Abstract
Androgens and estrogens, working together, promote prostate cancer (PRCA) initiation and progression, with androgens acting via androgen receptor (AR) and estrogens acting primarily through estrogen receptor-α (ERα). While the interplay between these steroid hormones has been established, the interaction between steroid hormone receptors in prostatic disease remains unstudied. The goal of this study was to objectively determine the incidence, stage specificity, and tissue/cell type specificity of AR and ERα expression, both independently and simultaneously, during the progression of PRCA. Using multiplexed immunohistochemistry and multispectral imaging analysis, AR, ERα, and smooth muscle α-actin (SMA) expression was detected and quantitated in benign prostate tissue (BPT), high-grade prostatic intraepithelial neoplasia (HGPIN), PRCA, and metastasis (MET) from patient specimens (n=340). Epithelial AR expression was significantly increased in HGPIN, PRCA, and MET compared to BPT, while ERα expression in epithelial and stromal cells was highest in HGPIN. With analysis of AR and ERα co-expression, we identified a unique population of double positive (AR+/ERα+) cells that increased in HGPIN specimens in both the stroma and the epithelium. Double negative (AR−/ERα−) cells significantly decreased across PRCA progression, from 65% in BPT to 30% in MET. Preliminary analysis of this AR+/ERα+ population indicates potential cell type specificity in SMA-negative stromal cells. This study demonstrates stage-, tissue-, and cell type-specific AR and ERα expression changes during PRCA progression, both independently and co-expressed. A more complete understanding of steroid hormones and their receptors in the initiation and progression of prostatic disease may elucidate improved strategies for PRCA prevention or therapy.
Keywords: androgens, estrogens, prostate cancer, stroma, epithelium
1. INTRODUCTION
In 2019, an estimated 174,650 men in the United States will be diagnosed with prostate cancer (PRCA) and nearly 32,000 men will die as a result 1. As PRCA is the most prevalent cancer diagnosed among men in the United States, understanding the molecular changes associated with disease initiation and progression are critical for better diagnosis and treatment of men with prostatic disease. For decades, it has been known that sex steroid hormones and their receptors play an important role in regulating the prostate, both in development and disease 2–4. While the interplay between hormone receptors has recently been recognized 5–7, the incidence, disease stage specificity, and cell type specificity of this interaction remains unresolved.
Androgens, acting via the androgen receptor (AR), play key roles in normal and neoplastic prostate growth 8, 9. While it is well accepted that AR is expressed in the nuclei of stromal and epithelial cells of the prostate 9, 10, there are discrepancies among reports of AR expression in PRCA progression 11–13. Estrogens have also been implicated in the development and progression of PRCA, though the role of estrogens in prostatic disease is not well understood 14–16. Unlike androgens, estrogens work primarily through two cognate receptors, estrogen receptor alpha (ERα) and estrogen receptor beta (ERβ). In the prostate, several lines of evidence support the concept that ERα promotes prostatic epithelial proliferation during carcinogenesis, whereas ERβ functions as a tumor suppressor 3, 4, 14, 17. As ERβ is considered to be lost during PRCA progression 14, 17, this study will focus on the role of ERα. Similar to AR, the literature on ERα expression during PRCA progression is inconsistent 12, 18–20. While there is some evidence of clinical success in targeting ERα for PRCA therapy or prevention 21, the interaction between AR and ERα in this context remains unresolved.
Recently, there has been an increasing interest in the synergy between steroid hormone receptors, and their combined role in PRCA development and progression. It has been established that both androgens and estrogens are required for malignant transformation of cells in early PRCA progression 9, 22, suggesting there may be receptor interaction during PRCA initiation. Others have identified a role for AR/ERα cooperation in androgen targeted therapy resistance 5, where prostate tumor heterogeneity underlying therapy resistance can be overcome by co-treatments targeting both AR and ERα signaling. Alternatively, cells that express AR and ERα may be intrinsically resistant to ADT due to sustained growth signaling through an ERαdependent mechanism. A recent study showed expression of canonical AR-target gene fusion TMPRSS-ERG is working through ERα, not AR 7. Yet another study suggests that AR and ERα co-localization in the nucleus occurred with estrogen, not androgen, stimulation, suggesting ADT may not be sufficient to prevent AR nuclear translocation 6. While these data introduce a potential interplay between AR and ERα in PRCA initiation or progression, the incidence and stage specificity of this interaction has not yet been identified.
Due to the inconsistencies in published data regarding AR and ERα expression in the prostate and the lack of information regarding co-expression of these receptors, there has been difficulty in discerning their combined significance in PRCA progression. In this study, we utilized an automated pathology analysis platform to quantitatively assess protein expression and localization of AR and ERα during PRCA progression, both simultaneously and independently 23. This objective machine learning based approach minimizes the limitations of previous studies (e.g. staining technique/inter-observer variability) and enables analysis of AR and ERα co-expression and co-localization in the same cell 23. Additionally, multiplexed staining with smooth muscle alpha-actin (SMA) allows preliminary analysis of cell type specificity of AR and ERα co-expression. With this technology, we aim to determine 1) the incidence of AR and ERα co-localization in human prostate, 2) the stage of prostatic disease at which AR and ERα are co-expressed, and 3) preliminary cell type specificity of AR and ERα co-expression. A more precise and quantitative understanding of AR and ERα localization and expression at different stages of PRCA could lead to a more complete picture of disease progression and potentially offer insight into new approaches for prevention or therapy.
2. MATERIALS AND METHODS
2.1. Immunohistochemistry
To assess AR and ERα protein localization and abundance during PRCA progression, we used a previously described tissue microarray (TMA) composed of duplicate tissue cores from prostates of different disease stages23. In accordance with the Institutional Review Board, informed consent was obtained for experimentation with human samples. Antibodies (AR: Biocare Medical ACI-109-A, 1:50; ERα: Thermo Scientific RM-9101, 1:400) were validated during optimization (Supplemental Figure S1). Tissue cores consisted of benign prostate tissue, not inclusive of BPH (BPT; n=101 cores, 52 patients), high-grade prostatic intraepithelial neoplasia (HGPIN; n=50 cores, 25 patients), primary tumor (PRCA; n=141 cores, 73 patients) and metastatic tumors (MET; n=44 cores, 22 patients) 23. Multiplexed immunohistochemistry (IHC) was performed to detect AR, ERα, and smooth muscle alpha-actin (SMA) as previously described 23, 24. SMA was used to designate stroma for stromal vs. epithelial analysis.
2.2. Image Analysis
For automatic image acquisition, we used the Vectra platform (Perkin Elmer, Waltham, MA) and InForm™ 1.4 software, (Perkin Elmer, Waltham, MA) for image analysis 23. Using InForm™ 1.4 software (Perkin Elmer, Waltham, MA), 18% of the total images (rendering approximately 97% accuracy) were trained by a genitourinary pathologist (WH) to segment nucleus from cytoplasm, and epithelium from stroma. This created an algorithm enabling automated cell and tissue segmentation for each tissue compartment within each core sample. The same algorithm and threshold was then applied to the entire TMA.
2.3. Staining Quantification
AR and ERα staining were quantified as the percentage of positive nuclei divided by the total number of nuclei in the respective tissue compartment (stroma and epithelium). To account for cells or tissue not already identified, we established a third compartment designated as “other”, which included artifact, edge-effect, nerves, red blood cells, and inflammatory cells. Co-localization of AR and ERα, within each cell, was assessed and used to quantify double positive cells (AR and ERα) in each tissue compartment 24. Using raw cell segmentation data exported from inForm v1.4 (PerkinElmer, Waltham, MA), and manual thresholding of AR (mean OD threshold=0.02), ERα (mean OD threshold=0.125), and SMA (mean OD threshold=0.08), we calculated the proportion of ERα+/AR+ double positive cells within the SMA-negative stromal compartment.
2.4. Statistical analysis
Graphpad Prism 5.04 (Graphpad Software, La Jolla, CA) was used for statistical analysis. We assessed differences among continuous variables with one-way ANOVA. Tukey’s Multiple Comparison Test was used to determine mean differences of BPT compared to HGPIN, PRCA, and MET. Data in bar graphs and tables show mean +/− SEM. For all analyses, p< 0.05 was considered statistically significant.
3. RESULTS
3.1. Tissue and cell segmentation
Using machine learning software, we objectively segmented tissue types (stroma, epithelia) and cell compartments (nucleus, cytoplasm) across disease progression in human prostate tissue (n=384). With this separation, we quantified cell number (based on nuclei) within each tissue type across PRCA progression, as shown in Table 1. The total number of epithelial cells increased from BPT to PRCA and from BPT to MET (p<0.001 for both), while the percentage of epithelial cells increased from BPT to HGPIN, PRCA, and MET (p<0.001 for all). There was a concordant decrease in the number of stromal cells from BPT to HGPIN (p<0.05), and the percentage of stromal cells decreased from BPT to HGPIN (p<0.01), PRCA, and MET (p<0.001 for both). As expected, the percent of tissue area increased for the epithelial compartment in PRCA and MET compared to BPT (p<0.001 for both), but decreased for the stromal compartment in MET vs. BPT (p<0.01). These segmentation techniques provide evidence of the sensitivity and accuracy of the software and serve as the basis for our analysis of AR and ERα positivity within the nucleus of stromal vs. epithelial tissue.
Table 1:
Cell quantification and tissue segmentation in prostate cancer
| BPT | HGPIN | PRCA | MET | |
|---|---|---|---|---|
| Number of Cells | ||||
| Total | 1153±36 | 1097±73 | 1498±37 c | 1827 ±109 c |
| Epithelial | 402±29 | 604±59 | 822±46 c | 1169±117 c |
| Stromal | 487±23 | 331±34 a | 436±25 | 375±89 |
| % of Cells | ||||
| Epithelial | 33±1.9 | 52±2.7 c | 52±2.1 c | 62±4.9 c |
| Stromal | 43±1.7 | 30±2.5 b | 30±1.8 c | 20±4.3 c |
| % Tissue Area | ||||
| Epithelial | 16±1.2 | 17±1.5 | 33±1.7 c | 44±3.8 c |
| Stromal | 35±1.8 | 33±3.0 | 29±1.8 | 23±4.5 b |
All values are presented as mean ± SEM per core
P < 0.05
P < 0.01
P <0.001 compared to BPT.
3.2. AR tissue localization and expression
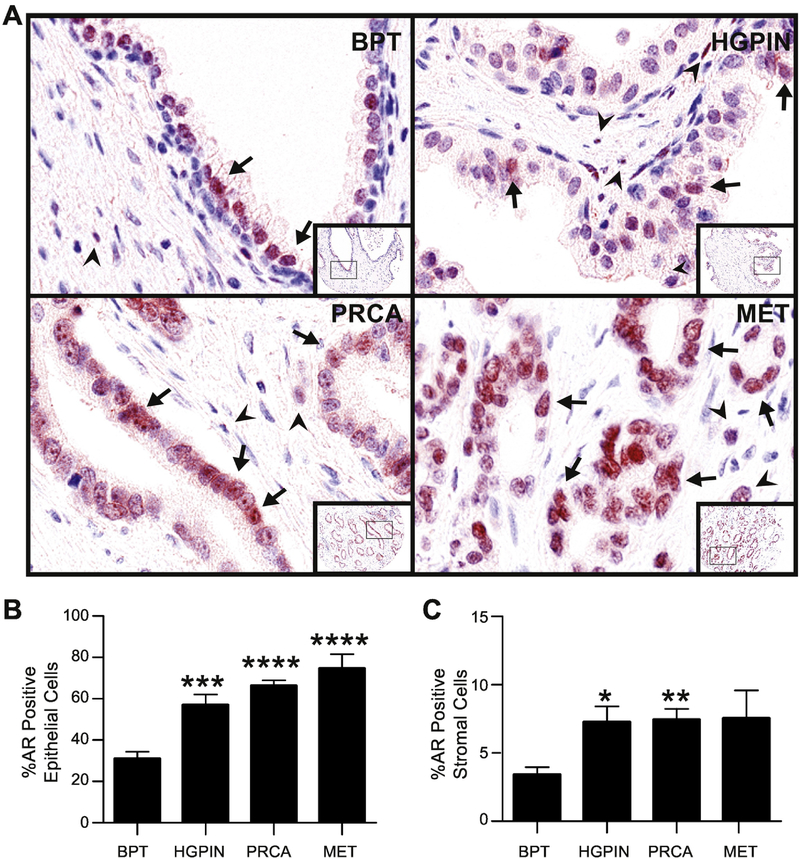
AR predominantly localized to nuclei of stromal and epithelial cells throughout all stages of PRCA progression. (Figure 1A). Using the tissue segmentation function of the machine learning software, we quantified AR positivity in both the epithelium and the stroma. In accordance with previously published data 10, 25, the percent of AR positivity in the epithelial compartment increased in HGPIN, PRCA, and MET compared to BPT (p< 0.001 for HGPIN, p<0.0001 for PRCA and MET; Figure 1B). Within the stroma, AR positivity was increased in HGPIN and PRCA compared to BPT (p<0.05, p<0.01, respectively) (Figure 1C). There was no difference in positivity between BPT and MET (p>0.05) (Figure 1C). In addition to expression trends through progression, we also noted both inter- and intra- sample heterogeneity of AR expression, which was categorized as high, medium or low AR expression (Supplemental Figure S2). Taken together, these data suggest that 1) AR is expressed in all stages of PRCA progression, though at different levels, and 2) AR in both the epithelium and the stroma may contribute to HGPIN and PRCA, while AR specifically in the epithelium may contribute to MET.
Fig. 1. Androgen receptor expression in prostate cancer progression.
A: Prostate composite image shows androgen receptor (AR, red) in nuclei of luminal epithelial cells (arrows) and stromal cells (arrowheads). AR positivity is seen more in the epithelium than stroma, and overall, expression increased in PRCA progression.
B: The percentage of AR positive cells significantly increased in the epithelium in HGPIN, PRCA and MET compared to BPT.
C: The percentage of AR positive cells in the stroma significantly increased in HGPIN and PRCA compared to BPT.
BPT = benign tissue prostate, HGPIN = high grade prostatic neoplasia, PRCA = prostate cancer, MET = metastasis. IHC 100X (20X inset). *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001 via post-hoc comparison to BPT.
3.3. ERα tissue localization and expression
ERα staining both supported and refuted previously published data 12, 17–20. As expected, ERα in BPT and HGPIN was predominantly nuclear and localized to basal epithelial and stromal cells (Figure 2A). Interestingly, in PRCA and MET, while ERα localized primarily to the stroma, there was a low percentage (2–5%) of positivity and in carcinoma cells. Segmenting stromal vs. epithelial staining revealed changes in percent positivity across PRCA progression within each tissue compartment. In the epithelium, ERα positivity was increased in HGPIN (p<0.0001) compared to BPT (Figure 3B). There was no change in ERα positivity between BPT and PRCA or METS (p>0.05) (Figure 2B). In the stroma, ERα positivity increased in HGPIN (p<0.05) and PRCA (p<0.01) compared to BPT (Figure 2C). There was no change in ERα positivity between BPT and METS (p>0.05) (Figure 2C). Taken together, these data show that ERα is expressed in both the epithelium and stroma in PRCA progression. Additionally, the highest percent positivity for ERα occurred in HGPIN for both tissue compartments, suggesting that this stage is where ERα contributes to prostatic disease.
Fig. 2. Estrogen receptor-alpha expression in prostate cancer progression.
A: Prostate composite image shows estrogen receptor-alpha (ERα, brown) in nuclei of epithelial (arrows) and stromal cells (arrow heads). In normal prostate epithelium, ERα was localized primarily in basal epithelial cells, while epithelial ERα expression in HGPIN and PRCA showed a marked increase.
B: The overall percentage of ERα positive cells in the epithelium is significantly increased in HGPIN compared to BPT.
C: The percentage of ERα positive stromal cells was increased in HGPIN and PRCA compared to BPT.
BPT = benign tissue prostate, HGPIN = high grade prostatic neoplasia, PRCA = prostate cancer, MET = metastasis. IHC 100X (20X inset). *P < 0.05, **P < 0.01, ****P < 0.0001 via post-hoc comparison to BPT.
Fig. 3. Co-localization of AR and ERα in normal and pathologic prostate.
A: Prostate composite image shows a small number of AR+/ERα+ double positive nuclei (yellow) seen within the epithelium (arrow) and stroma (arrowheads) in all stages of PRCA progression.
B: The percent positivity of AR+/ERα+ cells in the epithelium was increased in HGPIN only compared to BPT.
C: Double negative (AR−/ERα−) cells in the epithelium were significantly decreased in HGPIN, PRCA and MET compared to BPT.
D: In the stroma, AR+/ERα+ double positive cells were significantly higher in HGPIN compared to BPT.
E: Double negative (AR−/ERα−) cells in the stroma were significantly decreased in HGPIN and PRCA compared to BPT, but unchanged in MET compared to BPT.
BPT = benign tissue prostate, HGPIN = high grade prostatic neoplasia, PRCA = prostate cancer, MET = metastasis. IHC 100X (20X inset). **P < 0.01, ***P < 0.001, ****P < 0.0001 via post-hoc comparison to BPT.
3.4. Tissue co-expression and co-localization of AR and ERα
Because there may be synergistic effects between AR and ERα, we assessed the co-expression of these two receptors in PRCA progression (Figure 3A). In all stages of PRCA progression nuclear co-expression of these two receptors was detectable (Figure 3A). Within the epithelium, the proportion of AR+/ERα+ increased from BPT to HGPIN (p< 0.0001, Figure 3B), but did not change in PRCA or MET. Similarly, within the stroma, AR+/ERα+ double positivity increased from BPT to HGPIN (p< 0.01, Figure 3D), but did not change in PRCA or MET. These data show evidence of co-localization of AR and ERα in PRCA progression, in both the epithelial and stromal compartments; however, the incidence of this co-expression is relatively low, ranging from <1–5%. In addition to quantification of AR/ERα co-expression by double positivity, the percentage of cells within each tissue compartment that lack both receptors can be determined. The proportion of AR−/ERα− was lower in HGPIN, PRCA, and MET compared to BPT (p<0.0001 for all) for the epithelial compartment (Figure 3C). For the stromal compartment, there was a significant decrease in AR−/ERα− cells in HGPIN and PRCA compared to BPT (p<0.001, p<0.0001, respectively), but no change in MET compared to BPT (p>0.05) (Figure 3E). Taken together, these data support the idea that as PRCA progresses, there is increased expression of steroid hormone receptors in epithelial cells as seen by a decrease in double negativity through progression. Interestingly, while the incidence of AR/ERα co-expression is relatively low overall, there is a significant increase in AR and ERα co-expression in HGPIN, suggesting that steroid receptor interaction may contribute to disease initiation.
3.5. Localization and co-localization of AR and ERα in SMA-negative stroma
Prostatic stromal tissue is made up of a variety of different cell types: fibroblasts, myofibroblasts, endothelial cells, immune cells, etc 26. A preliminary assessment of cell type specificity for AR/ERα expression in the stroma was conducted using SMA as an initial criterion to identify differentiated smooth muscle cells and myofibroblasts. Specifically, we assessed AR/ERα expression, both separately and co-expressed, in SMA-negative stroma, which may represent cell types such as fibroblasts, immune cells, or endothelial cells. We observed an increase in the prevalence of AR positive cells in SMA-negative stroma in HGPIN and PRCA (p< 0.05 for both), but a decrease in AR positivity in SMA-negative stroma in MET (p< 0.01; Figure 4A). The number of ERα-positive SMA-negative stromal cells was increased in HGPIN compared to BPT (p<0.05), but no difference was observed in PRCA or MET (Figure 4B). The number of AR+/ERα+, SMA-negative cells was increased only in HGPIN (p<0.05; Figure 4C). Taken together, these data indicate that stromal AR and ERα expression in early PRCA progression may have cell type specificity that has not previously been described.
Fig. 4. AR and ERα expression and co-localization in smooth muscle α-actin negative stroma.
A: Prostate composite image shows localization of AR (red) and smooth muscle α-actin (SMA, turquoise) in PRCA progression. Quantification of AR in SMA-negative stroma showed a significant increase in HGPIN and PRCA compared to BPT, but a significant decrease in MET compared to BPT.
B: Prostate composite image shows localization of ERα (brown) and smooth muscle alpha-actin (SMA, turquoise) in PRCA progression. Quantification of AR in SMA-negative stroma showed a significant increase in HGPIN compared to BPT, and no significant change in PRCA or MET compared to BPT.
C: Prostate composite image shows co-localization of cells expressing both AR and ERα (yellow) and smooth muscle α-actin (SMA, turquoise) in PRCA progression. Quantification of AR+/ERα+ cells in SMA-negative stroma showed a significant increase in HGPIN compared to BPT, and no significant change in PRCA or MET compared to BPT.
BPT = benign tissue prostate, HGPIN = high grade prostatic neoplasia, PRCA = prostate cancer, MET = metastasis. IHC 100X (20X inset). *P < 0.05, **P < 0.01 via post-hoc comparison to BPT.
4. DISCUSSION
The interplay between androgens and estrogens is an aging event that has been established as a crucial component of PRCA initiation and progression 9, 22; however, the literature regarding the expression and localization of AR and ERα in PRCA progression is inconsistent 11–13, 18, 19. Therefore, the incidence, stage specificity, and cell type specificity of steroid hormone receptor interactions remains unresolved. Using multiplexed IHC followed by multispectral imaging, we showed that the independent expression of steroid hormone receptors AR and ERα changes through PRCA progression. These expression and localization findings support the conclusion that androgens are important at all stages of PRCA progression, while estrogens, working via ERα, likely play a role in early carcinogenesis. Interestingly, the staining pattern for AR also demonstrated the presence of AR low or negative cells throughout progression, which has previously been described as AR heterogeneity 27–29. In this study, there was both inter- and intra-sample heterogeneity of AR expression ranging from samples with very low AR expression to very high. While prostate luminal epithelial cells are generally considered to be AR-positive, we found a range of 20–70% of epithelial cells within a core that were considered AR-negative at all stages of PRCA progression. In PRCA, an average of 32% of cells within each core were considered AR low/negative in our study. Importantly, previous reports have correlated AR heterogeneity with poor patient outcomes 27–29. Though the potential role of AR negative cells in PRCA progression remains to be fully understood, this study provides evidence of AR heterogeneity in human PRCA specimens.
To our knowledge, no other studies have reported that AR and ERα are co-localized in the same cell and that the prevalence of these double positive cells changes with PRCA progression. This may be due, in part, to technical challenges associated with evaluating multiple markers with immunohistochemistry (IHC), especially with spatially overlapping targets. Additionally, steroid receptor positivity may vary based on focal areas (i.e. inflammation) and be unevenly distributed throughout the prostate. Conventional methods of analysis involve visually estimating staining intensity and evaluating hot spots. To circumvent limitations of current approaches, this study utilized an automated pathology analysis platform to quantify expression and localization of AR and ERα in PRCA progression using 340 patient specimens23. This platform allows for objective quantification of the prevalence of these hormone receptors within each tissue compartment (stroma, epithelium), both separately and together; importantly, the expression output reflects average optical density of each core as a whole, not a selected hot spot. While machine-learning based approaches to data analysis are more objective, one limitation of this technique is that results rely on the efficacy of the applied algorithm; in this study, nuclear segmentation was determined based on hematoxylin, which identifies nuclei. Therefore, cells that lack nuclei (i.e. platelets, red blood cells) were not included in the analysis23. Additionally, the tissues stained in the current study are duplicate cores taken from a larger tissue. Because of the high degree of heterogeneity within the prostate, these cores may not represent the prostate as a whole. Finally, these samples lack associated clinical data, resulting in a dearth of clinic-pathologic outcomes associated with AR/ERα co-expression. Despite the limitations to automated pathology platforms 23, 24, they remain a preferred approach due to efficiency and internal consistency.
Using this technology, we report the presence of AR/ERα double positive cells that may represent a subpopulation of cells mediating sex steroid synergy. Additionally, this study provides evidence that there are at least two distinct populations of hormone-responsive stromal cells (AR+ and ERα+) in PRCA. While this is an enticing area for future study, the implication of the present findings is that selective steroid receptor modulators directly targeting hormone receptors might be most effective when used for prevention of PRCA carcinogenesis. Taken together with former studies demonstrating stromal receptors are critical in PRCA progression 9, 24, therapies targeting stroma may be key to the prevention of PRCA development or progression. To evaluate AR and ERα in the stromal microenvironment, we used SMA to identify differentiated smooth muscle cells and myofibroblasts, as well as to aid in the identification of SMA-negative stroma. While this criterion does not definitively determine cell identity, due to the morphology and lack of SMA expression in these cells, it is likely that they are fibroblasts – an abundant cell type within the prostatic stroma 30, 31. Fibroblasts secrete growth factors, inflammatory cytokines, produce extracellular matrix proteins and can create a niche for cancer cells. In cancer, these cells are referred to as carcinoma-associated fibroblasts (CAFs), and play a role in evading immunosurveillance, promoting a chronic inflammatory environment, and altering the metabolism of the microenvironment towards a pro-tumorigenic state 26, 32. The present findings indicate that the stromal microenvironment of HGPIN contains an increased amount of double positive (AR+/ERα+) cells compared to BPT, which may be an important intersection in suppression or promotion of carcinogenesis. While this study provides preliminary evidence of cell type specificity for AR and ERα co-expression, a more definitive cell type identification is necessary to understand more completely the role of fibroblasts in this complex system.
Underscoring the importance of androgens and estrogens in PRCA, we demonstrate that AR and ERα expression in carcinoma cells and the stromal microenvironment changes with different stages of PRCA. Using an automated quantitative pathology approach, we report tissue specific expression and localization of AR, ERα, as well as cells expressing both receptors in PRCA progression. Our findings highlight the importance of disease stage-, tissue-, and cell type specificity of this co-localization, and provide a foundation for further investigation into the interaction of steroid hormone receptor in PRCA. A better understanding of hormone receptor expression through PRCA progression could increase our knowledge of events contributing to PRCA initiation or progression, and potentially provide rationale for co-targeting these receptors for PRCA prevention or therapy.
Supplementary Material
Supplemental Figure S1. Antibody Validation with Control Tissues
A: Androgen receptor (AR) expression in control tissues shows a lack of AR staining in human tonsil tissue (left panel) and strong nuclear staining (red) in prostate epithelial cells (right panel).
B: Estrogen receptor-alpha (ERα) expression in control tissues shows a lack of ERα staining in human kidney tissue (left panel) and strong nuclear staining (brown) in breast epithelial cells (right panel).
Supplemental Figure S2. Inter- and Intra-tumoral AR Heterogeneity
A: Prostate composite image shows androgen receptor (AR) in nuclei of luminal epithelial cells (red) in PRCA cores. There is inter-tumoral AR heterogeneity showing low, medium, and high AR expression.
B: The percentage of low, medium, and high AR expression was quantified among the PRCA cores (n=141), and represented in a pie chart. 57.45% of cores express high levels of AR, 29.78% expressed medium levels of AR, and 12.77% expressed low levels of AR.
C: An example prostate cancer core shows intra-tumoral AR heterogeneity, where AR expression is shown in red. The full core is shown on the left, and high magnification images are shown on the right. AR expression ranges from high (top, right) to low (bottom, right).
HIGHLIGHTS.
Steroid hormone receptor expression was objectively quantified in prostate cancer
Androgen receptor increased in cancer progression, yet AR-negative cells persist
Stromal estrogen receptor α expression increased in prostate cancer progression
Co-expression occurred early in progression in both stromal and epithelial cells
Stromal co-expression occurred predominantly in cells lacking smooth muscle α-actin
5. ACKNOWLEDGMENTS
The authors thank the University of Wisconsin TRIP laboratory, in part supported by the UW Department of Pathology and Laboratory Medicine and UWCCC, for use of its facilities and services. We also acknowledge the help of Laura Hogan for manuscript editing and Glen Leversen for bio-statistical assistance. TMN was a trainee in the Medical Scientist Training Program at the University of Rochester, and JEV is a trainee in the Cancer Biology Graduate Program at the University of Wisconsin-Madison.
GRANT SUPPORT: U54 DK104310 (WAR, WX, WH), R01 ES001332 (WAR, WT), P30 CA014520 (UWCCC), T32 GM07356 (TMN), F30 DK093173 (TMN), T32 CA009135 (JEV)
Footnotes
DISCLOSURE STATEMENT: The authors have nothing to disclose.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- [1].Siegel RL, Miller KD, Jemal A. Cancer statistics, 2019. CA: a cancer journal for clinicians. [DOI] [PubMed] [Google Scholar]
- [2].Prins GS, Korach KS. The role of estrogens and estrogen receptors in normal prostate growth and disease. Steroids 2008; 73:233–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Ricke WA, Wang Y, Cunha GR. Steroid hormones and carcinogenesis of the prostate. the role of estrogens. Differentiation 2007; 75:871–82. [DOI] [PubMed] [Google Scholar]
- [4].Ricke WA, McPherson SJ, Bianco JJ, Cunha GR, Wang Y, Risbridger GP. Prostatic hormonal carcinogenesis is mediated by in situ estrogen production and estrogen receptor alpha signaling. The FASEB journal 2008; 22:1512–20. [DOI] [PubMed] [Google Scholar]
- [5].Fujimura T, Takayama K, Takahashi S, Inoue S. Estrogen and androgen blockade for advanced prostate cancer in the era of precision medicine. Cancers 2018; 10:29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Ochiai I, Matsuda K-i, Nishi M, Ozawa H, Kawata M. Imaging analysis of subcellular correlation of androgen receptor and estrogen receptor α in single living cells using green fluorescent protein color variants. Molecular Endocrinology 2004; 18:26–42. [DOI] [PubMed] [Google Scholar]
- [7].Setlur SR, Mertz KD, Hoshida Y, et al. Estrogen-dependent signaling in a molecularly distinct subclass of aggressive prostate cancer. Journal of the National Cancer Institute 2008; 100:815–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Habib FK, Odoma S, Busuttil A, Chisholm GD. Androgen receptors in cancer of the prostate. Correlation with the stage and grade of the tumor. Cancer 1986; 57:2351–6. [DOI] [PubMed] [Google Scholar]
- [9].Ricke EA, Williams K, Lee Y-F et al. Androgen hormone action in prostatic carcinogenesis. stromal androgen receptors mediate prostate cancer progression, malignant transformation and metastasis. Carcinogenesis 2012; 33:1391–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Chodak GW, Kranc DM, Puy LA, Takeda H, Johnson K, Chang C. Nuclear localization of androgen receptor in heterogeneous samples of normal, hyperplastic and neoplastic human prostate. The Journal of Urology 1992; 147:798–803. [DOI] [PubMed] [Google Scholar]
- [11].Zhang SX, Bentel JM, Ricciardelli C, Horsfall DJ, Haagensen DE, Marshall VR, Tilley WD. Immunolocalization of apolipoprotein D, androgen receptor and prostate specific antigen in early stage prostate cancers. The Journal of urology 1998; 159:548–54. [DOI] [PubMed] [Google Scholar]
- [12].Brolin J, Skoog L, Ekman P. Immunohistochemistry and biochemistry in detection of androgen, progesterone, and estrogen receptors in benign and malignant human prostatic tissue. Prostate 1992; 20:281–95. [DOI] [PubMed] [Google Scholar]
- [13].Li R, Wheeler T, Dai H, Frolov A, Thompson T, Ayala G. High level of androgen receptor is associated with aggressive clinicopathologic features and decreased biochemical recurrence-free survival in prostate. cancer patients treated with radical prostatectomy. The American journal of surgical pathology 2004; 28:928–34. [DOI] [PubMed] [Google Scholar]
- [14].Bonkhoff H, Berges R. The evolving role of oestrogens and their receptors in the development and progression of prostate cancer. European urology 2009; 55:533–42. [DOI] [PubMed] [Google Scholar]
- [15].Carruba G Estrogen and prostate cancer. An eclipsed truth in an androgen-dominated scenario. Journal of cellular biochemistry 2007; 102:899–911. [DOI] [PubMed] [Google Scholar]
- [16].Härkönen PL, Mäkelä SI. Role of estrogens in development of prostate cancer. The Journal of steroid biochemistry and molecular biology 2004; 92:297–305. [DOI] [PubMed] [Google Scholar]
- [17].Bonkhoff H Estrogen receptor signaling in prostate cancer. implications for carcinogenesis and tumor progression. The Prostate 2018; 78:2–10. [DOI] [PubMed] [Google Scholar]
- [18].Hobisch A, Hittmair A, Daxenbichler G et al. Metastatic lesions from prostate cancer do not express oestrogen and progesterone receptors. The Journal of Pathology: A Journal of the Pathological Society of Great Britain and Ireland 1997; 182:356–61. [DOI] [PubMed] [Google Scholar]
- [19].Bonkhoff H, Fixemer T, Hunsicker I, Remberger K. Estrogen receptor expression in prostate cancer and premalignant prostatic lesions. The American journal of pathology 1999; 155:641–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Daniels G, Gellert LL, Melamed J, et al. Decreased expression of stromal estrogen receptor α and β in prostate cancer. American journal of translational research 2014; 6:140. [PMC free article] [PubMed] [Google Scholar]
- [21].Steiner MS, Raghow S, Neubauer BL. Selective estrogen receptor modulators for the chemoprevention of prostate cancer. Urology 2001; 57:68–72. [DOI] [PubMed] [Google Scholar]
- [22].Ricke WA, Ishii K, Ricke EA, Simko J, Wang Y, Hayward SW, Cunha GR. Steroid hormones stimulate human prostate cancer progression and metastasis. International journal of cancer: Journal international du cancer 2006; 118:2123–31. [DOI] [PubMed] [Google Scholar]
- [23].Huang W, Hennrick K, Drew S. A colorful future of quantitative pathology. validation of Vectra technology using chromogenic multiplexed immunohistochemistry and prostate tissue microarrays. Human pathology 2013; 44:29–38. [DOI] [PubMed] [Google Scholar]
- [24].Nicholson TM, Sehgal PD, Drew SA, Huang W, Ricke WA. Sex steroid receptor expression and localization in benign prostatic hyperplasia varies with tissue compartment. Differentiation 2013; 85:140–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Heinlein CA, Chang C. Androgen receptor in prostate cancer. Endocrine reviews 2004; 25:276–308. [DOI] [PubMed] [Google Scholar]
- [26].Kalluri R, Zeisberg M. Fibroblasts in cancer. Nature Reviews Cancer 2006; 6:392. [DOI] [PubMed] [Google Scholar]
- [27].De Winter JAR, Trapman J, Brinkmann AO et al. Androgen receptor heterogeneity in human prostatic carcinomas visualized by immunohistochemistry. The Journal of pathology 1990; 160:329–32. [DOI] [PubMed] [Google Scholar]
- [28].Magi-Galluzzi C, Xu X, Hlatky L et al. Heterogeneity of androgen receptor content in advanced prostate cancer. Modern pathology: an official journal of the United States and Canadian Academy of Pathology, Inc 1997; 10:839–45. [PubMed] [Google Scholar]
- [29].Takeda H, Akakura K, Masai M, Akimoto S, Yatani R, Shimazaki J. Androgen receptor content of prostate carcinoma cells estimated by immunohistochemistry is related to prognosis of patients with stage D2 prostate carcinoma. Cancer: Interdisciplinary International Journal of the American Cancer Society 1996; 77:934–40. [PubMed] [Google Scholar]
- [30].Dvorak HF. Tumors. wounds that do not heal. New England Journal of Medicine 1986; 315:1650–9. [DOI] [PubMed] [Google Scholar]
- [31].Gravina GL, Mancini A, Ranieri G et al. Phenotypic characterization of human prostatic stromal cells in primary cultures derived from human tissue samples. International journal of oncology 2013; 42:2116–22. [DOI] [PubMed] [Google Scholar]
- [32].Pavlides S, Whitaker-Menezes D, Castello-Cros R, et al. The reverse Warburg effect. aerobic glycolysis in cancer associated fibroblasts and the tumor stroma. Cell cycle 2009; 8:3984–4001. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental Figure S1. Antibody Validation with Control Tissues
A: Androgen receptor (AR) expression in control tissues shows a lack of AR staining in human tonsil tissue (left panel) and strong nuclear staining (red) in prostate epithelial cells (right panel).
B: Estrogen receptor-alpha (ERα) expression in control tissues shows a lack of ERα staining in human kidney tissue (left panel) and strong nuclear staining (brown) in breast epithelial cells (right panel).
Supplemental Figure S2. Inter- and Intra-tumoral AR Heterogeneity
A: Prostate composite image shows androgen receptor (AR) in nuclei of luminal epithelial cells (red) in PRCA cores. There is inter-tumoral AR heterogeneity showing low, medium, and high AR expression.
B: The percentage of low, medium, and high AR expression was quantified among the PRCA cores (n=141), and represented in a pie chart. 57.45% of cores express high levels of AR, 29.78% expressed medium levels of AR, and 12.77% expressed low levels of AR.
C: An example prostate cancer core shows intra-tumoral AR heterogeneity, where AR expression is shown in red. The full core is shown on the left, and high magnification images are shown on the right. AR expression ranges from high (top, right) to low (bottom, right).