Abstract
In Alzheimer’s disease (AD), amyloid β (Aβ) protein plays a detrimental role in neuronal injury and death. Recent in vitro and in vivo studies suggest that soluble oligomers of the Aβ peptide are neurotoxic. Structural properties of the oligomeric assembly, however, are largely unknown. Our present investigation established that the 40-residue-long Aβ peptide (Aβ40) became more helical, ordered, and compact in the oligomeric state, and both the helical and β-sheet components were found to increase significantly in the early event of oligomerization. The band-selective two-dimensional NMR analysis suggested that majority of the residues from sequence 12 to 22 gained a higher-ordered secondary structure in the oligomeric condition. The presence of a significant amount of helical conformation was confirmed by Raman bands at 1650 and 1336 cm–1. Other residues remained mostly in the extended polyproline II (PPII) and less compact β-conformation space. In the event of maturation of the oligomers into an amyloid fiber, both the helical content and the PPII-like structural components declined and ∼72% residues attained a compact β-sheet structure. Interestingly, however, some residues remained in the collagen triple helix/extended 2.51-helix conformation as evidenced by the amide III Raman signature band at 1272 cm–1. Molecular dynamics analysis using an optimized potential for liquid simulation force field with the peptide monomer indicated that some of the residues may have preferences for helical conformation and this possibly contributed in the event of oligomer formation, which eventually became a β-sheet-rich amyloid fiber.
Introduction
Alzheimer’s disease (AD) is a progressive dementia, and considerable evidence indicates that the disease is triggered by neurotoxic assemblies of the amyloid β (Aβ) peptide.1−4 Recent studies have shown that prefibrillar structures, such as oligomers5−11 and protofibril,12−15 of the Aβ peptide are key toxic species in AD. Soluble oligomers have been found to cause physical damage of synapses.16 Oligomers have also been implicated in memory failure in transgenic mice.17−19 Fibrillization of α-synuclein in Parkinson’s disease (PD) also involves the formation of similar oligomeric intermediates.20,21 Thus, prevention of oligomerization becomes an attractive therapeutic target for neuropathology of amyloid disease.4,22−26 However, very little is known about the protein/peptide conformation and the thermodynamics of the associated conformational changes during oligomer and fibril formation. The current investigation focused on the conformation and molecular details of the peptide in its oligomeric intermediate states. The results may clarify some of the fundamental questions regarding the molecular mechanism of amyloid formation, synaptic toxicity, and the nature of the intermolecular interactions that stabilize the oligomeric assembly of the peptide.
Early studies of amyloid formation of the Aβ peptide were largely centered on fibril morphology,27 and most of the structural investigations were focused on fibrillar assembly by solid-state NMR and Fourier transform infrared (FTIR) spectroscopic analyses. These studies suggested that in the fibrillar form the peptide attained a β-sheet conformation.28 Conformational studies of the monomeric Aβ peptide were often performed by circular dichroism (CD), FTIR, and NMR spectroscopic analyses.29,30 In vitro solution-phase NMR studies suggested that monomers of Aβ possessed no α-helical or β-sheet structure.31,32 Both Aβ40 and Aβ42 favored a predominately random and extended conformation.31 Following fibril formation, however, Aβ peptides attained a predominately β-sheet conformation28,33−35 within a repeating polymeric peptide assembly. Fiber X-ray diffraction analyses showed that Aβ fibrils contained cross-β structural motifs. In vitro solid-state NMR experiments on synthetic Aβ fibrils established a parallel β-sheet structure.35
The scarcity of structural information of oligomer intermediates arises due to their transient nature and the solubility problem. Their solubility and size render structural analysis by solution NMR problematic. X-ray crystallography is of no use, as crystals of the oligomeric intermediates cannot be obtained. Atomic force microscopy (AFM) provides morphological features of fibrils and some intermediates of Aβ7,36,37 and the morphology of other amyloidogenic aggregated proteins, including α-synuclein,38 lysozyme,39 and prion protein.40 However, the conformation of the protein backbone in the toxic oligomers has not been studied in detail. Lansbury et al. suggested that spherical oligomers of α-synuclein were rich in the β-sheet structure.41 Walsh et al. and Kirkitadze et al. provided limited structural information of the Aβ oligomeric intermediate on the basis of CD analysis.5,15,42,43
However, the detailed structural organizations starting from its monomeric state to the oligomer and fibril are less discussed and very little understood. In the current investigation, with a 40-residue-long Aβ40 peptide, we established that the peptide became more helical, ordered, and compact in the oligomeric state. Our analysis also suggested that the secondary structural fluctuation along with tertiary refolding stimulated the propagation of amyloid fiber formation and related aggregation. The band-selective-optimized-flip-angle short-transient (SOFAST)-heteronuclear multiple quantum coherence (HMQC) two-dimensional (2D) NMR measurements indicated that most of the residues in the sequence stretch from 12 to 22 in Aβ40 possessed strong propensities to acquire an ordered secondary structure in the very early event of oligomerization. In addition, analysis of the amide mode vibrations in the Raman spectra of the peptide confirmed the presence of a significant helical conformation in the oligomeric assembly structure.
Results and Discussion
Residue-Specific Conformation of Soluble Aβ Oligomer
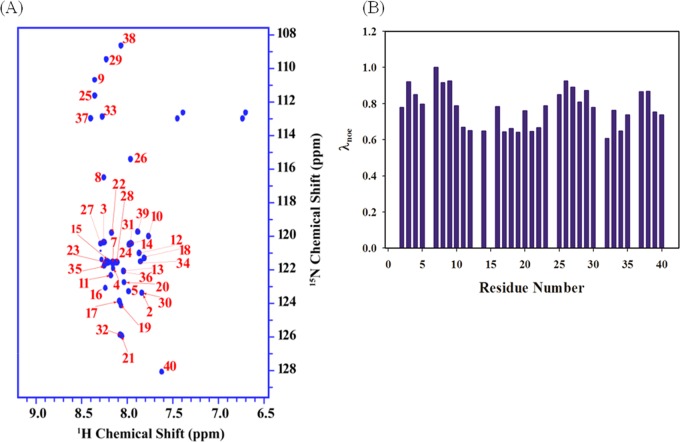
Aβ40 was dissolved (1.5 mg/mL) in a 10 mM phosphate buffer at pH 7.2, and both NMR and Raman spectroscopic measurements were made with this sample to derive structural information of the peptide. Two-dimensional 15N,1H heteronuclear correlation spectra of Aβ40 showed poor dispersion (∼1.1 ppm) of most of the backbone 1HN resonances and appeared between 7.5 and 8.4 ppm (Figure 1). In addition, the line widths were sharp, and very slow relaxation in the indirect dimension leads to a high acquisition time (t1 max), which is typical for unfolded macromolecules and indicated a lack of globular fold. To monitor the structural heterogeneity and compactness in a residue-specific manner, we have recorded one set of experiments. These experiments are based on the SOFAST-HMQC experiment, which was designed for rapid acquisition of heteronuclear correlation spectra of proteins. The hetero-SOFAST NMR experiments incorporate an additional band-selective inversion pulse at the beginning of the pulse sequence and the recycle period to act as the contact time.9,10 Two experiments are recorded with (excited) and without (reference) that specific selective pulse, and the ratio of peak volumes are taken. Among the different sets of experiments, this selective pulse differs. This selective pulse can be on aliphatic protons, which have a distinct chemical shift range (0–4 ppm), compared to that of the amide proton chemical shift (6–10 ppm for folded proteins). The experiment measures the nuclear Overhauser effect (NOE) between the perturbed aliphatic protons and the observed amide protons after a contact time of 200 ms.
Figure 1.
Solution-state NMR. (A) Two-dimensional [15N/1H] SOFAST-HMQC spectra of 15N-labeled Aβ40 (1.5 mg/mL) dissolved in a 20 mM phosphate buffer, pH 7.6. Residue numbers are marked in red. (B) Bar plot of λNOE (the ratio of effective spin diffusion of amide protons in the presence and absence of a selective pulse) against different residues of Aβ40 (details are given in Experimental Section).
The observed ratio (λNOE) probe the effective spin diffusion of that particular 1HN spin. This depends on the spin network around that particular spin and incorporates various factors such as number, density, and mobility of other close protons. This ratio thus can probe local structural propensity effectively in a residue-wise manner. For small unfolded peptides, the value can go above 0.8. For Aβ40, the average λNOE value was 0.76 and indicated that most of the peptide bond remained in the disordered region. However, the signal from residue stretches of 12–15, 17–19, and 21–22 showed a λNOE of less than 0.7, and these occurrences may be due to the proximity of the protons that can cause more spin diffusion (Figure 1). This observation suggested that these segments possessed some residual structure. In addition, the residues 32 and 34 also showed λNOE values of less than 0.7 and indicated that these two residues could be part of an ordered component.
The phenomenon of accessing alternate states could possibly act as an initiation point of mutual interactions as the residues 32 and 34 have been involved in making a tertiary fold, which was confirmed in the fibrillar state by various studies.44−46 Here also in the initiation phase of aggregation, our finding of accessing of alternate structures by residues 32 and 34 and the residue stretch of 12–22 provides a firm experimental support for the final topology of interaction and structural reorganization in the oligomeric state. We observed no new peak appearing in the HMQC spectra over time. As time progressed, peaks started disappearing. This implied the formation of oligomers of different sizes, which were large enough to be beyond the detection limit. From the NMR analysis, we probed the residues that access alternate structures in the initial phase of aggregation; however, an additional structural insight in a different state of aggregation was derived from the peptide bond vibration analysis via Raman spectroscopic measurement (Scheme S1).
Raman Spectra of Aβ40 and Quantification of Peptide Secondary Structure
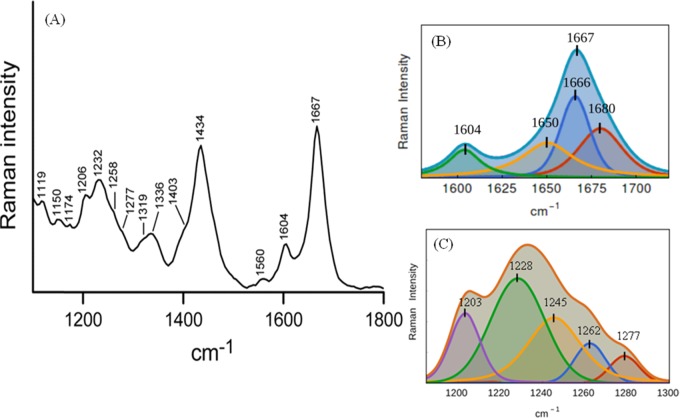
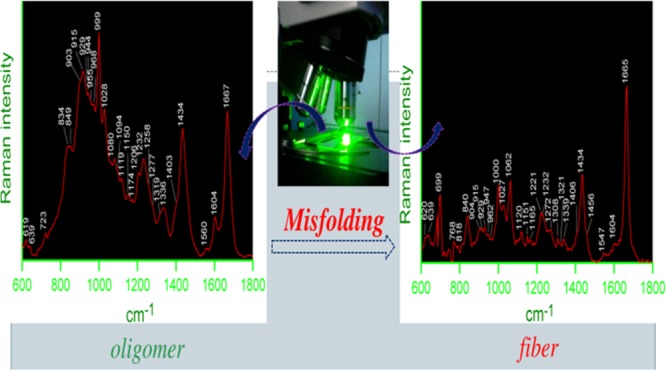
Figures 2 and 3, respectively, show the Raman spectra and transmission electron microscopy (TEM) images of Aβ40 oligomers. Figure S1 displays the Raman spectra of the Aβ40 monomer (upper trace) and matured fibril (bottom trace) in the frequency range of 1200–1800 cm–1. Table S1 provides assignments of some of the characteristic Raman bands. The amide I vibration bands often appear in the range of 1620–1700 cm–1, and it primarily represents C=O stretching; out-of-phase C–N motion also contributes to this band vibration. The peptide Raman spectrum in the monomeric state was dominated by a broad amide I band at 1675 cm–1. The full width at half-maximum (FWHM) was ∼35 cm–1. It was diagnostic of a highly fluctuating/dynamic secondary structure and could be a marker for a polyproline II (PPII)-like conformation.47−50 Thus, the amide I Raman band intensity data indicated that the secondary structural preference of the residues in Aβ40 as the monomer was mainly PPII-like conformation (77%) as defined in the Ramachandran plot. Table 1 shows the content of different conformational states of the monomer, oligomer, and fibrils of Aβ40. The values were obtained by fitting the broad amide band with different component bands. The area under each component band was proportionate to a particular kind of secondary structure (Figures 2B and S2).48,51 A component band centered at ∼1650 cm–1 (Figure S2A) that fitted well with the amide I Raman band of the peptide in the monomeric state could be linked to a short helical kink-like structure. Figure 2 displays the Raman spectra of the peptide in the oligomeric state. It contained a broad amide I band centered at 1667 cm–1 with an FWHM of ∼29 cm–1. The secondary structural distributions in the oligomeric state are shown in Figure 2B and listed in Table 1. In the oligomeric species, the helical content was 31% (component band at 1650 cm–1), whereas the β-strand/sheet content was 36% and was represented by a fitted band at 1666 cm–1, and the PPII-like conformation was 33% with a signature band at 1680 cm–1. In the state of amyloid β late assembly in the form of a fiber, the width of the amide I band was found to shrink; the value was close to 21 cm–1 and appeared at 1665 cm–1 (Figures S1 and S2). The narrow and sharp vibration band indicated a cross β-sheet conformation of the peptide in the fibrillar state (Figure S2B). A significant decrease of the PPII conformation was prominent, and the helical content was also declined; interestingly, however, the PPII-type signature was reduced maximally (Table 1 and Figure S2).
Figure 2.
Raman spectra of Aβ40 oligomers. (A) Raman spectra of Aβ40 oligomers in the frequency range of 1150–1800 cm–1 by a 532 nm laser. Oligomerized peptide solution was prepared in a 10 mM phosphate buffer, pH 7.2. The peptide solution (1.5 mg/mL) was incubated for 3 h at room temperature. To record the Raman spectrum, 20 μL of the sample solution was dropped onto a glass cover slip and the spectra were recorded at room temperature (25 °C). Laser power at the source was 30–35 mW, ∼2 mW at the sample. The recording scan time was 15 s, and the number of scans was 10. The displayed spectrum was the average of three/five such measurements. (B) Curve fitting of the amide I region of the Raman spectrum. The band fitting was based on a standard protocol as stated in Experimental Section. Three component bands that represent total amide I bands are shown separately. The violet line is the original spectrum, the red, blue, yellow, green lines are the individual component bands, and the cyan line is the sum of the bands. The fitted peak positions are also marked. 1604 cm–1 corresponds to vibration from the side chain residue. (C) Amide III region of the same spectrum and five component bands as obtained by curve fitting; their peak positions are marked.
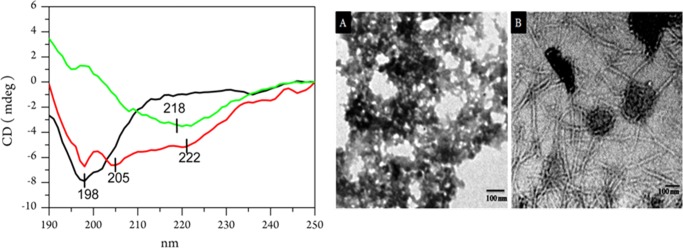
Figure 3.
CD spectra and TEM images of Aβ40 species. Left panel shows far-UV CD spectra of Aβ40 (1.5 mg/mL) dissolved in a 20 mM phosphate buffer of pH 7.2, at room temperature (25 °C). In the panel, the black line is the spectrum of the peptide monomer, recorded just after preparing the solution, the red line is for the oligomer, and the green line is for the fiber state of the peptide. Oligomer solution was prepared in a 20 mM phosphate buffer of pH 7.2, and the incubation time was 3 h. Aβ40 fiber was prepared in a 20 mM phosphate buffer of pH 7.2 at T = 25 °C, and the incubation time was 2 days (green line). The path length of the measuring cell was 0.1 cm, and each spectrum was an average of three scans. Right panel shows TEM images of Aβ40 oligomer (A) and Aβ40 fiber (B). The sample preparation condition was the same as above. Details of the recording conditions are given in the Methods section. Size of the scale bar is 100 nm.
Table 1. Secondary Structural Components of Aβ40 in Different States of Its Aggregation as a Percentage of the Total Band Areaa.
| peak no. | center (cm–1) | width (cm–1) | area (%) |
|---|---|---|---|
| Aβ40 Monomer | |||
| 1 | 1696 (3) | 26 (6) | 16 |
| 2 | 1677 (1) | 25 (11) | 61 |
| 3 | 1664 (3) | 24 (46) | 9 |
| 4 | 1650 (12) | 35 (12) | 14 |
| Oligomer | |||
| 1 | 1680 (4) | 29 (2) | 33 |
| 2 | 1666 (0) | 20 (7) | 36 |
| 3 | 1650 (6) | 35 (2) | 31 |
| Fiber | |||
| 1 | 1681 (4) | 20 (5) | 8 |
| 2 | 1665 (0) | 18 (2) | 72 |
| 3 | 1652 (2) | 30 (3) | 20 |
Results are obtained via curve fitting of the amide I band of Aβ40 in the Raman spectra of the peptide as a monomer, oligomer, and fiber. Values in parentheses are the standard errors.
Significant Number of Residues Attain an α-Helical Structure in the Oligomer
Figure 2C shows the amide III region (1200–1350 cm–1) in the Raman spectrum of oligomeric species, which also signified the results obtained from the amide I band. Weak bands at 1258 and 1319 cm–1 indicated the presence of a significant number of residues in the α-helical space.52−54 The Aβ40 oligomer also exhibited a distinct band at 1336 cm–1 (Figure 2A and Table S1). Thomas et al. suggested that the band near 1340–1345 cm–1 is a marker for α-helices and assigned it to the vibration localized specifically to the O=C–Cα–H linkage of the main peptide backbone.53 A signatory band near 1336 cm–1 in the Raman spectra of the Aβ40 oligomer (Figure 2A) suggested that the peptide bond geometry may be within the conformation of the α-helical structure.53,54 The band became much weaker in the fibrillar state (Figure S1) and confirmed the transition of the helical conformation into a β-sheet structure. The band at 1434 cm–1 in the oligomer state was assigned to CH2–, CH3– deformation and CH2– scissoring motion. The band was sharper than the corresponding band at 1448 cm–1 for the monomer (Figure S1). This again supported the fact that the peptide chain became more compact.
The peptide in a largely monomeric state in the aqueous buffer showed a significant negative signature at 198 nm in the CD spectra. It is often assigned to the n−π* transition and indicated that the residues mostly preferred the PPII-like conformation47 (Figure 3 left). However, the presence of helical content in the oligomerized peptide was imminent; an increase of the negative signal at 222 nm due to n−π* electronic transition was observed with ellipticity at 222 nm (Θ222nm was −0.91 mdeg) in the monomer, and the value became more negative (−4.96 mdeg) in the oligomeric state. Thus, the oligomeric population substantially inherited PPII and extended helix or combination of several short helical components (the absence of a strong positive signal for perpendicular π–π* transition at 192 nm indicated a lack of a very compact and large α-helical fold).
Ahmed et al. were able to prepare on-pathway oligomers under a low salt condition, keeping the temperature relatively low.55 They found no compact β-sheet secondary structure in Aβ42 oligomers. We also observed that the oligomers obtained in our preparation showed minimal binding to ThT (Figure S5). However, a very broad band at 1232 cm–1 indicated some residues with a β-conformation space. All these studies indicated that the peptide in the oligomeric state had less preference for a compact cross β-sheet structure. The fiber showed a major amide III signal (Figure S3) at 1221 cm–1 with a shoulder band at 1232 cm–1. The shift and compactness of the band indicated the presence of a β-sheet structure.48,51,54,56 The band became much sharper compared to that of the same band in the oligomeric state and effectively enhanced the ThT fluorescence (Figure S5). Wälti et al. in their recent article also suggested that in disease-relevant Aβ42 fibril, each layer of the fibril is composed of two molecules and the residues spanning between 15 and 42 adopt a horseshoe-like β-sheet conformation. The CD spectral signature of the amyloid fiber indicated that the peptide attained largely a β-sheet conformation and was devoid of the PPII conformation. It produced a positive signature at 196 nm (n−π* transition), suggesting a lack of large PPII contribution, and a negative minimum at 218 nm (π–π* transition) suggested a β-sheet structure.
Increase in the α-Helical Structure
Benevides et al. observed a very confirmative tertiary signature with a helix for the HK97 capsid assembly with a reliable Raman indicator at 933 cm–1.57,58 The 800–1200 cm–1 region of the Raman spectra of Aβ40 oligomers were overcrowded with signals at 929, 944 cm–1, and many others (Table S1 and Figure S4). These bands indicated the presence of a backbone tertiary helical structure and the conformational heterogeneity in the oligomeric assembly.54,56−58
Earlier Raman investigation with helical viral proteins also found a nonamide marker band for the helical secondary structure, and an associated tertiary fold was assigned at 1340 cm–1. The band was assigned to C–Cα–H valence angle bending. The isotope-edited study further confirmed that the O=C–Cα–H stretching motion contributes substantially to this vibration band. The band intensity was found to depend on the content of the α-helical structure, and least intensity suggests lesser amount of the α-helical fold. The band (intensity) also can guide the orientation (tertiary structure) of the α-helical segment in the protein. In our investigation, we observed the band at 1336 cm–1, which was found to substantially decrease in the fibrillar state and indicated a significant reorientation and decrease of the helical structure. With viral capsid proteins, it was suggested that α-helices may be short and connected by loops or disordered segments consisting of a few residues each.59 The helical signature band referenced at 1335–1340 cm–1 in viral proteins particularly was present in the intermediate capsid (procapsid) structure of viral coat proteins.52
N-Terminal Residues Preserve Collagen Triple Helix-Type/Extended 2.51-Helix Conformation
Majority of earlier investigations indicated that residues 1–14 may have some structural preferences.60 Our Raman spectroscopic analysis found minor amide III signals at 1275, 1277, and 1272 cm–1 for the monomer, oligomer, and fiber, respectively, and attributed it to the collagen triple helix type/extended 2.51-helix61,62 type of conformation. Asher et al. experimentally showed a stable 2.51-helix conformation in a model peptide (PGA) and found a signature of the conformation at 1271 cm–1.61 The structure was close to those in a PPII-like conformation; however, a larger separation distance appeared among the charge groups in the 2.51-helix conformation state. This region may be present in all the three states (monomer, oligomer, and fiber) of the peptide conformation. It is a highly extended PPII-like conformation and may be confined in the N-terminal residues (1–10) because the λNOE values were large (>0.75). These residues are highly polar and dynamic (MD calculations are provided in the supporting material). Petkova et al. also observed the extended structure of the region even in the fibrillar state.63,64
The intriguing fact in our observation was that upon oligomerization of the peptide, it became more helical, ordered, and compact. The existing literature from different experimental studies showed two significant points regarding the oligomer structure and the pathway of aggregation: (i) the amyloid oligomer mainly contained two well-defined secondary structures, β-strand and PPII; and (ii) the major structural change from monomer to oligomer proceeded via conversion of the PPII conformer to the β-strand, which was further extended to form a well-defined cross-β-sheet structure in the fibrillar state. However, our Raman analyses established a key information that the residues distributed in the helical and β-domain similar to a molten globule-like architecture65,66 before it transformed into a β-sheet-rich amyloid fiber. It was not a simple one-way structural conversion of PPII to the β-sheet structure. It is comparable to another disordered protein, α-synuclien,47 which also contained some signature of the helical structure in the oligomeric state.
Conformational Clustering of the Peptide and Ramachandran (ψ/Φ) Space of the Residues by Molecular Dynamics (MD) Simulation
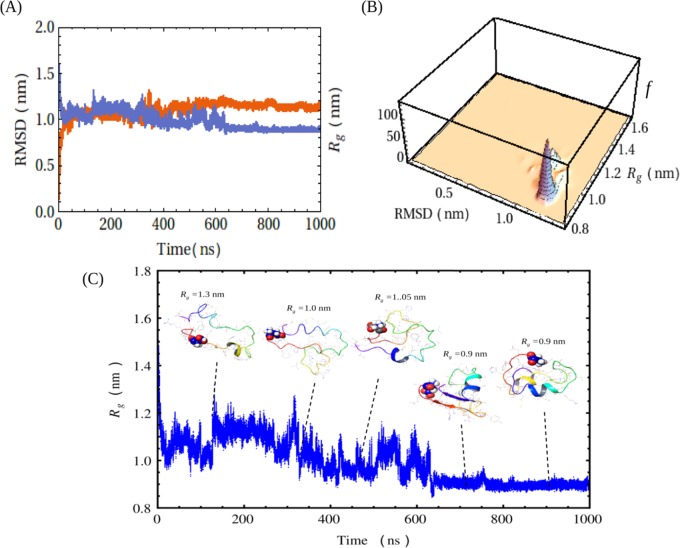
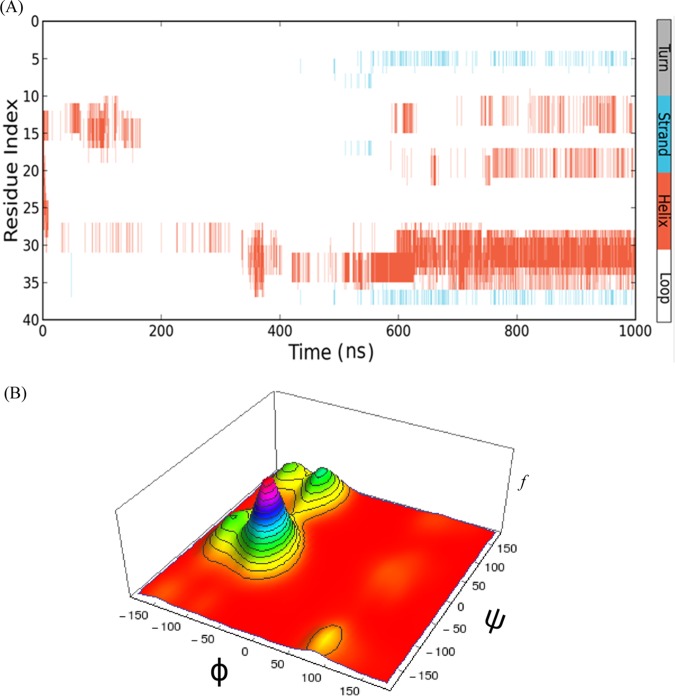
An early event of protein/peptide folding may govern the higher-order aggregation, such as amyloid fibril formation.65 To realize this folding event, often MD simulation is carried out as a powerful computational method.67−74 MD simulation was engaged in the current investigation to inspect early dynamics and overall conformational changes of Aβ40 as a monomer and dimer in a water system. The initial Aβ40 coordinates were taken from PDB entry 1BA4 and were subjected to simulation at 310 K for several nanoseconds. We analyzed the simulation trajectories, which include the measurements of root-mean-square deviations (RMSDs) of the α-carbons from its initial values, radius of gyration (Rg) of the peptide, and the secondary structure. Figure 4A shows the RMSDs of the Cα atoms and Rg values of the peptide against simulation time. In the beginning of the simulation, the RMSD was fluctuating and the values were in the range of 0.2–1.3 nm. However, after 400 ns of simulation, the fluctuation in the RMSD was minimized and an equilibrium conformation was reached. In the subsequent interval of ∼1000 ns, no significant fluctuation of RMSD was observed and it remained stable with average values of ∼1.2 nm. The Rg was also stabilized at about 600 ns, and the value was ∼0.9 nm. This suggested that the convergence of the simulation occurred and Aβ40 attained significant stability at this point of time. The conformational clustering (density distribution) of Aβ40 against RMSD and Rg is shown in Figure 4B. At an Rg value of 0.9 nm and an RMSD of 1.2 nm, the peptide gained significant stability and compactness. Figure 4C displays the snapshots of the conformation of the peptide at different time intervals. In the early state of the simulation, the peptide, for instance at 160 ns of simulation, was mostly disordered (Rg = 1.3 nm); however, with the increase in the simulation time, Aβ40 adopted more a compact and helical conformation with a decrease in the Rg value. At the end point (960 ns), we observed the presence of some helical content at residues 11–15 and 29–34.
Figure 4.
Conformational clustering and compactness of the peptide as obtained via MD analysis using Desmond algorithm. (A) Plot of RMSD and radius of gyration (Rg) against simulation time during a 1000 ns molecular dynamics simulation of Aβ40 at 310 K. Red listing indicates RMSD and blue listing indicates Rg. (B) Conformational clustering of the peptide by correlating the Rg with RMSD values obtained from simulation. The 3D histogram plot of Rg against RMSD shows the conformational clustering, where f is the frequency. (C) Structures generated at different time intervals of simulation and representative snapshots are shown. The simulation time and the Rg for each snapshot are given. The lower blue scattered plot shows the refinements in the radius of gyration of the Aβ40 backbone in the same time frame. N-Terminus of the peptide was marked by the Asp residue in the CPK model.
The residue-specific average secondary structural preference of the peptide is shown in Figure 5A. The analysis indicated helical preferences among residues 10–15, 17–21, and 28–36 segments, and a weak β-strand preference was observed of residues 37–38. These three different helical abundant amyloid segments might have a tendency to adopt a helical conformation during the early event of self-assembly as the conformational adaptation. This result was similar to the earlier NMR study of Aβ40 in which residue stretch 13–23 was shown to have a 310-helix conformation.75 Vivekanandan et al. showed that Aβ40 can adopt a partially folded structure.75 A collapsed coil structure was observed with the fragment of the peptide (residues from 10 to 35). It was also indicated in the CD spectral data and NMR analysis as discussed earlier.
Figure 5.
Structural analyses of Aβ40 and distribution of Ramachandran dihedral angles (ψ and Φ) in the full trajectory (1000 ns) of MD simulation using the Desmond algorithm. (A) Residue-specific average secondary structural preferences of the peptide. The color codes are as follows: white for loop, orange for helical conformation, and cyan for β-conformation. Y axis gives the residue number of the peptide as the residue index. (B) Three-dimensional plot of the distribution of Ramachandran dihedral angles (ψ and Φ) of all the residues present in the peptide. The z axis represents the frequency of occurrences.
The frequency of dihedral angles phi (Φ) and psi (Ψ) was monitored during the simulation time. The plot of the dihedral angle frequencies in a Ramachandran-like graph provides conformational preferences of the peptide. The major Ramachandran ψ/Φ angle distribution, as obtained by the MD analysis was found to peak at Ramachandran coordinates of ψ = −26° and Φ = −75° (Figure 5B). Three other comparatively week distributions were at (ψ = −140°, Φ = −24°), (ψ = +138°, Φ = −83°), and (ψ = +150°, Φ = −146°) in the Ramachandran (ψ, Φ) space. In the major ψ/Φ distribution, the Φ values were close to the right-handed α-helical space and the ψ values were close to the 310 helix.
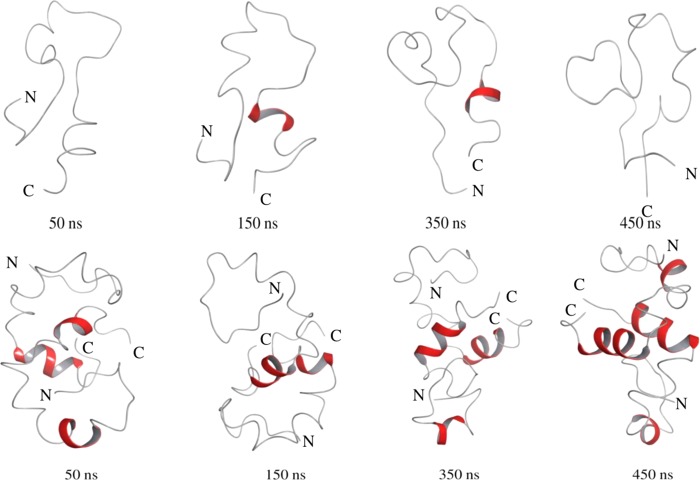
Possible conformational changes of the peptide as a dimer, which is believed to form in the early event of oligomerization of Aβ40, is shown in Figure 6 and compared with the conformational state of the monomer over an equal time of simulation. The total simulation time for the dimer was 450 ns. The backbone RMSD values for the dimer remained relatively low (1.1 nm), indicating their stability over the simulation time (Figure S6). It was found that a significant amount of the α-helix persisted for the dimer along with some residues with a turn conformation. Over a 450 ns simulation, a good number of residues attained the α-helix. The formation of some helix pattern in the sequence regions 13–16 and 28–36 was observed in the dimer (Figures 6 and S7).
Figure 6.
Snapshots of the conformational state of the Aβ40 peptide monomer (upper panel) and dimer (lower panel) over different time scales of MD simulations in water at 310 K using the Desmond algorithm. The peptide coloring schemes are as follows: red for helical conformation and gray for unstructured region.
The residue-specific interaction pattern (contact map) of the dimer of the Aβ40 peptide over a simulation period of 450 ns is shown in Figure S8. The contact probability map was made using all conformations generated at the full trajectory of the MD simulation. The residue segment, Leu17-Ala21, in one peptide and residues Gln15, Val18, and Ala21 in the other peptide were found to make high contact with each other. Contact probability in the Leu17-Ala21 of one peptide with Gly29-Met35 of the other peptide of the Aβ40 dimer was also observed. Many of these residues were in the segment that has the tendency to form a helical pattern. Tarus et al. examined the dimer of Aβ40 by molecular dynamics analysis and observed about 18% β-strand and 10% α helix residues. Their investigation also showed that one peptide segment with a β-sheet structure was stabilized by a peptide component with an α-helical structure.73
The results of the MD analysis largely depend on the accuracy and nature of the force fields embedded in the algorithm and the accompanying solvation models.76 However, in our current investigation, we used the optimized potential for liquid simulation (OPLS)/AA force field embedded in the Desmond algorithm. This was also used earlier to investigate the similar protein peptide dynamics. Earlier investigation using the OPLS force field found good agreement with the generated structures for Aβ40 and Aβ42 and their fragments with the experimental results.76,77 Gerben et al. found that OPLS-AA, GROMOS96 53A6, and GROMOS96 54A7 produce quite similar secondary structures (in terms of the helical and β-strand content) and matches well with the experimental results.77 Man et al. in their very recent investigation used four different force fields and observed different levels of helix content in the Aβ42 dimer; however, the helix content calculated by the OPLS-AA force field was comparable to CD data.70 When studying the effect of different force fields on peptide aggregation, Nguyen et al. also showed that OPLS/AA explored the most diverse conformations for the aggregation of the Aβ16–22 fragment in explicit water.78
Neumaier et al. investigated the helix–coil transition in helix-forming peptides and suggested that upon formation of the helix from the coil conformation a shrinkage of volume is eminent.80,79 Our simulation results also showed that compactness, as measured by the radius of gyration (Rg), decreases in the early event of folding as in the intermediate state. As a whole, MD simulation results indicated that the Aβ40 peptide had long-range residual preferences in unfoldedness; however, there was a strong helical tendency in aggregation-prone amyloidogenic regions. Thus, the observed compactness of the peptide as a monomer and its preferences of some of the residues for an ordered secondary structure may have a role in the processes of amyloid formation. We observed both the helical and β sheet content in the Raman spectroscopic measurement of the peptide in a different state of its aggregation, and our MD analysis indicated that the monomer itself may have attained some of this structure before its transformation into higher-order aggregates such as oligomer and fibril.
The C-terminal region of the peptide is more extended than the N-terminal region.81−84 Earlier molecular dynamics simulations predicted a partially structured monomeric state of the peptide.85−87 Our MD simulation analysis (based on the plot in Figure 5A) found that a maximum of 45% of the residues had helical propensity, ∼15% of which showed strong helical tenacity. We also observed that the λNOE for residue stretches of 12–15, 17–19, and 21–22 showed a low λNOE value for the amide protons, as stated earlier. A previous solid-state experimental study of Aβ40 by Petkova et al. showed that the main hydrophobic cross β-sheet core of the Aβ40 fiber (PDB: 2LMN) primarily consisted of LVFFAD (residues 17–22) and AIIGLM (residues 34–39).28 These segments were partially overlapping regions with our predicted most helicity-prone amyloid segment as obtained from MD simulation (Figure 5). The calculated grand averages of hydropathy of residue segments 17–21 and 28–36 are considerably high, and the values were 3.08 and 1.78, respectively. Thus, the regions may have a tendency to cause hydrophobic collapse in the processes of amyloid formation.
Our MD results and some of the previous investigations suggested that the peptide as a monomer itself has propensities to form some helical and β-sheet structure. The driving force of accelerated conversion of the monomer to early assembly formation of the oligomeric state could be the reconstruction of the highly energetic disordered peptide into two relatively stable ordered conformations, such as the helical fold and β-sheet/strand structures.88 Partial compact structures were also found for other amyloidogenic proteins. In the recent past, IR studies showed that the Josephin domain of ataxin-3 transformed into an amyloid fiber via formation of native oligomers, which possess both the random coil and α-helix conformations.89,90 These observations indicated that, like many other disordered proteins, the Aβ peptide may also contain a premolten globule state in its monomeric state just before the formation of oligomers, and some of these conformational states may be transmitted in the early stage of the soluble oligomer, which eventually transformed into the insoluble amyloid fiber.
Conclusions
In the current investigation, on the basis of mainly the Raman and CD spectroscopy data, we reported the content of a secondary structure of the amyloid β peptide (Aβ40) as a monomer and when it formed soluble oligomers and a highly insoluble amyloid fiber. It was found that ∼77% residues of the peptide as a monomer favored a disordered PPII-like conformation. However, in the oligomeric form the peptide contained residues distributed in the α-helical, PPII, and β-conformational spaces. Thus, the Raman signature suggested bifold secondary structural alteration (both helix and β conformation) in the processes of oligomerization of the peptide and disfavored the general assumption that the amyloid aggregation occurred through one direction, that is, mere β-conformational space transformation of the residues from its major PPII geometry in the monomeric state. However, in the fibrilar state the peptide mainly attained a β-sheet structure. Thus, the peptide secondary structure in the oligomeric state was very different from the fibrillar assembly. Like membrane fusion by a viral protein, the presence of helical segments in the oligomeric assembly may help the transmission of a toxic small oligomer from cell to cell. The observed result may thus be useful to understand the possible mechanism of Aβ aggregation and its link to disease formation and progression. This may also aid to design possible therapeutics targeting the region, particularly those show helical propensity, and to induce structural contact. Targeting completely disordered regions by a small molecule is very difficult due to the coexistence of an equivalent energetic ensemble of conformations or rather conformationally stable geometrical segments. In that sense, targeting residual segments, particularly in the sequence range of 12–22, may be more reliable compared to targeting highly disordered C-terminal residues to stop the formation of a soluble oligomer, which is believed now to be a potent neurotoxic species involving Aβ peptides in ADs.
Experimental Section
Sample Preparation
The peptide, full-length Aβ40, was purchased from American Peptide Company (777 E. Evelyn Ave., Sunnyvale, CA). Aβ40 was dissolved (1.5 mg/mL) in a 10 mM phosphate buffer of pH 7.2. The oligomerized peptide solution was prepared from the same monomeric solution by incubating for 3 h. The Aβ40 fibril was generated by dissolving the peptide (1.5 mg/mL) in the same buffer condition and incubating for 2 days at 25 °C.
Thioflavin T (ThT)-Binding Fluorescence Assay
A stock ThT dye was made (1 mM) in double-distilled water and stored in a cold and dark place. To monitor amyloid fibrillation, 600 μL of assay buffer [100 μM ThT in 20 mM PBS buffer (pH 7.2)] was mixed with 8 μL of incubated amyloid solution. The stock (1.5 mg/mL) was mixed properly before the fluorescence experiments. The mixed solution was taken in a 1 cm path length quartz cuvette, and the fluorescence measurement was performed using a Cary Eclipse fluorescence spectrophotometer. Emission spectra were acquired from 450 to 600 nm, excited at 440 nm, and the integration time was 1 s. Slit widths were 5 nm each. ThT in buffer without protein was also measured and used as the baseline.
CD
The far-UV CD spectra (190–250 nm) were recorded on a JASCO J-815 spectropolarimeter (Jasco) equipped with a Peltier temperature control unit. All of the CD measurements were performed at 25 °C with an accuracy of ±0.1 °C. The data acquisition interval value was 2 s. Spectra were recorded using a 0.1 cm path length cell with a 50 nm/min scan speed. For each sample, three scans were averaged, and the buffer background was subtracted. CD experimental studies at three different time intervals were performed using (1.5 mg/mL) Aβ40 in a 20 mM phosphate buffer (pH 7.2) at 25 °C. For CD analysis, 20 μL of the peptide sample (stock 1.5 mg/mL) was dissolved in 600 μL of the same PBS buffer to get a diluted solution of a concentration of about 10 μM.
Raman Spectroscopy
Raman spectra were recorded using a STR Raman spectrometer (model from AIRIX Corp.) equipped with a microscope (Olympus BX51 M, Japan) and 500 mm focal length triple grating monochromator connected with a charge-coupled device detector. The laser was focused through a 50× objective, and the scattered light from the sample was collected using the same objectives. The samples were excited with a 532 nm wavelength laser light, with 30–35 mW of radiant power at the source and ∼2 mW at the sample. The recording time was 15 s, and number of scans was 10. The wavenumbers of the Raman spectra were calibrated with the Raman band of silica wafer at 519 cm–1. Spectra were recorded from a spectral range of 500–1800 cm–1. Spectra were processed with the GRAMS/A1 software.
Curve Fitting of Amide I Band
The amide I band in the Raman spectra was used to investigating the changes of the secondary structure of the peptide. The band fitting process of the Raman amide I band was performed by the use of a Levenberg–Marquardt nonlinear least-squares process as applied in Curve Fit Ab routine in the GRAMS/AI 9.02 software.91 The Raman amide I mode (1580–1720 cm–1) was fitted assuming three or four symmetrical component bands that represent the different conformational states the peptide: α-helix (1650–1655 cm–1), β-sheet (1664–1670 cm–1), and polyproline II or extended β-strand (1670–1680 cm–1). During amide I band fitting, two different bands centered at 1604 and 1615 cm–1 for the ring vibrational mode of phenylalanine and tyrosine were also incorporated. A linear baseline correction was considered during the fitting processes. The standard error for both the peak positions and peak widths were ∼<5 cm–1, and uncertainty in the area measurement was ∼10% of the total area.
NMR Spectroscopy
All NMR data collection was done at 283 K on a BRUKER Avance NMR spectrometer equipped with a cryogenically cooled triple resonance probe. Freshly prepared full-length Aβ40 peptide sample was used. The concentration was the same as in the Raman experiments (1.5 mg/mL). Chemical shifts were calibrated with respect to standard 2,2-dimethyl-2-silapentane-5-sulfonate for proton, and 15N chemical shifts were calibrated indirectly. The sample had 10% 2H2O for deuterium locking. For efficient fast data acquisition, we have implemented SOFAST HSQC owing to the combined advantage of a small number of radio-frequency pulses, Ernst-angle excitation, and longitudinal relaxation optimization to perform the experiment with a high repetition rate to achieve a high signal to noise ratio using a small interscan delay period. Two-dimensional [15N,1H]-SOFAST-HMQC was recorded with the 1H carrier placed at the center of the amide region (8.5 ppm) and with the 15N carrier at 117.5 ppm. Selective excitation in the amide region was achieved with a 120° polychromatic pulse with 3 ms delay. For inversion, the R-SNOB pulse with 1 ms was used. Hetero-SOFAST NMR experiments incorporate an additional band-selective inversion pulse at the beginning of the pulse sequence, and 200 ms was used as the contact time. Two experiments are recorded with (excited) and without (reference) that specific inversion pulse, and the ratio of peak volumes are taken. Among the different sets of experiments, this selective pulse differs. This selective pulse (R-SNOB) was applied on aliphatic protons, which have a distinct chemicals shift range (0–4 ppm) compared to that of the amide proton chemical shift (6–10 ppm for folded proteins). Each experiment was acquired with 256 × 2048 complex points along the 15N and 1H dimensions, with an acquisition time of 63 ms and 107 ms, respectively. All of the data were processed with TOPSPIN 3.2 and were analyzed using CARA.
Transmission Electron Microscopy
Transmission Electron Microscopy has performed on a JEOL 1200EX transmission EM operating with an accelerating voltage of 120 kV. The protein sample (5 μL) was drop casted on 400 mesh copper grids (PST product, GSCu300C-50) for 5 min. Excess solution was adsorbed onto a filter paper. The grids were washed three times with distilled water and three times with filtered 1% uranyl acetate for negative staining and then dried for 20 min at room temperature.
MD Simulation (Desmond)
MD simulation analysis was performed using the Desmond Molecular Dynamics System 4.0. Wild-type Aβ40 was taken from PDB ID 1BA4.92 The Aβ40 peptide was solvated using the SPC water environment in a cubic box of 1 nm edge distance from any solute atom. For simulation of the dimer, we took two monomers with mostly an α-helical secondary structure in a cubic box and brought them into close proximity (∼3 nm). The total charge of the systems was neutralized with appropriate counterions. The OPLS 2005 force field parameter was used for this simulation.93 The initial system was subjected to energy minimization to 2000 steps of steepest descent integration to eliminate unfavorable contacts or steric clashes between the atoms. The pressure was regulated using the Martyna–Tobias–Klein method to 1 bar with isotropic coupling and a relaxation time of 2 ps, and a constant simulation temperature of 310 K was maintained using Nosé–Hoover thermostats.94−96 The equations of motion were integrated using the multistep RESPA integrator with bonded or near and far nonbonded interactions, at time steps of 2, 2, and 6 fs, respectively.97 Nonbonded interactions were subjected to a cutoff of 0.9 nm. Long-range electrostatics was treated with the smooth particle mesh Ewald method with a tolerance of 10–9. A five-step relaxation protocol was used starting with Brownian dynamics for 100 ps with restraints on solute heavy atoms at NVT (with T = 10 K), followed by 12 ps of dynamics with restraints at NVT (T = 10 K) and then at NPT (T = 10 K) using the Berendsen method. Then, the temperature was raised to 300 K for 12 ps, followed by a 24 ps relaxation step without restraints on the solute heavy atoms. The production MD was run at NPT with T = 310 K for 1000 ns. The molecular dynamics simulation trajectories were analyzed using a simulation event analysis tool, and all of the images were rendered using the Schrodinger Maestro Suite.
Acknowledgments
We sincerely thank Dr. Uttam Pal for his kind efforts in reading the manuscript and providing several meaningful suggestions that gave the manuscript its current form. Dr. K.C. acknowledges DST for financial support and project grant (DST-1323) as DST Inspire Faculty Fellowship. A.R. thanks UGC for the research fellowship, and S.D. also thanks CSIR-network projects BSC0113, BSC0115, and BSC0121 for funding and related support. The authors thank J. Mandal for CD measurement and also acknowledge the central instrumental facility, CSIR-IICB, and TIFR NMR facility, TIFR Mumbai. The Raman instrument was bought under DBT, New Delhi grant (GAP-299) to Dr. Nakul C. Maiti.
Supporting Information Available
The Supporting Information is available free of charge on the ACS Publications website at DOI: 10.1021/acsomega.7b00522.
Supporting Scheme S1, Table S1, and Figures S1–S8 (PDF)
The authors declare no competing financial interest.
Supplementary Material
References
- Golde T. E.; Estus S.; Younkin L. H.; Selkoe D. J.; Younkin S. G. Processing of the Amyloid Protein Precursor to Potentially Amyloidogenic Derivatives. Science 1992, 255, 728–730. 10.1126/science.1738847. [DOI] [PubMed] [Google Scholar]
- Hardy J. A.; Higgins G. A. Alzheimer’s Disease: The Amyloid Cascade Hypothesis. Science 1992, 256, 184–185. 10.1126/science.1566067. [DOI] [PubMed] [Google Scholar]
- Kirkitadze M. D.; Bitan G.; Teplow D. B. Paradigm Shifts in Alzheimer’s Disease and Other Neurodegenerative Disorders: The Emerging Role of Oligomeric Assemblies. J. Neurosci. Res. 2002, 69, 567–577. 10.1002/jnr.10328. [DOI] [PubMed] [Google Scholar]
- Selkoe D. J. Alzheimer Disease: Mechanistic Understanding Predicts Novel Therapies. Ann. Intern. Med. 2004, 140, 627. 10.7326/0003-4819-140-8-200404200-00010. [DOI] [PubMed] [Google Scholar]
- Walsh D. M.; Klyubin I.; Fadeeva J. V.; Cullen W. K.; Anwyl R.; Wolfe M. S.; Rowan M. J.; Selkoe D. J. Naturally Secreted Oligomers of Amyloid β Protein Potently Inhibit Hippocampal Long-Term Potentiation in Vivo. Nature 2002, 416, 535–539. 10.1038/416535a. [DOI] [PubMed] [Google Scholar]
- Klein W. L.; Stine W. B.; Teplow D. B. Small Assemblies of Unmodified Amyloid Beta-Protein Are the Proximate Neurotoxin in Alzheimer’s Disease. Neurobiol. Aging 2004, 25, 569–580. 10.1016/j.neurobiolaging.2004.02.010. [DOI] [PubMed] [Google Scholar]
- Chromy B. A.; Nowak R. J.; Lambert M. P.; Viola K. L.; Chang L.; Velasco P. T.; Jones B. W.; Fernandez S. J.; Lacor P. N.; Horowitz P.; et al. Self-Assembly of Abeta(1–42) into Globular Neurotoxins. Biochemistry 2003, 42, 12749–12760. 10.1021/bi030029q. [DOI] [PubMed] [Google Scholar]
- Lambert M. P.; Barlow A. K.; Chromy B. A.; Edwards C.; Freed R.; Liosatos M.; Morgan T. E.; Rozovsky I.; Trommer B.; Viola K. L.; et al. Diffusible, Nonfibrillar Ligands Derived from Aβ1–42 Are Potent Central Nervous System Neurotoxins. Proc. Natl. Acad. Sci. U.S.A. 1998, 95, 6448–6453. 10.1073/pnas.95.11.6448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kayed R.; Head E.; Thompson J. L.; McIntire T. M.; Milton S. C.; Cotman C. W.; Glabe C. G. Common Structure of Soluble Amyloid Oligomers Implies Common Mechanism of Pathogenesis. Science 2003, 300, 486–489. 10.1126/science.1079469. [DOI] [PubMed] [Google Scholar]
- Gong Y.; Chang L.; Viola K. L.; Lacor P. N.; Lambert M. P.; Finch C. E.; Krafft G. A.; Klein W. L. Alzheimer’s Disease-Affected Brain: Presence of Oligomeric Aβ Ligands (ADDLs) Suggests a Molecular Basis for Reversible Memory Loss. Proc. Natl. Acad. Sci. U.S.A. 2003, 100, 10417–10422. 10.1073/pnas.1834302100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi R. H.; Almeida C. G.; Kearney P. F.; Yu F.; Lin M. T.; Milner T. A.; Gouras G. K. Oligomerization of Alzheimer’s β-Amyloid within Processes and Synapses of Cultured Neurons and Brain. J. Neurosci. 2004, 24, 3592–3599. 10.1523/JNEUROSCI.5167-03.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harper J. D.; Wong S. S.; Lieber C. M.; Lansbury P. T. Assembly of Aβ Amyloid Protofibrils: An in Vitro Model for a Possible Early Event in Alzheimer’s Disease. Biochemistry 1999, 38, 8972–8980. 10.1021/bi9904149. [DOI] [PubMed] [Google Scholar]
- Harper J. D.; Lieber C. M.; Lansbury P. T. Atomic Force Microscopic Imaging of Seeded Fibril Formation and Fibril Branching by the Alzheimer’s Disease Amyloid-β Protein. Chem. Biol. 1997, 4, 951–959. 10.1016/S1074-5521(97)90303-3. [DOI] [PubMed] [Google Scholar]
- Harper J. D.; Lansbury P. T. Models of Amyloid Seeding in Alzheimer’s Disease and Scrapie: Mechanistic Truths and Physiological Consequences of the Time-Dependent Solubility of Amyloid Proteins. Annu. Rev. Biochem. 1997, 66, 385–407. 10.1146/annurev.biochem.66.1.385. [DOI] [PubMed] [Google Scholar]
- Walsh D. M.; Hartley D. M.; Kusumoto Y.; Fezoui Y.; Condron M. M.; Lomakin A.; Benedek G. B.; Selkoe D. J.; Teplow D. B. Amyloid Beta-Protein Fibrillogenesis. Structure and Biological Activity of Protofibrillar Intermediates. J. Biol. Chem. 1999, 274, 25945–25952. 10.1074/jbc.274.36.25945. [DOI] [PubMed] [Google Scholar]
- Mucke L.; Masliah E.; Yu G. Q.; Mallory M.; Rockenstein E. M.; Tatsuno G.; Hu K.; Kholodenko D.; Johnson-Wood K.; McConlogue L. High-Level Neuronal Expression of Abeta 1–42 in Wild-Type Human Amyloid Protein Precursor Transgenic Mice: Synaptotoxicity without Plaque Formation. J. Neurosci. 2000, 20, 4050–4058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morgan D.; Diamond D. M.; Gottschall P. E.; Ugen K. E.; Dickey C.; Hardy J.; Duff K.; Jantzen P.; DiCarlo G.; Wilcock D.; et al. A Beta Peptide Vaccination Prevents Memory Loss in an Animal Model of Alzheimer’s Disease. Nature 2000, 408, 982–985. 10.1038/35050116. [DOI] [PubMed] [Google Scholar]
- Dodart J.-C.; Bales K. R.; Gannon K. S.; Greene S. J.; DeMattos R. B.; Mathis C.; DeLong C. A.; Wu S.; Wu X.; Holtzman D. M.; et al. Immunization Reverses Memory Deficits without Reducing Brain Abeta Burden in Alzheimer’s Disease Model. Nat. Neurosci. 2002, 5, 452–457. 10.1038/nn842. [DOI] [PubMed] [Google Scholar]
- Kotilinek L. A.; Bacskai B.; Westerman M.; Kawarabayashi T.; Younkin L.; Hyman B. T.; Younkin S.; Ashe K. H. Reversible Memory Loss in a Mouse Transgenic Model of Alzheimer’s Disease. J. Neurosci. 2002, 22, 6331–6335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldberg M. S.; Lansbury P. T. Jr. Is There a Cause-and-Effect Relationship between α-Synuclein Fibrillization and Parkinson’s Disease?. Nat. Cell Biol. 2000, 2, E115–E119. 10.1038/35017124. [DOI] [PubMed] [Google Scholar]
- Lashuel H. A.; Petre B. M.; Wall J.; Simon M.; Nowak R. J.; Walz T.; Lansbury P. T. Alpha-Synuclein, Especially the Parkinson’s Disease-Associated Mutants, Forms Pore-like Annular and Tubular Protofibrils. J. Mol. Biol. 2002, 322, 1089–1102. 10.1016/S0022-2836(02)00735-0. [DOI] [PubMed] [Google Scholar]
- Manelli A. M.; Stine W. B.; Eldik L. J. V.; LaDu M. J. ApoE and Abeta1–42 Interactions: Effects of Isoform and Conformation on Structure and Function. J. Mol. Neurosci. 2004, 23, 235–246. 10.1385/JMN:23:3:235. [DOI] [PubMed] [Google Scholar]
- Kanemitsu H.; Tomiyama T.; Mori H. Human Neprilysin Is Capable of Degrading Amyloid β Peptide Not Only in the Monomeric Form but Also the Pathological Oligomeric Form. Neurosci. Lett. 2003, 350, 113–116. 10.1016/S0304-3940(03)00898-X. [DOI] [PubMed] [Google Scholar]
- Jabłonowska A.; Dadlez M. A Beta Peptide Oligomers - Potential Neurotoxic Agents in Alzheimer’s Disease. Polimery 2003, T. 48, 35–43. [Google Scholar]
- Lemere C. A.; Beierschmitt A.; Iglesias M.; Spooner E. T.; Bloom J. K.; Leverone J. F.; Zheng J. B.; Seabrook T. J.; Louard D.; Li D.; et al. Alzheimer’s Disease Abeta Vaccine Reduces Central Nervous System Abeta Levels in a Non-Human Primate, the Caribbean Vervet. Am. J. Pathol. 2004, 165, 283–297. 10.1016/S0002-9440(10)63296-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar S.; Hamilton A. D. α-Helix Mimetics as Modulators of Aβ Self-Assembly. J. Am. Chem. Soc. 2017, 139, 5744–5755. 10.1021/jacs.6b09734. [DOI] [PubMed] [Google Scholar]
- Murphy R. M. Peptide Aggregation in Neurodegenerative Disease. Annu. Rev. Biomed. Eng. 2002, 4, 155–174. 10.1146/annurev.bioeng.4.092801.094202. [DOI] [PubMed] [Google Scholar]
- Petkova A. T.; Buntkowsky G.; Dyda F.; Leapman R. D.; Yau W.-M.; Tycko R. Solid State NMR Reveals a pH-Dependent Antiparallel Beta-Sheet Registry in Fibrils Formed by a Beta-Amyloid Peptide. J. Mol. Biol. 2004, 335, 247–260. 10.1016/j.jmb.2003.10.044. [DOI] [PubMed] [Google Scholar]
- Filippov A. V.; Gröbner G.; Antzutkin O. N. Aggregation of Amyloid Abeta((1–40)) Peptide in Perdeuterated 2,2,2-Trifluoroethanol Caused by Ultrasound Sonication. Magn. Reson. Chem. 2010, 48, 427–434. 10.1002/mrc.2596. [DOI] [PubMed] [Google Scholar]
- Zagorski M. G.; Shao H.; Ma K.; Yang J.; Li H.; Zeng H.; Zhang Y.; Papolla M. Amyloid AB (1–40) and AB(1–42) Adopt Remarkably Stable, Monomeric, and Extended Structures in Water Solution at Neutral pH. Neurobiol. Aging 2000, 21, 10–11. 10.1016/S0197-4580(00)82724-4. [DOI] [Google Scholar]
- Shao H.; Jao S.; Ma K.; Zagorski M. G. Solution Structures of Micelle-Bound Amyloid Beta-(1–40) and Beta-(1–42) Peptides of Alzheimer’s Disease. J. Mol. Biol. 1999, 285, 755–773. 10.1006/jmbi.1998.2348. [DOI] [PubMed] [Google Scholar]
- Zhang S.; Iwata K.; Lachenmann M. J.; Peng J. W.; Li S.; Stimson E. R.; Lu Y.; Felix A. M.; Maggio J. E.; Lee J. P. The Alzheimer’s Peptide a Beta Adopts a Collapsed Coil Structure in Water. J. Struct. Biol. 2000, 130, 130–141. 10.1006/jsbi.2000.4288. [DOI] [PubMed] [Google Scholar]
- Kirschner D. A.; Abraham C.; Selkoe D. J. X-Ray Diffraction from Intraneuronal Paired Helical Filaments and Extraneuronal Amyloid Fibers in Alzheimer Disease Indicates Cross-Beta Conformation. Proc. Natl. Acad. Sci. U.S.A. 1986, 83, 503–507. 10.1073/pnas.83.2.503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirschner D. A.; Inouye H.; Duffy L. K.; Sinclair A.; Lind M.; Selkoe D. J. Synthetic Peptide Homologous to Beta Protein from Alzheimer Disease Forms Amyloid-like Fibrils in Vitro. Proc. Natl. Acad. Sci. U.S.A. 1987, 84, 6953–6957. 10.1073/pnas.84.19.6953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Antzutkin O. N.; Balbach J. J.; Leapman R. D.; Rizzo N. W.; Reed J.; Tycko R. Multiple Quantum Solid-State NMR Indicates a Parallel, Not Antiparallel, Organization of β-Sheets in Alzheimer’s β-Amyloid Fibrils. Proc. Natl. Acad. Sci. U.S.A. 2000, 97, 13045–13050. 10.1073/pnas.230315097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Legleiter J.; Czilli D. L.; Gitter B.; DeMattos R. B.; Holtzman D. M.; Kowalewski T. Effect of Different Anti-Abeta Antibodies on Abeta Fibrillogenesis as Assessed by Atomic Force Microscopy. J. Mol. Biol. 2004, 335, 997–1006. 10.1016/j.jmb.2003.11.019. [DOI] [PubMed] [Google Scholar]
- Moreno-Herrero F.; Pérez M.; Baró A. M.; Avila J. Characterization by Atomic Force Microscopy of Alzheimer Paired Helical Filaments under Physiological Conditions. Biophys. J. 2004, 86, 517–525. 10.1016/S0006-3495(04)74130-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoyer W.; Cherny D.; Subramaniam V.; Jovin T. M. Rapid Self-Assembly of α-Synuclein Observed by In Situ Atomic Force Microscopy. J. Mol. Biol. 2004, 340, 127–139. 10.1016/j.jmb.2004.04.051. [DOI] [PubMed] [Google Scholar]
- Arnaudov L. N.; de Vries R. Thermally Induced Fibrillar Aggregation of Hen Egg White Lysozyme. Biophys. J. 2005, 88, 515–526. 10.1529/biophysj.104.048819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aguzzi A.; Calella A. M. Prions: Protein Aggregation and Infectious Diseases. Physiol. Rev. 2009, 89, 1105–1152. 10.1152/physrev.00006.2009. [DOI] [PubMed] [Google Scholar]
- Rochet J. C.; Lansbury P. T. Amyloid Fibrillogenesis: Themes and Variations. Curr. Opin. Struct. Biol. 2000, 10, 60–68. 10.1016/S0959-440X(99)00049-4. [DOI] [PubMed] [Google Scholar]
- Walsh D. M.; Lomakin A.; Benedek G. B.; Condron M. M.; Teplow D. B. Amyloid Beta-Protein Fibrillogenesis. Detection of a Protofibrillar Intermediate. J. Biol. Chem. 1997, 272, 22364–22372. 10.1074/jbc.272.35.22364. [DOI] [PubMed] [Google Scholar]
- Kirkitadze M. D.; Condron M. M.; Teplow D. B. Identification and Characterization of Key Kinetic Intermediates in Amyloid Beta-Protein Fibrillogenesis. J. Mol. Biol. 2001, 312, 1103–1119. 10.1006/jmbi.2001.4970. [DOI] [PubMed] [Google Scholar]
- Nelson R.; Sawaya M. R.; Balbirnie M.; Madsen A. Ø.; Riekel C.; Grothe R.; Eisenberg D. Structure of the Cross-β Spine of Amyloid-like Fibrils. Nature 2005, 435, 773–778. 10.1038/nature03680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheidt H. A.; Morgado I.; Rothemund S.; Huster D.; Fändrich M. Solid-State NMR Spectroscopic Investigation of Aβ Protofibrils: Implication of a β-Sheet Remodeling upon Maturation into Terminal Amyloid Fibrils. Angew. Chem., Int. Ed. 2011, 50, 2837–2840. 10.1002/anie.201007265. [DOI] [PubMed] [Google Scholar]
- Schütz A. K.; Vagt T.; Huber M.; Ovchinnikova O. Y.; Cadalbert R.; Wall J.; Güntert P.; Böckmann A.; Glockshuber R.; Meier B. H. Atomic-Resolution Three-Dimensional Structure of Amyloid β Fibrils Bearing the Osaka Mutation. Angew. Chem., Int. Ed. Engl. 2015, 54, 331–335. 10.1002/anie.201408598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maiti N. C.; Apetri M. M.; Zagorski M. G.; Carey P. R.; Anderson V. E. Raman Spectroscopic Characterization of Secondary Structure in Natively Unfolded Proteins: Alpha-Synuclein. J. Am. Chem. Soc. 2004, 126, 2399–2408. 10.1021/ja0356176. [DOI] [PubMed] [Google Scholar]
- Apetri M. M.; Maiti N. C.; Zagorski M. G.; Carey P. R.; Anderson V. E. Secondary Structure of Alpha-Synuclein Oligomers: Characterization by Raman and Atomic Force Microscopy. J. Mol. Biol. 2006, 355, 63–71. 10.1016/j.jmb.2005.10.071. [DOI] [PubMed] [Google Scholar]
- Benevides J. M.; Bondre P.; Duda R. L.; Hendrix R. W.; Thomas G. J. Domain Structures and Roles in Bacteriophage HK97 Capsid Assembly and Maturation. Biochemistry 2004, 43, 5428–5436. 10.1021/bi0302494. [DOI] [PubMed] [Google Scholar]
- Thomas G. J. Jr. Raman Spectroscopy of Protein and Nucleic Acid Assemblies. Annu. Rev. Biophys. Biomol. Struct. 1999, 28, 1–27. 10.1146/annurev.biophys.28.1.1. [DOI] [PubMed] [Google Scholar]
- Huang K.; Maiti N. C.; Phillips N. B.; Carey P. R.; Weiss M. A. Structure-Specific Effects of Protein Topology on Cross-Beta Assembly: Studies of Insulin Fibrillation. Biochemistry 2006, 45, 10278–10293. 10.1021/bi060879g. [DOI] [PubMed] [Google Scholar]
- Xiong K.; Asher S. A. Circular Dichroism and UV Resonance Raman Study of the Impact of Alcohols on the Gibbs Free Energy Landscape of an α-Helical Peptide. Biochemistry 2010, 49, 3336–3342. 10.1021/bi100176a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsuboi M.; Suzuki M.; Overman S. A.; Thomas G. J. Intensity of the Polarized Raman Band at 1340–1345 Cm-1 as an Indicator of Protein Alpha-Helix Orientation: Application to Pf1 Filamentous Virus. Biochemistry 2000, 39, 2677–2684. 10.1021/bi9918846. [DOI] [PubMed] [Google Scholar]
- Tuma R. Raman Spectroscopy of Proteins: From Peptides to Large Assemblies. J. Raman Spectrosc. 2005, 36, 307–319. 10.1002/jrs.1323. [DOI] [Google Scholar]
- Ahmed M.; Davis J.; Aucoin D.; Sato T.; Ahuja S.; Aimoto S.; Elliott J. I.; Van Nostrand W. E.; Smith S. O. Structural Conversion of Neurotoxic Amyloid-β1–42 Oligomers to Fibrils. Nat. Struct. Mol. Biol. 2010, 17, 561–567. 10.1038/nsmb.1799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng R.; Zheng X.; Dong J.; Carey P. R. Proteins Can Convert to β-Sheet in Single Crystals. Protein Sci. 2004, 13, 1288–1294. 10.1110/ps.03550404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benevides J. M.; Bondre P.; Duda R. L.; Hendrix R. W.; Thomas G. J. Domain Structures and Roles in Bacteriophage HK97 Capsid Assembly and Maturation. Biochemistry 2004, 43, 5428–5436. 10.1021/bi0302494. [DOI] [PubMed] [Google Scholar]
- Benevides J. M.; Weiss M. A.; Thomas G. J. Design of the Helix-Turn-Helix Motif: Nonlocal Effects of Quaternary Structure in DNA Recognition Investigated by Laser Raman Spectroscopy. Biochemistry 1991, 30, 4381–4388. 10.1021/bi00232a003. [DOI] [PubMed] [Google Scholar]
- Overman S. A.; Thomas G. J. Amide Modes of the α-Helix: Raman Spectroscopy of Filamentous Virus Fd Containing Peptide 13C and 2H Labels in Coat Protein Subunits. Biochemistry 1998, 37, 5654–5665. 10.1021/bi972339c. [DOI] [PubMed] [Google Scholar]
- Wälti M. A.; Ravotti F.; Arai H.; Glabe C. G.; Wall J. S.; Böckmann A.; Güntert P.; Meier B. H.; Riek R. Atomic-Resolution Structure of a Disease-Relevant Aβ(1–42) Amyloid Fibril. Proc. Natl. Acad. Sci. U.S.A. 2016, 113, E4976–E4984. 10.1073/pnas.1600749113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mikhonin A. V.; Myshakina N. S.; Bykov S. V.; Asher S. A. UV Resonance Raman Determination of Polyproline II, Extended 2.51-Helix, and β-Sheet Ψ Angle Energy Landscape in Poly-L-Lysine and Poly-L-Glutamic Acid. J. Am. Chem. Soc. 2005, 127, 7712–7720. 10.1021/ja044636s. [DOI] [PubMed] [Google Scholar]
- Bonifacio A.; Sergo V. Effects of Sample Orientation in Raman Microspectroscopy of Collagen Fibers and Their Impact on the Interpretation of the Amide III Band. Vib. Spectrosc. 2010, 53, 314–317. 10.1016/j.vibspec.2010.04.004. [DOI] [Google Scholar]
- Petkova A. T.; Yau W.-M.; Tycko R. Experimental Constraints on Quaternary Structure in Alzheimer’s Beta-Amyloid Fibrils. Biochemistry 2006, 45, 498–512. 10.1021/bi051952q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petkova A. T.; Leapman R. D.; Guo Z.; Yau W.-M.; Mattson M. P.; Tycko R. Self-Propagating, Molecular-Level Polymorphism in Alzheimer’s ß-Amyloid Fibrils. Science 2005, 307, 262–265. 10.1126/science.1105850. [DOI] [PubMed] [Google Scholar]
- Kumar S.; Udgaonkar J. B. Mechanisms of Amyloid Fibril Formation by Proteins. Curr. Sci. 2010, 98, 639–656. [Google Scholar]
- Nath U.; Udgaonkar J. B. How Do Proteins Fold?. Curr. Sci. 1997, 72, 180–191. [Google Scholar]
- Adcock S. A.; McCammon J. A. Molecular Dynamics: Survey of Methods for Simulating the Activity of Proteins. Chem. Rev. 2006, 106, 1589–1615. 10.1021/cr040426m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karplus M.; McCammon J. A. Molecular Dynamics Simulations of Biomolecules. Nat. Struct. Biol. 2002, 9, 646–652. 10.1038/nsb0902-646. [DOI] [PubMed] [Google Scholar]
- MacKerell A. D.; Bashford D.; Bellott M.; Dunbrack R. L.; Evanseck J. D.; Field M. J.; Fischer S.; Gao J.; Guo H.; Ha S.; et al. All-Atom Empirical Potential for Molecular Modeling and Dynamics Studies of Proteins. J. Phys. Chem. B 1998, 102, 3586–3616. 10.1021/jp973084f. [DOI] [PubMed] [Google Scholar]
- Man V. H.; Nguyen P. H.; Derreumaux P. High-Resolution Structures of the Amyloid-β 1–42 Dimers from the Comparison of Four Atomistic Force Fields. J. Phys. Chem. B 2017, 121, 5977–5987. 10.1021/acs.jpcb.7b04689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Man V. H.; Nguyen P. H.; Derreumaux P. High-Resolution Structures of the Amyloid-β 1–42 Dimers from the Comparison of Four Atomistic Force Fields. J. Phys. Chem. B 2017, 121, 5977–5987. 10.1021/acs.jpcb.7b04689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Itoh S. G.; Okumura H. Oligomer Formation of Amyloid-β(29–42) from Its Monomers Using the Hamiltonian Replica-Permutation Molecular Dynamics Simulation. J. Phys. Chem. B 2016, 120, 6555–6561. 10.1021/acs.jpcb.6b03828. [DOI] [PubMed] [Google Scholar]
- Tarus B.; Tran T. T.; Nasica-Labouze J.; Sterpone F.; Nguyen P. H.; Derreumaux P. Structures of the Alzheimer’s Wild-Type Aβ1–40 Dimer from Atomistic Simulations. J. Phys. Chem. B 2015, 119, 10478–10487. 10.1021/acs.jpcb.5b05593. [DOI] [PubMed] [Google Scholar]
- Nguyen H. L.; Thi Minh Thu T.; Truong P. M.; Lan P. D.; Man V. H.; Nguyen P. H.; Tu L. A.; Chen Y.-C.; Li M. S. Aβ41 Aggregates More Like Aβ40 than Like Aβ42: In Silico and in Vitro Study. J. Phys. Chem. B 2016, 120, 7371–7379. 10.1021/acs.jpcb.6b06368. [DOI] [PubMed] [Google Scholar]
- Vivekanandan S.; Brender J. R.; Lee S. Y.; Ramamoorthy A. A Partially Folded Structure of Amyloid-Beta(1–40) in an Aqueous Environment. Biochem. Biophys. Res. Commun. 2011, 411, 312–316. 10.1016/j.bbrc.2011.06.133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith M. D.; Rao J. S.; Segelken E.; Cruz L. Force-Field Induced Bias in the Structure of Aβ21–30: A Comparison of OPLS, AMBER, CHARMM, and GROMOS Force Fields. J. Chem. Inf. Model. 2015, 55, 2587–2595. 10.1021/acs.jcim.5b00308. [DOI] [PubMed] [Google Scholar]
- Gerben S. R.; Lemkul J. A.; Brown A. M.; Bevan D. R. Comparing Atomistic Molecular Mechanics Force Fields for a Difficult Target: A Case Study on the Alzheimer’s Amyloid β-Peptide. J. Biomol. Struct. Dyn. 2014, 32, 1817–1832. 10.1080/07391102.2013.838518. [DOI] [PubMed] [Google Scholar]
- Nguyen P. H.; Li M. S.; Derreumaux P. Effects of All-Atom Force Fields on Amyloid Oligomerization: Replica Exchange Molecular Dynamics Simulations of the Aβ16–22 Dimer and Trimer. Phys. Chem. Chem. Phys. 2011, 13, 9778–9788. 10.1039/c1cp20323a. [DOI] [PubMed] [Google Scholar]
- Neumaier S.; Büttner M.; Bachmann A.; Kiefhaber T. Transition State and Ground State Properties of the Helix–coil Transition in Peptides Deduced from High-Pressure Studies. Proc. Natl. Acad. Sci. U.S.A. 2013, 110, 20988–20993. 10.1073/pnas.1317973110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imamura H.; Kato M. Effect of Pressure on Helix-Coil Transition of an Alanine-Based Peptide: An FTIR Study. Proteins 2009, 75, 911–918. 10.1002/prot.22302. [DOI] [PubMed] [Google Scholar]
- Lim K. H. A Weakly Clustered N Terminus Inhibits Aβ(1–40) Amyloidogenesis. ChemBioChem 2006, 7, 1662–1666. 10.1002/cbic.200600270. [DOI] [PubMed] [Google Scholar]
- Lim K. H.; Henderson G. L.; Jha A.; Louhivuori M. Structural, Dynamic Properties of Key Residues in Abeta Amyloidogenesis: Implications of an Important Role of Nanosecond Timescale Dynamics. Chembiochem Eur. J. Chem. Biol. 2007, 8, 1251–1254. 10.1002/cbic.200700194. [DOI] [PubMed] [Google Scholar]
- Yan Y.; Wang C. Aβ42 Is More Rigid than Aβ40 at the C Terminus: Implications for Aβ Aggregation and Toxicity. J. Mol. Biol. 2006, 364, 853–862. 10.1016/j.jmb.2006.09.046. [DOI] [PubMed] [Google Scholar]
- Yan Y.; Liu J.; McCallum S. A.; Yang D.; Wang C. Methyl Dynamics of the Amyloid-β Peptides Aβ40 and Aβ42. Biochem. Biophys. Res. Commun. 2007, 362, 410. 10.1016/j.bbrc.2007.07.198. [DOI] [PubMed] [Google Scholar]
- Vitalis A.; Caflisch A. Micelle-Like Architecture of the Monomer Ensemble of Alzheimer’s Amyloid-β Peptide in Aqueous Solution and Its Implications for Aβ Aggregation. J. Mol. Biol. 2010, 403, 148–165. 10.1016/j.jmb.2010.08.003. [DOI] [PubMed] [Google Scholar]
- Yang M.; Teplow D. B. Amyloid β-Protein Monomer Folding: Free-Energy Surfaces Reveal Alloform-Specific Differences. J. Mol. Biol. 2008, 384, 450–464. 10.1016/j.jmb.2008.09.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomaselli S.; Esposito V.; Vangone P.; van Nuland N. A. J.; Bonvin A. M. J. J.; Guerrini R.; Tancredi T.; Temussi P. A.; Picone D. The α-to-β Conformational Transition of Alzheimer’s Aβ-(1–42) Peptide in Aqueous Media Is Reversible: A Step by Step Conformational Analysis Suggests the Location of β Conformation Seeding. ChemBioChem 2006, 7, 257–267. 10.1002/cbic.200500223. [DOI] [PubMed] [Google Scholar]
- Zheng W.; Tsai M.-Y.; Chen M.; Wolynes P. G. Exploring the Aggregation Free Energy Landscape of the Amyloid-β Protein (1–40). Proc. Natl. Acad. Sci. U.S.A. 2016, 113, 11835–11840. 10.1073/pnas.1612362113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masino L.; Nicastro G.; De Simone A.; Calder L.; Molloy J.; Pastore A. The Josephin Domain Determines the Morphological and Mechanical Properties of Ataxin-3 Fibrils. Biophys. J. 2011, 100, 2033–2042. 10.1016/j.bpj.2011.02.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tayeb-Fligelman E.; Tabachnikov O.; Moshe A.; Goldshmidt-Tran O.; Sawaya M. R.; Coquelle N.; Colletier J.-P.; Landau M. The Cytotoxic Staphylococcus aureus PSMα3 Reveals a Cross-α Amyloid-like Fibril. Science 2017, 355, 831–833. 10.1126/science.aaf4901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marquardt D. W. An Algorithm for Least-Squares Estimation of Nonlinear Parameters. J. Soc. Ind. Appl. Math. 1963, 11, 431–441. 10.1137/0111030. [DOI] [Google Scholar]
- Coles M.; Bicknell W.; Watson A. A.; Fairlie D. P.; Craik D. J. Solution Structure of Amyloid Beta-peptide(1–40) in a Water-Micelle Environment. Is the Membrane-Spanning Domain Where We Think It Is?. Biochemistry 1998, 37, 11064–11077. 10.1021/bi972979f. [DOI] [PubMed] [Google Scholar]
- Banks J. L.; Beard H. S.; Cao Y.; Cho A. E.; Damm W.; Farid R.; Felts A. K.; Halgren T. A.; Mainz D. T.; Maple J. R.; et al. Integrated Modeling Program, Applied Chemical Theory (IMPACT). J. Comput. Chem. 2005, 26, 1752–1780. 10.1002/jcc.20292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martyna G. J.; Tobias D. J.; Klein M. L. Constant Pressure Molecular Dynamics Algorithms. J. Chem. Phys. 1994, 101, 4177–4189. 10.1063/1.467468. [DOI] [Google Scholar]
- Nosé S. A Unified Formulation of the Constant Temperature Molecular Dynamics Methods. J. Chem. Phys. 1984, 81, 511–519. 10.1063/1.447334. [DOI] [Google Scholar]
- Hoover W. G. Canonical Dynamics: Equilibrium Phase-Space Distributions. Phys. Rev. A 1985, 31, 1695–1697. 10.1103/PhysRevA.31.1695. [DOI] [PubMed] [Google Scholar]
- Tuckerman M.; Berne B. J.; Martyna G. J. Reversible Multiple Time Scale Molecular Dynamics. J. Chem. Phys. 1992, 97, 1990–2001. 10.1063/1.463137. [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.