Abstract
Mutations in the Isocitrate Dehydrogenase 1 (IDH1) gene occur in 70% of grade II and grade III gliomas, 10% of acute myeloid leukemia, as well as cholangiocarcinomas, melanomas, and chondrosarcomas. Numerous mechanisms have been proposed to illustrate the biological function of mutant IDH1. Most functional studies of mutant IDH1 have been conducted in exogenous overexpression systems with the IDH1 wild type background. This mini-review comments on recent publication by Wei et al, in which a highly efficient “single base editing” approach was employed to generate monoallelic IDH1 R132H mutation without the induction of a double strand break in the IDH1 gene.
Keywords: Glioma, Heterozygous IDH1 R132H Mutation, Single Base Editing, Yes-Associated Protein (YAP)
3. Introduction
Gliomas are the most prevalent type of tumors of the central nervous systems, accounting for up to 30% of all primary lesions and nearly 80% of all malignant forms [1,2]. Given their anatomical localization and locally infiltrative nature, these tumors are associated with high morbidity and mortality. Despite radical surgical resection coupled with chemo- and radiotherapy, these tumors often recur, leading to a dismal overall prognosis. With an approximate incident rate of 6.6 per 100,000 individuals annually in the USA, these malignancies result in a majority of deaths from primary brain tumors. Historically, gliomas have been classified based on their histological features and graded by their degree of anaplasia according to WHO criteria, serving as a “gold standard” for decades. However, in the case of low grade gliomas and particularly diffusely infiltrative gliomas, these methods are subject to intra observer variability. Thus, with the advent of molecular profiling, these tumors have now been further interrogated to identify diagnostically relevant alterations, including genomic, transcriptomic, and epigenetic variants, complementing the histological-based classification system [3–5].
3.1. IDH1 Mutation in Glioma
The recent identification of frequent mutations in the metabolic gene Isocitrate Dehydrogenase (IDH) 1 and 2 suggests the existence of different molecular subclasses of diffusely infiltrative gliomas with distinct biological and clinical attributes, prompting the WHO to propose revised classification guidelines [6]. Originally discovered in 2008 [7], it is now appreciated that 70–80% of grade II/III and 20% grade IV gliomas harbor mutations in IDH1; and that these alterations frequently coexist with TP53, ATRX mutations, and co-deletions of chromosome 1p and 19q [8,9]. Prior studies have identified mutations in IDH1 as one of the earliest events in gliomagenesis, possibly playing a significant role during tumor initiation and subsequent transformation [9–11]. The majority of IDH1 mutants contain heterozygous single amino acid missense mutation in its active site at arginine 132, altering its enzymatic activity that results in the neomorphic production of the oncometabolite 2-Hydroxyglutarate (2-HG) using α-ketoglutarate (α-KG) [12]. This aberrant production of 2-HG in turn inhibits α-KG-dependent dioxygenases, including histone demethylases and DNA demethylase Ten-Eleven Translocation 2 (TET 2) [13–15]. Consequently, IDH mutation is associated with global changes in DNA and histone methylation patterns as indicated by widespread hypermethylation of CpG islands [16]. Clinically, mutations in IDH1 prolong survival of glioma patients [8]. Given the pronounced frequency of IDH1 mutation in gliomas coupled with its impact on the biology and clinical progression of the disease, it is vital to further delineate the role of monoallelic IDH1 point mutations in gliomas.
3.2. Current Models for Mutant IDH1
While previous studies have investigated the biological function of mutant IDH1 in the context of tumorigenesis and tumor progression, these studies are often limited by the paucity of appropriate endogenous mutant IDH1 systems [17,18]. For instance, most prior studies have relied on the use of overexpression systems, which do not necessarily recapitulate the naturally occurring heterozygous IDH1 mutational status in this cancer [17]. Moreover, the underlying wild type IDH1 background in these exogenously overexpressing IDH1 mutant clonal cells may obscure the true biological and clinical impact of IDH1 mutation in this cancer. Although techniques to establish primary cultures carrying monoallelic IDH1 mutants from human tumor samples has been improved, it remains difficult to generate isogenic cellular models to study the function of mutant IDH1, especially during tumorigenesis [19]. Likewise, while orthotopic xenograft models are available, their utility is often limited [20]. Thus, it is important to establish clinically relevant cellular models that recapitulate the parental disease to methodically characterize the role of IDH1 mutation in this cancer. Such clinically representative in vitro disease models will enable systematic delineation of the molecular network driven by mutated IDH1, a prerequisite for effective therapeutic design.
3.3. An Efficient Approach to Create Heterozygous IDH1 R132H Mutation
To this end, we recently demonstrated the use of “Single base editing” method to generate isogenic cellular models carrying monoallelic IDH1 mutants [21]. Using a recently reported CRISPR-Cas9 technology which functions without the induction of a double strand break in IDH1 [22], we precisely introduced heterozygous IDH1 R132H point mutation in human astroglial cells with a successful rate of 20%. Compared with other nuclease and homology directed repair-based knock-in methods used to date [23–25], our work provides an efficient and easy approach to generate monoallelic IDH1 R132H mutation, and can be valuable to others in the field searching for models of endogenous heterozygous IDH1 mutation.
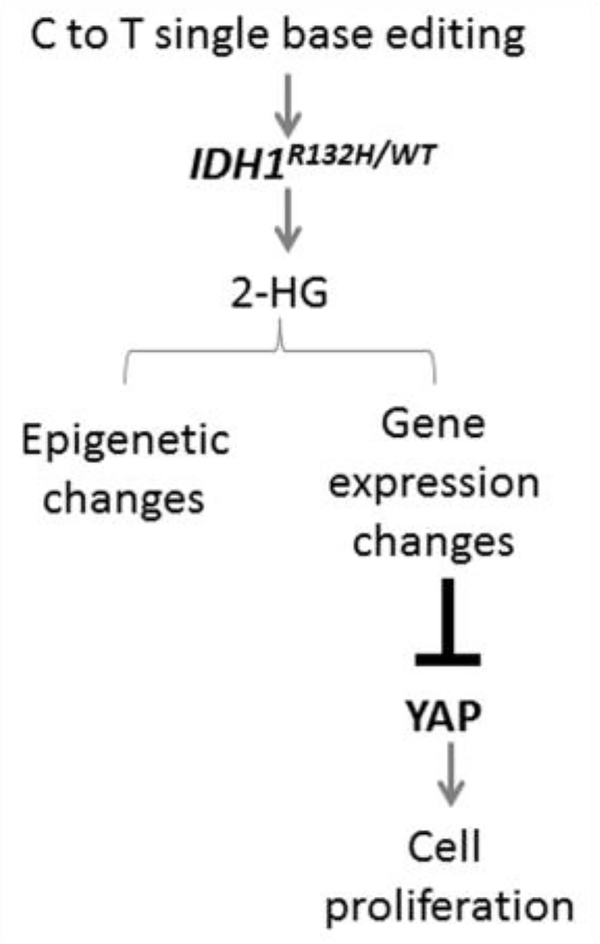
The monoallelic IDH1 mutants in our model displayed global alterations in DNA methylation and gene expression pattern coupled with dramatic changes in cellular behavior including decreased cell proliferation. Notably, we uncovered a previously unknown link between expression of YAP, an effector of the pro-growth Hippo pathway, and IDH1 mutation status (Figure 1). Specifically, our work revealed a Hippo-independent, 2-HG-dependent regulation of YAP expression in these monoallelic IDH1 mutant astroglial clones. The Hippo-YAP pathway has emerged as a critical network driving tumor growth and progression [26–28]. Thus, it is of interest to identify potent regulators of YAP and their role in cancer development. Our study suggests that YAP is responsive to changes in metabolic state, highlighting the intimate relationship between proto-oncogenes and cellular metabolism. While further mechanistic investigation is warranted to precisely elucidate the biological implication of YAP inhibition by IDH1 mutation, this study lays the groundwork in establishing a novel connection between oncometabolite production and activity of pro-growth signaling network in early disease development. Overall, this versatile and efficient “Single base gene editing” technique will permit thorough interrogation of the biological function of heterozygous IDH1 mutants in the context of glioma development and progression, and serve as a valuable model to test effective therapies for the management and treatment of gliomas.
Figure 1:
Schematic of the mechanistic details and functional effects of mutant IDH1-YAP signaling.
4.1.
Funding: Dr. Xia is supported by NIH R01NS091165. AQH was supported by the Mayo Clinic Professorship and a Clinician Investigator award as well as the NIH (R43CA221490, R01CA200399, R01CA183827, R01CA195503, R01CA216855).
Footnotes
Compliance with Ethical Standards
Conflict of Interest: The authors declare no conflict of interest.
Ethical approval: This article does not contain any studies with human participants or animals performed by any of the authors.
References
- 1.Dolecek TA, Propp JM, Stroup NE, Kruchko C (2012) CBTRUS statistical report: primary brain and central nervous system tumors diagnosed in the United States in 2005–2009. Neuro Oncol 14: v1–v49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ostrom QT, Gittleman H, Fulop J, Liu M, Blanda R, et al. (2015) CBTRUS Statistical Report: Primary Brain and Central Nervous System Tumors Diagnosed in the United States in 2008–2012. Neuro Oncol 17: iv1–iv62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Noushmehr H, Weisenberger DJ, Diefes K, Phillips HS, Pujara K, et al. (2010) Identification of a CpG island methylator phenotype that defines a distinct subgroup of glioma. Cancer Cell 17: 510–522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Phillips HS, Kharbanda S, Chen R, Forrest WF, Soriano RH, et al. (2006) Molecular subclasses of high-grade glioma predict prognosis, delineate a pattern of disease progression, and resemble stages in neurogenesis. Cancer Cell 9: 157–173. [DOI] [PubMed] [Google Scholar]
- 5.Verhaak RG, Hoadley KA, Purdom E, Wang V, Qi Y, et al. (2010) Integrated genomic analysis identifies clinically relevant subtypes of glioblastoma characterized by abnormalities in PDGFRA, IDH1, EGFR, and NF1. Cancer Cell 17: 98–110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hartmann C, Meyer J, Balss J, Capper D, Mueller W, et al. (2009) Type and frequency of IDH1 and IDH2 mutations are related to astrocytic and oligodendroglial differentiation and age: a study of 1,010 diffuse gliomas. Acta Neuropathol 118: 469–474. [DOI] [PubMed] [Google Scholar]
- 7.Parsons DW, Jones S, Zhang X, Lin JC, Leary RJ, et al. (2008) An integrated genomic analysis of human glioblastoma multiforme. Science 321: 1807–1812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yan H, Parsons DW, Jin G, McLendon R, Rasheed BA, et al. (2009) IDH1 and IDH2 mutations in gliomas. N Engl J Med 360: 765–773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Labussière M, Idbaih A, Wang XW, Marie Y, Boisselier B, et al. (2010) All the 1p19q codeleted gliomas are mutated on IDH1 or IDH2. Neurology 74: 1886–1890. [DOI] [PubMed] [Google Scholar]
- 10.Koivunen P, Lee S, Duncan CG, Lopez G, Lu G, et al. (2012) Transformation by the (R)-enantiomer of 2-hydroxyglutarate linked to EGLN activation. Nature 483: 484–488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Watanabe T, Nobusawa S, Kleihues P, Ohgaki H (2009) IDH1 mutations are early events in the development of astrocytomas and oligodendrogliomas. Am J Pathol 174: 1149–1153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dang L, White DW, Gross S, Bennett BD, Bittinger MA, et al. (2009) Cancer-associated IDH1 mutations produce 2-hydroxyglutarate. Nature 462: 739–744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Figueroa ME, Abdel-Wahab O, Lu C, Ward PS, Patel J, et al. (2010) Leukemic IDH1 and IDH2 mutations result in a hypermethylation phenotype, disrupt TET2 function, and impair hematopoietic differentiation. Cancer Cell 18: 553–567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lu C, Ward PS, Kapoor GS, Rohle D, Turcan S, et al. (2012) IDH mutation impairs histone demethylation and results in a block to cell differentiation. Nature 483: 474–478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Xu W, Yang H, Liu Y, Yang Y, Wang P, et al. (2011) Oncometabolite 2-hydroxyglutarate is a competitive inhibitor of alpha-ketoglutarate-dependent dioxygenases. Cancer Cell 19: 17–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Turcan S, Rohle D, Goenka A, Walsh LA, Fang F, et al. (2012) IDH1 mutation is sufficient to establish the glioma hypermethylator phenotype. Nature 483: 479–483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bralten LB, Kloosterhof NK, Balvers R, Sacchetti A, Lapre L, et al. (2011) IDH1 R132H decreases proliferation of glioma cell lines in vitro and in vivo. Ann Neurol 69: 455–463. [DOI] [PubMed] [Google Scholar]
- 18.Jin G, Pirozzi CJ, Chen LH, Lopez GY, Duncan CG, et al. (2012) Mutant IDH1 is required for IDH1 mutated tumor cell growth. Oncotarget 3: 774–782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Piaskowski S, Bienkowski M, Stoczynska-Fidelus E, Stawski R, Sieruta M, et al. (2011) Glioma cells showing IDH1 mutation cannot be propagated in standard cell culture conditions. Br J Cancer 104: 968–970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Luchman HA, Stechishin OD, Dang NH, Blough MD, Chesnelong C, et al. (2012) An in vivo patient-derived model of endogenous IDH1-mutant glioma. Neuro Oncol 14: 184–191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wei S, Wang J, Oyinlade O, Ma D, Wang S, et al. (2018) Heterozygous IDH1 (R132H/WT) created by “single base editing” inhibits human astroglial cell growth by downregulating YAP. Oncogene. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Komor AC, Kim YB, Packer MS, Zuris JA, Liu DR (2016) Programmable editing of a target base in genomic DNA without double-stranded DNA cleavage. Nature 533: p420–p424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Duncan CG, Barwick BG, Jin G, Rago C, Kapoor-Vazirani P, et al. (2012) A heterozygous IDH1R132H/WT mutation induces genome-wide alterations in DNA methylation. Genome Res 22: 2339–2355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ma S, Jiang B, Deng W, Gu ZK, Wu FZ, et al. (2015) D-2-hydroxyglutarate is essential for maintaining oncogenic property of mutant IDH-containing cancer cells but dispensable for cell growth. Oncotarget 6: 8606–8620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sulkowski PL, Corso CD, Robinson ND, Scanlon SE, Purshouse KR, et al. (2017) 2-Hydroxyglutarate produced by neomorphic IDH mutations suppresses homologous recombination and induces PARP inhibitor sensitivity. Sci Transl Med 9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Dong J, Feldmann G, Huang J, Wu S, Zhang N, et al. (2007) Elucidation of a universal size-control mechanism in Drosophila and mammals. Cell 130: 1120–1133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Moroishi T, Hansen CG, Guan KL (2015) The emerging roles of YAP and TAZ in cancer. Nat Rev Cancer 15: 73–79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Shah SR, David JM, Tippens ND, Mohyeldin A, Martinez-Gutierrez JC, et al. (2017) Brachyury-YAP Regulatory Axis Drives Stem ness and Growth in Cancer. Cell Rep 21: 495–507. [DOI] [PMC free article] [PubMed] [Google Scholar]