hybrids have been synthesized as antioxidants, Aβ1–42 aggregation inhibitors, and metal-chelating agents for Alzheimer’s disease (156). The hydroxyquinoline derivatives 5-((methylamino)methyl)-8-hydroxyquinoline (Q1) and 5-(morpholinomethyl)-8-hydroxyquinoline (Q4) (136), as well as polyhydroxyl coumarin, N-(1,3-dihydroxy-2-(hydroxymethyl)propan-2-yl)-2-(7-hydroxy-2-oxo-2H-chromen-4-yl) acetamide (CT51) (157), both exhibit potent free radical–neutralizing activity in vitro. Q1 has also been shown to protect dopaminergic neurons from cell death and oxidative stress in murine PD models (136). Mitochondria-targeted hexadentate (tricatechol-based) iron chelators linked to mitochondria-homing somatostatin-like peptides have demonstrated promise by alleviating mitochondrial iron overload in culture systems and protecting against UV radiation (158). DFO-conjugates, termed mtDFOs, which consist of DFO conjugated to molecules that can easily enter the mitochondria [TAT49–57, 1A, SS02, and SS20 (159) or to triphenylphosphonium (160)], have also been developed. Analogs of 2-pyridylcarboxaldehyde isonicotinoyl hydrazone have also shown promise in alleviating mitochondrial iron overload in model systems (161). Finally, a lipophilic small molecule, hinokitiol, a natural product isolated from the essential oil of the Chamaecyparis taiwanensis (Taiwan Hinoki), can autonomously perform transmembrane iron transport, including the delivery of iron to mitochondria (162), opening a new arena for small molecules that regulate mitochondrial iron transport. To summarize, the efficacy of most of these small molecules remains to be tested using in vivo models of disease. With these therapeutic approaches, great attention should be paid to not fully deplete mitochondrial iron stores to avoid undesirable consequences.
CONCLUSION
In conclusion, mitochondria are no longer considered as simple, discrete, kidney bean–shaped energy factories. Instead, they encompass a cell- and tissue-specific, organellar network that fuses, divides, and directs a vast array of functions central to cellular life, death, and differentiation. Maintaining iron homeostasis is an important contributory part of the essential nature of these dynamic organelles. Emerging evidence for the crucial role of mitochondrial iron storage, utilization, and export demonstrates that the mitochondrion is a distinct compartment of iron metabolism at the center of mitochondrial, cellular, and tissue iron homeostasis. Defects in mitochondrial function can lead to excess iron accumulation in mitochondria and consequent oxidative damage. Similarly, the overriding theme in mitochondrial iron–related diseases is dysfunctional mitochondrial Fe-S cluster synthesis or heme synthesis. This may lead to further downstream signals to increase mitochondrial iron import, giving rise to more oxidative damage. The mechanisms behind this increased mitochondrial iron uptake and how this uptake is regulated at the transporter level remain to be determined. For example, in red cells, MFRN1 works in a complex with ABCB10 and other heme synthesis enzymes to increase iron import (30–31). Yet, how MFRN transporter activity is regulated in other tissues or how other metal transporter activity is regulated is unknown. Future work in determining this regulation of activity (e.g., phosphorylation) may provide potential targeted therapies to reduce iron import into mitochondria under disease conditions.
Increased mitochondrial iron import in the face of defective Fe-S cluster synthesis or heme synthesis leads to the overriding problem in treating mitochondrial iron–based diseases, as treatments predominantly involve iron chelation therapies. These therapies will reduce iron delivery to mitochondria but will not repair defects in Fe-S cluster synthesis or heme synthesis. Hence, the development of efficacious precision-based approaches may involve using a “two-hit” approach encompassing the slowing down of iron delivery to the mitochondria (without depleting it), while at the same time repairing Fe-S cluster or heme synthesis. Alternatively, early interventions preventing excessive mitochondrial iron delivery or precisely targeting defective Fe-S cluster or heme synthesis may also be feasible. Although much remains to be learned about iron, mitochondria, and disease pathogenesis, evidence for dysregulated mitochondrial iron metabolism is clear in a range of cellular pathways, as well as in a plethora of disorders and diseases, thus confirming the importance of mitochondrial iron regulatory processes in human health and disease. Understanding how mitochondrial iron homeostasis is regulated will provide more insight into possible therapeutic targets of mitochondrial iron–based diseases.
ACKNOWLEDGMENTS
D.M.W. is supported by the US National Institutes of Health, National Institute of Diabetes and Digestive and Kidney Diseases (grants R01 DK030534, R01 DK052380), and the Friedreich’s Ataxia Research Alliance (grant 10047373). S.M.C. is supported by the US National Institutes of Health–National Heart, Lung, and Blood Institute (grant R00-HL125899).
Footnotes
DISCLOSURE STATEMENT
The authors are not aware of any affiliations, memberships, funding, or financial holdings that might be perceived as affecting the objectivity of this review.
LITERATURE CITED
- 1.Richardson DR, Lane DJ, Becker EM, Huang ML, Whitnall M, et al. 2010. Mitochondrial iron trafficking and the integration of iron metabolism between the mitochondrion and cytosol. PNAS 107:10775–82 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Paul BT, Manz DH, Torti FM, Torti SV. 2017. Mitochondria and iron: current questions. Expert Rev. Hematol 10:65–79 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Rauen U, Springer A, Weisheit D, Petrat F, Korth HG, et al. 2007. Assessment of chelatable mitochondrial iron by using mitochondrion-selective fluorescent iron indicators with different iron-binding affinities. ChemBioChem 8:341–52 [DOI] [PubMed] [Google Scholar]
- 4.Jhurry ND, Chakrabarti M, McCormick SP, Holmes-Hampton GP, Lindahl PA. 2012. Biophysical investigation of the ironome of human jurkat cells and mitochondria. Biochemistry 51:5276–84 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chevion M 1988. A site-specific mechanism for free radical induced biological damage: the essential role of redox-active transition metals. Free Radic. Biol. Med 5:27–37 [DOI] [PubMed] [Google Scholar]
- 6.Philpott CC. 2006. Iron uptake in fungi: a system for every source. Biochim. Biophys. Acta 1763:636–45 [DOI] [PubMed] [Google Scholar]
- 7.Khan A, Singh P, Srivastava A. 2017. Synthesis, nature and utility of universal iron chelator– siderophore: a review. Microbiol. Res 212–213:103–11 [DOI] [PubMed] [Google Scholar]
- 8.Bai L, Qiao M, Zheng R, Deng C, Mei S, Chen W. 2016. Phylogenomic analysis of transferrin family from animals and plants. Comp. Biochem. Physiol. D Genom. Proteom 17:1–8 [DOI] [PubMed] [Google Scholar]
- 9.Xiao G, Wan Z, Fan Q, Tang X, Zhou B. 2014. The metal transporter ZIP13 supplies iron into the secretory pathway in Drosophila melanogaster. eLife 3:e03191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Donovan A, Brownlie A, Zhou Y, Shepard J, Pratt SJ, et al. 2000. Positional cloning of zebrafish ferroportin1 identifies a conserved vertebrate iron exporter. Nature 403:776–81 [DOI] [PubMed] [Google Scholar]
- 11.Fleming RE, Ponka P. 2012. Iron overload in human disease. N. Engl. J. Med 366:348–59 [DOI] [PubMed] [Google Scholar]
- 12.Wilkinson N, Pantopoulos K. 2014. The IRP/IRE system in vivo: insights from mouse models. Front. Pharmacol 5:176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Shi H, Bencze KZ, Stemmler TL, Philpott CC. 2008. A cytosolic iron chaperone that delivers iron to ferritin. Science 320:1207–10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mancias JD, Wang X, Gygi SP, Harper JW, Kimmelman AC. 2014. Quantitative proteomics identifies NCOA4 as the cargo receptor mediating ferritinophagy. Nature 509:105–9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gunshin H, Mackenzie B, Berger UV, Gunshin Y, Romero MF, et al. 1997. Cloning and characterization of a mammalian proton-coupled metal-ion transporter. Nature 388:482–88 [DOI] [PubMed] [Google Scholar]
- 16.Jenkitkasemwong S, Wang CY, Mackenzie B, Knutson MD. 2012. Physiologic implications of metal-ion transport by ZIP14 and ZIP8. Biometals 25:643–55 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hamza I, Dailey HA. 2012. One ring to rule them all: trafficking of heme and heme synthesis intermediates in the metazoans. Biochim. Biophys. Acta 1823:1617–32 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kühlbrandt W 2015. Structure and function of mitochondrial membrane protein complexes. BMC Biol 13:89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhang AS, Sheftel AD, Ponka P. 2005. Intracellular kinetics of iron in reticulocytes: evidence for endosome involvement in iron targeting to mitochondria. Blood 105:368–75 [DOI] [PubMed] [Google Scholar]
- 20.Sheftel AD, Zhang AS, Brown C, Shirihai OS, Ponka P. 2007. Direct interorganellar transfer of iron from endosome to mitochondrion. Blood 110:125–32 [DOI] [PubMed] [Google Scholar]
- 21.Hamdi A, Roshan TM, Kahawita TM, Mason AB, Sheftel AD, Ponka P. 2016. Erythroid cell mitochondria receive endosomal iron by a “kiss-and-run” mechanism. Biochim. Biophys. Acta 1863:2859–67 [DOI] [PubMed] [Google Scholar]
- 22.Das A, Nag S, Mason AB, Barroso MM. 2016. Endosome-mitochondria interactions are modulated by iron release from transferrin. J. Cell Biol 214:831–45 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lambe T, Simpson RJ, Dawson S, Bouriez-Jones T, Crockford TL, et al. 2009. Identification of a Steap3 endosomal targeting motif essential for normal iron metabolism. Blood 113:1805–8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wolff NA, Garrick MD, Zhao L, Garrick LM, Ghio AJ, Thevenod F. 2018. A role for divalent metal transporter (DMT1) in mitochondrial uptake of iron and manganese. Sci. Rep 8:211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Foury F, Roganti T. 2002. Deletion of the mitochondrial carrier genes MRS3 and MRS4 sup-presses mitochondrial iron accumulation in a yeast frataxin-deficient strain. J. Biol. Chem 277:24475–83 [DOI] [PubMed] [Google Scholar]
- 26.Li L, Kaplan J. 2004. A mitochondrial-vacuolar signaling pathway in yeast that affects iron and copper metabolism. J. Biol. Chem 279:33653–61 [DOI] [PubMed] [Google Scholar]
- 27.Shaw GC, Cope JJ, Li L, Corson K, Hersey C, et al. 2006. Mitoferrin is essential for erythroid iron assimilation. Nature 440:96–100 [DOI] [PubMed] [Google Scholar]
- 28.Troadec MB, Warner D, Wallace J, Thomas K, Spangrude GJ, et al. 2011. Targeted deletion of the mouse Mitoferrin1 gene: from anemia to protoporphyria. Blood 117:5494–502 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chen W, Dailey HA, Paw BH. 2010. Ferrochelatase forms an oligomeric complex with mitoferrin-1 and Abcb10 for erythroid heme biosynthesis. Blood 116:628–30 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chen W, Paradkar PN, Li L, Pierce EL, Langer NB, et al. 2009. Abcb10 physically interacts with mitoferrin-1 (Slc25a37) to enhance its stability and function in the erythroid mitochondria. PNAS 106:16263–68 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Medlock AE, Shiferaw MT, Marcero JR, Vashisht AA, Wohlschlegel JA, et al. 2015. Identification of the mitochondrial heme metabolism complex. PLOS ONE 10:e0135896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Christenson ET, Gallegos AS, Banerjee A. 2018. In vitro reconstitution, functional dissection, and mutational analysis of metal ion transport by mitoferrin-1. J. Biol. Chem 293:3819–28 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ren Y, Yang S, Tan G, Ye W, Liu D, et al. 2012. Reduction of mitoferrin results in abnormal development and extended lifespan in Caenorhabditis elegans. PLOS ONE 7:e29666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chen YC, Wu YT, Wei YH. 2015. Depletion of mitoferrins leads to mitochondrial dysfunction and impairment of adipogenic differentiation in 3T3-L1 preadipocytes. Free Radic. Res 49:1285–95 [DOI] [PubMed] [Google Scholar]
- 35.Hung HI, Schwartz JM, Maldonado EN, Lemasters JJ, Nieminen AL. 2013. Mitoferrin-2-dependent mitochondrial iron uptake sensitizes human head and neck squamous carcinoma cells to photodynamic therapy. J. Biol. Chem 288:677–86 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Paradkar PN, Zumbrennen KB, Paw BH, Ward DM, Kaplan J. 2009. Regulation of mitochondrial iron import through differential turnover of mitoferrin 1 and mitoferrin 2. Mol. Cell. Biol 29:1007–16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wang C, Chen X, Zou H, Chen X, Liu Y, Zhao S. 2014. The roles of mitoferrin-2 in the process of arsenic trioxide-induced cell damage in human gliomas. Eur. J. Med. Res 19:49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Yoon H, Zhang Y, Pain J, Lyver ER, Lesuisse E, et al. 2011. Rim2, a pyrimidine nucleotide exchanger, is needed for iron utilization in mitochondria. Biochem. J 440:137–46 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Yan H, Hao S, Sun X, Zhang D, Gao X, et al. 2015. Blockage of mitochondrial calcium uniporter prevents iron accumulation in a model of experimental subarachnoid hemorrhage. Biochem. Biophys. Res. Commun 456:835–40 [DOI] [PubMed] [Google Scholar]
- 40.Abbate V, Reelfs O, Kong X, Pourzand C, Hider RC. 2016. Dual selective iron chelating probes with a potential to monitor mitochondrial labile iron pools. Chem. Commun 52:784–87 [DOI] [PubMed] [Google Scholar]
- 41.Lindahl PA, Moore MJ. 2016. Labile low-molecular-mass metal complexes in mitochondria: trials and tribulations of a burgeoning field. Biochemistry 55:4140–53 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Petrat F, de Groot H, Rauen U. 2001. Subcellular distribution of chelatable iron: a laser scanning microscopic study in isolated hepatocytes and liver endothelial cells. Biochem. J 356:61–69 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Shvartsman M, Cabantchik ZI. 2012. Intracellular iron trafficking: role of cytosolic ligands. Biometals 25:711–23 [DOI] [PubMed] [Google Scholar]
- 44.Ponka P, Sheftel AD, English AM, Bohle DS, Garcia-Santos D. 2017. Do mammalian cells really need to export and import heme? Trends Biochem. Sci 42:395–406 [DOI] [PubMed] [Google Scholar]
- 45.Verma A, Nye JS, Snyder SH. 1987. Porphyrins are endogenous ligands for the mitochondrial (peripheral-type) benzodiazepine receptor. PNAS 84:2256–60 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Krishnamurthy PC, Du G, Fukuda Y, Sun D, Sampath J, et al. 2006. Identification of a mammalian mitochondrial porphyrin transporter. Nature 443:586–89 [DOI] [PubMed] [Google Scholar]
- 47.Helias V, Saison C, Ballif BA, Peyrard T, Takahashi J, et al. 2012. ABCB6 is dispensable for erythropoiesis and specifies the new blood group system Langereis. Nat. Genet 44:170–73 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Braymer JJ, Lill R. 2017. Iron-sulfur cluster biogenesis and trafficking in mitochondria. J. Biol. Chem 292:12754–63 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Rouault TA, Maio N. 2017. Biogenesis and functions of mammalian iron-sulfur proteins in the regulation of iron homeostasis and pivotal metabolic pathways. J. Biol. Chem 292:12744–53 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kispal G, Csere P, Prohl C, Lill R. 1999. The mitochondrial proteins Atm1p and Nfs1p are essential for biogenesis of cytosolic Fe/S proteins. EMBO J 18:3981–89 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Bekri S, Kispal G, Lange H, Fitzsimons E, Tolmie J, et al. 2000. Human ABC7 transporter: gene structure and mutation causing X-linked sideroblastic anemia with ataxia with disruption of cytosolic iron–sulfur protein maturation. Blood 96:3256–64 [PubMed] [Google Scholar]
- 52.Li L, Miao R, Jia X, Ward DM, Kaplan J. 2014. Expression of the yeast cation diffusion facilitators Mmt1 and Mmt2 affects mitochondrial and cellular iron homeostasis: evidence for mitochondrial iron export. J. Biol. Chem 289:17132–41 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Drysdale J, Arosio P, Invernizzi R, Cazzola M, Volz A, et al. 2002. Mitochondrial ferritin: a new player in iron metabolism. Blood Cells Mol. Dis 29:376–83 [DOI] [PubMed] [Google Scholar]
- 54.Levi S, Corsi B, Bosisio M, Invernizzi R, Volz A, et al. 2001. A human mitochondrial ferritin encoded by an intronless gene. J. Biol. Chem 276:24437–40 [DOI] [PubMed] [Google Scholar]
- 55.Shi Y, Ghosh MC, Tong WH, Rouault TA. 2009. Human ISD11 is essential for both iron-sulfur cluster assembly and maintenance of normal cellular iron homeostasis. Hum. Mol. Genet 18:3014–25 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Corsi B, Cozzi A, Arosio P, Drysdale J, Santambrogio P, et al. 2002. Human mitochondrial ferritin expressed in HeLa cells incorporates iron and affects cellular iron metabolism. J. Biol. Chem 277:22430–37 [DOI] [PubMed] [Google Scholar]
- 57.Nie G, Sheftel AD, Kim SF, Ponka P. 2005. Overexpression of mitochondrial ferritin causes cytosolic iron depletion and changes cellular iron homeostasis. Blood 105:2161–67 [DOI] [PubMed] [Google Scholar]
- 58.Shi ZH, Nie G, Duan XL, Rouault T, Wu WS, et al. 2010. Neuroprotective mechanism of mitochondrial ferritin on 6-hydroxydopamine-induced dopaminergic cell damage: implication for neuroprotection in Parkinson’s disease. Antioxid. Redox Signal 13:783–96 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Wu WS, Zhao YS, Shi ZH, Chang SY, Nie GJ, et al. 2013. Mitochondrial ferritin attenuates β-amyloid-induced neurotoxicity: reduction in oxidative damage through the Erk/P38 mitogen-activated protein kinase pathways. Antioxid. Redox Signal 18:158–69 [DOI] [PubMed] [Google Scholar]
- 60.Chiabrando D, Marro S, Mercurio S, Giorgi C, Petrillo S, et al. 2012. The mitochondrial heme exporter FLVCR1b mediates erythroid differentiation. J. Clin. Investig 122:4569–79 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Ichikawa Y, Bayeva M, Ghanefar M, Potini V, Sun L, et al. 2012. Disruption of ATP-binding cassette B8 in mice leads to cardiomyopathy through a decrease in mitochondrial iron export. PNAS 109:4152–57 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Ichikawa Y, Ghanefar M, Bayeva M, Wu R, Khechaduri A, et al. 2014. Cardiotoxicity of doxorubicin is mediated through mitochondrial iron accumulation. J. Clin. Investig 124:617–30 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Xu W, Barrientos T, Andrews NC. 2013. Iron and copper in mitochondrial diseases. Cell Metab 17:319–28 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Santambrogio P, Dusi S, Guaraldo M, Rotundo LI, Broccoli V, et al. 2015. Mitochondrial iron and energetic dysfunction distinguish fibroblasts and induced neurons from pantothenate kinase-associated neurodegeneration patients. Neurobiol. Dis 81:144–53 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Tavsan Z, Ayar Kayali H. 2015. The variations of glycolysis and TCA cycle intermediate levels grown in iron and copper mediums of Trichoderma harzianum. Appl. Biochem. Biotechnol 176:76–85 [DOI] [PubMed] [Google Scholar]
- 66.Ristow M, Pfister MF, Yee AJ, Schubert M, Michael L, et al. 2000. Frataxin activates mitochondrial energy conversion and oxidative phosphorylation. PNAS 97:12239–43 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Lodi R, Cooper JM, Bradley JL, Manners D, Styles P, et al. 1999. Deficit of in vivo mitochondrial ATP production in patients with Friedreich ataxia. PNAS 96:11492–95 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Crooks DR, Maio N, Lane AN, Jarnik M, Higashi RM, et al. 2018. Acute loss of iron-sulfur clusters results in metabolic reprogramming and generation of lipid droplets in mammalian cells. J. Biol. Chem 293:8297–311 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Tyrakis PA, Yurkovich ME, Sciacovelli M, Papachristou EK, Bridges HR, et al. 2017. Fumarate hydratase loss causes combined respiratory chain defects. Cell Rep 21:1036–47 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Van Vranken JG, Jeong MY, Wei P, Chen YC, Gygi SP, et al. 2016. The mitochondrial acyl carrier protein (ACP) coordinates mitochondrial fatty acid synthesis with iron sulfur cluster biogenesis. eLife 5:e17828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Mishra P, Chan DC. 2014. Mitochondrial dynamics and inheritance during cell division, development and disease. Nat. Rev. Mol. Cell Biol 15:634–46 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Nikpour M, Pellagatti A, Liu A, Karimi M, Malcovati L, et al. 2010. Gene expression profiling of erythroblasts from refractory anaemia with ring sideroblasts (RARS) and effects of G-CSF. Br. J. Haematol 149:844–54 [DOI] [PubMed] [Google Scholar]
- 73.Baricault L, Segui B, Guegand L, Olichon A, Valette A, et al. 2007. OPA1 cleavage depends on decreased mitochondrial ATP level and bivalent metals. Exp. Cell Res 313:3800–8 [DOI] [PubMed] [Google Scholar]
- 74.Yoon YS, Yoon DS, Lim IK, Yoon SH, Chung HY, et al. 2006. Formation of elongated giant mitochondria in DFO-induced cellular senescence: involvement of enhanced fusion process through modulation of Fis1. J. Cell Physiol 209:468–80 [DOI] [PubMed] [Google Scholar]
- 75.Park J, Lee DG, Kim B, Park SJ, Kim JH, et al. 2015. Iron overload triggers mitochondrial fragmentation via calcineurin-sensitive signals in HT-22 hippocampal neuron cells. Toxicology 337:39–46 [DOI] [PubMed] [Google Scholar]
- 76.Jornayvaz FR, Shulman GI. 2010. Regulation of mitochondrial biogenesis. Essays Biochem 47:69–84 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Nam E, Han J, Suh JM, Yi Y, Lim MH. 2018. Link of impaired metal ion homeostasis to mitochondrial dysfunction in neurons. Curr. Opin. Chem. Biol 43:8–14 [DOI] [PubMed] [Google Scholar]
- 78.Mizumura K, Cloonan SM, Nakahira K, Bhashyam AR, Cervo M, et al. 2014. Mitophagy-dependent necroptosis contributes to the pathogenesis of COPD. J. Clin. Investig 124:3987–4003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Chu CT, Ji J, Dagda RK, Jiang JF, Tyurina YY, et al. 2013. Cardiolipin externalization to the outer mitochondrial membrane acts as an elimination signal for mitophagy in neuronal cells. Nat. Cell Biol 15:1197–205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Ishii KA, Fumoto T, Iwai K, Takeshita S, Ito M, et al. 2009. Coordination of PGC-1β and iron uptake in mitochondrial biogenesis and osteoclast activation. Nat. Med 15:259–66 [DOI] [PubMed] [Google Scholar]
- 81.Lin H, Magrane J, Rattelle A, Stepanova A, Galkin A, et al. 2017. Early cerebellar deficits in mitochondrial biogenesis and respiratory chain complexes in the KIKO mouse model of Friedreich ataxia. Dis. Model. Mech 10:1343–52 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Crooks DR, Natarajan TG, Jeong SY, Chen C, Park SY, et al. 2014. Elevated FGF21 secretion, PGC-1α and ketogenic enzyme expression are hallmarks of iron-sulfur cluster depletion in human skeletal muscle. Hum. Mol. Genet 23:24–39 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Schiavi A, Maglioni S, Palikaras K, Shaik A, Strappazzon F, et al. 2015. Iron-starvation-induced mitophagy mediates lifespan extension upon mitochondrial stress in C. elegans. Curr. Biol 25:1810–22 [DOI] [PubMed] [Google Scholar]
- 84.Esposito G, Vos M, Vilain S, Swerts J, De Sousa Valadas J, et al. 2013. Aconitase causes iron toxicity in Drosophila pink1 mutants. PLOS Genet 9:e1003478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Allen GF, Toth R, James J, Ganley IG. 2013. Loss of iron triggers PINK1/Parkin-independent mitophagy. EMBO Rep 14:1127–35 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Wilfinger N, Austin S, Scheiber-Mojdehkar B, Berger W, Reipert S, et al. 2016. Novel p53-dependent anticancer strategy by targeting iron signaling and BNIP3L-induced mitophagy. Oncotarget 7:1242–61 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Wang X, Yang H, Yanagisawa D, Bellier JP, Morino K, et al. 2016. Mitochondrial ferritin affects mitochondria by stabilizing HIF-1α in retinal pigment epithelium: implications for the pathophysiology of age-related macular degeneration. Neurobiol. Aging 47:168–79 [DOI] [PubMed] [Google Scholar]
- 88.Melber A, Haynes CM. 2018. UPRmt regulation and output: a stress response mediated by mitochondrial-nuclear communication. Cell Res 28:281–95 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Ciesielski SJ, Schilke B, Marszalek J, Craig EA. 2016. Protection of scaffold protein Isu from degradation by the Lon protease Pim1 as a component of Fe-S cluster biogenesis regulation. Mol. Biol. Cell 27:1060–68 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Tian Q, Li T, Hou W, Zheng J, Schrum LW, Bonkovsky HL. 2011. Lon peptidase 1 (LONP1)-dependent breakdown of mitochondrial 5-aminolevulinic acid synthase protein by heme in human liver cells. J. Biol. Chem 286:26424–30 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Kardon JR, Yien YY, Huston NC, Branco DS, Hildick-Smith GJ, et al. 2015. Mitochondrial ClpX activates a key enzyme for heme biosynthesis and erythropoiesis. Cell 161:858–67 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Guillon B, Bulteau AL, Wattenhofer-Donzé M, Schmucker S, Friguet B, et al. 2009. Frataxin deficiency causes upregulation of mitochondrial Lon and ClpP proteases and severe loss of mitochondrial Fe-S proteins. FEBS J 276:1036–47 [DOI] [PubMed] [Google Scholar]
- 93.Holmes-Hampton GP, Tong WH, Rouault TA. 2014. Biochemical and biophysical methods for studying mitochondrial iron metabolism. Methods Enzymol 547:275–307 [DOI] [PubMed] [Google Scholar]
- 94.Ma Y, Abbate V, Hider RC. 2015. Iron-sensitive fluorescent probes: monitoring intracellular iron pools. Metallomics 7:212–22 [DOI] [PubMed] [Google Scholar]
- 95.Abbate V, Reelfs O, Hider RC, Pourzand C. 2015. Design of novel fluorescent mitochondria-targeted peptides with iron-selective sensing activity. Biochem. J 469:357–66 [DOI] [PubMed] [Google Scholar]
- 96.Wei Y, Aydin Z, Zhang Y, Liu Z, Guo M. 2012. A turn-on fluorescent sensor for imaging labile Fe3+ in live neuronal cells at subcellular resolution. ChemBioChem 13:1569–73 [DOI] [PubMed] [Google Scholar]
- 97.Huang ML, Lane DJ, Richardson DR. 2011. Mitochondrial mayhem: the mitochondrion as a modulator of iron metabolism and its role in disease. Antioxid. Redox Signal 15:3003–19 [DOI] [PubMed] [Google Scholar]
- 98.Urrutia PJ, Aguirre P, Tapia V, Carrasco CM, Mena NP, Núñez MT. 2017. Cell death induced by mitochondrial complex I inhibition is mediated by Iron Regulatory Protein 1. Biochim. Biophys. Acta 1863:2202–9 [DOI] [PubMed] [Google Scholar]
- 99.Li-Harms X, Milasta S, Lynch J, Wright C, Joshi A, et al. 2015. Mito-protective autophagy is impaired in erythroid cells of aged mtDNA-mutator mice. Blood 125:162–74 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Chen ML, Logan TD, Hochberg ML, Shelat SG, Yu X, et al. 2009. Erythroid dysplasia, megaloblastic anemia, and impaired lymphopoiesis arising from mitochondrial dysfunction. Blood 114:4045–53 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Anso E, Weinberg SE, Diebold LP, Thompson BJ, Malinge S, et al. 2017. The mitochondrial respiratory chain is essential for haematopoietic stem cell function. Nat. Cell Biol 19:614–25 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Elorza A, Hyde B, Mikkola HK, Collins S, Shirihai OS. 2008. UCP2 modulates cell proliferation through the MAPK/ERK pathway during erythropoiesis and has no effect on heme biosynthesis. J. Biol. Chem 283:30461–70 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Khalil S, Holy M, Grado S, Fleming R, Kurita R, et al. 2017. A specialized pathway for erythroid iron delivery through lysosomal trafficking of transferrin receptor 2. Blood Adv 1:1181–94 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Puy H, Gouya L, Deybach JC. 2010. Porphyrias. Lancet 375:924–37 [DOI] [PubMed] [Google Scholar]
- 105.Chen FP, Risheg H, Liu Y, Bloomer J. 2002. Ferrochelatase gene mutations in erythropoietic protoporphyria: focus on liver disease. Cell Mol. Biol 48:83–89 [PubMed] [Google Scholar]
- 106.Whatley SD, Ducamp S, Gouya L, Grandchamp B, Beaumont C, et al. 2008. C-terminal deletions in the ALAS2 gene lead to gain of function and cause X-linked dominant protoporphyria without anemia or iron overload. Am. J. Hum. Genet 83:408–14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Wang Y, Langer NB, Shaw GC, Yang G, Li L, et al. 2011. Abnormal mitoferrin-1 expression in patients with erythropoietic protoporphyria. Exp. Hematol 39:784–94 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Fleming MD. 2011. Congenital sideroblastic anemias: iron and heme lost in mitochondrial translation. Hematol. Am. Soc. Hematol. Educ. Prog 2011:525–31 [DOI] [PubMed] [Google Scholar]
- 109.D’Hooghe M, Selleslag D, Mortier G, Van Coster R, Vermeersch P, et al. 2012. X-linked sideroblastic anemia and ataxia: a new family with identification of a fourth ABCB7 gene mutation. Eur. J. Paediatr. Neurol 16:730–35 [DOI] [PubMed] [Google Scholar]
- 110.Pondarre C, Campagna DR, Antiochos B, Sikorski L, Mulhern H, Fleming MD. 2007. Abcb7, the gene responsible for X-linked sideroblastic anemia with ataxia, is essential for hematopoiesis. Blood 109:3567–69 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Guernsey DL, Jiang H, Campagna DR, Evans SC, Ferguson M, et al. 2009. Mutations in mitochondrial carrier family gene SLC25A38 cause nonsyndromic autosomal recessive congenital sideroblastic anemia. Nat. Genet 41:651–53 [DOI] [PubMed] [Google Scholar]
- 112.Liu G, Guo S, Anderson GJ, Camaschella C, Han B, Nie G. 2014. Heterozygous missense mutations in the GLRX5 gene cause sideroblastic anemia in a Chinese patient. Blood 124:2750–51 [DOI] [PubMed] [Google Scholar]
- 113.Cazzola M, Invernizzi R. 2011. Ring sideroblasts and sideroblastic anemias. Haematologica 96:789–92 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Mallikarjun V, Sriram A, Scialo F, Sanz A. 2014. The interplay between mitochondrial protein and iron homeostasis and its possible role in ageing. Exp. Gerontol 56:123–34 [DOI] [PubMed] [Google Scholar]
- 115.Carvajal JJ, Pook MA, dos Santos M, Doudney K, Hillermann R, et al. 1996. The Friedreich’s ataxia gene encodes a novel phosphatidylinositol-4-phosphate 5-kinase. Nat. Genet 14:157–62 [DOI] [PubMed] [Google Scholar]
- 116.Campuzano V, Montermini L, Molto MD, Pianese L, Cossee M, et al. 1996. Friedreich’s ataxia: auto-somal recessive disease caused by an intronic GAA triplet repeat expansion. Science 271:1423–27 [DOI] [PubMed] [Google Scholar]
- 117.Puccio H, Simon D, Cossée M, Criqui-Filipe P, Tiziano F, et al. 2001. Mouse models for Friedreich ataxia exhibit cardiomyopathy, sensory nerve defect and Fe-S enzyme deficiency followed by intramitochondrial iron deposits. Nat. Genet 27:181–86 [DOI] [PubMed] [Google Scholar]
- 118.Cossee M, Puccio H, Gansmuller A, Koutnikova H, Dierich A, et al. 2000. Inactivation of the Friedreich ataxia mouse gene leads to early embryonic lethality without iron accumulation. Hum. Mol. Genet 9:1219–26 [DOI] [PubMed] [Google Scholar]
- 119.Koutnikova H, Campuzano V, Foury F, Dolle P, Cazzalini O, Koenig M. 1997. Studies of human, mouse and yeast homologues indicate a mitochondrial function for frataxin. Nat. Genet 16:345–51 [DOI] [PubMed] [Google Scholar]
- 120.Navarro JA, Botella JA, Metzendorf C, Lind MI, Schneuwly S. 2015. Mitoferrin modulates iron toxicity in a Drosophila model of Friedreich’s ataxia. Free Radic. Biol. Med 85:71–82 [DOI] [PubMed] [Google Scholar]
- 121.Sawicki KT, Shang M, Wu R, Chang HC, Khechaduri A, et al. 2015. Increased heme levels in the heart lead to exacerbated ischemic injury. J. Am. Heart Assoc 4:e002272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Chang HC, Wu R, Shang M, Sato T, Chen C, et al. 2016. Reduction in mitochondrial iron alleviates cardiac damage during injury. EMBO Mol. Med 8:247–67 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Wu W, Chang S, Wu Q, Xu Z, Wang P, et al. 2016. Mitochondrial ferritin protects the murine myocardium from acute exhaustive exercise injury. Cell Death Dis 7:e2475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.White K, Lu Y, Annis S, Hale AE, Chau BN, et al. 2015. Genetic and hypoxic alterations of the microRNA-210-ISCU1/2 axis promote iron-sulfur deficiency and pulmonary hypertension. EMBO Mol. Med 7:695–713 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Cloonan SM, Glass K, Laucho-Contreras ME, Bhashyam AR, Cervo M, et al. 2016. Mitochondrial iron chelation ameliorates cigarette-smoke induced bronchitis and emphysema in mice. Nat. Med 22:163–74 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Berg D, Hochstrasser H. 2006. Iron metabolism in Parkinsonian syndromes. Mov. Disord 21:1299–310 [DOI] [PubMed] [Google Scholar]
- 127.Querfurth HW, LaFerla FM. 2010. Alzheimer’s disease. N. Engl. J. Med 362:329–44 [DOI] [PubMed] [Google Scholar]
- 128.Kozlov S, Afonin A, Evsyukov I, Bondarenko A. 2017. Alzheimer’s disease: as it was in the beginning. Rev. Neurosci 28:825–43 [DOI] [PubMed] [Google Scholar]
- 129.Lovell MA, Robertson JD, Teesdale WJ, Campbell JL, Markesbery WR. 1998. Copper, iron and zinc in Alzheimer’s disease senile plaques. J. Neurol. Sci 158:47–52 [DOI] [PubMed] [Google Scholar]
- 130.Smith MA, Harris PL, Sayre LM, Perry G. 1997. Iron accumulation in Alzheimer disease is a source of redox-generated free radicals. PNAS 94:9866–68 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Yang H, Guan H, Yang M, Liu Z, Takeuchi S, et al. 2015. Upregulation of mitochondrial ferritin by proinflammatory cytokines: implications for a role in Alzheimer’s disease. J. Alzheimers Dis 45:797–811 [DOI] [PubMed] [Google Scholar]
- 132.Forno LS. 1996. Neuropathology of Parkinson’s disease. J. Neuropathol. Exp. Neurol 55:259–72 [DOI] [PubMed] [Google Scholar]
- 133.Muñoz Y, Carrasco CM, Campos JD, Aguirre P, Núñez MT. 2016. Parkinson’s disease: the mitochondria-iron link. Parkinsons Dis 2016:7049108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Mizuno Y, Ikebe S, Hattori N, Nakagawa-Hattori Y, Mochizuki H, et al. 1995. Role of mitochondria in the etiology and pathogenesis of Parkinson’s disease. Biochim. Biophys. Acta 1271:265–74 [DOI] [PubMed] [Google Scholar]
- 135.Carroll CB, Zeissler ML, Chadborn N, Gibson K, Williams G, et al. 2011. Changes in iron-regulatory gene expression occur in human cell culture models of Parkinson’s disease. Neurochem. Int 59:73–80 [DOI] [PubMed] [Google Scholar]
- 136.Mena NP, García-Beltrán O, Lourido F, Urrutia PJ, Mena R, et al. 2015. The novel mitochondrial iron chelator 5-((methylamino)methyl)-8-hydroxyquinoline protects against mitochondrial-induced oxidative damage and neuronal death. Biochem. Biophys. Res. Commun 463:787–92 [DOI] [PubMed] [Google Scholar]
- 137.Mastroberardino PG, Hoffman EK, Horowitz MP, Betarbet R, Taylor G, et al. 2009. A novel transferrin/TfR2-mediated mitochondrial iron transport system is disrupted in Parkinson’s disease. Neurobiol. Dis 34:417–31 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Guan H, Yang H, Yang M, Yanagisawa D, Bellier JP, et al. 2017. Mitochondrial ferritin protects SH-SY5Y cells against H2O2-induced oxidative stress and modulates α-synuclein expression. Exp. Neurol 291:51–61 [DOI] [PubMed] [Google Scholar]
- 139.Matak P, Matak A, Moustafa S, Aryal DK, Benner EJ, et al. 2016. Disrupted iron homeostasis causes dopaminergic neurodegeneration in mice. PNAS 113:3428–35 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.MacDonald ME, Ambrose CM, Duyao MP, Myers RH, Lin C, et al. 1993. A novel gene containing a trinucleotide repeat that is expanded and unstable on Huntington’s disease chromosomes. Cell 72:971–83 [DOI] [PubMed] [Google Scholar]
- 141.Vonsattel JP, DiFiglia M. 1998. Huntington disease. J. Neuropathol. Exp. Neurol 57:369–84 [DOI] [PubMed] [Google Scholar]
- 142.Jansen AH, van Hal M, Op den Kelder IC, Meier RT, de Ruiter AA, et al. 2017. Frequency of nuclear mutant huntingtin inclusion formation in neurons and glia is cell-type-specific. Glia 65:50–61 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Gu M, Gash MT, Mann VM, Javoy-Agid F, Cooper JM, Schapira AH. 1996. Mitochondrial defect in Huntington’s disease caudate nucleus. Ann. Neurol 39:385–89 [DOI] [PubMed] [Google Scholar]
- 144.Agrawal S, Fox J, Thyagarajan B, Fox JH. 2018. Brain mitochondrial iron accumulates in Huntington’s disease, mediates mitochondrial dysfunction, and can be removed pharmacologically. Free Radic. Biol. Med 120:317–29 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Danielpur L, Sohn YS, Karmi O, Fogel C, Zinger A, et al. 2016. GLP-1-RA corrects mitochondrial labile iron accumulation and improves β-cell function in type 2 Wolfram Syndrome. J. Clin. Endocrinol. Metab 101:3592–99 [DOI] [PubMed] [Google Scholar]
- 146.Mochel F, Knight MA, Tong WH, Hernandez D, Ayyad K, et al. 2008. Splice mutation in the iron-sulfur cluster scaffold protein ISCU causes myopathy with exercise intolerance. Am. J. Hum. Genet 82:652–60 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Stehling O, Wilbrecht C, Lill R. 2014. Mitochondrial iron-sulfur protein biogenesis and human disease. Biochimie 100:61–77 [DOI] [PubMed] [Google Scholar]
- 148.Porter JB, Rafique R, Srichairatanakool S, Davis BA, Shah FT, et al. 2005. Recent insights into interactions of deferoxamine with cellular and plasma iron pools: implications for clinical use. Ann. N. Y. Acad. Sci 1054:155–68 [DOI] [PubMed] [Google Scholar]
- 149.Sohn YS, Breuer W, Munnich A, Cabantchik ZI. 2008. Redistribution of accumulated cell iron: a modality of chelation with therapeutic implications. Blood 111:1690–99 [DOI] [PubMed] [Google Scholar]
- 150.Strawser C, Schadt K, Hauser L, McCormick A, Wells M, et al. 2017. Pharmacological therapeutics in Friedreich ataxia: the present state. Expert Rev. Neurother 17:895–907 [DOI] [PubMed] [Google Scholar]
- 151.Kakhlon O, Breuer W, Munnich A, Cabantchik ZI. 2010. Iron redistribution as a therapeutic strategy for treating diseases of localized iron accumulation. Can. J. Physiol. Pharmacol 88:187–96 [DOI] [PubMed] [Google Scholar]
- 152.Velasco-Sánchez D, Aracil A, Montero R, Mas A, Jiménez L, et al. 2011. Combined therapy with idebenone and deferiprone in patients with Friedreich’s ataxia. Cerebellum 10 10.1007/s12311-010-0212-7 [DOI] [PubMed] [Google Scholar]
- 153.Arpa J, Sanz-Gallego I, Rodriguez-de-Rivera FJ, Dominguez-Melcón FJ, Prefasi D, et al. 2014. Triple therapy with deferiprone, idebenone and riboflavin in Friedreich’s ataxia—open-label trial. Acta Neurol. Scand 129:32–40 [DOI] [PubMed] [Google Scholar]
- 154.Prasanthi JR, Schrag M, Dasari B, Marwarha G, Dickson A, et al. 2012. Deferiprone reduces amyloid-β and tau phosphorylation levels but not reactive oxygen species generation in hippocampus of rabbits fed a cholesterol-enriched diet. J. Alzheimers Dis 30:167–82 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Martin-Bastida A, Ward RJ, Newbould R, Piccini P, Sharp D, et al. 2017. Brain iron chelation by deferiprone in a phase 2 randomised double-blinded placebo controlled clinical trial in Parkinson’s disease. Sci. Rep 7:1398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Xu P, Zhang M, Sheng R, Ma Y. 2017. Synthesis and biological evaluation of deferiprone-resveratrol hybrids as antioxidants, Aβ1–42 aggregation inhibitors and metal-chelating agents for Alzheimer’s disease. Eur. J. Med. Chem 127:174–86 [DOI] [PubMed] [Google Scholar]
- 157.García-Beltrán O, Mena NP, Aguirre P, Barriga-González G, Galdámez A, et al. 2017. Development of an iron-selective antioxidant probe with protective effects on neuronal function. PLOS ONE 12:e0189043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Reelfs O, Abbate V, Hider RC, Pourzand C. 2016. A powerful mitochondria-targeted iron chelator affords high photoprotection against solar ultraviolet A radiation. J. Investig. Dermatol 136:1692–700 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Alta RY, Vitorino HA, Goswami D, Liria CW, Wisnovsky SP, et al. 2017. Mitochondria-penetrating peptides conjugated to desferrioxamine as chelators for mitochondrial labile iron. PLOS ONE 12:e0171729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Alta RYP, Vitorino HA, Goswami D, Teresa Machini M, Esposito BP. 2017. Triphenylphosphonium-desferrioxamine as a candidate mitochondrial iron chelator. Biometals 30:709–18 [DOI] [PubMed] [Google Scholar]
- 161.Richardson DR. 2003. Friedreich’s ataxia: iron chelators that target the mitochondrion as a therapeutic strategy? Expert Opin. Investig. Drugs 12:235–45 [DOI] [PubMed] [Google Scholar]
- 162.Grillo AS, SantaMaria AM, Kafina MD, Cioffi AG, Huston NC, et al. 2017. Restored iron transport by a small molecule promotes absorption and hemoglobinization in animals. Science 356:608–16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Rademakers LH, Koningsberger JC, Sorber CW, Baart de la Faille H, Van Hattum J, Marx JJ. 1993. Accumulation of iron in erythroblasts of patients with erythropoietic protoporphyria. Eur. J. Clin. Investig 23:130–38 [DOI] [PubMed] [Google Scholar]
- 164.Yamamoto M, Arimura H, Fukushige T, Minami K, Nishizawa Y, et al. 2014. Abcb10 role in heme biosynthesis in vivo: Abcb10 knockout in mice causes anemia with protoporphyrin IX and iron accumulation. Mol. Cell. Biol 34:1077–84 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Kaneko K, Kubota Y, Nomura K, Hayashimoto H, Chida T, et al. 2018. Establishment of a cell model of X-linked sideroblastic anemia using genome editing. Exp. Hematol 65:57–68.e2 [DOI] [PubMed] [Google Scholar]
- 166.Cavadini P, Biasiotto G, Poli M, Levi S, Verardi R, et al. 2007. RNA silencing of the mitochondrial ABCB7 transporter in HeLa cells causes an iron-deficient phenotype with mitochondrial iron overload. Blood 109:3552–59 [DOI] [PubMed] [Google Scholar]
- 167.Sato T, Chang HC, Bayeva M, Shapiro JS, Ramos-Alonso L, et al. 2018. mRNA-binding protein tristetraprolin is essential for cardiac response to iron deficiency by regulating mitochondrial function. PNAS 115:E6291–300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Navarro-Sastre A, Tort F, Stehling O, Uzarska MA, Arranz JA, et al. 2011. A fatal mitochondrial disease is associated with defective NFU1 function in the maturation of a subset of mitochondrial Fe-S proteins. Am. J. Hum. Genet 89:656–67 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Alhawaj R, Patel D, Kelly MR, Sun D, Wolin MS. 2015. Heme biosynthesis modulation via δ-aminolevulinic acid administration attenuates chronic hypoxia-induced pulmonary hypertension. Am. J. Physiol. Lung Cell. Mol. Physiol 308:L719–28 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.DeMeo DL, Mariani T, Bhattacharya S, Srisuma S, Lange C, et al. 2009. Integration of genomic and genetic approaches implicates IREB2 as a COPD susceptibility gene. Am. J. Hum. Genet 85:493–502 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Chang NC, Nguyen M, Bourdon J, Risse PA, Martin J, et al. 2012. Bcl-2-associated autophagy regulator Naf-1 required for maintenance of skeletal muscle. Hum. Mol. Genet 21:2277–87 [DOI] [PubMed] [Google Scholar]
- 172.Nordin A, Larsson E, Thornell LE, Holmberg M. 2011. Tissue-specific splicing of ISCU results in a skeletal muscle phenotype in myopathy with lactic acidosis, while complete loss of ISCU results in early embryonic death in mice. Hum. Genet 129:371–78 [DOI] [PubMed] [Google Scholar]
- 173.Olsson A, Lind L, Thornell LE, Holmberg M. 2008. Myopathy with lactic acidosis is linked to chromosome 12q23.3–24.11 and caused by an intronmutation in the ISCU gene resulting in a splicing defect. Hum. Mol. Genet 17:1666–72 [DOI] [PubMed] [Google Scholar]
- 174.Saha PP, Kumar SK, Srivastava S, Sinha D, Pareek G, D’Silva P. 2014. The presence of multiple cellular defects associated with a novel G50E iron-sulfur cluster scaffold protein (ISCU) mutation leads to development of mitochondrial myopathy. J. Biol. Chem 289:10359–77 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Spiegel R, Saada A, Halvardson J, Soiferman D, Shaag A, et al. 2014. Deleterious mutation in FDX1L gene is associated with a novel mitochondrial muscle myopathy. Eur. J. Hum. Genet 22:902–6 [DOI] [PMC free article] [PubMed] [Google Scholar]