Supplemental Digital Content is available in the text
Keywords: childhood, cord blood, DNA methylation, hepatocyte nuclear factor 4 alpha, melanocortin 4 receptor, triglyceride
Abstract
Although the changes in DNA methylation are assumed to be due to the association between adverse intrauterine conditions and adult metabolic health, evidence from human studies is rare. Little is known about the changes in DNA methylation present at birth that affect metabolic profiles in childhood. Previous studies have shown that the melanocortin 4 receptor (MC4R) and hepatocyte nuclear factor 4 alpha (HNF4α) genes are associated with obesity and metabolic disorders. Thus, we investigated the associations of the DNA methylation statuses of MC4R and HNF4α in cord blood with metabolic profiles in childhood.
We collected data from 90 children 7 to 9 years of age included in the Ewha Birth & Growth Cohort Study in Korea. DNA methylation was analyzed by pyrosequencing. The children were split into 2 groups according to the cutoff triglyceride (TG) levels (<110 and ≥110 mg/dL).
The methylation statuses of MC4R and HNF4α at birth were significantly associated with the TG level in childhood (P < .05). It was interesting to note that the methylation statuses of MC4R and HNF4α in cord blood were significantly decreased, whereas childhood body mass index was significantly increased, in children with high TG levels compared with children with low TG levels (P < .05).
Our findings show that the methylation statuses of MC4R and HNF4α at birth are associated with metabolic profiles in childhood. These epigenetic modifications occurring in early life may contribute to subsequent metabolic-related disorders. Thus, we suggest that DNA methylation status in cord blood may be predictive of the risk of developing metabolic syndrome.
1. Introduction
Metabolic syndrome (MetS) is a complex disorder caused by a cluster of interrelated risk factors for the development of type 2 diabetes and cardiovascular disease.[1,2] Globally, MetS is becoming increasingly prevalent and a major public health concern. In Korea, the rate of MetS in children and adolescents is lower (3%) compared with that in adults aged 30 years (28.8%),[3] but MetS-related traits continue into adulthood, increasing the risk of adult MetS.[4,5] Although the molecular mechanisms underlying the etiology of MetS remain largely unknown, both genetic and environmental factors play important roles in the pathogenesis of MetS.
The melanocortin 4 receptor gene (MC4R), which is involved in the regulation of energy homeostasis, and hepatocyte nuclear factor 4 alpha gene (HNF4α), a transcription factor involved in gluconeogenesis and lipid homeostasis, are associated with obesity and MetS.[6–12] Humans with an MC4R deficiency have a higher risk of early onset obesity and hyperphagia due to haploinsufficiency.[6,7] Similarly, MC4R-null mice showed elevated triglyceride (TG) levels and abnormal expression of genes related to fatty acid synthesis.[8,9] In humans, mutations in the 2 promoters of HNF4α, P1 and P2, led to impaired insulin secretion and, consequently, maturity-onset diabetes in young.[10–12] In addition, HNF4α-deficient mice showed altered TG, total cholesterol, and high-density lipoprotein cholesterol levels and abnormal expression of genes associated with lipid transport and metabolism.[13]
Epigenetic modifications induced in early life influence individual susceptibility to MetS in adults, although the underlying mechanism in humans remains unclear.[14–16] Indeed, maternal diet can affect the epigenome of offspring, resulting in obesity and MetS.[17,18] In our previous study, there were significant associations between the pro-opiomelanocortin methylation status at birth and increased TG and insulin levels in childhood.[19] In addition, MC4R and HNF4α methylation contributed to elevated TG levels in the cord blood of preterm babies.[20] However, DNA methylation of some genes is relatively stable following birth and may persistently affect long-term health outcomes, but that of other genes is temporally reversible.[21,22] To date, little is known about the epigenetic modifications of metabolism-related genes that affect metabolic profiles in childhood.
In this study, we measured methylation of MC4R and HNF4α in DNA extracted from cord blood and the blood of children included in a prospective cohort. Next, we investigated the associations between the methylation statuses of MC4R and HNF4α at birth and metabolic profiles in childhood.
2. Methods
2.1. Study design
This study was conducted as part of the Ewha Birth and Growth Cohort Study, which is a birth cohort established at the Ewha Womans University Hospital, Seoul, Korea, between 2001 and 2006. This cohort study had a single hospital-based cohort design and involved longitudinal observations of child growth and health from early life. Methodologic details of the cohort are described elsewhere.[19,23] Briefly, pregnant women who received prenatal care between 24 and 28 weeks’ gestation at the hospital were recruited (baseline n = 940). The children were followed up at 3, 5, and annually after 7 years of age. In 2011, we contacted 344 consenting mothers whose children were 7 to 9 years old, and 260 subjects participated in the follow-up examination. During follow-up, anthropometric data and blood samples were collected from the participants. We excluded subjects who had a congenital defect (n = 2), who had not given a blood sample (n = 1), or whose mother had a history of preeclampsia, gestational diabetes mellitus, or chronic disease at baseline (n = 24). Of the 260 participants, 90 had stored cord blood and were included in the final analysis. All participants gave their written informed consent before enrollment in this study. This study was approved by the Institutional Review Board of the Ewha Womans University Hospital (ECT 13-01A-13).
2.2. Anthropometric measurements
Trained researchers collected data on maternal and offspring characteristics, including gestational age (GA), birth length, birth weight, and maternal features at delivery using medical charts, and mother's education and family's income using questionnaires. GA was calculated based on the last menstrual period.[20] The Ponderal index was calculated as weight in kilograms divided by the cube of height in meters (kg/m3). Children's height and weight were measured to the nearest 0.1 cm and 0.1 kg using an automatic electronic scale (Dong Sahn Jenix Co Ltd, Seoul, Korea). Body mass index (BMI) was calculated as weight divided by height squared (kg/m2). Mother's education level was divided into 2 groups: graduated from high school (<12 years) and graduated from university or higher (≥12 years). Family income was categorized into 3 groups by monthly income: <3 million KRW, 3.0 to 4.9 million KRW, and >5 million KRW (KRW, South Korean Won).[24]
2.3. Biochemical assessments
Cord blood samples were collected at birth and stored at –80°C until analysis. Fasting blood samples at follow-up examinations were drawn from the antecubital veins of the children into Vacutainer tubes containing EDTA or serum tubes (BD Biosciences, San Jose, CA). The blood samples were centrifuged at 3000 rpm for 10 minutes, and the plasma was stored at –80°C. Levels of glucose, TG, total cholesterol, and high-density lipoprotein cholesterol were analyzed using a model 7180 automatic analyzer (Hitachi, Tokyo, Japan). Insulin was analyzed using an immunoradiometric assay kit (MyBiosource, San Diego, CA) according to the manufacturer's protocol. Insulin resistance was determined by the widely used homeostasis model assessment of insulin resistance (HOMA-IR) method, which was calculated as (plasma glucose [mmol/L] × insulin [μIU/mL])/22.5.[25]
2.4. Analysis of DNA methylation at CpG site by pyrosequencing
Genomic DNA samples were isolated from the cord blood and blood samples of children using the DNeasy Blood and Tissue kit (Qiagen, Hilden, Germany), according to the manufacturer's methods. DNA concentration and purity were measured using a spectrophotometer (Nanodrop Technologies, Wilmington, DE).
Analysis of DNA methylation at CpG sites within the MC4R and HNF4α genes was performed by pyrosequencing. To identify the CpG islands in promoter regions, the areas within 2000 bp upstream from the transcriptional start sites (TSSs) of MC4R and the P1 promoter of HNF4α were analyzed using PSQ Assay Design software (Biotage AB, Uppsala, Sweden). The P2 promoter of HNF4α was designed based on previous studies and using UCSC Genome Browser.[26–28] Three CpG sites located within the promoter regions (–800, –788, and –783 bp upstream of the TSS) of MC4R and 8 CpG sites located within the P1 promoter regions (–119, –103, –100, and –92 bp upstream of the TSS) and P2 promoter regions (–44,924, –44,921, –44,919, and –44,907 bp upstream of the TSS) of HNF4α were amplified by a primer set designed (Supplemental Digital Content: Figure S1 and Table S1). Among them, the CpG2 site in the P2 promoter of HNF4α was mostly methylated (>99%). Therefore, it was excluded from the final analysis. Methodologic details of the DNA methylation analysis have been provided in our previous study.[19]
2.5. Statistical analysis
Continuous variables are expressed as mean ± standard deviation or medians (interquartile range), and categorical variables are expressed as numbers (percentages). The TG level, insulin level and HOMA-IR values were log-transformed to satisfy normality. To explore the persistence of methylation status from birth to 7 to 9 years of age, Spearman correlation was used to assess the relationships between DNA methylation of cord blood and of children's blood. In addition, a partial correlation analysis was performed to estimate the relationships between the methylation status of MC4R or HNF4α at birth and metabolic profiles in children after adjustment for sex, child's age, and BMI.
Multiple regression analyses were used to assess the influence of MC4R and HNF4α methylation in cord blood on the metabolic profiles in children. Each CpG site in MC4R and HNF4α showed a nonsignificant correlation with the metabolic profiles in children. Therefore, all were included as independent variables in the multiple regression analyses. An adjusted P-value for multiple testing involving 10 CpG sites was estimated using the Benjamini–Hochberg procedure to control the false discovery rate.
Further, to identify the influences of site-specific CpG methylation in MC4R and HNF4α in cord blood and BMI in childhood on abnormal metabolic levels, an abnormal TG cutoff level in children was defined (110 mg/dL) according to the National Cholesterol Education Program Adult Treatment Panel III criteria, and the subjects were divided into 2 groups (<110 and ≥110 mg/dL).[29] Accordingly, we compared the methylation statuses of MC4R and HNF4α at birth with BMI in childhood according to the childhood TG level by analysis of covariance, to examine whether the DNA methylation status is an early predictive marker of metabolic abnormalities.
Confounding factors that could influence the relationship between DNA methylation and metabolic profiles, including maternal age, prepregnancy BMI, mother's education, GA, sex, birth weight, child's age, child's BMI, and methylation levels in children, have been reported in previous studies.[19,30–33] There was no multicollinearity problem. All statistical analyses were conducted using SAS software (ver. 9.3; SAS Institute Inc, Cary, NC). All analyses were 2-tailed, and P < .05 was considered to indicate statistical significance.
3. Results
3.1. General characteristics of the study subjects
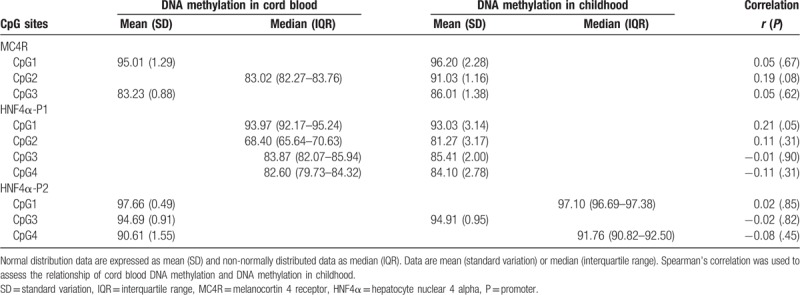
Ninety subjects were included in this study, of whom 56.7% were girls. At birth, the mean GA and birth weight were 39.4 weeks and 3290 g, respectively. The incidence of preterm birth (GA <37 weeks) was 2.2%. At 7 to 9 (mean 7.8) years of age, the mean BMI and percent body fat mass of the children were 16.62 kg/m2 and 22.83%, respectively (Table 1). The average methylation levels of MC4R and HNF4α in cord blood and blood from the children are shown in Table 2. The DNA methylation levels of CpG sites in MC4R and HNF4α mostly increased from birth to 7 to 9 years of age, with the exception of HNF4α-CpG1. The methylation status of the CpG1 site in the P1 promoter of HNF4α at birth was significantly correlated with that in childhood (r = 0.21, P = .05). By contrast, no correlation was observed in the methylation statuses of the CpG sites at birth and the children.
Table 1.
Basic characteristics of the study subjects.
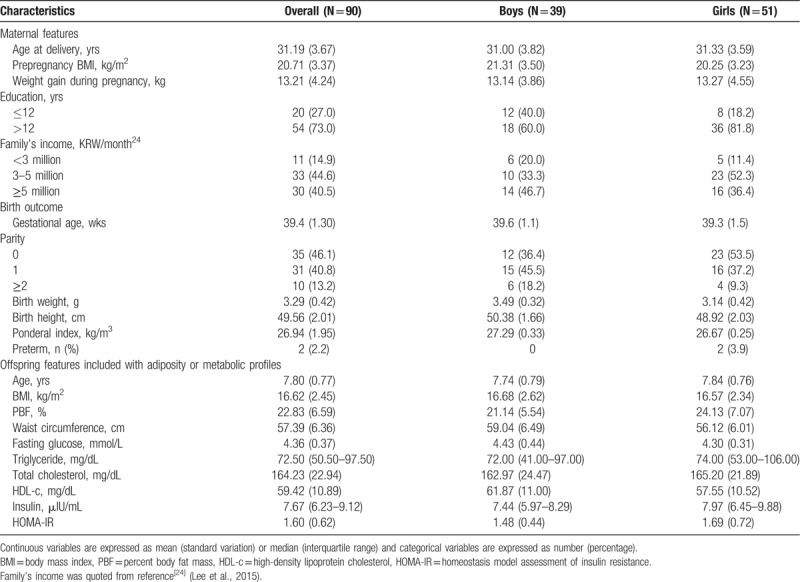
Table 2.
DNA methylation levels in cord blood and blood in childhood.
3.2. Associations between DNA methylation statuses in cord blood and TG levels in children
To access the correlation between methylation at birth and metabolic indices in children aged 7 to 9 years, we analyzed the linear relationships between the MC4R and HNF4α methylation statuses in cord blood and metabolic parameters in childhood. Especially, TG levels were significantly correlated with the methylation statuses of MC4R-CpG1 and -CpG2 and HNF4α-CpG2, -CpG3, and -CpG4 in the P1 promoter after adjusting for sex, age, and BMI (P < .05, Fig. 1).
Figure 1.
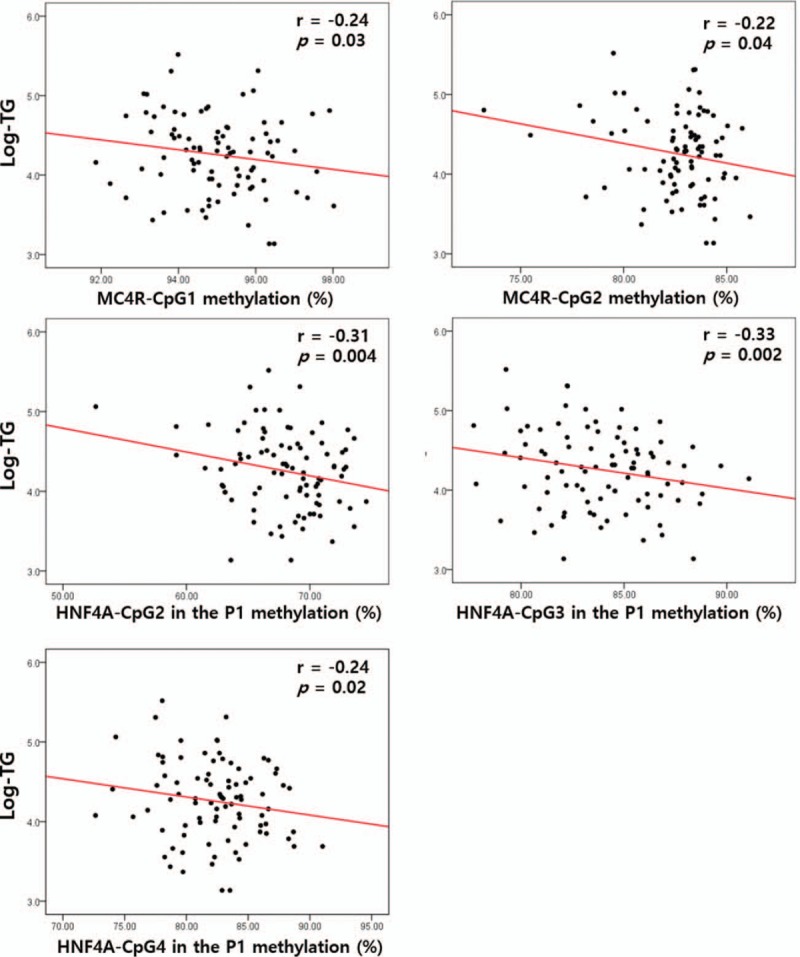
Analysis of the associations between triglyceride (TG) levels and the methylation statuses of the melanocortin 4 receptor (MC4R) and hepatocyte nuclear factor 4 alpha (HNF4a) promoters in the blood of children, after adjustment for sex, child's age, and child's body mass index.
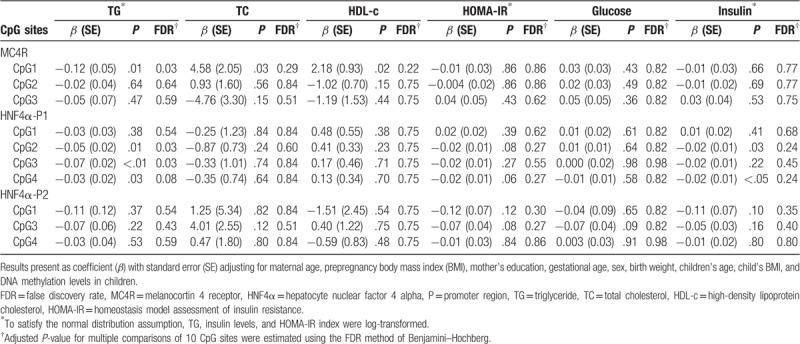
Additionally, the MC4R and HNF4α methylation statuses in cord blood were related to the metabolic profiles of children after controlling for multiple comparisons of 10 CpG sites (adjusted P-value) and potential risk factors to evaluate the influence of methylation status at birth on metabolic profiles in childhood (Table 3). The methylation statuses of MC4R-CpG1, HNF4α-CpG2, and HNF4α-CpG3 in the P1 promoter in cord blood were significantly and negatively associated with a higher TG level in childhood, after adjusting for maternal age, prepregnancy BMI, mother's education, GA, sex, birth weight, child's age, child's BMI, and methylation levels in childhood (β = –0.12, adjusted P = .03; β = –0.05, adjusted P = .03; β = –0.07, adjusted P = .03, respectively). Additionally, the methylation status of HNF4α-CpG4 in the P1 promoter in cord blood was marginally but significantly associated with a higher TG level in childhood (β = –0.03, adjusted P = .08). No significant associations between the methylation status and other metabolic profiles were observed.
Table 3.
Association between DNA methylation of MC4R and HNF4α in cord blood and metabolic indices in children.
3.3. Associations between DNA methylation statuses in cord blood and BMI in childhood according to TG level
To evaluate whether methylation status is an early marker of metabolic abnormalities, we compared the methylation statuses of MC4R and HNF4α in cord blood with BMI in childhood according to the TG level in childhood (Fig. 2). Four CpG sites were significantly associated with the methylation status and TG level (Table 3). The DNA methylation statuses of MC4R-CpG1 and HNF4α-CpG2, -CpG3, and -CpG4 in the P1 promoter were significantly lower in children with a high TG level after adjusting for maternal age, prepregnancy BMI, mother's education, GA, sex, birth weight, child's age, child's BMI, and methylation levels in children (LS means: 95.43 vs 94.60%, P = .04; 69.77 vs 66.61%, P = .003; 84.72 vs 82.28%, P = .001; 83.42 vs 80.86%, P = .03, respectively), whereas BMI was significantly higher in children with high TG levels than in those with low TG levels, after adjusting for maternal age, prepregnancy BMI, mother's education, GA, sex, birth weight, child's age, and methylation levels in cord blood (LS means: 17.49 vs 19.10 kg/m2, P = .04; 17.36 vs 19.14 kg/m2, P = .02; 17.39 vs 19.26 kg/m2, P = .02; 17.41 vs 19.15 kg/m2, P = .02, respectively).
Figure 2.
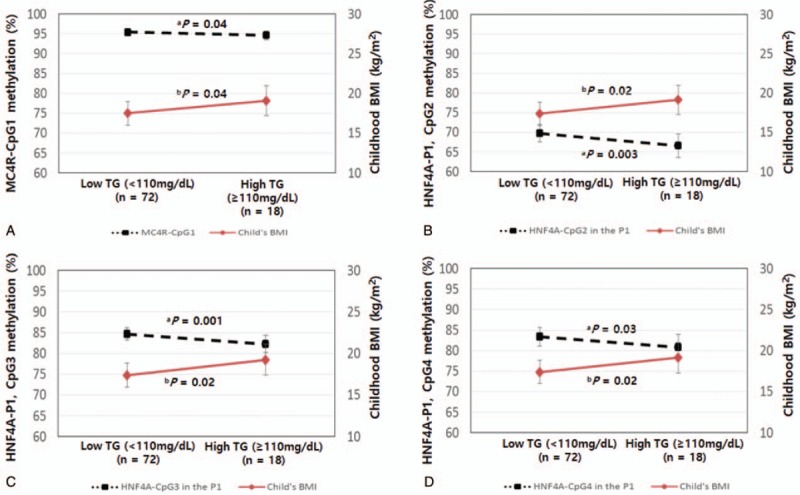
DNA methylation statuses of melanocortin 4 receptor (MC4R) and hepatocyte nuclear factor 4 alpha (HNF4α) at CpG sites in cord blood and child's body mass index (BMI) according to triglyceride (TG) levels in children. (A) P-values were calculated by analysis of covariance (ANCOVA), adjusting for maternal age, prepregnancy BMI, mother's education, gestational age (GA), sex, birth weight, child's age, child's BMI, and DNA methylation levels in children. (B) P-values were calculated by ANCOVA, adjusting for maternal age, prepregnancy BMI, mother's education, GA, sex, birth weight, child's age, and DNA methylation levels in cord blood. Dotted lines (----) represent differences in the methylation status between low TG and high TG levels. Solid lines (—) represent differences in BMI in childhood between low and high TG levels.
4. Discussion
In this study, we conducted follow-up evaluations on the growth of children from birth to 7 to 9 years of age via a prospective cohort study. Interestingly, we identified associations of the MC4R and HNF4α methylation statuses in cord blood with metabolic profiles in children, especially with the TG level. These observations suggest that an altered DNA methylation status early in life can continue to influence metabolic profiles in childhood.
We found that reduced methylation of MC4R at birth was significantly associated with an increased TG level in childhood (Table 3). In particular, the methylation status of MC4R-CpG1 was significantly lower, whereas BMI was significantly higher, in children with high TG levels than in those with low TG levels (Fig. 2). Reduced methylation of MC4R was associated with obesity in mice fed a high-fat diet; however, to our knowledge, no previous studies have examined the associations between MC4R methylation and metabolic profiles in humans.[34] We recently showed that reduced methylation of MC4R was significantly associated with higher TG levels in the cord blood of preterm infants.[20] Based on this finding, we speculate that a lower level of MC4R methylation at birth is involved in obesity and MetS in childhood. Kooijman et al found that inhibition of the MC3/4R synthetic antagonist reduces very-low-density lipoprotein and TG production, resulting in an increased TG level in white adipose tissue.[35] However, another study found that blocking the MC4R antagonist did not increase TG levels in pair-fed rats.[36] MC4R activation leads to increases in arterial pressure and heart rate despite a decrease in food intake.[37] Thus, we suggest that altered DNA methylation levels of MC4R established in utero might contribute to TG dysregulation in childhood and thus influence the likelihood of developing obesity and MetS later in life. This underscores the need for a long-term follow-up study of our cohort.
We also found that the methylation statuses of HNF4α-CpG2 and -CpG3 in the P1 promoter at birth were significantly and negatively associated with the TG level in childhood (Table 3). In humans, differences in DNA methylation, particularly in HNF4α, due to intrauterine growth restriction have been identified in cord blood.[38] Additionally, HNF4α methylation in adipose tissue is elevated in patients with type 2 diabetes.[39] Recently, we reported that an altered methylation status of HNF4α results in increased TG levels in the cord blood of preterm infants.[20] Overexpression of HNF4α caused increases in TG-rich lipoproteins.[40] Of the 2 promoters of HNF4α (P1 and P2),[12] genetic variation in the P1 promoter is associated with a risk of MetS in children and adolescents.[41] Genetic variation in the P2 promoter is related to insulin levels and BMI in adults.[42] The mechanisms underlying these associations are unknown, but in this study, the P1 promoter of HNF4α may have had a greater contribution than that of the P2 promoter to TG levels in childhood. Collectively, these findings suggest that the altered methylation status of HNF4α in the P1 promoter in cord blood is closely related to the TG level in children. Although the biologic effects of small changes in methylation levels are unknown, there is concern that the epigenetic patterns induced in early life continue to influence physical health throughout life. Thus, we suggest that the methylation status at birth is predictive of obesity and MetS in later life. Longitudinal and follow-up studies are needed to confirm these associations.
Regarding the persistence of DNA methylation patterns from birth to 7 to 9 years of age, we found that the methylation statuses of MC4R and HNF4α at birth were not significantly correlated with those in childhood, except for the HNF4α-CpG1 methylation status in the P1 promoter (Table 2). Cord blood and blood from 7- to 9-year-old children differed in terms of lymphocyte composition.[43,44] In addition, DNA methylation status may differ according to blood cell count and type.[45,46] This study measured DNA methylation levels in whole blood, which contains heterogeneous cell types. Previous studies have reported that the DNA methylation status of some CpG loci in buccal cells, which are associated with neurologic disorders, differ markedly in twins from age 5 to 10 years.[47] Furthermore, DNA methylation levels increase from infancy to puberty.[48] These epigenetic changes during infancy and childhood may be due to environmental influences rather than heritable factors. Therefore, we analyzed the association between DNA methylation in cord blood and metabolic profiles in childhood after adjusting for DNA methylation levels in children aged 7 to 9 years.
To the best of our knowledge, this is the 1st study to explore the associations of the MC4R and HNF4α methylation statuses in cord blood with metabolic profiles in the context of health outcomes in healthy children with a normal BMI. We did not assess paternal factors such as diet, nor did we analyze mRNA levels of the genes because of insufficient samples. However, we did verify the associations of gene methylation statuses with metabolic profiles in normal weight and obese children aged 7 to 9 years in a nested case–control study[49] and in term and preterm infants at birth in a case–control study.[20] Although this study did not measure gene expression levels, our previous study identified that MC4R and HNF4α methylation statuses were significantly lower, but that expression levels were significantly higher, in preterm than in term infants.[20] Thus, epigenetic changes established early in life may affect gene function and eventually contribute to the development of MetS later in life. Further studies with long-term follow-up are needed to determine whether changes in the methylation status at birth contribute to the development of MetS in adults.
Our findings indicate that TG levels in childhood may be altered by the methylation status of MC4R or HNF4α in cord blood. Hence, epigenetic modifications at birth may influence metabolic profiles in childhood, thereby contributing to the development of MetS in adulthood. Thus, we suggest that DNA methylation in cord blood is predictive of MetS later in life.
Author contributions
Conceptualization: Eun Jin Kwon, Young Ju Kim.
Data curation: Hye Ah Lee, Hyesook Park, Eun Ae Park, Eun Hee Ha.
Formal analysis: Eun Jin Kwon, Hye Ah Lee, Young-Ah You, Jae Young Yoo, Hyesook Park.
Funding acquisition: Young Ju Kim.
Investigation: Eun Jin Kwon, Jae Young Yoo, Eun Ae Park, Eun Hee Ha.
Supervision: Young Ju Kim.
Validation: Young-Ah You.
Writing – original draft: Eun Jin Kwon.
Writing – review & editing: Hye Ah Lee, Young-Ah You, Jae Young Yoo, Eun Hee Ha, Young Ju Kim.
Supplementary Material
Footnotes
Abbreviations: BMI = body mass index, GA = gestational age, HNF4α = hepatocyte nuclear factor 4 alpha, LDL-c = low-density lipoprotein cholesterol, MC4R = melanocortin 4 receptor, MetS = metabolic syndrome, TG = triglyceride.
This study was supported by the Basic Science Research Program through the National Research Foundation of Korea funded by the Ministry of Education (NRF-2016 R1D1A1A09918620) and by the Ministry of Health & Welfare, Republic of Korea (HI14C0306, HI15C2059) through the Korea Health Industry Development Institute.
The authors have no conflicts of interest to disclose.
Supplemental Digital Content is available for this article.
References
- [1].Alberti KG, Eckel RH, Grundy SM, et al. Harmonizing the metabolic syndrome: a joint interim statement of the International Diabetes Federation Task Force on Epidemiology and Prevention; National Heart, Lung, and Blood Institute; American Heart Association; World Heart Federation; International Atherosclerosis Society; and International Association for the Study of Obesity. Circulation 2009;120:1640–5. [DOI] [PubMed] [Google Scholar]
- [2].Kassi E, Pervanidou P, Kaltsas G, et al. Metabolic syndrome: definitions and controversies. BMC Med 2011;9:48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Ministry of Health and Welfare. Korea Centers for Disease Control and, Prevention. Korea Health Statistics 2010: Korea National Health and Nutrition Examination Survey, (KNHANESV-1). Cheongwon: Ministry of Health and Welfare, 2011 [Google Scholar]
- [4].Eisenmann JC, Welk GJ, Wickel EE, et al. Stability of variables associated with the metabolic syndrome from adolescence to adulthood: the aerobics center longitudinal study. Am J Hum Biol 2004;16:690–6. [DOI] [PubMed] [Google Scholar]
- [5].Katzmarzyk PT, Perusse L, Malina RM, et al. Stability of indicators of the metabolic syndrome from childhood and adolescence to young adulthood: the Quebec Family Study. J Clin Epidemiol 2001;54:190–5. [DOI] [PubMed] [Google Scholar]
- [6].Farooqi IS, Keogh JM, Yeo GS, et al. Clinical spectrum of obesity and mutations in the melanocortin 4 receptor gene. N Engl J Med 2003;348:1085–95. [DOI] [PubMed] [Google Scholar]
- [7].Yeo GS, Lank EJ, Farooqi IS, et al. Mutations in the human melanocortin-4 receptor gene associated with severe familial obesity disrupts receptor function through multiple molecular mechanisms. Hum Mol Genet 2003;12:561–74. [DOI] [PubMed] [Google Scholar]
- [8].Albarado DC, McClaine J, Stephens JM, et al. Impaired coordination of nutrient intake and substrate oxidation in melanocortin-4 receptor knockout mice. Endocrinology 2004;145:243–52. [DOI] [PubMed] [Google Scholar]
- [9].Sutton GM, Trevaskis JL, Hulver MW, et al. Diet-genotype interactions in the development of the obese, insulin-resistant phenotype of C57BL/6J mice lacking melanocortin-3 or -4 receptors. Endocrinology 2006;147:2183–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Saif-Ali R, Harun R, Kamaruddin NA, et al. Association of hepatocyte nuclear factor 4 alpha polymorphisms with type 2 diabetes with or without metabolic syndrome in Malaysia. Biochem Genet 2012;50:298–308. [DOI] [PubMed] [Google Scholar]
- [11].Lehto M, Bitzén PO, Isomaa B, et al. Mutation in the HNF-4alpha gene affects insulin secretion and triglyceride metabolism. Diabetes 1999;48:423–5. [DOI] [PubMed] [Google Scholar]
- [12].Love-Gregory LD, Wasson J, Ma J, et al. A common polymorphism in the upstream promoter region of the hepatocyte nuclear factor-4 ( gene on chromosome 20q is associated with type 2 diabetes and appears to contribute to the evidence for linkage in an Ashkenazi Jewish population. Diabetes 2004;53:1134–40. [DOI] [PubMed] [Google Scholar]
- [13].Hayhurst GP, Lee YH, Lambert G, et al. Hepatocyte nuclear factor 4-alpha (nuclear receptor 2A1) is essential for maintenance of hepatic gene expression and lipid homeostasis. Mol Cell Biol 2001;21:1393–403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Vickers MH. Early life nutrition, epigenetics and programming of later life disease. Nutrients 2014;6:2165–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Martínez JA, Cordero P, Campión J, et al. Interplay of early-life nutritional programming on obesity, inflammation and epigenetic outcomes. Proc Nutr Soc 2012;71:276–83. [DOI] [PubMed] [Google Scholar]
- [16].Kwon EJ, Kim YJ. What is fetal programming?: a lifetime health is under the control of in utero health. Obstet Gynecol Sci 2017;60:506–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Burdge GC, Slater-Jefferies J, Torrens C, et al. Dietary protein restriction of pregnant rats in the F0 generation induces altered methylation of hepatic gene promoters in the adult male offspring in the F1 and F2 generations. Br J Nutr 2007;97:435–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Lee HS. Impact of maternal diet on the epigenome during in utero life and the developmental programming of diseases in childhood and adulthood. Nutrients 2015;17:9492–507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Yoo JY, Lee S, Lee HA, et al. Can proopiomelanocortin methylation be used as an early predictor of metabolic syndrome? Diabetes Care 2014;37:734–9. [DOI] [PubMed] [Google Scholar]
- [20].Kwon EJ, Lee HA, You YA, et al. DNA methylations of MC4R and HNF4 ( are associated with increased triglyceride levels in cord blood of preterm infants. Medicine 2016;95:e4590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Parets SE, Conneely KN, Kilaru V, et al. Fetal DNA methylation associates with early spontaneous preterm birth and gestational age. PLoS One 2013;8:e67489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Bishop KS, Ferguson LR. The interaction between epigenetics, nutrition and the development of cancer. Nutrients 2015;7:922–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Min JW, Kong KA, Park BH, et al. Effect of postnatal catch-up growth on blood pressure in children at 3 years of age. J Hum Hypertens 2007;21:868–74. [DOI] [PubMed] [Google Scholar]
- [24].Lee HA, Kim YJ, Lee H, et al. The preventive effect of breast-feeding for longer than 6 months on early pubertal development among children aged 7-9 years in Korea. Public Health Nutr 2015;18:3300–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Song Y, Manson JE, Tinker L, et al. Insulin sensitivity and insulin secretion determined by homeostasis model assessment and risk of diabetes in a multiethnic cohort of women: the Women's Health Initiative Observational Study. Diabetes Care 2007;30:1747–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Eeckhoute J, Moerman E, Bouckenooghe T, et al. Hepatocyte nuclear factor 4 alpha isoforms originated from the P1 promoter are expressed in human pancreatic beta-cells and exhibit stronger transcriptional potentials than P2 promoter-driven isoforms. Endocrinology 2003;144:1686–94. [DOI] [PubMed] [Google Scholar]
- [27].Thomas H, Jaschkowitz K, Bulman M, et al. A distant upstream promoter of the HNF-4alpha gene connects the transcription factors involved in maturity-onset diabetes of the young. Hum Mol Genet 2001;10:2089–97. [DOI] [PubMed] [Google Scholar]
- [28].Kent WJ, Sugnet CW, Furey TS, et al. The human genome browser at UCSC. Genome Res 2002;12:996–1006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Rodríguez-Moran M, Guerrero-Romero F. Low birthweight and elevated levels of lipoprotein (a) in prepubertal children. J Paediatr Child Health 2014;50:610–4. [DOI] [PubMed] [Google Scholar]
- [30].Godfrey KM, Sheppard A, Gluckman PD, et al. Epigenetic gene promoter methylation at birth is associated with child's later adiposity. Diabetes 2011;60:1528–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Agha G, Hajj H, Rifas-Shiman SL, et al. Birth weight-for-gestational age is associated with DNA methylation at birth and in childhood. Clin Epigenetics 2016;8:118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Engel SM, Joubert BR, Wu MC, et al. Neonatal genome-wide methylation patterns in relation to birth weight in the Norwegian Mother and Child Cohort. Am J Epidemiol 2014;179:834–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Relton CL, Groom A, St Pourcain B, et al. DNA methylation patterns in cord blood DNA and body size in childhood. PLoS One 2012;7:e31821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Widiker S, Karst S, Wagener A, et al. High-fat diet leads to a decreased methylation of the Mc4r gene in the obese BFMI and the lean B6 mouse lines. J Appl Genet 2010;51:193–7. [DOI] [PubMed] [Google Scholar]
- [35].Kooijman S, Boon MR, Parlevliet ET, et al. Inhibition of the central melanocortin system decreases brown adipose tissue activity. J Lipid Res 2014;55:2022–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Baran K, Preston E, Wilks D, et al. Chronic central melanocortin-4 receptor antagonism and central neuropeptide-Y infusion in rats produce increased adiposity by divergent pathways. Diabetes 2002;51:152–8. [DOI] [PubMed] [Google Scholar]
- [37].Tao YX. The melanocortin-4 receptor: physiology, pharmacology, and pathophysiology. Endocr Rev 2010;3:506–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Einstein F, Thompson RF, Bhagat TD, et al. Cytosine methylation dysregulation in neonates following intrauterine growth restriction. PLoS One 2010;5:e8887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Ribel-Madsen R, Fraga MF, Jacobsen S, et al. Genome-wide analysis of DNA methylation differences in muscle and fat from monozygotic twins discordant for type 2 diabetes. PLoS One 2012;7:e51302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Krapivner S, Iglesias MJ, Silveira A, et al. DGAT1 participates in the effect of HNF4A on hepatic secretion of triglyceride-rich lipoproteins. Arterioscler Thromb Vasc Biol 2010;30:962–7. [DOI] [PubMed] [Google Scholar]
- [41].Marcil V, Amre D, Seidman EG, et al. Hepatocyte nuclear factor 4 alpha polymorphisms and the metabolic syndrome in French-Canadian youth. PLoS One 2015;10:e0117238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Saif-Ali R, Harun R, Al-Jassabi S, et al. Hepatocyte nuclear factor 4 alpha P2 promoter variants associate with insulin resistance. Acta Biochim Pol 2011;58:179–86. [PubMed] [Google Scholar]
- [43].López MC, Palmer BE, Lawrence DA. Phenotypic differences between cord blood and adult peripheral blood. Cytometry B Clin Cytom 2009;76:37–46. [DOI] [PubMed] [Google Scholar]
- [44].Orkin SHND, Ginsburg D, Look AT, et al. Nathan and Oski's Hematology of Infancy and Childhood. Philadelphia, PA: Saunders Elsevier; 2009. [Google Scholar]
- [45].Wu HC, Wang Q, Delgado-Cruzata L, et al. Genomic methylation changes over time in peripheral blood mononuclear cell DNA: differences by assay type and baseline values. Cancer Epidemiol Biomarkers Prev 2012;21:1314–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].Herbstman JB, Wang S, Perera FP, et al. Predictors and consequences of global DNA methylation in cord blood and at three years. PLoS One 2013;8:e72824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].Wong CC, Caspi A, Williams B, et al. A longitudinal study of epigenetic variation in twins. Epigenetics 2010;5:516–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [48].Szyf M. The dynamic epigenome and its implications in toxicology. Toxicol Sci 2007;100:7–23. [DOI] [PubMed] [Google Scholar]
- [49].Kwon EJ, You YA, Park B, et al. Association between the DNA methylations of POMC, MC4R, and HNF4A and metabolic profiles in the blood of children aged 7-9 years. BMC Pediatr 2018;18:121. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


