Abstract
The splicing factor RBM10 and the close homologues RBM5 and RBM6 govern the splicing of oncogenes such as Fas, NUMB and Bcl-X. The molecular architecture of these proteins includes zinc fingers (ZnF) and RNA recognition motifs (RRM). Three of these domains in RBM10, which constitute the RNA-binding part of this splicing factor, were found to individually bind RNAs with μM affinities. It was thus of interest to further investigate the structural basis of the well-documented high affinity RNA-binding by RMB10. Here, we investigated RNA-binding by combinations of two or three of these domains, and we thus discovered that a polypeptide containing RRM1, ZnF1 and RRM2 connected by their natural linkers recognizes specific sequences of the Fas exon 6 mRNA with 20 nM affinity. NMR structures of the RBM10 domains RRM1, ZnF1, and the V354del variant of RRM2 further confirmed that the interactions with RNA are driven by canonical RNA recognition elements. The well-known high-fidelity RNA splice site recognition by RBM10, and probably by RBM5 and RBM6, can thus be largely rationalized by a cooperative action of RRM and ZnF domains.
Keywords: Splicing factor, protein: RNA interactions, NMR structure determination
INTRODUCTION
The splicing factor RBM10 modulates the cellular isoform rates of multiple apoptotic genes, such as Fas1, NUMB2 and Bcl-x,3, 4 and has been linked to the onset of multiple types of cancer.5, 6 In their physiological functions, RBM5, RBM10 and the more distant homologue RBM6 mediate exon skipping as well as inclusion events, and participate in the processing of multiple oncogenes. While in some alternative splicing events the involvement of the three RBM proteins favors the same gene product, the individual splicing factors may also lead to the production of different isoforms.2 For example, in the alternative splicing of Fas, RBM5 and RBM10 behave similarly, by promoting exon 6 skipping and thus increasing the yield of an antiapoptotic isoform. In contrast, in the processing of NUMB, RBM10 induces exon 9 inclusion, while RBM5 increases skipping.1, 2, 4, 7–9
RBM10, RBM5 and RBM6 share similar domain architectures, which include an N-terminal RNA recognition region consisting of a zinc finger (ZnF1) flanked by two RRMs (RRM1 and RRM2),10–12 and a C-terminal protein-interacting region containing a second zinc finger (ZnF2), an OCRE sequence motif13, 14 and a G-patch motif (Fig. 1A). The RNA recognition domains in RBM5 and RBM10 have about 60% sequence identity, recognize similar splice sites, and together with the OCRE domain15–17 govern Fas isoform ratios. In this manuscript we report on recognition of the exon 6 in Fas by cooperative action of the RRM1, ZnF1 and RRM2 domains of RBM10, and present NMR structures of the individual RBM10 RNA-binding domains.
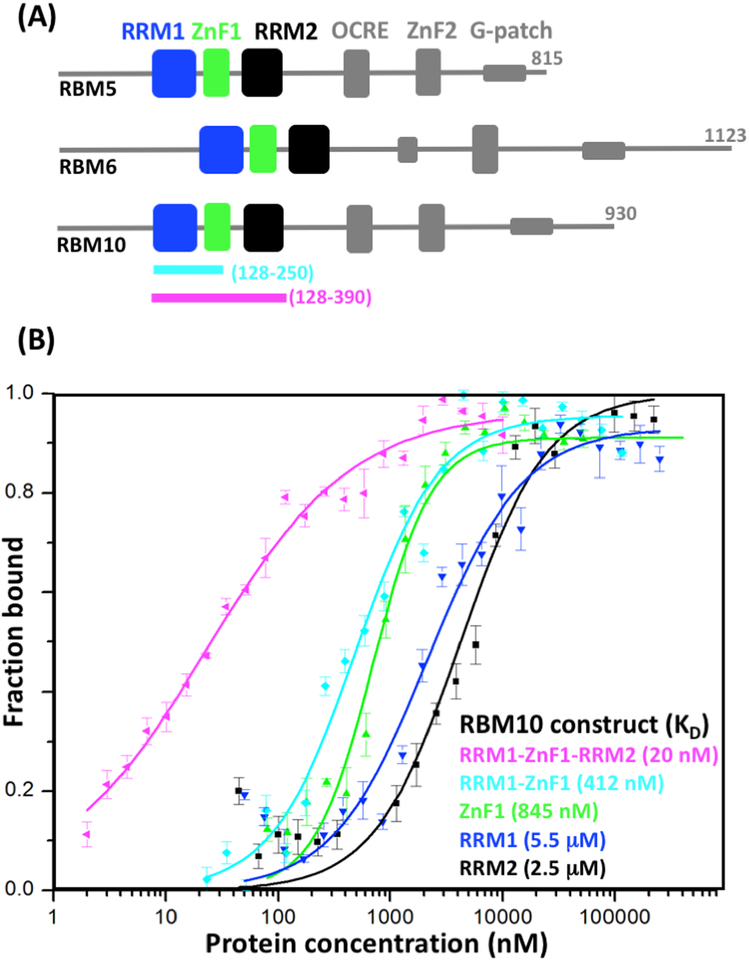
Figure 1. RBM5/6/10 domain architectures and recognition of exon 6 from Fas by RBM10 fragments.
(A) Schematic representation of the domain architectures of the three proteins. The three globular domains studied here are highlighted in color. At the bottom, the RRM1–ZnF1 and RRM1–ZnF1–RRM2 polypeptide fragments of RBM10 used for RNA binding assays are indicated by thick colored lines and indication of the chain ends. (B) Protein–RNA association curves with a 22-nucleotide RNA sequence from exon 6 of Fas, UAAUUGUUUGGGGUAAGUUCUU. The data were obtained using nitrocellulose binding assays with the RBM10 polypeptide fragments indicated in the figure. The same color code is used as in panel (A). Average values from three independent experiments are shown as geometric symbols, and the standard deviations are represented as vertical bars. KD values obtained with Hill-equation fitting are indicated in the lower right.
RESULTS
The biological functions of RBM5, RMB6 and RBM10 indicate that these splicing factors bind to mRNA with high affinity.2, 4, 7, 9, 17–19 Here, we set out to investigate the structural basis of these implicated interactions. Working with RBM10, we measured the binding affinities for RNA fragments from exon 6 of Fas of the individual RRMs, the zinc finger, and of combinations of two and three of these globular domains. We also determined NMR structures of the individual domains, which may support future studies of the general mechanisms of action by this class of splicing factors.
Multidomain RNA recognition by the splicing factor RBM10
In order to investigate the impact of synergies between multiple RNA binding domains in RBM10 during Fas recognition, we prepared RBM polypeptide fragments of variable lengths and evaluated their affinities for the 22-nucleotide sequence UAAUUGUUUGGGGUAAGUUCUU found in exon 6 of Fas (Fig. 1B). High affinity was observed for a three-domain construct containing RRM1, ZnF1 and RRM2 connected by the natural linkers in RBM10 (RRM1–ZnF1–RRM2), with KD = 20 nM, as compared to 2.5 μM and 5.5 μM, respectively, for the individual domains RRM1 and RRM2, and 845 nM for ZnF1 (Fig. 1B). The KD value for ZnF1 is similar to the affinites observed for other members of the ZRANB2 family.11, 20 A construct of residues 128–250 containing RRM1 and ZnF1 (RRM1–ZnF1) was found to bind with intermediate affinity (KD = 412 nM; Fig. 1B). Overall, the data of Fig. 1B show that synergies among three RBM10 domains can afford high-affinity recognition of a sequence Fas mRNA.
NMR Structures of the RBM10 domains RRM1, ZnF1 and RRM2[V354del].
For the NMR structure determinations we followed the J-UNIO protocol with non-uniform sampling of the 3D heteronuclear-resolved [1H,1H]-NOESY data sets21–23, high-quality structures were thus obtained, as indicated by the statistics presented in Table 1.
Table 1.
Input for the structure calculations and characterization of bundles of 20 energy-minimized CYANA conformers representing the NMR structures of the RBM10 domains RRM1, ZnF1 and RRM2[V354del].
| Quantitya | RRM1 | ZnF1 | RRM2[V354del] |
| NOE upper distance limits | 1960 | 420 | 1693 |
| Intraresidual | 457 | 135 | 442 |
| Short range | 540 | 109 | 556 |
| Medium range | 376 | 77 | 278 |
| Long range | 587 | 99 | 417 |
| Dihedral angle constraints | 385 | 179 | 336 |
| Residual target function value (Å2) | 1.66 ± 0.30 |
0.34 ± 0.05 | 1.44 ± 0.24 |
| Residual NOE violations | |||
| Number ≥ 0.1 Å | 5 ± 2 |
1 ± 1 |
6 ± 1 |
| Maximum (Å) | 0.14 | 0.12 | 0.13 |
| Residual dihedral angle violations | |||
| Number ≥ 2.5° | 1 ± 1 |
1 ± 1 |
0 ± 0 |
| Maximum (°) | 1.56 |
1.78 | 0.87 |
| Amber energies (kcal/mol) | |||
| Total | -3462 ± 105 |
-1117 ± 87 |
- 3801 ± 57 |
| Van der Waals | -279 ± 18 |
-208 ± 15 |
- 223 ± 18 |
| Electrostatic | - 3994 ± 92 |
- 1519 ± 112 |
- 3291 ± 68 |
| RMSD from ideal geometry | |||
| Bond lengths (Å) | 0.0091 |
0.0081 6 |
0.0078 |
| Bond angles (°) |
1.48 |
1.31 |
1.44 |
| RMSD to the mean co-ordinatesb (Å)b | |||
| bb | 0.46 ± 0.06 | 0.41 ± 0.07 | 0.68 ± 0.08 |
| ha | 0.93 ± 0.09 | 1.01 ± 0.12 | 1.04 ± 0.09 |
| Ramachandran plot statistics (%)c c | |||
| Most favoured regions | 83.5 | 85.3 | 74.5 |
| Additional allowed regions | 13.9 | 13.3 | 21.3 |
| Generously allowed regions | 2.6 | 0.9 | 3.1 |
| Disallowed regions | 0 | 0.5 | 1.1 |
Except for the top six entries, which describe the input generated in the final cycle of the ATNOS/CANDID/CYANA calculation,36–38 the last entries refer to the 20 best CYANA conformers after energy minimization with OPALp (see text). Where applicable, the average value for the bundle of 20 conformers and the standard deviation are given.
bb indicates the backbone atoms N, Cα, and C’; ha stands for all heavy atoms.
As determined by PROCHECK.
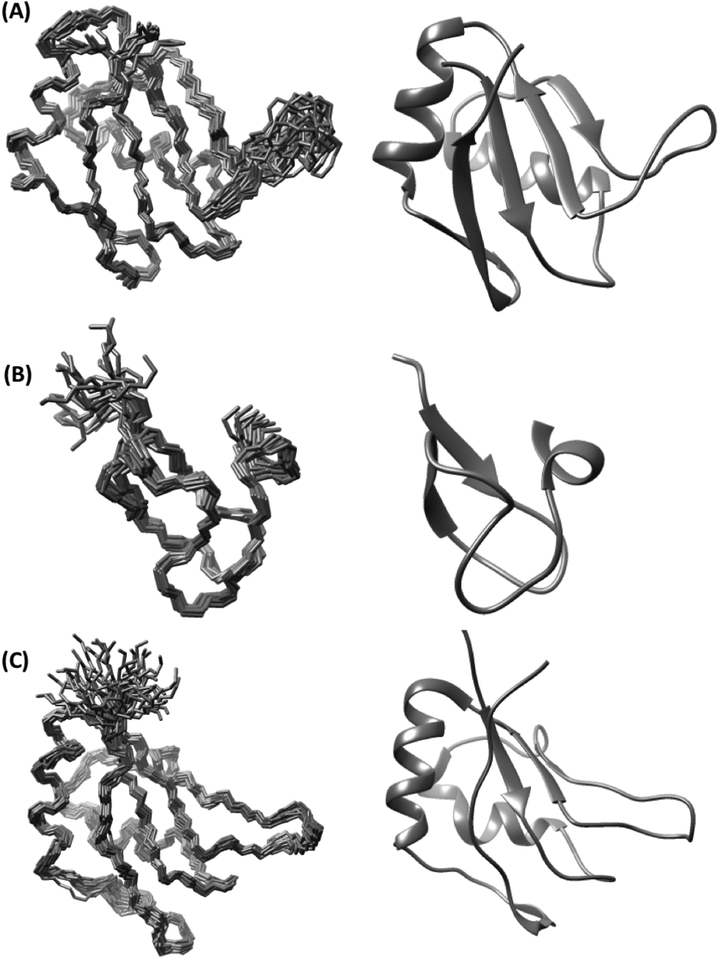
RRM1 shows a variation of the canonical RRM architecture, with a four-stranded antiparallel β-sheet and two α-helices (Fig. 2A).24, 25 The helices α1 and α2 contain the residues 141–153 and 181–191, and β-strands are formed by the polypeptide segments 128–132 (β1), 157–162 (β2), 174–178 (β3), 195–198 (β3’) and 201–205 (β4). The unique feature of RRM1 is that the linker between α2 and β4 forms an additional strand, β3’, which is not part of the canonical RRM β-sheet.10, 24, 25
Figure 2. NMR structures of RBM10 domains.
The RBM10 domains RRM1(A), ZnF1(B) and RRM2[V354del] (C) are represented by bundles of 20 conformers (left) and ribbon presentations of the conformer closest to the mean coordinates (right). The chain ends are identified by N and C.
The ZnF1 domain adopts a ZRANB2-like structure (Fig. 2B)11, 20 containing an α-turn with residues 240–244 and four short β-strands of residues 315–318, 324–328, 336–338 and 340–344. Zn2+ is coordinated by four cysteines in positions 319, 322, 333 and 336, which are located within or near the four β-strands.
The RRM2[V354del] structure contains the helices α1 and α2 with residues 310–324 and 351–359, and the four β-strands are formed by the polypeptide segments 301–305 (β1), 329–334 (β2), 344–349 (β3) and 378– 381 (β4) (Fig. 2C). RRM2[V254del] thus adopts a canonical RRM structure, and therefore it did not come as a surprise that this deletion did not have a significant effect on RNA binding (Fig. 3). In view of the near-identity of these domain structures with those for which detailed studies of RNA-complexation have been reported10,24,25, we hypothesize that they also have similar patterns of contacts with RNAs.
Figure 3. Association curves of RRM2 and RRM2[V354del] with the 22-nucleotide RNA fragment UAAUUGUUUGGGGUAAGUUCUU from exon 6 of Fas.
Same measurement details and presentation as in Fig. 1(B).
DISCUSSION
The function of RBM10 in the regulation of Fas alternative splicing has been shown to be based on both, specific recognition of oligonucleotide motifs in the mRNA and interactions with supplementary splicing factors1,10. While the C-terminal region of RBM10 is involved in protein recruitment, the N-terminal region with the three RNA binding domains RRM1, ZnF1 and RRM2 ensures recognition and binding of specific splice sites on the mRNA, which is the focus of the present work. Previous studies with tandem RRM constructs showed enhanced affinities when compared with respect to the individual RRMs.24–29 RBM10 now provided an opportunity to investigate possible cooperativity between RRMs and a zinc-finger domain, which exhibits a different structure and a different mode of RNA recognition. Introduction of non-RRM domains into splicing factors obviously carries the promise to diversify RNA recognition, which has found extensive use in nature (Fig. 1A)
Individually, the RRMs and ZnF1 in RBM10 have nearly identical structural properties to corresponding domains in other splicing factors,10, 12, 24–26, 30–33 with the sole exception that RBM10-RRM1 includes an additional, non-canonical regular secondary structure element, β3’ (Fig. 2A). We now showed that recognition of exon 6 in Fas by the combination of the three RNA binding domains of RBM10 results in low nM binding affinities, although the individual domains have weak affinities for RNA binding (Fig. 1B).
Complementarity of the biological functions of the splicing factors RBM5 and RBM10 has previously been extensively investigated.2, 4, 7, 17–19, 34, 35 The high sequence homology and the close similarity of the three-dimensional structures of their RNA binding domains, in as far as they are available,11, 12 suggest a homologous RNA recognition mode, which coincides with evidence that the two splicing factors may target closely related mRNA motifs.2, 32, 35
Acknowledgements
Part of this work was supported by the Joint Center for Structural Genomics (JCSG; www.jcsg.org, grant number U54 GM094586).
Abbreviations:
- APSY
automated projection spectroscopy
- ASCAN
software for automated side-chain resonance assignment
- ATNOS
software for automated NMR peak picking
- CANDID
software for automated NOE assignment
- CYANA
software for NMR structure calculation
- EDTA
ethylenediaminetetraacetic acid
- HSQC
heteronuclear single-quantum coherence spectroscopy
- J-UNIO
protocol for automated determination of NMR structures of proteins
- MATCH
software used for backbone NMR chemical shift assignments
- NOE
nuclear Overhauser effect
- NOESY
nuclear Overhauser effect spectroscopy
- PDB
protein data bank
- RMSD
root-mean-square deviation
- TEV
tobacco etch virus
References
- [1].Bonnal S, Martinez C, Forch P, Bachi A, Wilm M, and Valcarcel J (2008) RBM5/Luca-15/H37 regulates Fas alternative splice site pairing after exon definition. Mol Cell 32, 81–95. [DOI] [PubMed] [Google Scholar]
- [2].Bechara EG, Sebestyen E, Bernardis I, Eyras E, and Valcarcel J (2013) RBM5, 6, and 10 differentially regulate NUMB alternative splicing to control cancer cell proliferation. Mol Cell 52, 720–733. [DOI] [PubMed] [Google Scholar]
- [3].Bielli P, Bordi M, Di Biasio V, and Sette C (2014) Regulation of BCL-X splicing reveals a role for the polypyrimidine tract binding protein (PTBP1/hnRNP I) in alternative 5’ splice site selection. Nucleic Acids Res 42, 12070–12081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Inoue A, Yamamoto N, Kimura M, Nishio K, Yamane H, and Nakajima K (2014) RBM10 regulates alternative splicing. FEBS Lett 588, 942–947. [DOI] [PubMed] [Google Scholar]
- [5].Rintala-Maki ND, and Sutherland LC (2004) LUCA-15/RBM5, a putative tumor suppressor, enhances multiple receptor-initiated death signals. Apoptosis 9, 475–484. [DOI] [PubMed] [Google Scholar]
- [6].Imielinski M, Berger AH, Hammerman PS, Hernandez B, Pugh TJ, Hodis E, Cho J, Suh J, Capelletti M, Sivachenko A, Sougnez C, Auclair D, Lawrence MS, Stojanov P, Cibulskis K, Choi K, de Waal L, Sharifnia T, Brooks A, Greulich H, Banerji S, Zander T, Seidel D, Leenders F, Ansen S, Ludwig C, Engel-Riedel W, Stoelben E, Wolf J, Goparju C, Thompson K, Winckler W, Kwiatkowski D, Johnson BE, Janne PA, Miller VA, Pao W, Travis WD, Pass HI, Gabriel SB, Lander ES, Thomas RK, Garraway LA, Getz G, and Meyerson M (2012) Mapping the hallmarks of lung adenocarcinoma with massively parallel sequencing. Cell 150, 1107–1120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Loiselle JJ, and Sutherland LC (2014) Differential downregulation of Rbm5 and Rbm10 during skeletal and cardiac differentiation. In Vitro Cell Dev Biol Anim 50, 331–339. [DOI] [PubMed] [Google Scholar]
- [8].Ray D, Kazan H, Cook KB, Weirauch MT, Najafabadi HS, Li X, Gueroussov S, Albu M, Zheng H, Yang A, Na H, Irimia M, Matzat LH, Dale RK, Smith SA, Yarosh CA, Kelly SM, Nabet B, Mecenas D, Li W, Laishram RS, Qiao M, Lipshitz HD, Piano F, Corbett AH, Carstens RP, Frey BJ, Anderson RA, Lynch KW, Penalva LO, Lei EP, Fraser AG, Blencowe BJ, Morris QD, and Hughes TR (2013) A compendium of RNA-binding motifs for decoding gene regulation. Nature 499, 172–177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Wang Y, Gogol-Doring A, Hu H, Frohler S, Ma Y, Jens M, Maaskola J, Murakawa Y, Quedenau C, Landthaler M, Kalscheuer V, Wieczorek D, Wang Y, Hu Y, and Chen W (2013) Integrative analysis revealed the molecular mechanism underlying RBM10-mediated splicing regulation. EMBO Mol Med 5, 1431–1442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Maris C, Dominguez C, and Allain FH (2005) The RNA recognition motif, a plastic RNA-binding platform to regulate post-transcriptional gene expression. FEBS J 272, 2118–2131. [DOI] [PubMed] [Google Scholar]
- [11].Nguyen CD, Mansfield RE, Leung W, Vaz PM, Loughlin FE, Grant RP, and Mackay JP (2011) Characterization of a family of RanBP2-type zinc fingers that can recognize single-stranded RNA. J Mol Biol 407, 273–283. [DOI] [PubMed] [Google Scholar]
- [12].Song Z, Wu P, Ji P, Zhang J, Gong Q, Wu J, and Shi Y (2012) Solution structure of the second RRM domain of RBM5 and its unusual binding characters for different RNA targets. Biochemistry 51, 6667–6678. [DOI] [PubMed] [Google Scholar]
- [13].Callebaut I, and Mornon JP (2005) OCRE: a novel domain made of imperfect, aromatic-rich octamer repeats. Bioinformatics 21, 699–702. [DOI] [PubMed] [Google Scholar]
- [14].Inoue A, Takahashi K, Kimura M, Watanabe T, and Morisawa S (1996) Molecular Cloning of a RNA Binding Protein, S1–1. Nucleic Acids Res 24, 2990–2997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Mourão A, Bonnal S, Komal S, Warner L, Bordonné R, Valcárcel J, and Sattler M (2016) Structural basis for the recognition of spliceosomal SmN/B/B’ proteins by the RBM5 OCRE domain in splicing regulation. eLIFE eLife 2016;5:e14707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Martin BT, Serrano P, Geralt M, and Wüthrich K (2016) Nuclear Magnetic Resonance Structure of a Novel Globular Domain in RBM10 Containing OCRE, the Octamer Repeat Sequence Motif. Structure 24, 158–164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Xiao SJ, Wang LY, Kimura M, Kojima H, Kunimoto H, Nishiumi F, Yamamoto N, Nishio K, Fujimoto S, Kato T, Kitagawa S, Yamane H, Nakajima K, and Inoue A (2013) S1–1/RBM10: multiplicity and cooperativity of nuclear localisation domains. Biol Cell 105, 162–174. [DOI] [PubMed] [Google Scholar]
- [18].Hernández J, Bechara E, Schlessinger D, Delgado J, Serrano L, and Valcárcel J (2016) Tumor suppressor properties of the splicing regulatory factor RBM10. RNA Biology 13, 466–672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Tessier S, Loiselle J, McBain A, Pullen C, Koenderink B, Roy J, and Sutherland L (2015) Insight into the role of alternative splicing within the RBM10v1 exon 10 tandem donor site. BMC Research Notes 8, DOI: 10.1186/s13104-13015-10983-13105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Loughlin FE, Mansfield RE, Vaz PM, McGrath AP, Setiyaputra S, Gamsjaeger R, Chen ES, Morris BJ, Guss JM, and Mackay JP (2009) The zinc fingers of the SR-like protein ZRANB2 are single-stranded RNA-binding domains that recognize 5’ splice site-like sequences. Proc Natl Acad Sci U S A 106, 5581–5586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Serrano P, Pedrini B, Mohanty B, Geralt M, Herrmann T, and Wüthrich K (2012) The J-UNIO protocol for automated protein structure determination by NMR in solution. J Biomol NMR 53, 341–354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Didenko T, Proudfoot A, Dutta SK, Serrano P, and Wüthrich K (2015) Non-Uniform Sampling and J-UNIO Automation for Efficient Protein NMR Structure Determination. Chemistry 21, 12363–12369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Dutta SK, Serrano P, Proudfoot A, Geralt M, Pedrini B, Herrmann T, and Wüthrich K (2015) APSY-NMR for protein backbone assignment in high-throughput structural biology. J Biomol NMR 61, 47–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Daubner GM, Clery A, and Allain FHT (2013) RRM-RNA recognition: NMR or crystallography…and new findings. Curr Opin Struct Biol 23, 100–108. [DOI] [PubMed] [Google Scholar]
- [25].Cléry A, Blatter M, and Allain FHT (2008) RNA recognition motifs: boring? Not quite. Curr Opin Struc Biol 18, 290–298. [DOI] [PubMed] [Google Scholar]
- [26].Wang H, Zeng F, Liu Q, Liu H, Liu Z, Niu L, Teng M, and Li X (2013) The structure of the ARE-binding domains of Hu antigen R (HuR) undergoes conformational changes during RNA binding. Acta Crystallogr D Biol Crystallogr 69, 373–380. [DOI] [PubMed] [Google Scholar]
- [27].de Mollerat XJ (2003) A genomic rearrangement resulting in a tandem duplication is associated with split hand-split foot malformation 3 (SHFM3) at 10q24. Hum Mol Genet 12, 1959–1971. [DOI] [PubMed] [Google Scholar]
- [28].Conte M, Grüne T, Ghuman J, Kelly G, Ladas A, Matthews S, and Curry S (2000) Structure of tandem RNA recognition motifs from polypyrimidine tract binding protein reveals novel features of the RRM fold. EMBO J 19, 3132–3141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Dominguez C, Schubert M, Duss O, Ravindranathan S, and Allain FHT (2011) Structure determination and dynamics of protein-RNA complexes by NMR spectroscopy. Prog Nucl Magn Reson Spectrosc 58, 1–61. [DOI] [PubMed] [Google Scholar]
- [30].Wang I, Hennig J, Jagtap PK, Sonntag M, Valcarcel J, and Sattler M (2014) Structure, dynamics and RNA binding of the multi-domain splicing factor TIA-1. Nucleic Acids Res 42, 5949–5966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Tripsianes K, Friberg A, Barrandon C, Brooks M, van Tilbeurgh H, Seraphin B, and Sattler M (2014) A novel protein-protein interaction in the RES (REtention and Splicing) complex. J Biol Chem 289, 28640–28650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Sutherland LC, Rintala-Maki ND, White RD, and Morin CD (2005) RNA binding motif (RBM) proteins: a novel family of apoptosis modulators? J Cell Biochem 94, 5–24. [DOI] [PubMed] [Google Scholar]
- [33].Kenan DJ, Query CC, and Keene JD (1991) RNA recognition: towards identifying determinants of specificity. Trends Biochem Sci 16, 214–220. [DOI] [PubMed] [Google Scholar]
- [34].Wang K, Bacon ML, Tessier JJ, Rintala-Maki ND, Tang V, and Sutherland LC (2012) RBM10 Modulates Apoptosis and Influences TNF-alpha Gene Expression. J Cell Death 5, 1–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Sutherland LC, Wang K, and Robinson AG (2010) RBM5 as a putative tumor suppressor gene for lung cancer. J Thorac Oncol 5, 294–298. [DOI] [PubMed] [Google Scholar]
- [36].Herrmann T, Güntert P, and Wüthrich K (2002) Protein NMR structure determination with automated NOE-identification in the NOESY spectra using the new software ATNOS. J Biomol NMR 24, 171–189. [DOI] [PubMed] [Google Scholar]
- [37].Herrmann T, Güntert P, and Wüthrich K (2002) Protein NMR structure determination with automated NOE assignment using the new software CANDID and the torsion angle dynamics algorithm DYANA. J Mol Biol 319, 209–227. [DOI] [PubMed] [Google Scholar]
- [38].Güntert P, Mumenthaler C, and Wüthrich K (1997) Torsion angle dynamics for NMR structure calculation with the new program DYANA. J Mol Biol 273, 283–298. [DOI] [PubMed] [Google Scholar]