Abstract
About one fourth of all newly identified cases of breast carcinoma are diagnoses of breast ductal carcinoma in situ (DCIS). Since we cannot yet distinguish DCIS cases that would remain indolent from those that may progress to life-threatening invasive ductal carcinoma (IDC), almost all women undergo aggressive treatment. In order to allow for more rational individualized treatment, we and others are developing in vitro models to identify and validate druggable pathways that mediate the transition of DCIS to IDC. These models range from conventional two-dimensional (2D) monolayer cultures on plastic to 3D cultures in natural or synthetic matrices. Some models consist solely of DCIS cells, either cell lines or primary cells. Others are co-cultures that include additional cell types present in the normal or cancerous human breast. The 3D co-culture models more accurately mimic structural and functional changes in breast architecture that accompany the transition of DCIS to IDC. Mechanistic studies of the dynamic and temporal changes associated with this transition are facilitated by adapting the in vitro models to engineered microfluidic platforms. Ultimately, the goal is to create in vitro models that can serve as a reproducible preclinical screen for testing therapeutic strategies that will reduce progression of DCIS to IDC. This review will discuss the in vitro models that are currently available, as well as the progress that has been made using them to understand DCIS pathobiology.
Keywords: 2D cultures, 3D cultures, Co-cultures, Tumor microenvironment, Natural matrices, Synthetic matrices, Engineered microfluidic platforms, Preclinical screens
Introduction
Last year roughly one out of every four newly diagnosed cases of breast carcinoma was ductal carcinoma in situ (DCIS) [1]. Many women diagnosed with DCIS will receive unnecessary care because we currently lack biomarkers to help differentiate between indolent pre-invasive lesions and those which may progress to invasive ductal carcinoma (IDC). There is an unmet need to identify biologically important markers in DCIS [2]. In order to help define these markers and allow for more rational, individualized treatment, models that accurately recapitulate disease are needed [3]. We and others are developing such systems in order to identify and validate pathways that play key roles in the transition of DCIS to IDC. The systems include mouse models, conventional two-dimensional (2D) monolayer cultures on plastic, and three-dimensional (3D) cultures in natural or synthetic matrices. Some culture models are comprised solely of DCIS cells (either cell lines or primary cells). Others are co-cultures, in which additional cell types found in the normal or cancerous human breast are added. 3D co-culture models can be particularly informative because they have been shown to accurately mimic the structural and functional changes in the architecture of the cancerous breast as shown elegantly by Bissell and colleagues (for review, see [4–6]). In this review article, we will present studies demonstrating that 3D co-culture models also can mimic the transition of DCIS to IDC. Cell:cell interactions that enhance this transition can be observed to occur over time. Adapting these in vitro models to engineered microfluidic platforms further facilitates the study of these cultures over more extended periods, providing another means of interrogating disease pathophysiology. Each model system has its own merits and can be a useful tool for answering specific questions. The advantages and disadvantages of in vivo modeling have been discussed in another review (Machado and Behbod). In this review we will focus on in vitro systems, their ability to recapitulate disease biology, and how they may serve as a reproducible and quantifiable pre-clinical screen for testing therapeutic strategies.
DCIS Cell Lines
Our understanding of cell culture has changed dramatically over the years. As a community, we now know that tumor cells do not “dedifferentiate” immediately upon being placed in culture; rather that early methods gave certain stromal components (e.g., fibroblasts) the edge they needed to take over the sample [7]. Similarly, our understanding of how to apply this technique to biomedical research has also evolved.
The maintenance of cells isolated from a single source on plastic, also known as 2D culture, acts as the data workhorse of many research labs around the world. This is especially true in the broad field of cancer research [8], where a plethora of cell lines are available and provide a relatively cheap, renewable source of biological material for the characterization of various tumor types. Generally speaking, when cultured in the absence of other cell types (mono-culture), these cell lines reliably express the hallmarks of cancer [9] with the exceptions of angiogenesis and evading immune destruction, which require endothelial and immune cells, respectively. This makes them ideal for identifying important oncogenes and tumor suppressors, discerning the mechanism of action for drugs, as well as understanding cell signaling pathways and how they may contribute to pathogenesis in a particular context. Of course, the relevance of any cell line is dependent on how closely it resembles the state that it is supposed to model, and with so many cell lines available some are inevitably less beneficial than others. Furthermore, the patient-to-patient heterogeneity of these diseases means that no single-source model will ever accurately represent all cases. Therefore, validating research findings using models comprised of cells from more than one line or patient will continue to be an important practice. This and other factors (reviewed in more detail here [10, 11]) have made the usefulness of cell lines a well-debated topic for many decades. Encouragingly, these matters have not only put a greater emphasis on careful study design and the proper handling of chosen lines, they have also led to the creation of more applicable model systems.
There are many human breast cancer cell lines available that can be employed to better understand various aspects of this malignancy. However, despite the fact that DCIS comprises such a high proportion of newly diagnosed cases of breast cancer, the same cannot be said for DCIS cell lines. This is likely due to the relatively small size of DCIS lesions as compared to established, invasive breast tumors. With less tissue, it is difficult to establish cell lines. As a result, the handful of established DCIS cell lines that do exist (MCF10DCIS.com in the MCF10 series, S3 in the HMT-3522 series, 21NT in the 21T series, h.DCIS.01, SUM102PT, and SUM225) have proved to be invaluable to the field, and represent a variety of breast cancer subtypes. Even so, established cell lines that are cultured over long periods of time can experience changes in their molecular profiles. A potential solution is the generation of new cell lines from patient-derived xenograft models or tumor cells, which should more closely resemble the original tumor.
HER2-Negative DCIS Cell Lines
MCF10DCIS.com is an ER-negative/PR-negative/HER2-negative basal-like cell line that recapitulates DCIS structures when grown in mice [12–14] and under 3D culture conditions [14, 15]. As the naming convention suggests, this line is part of the MCF10 series of breast cancer progression which also includes non-transformed breast epithelial cells (MCF10A), premalignant variants (MCF10AT), as well as malignant variants (MCF10CA). These isogenic lines originated from tissue provided by a 36 year-old woman who underwent a reduction mammoplasty for fibrocystic breast disease [16]. MCF10A cells immortalized spontaneously and do not form xenografts in nude mice. Cell lines derived from MCF10A, which contain a stably transfected T24 H-Ras oncogene, do grow as xenografts [17]. The MCF10DCIS.com line, cloned from an MCF10AT xenograft after it had undergone two successive trocar passages, forms predominantly high-grade comedo DCIS in vivo [12]. At 3 weeks these lesions are not invasive, however, some have been observed to progress to IDC after 9 weeks. Why some of the DCIS lesions progress to IDC is not known; it is however intriguing that this model recapitulates the heterogeneity observed in patients. Of potential relevance is that Natrajan and colleagues have reported that the MCF10 series harbors relevant driver alterations that are also seen in primary breast cancers (e.g., PIK3CA) [18]. Despite the infrequency of HRAS mutation in primary breast lesions, they conclude that these cell lines represent a good model for studying disease progression and evaluating potential biomarkers or therapeutic targets. At the same time, functional activation of wild-type Ras in breast cancers through overexpressed growth factors and their receptors [19] is thought to be a more common occurrence, and thus therapeutics that target Ras may be useful in disease treatment [20].
Another ER-negative/PR-negative/HER2-negative DCIS line is S3 of the HMT-3522 human breast epithelial cell line series (S1, S2, S3, T4-2) [21]. The HMT-3522 cell lines like the MCF10 cell lines are derived from a patient who underwent reduction mammoplasty for fibrocystic breast disease. S1 cells immortalized spontaneously and S2 cells were established through continuous cell passaging in defined medium. The S2 cells gave rise to both the pre-invasive S3 and invasive T4-2 cells through 3D culture in laminin-rich basement membrane or implantation in nude mice, respectively. In Boyden chamber assays, S1, S2, and S3 cell lines do not invade whereas T4-2 cells do. However, when S3 cells are exposed to conditioned medium from T4-2 cells they become invasive, with the S3-C variant having the highest invasive potential. Importantly, orthotopic transplantation of the series into the mammary fat pad results in increases in tumor frequency from S1 (no tumors) to T4-2 (many tumors) that parallel the rates of invasion in Boyden chamber assays. The HMT series is well-established [22] and models metaplastic disease, with the S3 line operating similarly to MCF10DCIS.com as a DCIS that is poised to invade [21].
SUM102PT is also an ER-negative/PR-negative/HER2-negative DCIS cell line (https://sumlineknowledgebase.com/?page_id=1181). The origin of this cell line differs from the two examples above as it was derived from a 56 year-old woman who had undergone neo-adjuvant chemotherapy followed by modified radical mastectomy for a minimally invasive apocrine adenocarcinoma with extensive DCIS [23]. This line is dependent on epidermal growth factor receptor (EGFR) signaling for growth, and over-expresses the receptor without gene amplification. SUM102PT is classified as a basal-like model of DCIS with micro-invasion [24] and has been studied in both 3D culture [14, 25] and nude mice [26].
Another example of an ER-negative/PR-negative/HER2-negative cell line is h.DCIS.01. This line was derived from a primary culture of hyperplastic breast epithelial cells (columnar cell hyperplasia) and form lesions in vivo that are similar to those formed by MCF10DCIS.com At later passages (>30) these cells spontaneously transformed in culture into a cell line exhibiting a DCIS-like phenotype [27].
HER2-Positive DCIS Cell Lines
HER2-positive DCIS lesions have a significantly higher risk for local in situ recurrence [28]. This positivity is also reported to increase the rate of upstaging to invasive breast cancer as compared to HER2-negative DCIS [29]. Therefore, cell lines that are HER2-positive are needed to model DCIS. To date, there are only two cell lines that fit this description: SUM225 and 21NT from the 21T series, both of which exhibit HER2 amplification. SUM225 was isolated through culturing a chest wall recurrence from a patient with DCIS treated by mastectomy [24]. Notably, SUM225 forms dysplastic structures in both 3D culture models [25] and in vivo in an intraductal mouse model [13]. The 21T series is comprised of four cell lines: 21PT, 21NT, 21MT-1, and 21MT-2. These were obtained from a 36 year-old woman who was first diagnosed with infiltrating and intraductal carcinoma, then developed lung metastases a year later [30]. 21PT and 21NT were collected from separate primary lesions and serve as models of atypical ductal hyperplasia and DCIS, respectively. 21MT-1 and 21MT-2 were both isolated from a metastatic pleural effusion after mastectomy and chemotherapy, with the former more closely resembling the primary tumor and the latter displaying a much more aggressive profile [30]. In 3D, 21PT cells, in contrast to 21NT cells, formed acinar structures with lumens [31]. The most striking difference between the 21PT and 21NT cells is their ability to form tumors in nude mice (21NT cells can form tumors, but 21PT cannot). 21NT xenografts mimic comedo DCIS with neoplastic cells filling the entire mammary fat pad duct, often with accompanying central necrosis.
ER-Positive/PR-Positive Cell Lines
There are few ER-positive/PR-positive models of DCIS. Lee and colleagues reported the creation of five ER-positive/PR-positive DCIS cell lines in 2014 [32]. All five cell lines derive from a single source, i.e., a pre-invasive DCIS excised from a 47 year-old Chinese woman. These cells were transfected with a pGRN145 vector containing hTERT and antibiotic resistance sequences. Single cell colonies were then allowed to grow under selection for at least 2 weeks, leading to the generation of ETCC006, ETCC007, ETCC008, ETCC010, and ETCC011 lines. Of these, ETCC006 (and to a lesser degree ETCC010) resembled the original lesion the closest with regard to expression of pan-cytokeratin, cytokeratin-19, vimentin, estrogen receptor α/β, and progesterone receptor. HER2 expression was indirectly assessed through treatment with trastuzumab/Herceptin and no response was reported. The parental line and ETCC008 were non-tumorigenic over a 90-day period as assessed by subcutaneous transplantation into severe combined immunodeficiency female mice. In contrast, ETCC006, ETCC007, ETCC010 and ETCC011 all showed tumor growth after 50 days. To date these lines have not been studied by other laboratories.
2D DCIS Mono-cultures as a Tool for Understanding Disease Biology
Some progress has been made in understanding how DCIS relates to invasive breast cancer using 2D mono-cultures. Porter and colleagues used MCF10DCIS.com 2D mono-cultures, xenografts, and patient tissue to elucidate the role of singleminded-2s (SIM2s) in malignant progression [33]. Their previous work had suggested that SIM2s is a tumor suppressor and that, by inhibiting epithelial to mesenchymal transition signaling and promoting differentiation, SIM2s maintains the integrity of the epithelium [34–38]. In a more recent paper [33], they analyzed the effect of modulating SIM2s expression on DCIS progression. SIM2s expression in patient tissues decreases from normal to DCIS to IDC and in parallel with malignancy of breast cell lines. Schribner et al. [33] found that stable knockdown of SIM2s in MCF10DCIS.com cells results in an increase in proliferation and invasiveness. In contrast, over-expression of SIM2s inhibits proliferation, yet has no effect on invasiveness. Contradictory changes in expression of several matrix metal-loproteinases (MMPs) are observed in xenografts generated from the cells in which SIM2s expression had been manipulated. MMP expression was analyzed in the xenografts, but not in the cells in mono-culture. Therefore, it is not possible to assess whether the contradictory changes in expression of MMPs reflect changes induced in host cells that have infiltrated into the xenografts, changes in the MCF10DCIS.com cells or a combination of the two. Porter and colleagues concluded that SIM2s may play a role in breast cancer progression, and that expression of the protein can promote tumor differentiation [33]. MCF10DCIS.com 2D monocultures have also been used to assess other possible treatment strategies that will induce DCIS differentiation, in this case of peroxisome proliferator-activated receptor gamma (PPARγ) [39]. PPARγ signaling is known to affect multiple cellular processes, including differentiation, proliferation, and apoptosis in both normal and cancerous tissue [40]. Ory et al. [39] tested effects of a third-generation PPARγ agonist, efatutazone. In 2D mono-cultures, they observed increased expression of luminal epithelial markers and decreased expression of basal epithelial markers, consistent with an effect on differentiation. They also analyzed effects on 3D cultures of MCF10DCIS.com cells grown in reconstituted basement membrane (rBM) and on xenografts. Tumorsphere or mammosphere formation by 3D rBM cultures was reduced as was invasiveness of the xenografts, both consistent with efatutazone treatment inducing differentiation. Thus, by using 2D mono-cultures of DCIS cells, two pathways that can induce differentiation and delay progression of DCIS have been identified. In both cases, the results have been validated in xenograft models. The complex changes in MMPs in the study on SIM2s and the changes in expression of inflammatory response pathway genes in the study on PPARγ suggest a need to consider possible effects on the microenvironment as well as on DCIS cells.
2D DCIS Cultures: Analyses of Interactions with Cellular and Pathochemical Tumor Microenvironment
Many studies have looked at the potential role of the microenvironment in DCIS progression using 3D cultures, yet only a few of these have used 2D cultures of DCIS cells with other cells present or in pathochemical conditions present in the microenvironment of breast cancers.
2D DCIS:Fibroblast Co-cultures
Boerner and colleagues [41] primarily investigated resistance to inhibitors of epidermal growth factor receptor (EGFR) in triple-negative breast cancers (TNBCs), which frequently over-express EGFR [42, 43]. They did, however, compare results to those for the SUM102PT DCIS cell line due to its reliance on EGFR and its being basal-like as are many TNBCs. EGFR can be activated not only by its ligand, EGF, but also through crosstalk with the MET signaling transduction pathway [44, 45]. MET signaling is commonly activated when the ligand for MET, hepatocyte growth factor (HGF), is present at high levels [46]. Therefore, if MET signaling is active in the presence of an EGFR inhibitor, this crosstalk could negate the therapeutic effects of the EGFR inhibitor. HGF is not expressed in epithelial cells, but is secreted by fibroblasts in the tumor stroma [41]. To recapitulate this environment, SUM102PT was co-cultured with HGF-expressing fibroblasts. This resulted in Met phosphorylation and DNA synthesis in the presence of the EGFR inhibitor gefitinib. Fibroblast-conditioned medium, which contains HGF, also stimulates clonogenic survival in the presence of gefitinib. Mueller et al. [41] concluded that EGFR/Met crosstalk, mediated by tumor-stromal interactions, may play a role in the resistance of breast cancer patients to tyrosine kinase inhibitors. Importantly, this work also suggests that resistance mediated by crosstalk between breast cancer cells and fibroblasts can occur in premalignant disease as well as IDC.
2D DCIS:Adipocyte Co-cultures
Zhou and colleagues have reported that co-culturing adipocytes with either of the two DCIS cell lines, MCF10DCIS.com or SUM102PT, results in increased migration [47]. They also observed a more aggressive phenotype of MCF10DCIS.com, characterized by enhanced invasiveness and increased tumor growth in vivo. In a subsequent paper, they examined how inhibiting exosome trafficking from pre-adipocytes influenced the ability of MCF10DCIS.com to migrate in 2D culture and grow as tumors in nude mice [48]. Their rationale for this study was that tumor cells can actively influence certain cell types or cellular processes in their immediate vicinity to create a more hospitable niche, and this process promotes their continued growth and survival [49]. The secretion of exosomes, then, is one way that tumor cells are able to influence their microenvironment. This rationale appeared correct, as migration of MCF10DCIS.com cells did increase when the cells were co-cultured with exosomes [48]. Conversely, when pre-adipocytes are treated with shikonin (an anti-tumor compound that induces necroptosis and also inhibits cancer cell glycolysis [50]) before co-culturing with the DCIS cells, the migration of DCIS cells is less than observed in co-cultures with untreated pre-adipocytes. Importantly, the authors observed a similar trend when tumor growth was assessed in vivo: exosome co-culture increases tumor growth, and treating pre-adipocytes with shikonin before incubating the DCIS cells with adipocyte exosomes partially reverses this effect. From these results they concluded that intercellular signaling mediated by adipocyte exosomes in the tumor microenvironment is important for the regulation of breast tumorigenesis and disease progression.
2D DCIS Cultures and Acidosis
Gillies and colleagues have investigated whether chronic acidosis, which is characteristic of the pathochemical microenvironment of solid tumors, has an effect on malignant progression of DCIS [51]. They had previously demonstrated that the acidic microenvironment of solid tumors promotes tumor progression by stimulating invasion and metastasis (as reviewed in [52–54]), as well as by degrading/remodeling the extracellular matrix (ECM) to create an environment that is toxic to normal cells [55]. Interestingly, they propose that these changes may arise from the tumor selecting for phenotypes that can overcome the proliferative barriers created by the microenvironment [52]. However, the mechanisms by which cancer cells survive these conditions are not well understood. Through proteomic screening of acid-adapted and acid-naïve MCF-7 breast cancer cells [51], they found that lysosome-associated membrane protein-2 (LAMP2) expression is significantly increased with adaption. They confirmed these findings in other breast cell lines and also in MCF10DCIS.com cells cultured in 2D under acidic conditions. The increase in LAMP2 expression in MCF10DCIS.com cells and an increase in LAMP2 expression in DCIS patient samples compared to normal breast tissue samples suggests that this change precedes the transition to IDC. Further examination of stained patient samples revealed that LAMP2 expression is high in areas of DCIS microinvasion—a location in which acidification potentially promotes disease progression. Gillies and colleagues propose that under evolutionary pressure cancer cells adapt to a chronically acidic environment by increasing lysosomal turnover. This, in turn, leads to an increase of LAMP2 at the plasma membrane.
As indicated by the studies above, the microenvironment has a strong influence on the behavior of a tumor. Factors secreted from surrounding cellular components are not only able to promote migration and growth, they are able to influence drug resistance. Moreover, when the local environment is chronically acidic, tumor cells can adapt by increasing lysosomal turnover to create a survival advantage. These observations of environmental adaptation also offer an explanation for the heterogeneity commonly observed in the tumor mass (i.e., there are many genomic paths that lead to a common phenotype).
3D DCIS Mono-cultures as a Tool for Understanding Disease Biology
Culturing tumor cells in 2D, even with the addition of other relevant cell types, only provides a small part of the biological picture because many of the checks and balances are missing. 3D cultures in which tumor cells grow in an architecture resembling the cell:cell interactions and cell:ECM interactions that are found in vivo are more relevant (see Table 1 for comparisons of benefits and limitations of culture systems) and are proving to be a preclinical model that can predict drug efficacy and toxicity (for review, see [75–78]). This has been shown in studies using DCIS cell lines and cells isolated from patient samples by Bundred and colleagues [79–82]. The DCIS cells are grown in non-adherent 3D cultures as mammospheres, an assay that selects for cells that exhibit stem-like properties and express E-cadherin on their surface [83]. The only matrix in the mammosphere cultures is that produced by the DCIS cells themselves. Formation of mammospheres by DCIS cells is reduced by inhibiting EGFR or Notch signaling pathways with gefitinib and Notch 4-neutralizing antibody, respectively [79]. Drug combinations that inhibit EGFR (e.g., gefitinib) or Her2/neu (e.g., lapatinib) and Notch indirectly (e.g., DAPT (N-[N-(3,5-difluorophenacetyl-L-alanyl)]-S-phenylglycine t-butyl ester), a gamma-secretase inhibitor) are more effective [80]. Formation of mammospheres by DCIS cells also can be reduced by targeting a novel focal adhesion kinase-Wnt/beta-catenin signaling pathway that regulates DCIS stem cell activity [82]. The mammosphere studies thus support the potential of 3D DCIS mono-cultures as pre-clinical models for drug discovery.
Table 1.
Comparison of the advantages and disadvantages of 2D culture systems vs. 3D mono-culture, co-culture, and 3D chamber systems relevant to breast cancer [18, 22, 56–7,3]
| Model system | Benefits | Limitations |
|---|---|---|
| 2D monoculture | • Relatively fast data acquisition [61] • Cost effective • Discover new mechanisms and pathways in cells • Provides a controlled environment • Can perform certain types of experiments (e.g., cellular fractionation, pull-downs) • Can be applied to high-throughput applications |
• No interactions between cells and the ECM or other aspects of the tissue microenvironment [63, 74] • Not necessarily reflective of patients |
| 3D culture in synthetic matrix (e.g., agarose) | • Discover new mechanisms and pathways in cells • Provides a controlled environment • Scalable to different plate format • Can be applied to high-throughput applications • Can also accommodate co-culture techniques • Can study impact of microenvironment [64] • Can mine for genetic drivers in malignant progression [18] • Can simulate physical environment [65] |
• Acquisition of data takes time [66] • Can be expensive • May require specialized devices and software • Limits techniques due to the presence of matrix • Long-term culture is difficult to achieve • Difficult to apply to high-throughput applications |
| 3D rBM monoculture | • More accurately represents gene [67] and protein expression [68], as well as drug [60, 68] and radiation response [69] than 2D culture • Better recapitulates changes in cellular polarity [70], responses to environmental cues [22], and presents a more accurate cell signalling profile [71] than 2D culture or synthetic matrices. • Facilitates study and characterization of proteases involved in ECM remodeling [56, 72] |
• Acquisition of data takes time • Expensive • Limits techniques due to the presence of matrix • Matrix may differ from lot to lot (leading to inconsistencies in cell behavior) • Long-term culture is difficult to achieve • Difficult to apply to high-throughput applications |
| 3D rBM co-culture | • Incorporates multiple cell types present in the microenvironment such as fibroblasts [57] and myoepithelial cells [58] for a more accurate depiction of tissue niche • Direct and indirect methods can elucidate the need for physical cellular interation |
• Acquisition of data takes time • Expensive • Limits techniques due to the presence of matrix • Matrix may differ from lot to lot (leading to inconsistencies in cell behavior) • Long-term culture is difficult to achieve • Multiple cell types need to be distinguishable from one another (e.g., fluorescent tags for microscopy) • Difficult to apply to high-througput applications • Ratio of cell types plays a role in the outcome |
| 3D chamber sytems | • Facilitates analysis over long periods of time [73] • Facilitates investigation of environmental factors of the invasion of ductal carcinoma [59] |
• Acquisition of data takes time • Expensive • Limits techniques due to the presence of matrix • Requires specialized devices • Difficult to apply to high-throughput applications |
Mammosphere studies might be interpreted as evidence that interactions of DCIS cells with surrounding matrices are of little importance for drug discovery. Nonetheless, there are matrix-related risk factors for breast cancer development. These include increases in mammographic density due to elevated collagen deposition [84]; increases in matrix stiffness due to changes in the mechanical properties of collagen (for review, see [85]); and changes in matrix topography due to realignment of collagen fibers [86] all are risk factors for breast cancer development. Changes in collagen I have been observed in DCIS in patients and in mouse models. For example, there are increases in fibrillar collagen in DCIS in a mouse model of postpartum breast cancer [87]. At the sites of stromal reactions in DCIS lesions, two proteins that will increase matrix stiffness, the collagen cross-linker lysyl oxidase and a lysyl oxidase-like protein, colocalize [88]. In addition, increased expression of focal adhesion kinase, a downstream pathway in stimulation of collagen I production [89, 90], is observed in DCIS lesions [91]. Wei et al. [92] have evaluated the effect of matrix stiffness on MCF10DCIS.com cells grown in 3D culture. In this case, the cells were grown in a complex system comprised of rBM overlay cultures plated on collagen I-coated glass slides of calibrated Pascals of stress. Increased matrix stiffness induced nuclear translocation of the transcription factor TWIST1 in the DCIS cells. This response to increased matrix stiffness is one conserved in breast epithelial cells. When the cytoplasmic partner of TWIST1, the Ras GTPase-activating protein-binding protein 2 or G3BP2 was knocked down in the MCF10DCIS.com cells, the TWIST1-G3BP2 mechanotransduction pathway was activated, inducing epithelial to mesenchymal transition and invasiveness. These results, however, are inconsistent with those of Gupta et al. [93] who reported that high expression of G3BP2, which regulates initiation of breast cancer, is associated with poor survival of breast cancer patients.
Studies on DCIS progression have focused on two different hypotheses for the acquisition of invasiveness that characterizes the transition from DCIS to IDC: 1) molecular and genetic alterations in the DCIS cells or 2) alterations resulting from cross-talk between DCIS cells and their microenvironment including matrices as discussed above or stromal cells and pathochemical factors as discussed below (see “3D DCIS cultures: analyses of interactions with tumor microenvironment”). Profiling of DCIS patient samples has revealed significant changes in gene expression in association with the transition to IDC [27, 94–97], similarities in gene expression and mutation in epithelial cells from in situ and invasive regions [98–101], and both changes and similarities [99, 102]. These disparate results may be due in part to difficulties in obtaining DCIS lesions from patients that do not contain contaminating cells even when using laser capture microdissection techniques [103, 104]. Another approach is to isolate RNA from cultured DCIS cells [25, 105].
We have performed next-generation sequencing of RNA isolated from 3D rBM mono-cultures of three DCIS cell lines and the MCF10A non-transformed human breast epithelial cell line. Our transcriptome DCIS signature [25] (NCBI: GSE 36863) was validated by the identification of genes previously associated with DCIS, i.e., transforming growth factor beta 1 (TGFB1) [106], dystonin (DST) [27], high-temperature requirement a serine peptidase 1 (HTRA1) [107], and gap junction protein beta 2 (GJB2) [108]. Using the Genomatix Pathway System we identified two highly-enriched common frameworks (336-fold and 254-fold) in the promoters of 3 and 4 genes, respectively, in the human genome. Pursuing these potential target genes has led to some intriguing findings, not only for those that appear to play a role in DCIS progression, such as Rap 1Gap [109], but also for those which can influence the invasive capability of malignant breast cancer cells. An example of the latter is the gene encoding for C-C motif chemokine ligand 20 (CCL20), also known as macrophage inflammatory protein-3 alpha (MIP-3α). Secretion of CCL20/MIP-3α increases in 3D rBM mono-cultures of MCF10 variants from atypical hyperplastic to DCIS and remains high in the isogenic IDC cell line. Furthermore, when highly malignant MDA-MB-231 cells are grown under the same conditions but in the presence of a neutralizing antibody to human CCL20, we observed a reduction in both invasion and proteolysis (Osuala and Sloane, unpublished data). Validating the functional roles of targets such as those found in our screen may identify biomarkers for discriminating indolent lesions from premalignant lesions and also may reveal new therapeutic targets.
3D DCIS Cultures: Analyses of Interactions with Tumor Microenvironment
The microenvironment makes important contributions to modulating DCIS progression to IDC ([56, 57, 97, 110]; see also review by Nelson et al.). 3D cultures that model interactions between DCIS and its microenvironment have been used to analyze the mechanisms inducing transition of pre-invasive DCIS to an invasive phenotype [56–59, 111, 112].
3D DCIS:Fibroblast Co-cultures
Carcinoma-associated fibroblasts (CAFs) are the most abundant cell component of the tumor microenvironment and communicate with cancer cells via secretion of growth factors and chemokines (for recent comprehensive reviews, see [113, 114]). They play a variety of roles in promoting: 1) cancer cell proliferation and survival; 2) metastasis; and 3) resistance to therapy. Studies using 3D rBM co-culture models of DCIS cells and CAFs have shown that proteases and cytokines are among the mechanisms that promote the DCIS-to-IDC transition (Fig. 2), findings also validated in in vivo studies [14, 56, 57, 118]. Hu et al. [118] showed that DCIS cells in co-culture with CAFs exhibit increased invasiveness. There are also increases in cyclooxygenase-2 in the DCIS cells and in secretion of MMP-9 and −1 from the DCIS cells. We demonstrated that secretion of interleukin-6 by CAFs in co-culture with DCIS cells results in increased growth and invasiveness of the DCIS cells, in part through ECM degradation by the cysteine protease cathepsin B [57]. Gascard and Tlsty [114] cited the latter study for extending the roles of CAFs and reactive stroma to earlier in malignant progression, i.e., during the transition from DCIS to IDC.
3D DCIS:Myoepithelial Cell Co-cultures
Myoepithelial cells (MEPs) play a role in: 1) polarity of the epithelial cells in normal mammary acini, 2) synthesis and maintenance of basement membrane and 3) lactation [119]. Loss of the myoepithelial cell layer, degradation of the underlying basement membrane, and invasion of tumor cells into the stroma are hallmarks of DCIS-to-IDC progression. Moreover, MEPs exert an inhibitory effect on cancer growth and invasion by secretion of protease inhibitors (e.g., myoepithelium-derived serine proteinase inhibitor [120]; TIMP-1 [121]; secretion of anti-angiogenic mediators [121] (e.g., thrombospondin-1, plasminogen, and prolactin); and expression of potential tumor suppressor proteins such as relax-in, activin, connexin-43, and neogenin [121, 122]. In 3D rBM co-cultures with DCIS cells, MEPs can be shown to exert inhibitory effects on growth, invasiveness, and proteolysis of the DCIS cells, reliant on high levels of secretion of plasminogen activator inhibitor-1, an inhibitor of urokinase plasminogen activator [56]. Duivenvoorden et al. [123] have shown that another protease inhibitor, i.e., stefin A, an inhibitor of cysteine cathepsins, is abundant in MEPs and that stefin A suppresses DCIS invasion, an inhibitory effect reliant on cathepsin B. Targeted downregulation of stefin A in MEPs promotes invasion [123].
3D DCIS:Myoepithelial Cell:Fibroblast Co-cultures
We have also used a 3D tri-culture model to more closely mimic the in vivo architecture of breast tumors in the context of their microenvironment [56]. By live-cell confocal imaging we observed and quantified changes in the malignant phenotype and proteolytic activity of pre-invasive MCF10DCIS.com cells co-cultured with CAFs, MEPs or both cell types. We analyzed tri-cultures in which the CAFs were embedded in type I collagen below the breast epithelial cells and MEPs in rBM (Fig. 1a). These multi-matrix layered co-cultures mimicked the in vivo location of CAFs in surrounding stroma prior to their infiltration into the DCIS lesions. They also allowed us to observe dynamic and temporal changes in migration of CAFs toward, and their invasion into, DCIS structures over time in culture. The studies in the multi-matrix layered co-cultures were compared to ones in rBM mixed co-cultures (Fig. 1b), the latter mimicking DCIS lesions into which CAFs have infiltrated. By spiking the matrices used in these co-cultures with 1% dye quenched (DQ)-collagen I and 0.2% DQ-collagen IV in collagen I and rBM, respectively, we can image and quantify degradative activity [56]. Representative examples of the two types of co-cultures show that CAFs increase degradative activity and invasiveness of DCIS cells, in association with migration of CAFs toward DCIS structures (Fig. 2a and b, top). MEPs suppress the CAF-induced degradative activity and invasiveness of DCIS structures in the multi-matrix layered co-cultures at both 8-and 21-days (Fig. 2a, bottom). In contrast, MEPs do not suppress degradative activity in rBM mixed co-cultures in which CAFs are in direct contact with the DCIS structures (Fig. 2b, bottom). Our data thus demonstrate that 3D co-cultures can be used to evaluate dynamic and temporal interactions among DCIS cells, CAFs, and MEPs, and should be informative about therapeutic approaches to reduce progression of DCIS to IDC. Similar models are likely to be able to assess cross-talk between DCIS cells and other cell types (e.g., adipocytes and immune cells) in the transition to IDC as well as to understand effects on progression induced by pathochemical aspects of the microenvironment.
Fig. 1. 3D co-culture models for analysis of DCIS transition to invasive phenotype.
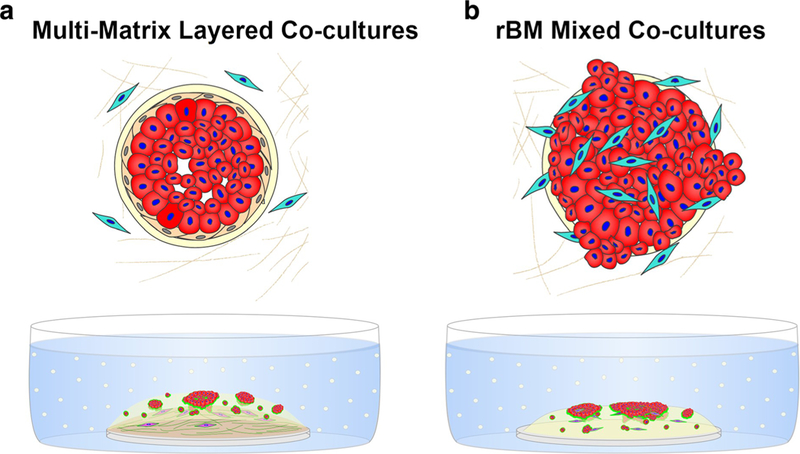
Top: a Architecture of pre-invasive DCIS in which the basement membrane (cream) and layer of myoepithelial cells (MEPs; beige) surrounding DCIS cells (red) are intact. Fibroblasts (cyan) are present in surrounding stroma. b Architecture of microinvasive DCIS in which areas of basement membrane (cream) are disrupted, MEPs (beige) lost, and fibroblasts (cyan) have infiltrated into lesion. Bottom: Pathomimetic 3D co-cultures to model pre-invasive (a) and microinvasive (b) DCIS. a In multi-matrix layered co-cultures, DCIS cells and fibroblasts are cultured in relevant matrix: fibroblasts (fuchsia)in lower layer of type I collagen and DCIS cells (red) ± breast myoepithelial cells (unlabeled) in a top layer of rBM as in a rBM overlay culture. To follow proteolysis, quenched fluorescent substrates (dyequenched (DQ)-collagens IV and I) are mixed with rBM and collagen I, respectively. Green represents the fluorescent cleavage products of these substrates. b In the rBM mixed co-cultures, a mixture of DCIS cells ± MEPs and fibroblasts are plated in a rBM overlay culture. To follow proteolysis, quenched fluorescent substrate (DQ-collagen IV) is mixed with rBM. Green represents the fluorescent cleavage products of these substrates
Fig. 2. Images of 3D co-cultures illustrating increases in DCIS invasive phenotype induced by co-culture with CAFs and decreases induced by co-culture with MEPs.
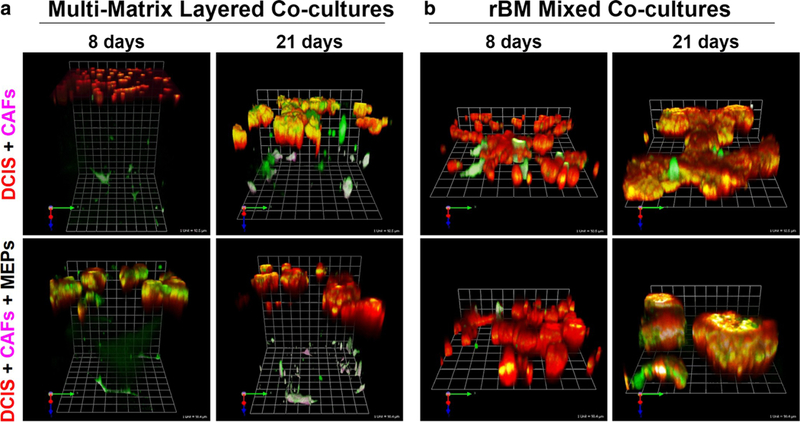
Multi-matrix layered co-cultures (a) and rBM mixed co-cultures (b), comprised of MCF10DCIS.com-lenti- RFP cells (red), WS-12Ti-lenti-YFP CAFs (pseudocolored fuchsia) and/or MEPs (unlabeled), are described in legend to Fig. 1. Structures and interactions were imaged live by confocal microscopy and proteolysis was assessed by a live-cell proteolysis assay developed in our laboratory [115]. Green fluorescence in A represents degradation products of DQ-collagen IV in the top layer of rBM and DQ-collagen I in the bottom layer of type I collagen. Green fluorescence in B represents degradation products of DQ-collagen IV in rBM. Degradation at surface of structures or cells appears pink-white on CAFs and yellow-white on DCIS cells. Representative angled views of 8- and 21-day co-cultures are tiled from 16 contiguous fields of optical sections taken through the entire depth of the 3D co-cultures and reconstructed in 3D with Volocity software; one grid unit = 92 μm (top) and 90 μm (bottom). For details on assembling and imaging the co-cultures and detailed methodologies for live-cell proteolysis assay and quantification of cell numbers, growth in 3D, degradation products and invasiveness, see [56, 115–117]
3D DCIS:Macrophage Co-cultures
Inflammatory cells, such as macrophages and lymphocytes, are recruited into tumors, where they play a critical role in tumor-associated inflammation (see reviews, [124, 125]. Macrophages infiltrating into tumors, termed tumor-associated macrophages, are composed of multiple subpopulations with overlapping phenotypes [126]. Thus, tumor-associated macrophages contribute to: 1) tumor progression through production of IL-10 and CCL17, 18, 22 andmannose receptor and 2) tumor suppression through secretion of IL-1, −12, and TNF-alpha [126, 127]. In 3D rBM co-cultures, bone marrow-derived macrophages have been shown to induce an invasive phenotype in pre-malignant cells: non-invasive murine mammary epithelial cells [128] and pre-invasive PN1a murine ductal hyperplastic cells [129]. In the study of non-invasive epithelial cells, migration and invasion were promoted by the macrophage-derived chemokine receptor CXCR2 [128]. CXCR2 is implicated in breast cancer progression as targeting CXCR2 in mouse mammary carcinoma models enhances responses to chemotherapy and inhibits tumor growth, angiogenesis, and lung metastasis [130].
3D DCIS Cultures in Engineered Microfluidic Platforms
Despite the advantages of 3D culture models, they only partially replicate in vivo tissue:tissue interactions and tissue geometries/structures that are of pathophysiological relevance [131, 132], and they do not allow for analyses of cell:cell interactions and paracrine signaling over extended times in culture. Engineered microfluidic devices have emerged recently that can support models recapitulating tumor progression, including dynamic models in which one can observe spatiotemporal changes in cell:cell interactions that accompany the transition of DCIS to invasiveness (e.g., see [111]) and interactions with other aspects of the tumor microenvironment such as acidosis, hypoxia, and concentration gradients of growth factors, cytokines, etc.
Microfluidic 3D culture systems can be designed to meet the requirements of specific studies. This can include multiple compartments or channels in which to grow individual cell types such as DCIS cells and fibroblasts [59], thus generating a phenotype in vitro that more closely resembles the in vivo structure. Alternatively, distinct physical compartments fabricated from silicon-based organic polymers such as polydimethylsiloxane can be used to separate cell types (e.g., the microchannels in Bischel et al. [59] and Choi et al. [112]). Separation of cell types may be achieved by layering of matrices as shown in Fig. 1 and in Choi et al. [112]. Formation of structures resembling mammary acini and ducts may be achieved by plating breast epithelial cells and breast myoepithelial cells sequentially in rBM [56] or by viscous finger patterning of lumens using hydrogels that differ in viscosity [59]. This variety of techniques can generate platforms with well-defined spatial control of specific cell types and channels that can be used for temporal control of additions such as culture medium or cells.
Beebe and colleagues have developed a series of microfluidic devices for studying the transition of DCIS to IDC. In one for co-culture of MCF10DCIS.com cells and fibroblasts [111], they provide spatial control of the two cell types by loading them simultaneously in two compartments by pumping. They observed transition of the DCIS cells to an invasive phenotype in regions where there is physical contact between the two cell types, consistent with observations in xenografts [14]. The pumping system in this device can also be used for analyses of temporal control by sequential loading of the MCF10DCIS.com cells and fibroblasts, (i.e., loading the DCIS cells and allowing them to form structures for 6 days prior to loading the fibroblasts). Here, Beebe and colleagues found that the invasive transition of DCIS occurs in regions where the two cell types are close. In contrast, DCIS further away from the fibroblasts retains a non-invasive phenotype. Truong et al. [133] have developed a dual chamber/dual matrix microfluidic device for quantitative analysis of single cell invasion and the invasive front of breast cancer cells, but to our knowledge this has not been used for studies on DCIS cells. Choi et al. [112] have developed a breast cancer-on-a-chip microdevice for use as a predictive tool in DCIS diagnosis as well as a drug screening platform. This model uses a compartmentalized 3D platform in which two channels are separated by a vitrified collagen membrane. Human primary fibroblasts are cultured in the lower chamber in a collagen gel. The collagen membrane is coated with rBM on which a non-transformed breast epithelial cell line is grown to confluence. Spheroids of MCF10DCIS.com cells are then generated on a hanging drop plate and placed on top of the epithelial cells. Over a culture period of 2–7 days, the DCIS spheroids incorporate into the epithelial layer and the fibroblasts retain a spindle-shaped morphology in the collagen gel. An advantage of this DCIS-on-a-chip model is that drugs can be introduced to the lower channel to simulate intravenous administration in patients. For example, introduction of the taxane paclitaxel inhibits the growth of the DCIS spheroids, supporting the potential of this model for testing of drug efficacy and toxicity. DCIS, however, do not grow as spheroids in vivo, but form irregularly shaped structures that often include areas of microinvasion. The imposition of a spherical architecture may limit the ability of this DCIS-on-a-chip model to accurately predict drug efficacies.
2D cultures have been used to model drug resistance by adding successively higher concentrations of drugs as the cells are passaged [134]. In patients, however, resistant subpopulations of tumor cells may arise over the course of drug administration at a single concentration. As we have shown for DCIS and IDC cells [60, 116], 3D cell cultures are more predictive of drug responses than are 2D monolayer cultures. This is hypothesized to reflect the architecture of cell:cell and cell:ECM interactions in 3D cultures [135]. One potential advantage of 3D cultures that is not yet routinely exploited is that drug treatment can be initiated either early or during the course of the development of structures, allowing results to be compared with other preclinical models (e.g., animal studies where drug treatment is initiated after the tumors are well established) [116]. In 3D DCIS co-cultures, the effects of drugs on the complex interplay between DCIS cells and other cell types and resistance mechanisms can be assessed. This interplay will vary with time and may include changes in cytokine secretion, proteolytic activity, and morphology such as formation of multicellular invasive outgrowths. Quantitative assays will be needed such as those that can be performed with cultures growing in engineered microfluidic platforms (e.g., platforms that allow regulated delivery of drugs at specific time points [136] and platforms designed for direct or indirect interactions among the cell types [111, 136]).
Transition of DCIS to IDC involves changes in various pathophysiological processes such as matrix remodeling and paracrine signaling which leads to local invasion and eventually metastasis to distant organs. These processes occur over extended periods of time that are not readily recapitulated in vitro. Therefore, we have fabricated and tested microfluidic platforms that will allow us to image and quantify cell:cell and cell:matrix interactions in 3D cultures over long periods of time and over these same periods collecting conditioned media for analysis of the secretome without disturbing the cultures. The culture wells in our platforms are fabricated from polymers of acrylates [136]. We chose acrylic due to concerns about leaching and absorption in devices made of polydimethylsiloxane (for discussion, see [74,137,138], which will be of higher concern for devices intended for long-term cultures. Using our platforms, we have cultured MDA-MB-231 human breast cancer cells alone and with human breast CAFs and normal fibroblasts for up to 70 days (Ji and Sloane, unpublished data). We are using our platforms for studies of how interactions of breast cancer cells, including MCF10DCIS.com cells, are affected by interactions with other cell types associated with malignant progression (e.g., microvascular endothelial cells of lymphatic and vascular origin), by acidification of the tumor microenvironment, and by a variety of therapeutic approaches designed to target cell:cell interactions and acidosis.
Conclusions
In vitro models to distinguish indolent DCIS from those lesions that will progress to IDC are needed to identify and validate druggable pathways that mediate the transition of DCIS to IDC. Such studies are limited in 2D monolayer cultures in part because 2D cultures do not include cell:cell and cell:ECM interactions comparable to those present in vivo. DCIS in vivo models provide valuable information by capturing the complexity and physiological pathways present in the body; however, it is difficult, expensive, and time-consuming to perform mechanistic and quantitative studies in these models. Recent advances in 3D culture models in combination with tissue engineering and microfabrication techniques are overcoming the limits of traditional 2D cultures and better mimic tissue architecture in vivo. More pathophysiologically relevant 3D models are being generated by co-culturing DCIS cells with other cell types that positively or negatively impact progression to IDC as well as under pathochemical conditions such as acidosis that impact progression. These models are proving to be powerful tools for identifying pathways that mediate progression and for testing drugs that target those pathways. Adapting such models to engineered microfluidic platforms will increase their reproducibility, facilitate mechanistic studies of dynamic and temporal changes associated with the DCIS to IDC transition, and increase their use as preclinical screens of therapies to reduce progression of DCIS to IDC.
Abbreviations
- 2D
Two-dimensional
- 3D
Three-dimensional
- DCIS
Ductal carcinoma in situ
- IDC
Invasive ductal carcinoma
- ER
Estrogen receptor
- PR
Progesterone receptor
- HER2
Human epidermal growth factor receptor 2
- SIM2s
Singleminded-2s
- TNBCs
Triple-negative breast cancers
- EGF
Epidermal growth factor
- EGFR
Epidermal growth factor receptor
- HGF
Hepatocyte growth factor
- MET
Hepatocyte growth factor receptor
- PPARγ
Peroxisome proliferator-activated receptor gamma
- rBM
Reconstituted basement membrane
- MMP
Matrix metalloproteinase
- ECM
Extracellular matrix
- LAMP2
Lysosome-associated membrane protein-2
- CCL20
C-C motif chemokine ligand 20
- CAF
Carcinoma-associated fibroblast
- MEPs
Myoepithelial cells
- DQ
Dye quenched
References
- 1.Siegel RL, Miller KD, Jemal A. Cancer statistics, 2018. CA Cancer J Clin. 2018;68(1):7–30. [DOI] [PubMed] [Google Scholar]
- 2.Lari SA, Kuerer HM. Biological markers in DCIS and risk of breast recurrence: a systematic review. J Cancer. 2011;2:232–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cowell CF, Weigelt B, Sakr RA, Ng CK, Hicks J, King TA, et al. Progression from ductal carcinoma in situ to invasive breast cancer: revisited. Mol Oncol. 2013;7(5):859–69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Weigelt B, Bissell MJ. Unraveling the microenvironmental influences on the normal mammary gland and breast cancer. Semin Cancer Biol. 2008;18(5):311–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Weigelt B, Ghajar CM, Bissell MJ. The need for complex 3D culture models to unravel novel pathways and identify accurate biomarkers in breast cancer. Adv Drug Deliv Rev. 2014;69–70: 42–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Schmeichel KL, Bissell MJ. Modeling tissue-specific signaling and organ function in three dimensions. J Cell Sci. 2003;116(Pt 12):2377–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sato G Tissue culture: the unrealized potential. Cytotechnology. 2008;57(2):111–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sharma SV, Haber DA, Settleman J. Cell line-based platforms to evaluate the therapeutic efficacy of candidate anticancer agents. Nat Rev Cancer. 2010;10(4):241–53. [DOI] [PubMed] [Google Scholar]
- 9.Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell. 2011;144(5):646–74. [DOI] [PubMed] [Google Scholar]
- 10.van Staveren WC, Solis DY, Hebrant A, Detours V, Dumont JE, Maenhaut C. Human cancer cell lines: experimental models for cancer cells in situ? For cancer stem cells? Biochim Biophys Acta. 2009;1795(2):92–103. [DOI] [PubMed] [Google Scholar]
- 11.Gillet JP, Varma S, Gottesman MM. The clinical relevance of cancer cell lines. J Natl Cancer Inst. 2013;105(7):452–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Miller FR, Santner SJ, Tait L, Dawson PJ. MCF10DCIS.com xenograft model of human comedo ductal carcinoma in situ. J Natl Cancer Inst. 2000;92(14):1185–6. [DOI] [PubMed] [Google Scholar]
- 13.Behbod F, Kittrell FS, LaMarca H, Edwards D, Kerbawy S, Heestand JC, et al. An intraductal human-in-mouse transplantation model mimics the subtypes of ductal carcinoma in situ. Breast Cancer Res. 2009;11(5):R66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jedeszko C, Victor BC, Podgorski I, Sloane BF. Fibroblast hepatocyte growth factor promotes invasion of human mammary ductal carcinoma in situ. Cancer Res. 2009;69(23):9148–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Li Q, Mullins SR, Sloane BF, Mattingly RR. p21-activated kinase 1 coordinates aberrant cell survival and pericellular proteolysis in a three-dimensional culture model for premalignant progression of human breast cancer. Neoplasia. 2008;10(4):314–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Soule HD, Maloney TM, Wolman SR, Peterson WD Jr, Brenz R, McGrath CM, et al. Isolation and characterization of a spontaneously immortalized human breast epithelial cell line, MCF-10. Cancer Res. 1990;50(18):6075–86. [PubMed] [Google Scholar]
- 17.Basolo F, Elliott J, Tait L, Chen XQ, Maloney T, Russo IH, et al. Transformation of human breast epithelial cells by c-Ha-ras oncogene. Mol Carcinog. 1991;4(1):25–35. [DOI] [PubMed] [Google Scholar]
- 18.Maguire SL, Peck B, Wai PT, Campbell J, Barker H, Gulati A, et al. Three-dimensional modelling identifies novel genetic dependencies associated with breast cancer progression in the isogenic MCF10 model. J Pathol. 2016;240(3):315–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Eckert LB, Repasky GA, Ulku AS, McFall A, Zhou H, Sartor CI, et al. Involvement of Ras activation in human breast cancer cell signaling, invasion, and anoikis. Cancer Res. 2004;64(13):4585–92. [DOI] [PubMed] [Google Scholar]
- 20.Li T, Sparano JA. Inhibiting Ras signaling in the therapy of breast cancer. Clin Breast Cancer. 2003;3(6):405–16. discussion 17–20 [DOI] [PubMed] [Google Scholar]
- 21.Rizki A, Weaver VM, Lee SY, Rozenberg GI, Chin K, Myers CA, et al. A human breast cell model of preinvasive to invasive transition. Cancer Res. 2008;68(5):1378–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wang F, Weaver VM, Petersen OW, Larabell CA, Dedhar S, Briand P, et al. Reciprocal interactions between |31-integrin and epidermal growth factor receptor in three-dimensional basement membrane breast cultures: a different perspective in epithelial biology. Proc Natl Acad Sci. 1998;95(25):14821–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sartor CI, Dziubinski ML, Yu CL, Jove R, Ethier SP. Role of epidermal growth factor receptor and STAT-3 activation in autonomous proliferation of SUM-102PT human breast cancer cells. Cancer Res. 1997;57(5):978–87. [PubMed] [Google Scholar]
- 24.Forozan F, Veldman R, Ammerman CA, Parsa NZ, Kallioniemi A, Kallioniemi OP, et al. Molecular cytogenetic analysis of 11 new breast cancer cell lines. Br J Cancer. 1999;81(8):1328–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kaur H, Mao S, Li Q, Sameni M, Krawetz SA, Sloane BF, et al. RNA-Seq of human breast ductal carcinoma in situ models reveals aldehyde dehydrogenase isoform 5A1 as a novel potential target. PLoS One. 2012;7(12):e50249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Eck SM, Cote AL, Winkelman WD, Brinckerhoff CE. CXCR4 and matrix Metalloproteinase-1 are elevated in breast carcinoma- associated fibroblasts and in normal mammary fibroblasts exposed to factors secreted by breast cancer cells. Mol Cancer Res. 2009;7(7):1033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lee S, Stewart S, Nagtegaal I, Luo J, Wu Y, Colditz G, et al. Differentially expressed genes regulating the progression of ductal carcinoma in situ to invasive breast cancer. Cancer Res. 2012;72(17):4574–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Curigliano G, Disalvatore D, Esposito A, Pruneri G, Lazzeroni M, Guerrieri-Gonzaga A, et al. Risk of subsequent in situ and invasive breast cancer in human epidermal growth factor receptor 2-positive ductal carcinoma in situ. Ann Oncol. 2015;26(4):682–7. [DOI] [PubMed] [Google Scholar]
- 29.Mustafa RE, DeStefano LM, Bahng J, Yoon-Flannery K, Fisher CS, Zhang PJ, et al. Evaluating the risk of upstaging HER2-positive DCIS to invasive breast Cancer. Ann Surg Oncol. 2017;24(10):2999–3003. [DOI] [PubMed] [Google Scholar]
- 30.Band V, Zajchowski D, Swisshelm K, Trask D, Kulesa V, Cohen C, et al. Tumor progression in four mammary epithelial cell lines derived from the same patient. Cancer Res. 1990;50(22):7351–7. [PubMed] [Google Scholar]
- 31.Souter LH, Andrews JD, Zhang GH, Cook AC, Postenka CO, Al-Katib W, et al. Human 21T breast epithelial cell lines mimic breast cancer progression in vivo and in vitro and show stage-specific gene expression patterns. Lab Investig. 2010;90(8):1247–58. [DOI] [PubMed] [Google Scholar]
- 32.Yong JW, Choong ML, Wang S, Wang Y, Lim SQ, Lee MA. Characterization of ductal carcinoma in situ cell lines established from breast tumor of a Singapore Chinese patient. Cancer Cell Int. 2014;14(1):94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Scribner KC, Behbod F, Porter WW. Regulation of DCIS to invasive breast cancer progression by Singleminded-2s (SIM2s). Oncogene. 2013;32(21):2631–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Scribner KC, Wellberg EA, Metz RP, Porter WW. Singleminded-2s (Sim2s) promotes delayed involution of the mouse mammary gland through suppression of Stat3 and NF kB. Mol Endocrinol. 2011;25(4):635–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gustafson TL, Wellberg E, Laffin B, Schilling L, Metz RP, Zahnow CA, et al. Ha-Ras transformation of MCF10A cells leads to repression of Singleminded-2s through NOTCH and C/EBPβ. Oncogene. 2009;28:1561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Laffin B, Wellberg E, Kwak HI, Burghardt RC, Metz RP, Gustafson T, et al. Loss of singleminded-2s in the mouse mammary gland induces an epithelial-mesenchymal transition associated with up-regulation of slug and matrix metalloprotease 2. Mol Cell Biol. 2008;28(6):1936–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kwak HI, Gustafson T, Metz RP, Laffin B, Schedin P, Porter WW. Inhibition of breast cancer growth and invasion by single-minded 2s. Carcinogenesis. 2007;28(2):259–66. [DOI] [PubMed] [Google Scholar]
- 38.Wellberg E, Metz RP, Parker C, Porter WW. The bHLH/PAS transcription factor singleminded 2s promotes mammary gland lactogenic differentiation. Development. 2010;137(6):945–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ory V, Kietzman WB, Boeckelman J, Kallakury BV, Wellstein A, Furth PA, et al. The PPARgamma agonist efatutazone delays invasive progression and induces differentiation of ductal carcinoma in situ. Breast Cancer Res Treat. 2018;169(1):47–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Michalik L, Desvergne B, Wahli W. Peroxisome-proliferator-activated receptors and cancers: complex stories. Nat Rev Cancer. 2004;4(1):61–70. [DOI] [PubMed] [Google Scholar]
- 41.Mueller KL, Madden JM, Zoratti GL, Kuperwasser C, List K, Boerner JL. Fibroblast-secreted hepatocyte growth factor mediates epidermal growth factor receptor tyrosine kinase inhibitor resistance in triple-negative breast cancers through paracrine activation of met. Breast Cancer Res. 2012;14(4):R104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Baselga J, Albanell J, Ruiz A, Lluch A, Gascon P, Guillem V, et al. Phase II and tumor pharmacodynamic study of gefitinib in patients with advanced breast cancer. J Clin Oncol. 2005;23(23):5323–33. [DOI] [PubMed] [Google Scholar]
- 43.Tan AR, Yang X, Hewitt SM, Berman A, Lepper ER, Sparreboom A, et al. Evaluation of biologic end points and pharmacokinetics in patients with metastatic breast cancer after treatment with erlotinib, an epidermal growth factor receptor tyrosine kinase inhibitor. J Clin Oncol. 2004;22(15):3080–90. [DOI] [PubMed] [Google Scholar]
- 44.Linklater ES, Tovar EA, Essenburg CJ, Turner L, Madaj Z, Winn ME, et al. Targeting MET and EGFR crosstalk signaling in triple-negative breast cancers. Oncotarget. 2016;7(43):69903–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Maroun CR, Rowlands T. The met receptor tyrosine kinase: a key player in oncogenesis and drug resistance. Pharmacol Ther. 2014;142(3):316–38. [DOI] [PubMed] [Google Scholar]
- 46.Liu X, Newton RC, Scherle PA. Developing c-MET pathway inhibitors for cancer therapy: progress and challenges. Trends Mol Med. 2010;16(1):37–45. [DOI] [PubMed] [Google Scholar]
- 47.Li Q, Xia J, Yao Y, Gong DW, Shi H, Zhou Q. Sulforaphane inhibits mammary adipogenesis by targeting adipose mesenchymal stem cells. Breast Cancer Res Treat. 2013;141(2):317–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Gernapudi R, Yao Y, Zhang Y, Wolfson B, Roy S, Duru N, et al. Targeting exosomes from preadipocytes inhibits preadipocyte to cancer stem cell signaling in early-stage breast cancer. Breast Cancer Res Treat. 2015;150(3):685–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Barcellos-Hoff MH, Lyden D, Wang TC. The evolution of the cancer niche during multistage carcinogenesis. Nat Rev Cancer. 2013;13(7):511–8. [DOI] [PubMed] [Google Scholar]
- 50.Chen J, Xie J, Jiang Z, Wang B, Wang Y, Hu X. Shikonin and its analogs inhibit cancer cell glycolysis by targeting tumor pyruvate kinase-M2. Oncogene. 2011;30(42):4297–306. [DOI] [PubMed] [Google Scholar]
- 51.Damaghi M, Tafreshi NK, Lloyd MC, Sprung R, Estrella V, Wojtkowiak JW, et al. Chronic acidosis in the tumour microenvironment selects for overexpression of LAMP2 in the plasma membrane. Nat Commun. 2015;6:8752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Gatenby RA, Gillies RJ. A microenvironmental model of carcino-genesis. Nat Rev Cancer. 2008;8(1):56–61. [DOI] [PubMed] [Google Scholar]
- 53.Gatenby RA, Gillies RJ. Why do cancers have high aerobic glycolysis? Nat Rev Cancer. 2004;4(11):891–9. [DOI] [PubMed] [Google Scholar]
- 54.Damaghi M, Wojtkowiak JW, Gillies RJ. pH sensing and regulation in cancer. Front Physiol. 2013;4(370):370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Estrella V, Chen T, Lloyd M, Wojtkowiak J, Cornnell HH, Ibrahim-Hashim A, et al. Acidity generated by the tumor microenvironment drives local invasion. Cancer Res. 2013;73(5):1524–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Sameni M, Cavallo-Medved D, Franco OE, Chalasani A, Ji K, Aggarwal N, et al. Pathomimetic avatars reveal divergent roles of microenvironment in invasive transition of ductal carcinoma in situ. Breast Cancer Res. 2017;19(1):56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Osuala KO, Sameni M, Shah S, Aggarwal N, Simonait ML, Franco OE, et al. Il-6 signaling between ductal carcinoma in situ cells and carcinoma-associated fibroblasts mediates tumor cell growth and migration. BMC Cancer. 2015;15(1):584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Carter EP, Gopsill JA, Gomm JJ, Jones JL, Grose RP. A 3D in vitro model of the human breast duct: a method to unravel myoepithelial-luminal interactions in the progression of breast cancer. Breast Cancer Res. 2017;19(1):50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Bischel LL, Beebe DJ, Sung KE. Microfluidic model of ductal carcinoma in situ with 3D, organotypic structure. BMC Cancer. 2015;15(1):12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Li Q, Chow AB, Mattingly RR. Three-dimensional overlay culture models of human breast cancer reveal a critical sensitivity to mitogen-activated protein kinase kinase inhibitors. J Pharmacol Exp Ther. 2010;332(3):821–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Bissell MJ, Rizki A, Mian IS. Tissue architecture: the ultimate regulator of breast epithelial function. Curr Opin Cell Biol. 2003;15(6):753–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Cukierman E, Pankov R, Stevens DR, Yamada KM. Taking cell-matrix adhesions to the third dimension. Science. 2001;294(5547):1708–12. [DOI] [PubMed] [Google Scholar]
- 63.Fennema E, Rivron N, Rouwkema J, van Blitterswijk C, de Boer J. Spheroid culture as a tool for creating 3D complex tissues. Trends Biotechnol. 2013;31(2):108–15. [DOI] [PubMed] [Google Scholar]
- 64.Kimlin LC, Casagrande G, Virador VM. In vitro three-dimensional (3D) models in cancer research: an update. Mol Carcinog. 2013;52(3):167–82. [DOI] [PubMed] [Google Scholar]
- 65.Ghaffarizadeh A, Heiland R, Friedman SH, Mumenthaler SM, Macklin P. PhysiCell: an open source physics-based cell simulator for 3-D multicellular systems. PLoS Comput Biol. 2018;14(2): e1005991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Benien P, Swami A. 3D tumor models: history, advances and future perspectives. Future Oncol. 2014;10(7):1311–27. [DOI] [PubMed] [Google Scholar]
- 67.Zschenker O, Streichert T, Hehlgans S, Cordes N. Genome-wide gene expression analysis in cancer cells reveals 3D growth to affect ECM and processes associated with cell adhesion but not DNA repair. PLoS One. 2012;7(4):e34279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Myungjin Lee J, Mhawech-Fauceglia P, Lee N, Cristina Parsanian L, Gail Lin Y, Andrew Gayther S, et al. A three-dimensional microenvironment alters protein expression and chemosensitivity of epithelial ovarian cancer cells in vitro. Lab Investig. 2013;93: 528. [DOI] [PubMed] [Google Scholar]
- 69.Eke I, Cordes N. Radiobiology goes 3D: how ECM and cell morphology impact on cell survival after irradiation. Radiother Oncol. 2011;99(3):271–8. [DOI] [PubMed] [Google Scholar]
- 70.Schmeichel KL, Weaver VM, Bissell MJ. Structural cues from the tissue microenvironment are essential determinants of the human mammary epithelial cell phenotype. J Mammary Gland Biol Neoplasia. 1998;3(2):201–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Gangadhara S, Smith C, Barrett-Lee P, Hiscox S. 3D culture of Her2+ breast cancer cells promotes AKT to MAPK switching and a loss of therapeutic response. BMC Cancer. 2016;16(1):345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Moin K, Sameni M, Victor BC, Rothberg JM, Mattingly RR, Sloane BF. Chapter ten - 3D/4D functional imaging of tumor-associated proteolysis: impact of microenvironment In: Conn PM, editor. Methods in enzymology, vol. 506 San Diego: Academic Press; 2012. p. 175–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Ji K, Zhao Z, Moin K, Xu Y, Sloane BF. Abstract B65: live-cell imaging of 3D/4D parallel co-cultures of breast carcinoma cells and breast fibroblasts in tissue architecture and microenvironment engineering (TAME) chambers. Mol Cancer Res. 2016;14(2 Supplement):B65. [Google Scholar]
- 74.Becker H Mind the gap! Lab Chip. 2010;10(3):271–3. [DOI] [PubMed] [Google Scholar]
- 75.Katt ME, Placone AL, Wong AD, Xu ZS, Searson PC. In vitro tumor models: advantages, disadvantages, variables, and selecting the right platform. Front Bioeng Biotechnol. 2016;4(12):12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Ravi M, Ramesh A, Pattabhi A. Contributions of 3D cell cultures for cancer research. J Cell Physiol. 2017;232(10):2679–97. [DOI] [PubMed] [Google Scholar]
- 77.Maddaly R, Subramaniyan A, Balasubramanian H. Cancer cytokines and the relevance of 3D cultures for studying those implicated in human cancers. J Cell Biochem. 2017;118(9):2544–58. [DOI] [PubMed] [Google Scholar]
- 78.Lv D, Hu Z, Lu L, Lu H, Xu X. Three-dimensional cell culture: a powerful tool in tumor research and drug discovery. Oncol Lett. 2017;14(6):6999–7010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Farnie G, Clarke RB, Spence K, Pinnock N, Brennan K, Anderson NG, et al. Novel cell culture technique for primary ductal carcinoma in situ: role of Notch and epidermal growth factor receptor signaling pathways. J Natl Cancer Inst. 2007;99(8):616–27. [DOI] [PubMed] [Google Scholar]
- 80.Farnie G, Willan PM, Clarke RB, Bundred NJ. Combined inhibition of ErbB½ and Notch receptors effectively targets breast ductal carcinoma in situ (DCIS) stem/progenitor cell activity regardless of ErbB2 status. PLoS One. 2013;8(2):e56840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Farnie G, Johnson RL, Williams KE, Clarke RB, Bundred NJ. Lapatinib inhibits stem/progenitor proliferation in preclinical in vitro models of ductal carcinoma in situ (DCIS). Cell Cycle. 2014;13(3):418–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Williams KE, Bundred NJ, Landberg G, Clarke RB, Farnie G. Focal adhesion kinase and Wnt signaling regulate human ductal carcinoma in situ stem cell activity and response to radiotherapy. Stem Cells. 2015;33(2):327–41. [DOI] [PubMed] [Google Scholar]
- 83.Manuel Iglesias J, Beloqui I, Garcia-Garcia F, Leis O, Vazquez-Martin A, Eguiara A, et al. Mammosphere formation in breast carcinoma cell lines depends upon expression of E-cadherin. PLoS One. 2013;8(10):e77281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Alowami S, Troup S, Al-Haddad S, Kirkpatrick I, Watson PH. Mammographic density is related to stroma and stromal proteoglycan expression. Breast Cancer Res. 2003;5(5):R129–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Butcher DT, Alliston T, Weaver VM. A tense situation: forcing tumour progression. Nat Rev Cancer. 2009;9(2):108–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Riching KM, Cox BL, Salick MR, Pehlke C, Riching AS, Ponik SM, et al. 3D collagen alignment limits protrusions to enhance breast cancer cell persistence. Biophys J. 2014;107(11):2546–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Lyons TR, O’Brien J, Borges VF, Conklin MW, Keely PJ, Eliceiri KW, et al. Postpartum mammary gland involution drives progression of ductal carcinoma in situ through collagen and COX-2. Nat Med. 2011;17(9):1109–U116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Decitre M, Gleyzal C, Raccurt M, Peyrol S, Aubert-Foucher E, Csiszar K, et al. Lysyl oxidase-like protein localizes to sites of de novo fibrinogenesis and in the early stromal reaction of ductal breast carcinomas. 1998. 143–51 p. [PubMed] [Google Scholar]
- 89.Zhao XK, Cheng Y, Liang Cheng M, Yu L, Mu M, Li H, et al. Focal adhesion kinase regulates fibroblast migration via integrin beta-1 and plays a central role in fibrosis. Sci Rep. 2016;6:19276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Wong VW, Rustad KC, Akaishi S, Sorkin M, Glotzbach JP, Januszyk M, et al. Focal adhesion kinase links mechanical force to skin fibrosis via inflammatory signaling. Nat Med. 2011;18(1): 148–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Cance WG, Harris JE, Iacocca MV, Roche E, Yang X, Chang J, et al. Immunohistochemical analyses of focal adhesion kinase expression in benign and malignant human breast and colon tissues: correlation with preinvasive and invasive phenotypes. Clin Cancer Res. 2000;6(6):2417–23. [PubMed] [Google Scholar]
- 92.Wei SC, Fattet L, Tsai JH, Guo Y, Pai VH, Majeski HE, et al. Matrix stiffness drives epithelial-mesenchymal transition and tumour metastasis through a TWIST1-G3BP2 mechanotransduction pathway Nat Cell Biol. 2015;17:678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Gupta N, Badeaux M, Liu Y, Naxerova K, Sgroi D, Munn LL, et al. Stress granule-associated protein G3BP2 regulates breast tumor initiation. Proc Natl Acad Sci USA. 2017;114(5):1033–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Muggerud AA, Hallett M, Johnsen H, Kleivi K, Zhou W, Tahmasebpoor S, et al. Molecular diversity in ductal carcinoma in situ (DCIS) and early invasive breast cancer. Mol Oncol. 2010;4(4):357–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Knudsen ES, Ertel A, Davicioni E, Kline J, Schwartz GF, Witkiewicz AK. Progression of ductal carcinoma in situ to invasive breast cancer is associated with gene expression programs of EMT and myoepithelia. Breast Cancer Res Treat. 2012;133(3): 1009–24. [DOI] [PubMed] [Google Scholar]
- 96.Burkhardt L, Grob TJ, Hermann I, Burandt E, Choschzick M, Janicke F, et al. Gene amplification in ductal carcinoma in situ of the breast. Breast Cancer Res Treat. 2010;123(3):757–65. [DOI] [PubMed] [Google Scholar]
- 97.Hu M, Yao J, Carroll DK, Weremowicz S, Chen H, Carrasco D, et al. Regulation of in situ to invasive breast carcinoma transition. Cancer Cell. 2008;13(5):394–406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Petridis C, Brook MN, Shah V, Kohut K, Gorman P, Caneppele M, et al. Genetic predisposition to ductal carcinoma in situ of the breast. Breast Cancer Res. 2016;18(1):22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Ma XJ, Salunga R, Tuggle JT, Gaudet J, Enright E, McQuary P, et al. Gene expression profiles of human breast cancer progression. Proc Natl Acad Sci USA. 2003;100(10):5974–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Schuetz CS, Bonin M, Clare SE, Nieselt K, Sotlar K, Walter M, et al. Progression-specific genes identified by expression profiling of matched ductal carcinomas in situ and invasive breast tumors, combining laser capture microdissection and oligonucleotide microarray analysis. Cancer Res. 2006;66(10):5278–86. [DOI] [PubMed] [Google Scholar]
- 101.Vincent-Salomon A, Lucchesi C, Gruel N, Raynal V, Pierron G, Goudefroye R, et al. Integrated genomic and transcriptomic analysis of ductal carcinoma in situ of the breast. Clin Cancer Res. 2008;14(7):1956–65. [DOI] [PubMed] [Google Scholar]
- 102.Hernandez L, Wilkerson PM, Lambros MB, Campion-Flora A, Rodrigues DN, Gauthier A, et al. Genomic and mutational profiling of ductal carcinomas in situ and matched adjacent invasive breast cancers reveals intra-tumour genetic heterogeneity and clonal selection. J Pathol. 2012;227(1):42–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Luzzi V, Holtschlag V, Watson MA. Expression profiling of ductal carcinoma in situ by laser capture microdissection and high-density oligonucleotide arrays. Am J Pathol. 2001;158(6):2005–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Perou CM, Sorlie T, Eisen MB, van de Rijn M, Jeffrey SS, Rees CA, et al. Molecular portraits of human breast tumours. Nature. 2000;406(6797):747–52. [DOI] [PubMed] [Google Scholar]
- 105.Kaur H, Mao S, Shah S, Gorski DH, Krawetz SA, Sloane BF, et al. Next-generation sequencing: a powerful tool for the discovery of molecular markers in breast ductal carcinoma in situ. Expert Rev Mol Diagn. 2013;13(2):151–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Calaf GM, Echiburu-Chau C, Zhao YL, Hei TK BigH3 protein expression as a marker for breast cancer. Int J Mol Med. 2008;21(5):561–8. [PubMed] [Google Scholar]
- 107.Wang N, Eckert KA, Zomorrodi AR, Xin P, Pan W, Shearer DA, et al. Down-regulation of HtrA1 activates the epithelial-mesenchymal transition and ATM DNA damage response pathways. PLoS One. 2012;7(6):e39446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Castellana B, Escuin D, Peiro G, Garcia-Valdecasas B, Vazquez T, Pons C, et al. ASPN and GJB2 are implicated in the mechanisms of invasion of ductal breast carcinomas. J Cancer. 2012;3:175–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Shah S, Brock EJ, Jackson RM, Ji K, Boerner JL, Sloane BF, Mattingly RR. Downregulation of Rap1Gap: A switch from DCIS to invasive breast carcinoma via ERK/MAPK activation. Neoplasia. 2018; (in press). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Schnitt SJ. The transition from ductal carcinoma in situ to invasive breast cancer: the other side of the coin. Breast Cancer Res. 2009;11(1):101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Sung KE, Yang N, Pehlke C, Keely PJ, Eliceiri KW, Friedl A, et al. Transition to invasion in breast cancer: a microfluidic in vitro model enables examination of spatial and temporal effects. Integr Biol (Camb). 2011;3(4):439–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Choi Y, Hyun E, Seo J, Blundell C, Kim HC, Lee E, et al. A microengineered pathophysiological model of early-stage breast cancer. Lab Chip. 2015;15(16):3350–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Kalluri R The biology and function of fibroblasts in cancer. Nat Rev Cancer. 2016;16(9):582–98. [DOI] [PubMed] [Google Scholar]
- 114.Gascard P, Tlsty TD. Carcinoma-associated fibroblasts: orchestrating the composition ofmalignancy. Genes Dev. 2016;30(9):1002–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Chalasani A, Ji K, Sameni M, Mazumder SH, Xu Y, Moin K, et al. Live-cell imaging of protease activity: assays to screen therapeutic approaches. Methods Mol Biol. 2017;1574:215–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Sameni M, Tovar EA, Essenburg CJ, Chalasani A, Linklater ES, Borgman A, et al. Cabozantinib (XL184) inhibits growth and invasion ofpreclinical TNBC models. Clin Cancer Res. 2016;22(4): 923–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Sameni M, Anbalagan A, Olive MB, Moin K, Mattingly RR, Sloane BF. MAME models for 4D live-cell imaging of tumor: microenvironment interactions that impact malignant progression. J Vis Exp. 2012; (60):e3661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Hu M, Peluffo G, Chen H, Gelman R, Schnitt S, Polyak K. Role of COX-2 in epithelial-stromal cell interactions and progression of ductal carcinoma in situ of the breast. Proc Natl Acad Sci. 2009;106(9):3372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Sternlicht MD, Barsky SH. The myoepithelial defense: a host defense against cancer. Med Hypotheses. 1997;48(1):37–46. [DOI] [PubMed] [Google Scholar]
- 120.Xiao G, Liu YE, Gentz R, Sang QA, Ni J, Goldberg ID, et al. Suppression of breast cancer growth and metastasis by a serpin myoepithelium-derived serine proteinase inhibitor expressed in the mammary myoepithelial cells. Proc Natl Acad Sci U S A. 1999;96(7):3700–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Barsky SH, Karlin NJ. Myoepithelial cells: autocrine and paracrine suppressors of breast cancer progression. J Mammary Gland Biol Neoplasia. 2005;10(3):249–60. [DOI] [PubMed] [Google Scholar]
- 122.Pandey PR, Saidou J, Watabe K. Role of myoepithelial cells in breast tumor progression. Front Biosci. 2010;15:226–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Duivenvoorden HM, Rautela J, Edgington-Mitchell LE, Spurling A, Greening DW, Nowell CJ, et al. Myoepithelial cell-specific expression of stefin a as a suppressor of early breast cancer invasion. J Pathol. 2017;243(4):496–509. [DOI] [PubMed] [Google Scholar]
- 124.Coussens LM, Werb Z. Inflammation and cancer. Nature. 2002;420(6917):860–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Grivennikov SI, Greten FR, Karin M. Immunity, inflammation, and cancer. Cell. 2010;140(6):883–99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Quatromoni JG, Eruslanov E. Tumor-associated macrophages: function, phenotype, and link to prognosis in human lung cancer. Am J Transl Res. 2012;4(4):376–89. [PMC free article] [PubMed] [Google Scholar]
- 127.Sica A, Schioppa T, Mantovani A, Allavena P. Tumour-associated macrophages are a distinct M2 polarised population promoting tumour progression: potential targets of anti-cancer therapy. Eur J Cancer. 2006;42(6):717–27. [DOI] [PubMed] [Google Scholar]
- 128.Bohrer LR, Schwertfeger KL. Macrophages promote fibroblast growth factor receptor-driven tumor cell migration and invasion in a CXCR2-dependent manner. Mol Cancer Res. 2012;10(10): 1294–305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Carron EC, Homra S, Rosenberg J, Coffelt SB, Kittrell F, Zhang Y, et al. Macrophages promote the progression of premalignant mammary lesions to invasive cancer. Oncotarget. 2017;8(31):50731–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Sharma B, Nawandar DM, Nannuru KC, Varney ML, Singh RK. Targeting CXCR2 enhances chemotherapeutic response, inhibits mammary tumor growth, angiogenesis, and lung metastasis. Mol Cancer Ther. 2013;12(5):799–808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Nelson CM, Bissell MJ. Of extracellular matrix, scaffolds, and signaling: tissue architecture regulates development, homeostasis, and cancer. Annu Rev Cell Dev Biol. 2006;22(1):287–309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Bischel LL, Sung KE, Jimenez-Torres JA, Mader B, Keely PJ, Beebe DJ. The importance of being a lumen. FASEB J. 2014;28(11):4583–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Truong D, Puleo J, Llave A, Mouneimne G, Kamm RD, Nikkhah M. Breast cancer cell invasion into a three dimensional tumor-stroma microenvironment. Sci Rep. 2016;6:34094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Mattingly RR, Milstein ML, Mirkin BL. Down-regulation of growth factor-stimulated MAP kinase signaling in cytotoxic drug-resistant human neuroblastoma cells. Cell Signal. 2001;13(7):499–505. [DOI] [PubMed] [Google Scholar]
- 135.Horning JL, Sahoo SK, Vijayaraghavalu S, Dimitrijevic S, Vasir JK, Jain TK, et al. 3-D tumor model for in vitro evaluation of anticancer drugs. Mol Pharm. 2008;5(5):849–62. [DOI] [PubMed] [Google Scholar]
- 136.Ji K, Heyza J, Cavallo-Medved D, Sloane BF. Pathomimetic cancer avatars for live-cell imaging of protease activity. Biochimie. 2016;122:68–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Regehr KJ, Domenech M, Koepsel JT, Carver KC, Ellison-Zelski SJ, Murphy WL, et al. Biological implications of polydimethylsiloxane-based microfluidic cell culture. Lab Chip. 2009;9(15):2132–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Berthier E, Young EW, Beebe D. Engineers are from PDMS-land, biologists are from polystyrenia. Lab Chip. 2012;12(7):1224–37. [DOI] [PubMed] [Google Scholar]


