Abstract

It was previously reported that two naphthyl-based trans-stilbene probes, (E)-4-(2-(naphthalen-1-yl)vinyl)benzene-1,2-diol (1) and (E)-4-(2-(naphthalen-2-yl)vinyl)benzene-1,2-diol (3), can bind to both native transthyretin (TTR) and misfolded protofibrillar TTR at physiological concentrations, displaying distinct emission maxima bound to the different conformational states (>100 nm difference). To further explore this amyloid probe scaffold to obtain extended fluorescence lifetimes, two new analogues with expanded aromatic ring systems (anthracene and pyrene), (E)-4-(2-(anthracen-2-yl)vinyl)benzene-1,2-diol (4) and (E)-4-(2-(pyren-2-yl)vinyl)benzene-1,2-diol (5), were synthesized employing the palladium-catalyzed Mizoroki–Heck reaction. (E)-4-Styrylbenzene-1,2-diol (2), 3, 4, and 5 were investigated with respect to their photophysical properties in methanol and when bound to insulin, lysozyme, and Aβ1-42 fibrils, including time-resolved fluorescence measurements. In conclusion, 4 and 5 can bind to both native and fibrillar TTR, becoming highly fluorescent. Compounds 2–5 bind specifically to insulin, lysozyme, and Aβ1-42 fibrils with an apparent fluorescence intensity increase and moderate binding affinities. The average fluorescence lifetimes of the probes bound to Aβ1-42 fibrils are 1.3 ns (2), 1.5 ns (3), 5.7 ns (4), and 29.8 ns (5). In summary, the variable aromatic moieties of the para-positioned trans-stilbenoid vinyl-benzene-1,2-diol with benzene, naphthalene, anthracene, and pyrene showed that the extended conjugated systems retained the amyloid targeting properties of the probes. Furthermore, both the anthracene and pyrene moieties extensively enhanced the fluorescence intensity and prolonged lifetimes. These attractive probe properties should improve amyloid detection and characterization by fluorescence-based techniques.
Introduction
Many aspects of amyloid fibrils have been studied owing to their association with a variety of human diseases, such as Alzheimer’s disease, type II diabetes, amyloidosis, and others.1−7 Amyloid fibrils form when native proteins misfold and self-assemble into insoluble aggregates,8,9 which are deposited in organs or tissues where the aggregates have a pathogenic effect.10 Up to date, several characteristics of the amyloid fibril structure are widely accepted, for example, (1) mature fibrils are composed of parallel β-strands, which are oriented perpendicular to the long fibril axis,3,7,11,12 (2) mature fibrils have variable lengths up to several micrometers with a width between 8 and 10 nm and are often twisted and are generally unbranched.11−13 It has been recently reported that most proteins could form amyloid fibrils under specific conditions14,15 and that fibril formation is an intrinsic property of the polypeptide backbone.3
Characterization of amyloid fibrils has been performed by using various spectroscopic and microscopic techniques, such as atomic force and electron microscopy, circular dichroism, Fourier transform infrared, luminescence, and others.16−18 The most commonly used, fast, and reliable method for spectroscopic determination of amyloid fibril formation is by using fluorescent dyes, such as luminescent conjugated poly-, and oligothiophenes19,20 (LCP/LCO), Congo red (CR), or thioflavin T (ThT). Furthermore, upon binding to amyloid fibrils, CR generates a characteristic apple-green birefringence when viewed through crossed polarizers, and a characteristic red shift in absorbance maximum from about 490 nm to about 540 nm.21−24 However, it has been reported that CR can bind to native proteins.24 ThT, a charged molecular rotor molecule, shows an apparent fluorescence intensity increase upon binding to fibrils and its binding property is different at acidic and neutral pH,25−28 and it has been reported to bind poorly to transthyretin (TTR) fibrils at an acidic pH.29 In addition, both LCOs and ThT have a relatively short fluorescence lifetime with average t1/2 < 1 ns.29 We have previously demonstrated the power of time-resolved fluorescence for the characterization of oligomerization of amyloid proteins, allowing both real-time detection of oligomerization and size distribution analysis.17,29 Especially, a noncovalent small-molecule amyloid targeting probe with an extended lifetime is desirable in this regard. Here, there is ample room for current probe improvement to facilitate detection and characterization of amyloid protein structures. Such probes that utilize both spectral and temporal differences open up alternative modes of detection, for example, fluorescence lifetime imaging and related methods.30,31
In recent years, various trans-stilbene-based probes for the detection of amyloid fibrils and amyloidogenic proteins have been reported (Chart 1). The Kelly group has reported that resveratrol can bind native TTR and becomes fluorescent.32,33 [11C]BF-227 and 18F-florbetapir containing a trans-stilbenoid structure have been successfully used as positron emission tomography ligands to trace cardiac TTR amyloid.34,35 Previously, we successfully developed two naphthyl-based trans-stilbene probes, (E)-4-(2-(naphthalen-1-yl)vinyl)benzene-1,2-diol (1) and (E)-4-(2-(naphthalen-2-yl)vinyl)benzene-1,2-diol (3), that can bind both native TTR and misfolded protofibrillar TTR at physiological concentrations.36 These probes displayed distinct shift in emission maxima (>100 nm) when bound to the two different TTR conformations, allowing conformational discrimination by fluorescence spectroscopy. Hence, this class of compounds having a trans-stilbene or trans-stilbenoid structure are indeed a promising fluorescent probe scaffold for amyloid research. Accordingly, we designed and prepared two new analogues (4 and 5, see the structures in Chart 1), where the naphthyl was replaced by anthracenyl or pyrenyl to investigate their spectral discrimination of binding native and protofibrillar TTR as well as three other canonical amyloid proteins. Subsequently, the emission properties of these analogues (2–5) with different aromatic ring moieties were investigated using steady-state and time-resolved fluorescence spectroscopies, which demonstrate the feasibility of these new scaffolds for amyloid fibril characterization.
Chart 1. Chemical Structures of trans-Stilbenoidsa.
a 1, 2, and 3 have been reported previously by our group.364 and 5 were synthesized in this paper.
Experimental Section
Materials and Methods
All chemicals were obtained from conventional commercial sources and used without further purification unless otherwise noted. The synthesis of compound 2-(Bpin)pyrene (6) was performed according to the literature.37−39 The synthesis of compound 2-bromopyrene (7) was carried out according to the literature.39−41 3,4-Dimethoxystyrene (9) was synthesized according to the literature.42,43 The synthetic method of compounds 4 and 5 is shown in the Supporting Information. Thin layer chromatography was performed on Merck silica gel 60F254 glass-backed plates. Flash chromatography was carried out with silica gel 60. Nuclear magnetic resonance (NMR) spectra were recorded on a Varian AVANCE 300 MHz instrument with solvent indicated. The 1H NMR and 13C NMR chemical shifts are reported in parts per million relative to tetramethylsilane or the residual solvent protons.
Amyloid Fibril Formation
WT TTR was isolated and purified, as previously described.44 The TTR protein concentration was determined using the molar extinction coefficient (ε) of the monomeric TTR 18 289 M–1 cm–1 at 280 nm.45 The fibrillation of native TTR in dH2O was induced by adding a stock solution of 1 M NaAc and 2 M NaCl to a final concentration of 50 mM NaAc and 100 mM NaCl (final pH 3.0). Concentration of the insulin was determined by using ε = 5840 M–1 cm–1 at 277 nm. Bovine insulin fibril formation was initiated by incubating 200 μM in 25 mM HCl (pH 1.6) at 65 °C.46 The chicken egg-white lysozyme protein concentration was determined using the ε = 37 752 M–1 cm–1 at 280 nm, and fibril formation was initiated using 200 μM at 65 °C in 25 mM HCl (pH 1.6).46,47 Insulin and lysozyme fibrils were dialyzed against 50 mM NaAc and 100 mM NaCl (final pH 3.0) prior to measurements. Recombinant Aβ1-42 was expressed and purified using the system described by Linse and co-workers,48 containing an additional methionine in the N-terminus. Before the final step, the lyophilized Aβ1-42 sample was solubilized in 6 M GuHCl before gel filtration on a Superdex 75 10/300 column (GE-Healthcare) in phosphate-buffered saline (PBS) buffer pH 7.5. Freshly monomeric gel filtered Aβ1-42 was used in all experiments. Peptide Aβ1-42 (Atlanta, GA, USA) without N-terminal methionine from HFIP was also dissolved in 6 M GuHCl and gel filtered to make comparable preparations. Aβ1-42 protein concentration was calculated using ε = 1490 M–1 cm–1 at 277 nm. The Aβ1-42 fibrils were generated through incubation of 10 μM Aβ1-42 at 37 °C in PBS buffer (pH 7.5) for 24 h under quiescent conditions. Fibril formation of TTR, insulin, lysozyme, and Aβ1-42 was verified using the ThT (neutral pH) and X-34 (acidic pH) fluorescence assays.
UV–Vis Absorption Measurements
The ultraviolet–visible (UV–vis) absorption measurements were carried out employing a Cary winUV 100 spectrophotometer, using a 1 cm quartz cuvette at room temperature. The concentration of each probe was 20 μM in methanol.
Fluorescence Spectroscopy Measurements
All fluorescence spectra for each probe in methanol (20 μM) were recorded in a Tecan Safire 2 Microplate reader (Tecan, Männerdorf, Switzerland) or a Tecan infinite M1000 instrument and collected between 350 and 600 nm with the excitation wavelength at 330 nm at 1 nm spectral resolution. Fluorescence analysis for each probe upon binding to native protein or amyloid fibrils was carried out by mixing 1.0 μM of each molecule from a dimethyl sulfoxide (DMSO) stock with 5 μM proteins or fibrils (on a monomer basis) in PBS buffer (pH 7.5) or HOAc/NaAc buffer (pH 3.0) for native proteins or fibrils, except for both native (freshly dissolved monomeric) Aβ1-42 and Aβ1-42 fibrils in PBS buffer.
To avoid solubility problems for the probes, we used variable fibril concentrations to a fixed low concentration of probe to determine the binding affinity. A mixture (100 μL) containing each probe (final concn 1 or 0.5 μM for Aβ1-42 fibrils) and fibrils (final concn 0–20 μM on a monomer basis) was incubated at room temperature for 30 min to achieve equilibrium. To assess the binding interactions in supermolecular chemistry is not trivial.49 Especially, this is true for amyloid fibrils that are known to be structurally heterogenous and may contain several binding pockets for each fibril type.50 For this assay, we assumed that the fluorescence increase in the complex between the fibril and the compound reports on binding. A high-resolution probe binding pocket within model amyloid fibrils has been defined,51 where CR (an analogous molecule to our compounds) needs a defined pocket of sufficient order lined by side chains to bind. From this model, it is observed that a molecule of CR binds across six β-strands. In comparison, the compounds of this study are half the size of CR and hence should span three β-strands. From the experimental data, we assumed the simplistic model of one saturable binding site for each fibril protein. The fluorescence spectra were collected between 350 and 700 nm with excitation at 330 nm. The dissociation constant (Kd) of amyloid fibrils to each probe was calculated by GraphPad Prism 7.01 using nonlinear fit with one binding site and a fixed Bmax adjusting for the signal from the unbound compound. All binding curves and fits are shown in the Supporting Information.
Fluorescence Lifetime Measurements
Fluorescence decays were performed using an IBH time-correlated single photon counting (TCSPC) spectrometer. The solution of each probe in methanol was degassed with an argon gas, whereas the solution of each probe incubated with amyloid fibrils was not. After the sample solution was excited at 337 nm, the fluorescence decays at each respective emission maximum for each probe either free in methanol or bound to fibrils were monitored at room temperature. The instrument response function was determined with a fibril solution. Decays were fitted with a sum of exponentials (eq 1), and the quality of the fit was determined from the χ2 parameter, the random distribution of the residuals, and the autocorrelation of the residuals.
| 1 |
where αi is the pre-exponential factor associated with the decay time τi. The average lifetime of fluorescence decay was calculated according to the following equation
| 2 |
Results and Discussion
Synthesis and Optical Characterization of trans-Stilbenoid Derivatives
Our approach for the synthesis of the trans-stilbene derivatives 4 and 5 was to apply the palladium-catalyzed Mizoroki–Heck reaction of 9 to the corresponding 2-bromopyrene (7) and 2-bromoanthracene (10). Marder et al. have reported the synthesis of 2- and 2,7-functionalized pyrene derivatives including compounds 6 and 7 (Scheme 1).37−41 Employing the catalyst that was prepared in situ by the reaction of [Ir(u-OMe)cod]2 with 4,4′-di-tert-butyl-2,2′-dipyridyl (dtbpy), the product 6 was obtained in 62% yield. Then, the reaction of compound 6 with 3 equiv of CuBr2 in methanol/H2O (1:1) gave 7 in 78% yield. From 3,4-dihydroxybenzaldehyde (8), 9 was obtained by a witting reaction in 59% isolated yield, in which the Mizoroki–Heck reaction with 10 and 7 yielded the target molecules 4 and 5 in 35 and 28% isolated yields, respectively. Scheme 1 depicts the synthesis of the target molecules.
Scheme 1. Synthesis of Compounds 4 and 5.
Reagents and conditions: (a) B2Pin2 (1.1 equiv), [Ir(u-OMe)cod]2 (1 mol %), dtbpy (2 mol %), hexane, 80 °C, overnight, 60%. (b) CuBr2 (3 equiv), methanol/H2O (1:1), 90 °C, overnight, 75%. (c) Ph3PCH3Br (1.5 equiv), t-BuOK (4 equiv), THF, rt, 6 h, 58%. (d) Pd(OAc)2 (20% mol), triethanolamine (4 equiv), dry DMF, 100 °C, 24 h. [Ir(μ-OMe)cod]2 = (1,5-cyclooctadiene)(methoxy)iridium(I) dimer, dtbpy = 4,4′-di-tert-butyl-2,2′-dipyridyl.
After synthesis and purification, the photophysical properties of the probes (2–5), including absorption and emission spectra in methanol, and extinction coefficients were determined (Figure 1, Table 1, and Figure S1 in the Supporting Information). Methanol was a good solvent for the probes where absorbance showed clear linearity over a concentration range up to 50 μM. In water, the absorbance for 2 also showed a clear linearity over a concentration range up to 50 μM or more, 3 (up to 10 μM), 4 (up to 5 μM), and 5 (up to 2 μM) (Figure S1). Deviation from linearity above these concentrations shows that aggregation or micelle formation occurs above this concentration for each probe. As shown in Figure 1 and Table 1, the absorbance maxima are quite similar and centered around 330 nm for the whole series except for 4 that has a double-peak shoulder extending toward 400 nm. The absorbance peak for 2 was at 325 nm, 3 at 334 nm, and 4 at 335 nm. The lengthening of the π-conjugated system leads to a progressive red shift of the emission maximum from 386 to 483 nm. Compound 5 was an exception with the emission maximum at 415 nm. The progressive shift is ascribed to the lengthening of the π-conjugation from the central moiety over the substituent, whereas the emission at 415 nm for the pyrene variant suggests a weak electronic interaction between the pyrene center and the dihydroxyphenyl substituent.37,52 In conclusion, these probes showed Stokes shifts ranging from 61 to 148 nm (Table 1) using methanol as the solvent.
Figure 1.

Absorption (solid lines) and normalized fluorescence (dashed lines) spectra of compounds 2–5 dissolved in methanol [20 μM] at 25 °C. 2 (black); 3 (red); 4 (green); and 5 (blue).
Table 1. Spectroscopic Properties of 2, 3, 4, and 5.
| probe | λmax (abs) [nm] | ε [L·M–1 cm–1]a | λmax (em)b [nm] | Stokes shift [nm] |
|---|---|---|---|---|
| 2 | 325 | 25 038 | 386 | 61 |
| 3 | 334 | 30 170 | 420 | 86 |
| 4 | 335 | 27 070 | 483 | 148 |
| 5 | 337 | 47 668 | 415 | 78 |
Absorption spectra acquired at four different concentrations in methanol (2, 3, and 4 from 50 to 5 μM and 5 from 10 to 2 μM).
Excitation wavelength: 330 nm, [probe] = 20 μM.
ThT Fluorescence is pH-Dependent
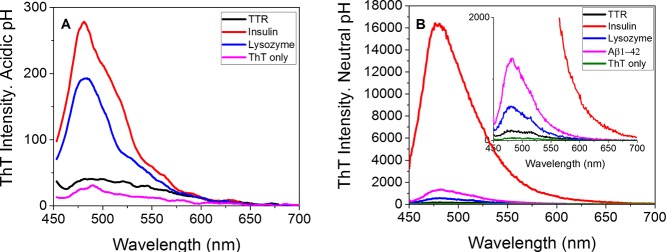
To benchmark our new probes with established amyloid probes, we used ThT. ThT fluorescence when bound to fibrils is known to be pH dependent.53 The fluorescence intensity of ThT when bound to different amyloid fibrils at acidic pH (pH 3.0) or at neutral pH (pH 7.5) was compared. Herein, TTR, insulin, lysozyme, and Aβ1-42 fibrils were measured. As shown in Figure 2A,B, the fluorescence of ThT bound to amyloid fibrils is very weak at pH 3.0 probably because of the electrostatic repulsion of the dye by an increase in the positive charge. In general, the fluorescence intensity of ThT upon binding to fibrils increased greatly when comparing at acidic pH and neutral pH: TTR (4.3-fold), insulin (57.1-fold), and lysozyme (3.1-fold), in agreement with our previous paper.53
Figure 2.
ThT shows pH-dependent fluorescence when bound to amyloid fibrils. (A) ThT fluorescence spectra of different amyloid fibrils in HOAc/NaAc buffer (pH 3.0). (B) ThT fluorescence spectra of different amyloid fibrils in PBS buffer (pH 7.5). All spectra were obtained at 5 μM fibrils (on a monomer basis) + 1 μM ThT with identical settings.
trans-Stilbenoids 4 and 5 Bind to Native and Fibrillar TTR
Previously, compounds 1 and 3 were reported to bind to native and misfolded fibrillar TTR, providing distinguishable emission spectra that shifted more than 100 nm for the two conformational states.36 Here, the binding response of the series 2–5 to native TTR and TTR fibrils was also determined by using fluorescence spectroscopy (excitation: 330 nm, emission: 350–600 nm). Typical results are shown in Figure 3. Upon binding to native TTR, 4 generated two emission peaks at 449 and 476 nm, with a total fluorescence intensity increase of about 6.1 times the intensity of the probe in buffer (Figure 3A). Upon binding to fibrillar TTR, there were also two emission peaks at 451 and 475 nm; however, the dominating contribution was shifted toward the shorter wavelength (Figure 3A). Here, the apparent increase in fluorescence intensity was 8.8-fold. As found from our results of 3,36 the spectrum of 4 bound to fibrils was blue-shifted compared with native TTR (Figure 3A). As shown in Figure 3B, probe 5, upon binding to either native or fibrillar TTR, showed very similar emission spectra with peaks at 420, 445, and 475 nm. The difference here is the apparent fluorescence intensity increase: 15.8-fold for native TTR compared with 6.3-fold for fibrillar TTR (Figure 3B). These two conformational states of TTR are vastly different as shown by us using a large collection of biophysical techniques.36 We noted intense fluorescence of 5 bound to the native tetrameric TTR. The determined Kd value for 5 binding to the high-fluorescence site for native TTR was 0.2 ± 0.01 μM (Figure S2). The intensity increase of ThT bound to TTR fibrils was a mere 1.8, indicating that these new probes are more sensitive for the detection of TTR fibrils in vitro at acidic pH (pH 3.0).
Figure 3.
Fluorescence emission spectra of 4 and 5. (A) 4, upon binding to either native or fibrillar TTR and (B) 5, upon binding to either native or fibrillar TTR. [native TTR] = [TTR fibrils] = 5 μM (on a monomer basis), [probe] = 1 μM, excitation wavelength: 330 nm. Identical settings were used in all measurements.
We further investigated the titration of TTR fibrils on a fixed concentration of probe (1.0 μM) to determine the Kd values for the whole series. As found previously with 1 and 3, two separate binding events were detected: 1 (Kd1 = 0.11 μM and Kd2 > 10 μM) and 3 (Kd1 = 0.07 μM and Kd2 > 10 μM).36 The high-affinity binding site showed minor fluorescence contribution; hence, we herein for fluorescence discrimination compare all probes for the high-fluorescence binding site. The Kd values were 15.1 μM for 2, 12.3 μM for 3, 7.4 μM for 4, and 6.0 μM for 5, indicating that the affinity was in the following order: 5 > 4 > 3 > 2, for details see Table 2 and Figures S3–S6 in the Supporting Information.
Table 2. Spectroscopic and Binding Data of 2, 3, 4, and 5.
| TTR
fibrils |
insulin
fibrils |
lysozyme
fibrils |
Aβ1-42 fibrils |
||||||
|---|---|---|---|---|---|---|---|---|---|
| probe | λem1a [nm] | Intb (λem2) | Kdc [μM] | Intb (λem2) | Kdc [μM] | Intb (λem2) | Kdc [μM] | Intb (λem2) | Kdc [μM] |
| 2 | --- | 1.7 (395) | 15.1 ± 0.9 | 7.5 (382) | 8.0 ± 1.6 | 3.8 (382) | 7.0 ± 1.6 | 2.6 (384) | 4.3 ± 0.2 |
| 3 | --- | 3.1 (395) | 12.3 ± 1.0 | 13.7 (410) | 4.5 ± 0.4 | 13.1 (410) | 2.9 ± 0.2 | 6.9 (408) | 2.2 ± 0.4 |
| 4 | --- | 8.8 (451) | 7.4 ± 0.5 | 91.4 (451) | 3.7 ± 0.6 | 37.5 (451) | 2.8 ± 0.4 | 10.3 (451) | 1.6 ± 0.1 |
| 5 | 420 | 6.3 (420) | 6.0 ± 0.3 | 23.2 (420) | 2.3 ± 0.4 | 8.9 (420) | 1.6 ± 0.3 | 11.1 (421) | 1.3 ± 0.1 |
The emission maxima of probes measured in HOAc/NaOAc buffer at pH 3.0 or PBS buffer at pH 7.5.
The fold increase in the fluorescence intensity upon binding to amyloid fibrils. The values in parentheses indicate the emission maxima of probes bound to amyloid fibrils measured in NaOAc buffer or PBS buffer (only for Aβ1-42 fibrils).
Dissociation constant: amyloid fibrils’ titration on fixed concentration of probes (1 μM for TTR fibrils, insulin fibrils, and lysozyme fibrils. 0.5 μM for Aβ1-42 fibrils). ± denotes the standard deviation from Kd values obtained from two independent binding series. --- no apparent peak.
We also experimentally addressed the finding of nearly identical spectra for compound 5 when bound to protofibrillar and native tetrameric TTR. Although the fluorescence intensity correlates well with the high intensity found in toluene, the spectral Stokes shift of probe 5 appears insensitive toward polarity of the binding site using four different solvents (PBS buffer, methanol, DMSO, and toluene) (Figure S7A). Notably, the relative intensities of three emission peaks do change, but the peak positions remain (Figure S7B). Our conclusion from the observation that the probe spectra bound to TTR are similar to that of 5 dissolved in toluene shows that both binding sites in the different conformational states are highly hydrophobic. Because the Kd toward native TTR is significantly lower than that of TTR protofibrils (0.2 μM compared with 6 μM), the occupancy of 5 on protofibrils is lower compared with native TTR under the conditions described in Figure 3, explaining the difference in intensity. This large Kd difference underlines the difference of a highly ordered binding site in the tetramer (T4 binding pocket) versus a more dynamic and fluid binding site on protofibrils.36
trans-Stilbenoids 2, 3, 4, and 5 Bind to Insulin Amyloid Fibrils
Similar experiments as discussed in the previous section were carried out to evaluate the probe series (Chart 1) for detecting insulin fibrils and as a control to native monomeric/dimeric insulin at pH 3.0. As shown in Figure 4 and Table 2, when these probes were incubated with native insulin, we found no significant change in fluorescence, indicating that there were only weak interactions between the probes and native insulin. By contrast, upon binding to insulin fibrils, 2 showed an emission peak at 382 nm and an apparent fluorescence intensity increase (7.5-fold over free probe); 3 displayed an emission peak at 410 nm and an apparent fluorescence intensity increase (13.7-fold); 4 had an apparent emission peak at 451 nm, a distinct fluorescence intensity increase (91.4-fold), and an additional peak at 471 nm; and 5 had emission peaks at 420, 445, and 475 nm and an obvious fluorescence intensity increase (23.2-fold over free probe for the peak at 420 nm). An apparent lower background fluorescence was observed from 2 to 5, especially 4 and 5 very little fluorescence was given in either buffer, suggesting that these probes formed aggregates or micelles in the absence of fibrils. Compared with ThT at pH 3.0, the intensity increase upon bound to insulin fibrils was in the following order: 4 > 5 > 3 > ThT > 2, suggesting that 3, 4, and 5 are more sensitive probes to detect insulin fibrils in vitro at acidic pH (pH 3.0).
Figure 4.
Fluorescence emission spectra of (A) 2, (B) 3, (C) 4, and (D) 5 bound to either native insulin or insulin fibrils. [native insulin] = [insulin fibrils] = 5 μM (on a monomer basis), [probe] = 1 μM, excitation wavelength: 330 nm. Identical settings were used in all measurements.
To quantitatively evaluate the binding affinities of these probes to insulin fibrils, we measured the change in the fluorescence intensity by titrating a fixed low concentration of probes (1.0 μM) with insulin fibrils. All probes mainly showed one distinct binding site with Kd = 8.0 μM for 2, 4.5 μM for 3, 3.7 μM for 4, and 2.3 μM for 5 (for details see Table 2 and Figures S8–S11 in the Supporting Information). Groenning et al54 have reported that the Kd value of ThT binding to insulin fibrils was 10.4 μM with one binding site. Therefore, the affinity of insulin fibrils to probes was in the following order: 5 > 4 > 3 > 2 > ThT. These results indicate that the probes can specifically bind to insulin amyloid fibrils with an obvious fluorescence intensity increase (from 7.5-fold to 91.4-fold) and moderate binding affinities.
trans-Stilbenoids 2, 3, 4, and 5 Bind to Lysozyme Amyloid Fibrils
The probe fluorescence response toward native lysozyme and lysozyme amyloid fibrils was assessed. As shown in Figure 5 and Table 2, there was essentially no fluorescence change of any probe with native lysozyme. On the contrary, upon binding to lysozyme fibrils, there is an increase in fluorescence intensity: 3.8-, 13.1-, 37.5-, and 8.9-fold for 2, 3, 4, and 5, respectively, as compared to the free probe. The emission spectra recorded for each probe upon binding to lysozyme fibrils were similar to those recorded upon binding to insulin fibrils, which means that the emission spectra for each probe occur from the same excited state. We also measured the Kd values: 7.0 μM for 2, 2.9 μM for 3, 2.8 μM for 4, and 1.6 μM for 5 (for details see Table 2 and Figures S12–S15 in the Supporting Information). Sulatskaya et al.55 reported that ThT binds to lysozyme fibrils with two binding sites: Kd1 = 0.13 μM and Kd2 = 17.86 μM, which suggested that the affinity of lysozyme fibrils to probes was in the order: 5 > 4 ≈ 3 > 2 > ThT (Kd2 = 17.86 μM). The fluorescence intensity was found to increase for probes 2, 4, and 5 but not to the same extent during binding to lysozyme fibrils as found for insulin fibrils. Probe 3 increased similarly (13.7-fold for insulin fibrils and 13.1-fold for lysozyme fibrils) for both fibril systems. Conclusively, these probes can specifically bind to lysozyme fibrils concomitant with the fluorescence intensity increase and overall with a higher affinity than that found for insulin fibrils.
Figure 5.
Fluorescence emission spectra of (A) 2, (B) 3, (C) 4, and (D) 5 bound to either native lysozyme or lysozyme fibrils. [native lysozyme] = [lysozyme fibrils] = 5 μM (on a monomer basis), [probe] = 1 μM, excitation wavelength: 330 nm. Identical settings were used in all measurements.
trans-Stilbenoids 2, 3, 4, and 5 Bind to Aβ1-42 Amyloid Fibrils
After specific detection of TTR, insulin, and lysozyme amyloid fibrils by these probes, we further extended the use of the series of probes in identifying Aβ1-42 amyloid fibrils at neutral pH (pH 7.5). As shown in Figure 6, 2 binds to Aβ1-42 fibrils emitting fluorescence with a wavelength maximum at 384 nm and a 2.6-fold fluorescence intensity increase. 3 showed a 6.9-fold increased fluorescence intensity, at an emission maximum at 408 nm. 4 and 5, both bind to Aβ1-42 fibrils with an apparent 10.3- and 11.1-fold fluorescence intensity increase and emission maxima at around 450 and 421 nm, respectively. Neither 2, 4, or 5 fluoresced appreciably in the presence of freshly prepared native Aβ1-42, whereas 3 showed some binding to fresh Aβ1-42 (Figure 6B). The measured binding constant to Aβ1-42 fibrils is summarized in Table 2 and Figures S14–S17 in the Supporting Information. The affinity for Aβ1-42 fibrils was stronger for 2 (Kd = 4.3 μM), 3 (Kd = 2.2 μM), 4 (Kd = 1.6 μM), and 5 (Kd = 1.3 μM), compared with TTR, insulin, and lysozyme fibrils. The stronger binding affinities to Aβ1-42 fibrils (performed at pH 7.5) compared with the other fibril types could indicate higher affinity at this pH. Furthermore, it was found that the increase in the fluorescence intensity of 2, 3, and 4 bound to Aβ1-42 fibrils was lower than that detected for insulin and lysozyme fibrils. Conclusively, all of the probes can specifically bind to Aβ1-42 fibrils, followed by an increase in the fluorescence intensity (from 2.6-fold to 11.1-fold compared to the free probe) and with higher affinity compared with TTR, insulin, and lysozyme fibrils. For Aβ1-42 fibrils, the ThT fluorescence increase (36.9-fold) is even higher than that for 5 mainly because ThT is almost completely quenched in the free state. Various Kd values for ThT have been reported toward Aβ fibrils in the 2–30 μM range.26,50
Figure 6.
Fluorescence emission spectra of (A) 2, (B) 3, (C) 4, and (D) 5 bound to either freshly dissolved Aβ1-42 or Aβ1-42 fibrils as well as recombinant Aβ1-42 with an additional methionine in the N-terminus (Aβ M1-42). Concentrations: [native Aβ1-42] = [Aβ1-42 fibrils] = 5 μM (on a monomer basis), [probe] = 1 μM, excitation wavelength: 330 nm. Identical settings were used in all measurements.
The majority of data in our study was collected on recombinant Aβ1-42 with an extra methionine in the N-terminus (AβM1-42). The extra methionine has been reported not to influence kinetics or fibril morphology48 compared with the wild-type Aβ1-42. Importantly, two high-resolution structures were recently published where one used this construct56 and the other study used a construct lacking the N-terminal methionine.57 Both fibril structures were found to be nearly identical. To verify this similar structure for our probe spectra, we prepared fibrils for both types. Nearly identical spectral profiles were observed (Figure 6).
Fluorescence Lifetimes of trans-Stilbenoids 2, 3, 4, and 5
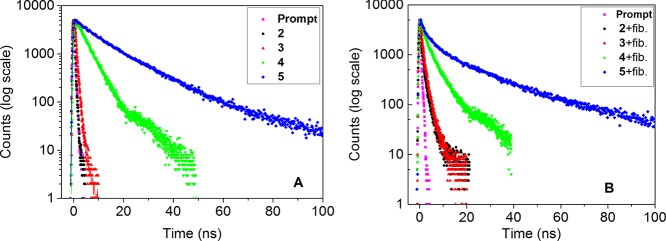
The probes can specifically bind to four different amyloid fibrils, with a substantial fluorescence intensity increase and similar emission maxima for each fibril type. No appreciable binding was detected for native insulin, lysozyme, and Aβ1-42. Only native TTR showed probe binding as expected from previous data, likely reflecting binding to the T4 binding site.36 Herein, we chose Aβ1-42 fibrils as model fibrils to investigate the emission properties of the probes bound to amyloid fibrils from the viewpoint of the probe structures and the interaction modes between the probes and fibrils. We conducted time-resolved fluorescence measurements using the TCSPC method. The probes showed very weak and quenched fluorescence when dissolved without amyloid using the water-based buffers applied for the amyloid or protein association experiments. A prerequisite for the assessment of fluorescence lifetimes is to have sufficient signal. Therefore, the intrinsic free lifetimes of the probes were determined using degassed methanol. In methanol, the solubility is high and by removing solvent oxygen, the parasitic quenching by oxygen, being important especially for probes with long lifetimes, was removed. Figure 7A shows the decay curves of the probes dissolved in methanol, in which oxygen from air has been removed by flushing the sample with an argon gas. Thus, in degassed methanol solution, where emission strictly occurs from the free dyes, the fluorescence decay of the excited states of 2 is very fast (<0.05 ns). The fluorescence decay of 3 and 4 is single exponential with decay times of 0.52 and 4.23 ns, respectively. The emission decay at 415 nm for 5 is double exponential in character with the τavg = 19.11 ns (τ1 = 11.00 ns and τ2 = 29.30 ns, with the corresponding pre-exponential factors α1 = 0.77 and α2 = 0.23, respectively) (Table 3). As expected for a pyrene fluorophore, probe 5 shows the longest lifetime. Consequently, the fluorescence lifetimes of the probes in degassed methanol are in the following order: 2 < < 3 < 4 < 5, indicating that the different aromatic rings (benzene, naphthyl, antracene, and pyrene) conjugated to the stilbene are promoting extended excited-state lifetimes.
Figure 7.
Fluorescence decays for excitation prompt (magenta), 2 (black), 3 (red), 4 (green), and 5 (blue) in degassed methanol (A) or bound to Aβ1-42 fibrils (B).
Table 3. Fluorescence Lifetimes for the Probes in Degassed Methanol or Bound to Aβ1-42 Fibrils and Normalized Pre-Exponential Factors.
| probe | a | λem1b [nm] | τ1(α1)c [ns] | τ2(α2)c [ns] | τ3(α3)c [ns] | τavg [ns] |
|---|---|---|---|---|---|---|
| 2 | A | 395 | <0.05(1) | <0.05 | ||
| B | 410 | 0.28(0.86) | 2.31(0.14) | 1.33 | ||
| 3 | A | 415 | 0.52(1) | 0.52 | ||
| B | 420 | 0.23(0.54) | 1.65(0.46) | 1.45 | ||
| 4 | A | 475 | 4.23(1) | 4.23 | ||
| B | 460 | 0.64(0.38) | 6.06(0.62) | 5.73 | ||
| 5 | A | 425 | 11.00(0.77) | 29.30(0.23) | 19.11 | |
| B | 430 | 0.83(0.11) | 8.89(0.42) | 34.70(0.47) | 29.76 |
(A) Measured in degassed methanol, [probe] = 5 μM; (B) bound to Aβ1-42 fibrils in PBS buffer, [probe] = 5 μM and [Aβ1-42 fibrils] = 20 μM.
The emission maxima measured in PBS buffer upon binding to Aβ1-42 fibrils.
Lifetime (τ) and corresponding amplitude (α). The uncertainties of fluorescence lifetimes are within 10%.
To obtain sufficient signal to noise and to work under saturated conditions for the bound probe, the fluorescence decays for the probes bound to Aβ1-42 fibrils were investigated under the following conditions: [probe] = 5 μM, [Aβ1-42 fibrils] = 20 μM (on a monomer basis), and PBS buffer, pH 7.5, at 25 °C. By the law of mass action and the determined Kd values of the probes bound to Aβ1-42 fibrils applied at a 10-fold concentration above the Kd for 3, 4, and 5, we expect >90% of the signal to originate from the fibril-bound probe. Figure 7B shows the decay curves of the probes bound to Aβ1-42 fibrils. In general, there are small changes in the decay upon binding, but one can note a more pronounced contribution from longer decay time components, generating a “bent”, multicomponent decay in all cases. Specifically, the fluorescence decay curves for 2, 3, and 4 are successfully analyzed using the double-exponential kinetic law with good χ2 values, whereas a three-component decay is needed to accurately reproduce the decay of 5 with fibrils (see details in Table 3). Multiexponential decays can arise from populations of different binding sites in the aggregated protein, thus giving different lifetimes due to nearby quenching amino acid residues and/or solvent water. Multiexponential decays can also arise from mechanisms such as fluorescence resonance energy transfer, dynamic or static collision quenching, or combinations thereof, each model requiring a thorough analysis and definition of the averaging procedure.58−60 To compare the different probes side-by-side, the multiexponential decays were summarized as intensity averaged lifetimes (see Experimental Section, Table 3). The average lifetime reflects the state population of each contributing lifetime, which we assume to best describe the binding sites on the amyloid fibril. The associated time constants are presented with the shortest first and the longest last (Table 3). As can be seen in Table 3, each probe, when bound to Aβ1-42 fibrils, showed a longer average lifetime than for the free probe in degassed methanol. For 2, 3, and 4, the existence of a long lifetime component (τ2) population resulted from the probe incubated with Aβ1-42 fibrils; the origin of a short phase (τ1) can possibly be attributed to the small fraction of the free probe unbound to Aβ1-42 fibrils. Another possible origin of the short phase is the light-scattering effect from fibrils in PBS solution, although a spectral filter was used in the setup to minimize the effect of scatter from the excitation wavelength. For 5, the lifetime component τ3 most likely resulted from the probe bound to Aβ1-42 fibrils. The pre-exponential factors (amplitude weights) of the multicomponent decays also show some systematic patterns. For 2, the pre-exponential factors of τ1 are found to be larger than that of τ2, indicating that the shorter decay of the free probe is predominant in the excited state. For 3, the pre-exponential factors of τ1 and τ2 are almost the same, meaning that the free probe and bound probe are nearly equal in the excited state. For 4, the pre-exponential factors of τ1 are smaller than that of τ2, demonstrating that the bound probe is predominant in the excited state. Conclusively, the binding of the series of new probes to Aβ1-42 amyloid fibrils results in a fluorescent decay time that is substantially longer than the corresponding lifetime found in a good solvent (methanol).
Conclusions
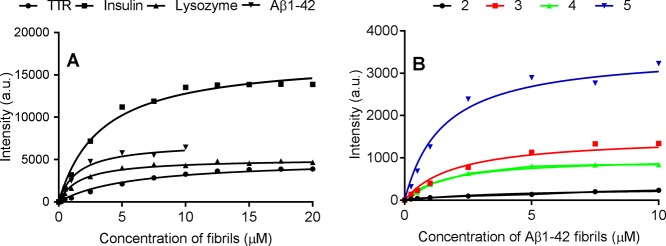
Inspired by previous probe developments for TTR conformational states,36 we have herein successfully synthesized two new trans-stilbene-based fluorescent molecules 4 and 5 that can bind to both native and protofibrillar TTR. Both probes become highly fluorescent in the bound state. The amyloid-binding propensity for the series of four analogous trans-stilbenoids (two previously reported: 2 and 3, two new: 4 and 5) with different aromatic ring moieties was compared. Because of the common overall fold and core structure of amyloid fibrils, we expected these probes to bind to several amyloid fibrils while being refractive to native monomeric protein conformations. Importantly, all studied probes can bind specifically to amyloid fibrils of vastly heterologous proteins (insulin, lysozyme, TTR, and Aβ1-42 fibrils) with apparent fluorescence intensity augmentation and moderate affinities. The assays of fibrils were performed at the preferred pH for formation and stability, that is, pH 3.0 for insulin, lysozyme, and TTR and at pH 7.5 for Aβ1-42. To exemplify the binding response of the probes versus various amyloid fibrils, Figure 8A shows the fluorescence response of 5 versus various amyloid fibril concentrations at a fixed concentration of probe. A corresponding probe comparison for 2–5 is shown for Aβ1-42 fibrils (Figure 8B). Both datasets display the various fluorescence responses being both protein- and probe-dependent.
Figure 8.
Representative saturation binding curves of amyloid fibrils to probes. (A) TTR, insulin, lysozyme, and Aβ1-42 fibrils to 5. (B) Aβ1-42 fibrils to 2, 3, 4, and 5.
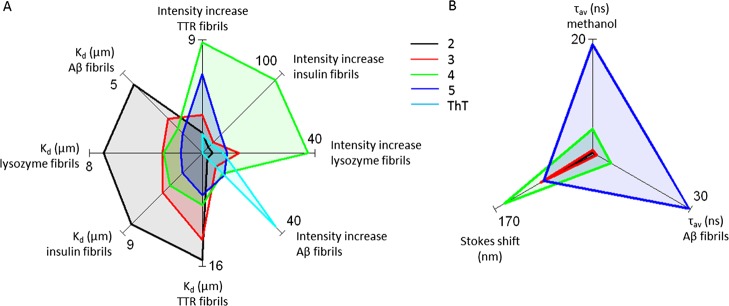
The fluorescence lifetime of ThT is rather short: 80% is <1 ns and 20% 3.9 ns.29 One specific aim of the current study was to develop long lifetime fluorescent probes to allow measurements of solution dynamics of amyloid species. To our knowledge, no small-molecule amyloid targeting probes have been reported with pyrene or anthracene directly binding to the amyloid fibril binding sites. In previous work, we have shown the virtue of pyrene-based cysteine-specific probes covalently linked to TTR for size determination of TTR oligomers up to 20 mers,61 and we hypothesized on the applicability of extended lifetime noncovalent amyloid targeting probes described herein.17 The fluorescence lifetimes of the amyloid probes described in this work in degassed methanol are ranked as (2 < 0.05 ns) < 3 = 0.52 ns < 4 = 4.23 ns < 5 = 19.11 ns, indicating that the different aromatic ring moieties determine the respective probe residence time in the excited state. When bound to Aβ1-42 fibrils, the lifetimes are 1.3 ns (2), 1.5 ns (3), 5.7 ns (4), and 29.8 ns (5). Hence, each probe bound to Aβ1-42 fibrils showed longer lifetime than in degassed methanol, suggesting that the probe binding sites are hydrophobic and well-shielded from the surrounding water solvent. Obviously, the pyrene-based 5 showed the longest fluorescence lifetime when bound to Aβ1-42 fibrils, opening the opportunity for size determination of the oligomerization process by dynamic fluorescence spectroscopy. To make a palpable overall comparison of the probes against various amyloid fibrils, we generated a starplot (Figure 9A) to compare various Kd values and fluorescence increase for the respective probe. This graphical representation illustrates that 4 shows the strongest fluorescence increase for fibrils at acidic pH and that the affinity (low Kd) generally increases as the hydrophobicity of the probe increases, with the strongest binding for 5. Furthermore, a starplot representation of the photophysical properties, Stokes shift, and fluorescence lifetimes clearly display a similar separation with highest Stokes shift for 4 and the longest fluorescence lifetimes for 5 (Figure 9B). We foresee that this class of probes will be useful for amyloid fibril research.
Figure 9.
Starplot representation to compare all probe parameters in graphical format. (A) Binding properties represented by dissociation constants (Kd in μM) of the probes against different fibril types and the relative fold fluorescence intensity increase of the fibril-bound probe compared with the free probe. (B) Starplot representation of the photophysical properties, Stokes shift (nm), and average fluorescence lifetime (τavg in ns) of the probes.
Acknowledgments
We thank Maria Thörnkvist for assistance with preparation of TTR. This work was supported by the Swedish Research Council, the Swedish Alzheimer Foundation, the Swedish Brain Foundation, LiU-Neuro, and Göran Gustafsson Foundation. M.L. is grateful to Linköping University for a guest professor tenure during which a part of this work was carried out.
Supporting Information Available
The Supporting Information is available free of charge on the ACS Publications website at DOI: 10.1021/acsomega.7b00535.
Synthesis of compounds 4 and 5; absorbance spectra of all compounds; emission spectra in various solvents for compound 5 and binding of 5 toward native TTR; and saturation binding curves of all compounds to different fibrils: TTR, insulin, lysozyme, and Aβ1-42 fibrils (PDF)
The authors declare no competing financial interest.
Supplementary Material
References
- Chiti F.; Dobson C. M. Protein Misfolding, Functional Amyloid, and Human Disease. Annu. Rev. Biochem. 2006, 75, 333–366. 10.1146/annurev.biochem.75.101304.123901. [DOI] [PubMed] [Google Scholar]
- Ross C. A.; Poirier M. A. Protein Aggregation and Neurodegenerative Disease. Nat. Med. 2004, 10, S10–S17. 10.1038/nm1066. [DOI] [PubMed] [Google Scholar]
- Dobson C. M. Protein Folding and Misfolding. Nature 2003, 426, 884–890. 10.1038/nature02261. [DOI] [PubMed] [Google Scholar]
- Ban T.; Hamada D.; Hasegawa K.; Naiki H.; Goto Y. Direct Observation of Amyloid Fibril Growth Monitored by Thioflavin T Fluorescence. J. Biol. Chem. 2003, 278, 16462–16465. 10.1074/jbc.c300049200. [DOI] [PubMed] [Google Scholar]
- Harper J. D.; Lansbury P. T. Models of Amyloid Seeding in Alzheimier’s Disease and Scrapie: Mechanistic Truths and Physiological Consequences of the Time-Dependent Solubility of Amyloid Proteins. Annu. Rev. Biochem. 1997, 66, 385–407. 10.1146/annurev.biochem.66.1.385. [DOI] [PubMed] [Google Scholar]
- Bulic B.; Pickhardt M.; Schmidt B.; Mandelkow E.-M.; Waldmann H.; Mandelkow E. Development of Tau Aggregation Inhibitors for Alzheimer’s Disease. Angew. Chem., Int. Ed. 2009, 48, 1740–1752. 10.1002/anie.200802621. [DOI] [PubMed] [Google Scholar]
- Selkoe D. J. Folding Proteins in Fatal Ways. Nature 2003, 426, 900–904. 10.1038/nature02264. [DOI] [PubMed] [Google Scholar]
- Sipe J. D. Amyloidosis. Annu. Rev. Biochem. 1992, 61, 947–975. 10.1146/annurev.bi.61.070192.004503. [DOI] [PubMed] [Google Scholar]
- Eva Ž. Amyloid-Fibril Formation. Eur. J. Biochem. 2002, 269, 3362–3371. 10.1046/j.1432-1033.2002.03024.x. [DOI] [PubMed] [Google Scholar]
- Pepys M. B. Pathogenesis, Diagnosis and Treatment of Systemic Amyloidosis. Philos. Trans. R. Soc., B 2001, 356, 203–211. 10.1098/rstb.2000.0766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sunde M.; Serpell L. C.; Bartlam M.; Fraser P. E.; Pepys M. B.; Blake C. C. F. Common Core Structure of Amyloid Fibrils by Synchrotron X-Ray Diffraction. J. Mol. Biol. 1997, 273, 729–739. 10.1006/jmbi.1997.1348. [DOI] [PubMed] [Google Scholar]
- Makin O. S.; Serpell L. C. Structures for Amyloid Fibrils. FEBS J. 2005, 272, 5950–5961. 10.1111/j.1742-4658.2005.05025.x. [DOI] [PubMed] [Google Scholar]
- Serpell L. C.; Sunde M.; Benson M. D.; Tennent G. A.; Pepys M. B.; Fraser P. E. The Protofilament Substructure of Amyloid Fibrils. J. Mol. Biol. 2000, 300, 1033–1039. 10.1006/jmbi.2000.3908. [DOI] [PubMed] [Google Scholar]
- Krebs M. R. H.; Bromley E. H. C.; Donald A. M. The Binding of Thioflavin-T to Amyloid Fibrils: Localisation and Implications. J. Struct. Biol. 2005, 149, 30–37. 10.1016/j.jsb.2004.08.002. [DOI] [PubMed] [Google Scholar]
- Hwang W.; Zhang S.; Kamm R. D.; Karplus M. Kinetic Control of Dimer Structure Formation in Amyloid Fibrillogenesis. Proc. Natl. Acad. Sci. U.S.A. 2004, 101, 12916–12921. 10.1073/pnas.0402634101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pedersen J. S.; Dikov D.; Flink J. L.; Hjuler H. A.; Christiansen G.; Otzen D. E. The Changing Face of Glucagon Fibrillation: Structural Polymorphism and Conformational Imprinting. J. Mol. Biol. 2006, 355, 501–523. 10.1016/j.jmb.2005.09.100. [DOI] [PubMed] [Google Scholar]
- Lindgren M.; Hammarström P. Amyloid Oligomers: Spectroscopic Characterization of Amyloidogenic Protein States. FEBS J. 2010, 277, 1380–1388. 10.1111/j.1742-4658.2010.07571.x. [DOI] [PubMed] [Google Scholar]
- Nilsson M. R. Techniques to Study Amyloid Fibril Formation in Vitro. Methods 2004, 34, 151–160. 10.1016/j.ymeth.2004.03.012. [DOI] [PubMed] [Google Scholar]
- Nilsson K. P. R.; Hammarström P. Luminescent Conjugated Polymers: Illuminating the Dark Matters of Biology and Pathology. Adv. Mater. 2008, 20, 2639–2645. 10.1002/adma.200800663. [DOI] [Google Scholar]
- LeVine H. III; Peter K.; Nilsson R.; Hammarström P.. Reporters of Amyloid Structural Polymorphism. In Bio-Nanoimaging: Protein Misfolding and Aggregation; Elsevier: London, 2014; Chapter 7. [Google Scholar]
- Ban T.; Hamada D.; Hasegawa K.; Naiki H.; Goto Y. Direct Observation of Amyloid Fibril Growth Monitored by Thioflavin T Fluorescence. J. Biol. Chem. 2003, 278, 16462–16465. 10.1074/jbc.c300049200. [DOI] [PubMed] [Google Scholar]
- McParland V. J.; Kad N. M.; Kalverda A. P.; Brown A.; Kirwin-Jones P.; Hunter M. G.; Sunde M.; Radford S. E. Partially Unfolded States of β2-Microglobulin and Amyloid Formation in Vitro. Biochemistry 2000, 39, 8735–8746. 10.1021/bi000276j. [DOI] [PubMed] [Google Scholar]
- Kim Y.-S.; Randolph T. W.; Manning M. C.; Stevens F. J.; Carpenter J. F. Congo Red Populates Partially Unfolded States of an Amyloidogenic Protein to Enhance Aggregation and Amyloid Fibril Formation. J. Biol. Chem. 2003, 278, 10842–10850. 10.1074/jbc.m212540200. [DOI] [PubMed] [Google Scholar]
- Khurana R.; Uversky V. N.; Nielsen L.; Fink A. L. Is Congo Red an Amyloid-Specific Dye?. J. Biol. Chem. 2001, 276, 22715–22721. 10.1074/jbc.m011499200. [DOI] [PubMed] [Google Scholar]
- LeVine H. III [18] Quantification of β-Sheet Amyloid Fibril Structures with Thioflavin T. Methods Enzymol. 1999, 309, 274–284. 10.1016/s0076-6879(99)09020-5. [DOI] [PubMed] [Google Scholar]
- Levine H. Thioflavine T Interaction with Synthetic Alzheimer’s Disease β-Amyloid Peptides: Detection of Amyloid Aggregation in Solution. Protein Sci. 2008, 2, 404–410. 10.1002/pro.5560020312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maskevich A. A.; Stsiapura V. I.; Kuzmitsky V. A.; Kuznetsova I. M.; Povarova O. I.; Uversky V. N.; Turoverov K. K. Spectral Properties of Thioflavin T in Solvents with Different Dielectric Properties and in a Fibril-Incorporated Form. J. Proteome Res. 2007, 6, 1392–1401. 10.1021/pr0605567. [DOI] [PubMed] [Google Scholar]
- Stsiapura V. I.; Maskevich A. A.; Kuzmitsky V. A.; Uversky V. N.; Kuznetsova I. M.; Turoverov K. K. Thioflavin T as a Molecular Rotor: Fluorescent Properties of Thioflavin T in Solvents with Different Viscosity. J. Phys. Chem. B 2008, 112, 15893–15902. 10.1021/jp805822c. [DOI] [PubMed] [Google Scholar]
- Lindgren M.; Sörgjerd K.; Hammarström P. Detection and Characterization of Aggregates, Prefibrillar Amyloidogenic Oligomers, and Protofibrils Using Fluorescence Spectroscopy. Biophys. J. 2005, 88, 4200–4212. 10.1529/biophysj.104.049700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arja K.; Sjölander D.; Åslund A.; Prokop S.; Heppner F. L.; Konradsson P.; Lindgren M.; Hammarström P.; Åslund K. O. A.; Nilsson K. P. R. Enhanced Fluorescent Assignment of Protein Aggregates by an Oligothiophene-Porphyrin-Based Amyloid Ligand. Macromol. Rapid Commun. 2013, 34, 723–730. 10.1002/marc.201200817. [DOI] [PubMed] [Google Scholar]
- Berezin M. Y.; Achilefu S. Fluorescence Lifetime Measurements and Biological Imaging. Chem. Rev. 2010, 110, 2641–2684. 10.1021/cr900343z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klabunde T.; Petrassi H. M.; Oza V. B.; Raman P.; Kelly J. W.; Sacchettini J. C. Rational Design of Potent Human Transthyretin Amyloid Disease Inhibitors. Nat. Struct. Mol. Biol. 2000, 7, 312–321. 10.1038/74082. [DOI] [PubMed] [Google Scholar]
- Hammarström P.; Jiang X.; Hurshman A. R.; Powers E. T.; Kelly J. W. Sequence-Dependent Denaturation Energetics: A Major Determinant in Amyloid Disease Diversity. Proc. Natl. Acad. Sci. U.S.A. 2002, 99, 16427–16432. 10.1073/pnas.202495199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dorbala S.; Vangala D.; Semer J.; Strader C.; Bruyere J. R. Jr.; Di Carli M. F.; Moore S. C.; Falk R. H. Imaging Cardiac Amyloidosis: A Pilot Study Using 18F-Florbetapir Positron Emission Tomography. Eur. J. Nucl. Med. Mol. Imaging 2014, 41, 1652–1662. 10.1007/s00259-014-2787-6. [DOI] [PubMed] [Google Scholar]
- Furukawa K.; Ikeda S.-i.; Okamura N.; Tashiro M.; Tomita N.; Furumoto S.; Iwata R.; Yanai K.; Kudo Y.; Arai H. Cardiac Positron-Emission Tomography Images with an Amyloid-Specific Tracer in Familial Transthyretin-Related Systemic Amyloidosis. Circulation 2012, 125, 556–557. 10.1161/circulationaha.111.045237. [DOI] [PubMed] [Google Scholar]
- Campos R. I.; Wu X.; Elgland M.; Konradsson P.; Hammarström P. Novel Trans-Stilbene-Based Fluorophores as Probes for Spectral Discrimination of Native and Protofibrillar Transthyretin. ACS Chem. Neurosci. 2016, 7, 924–940. 10.1021/acschemneuro.6b00062. [DOI] [PubMed] [Google Scholar]
- Wanninger-Weiß C.; Wagenknecht H.-A. Synthesis of 5-(2-Pyrenyl)-2′-Deoxyuridine as a DNA Modification for Electron-Transfer Studies: The Critical Role of the Position of the Chromophore Attachment. Eur. J. Org. Chem. 2008, 64–71. 10.1002/ejoc.200700818. [DOI] [Google Scholar]
- Coventry D. N.; Batsanov A. S.; Goeta A. E.; Howard J. A. K.; Marder T. B.; Perutz R. N. Selective Ir-Catalysed Borylation of Polycyclic Aromatic Hydrocarbons: Structures of Naphthalene-2,6-Bis(Boronate), Pyrene-2,7-Bis(Boronate) and Perylene-2,5,8,11-Tetra(Boronate) Esters. Chem. Commun. 2005, 2172–2174. 10.1039/b501778e. [DOI] [PubMed] [Google Scholar]
- Crawford A. G.; Liu Z.; Mkhalid I. A. I.; Thibault M.-H.; Schwarz N.; Alcaraz G.; Steffen A.; Collings J. C.; Batsanov A. S.; Howard J. A. K.; Marder T. B. Synthesis of 2- and 2,7-Functionalized Pyrene Derivatives: An Application of Selective C–H Borylation. Chem.—Eur. J. 2012, 18, 5022–5035. 10.1002/chem.201103774. [DOI] [PubMed] [Google Scholar]
- Yamato T.; Noda K.; Tsuzuki H. Medium-Sized Cyclophanes. Part 56. 8-Substituted 5-Tert-Butyl[2.2]Metaparacyclophane-1,9-Dienes. Preparation, X-Ray Diffraction Study and Their Treatment with Lewis and Protic Acids. New J. Chem. 2001, 25, 721–727. 10.1039/b009932p. [DOI] [Google Scholar]
- Harvey R. G.; Schmolka S.; Cortez C.; Lee H. Syntheses of 2-Bromopyrene and 2-Hydroxypyrene. Synth. Commun. 1988, 18, 2207–2209. 10.1080/00397918808082362. [DOI] [Google Scholar]
- Albert S.; Horbach R.; Deising H. B.; Siewert B.; Csuk R. Synthesis and Antimicrobial Activity of (E) Stilbene Derivatives. Bioorg. Med. Chem. 2011, 19, 5155–5166. 10.1016/j.bmc.2011.07.015. [DOI] [PubMed] [Google Scholar]
- Fisher A. J.; Kerrigan F. A New Convenient Synthesis of 1-(3-Hydroxy-4-Methoxyphenyl)Ethane-1,2-Diol (Iso-Mhpg) and Its Enantiomers. Synth. Commun. 1998, 28, 2959–2968. 10.1080/00397919808004875. [DOI] [Google Scholar]
- Groenning M.; Campos R. I.; Fagerberg C.; Rasmussen A. A.; Eriksen U. H.; Powers E. T.; Hammarström P. Thermodynamic Stability and Denaturation Kinetics of a Benign Natural Transthyretin Mutant Identified in a Danish Kindred. Amyloid 2011, 18, 35–46. 10.3109/13506129.2011.560215. [DOI] [PubMed] [Google Scholar]
- Raz A.; Goodman D. S. The Interaction of Thyroxine with Human Plasma Prealbumin and with the Prealbumin-Retinol-Binding Protein Complex. J. Biol. Chem. 1969, 244, 3230–3237. [PubMed] [Google Scholar]
- Krebs M. R. H.; Morozova-Roche L. A.; Daniel K.; Robinson C. V.; Dobson C. M. Observation of Sequence Specificity in the Seeding of Protein Amyloid Fibrils. Protein Sci. 2004, 13, 1933–1938. 10.1110/ps.04707004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mishra R.; Sörgjerd K.; Nyström S.; Nordigården A.; Yu Y.-C.; Hammarström P. Lysozyme Amyloidogenesis Is Accelerated by Specific Nicking and Fragmentation but Decelerated by Intact Protein Binding and Conversion. J. Mol. Biol. 2007, 366, 1029–1044. 10.1016/j.jmb.2006.11.084. [DOI] [PubMed] [Google Scholar]
- Walsh D. M.; Thulin E.; Minogue A. M.; Gustavsson N.; Pang E.; Teplow D. B.; Linse S. A Facile Method for Expression and Purification of the Alzheimer’s Disease-Associated Amyloid β-Peptide. FEBS J. 2009, 276, 1266–1281. 10.1111/j.1742-4658.2008.06862.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thordarson P. Determining Association Constants from Titration Experiments in Supramolecular Chemistry. Chem. Soc. Rev. 2011, 40, 1305–1323. 10.1039/c0cs00062k. [DOI] [PubMed] [Google Scholar]
- Lockhart A.; Ye L.; Judd D. B.; Merritt A. T.; Lowe P. N.; Morgenstern J. L.; Hong G.; Gee A. D.; Brown J. Evidence for the Presence of Three Distinct Binding Sites for the Thioflavin T Class of Alzheimer’s Disease Pet Imaging Agents on β-Amyloid Peptide Fibrils. J. Biol. Chem. 2005, 280, 7677–7684. 10.1074/jbc.m412056200. [DOI] [PubMed] [Google Scholar]
- Schütz A. K.; Soragni A.; Hornemann S.; Aguzzi A.; Ernst M.; Böckmann A.; Meier B. H. The Amyloid-Congo Red Interface at Atomic Resolution. Angew. Chem., Int. Ed. 2011, 50, 5956–5960. 10.1002/anie.201008276. [DOI] [PubMed] [Google Scholar]
- Crawford A. G.; Dwyer A. D.; Liu Z.; Steffen A.; Beeby A.; Pålsson L.-O.; Tozer D. J.; Marder T. B. Experimental and Theoretical Studies of the Photophysical Properties of 2- and 2,7-Functionalized Pyrene Derivatives. J. Am. Chem. Soc. 2011, 133, 13349–13362. 10.1021/ja2006862. [DOI] [PubMed] [Google Scholar]
- Mishra R.; Sjölander D.; Hammarström P. Spectroscopic Characterization of Diverse Amyloid Fibrilsin Vitro by the Fluorescent Dye Nile Red. Mol. BioSyst. 2011, 7, 1232–1240. 10.1039/c0mb00236d. [DOI] [PubMed] [Google Scholar]
- Groenning M.; Norrman M.; Flink J. M.; van de Weert M.; Bukrinsky J. T.; Schluckebier G.; Frokjaer S. Binding Mode of Thioflavin T in Insulin Amyloid Fibrils. J. Struct. Biol. 2007, 159, 483–497. 10.1016/j.jsb.2007.06.004. [DOI] [PubMed] [Google Scholar]
- Sulatskaya A. I.; Kuznetsova I. M.; Turoverov K. K. Interaction of Thioflavin T with Amyloid Fibrils: Fluorescence Quantum Yield of Bound Dye. J. Phys. Chem. B 2012, 116, 2538–2544. 10.1021/jp2083055. [DOI] [PubMed] [Google Scholar]
- Colvin M. T.; Silvers R.; Ni Q. Z.; Can T. V.; Sergeyev I.; Rosay M.; Donovan K. J.; Michael B.; Wall J.; Linse S.; Griffin R. G. Atomic Resolution Structure of Monomorphic Aβ42 Amyloid Fibrils. J. Am. Chem. Soc. 2016, 138, 9663–9674. 10.1021/jacs.6b05129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wälti M. A.; Ravotti F.; Arai H.; Glabe C. G.; Wall J. S.; Böckmann A.; Güntert P.; Meier B. H.; Riek R. Atomic-resolution structure of a disease-relevant Aβ(1-42) amyloid fibril. Proc. Natl. Acad. Sci. U.S.A. 2016, 113, E4976–E4984. 10.1073/pnas.1600749113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Webber S. E. The Role of Time-Dependent Measurements in Elucidating Static Versus Dynamic Quenching Processes. Photochem. Photobiol. 1997, 65, 33–38. 10.1111/j.1751-1097.1997.tb01874.x. [DOI] [Google Scholar]
- Sillen A.; Engelborghs Y. The Correct Use of “Average” Fluorescence Parameters. Photochem. Photobiol. 1998, 67, 475–486. 10.1111/j.1751-1097.1998.tb09443.x. [DOI] [Google Scholar]
- Lakowicz J. R.Principles of Fluorescence Spectroscopy; Springer: New York, 2010. [Google Scholar]
- Sörgjerd K.; Klingstedt T.; Lindgren M.; Kågedal K.; Hammarström P. Prefibrillar Transthyretin Oligomers and Cold Stored Native Tetrameric Transthyretin Are Cytotoxic in Cell Culture. Biochem. Biophys. Res. Commun. 2008, 377, 1072–1078. 10.1016/j.bbrc.2008.10.121. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.