Abstract
Pyrroline-5-carboxylate reductase 1 (PYCR1) is an enzyme involved in cell metabolism and is upregulated in cancer. However, the correlations of PYCR1 expression with the clinicopathological features and prognosis of renal cell carcinoma (RCC) remain unclear. The purpose of this study was to identify the expression of PYCR1 and its clinical relevance in RCC patients.
PYCR1 mRNA expression differences between RCC and the adjacent normal renal tissues were assessed using the Cancer Genome Atlas database (TCGA). Subsequently, the expression of PYCR1 mRNA and protein were evaluated by quantitative real-time polymerase chain reaction, Western blot, and immunochemistry using 30 paired frozen samples of RCC and the adjacent normal renal tissues. The protein expression of PYCR1 was evaluated by immunostaining formalin-fixed, paraffin-embedded sections of RCC samples from 96 patients who underwent radical nephrectomy, and its relationship with clinical features were analyzed. Nonpaired t tests were used to statistically analyze the differences between the 2 groups. Cox univariable and multivariable analyses of overall survival (OS) among RCC patients were performed.
The expression of PYCR1 mRNA was significantly upregulated in RCC tissues compared to adjacent normal renal tissues in the TCGA database (P < .01). The area under the receiver operating characteristic curve value was 0.748. The expression of PYCR1 mRNA and protein was significantly upregulated in RCC compared with that in paired normal renal tissues (P < .01). Higher PYCR1 levels were associated with metastasis (P < .01). Kaplan–Meier survival curves indicated that higher PYCR1 expression was correlated with poorer OS. Therefore, PYCR1 may act as a novel prognostic marker and therapeutic target in the diagnosis and treatment of RCC.
Keywords: clinical significance, PYCR 1, renal cell carcinoma
1. Introduction
Renal cell carcinoma (RCC) is a common malignant tumor in the genitourinary system. It is estimated that approximately 62,700 patients are expected to be diagnosed with RCC in 2016, and 14,260 deaths are predicted to be related to this disease.[1] Currently, RCC has no specific tumor markers for diagnosis. Therefore, early diagnosis is difficult. 25% of RCC patients have been diagnosed with advanced or metastatic disease.[2] Current treatments have shown limited efficacy and metastatic RCC remains incurable.[3] In patients with metastatic disease, the 5-year survival rates are less than 10%.[4] It is very important to make early diagnoses and to find new targets for the treatment of RCC.
Tumor cell metabolism is closely related to rapid cell proliferation and maintenance of redox homeostasis. A variety of oncogenes and tumor suppressors are also involved in the regulation of tumor cell metabolism. Proline is an important amino acid in the cellular microenvironment. The metabolism and synthesis of proline are involved in the cellular tricarboxylic acid cycle, the urea cycle, and the pentose phosphate pathway.[5,6] Proline dehydrogenase/oxidase is the first step in the catabolism of proline, and pyrrole-5-carboxylate reductase (PYCR) is the last key enzyme that catalyzes the synthesis of proline, including PYCR1, PYCR2, and PYCR like.[7] PYCR1 is mutated in patients with autosomal recessive dermatitis type 2, which suggests that PYCR1 plays an important role in the normal development of cells.[8]
Accumulated studies have demonstrated that PYCR1 may play an important role in tumorigenesis. PYCR1 is overexpressed in breast cancer, prostate cancer, and lung cancer cells.[9–11] Silencing PYCR1 expression can suppress tumor cell proliferation and survival.[9,11] The roles of PYCR1 in RCC are still unclear. In this study, we have compared the expression of PYCR1 between RCC and adjacent normal renal tissues, analyzed the relationships between PYCR1 expression and clinicopathological features and prognosis of RCC, and explored the potential roles of PYCR1 in the development of RCC.
2. Materials and methods
2.1. Analysis of the Cancer Genome Atlas database
The data of PYCR1 mRNA and the clinicopathological features of RCC patients were downloaded from the Cancer Genome Atlas database (TCGA) to integrate the data (http://tcga-data.nci.nih.gov/tcga/). As described in the UCSC Xena website, the gene expression profile was measured experimentally using the Illumina HiSeq 2000 RNA Sequencing platform by the University of North Carolina TCGA genome characterization center. RNA-seq data were quantified using RNA-seq by expectation-maximization as described previously.[12] Associations between the expression of PYCR1 and clinicopathological features of RCC patients were investigated.
2.2. Patients and samples
Human RCC tissue samples (n = 30) and the adjacent normal renal tissues (n = 30) were collected from patients undergoing radical nephrectomy at the First Affiliated Hospital of GuangXi Medical University between January 1, 2016 and December 31, 2017. None of the patients received chemotherapy or radiotherapy before surgery. The RCC and the adjacent normal renal tissues were stored in liquid nitrogen. Specimens were stored frozen until RNA and protein were extracted. The partial specimens were embedded in paraffin for immunohistochemical staining.
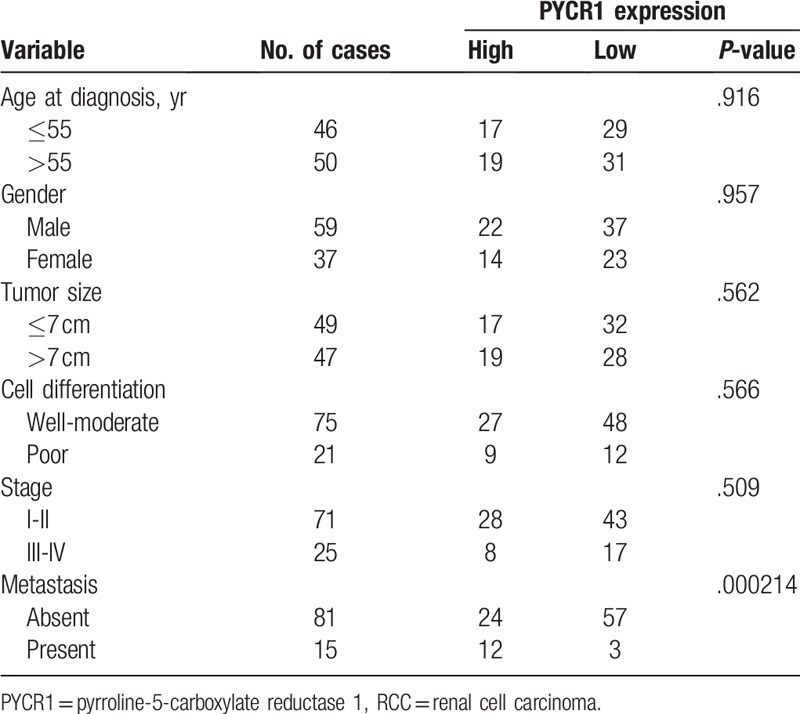
All patients were confirmed as clear renal cell carcinoma (cRCC) by pathology. In addition, paraffin-embedded specimens from 96 cRCC patients who underwent radical nephrectomy between January 1, 2009 and December 31, 2013 were analyzed by immunohistochemical staining by the Department of Pathology, First Affiliated Hospital of GuangXi Medical University. The study was approved by the Ethics Committee of the First Affiliated Hospital of GuangXi Medical University. Informed consent was collected from each patient. Clinical staging was performed according to the AJCC 7th edition, and cell grading was evaluated according to the Furhman criteria. The clinicopathological data are detailed in Table 1.
Table 1.
Relationship of PYCR1 expression and clinicopathological parameters in RCC patients.
2.3. Quantitative real-time PCR
Total RNA was extracted from collected 30 paired RCC and adjacent normal renal tissues according to manufacturer instructions. Reverse transcription was performed according to TaKaRa's manufacturer instructions. Quantitative Real-time polymerase chain reaction (qRT-PCR) was performed on the StepOne III system. The primers used for PCR amplification were 5′-GAAGATGGGGGTGAAGTTGA-3′ (forward) and 5′-CTCAATGGAGCTGATGGTGA-3′ (reverse) for PYCR1; 5′-CTCCATCCTGGCCTCGCTGT-3′ (forward) and 5′-GCTGTCACCTTCACCGTTCC-3′ (reverse) for β-actin. Conditions used for PCR were as follows: an initial 10 minutes 95°C period followed by 40 cycles of 95°C for 30 seconds and 60°C for 60 seconds. The relative mRNA expression levels were evaluated by the 2−ΔΔCQ method. Each experiment was repeated 3 times.
2.4. Western blot assay
Total protein was extracted from tissue lysate and the protein concentration was measured. An acrylamide gel was prepared at 10% for separation and 5% for loading, 40 μg of protein per well was subjected to sodium dodecyl sulfate-polyacrylamide gel electrophoresis, then transferred to a polyvinylidene difluoride membrane, blocked, and exposed to rabbit anti-goat PYCR1 polyclonal antibody (ab103314; Abcam Inc, Cambridge, UK) overnight at 4°C. The secondary antibody was incubated for 2 hours at room temperature. β-actin was used as an internal reference. The relative expression of PYCR1 protein was calculated by using a biological imaging system. Each experiment was repeated 3 times.
2.5. Immunohistochemistry
All tissues were fixed in 10% formalin, routinely dehydrated, dipped in wax, embedded in paraffin, and sliced to a thickness of 4 μm. Phosphate-buffered saline was used instead of primary antibody as a negative control. The primary antibody was PYCR1 (polyclonal rabbit antibody, ab103314; Abcam Inc, concentration: 1:100), dewaxing using xylene, dephenylation using ethanol, 3% dehydrogenation of endogenous peroxidase by hydrogen peroxide, repaired by high pressure of citric acid, and overnight refrigeration at 4°C. The secondary antibody (goat anti-rabbit antibody, SP-9000; Beijing Zhongshan Jinqiao Biotechnology Co, Ltd, Beijing, China) was incubated in a 37% water bath for 25 minutes, followed by DAB mean (3,3′-diaminobenzidine, DAB) coloration, hematoxylin staining, and sealing. PYCR1 protein expression was shown by brown staining mainly in the cytoplasm. PYCR1 expression was evaluated by 2 independent pathologists.
The expression of PYCR1 was measured by staining index. Scoring was based on the following criteria:
-
(1)
number of positive staining cells, where 0 indicates ≦5%, 1 indicates 6% to 20%, 2 indicates 21% to 50%, and 3 indicates >51%;
-
(2)
cell staining intensity, where 0 indicates no staining of cells, 1 indicates weakly positive (light yellow), 2 indicates positive (brown), and 3 indicates strongly positive (tan).
Staining index = positive staining cells + cell staining intensity, score (0–12). A staining index of >3 suggests a high expression, a staining index of ≦ 3 for low expression.[9,11]
3. Statistical analysis
All statistical analyses were performed using SPSS statistical software (version 21.0) and GraphPad Prism (version 6.0). Each experiment was repeated 3 times, all results were presented as the mean ± standard deviation (x ± s). Nonpaired t test was employed to analyze the difference in 2 groups regarding the expression of PYCR1 in RCC and adjacent normal renal tissues, Chi-square test was used to evaluate the correlations between PYCR1 expression and the clinicopathologic features of RCC patients. Additionally, log-rank test was used to analyze the overall survival (OS) in patients with RCC. Kaplan–Meier plots were generated for the survival analysis. Cox univariable and multivariable analyses of OS among RCC patients were performed. P < .05 was considered statistically significant.
4. Results
4.1. Analysis of PYCR1 expression in the TCGA dataset
The data of PYCR1 expression at the mRNA level were obtained from 606 tissues (534 RCC and 72 adjacent normal renal tissues) in the TCGA database. As shown in Figure 1A and B, the expression of PYCR1 was significantly upregulated in RCC tissues compared to adjacent normal renal tissues (7.1315 ± 1.5574 vs 5.9646 ± 0.8031, P < .01). Significant association was observed between high expression of PYCR1 and shorter OS (P < .05, Fig. 1C).
Figure 1.

The expression levels of PYCR1 mRNA in RCC and normal renal tissues from TCGA database. (A and B) RSEM of PYCR1 mRNA. Significant differences were observed between RCC tissues (534 cases) and normal renal tissues (72 cases). (C) Associations between PYCR1 mRNA expression and overall survival from TCGA database. (D) ROC curve of PYCR1. ∗∗∗P < .001. mRNA = messenger RNA, PYCR1 = pyrroline-5-carboxylate reductase 1, RCC = renal cell carcinoma, ROC = receiver operating characteristic, RSEM = RNA-seq by expectation-maximization, TCGA = the cancer genome atlas.
To further evaluate the diagnostic significance of PYCR1, an ROC curve was constructed by plotting sensitivity versus specificity (Fig. 1D). The value for AUC of PYCR1 was 0.748 (P < .01). Sensitivity and specificity for predicting RCC were calculated to be 54.3% and 90.3%, respectively.
4.2. Expression of PYCR1 mRNA and protein in RCC tissues
The results of qRT-PCR and Western blot have shown that PYCR1 mRNA and protein expression was upregulated in 30 RCC samples compared with the paired adjacent renal tissues. The mRNA and protein expression levels of PYCR1 in RCC tissues were significantly higher than those in adjacent normal renal tissues (Fig. 2A–C, P < .01). Immunohistochemistry was used to evaluate PYCR1 expression in RCC and adjacent normal renal tissues. The results revealed that 70% (21/30) of patients demonstrated high PYCR1 protein expression in RCC tissues (Fig. 2E) compared with 33.3% (10/30) of patients in adjacent normal renal tissues (Fig. 2D, P < .01).
Figure 2.

Expression of PYCR1 mRNA and protein in RCC and paired adjacent normal renal tissues. (A) qRT-PCR analysis showed the higher expression of PYCR1 mRNA in RCC tissues compared with adjacent normal renal tissues. (B) Expression of PYCR1 protein in 4 representative pairs of RCC tissues is presented. (C) The PYCR1 protein expression was higher in RCC tissues than in adjacent normal renal tissues. (D) Immunohistochemical analysis showed weak staining of PYCR1 in adjacent normal renal tissues. (E) Immunohistochemical analysis showed strong staining of PYCR1 in RCC tissues. C: RCC tissues, N: adjacent normal renal tissues, ∗∗P < .01. mRNA = messenger RNA, PYCR1 = pyrroline-5-carboxylate reductase 1, qRT-PCR = quantitative real-time polymerase chain reaction, RCC = renal cell carcinoma.
4.3. The correlation between PYCR1 expression and clinical characteristics of RCC patients
To investigate the correlation between PYCR1 expression and clinical characteristics of RCC patients, the clinical data of 96 patients were collected, specifically age, gender, tumor size, cell differentiation, metastasis, and stage. The results showed that PYCR1 expression in RCC tissues was significantly associated with metastasis (P < .001). These results suggested that high expression of PYCR1 might be associated with tumor aggression and progression of RCC.
4.4. The correlation between PYCR1 expression and prognosis of RCC patients
Kaplan–Meier method found that the median OS for the high expression PYCR1 group was 55.69 months, whereas the median OS for low expression patients was 74.8 months. Significant association was observed between high expression of PYCR1 and shorter OS (P < .05, hazard ratio = 0.2092, 95% confidence interval: 0.09–0.4862, Fig. 3).
Figure 3.

Associations between PYCR1 expression and patients’ overall survival. PYCR1 = pyrroline-5-carboxylate reductase 1.
4.5. Cox univariable and multivariable analyses of OS among RCC patients
Cox univariable analyses showed that gender (P = .013), cell differentiation (P = .039), stage (P = .017), and PYCR1 expression (P = .004) all influence OS in RCC patients. Subsequently, the cox multivariable analyses suggested that PYCR1 expression (P = .001) was an independent risk factor of OS in RCC patients (Table 2).
Table 2.
Univariate and multivariate analysis of prognostic factors in 96 patients with RCC.

5. Discussion
Disorder of glutamine metabolism is related to the development of tumors, whereas glutamic acid is one of the sources of proline synthesis. Therefore, attention has been drawn to the role of proline synthesis in the development and progression of tumors.[13,14] Pyrroline-5-carboxylate (P5C) is a glutamate intermediate formed by P5C synthase that is then converted into proline by PYCR. PYCR1 is an enzyme of PYCR.[7] PYCR1 is overexpressed in several cancer tissues, suggesting that it may be closely related to the development and progression of cancer.[9,10,15] However, the expression and related functions of PYCR1 in RCC are still unclear.
Our study first analyzed the TCGA database and found that PYCR1 mRNA was highly expressed in RCC. Then, qRT-PCR and Western blot were used to verify that PYCR1 mRNA and protein were highly expressed in RCC compared with adjacent normal renal tissues, consistent with the findings in the TCGA database. Immunohistochemistry showed the high expression rate of PYCR1 in RCC tissues at 70%, which was higher than that in adjacent normal renal tissues (33.3%). It is confirmed that PYCR1 is overexpressed in RCC, suggesting that it may play an important role in the tumorigenesis and development of RCC.
Further analysis showed that the expression of PYCR1 was associated with metastasis of RCC. These results suggested that high expression of PYCR1 might be associated with tumor aggression and progression of RCC. This study found that the median OS time for the high expression PYCR1 group was 55.69 months, whereas the median OS time for low expression patients was 74.8 months. Poor prognosis may be associated with positive expression of PYCR1. Cox multivariable analyses demonstrated that PYCR1 was an independent predictor of the presence of RCC in patients. It is suggested that PYCR1 expression may be closely related to the occurrence and development of RCC.
However, there are no cytological evidences that the expression of PYCR1 is directly related to the malignant transformation of RCC and its mechanism. The next step is to verify the effect of PYCR1 on renal carcinoma cells in vitro and in vivo and to explore its specific mechanisms of action. Furthermore, we will investigate which pathway is involved in PYCR1 function in RCC.
6. Conclusion
PYCR1 is overexpressed in RCC, which is associated with tumor metastasis. The prognosis of high expression of PYCR1 is significantly worse than that of low expression among RCC patients. It is suggested that PYCR1 may play an important role in the development of RCC. PYCR1 is expected to be a valuable biomarker for RCC progression and a potential target for RCC therapy.
Author contributions
Conceptualization: Fu Weijin, Cheng Jiwen.
Data curation: Fu Weijin, Xie Zhibin, Liu Deyun.
Formal analysis: Zheng Shengfeng, Ding Qijian, Liu Jiayi.
Funding acquisition: Fu Weijin.
Investigation: Chen Yilong, Yang Xiaoli, Mi Hua.
Supervision: Fu Weijin, Xie Zhibin, Liang Qiumei.
Writing – original draft: Fu Weijin, Xie Zhibin.
Writing – review and editing: Fu Weijin, Xie Zhibin.
Footnotes
Abbreviations: cRCC = clear renal cell carcinoma, OS = overall survival, PYCR1 = pyrroline-5-carboxylate reductase 1, RCC = renal cell carcinoma, TCGA = the Cancer Genome Atlas database.
WF, ZX, and SZ contributed equally to this study.
The research was supported by 2018 Guangxi Scholarship Fund of Guangxi Education Department.
The authors have no conflicts of interest to disclose.
References
- [1].Siegel RL, Miller KD, Jemal A. Cancer statistics, 2016. CA Cancer J Clin 2016;66:7–30. [DOI] [PubMed] [Google Scholar]
- [2].Shuch B, Amin A, Armstrong AJ, et al. Understanding pathologic variants of renal cell carcinoma: distilling therapeutic opportunities from biologic complexity. Eur Urol 2015;67:85–97. [DOI] [PubMed] [Google Scholar]
- [3].Capitanio U, Montorsi F. Renal cancer. Lancet 2016;387:894–906. [DOI] [PubMed] [Google Scholar]
- [4].Minardi D, Quaresima L, Santoni M, et al. Recent aspects of sunitinib therapy in patients with metastatic clear-cell renal cell carcinoma: a systematic review of the literature. Curr Urol Rep 2015;16:3. [DOI] [PubMed] [Google Scholar]
- [5].Phang JM, Liu W. Proline metabolism and cancer. Front Biosci 2012;17:1835–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Phang JM, Liu W, Hancock CN, et al. Proline metabolism and cancer: emerging links to glutamine and collagen. Curr Opin Clin Nutr Metab Care 2015;18:71–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Kuo ML, Lee MB, Tang M, et al. PYCR1 and PYCR2 interact and collaborate with RRM2B to protect cells from overt oxidative stress. Sci Rep 2016;6:18846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Guernsey DL, Jiang H, Evans SC, et al. Mutation in pyrroline-5-carboxylate reductase 1 gene in families with cutis laxa type 2. Am J Hum Genet 2009;85:120–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Ding J, Kuo ML, Su LP, et al. Human mitochondrial pyrroline-5-carboxylate reductase 1 promotes invasiveness and impacts survival in breast cancers. Carcinogenesis 2017;38:519–31. [DOI] [PubMed] [Google Scholar]
- [10].Zeng T, Zhu L, Liao M, et al. Knockdown of PYCR1 inhibits cell proliferation and colony formation via cell cycle arrest and apoptosis in prostate cancer. Med Oncol 2017;34:27. [DOI] [PubMed] [Google Scholar]
- [11].Cai F, Miao Y, Liu C, et al. Pyrroline-5-carboxylate reductase 1 promotes proliferation and inhibits apoptosis in non-small cell lung cancer. Oncol Lett 2018;15:731–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Lilljebjorn H, Henningsson R, Hyrenius-Wittsten A, et al. Identification of ETV6-RUNX1-like and DUX4-rearranged subtypes in paediatric B-cell precursor acute lymphoblastic leukaemia. Nat Commun 2016;7:11790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].De Ingeniis J, Ratnikov B, Richardson AD, et al. Functional specialization in proline biosynthesis of melanoma. PloS One 2012;7:e45190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Hu CA, Bart Williams D, Zhaorigetu S, et al. Functional genomics and SNP analysis of human genes encoding proline metabolic enzymes. Amino acids 2008;35:655–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Ernst T, Hergenhahn M, Kenzelmann M, et al. Decrease and gain of gene expression are equally discriminatory markers for prostate carcinoma: a gene expression analysis on total and microdissected prostate tissue. Am J Pathol 2002;160:2169–80. [DOI] [PMC free article] [PubMed] [Google Scholar]


