Abstract
Systemic lupus (SLE) is characterized by a break of B cell tolerance that plays a central role in disease pathophysiology. An early checkpoint defect occurs at the transitional stage leading to the survival of autoreactive B cells and consequently the production of pathogenic autoantibodies. The main purpose of our work was to determine whether transitional B cells, as the most immature naïve B cell subset upstream of pathogenic B cells, display specific features compared to healthy non SLE subjects. Through extensive analysis of transitional B cells from untreated or low treated, mostly Caucasian, SLE patients, we demonstrated that transitional (T1 and T2) B cell frequencies were increased in SLE and positively correlated with disease activity. SLE transitional B cells displayed defects in two closely inter-related molecules (i.e. TLR9 defective responses and CD19 downregulation). RNA sequencing of sorted transitional B cells from untreated patients revealed a predominant overexpression of interferon stimulated genes (ISGs) even out of flares. In addition, early transitional B cells from the bone marrow displayed the highest interferon score, reflecting a B cell interferon burden of central origin. Hence, the IFN signature in transitional B cells is not confined to African American SLE patients and exists in quiescent disease since the medullary stage. These results suggest that in SLE these 3 factors (i.e. IFN imprintment, CD19 downregulation and TLR9 responses impairment) could take part at the early transitional B cell stage in B cell tolerance by-pass, ultimately leading in periphery to the expansion of autoantibodies-secreting cells.
Keywords: Systemic lupus erythematosus, Transitional B cells, Interferon, CD19, TLR9
1. INTRODUCTION
Systemic lupus erythematosus (SLE) is a severe autoimmune disease characterized by flares and mostly affecting young women. Patients produce pathogenic autoantibodies, specifically against double-stranded DNA (dsDNA) but also against ribonucleoproteins (SSA/Ro52, Sm) and phospholipids. The etiology of SLE is still not well understood but B cells and their autoantibodies play an essential role in disease pathophysiology [1,2]. There is currently a debate on SLE B cell fate and on the origin of ANA+ IgG+ cells [3]. Sequencing studies in young SLE patients and in the NZB/W lupus prone mice recorded greater percentages of ANA+ cells in naïve or new emigrant/transitional B cells, and an early breakdown of B tolerance [4,5]. Other studies argue for a late loss of control, as demonstrated by the expansion of IgG+ plasma cells in some patients with SLE and in the MRL/lpr model [5].
Strong data support the “early or naïve B cell model” in SLE. Autoreactive B cells including anti-nuclear and anti-ds DNA B cells are known to be controlled at the bone marrow (BM) level mainly by deletion or editing [6–8]. There is in patients affected with SLE an increased frequency of ANA+ and anti-dsDNA+ in naïve B cells compared to immature B cells suggesting defective selection at the transitional stage [4]. SLE mature naïve B cells display a specific decrease in the expression of CD19, which is necessary for efficient B cell signaling [9], and an altered in vitro toll like receptor 9 (TLR9) responses [10]. The latter observations are in agreement with disease worsening in TLR9 deficient lupus-prone murine models, and with TLR9 preventing the survival of autoreactive naive B cells when co-engaged with B cell receptor (BCR) via nucleic acid-autoantibodies complexes [11].
Nucleic acid-containing immune complexes also activate myeloid and plasmacytoid dendritic cells (pDCs) leading to the secretion of type I and II interferons (IFN), key cytokines associated with SLE pathogenesis [12,13]. Mei Liu et al. showed that plasma from Chinese patients enhance transitional B cell survival in a type I IFN dependent manner [14] In addition, increased levels of IFN-β were particularly detected in transitional B cells from African American SLE patients with nephritis and high levels of anti-ribonucleoproteins autoantibodies [15]. Hence, transitional B cells could be early key actors in SLE pathogenesis.
Considering these critical issues and the fact that transitional B cells are often increased in the blood of SLE patients independently of their ethnicity [10,16,17], we explored in depth transitional B cells from untreated or low treated, mostly Caucasian, SLE patients in quiescent or active phase of their disease. The main purpose of our work was to determine whether transitional B cells, as a potential checkpoint of tolerance and as the most immature naïve B cell subset upstream of pathogenic B cells, display specific features compared to healthy non SLE subjects. The question at hand is very important considering the needs in SLE for targeted or personalized therapy.
We confirm herein the increased frequency of transitional B cells in SLE, and more specifically of T1 and T2 subsets. This increase does not result from a proliferation or apoptosis defect but likely output from the BM. SLE transitional B cells display an abnormal phenotype with a CD19 reduced expression and an impaired response to TLR9 stimuli. In addition, a strong IFN signature, of central origin, is present at this stage, even in quiescent patients. We suggest that these 3 factors (i.e. IFN imprintment, CD19 downregulation and TLR9 response impairment) could take part at the early transitional B cell stage in B cell tolerance by-pass.
2. MATERIALS AND METHODS
2.1. Study approval
The study was conducted in accordance with the principles of Helsinki declaration. Informed consents were obtained from all patients and healthy donors (HDs). The study was approved by the Clinical Research Ethic Committee of Strasbourg’s University Hospital and the Human Subjects Institutional Review Board of the University of Rochester Medical Center (URMC).
2.2. Patients and samples
For blood samples, 23 patients (21 females and 2 males; 17 quiescent and 6 active patients) aged from 29 to 68 years with the diagnosis of systemic lupus erythematosus (SLE) were selected. The main characteristics of SLE patients are listed in Supplementary Table 1. All patients fulfilled the ACR SLE classification criteria [18]. Disease activity was measured by the modified Systemic Lupus Erythematosus Disease Activity Index (SLEDAI-2K) [19]. A SLEDAI-2K below 5 defines the quiescent patients associated with no activity in major organ systems, no vasculitis, no fever, no haemolytic anaemia, no new features of lupus activity or increase in treatment [20]. Only patients with no treatment (i.e. no immunosuppressive drugs, steroids and hydroxychloroquine within the last 6 months), or treated with low-dose steroids (<10 mg per day) with or without hydroxychloroquine were included. Patients treated with immunosuppressive drugs within the previous 6 months were systematically excluded. For transcriptomic analysis, only untreated patients were included, as hydroxychloroquine and corticosteroid therapy modify the lymphocyte transcriptomic profile [13]. Routine measurements were performed to determine ANAs (indirect immunofluorescence with Hep-2 cells, Zeus Scientific, USA) and anti-dsDNA titers (screened by ELISA; Kallestad anti-DNA microplate EIA; BioRad, Hercules, CA, USA). ANAs were considered positive when > 1/160. SSA/Ro60, SSB/La, and Sm antibodies identification was performed using enzyme immunoassays (EuroImmun, Lubeck, Germany) according to manufacturer’ specifications. Anticoagulated venous blood was drawn from SLE patients and from ethnicity, age and sex matched HDs and was subjected to density gradient centrifugation to get peripheral blood mononuclear cells (PBMC) and remove dead cells.
For bone marrow (BM) samples, 4 patients (4 females, all quiescent) aged from 24 to 55 years with the diagnosis of systemic lupus erythematosus (SLE) were selected. The main characteristics of SLE patients are listed in Supplementary Table 1. Research BM aspirates from SLE patients and HDs were obtained (i) at URMC by aspiration from the iliac crest (n=3) and (ii) at Strasbourg University Hospital by aspiration from the sternum (n=1). Paired peripheral blood samples were obtained for 3 SLE patients and 3 HDs (2 at URMC and 1 in Strasbourg). BM aspirate were subjected to density gradient centrifugation to get BM mononuclear cells and remove dead cells.
2.3. Flow Cytometry
Cell viability was assessed by incubation of cells with Fixable Viability Dye eFluor® 780 (eBioscience, San Diego, CA, USA) following the manufacturer’s protocol. Samples were incubated with antibodies for 15 minutes at 4°C. Intracellular staining was performed, for Ki-67 using True Nuclear™ Transcription Factor Fix (BioLegend, San Diego, CA, USA) following the manufacturer’s recommendations. The cells were washed twice in PBE (PBS, 0.5% Bovine Serum Albumin, 2mM EDTA) and analyzed on a Gallios flow cytometer (Beckman Coulter, Villepinte, France). For the analysis of protein expression, level of the geometric mean fluorescence intensity (MFI) was used. Fluorescence Minus One (FMO) controls were performed. Data analysis was performed using Kaluza software (Beckman Coulter) with “logicle” compensation [21]. MFI ratio is determined as follow: [MFI of the considered marker]/[mean of the MFI of the considered marker in HD group]. The following monoclonal anti-human antibodies were used: CD10 (HI10a), CD19 (J3–119) and CD27 (B12701) from Beckman Coulter, Ki67 (c35/ki-67), CD3 (UCHT1), CD19 (HIB19), CD21 (B-ly4), CD24 (ML5), CD27 (M-T271), CD38 (HIT2), CD69 (H1.2F3), CD86 (2331/FUN-1), IgD (HI10a) and IgM (G20–127) from BD Biosciences.
2.4. Apoptosis assay
B cells were purified with magnetic separation with EasySep™ Human B Cell Isolation Kit (StemCell™ Technologies, Vancouver, Canada). The resultant post-sort purity was > 95%. Sorted B cells were plated at 100 000 cells/well in a 96 well-plate in RPMI 2% FBS for 8 hours, with DMSO 5%, positive control) or left untreated. Classical surface staining, to identify transitional B cells, and annexin V labeling (BD Bioscience), following the manufacturer’s recommendations, were performed. Cells were resuspended in a PBS solution containing DAPI and analyzed on a Gallios flow cytometer (Beckman Coulter).
2.5. B-cell stimulation
B cells from SLE patients without hydroxychloroquine treatment were purified with magnetic separation with EasySep™ Human B Cell Isolation Kit (StemCell™ Technologies). The resultant post-sort purity was >95%. Sorted B cells were plated at 100 000 cells/well in a 96 well-plate in RPMI 10% FBS and gentamycine 10µ g/ml with 2µ g/ml class B CpG (TLR9 agonist, ODN 2006, InvivoGen, San Diego, CA, USA), 1µ g/ml Gardiquimod (TLR7 agonists, InvivoGen) or 1µ g/ml CD40 ligand (R&D System®, Mineapolis, MN, USA). Expression of surface activation markers was analyzed after 48 hours with Gallios flow cytometer (Beckman Coulter).
2.6. Quantitative real-time RT-PCR analysis
For blood samples, B cells were purified, from PBMC of HDs and of SLE patients, with magnetic separation with EasySep™ Human B Cell Isolation Kit (StemCell™ Technologies). Then, transitional B cells (CD19+CD27−IgM+IgD+CD24++CD38++) and mature naïve (CD19+CD27−IgM+IgD+CD24+CD38+) populations were sorted (FACSAria II, BD Biosciences). For BM samples, pre/proB (CD19+CD24++CD38++IgM−IgD−), immature (CD19+CD24++CD38++IgM+IgD−) and early transitional (CD19+CD24++CD38++IgM+IgD+) populations were sorted. Cell viability was assessed with DAPI (Sigma-Aldrich) and the resultant post-sort purity was > 95%. RNA was isolated using RNeasy Microkit (Qiagen, Valencia, CA, USA) following the manufacturer’s protocol, including a DNase digestion. Reverse transcription was performed on a T100™ Thermal cycler (BioRad) using High Capacity cDNA Reverse transcription Kit (Applied Biosystems, Foster City, CA) following the manufacturer’s protocol. 5ng of cDNA were PCR-preamplified for 14 cycles using TaqMan® PreAmp Master Mix Kit (Applied Biosystems). Quantitative real-time PCR reactions were performed on a StepOnePlus realtime thermal cycler (Applied Biosystems) using the TaqMan® Gene Expression Master Mix (Applied Biosystems) following the manufacturer’s protocol. Assays were plated in triplicate. Relative expression levels were calculated with the comparative Ct method using the mean of the Ct between GAPDH and HPRT1 housekeeping gene for normalization. As defined by Rice et al. [22], the median fold change of the 6 interferon-stimulated genes IFI27, IFI44L, IFIT1, ISG15, RSAD2, SIGLEC1 were used to create an interferon score for each individual. The following list of TaqMan® probes (Applied Biosystems) was used: GAPDH (Hs99999905_m1), HERC5 (Hs00180943_m1), HERC6 (Hs00215555_m1), HPRT1 (Hs01003267_m1), IFI27 (Hs01086370_m1), IFI44L (Hs00199115_m1), IFIT1 (Hs01675197_m1), ISG15 (Hs00192713_m1), RSAD2 (Hs01057264_m1) and SIGLEC1 (Hs00988063_m1).
2.7. Whole transcriptome sequencing (RNA-seq)
B cells were purified, from PBMC of quiescent (n=3) and active (n=3) SLE patients and from PBMC of healthy donors (n=6), with magnetic separation with EasySep™ Human B Cell Isolation Kit (StemCell™ Technologies). Then, transitional B cells (CD3−CD19+CD27−IgM+IgD+CD24++CD38++) populations were sorted (FACSAria II, BD Biosciences). Cell viability was assessed with DAPI (Sigma-Aldrich) and the resultant post-sort purity was >95%. RNA was isolated using RNeasy Microkit (Qiagen) following the manufacturer’s protocol, including a DNase digest. Sample quality was assessed using Agilent 2100 bioanalyzer (Agilent Technologies, Palo Alto, CA, USA). RNA-seq libraries preparation and sequencing were performed in the GenomEast platform (Strasbourg, France). Full length cDNA was generated from 1 ng of total RNA using Clontech SMART-Seq v4 Ultra Low Input RNA kit for Sequencing (Takara Bio Europe, Saint Germain en Laye, France) according to ma’ufacturer’s instructions, with 12 cycles of PCR for cDNA amplification by Seq-Amp polymerase. 600 pg of pre-amplified cDNA were then used as input for Tn5 transposon tagmentation using the Nextera XT DNA Library Preparation Kit (Illumina, San Diego, CA) followed by 12 cycles of library amplification. Following purification with Agencourt AMPure XP beads (Beckman-Coulter), the size and concentration of libraries were assessed by capillary electrophoresis. Sequencing was performed on an Illumina HiSeq 4000 with 50 bp single-end reads. Image analysis and base calling were performed using RTA 2.7.3 and bcl2fastq 2.17.1.14. Adapters and low-quality sequences (Phred score < 20) were removed using cutadapt v1.10. After this preprocessing, reads shorter than 40 bases and reads mapping to rRNA sequences were discarded for further analysis. Reads were mapped onto the hg38 assembly of the human genome using STAR v2.5.3a [23]. Gene expression was quantified using HTSeq v0.6.1p1 [24] and gene annotations from Ensembl release 91. Gene expression comparisons between quiescent SLE patients and HDs, active SLE patients and HDs and active versus quiescent SLE patients were performed using the method proposed by Love et al. [25] implemented in the DESeq2 Bioconductor library (v1.16.1). Adjustment for multiple testing was performed with the Benjamini and Hochberg method [26]. Results were interpreted using Ingenuity Pathway Analysis 2.3 (Qiagen) and the second generation of the modular transcriptional framework developed by Chaussabel and Baldwin [27]. Considering transcripts with a sum of normalized read count across all samples >150, a 2-fold change cut-off with an adjusted p-value<0.01 was set to identify differentially expressed genes. Variant calling was performed according to The GATK Best Practices workflow for variant calling on RNA-seq data [28]. Duplicate reads were marked using MarkDuplicates from Picard version 1.122. Further analyses were performed using GATK version 3.4–46: reads were split into exon segments and sequences overhanging intronic regions were hard-clipped using SplitNCigarReads, variant calling was then performed using HaplotypeCaller. Resulting variants were filtered according to GATK recommendations for RNA-seq data: clusters of at least 3 SNPs within a window of 35 bases were filtered and filtering based on Fisher Strand values (> 30) and Quality by Depth (< 2) values were also performed. Variants were annotated using GATK, SnpSift [29] and SnpEff [30]. ClinVar database (https://www.ncbi.nlm.nih.gov/clinvar/) was used to assess if variants involved in monogenic forms of SLE, primary immunodeficiencies with SLE-like features or interferonopathies were found (clinical significance value: “pathogenic” or “risk factor”, 28 genes checked).
2.8. Statistical analysis
Data were analyzed using GraphPad Prism version 7 (GraphPad software Inc, San Diego, CA, USA). Statistical significances were calculated with two-tailed unpaired Mann-Whitney U-test or two-tailed Spearman rank correlation test. All values are mean ±SEM. P values lower than 0.05 were considered statistically significant, or lower than 0.01 for RNA-seq.
3. RESULTS
3.1. Transitional B cell expansion in peripheral blood in SLE
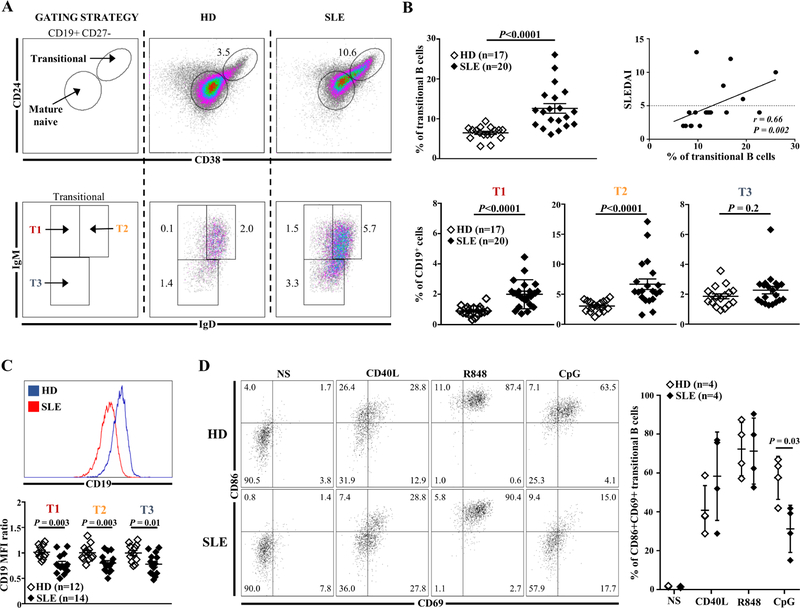
Transitional B cells represent a central developmental stage for B cells, reflecting ontogenesis in the BM before peripheral differentiation and selection into long-lived mature B cells. The classification of human transitional B cells into immature T1, intermediate T2 and T3 subsets is based on the expression of CD24, CD38, IgM, IgD markers in the CD19+CD27− lymphocyte gate, as described by Simons et al. and others (Fig. 1A) [17,31,32]. Immature T1 is the predominant subset in the BM while T2 is predominant in the peripheral blood [32]. We carried out detailed phenotypic analysis of these populations in the blood of 20 low treated or untreated SLE patients. All except one patient were Caucasian and their main clinical characteristics are presented in Supplementary Table 1. Most patients were female with at least 4 ACR criteria (see methods) but with different clinical and biological presentations of SLE. All patients were positive for anti-dsDNA antibodies. The frequency of transitional B cells (CD19+CD27−CD24++CD38++) was significantly higher in SLE patients than in healthy donors (HDs) (12.6%±1.2 versus 6.5%±0.3, p<0.0001) and correlated with the SLEDAI-2K score (r=0.66, p<0.01) (Fig. 1A, 1B). Transitional B cell increase in SLE patients was mostly the consequence of T1 and T2 subsets expansion (2.0±0.9% vs 0.9%±0.3 and 6.7%±3.9 vs 3.0%±0.9, respectively; p<0.0001) (Fig. 1B and Supplementary Fig. 1). Results were comparable considering either quiescent or active patients (Supplementary Fig. 2).
Fig. 1.
(A) Representative dot plots of transitional B cells and their subsets in one HD and one SLE patient. (B) Frequency of transitional B cells (top left panel) and their subsets (bottom panel) (% of CD19+) in HDs compared to SLE patients. Correlation between transitional B cells (% of CD19+) and disease activity assessed by the SLEDAI-2K score (top right panel). (C) MFI of CD19 in transitional B cells subsets from HDs and SLE patients. (D) Frequency of CD86+CD69+ transitional B cells from HDs or SLE patients after no stimulation (NS) or in vitro stimulation with CD40L (CD40 ligand), R848 (TLR7 agonist) or CpG (TLR9 agonist) for 2 days. HDs: healthy donors; MFI: mean of fluorescence intensity; SLE: Systemic lupus erythematosus.
3.2. Defect in transitional B cell responses to TLR9
Based on our previous recent results [10], we hypothesized that transitional B cells present a lower expression of CD19, concomitantly with an impaired TLR9 response that could influence their selection. As shown in Fig. 1C, SLE transitional T1, T2 and T3 B cells displayed a decrease in CD19 expression. This downregulation was present in transitional B cells from quiescent and active SLE patients (Supplementary Fig. 3). In addition, we further tested TLR9 responses in rare non-treated SLE patients to circumvent hydroxychloroquine interference [10]. We confirmed that the induction of CD86 and CD69 activation markers on transitional B cells was defective after TLR9 activation when compared to HDs (31.9%±5.5 vs 57.3%±6.1; p<0.05), while their responses after TLR7/TLR8 and CD40 stimulation were similar to HD counterparts (Fig. 1D).
3.3. IFN signature in SLE PB transitional B cells
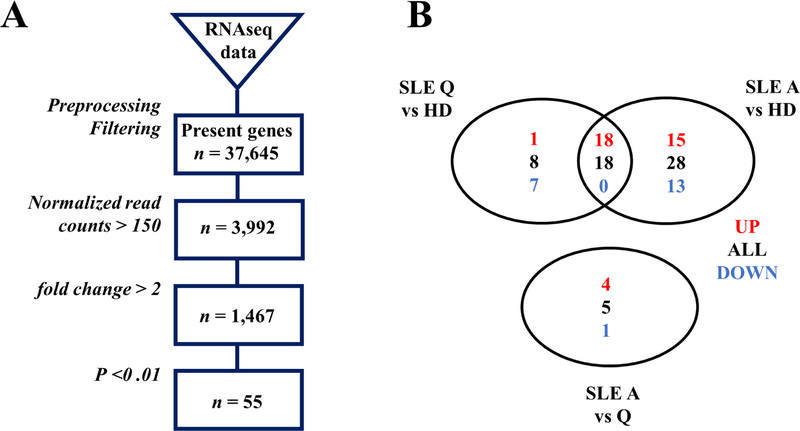
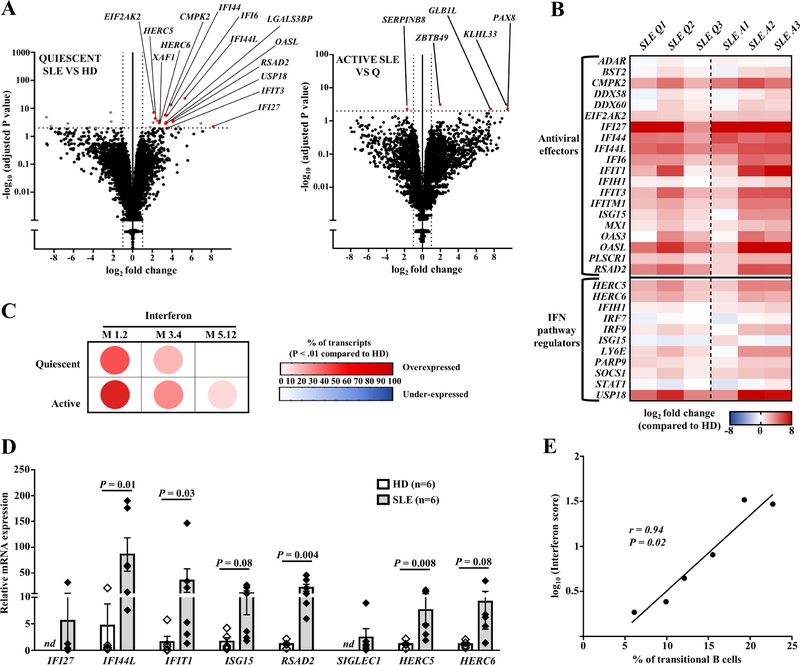
These data prompted us to explore if SLE transitional B cells could harbor a specific transcriptomic signature, to explain their increased number, their phenotype, and more generally a breakdown of tolerance at this stage. We performed RNA-sequencing analysis on sorted transitional B cells (CD19+CD27−CD24++CD38++) from 12 individuals including 3 SLE quiescent patients (mean SLEDAI-2K 3.3), 3 active patients (mean SLEDAI-2K 10) and 6 HDs (see Supplementary Table 1 for the main characteristics of these SLE patients). For transcriptomic analysis, only untreated patients (for the last six months) were included, as even hydroxychloroquine or corticosteroids can modify the lymphocyte transcriptomic profile [13]. All SLE patients selected for this analysis displayed a high percentage of transitional B cells and lower CD19 levels on these cells than HDs. Genes with a minimum of 2-fold change and an adjusted p-value<0.01 were considered as differentially expressed genes (see Materials and Methods section). Comparisons were performed between quiescent SLE patients versus HDs (19 upregulated genes, 7 downregulated genes), active SLE patients versus HDs (33 upregulated genes, 13 downregulated genes), and in active versus quiescent SLE patients (4 upregulated genes, 1 downregulated gene) (Fig. 2 and Supplementary Tables 2–4). No difference was detected between SLE and HD transitional B cells considering the expression of genes involved in TLR7 or TLR9 signaling, of CD19, CD21 and PAX5 (which encodes a regulator of CD19), or of genes encoding molecules involved in BCR signaling cascade, namely PI3K, BTK, or AKT. These results were consistent with previous q-RT PCR analysis of sorted naive SLE B cells [10]. This approach identified interferon stimulated genes (ISGs) as the most overexpressed genes in SLE patients compared to controls, i.e. 14 and 25 ISGs in sorted transitional B cells from quiescent and active SLE patients compared to HDs, respectively (Fig. 3A–B). Results were analyzed using Ingenuity Pathway Analysis, and the modular transcriptional repertoire developed by Chaussabel et al. [27], which describes 260 modules of genes sets in blood in various immunological conditions. The three IFN-related modules (M 1.2, M 3.4, and M 5.12) driven by type I IFN but also type II IFN (for M 3.4 and M 5.12), appeared up-regulated in SLE transitional B cells (Fig. 3C). IFN signature was already present in quiescent patients and higher in active patients. The analysis of RNA sequences did not highlight deleterious mutations in genes (SNPs and indels called as described in Materials and Methods section) related to monogenic interferonopathies or to IFN pathway (32 genes, including TREX1, TMEM173, RIG-1, CECR1, RNASEH2A, RNASEH2B, RNASEH2C) [22,33]. Genes coding for IFN were not over-expressed, neither genes coding for IFN receptors compared to controls. We did not detect significant transcriptomic signatures for other cytokines.
Fig. 2.
(A) Flow chart of RNA Sequencing analysis. (B) Venn Diagram representing genes significantly up (in red) or down-regulated (in blue) between quiescent SLE patients, active SLE patients and HDs. A: active; HDs: healthy donors; Q: quiescent; SLE: systemic lupus erythematosus.
Fig. 3.
(A) Volcano plot representation of RNA-Seq data comparing transitional B cells from HDs (n=6) versus untreated quiescent SLE patients (n=3) (left panel) and transitional B cells from untreated quiescent versus active patients (n=3) (right panel). (B) Hierarchical representation of interferon stimulated genes (ISGs) expression in transitional B cells from each patient compared to HDs. (C) Modular analysis of RNA-Seq data representing the % of transcripts over/under-expressed (P<0.01) in untreated SLE patients (quiescent, n=3; active, n=3) compared to HDs (n=6). (D) qRT-PCR of ISGs in sorted transitional B cells of quiescent SLE patients compared to HDs. (E) Correlation between transitional B cells (% of CD19+ cells) and the interferon score. HDs: healthy donors; ND: not detected; SLE: Systemic lupus erythematosus.
IFN signature in whole PBMCs from SLE patients has been previously associated to disease activity [12,13]. To confirm that IFN signature was reproducible in the transitional B cells of quiescent SLE patients (i.e. out of flares), we tested on sorted cells from additional quiescent SLE patients (n=6, mean SLEDAI-2K 3.3; see Supplementary Table 1 for the main characteristics of these SLE patients) the relative expression of eight ISGs, some of which (namely IFI27, IFI44L, IFIT1, ISG15, RSAD2, SIGLEC1) were previously used to determine an interferon score in inflammatory diseases [20]. HERC5 and HERC6 genes were added as they encode positive regulators of IFN pathways [34] and were found over-expressed in our RNA-Seq analysis. Transcripts coding for all these genes were very significantly amplified by RT-qPCR in sorted transitional B cells from quiescent SLE patients compared to HDs (Fig. 3D). Of note, the IFN-score correlated with the frequency of transitional B cells (r = 0.94, p<0.01) (Fig. 3E).
3.4. IFN signature in BM B cells
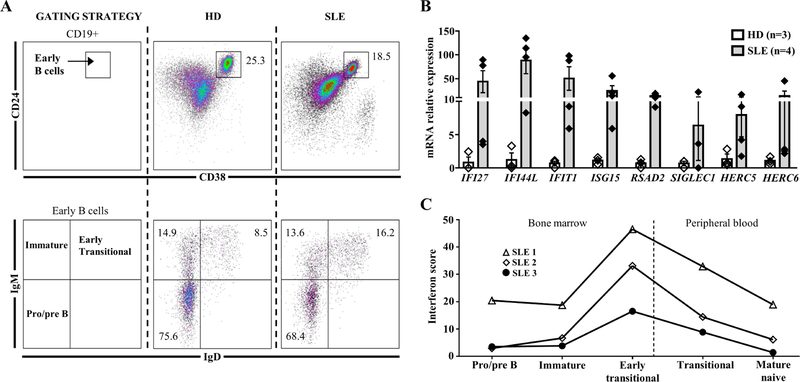
The IFN signature in transitional B cells from SLE patients could be the consequence of higher levels of IFN in the peripheral blood or affecting the early transitional stage in the BM [35]. To assess this point, and despite the rarity of the samples, the IFN score of sorted BM B cells was compared to sorted peripheral B cells subsets in the same SLE patients (Fig. 4A). BM early transitional B cells displayed the highest IFN score, followed by a progressive decrease in the peripheral subsets, revealing a B cell IFN burden of central origin (Fig. 4B–C).
Fig. 4.
(A) Gating strategy for BM analysis of pre/proB (CD19+CD24++CD38++IgM−IgD−), immature (CD19+CD24++CD38++IgM+IgD−) and early transitional B cells (CD19+CD24++CD38++IgM+IgD+). (B) qRT-PCR of eight interferon stimulated genes in sorted early transitional B cells from BM of SLE patients compared to control. (C) Representation of the interferon score in BM and peripheral B cell subpopulations in three SLE patients. BM: Bone marrow; ND: not detected; SLE: Systemic lupus erythematosus.
3.5. Activation and proliferation status
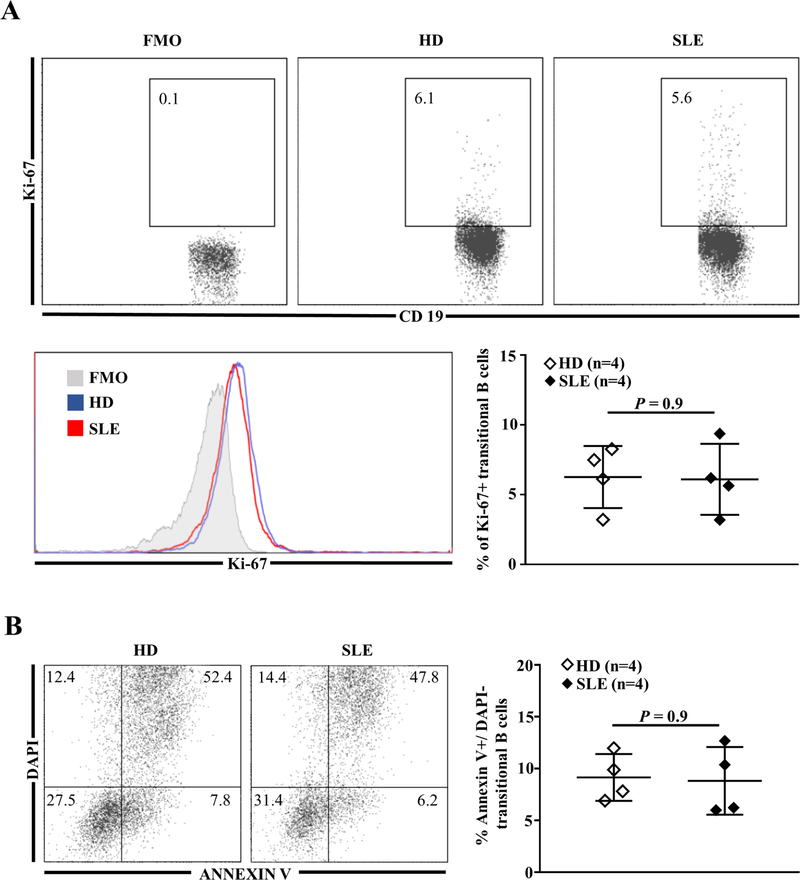
Finally, RNA-sequencing analysis of peripheral transitional B cells showed an over-expression of genes related to the regulation of cell-cycle and coding for pro- or anti-apoptotic proteins (Supplementary Tables 2–4). Per se transitional B cell activation could not explain such gene regulation as the frequency of CD86+ B cells and the intensity of HLA-DR expression in SLE patients were comparable to that of HDs (Supplementary Fig. 4). We tested the hypothesis that SLE transitional B cells proliferate more or die less than HDs transitional B cells. However, flow cytometric analysis did not highlight any difference in proliferation (Fig. 5A) or early apoptosis in freshly isolated transitional SLE B cells (Fig. 5B), arguing for an accelerate differentiation from immature B cells to transitional stage in SLE patients.
Fig. 5.
(A) Representative dot plots of Ki-67+ cells (top panel) and Ki-67 MFI (bottom left panel) in gated transitional B cells from one HD and one SLE patient; frequency of Ki-67+ transitional B cells from HDs and SLE patients (bottom right panel). (B) Frequency of early apoptotic (DAPI− AnnexinV+) transitional B cells in HDs and SLE patients. FMO: Fluorescence Minus One; HDs: healthy donors; MFI: mean of fluorescence intensity; SLE: systemic lupus erythematosus.
4. DISCUSSION
Organ damage in SLE is driven by immune complexes deposition and anti-nuclear antibodies [2]. Hence, lots of work to elucidate SLE pathogenesis focused on the behavior of autoreactive B cells to explain how pathogenic B cells producing high affinity auto-antibodies can arise. Autoreactive B cells are submitted to stringent selection in the BM, via editing and deletion [36–39], and during B cell differentiation their percentage dropped significantly in HDs between the transitional stage and the mature naive stage [6]. However, a percentage of ignorant or anergic autoreactive B cells remain and they will be ultimately control in the periphery [40].
Herein, we describe that transitional B cells in SLE patients displayed, as naive B cells [10], an impairment in TLR9 response, and low levels of CD19 concerning all 3 subsets. TLR9 deficiency in lupus-prone mice leads to disease worsening, and Sindhava et al. showed in humans the crucial role of TLR9 in controlling the survival of autoreactive naive B cells when co-engaged with BCR [11]. CD19 downregulation could participate for diminished TLR9 response in early SLE B cells [9,10]. It may also favor per se abnormalities in selection at the BM level, considering autoreactive B cell fate in CD19 deficient patients [41], in CD19 deficient transgenic cell lines [42] or in mice [43]. Thus, the defect in transitional B cells of two closely inter-related molecules (i.e. CD19 and TLR9) may favor the B-cell tolerance breakdown described in SLE at this stage.
We detected an IFN signature in sorted transitional B cells from both peripheral blood and BM. Domeier et al. recently reported in mice, that type I IFN maintains BCR signaling beyond the threshold required for effective tolerance, driving autoreactive B cell development into the germinal center (GC) pathway [44]. Since IFN influence appears in the BM at the early transitional stage, it could affect the BCR signaling threshold also at the medullary B-cell stages and not only in the periphery.
IFN signature and abnormalities in CD19/TLR9 signaling could have different causes. CD19/TLR9 defects, are undoubtedly present in quiescent SLE patients. Munroe et al. were able to examine type I and type II interferon activities, autoantibodies and cytokines levels in samples collected before and after diagnosis of SLE and it appears that IFN activities precede the disease [45]. Studies in ANA+ healthy subjects or in other autoimmune conditions may answer about the causal/relationship of our findings in SLE and in autoantibody mediated diseases. For example, transitional B cells are expanded in Sjögren syndrome [17], although the expression of CD19 remains normal [10]. Thus, investigations in other conditions and in prospective longitudinal cohorts could help to precise if the frequency of transitional B cells is a reflection of IFN burden without real disease specificity and whether or not it can predict the occurrence of some of them. We confirmed that IFN imprintment in SLE is not confined to African American patients, especially to those with nephritis and anti-Ro antibodies [15] but also concerns Caucasian patients with anti-dsDNA. Transitional B cells from African American SLE patients displayed an autocrine production of IFN-β [15]. However, we did not detect overexpression of genes coding for type I or type II IFN in our cohort, suggesting an extrinsic influence in Caucasian patients, i.e. IFN secretion by other cells. This is consistent with the low transcription of genes coding for type I IFN in another group of European American SLE patients compared to African American SLE in sorted transitional B cells [15]. In this view, Palanichamy et al. described in SLE a significant production of IFN-α and β by resident BM cells, associated with a reduction in the fraction of precursor B cells and a T1 and T2 B cell subsets expansion [35]. The correlation of the IFN score with the percentage of transitional B cells, together with the normal status of peripheral transitional B cells in terms of activation, proliferation and survival, argue for the influence of IFN on early BM differentiation. We cannot rule out that for some reasons, transitional B cells in SLE are more sensitive to IFN although transcriptional levels of IFN receptors did not differ in our analysis.
In our previous work, we correlated the CD19 low expression on B cells to the presence of anti-dsDNA antibodies, suggesting that apoptotic bodies and immune complexes containing DNA participate to CD19 and TLR9 regulation in B cells in the BM. Immune complexes could also trigger IFN production via TLR7, or directly via DNA-sensors in myeloid cells [46], and favor the output of transitional B cells, interfering with the elimination of autoreactive B cells at this stage [47]. Interestingly, TLR7 can escape X chromosome inactivation in immune cells and its expression is higher after puberty in female and correlates with sex hormones levels [48]. However, we did not observe significant differences in responses of SLE transitional B cells to TLR7/TLR8 agonist compared to controls, neither differences in TLR7/TLR9 transcription in transitional B cells via RNA sequencing analysis. It would be however of great interest to focus on this point.
SLE differs from one patient to another, depending of genetic factors and environment. The idea that central tolerance and early escape of autoreactive B cells participate to the genesis of SLE has been harmed by elegant studies demonstrating the extra-follicular activation of autoreactive B cells and of plasmocytes in periphery [5]. In view of our results, early B cells are abnormal likely since the BM, considering frequency or signaling, in many SLE patients even out of flares. Thus, both early and late mechanisms could interfere with survival and activation of autoreactive B cells in SLE patients and further analysis should clarify at what point it is in a personalized way.
Supplementary Material
HIGHLIGHTS.
SLE transitional B cells display CD19 downregulation and impaired TLR9 response
IFN signature exists since the B-cell medullary stage, even out of flares
IFN signature in transitional B cells is not confined to African American SLE
IFN score correlates with the increase in peripheral transitional B cells
ACKNOWLEDGMENTS
We thank C. Herouard-Molina (IGBMC, Illkirch, France) for expert technical assistance for the whole transcriptome sequencing. Sequencing was performed by the GenomEast platform, a member of the “France Génomique” consortium (ANR-10-INBS-0009). We thank Dr L. Chiche (Department of Internal Medicine, Hôpital Européen, Marseille, France) and Dr R. Carapito (Genomax Plateform, INSERM 1109, Université de Strasbourg, Strasbourg, France) for assistance with the RNA-Seq analysis.
A.K.B. has been supported by a National Institutes of Health Clinical and Translational Science Award (5UL1TR000042–10) Trainee Pilot and a Center for Musculoskeletal Research Training Grant (2 T32AR053459).
J.L.B. has been supported by a Rheumatology Research Foundation (RRF) Scientist Development Award.
A.S.K. is supported by grants from the French Ministry of Health (PHRC IR 2011), from the Société Nationale Française de Médecine Interne (SNFMI), from the Hôpitaux Universitaires de Strasbourg (HUS) and from the EU-funded (ERDF) project INTERREG V “RARENET”.
J.H.A. is supported by AI563262, AI078907, AR071670, the NIAMS Accelerated Medicines Partnership (1UH2AR067690), and the Bertha and Louis Weinstein research fund.
Footnotes
CONFLICT-OF-INTEREST DISCLOSURE
The authors declare no competing financial interests.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- [1].Arbuckle MR, McClain MT, Rubertone MV, Scofield RH, Dennis GJ, James JA, Harley JB, Development of autoantibodies before the clinical onset of systemic lupus erythematosus, N. Engl. J. Med 349 (2003) 1526–1533. doi: 10.1056/NEJMoa021933. [DOI] [PubMed] [Google Scholar]
- [2].Tsokos GC, Lo MS, Reis P. Costa, Sullivan KE, New insights into the immunopathogenesis of systemic lupus erythematosus, Nat. Rev. Rheumatol 12 (2016) 716–730. doi: 10.1038/nrrheum.2016.186. [DOI] [PubMed] [Google Scholar]
- [3].Tsokos GC, A new checkpoint in lupus, J. Allergy Clin. Immunol 143 (2019) 1351–1352. doi: 10.1016/j.jaci.2018.12.996. [DOI] [PubMed] [Google Scholar]
- [4].Yurasov S, Wardemann H, Hammersen J, Tsuiji M, Meffre E, Pascual V, Nussenzweig MC, Defective B cell tolerance checkpoints in systemic lupus erythematosus, J. Exp. Med 201 (2005) 703–711. doi: 10.1084/jem.20042251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Suurmond J, Atisha-Fregoso Y, Marasco E, Barlev AN, Ahmed N, Calderon SA, Wong MY, Mackay MC, Aranow C, Diamond B, Loss of an IgG plasma cell checkpoint in patients with lupus, J. Allergy Clin. Immunol 143 (2019) 1586–1597. doi: 10.1016/j.jaci.2018.10.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Wardemann H, Yurasov S, Schaefer A, Young JW, Meffre E, Nussenzweig MC, Predominant autoantibody production by early human B cell precursors, Science 301 (2003) 1374–1377. doi: 10.1126/science.1086907. [DOI] [PubMed] [Google Scholar]
- [7].Li H, Jiang Y, Prak EL, Radic M, Weigert M, Editors and editing of anti-DNA receptors, Immunity 15 (2001) 947–957. [DOI] [PubMed] [Google Scholar]
- [8].Goodnow CC, Sprent J, de St Groth B. Fazekas, Vinuesa CG, Cellular and genetic mechanisms of self tolerance and autoimmunity, Nature 435 (2005) 590–597. doi: 10.1038/nature03724. [DOI] [PubMed] [Google Scholar]
- [9].Morbach H, Schickel J-N, Cunningham-Rundles C, Conley ME, Reisli I, Franco JL, Meffre E, CD19 controls Toll-like receptor 9 responses in human B cells, J. Allergy Clin. Immunol 137 (2016) 889–898. 10.1016/j.jaci.2015.08.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Gies V, Schickel J-N, Jung S, Joublin A, Glauzy S, Knapp A-M, Soley A, Poindron V, Guffroy A, Choi J-Y, Gottenberg J-E, Anolik JH, Martin T, Soulas-Sprauel P, Meffre E, Korganow A-S, Impaired TLR9 responses in B cells from patients with systemic lupus erythematosus, JCI Insight 3 (2018). doi: 10.1172/jci.insight.96795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Sindhava VJ, Oropallo MA, Moody K, Naradikian M, Higdon LE, Zhou L, Myles A, Green N, Nündel K, Stohl W, Schmidt AM, Cao W, Dorta-Estremera S, Kambayashi T, Marshak-Rothstein A, Cancro MP, A TLR9-dependent checkpoint governs B cell responses to DNA-containing antigens, J. Clin. Invest 127 (2017) 1651–1663. doi: 10.1172/JCI89931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Chiche L, Jourde-Chiche N, Whalen E, Presnell S, Gersuk V, Dang K, Anguiano E, Quinn C, Burtey S, Berland Y, Kaplanski G, Harle J-R, Pascual V, Chaussabel D, Modular transcriptional repertoire analyses of adults with systemic lupus erythematosus reveal distinct type I and type II interferon signatures, Arthritis Rheumatol. Hoboken NJ 66 (2014) 1583–1595. doi: 10.1002/art.38628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Banchereau R, Hong S, Cantarel B, Baldwin N, Baisch J, Edens M, Cepika A-M, Acs P, Turner J, Anguiano E, Vinod P, Kahn S, Obermoser G, Blankenship D, Wakeland E, Nassi L, Gotte A, Punaro M, Liu Y-J, Banchereau J, Rossello-Urgell J, Wright T, Pascual V, Personalized Immunomonitoring Uncovers Molecular Networks that Stratify Lupus Patients, Cell 165 (2016) 551–565. doi: 10.1016/j.cell.2016.03.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Liu M, Guo Q, Wu C, Sterlin D, Goswami S, Zhang Y, Li T, Bao C, Shen N, Fu Q, Zhang X, Type I interferons promote the survival and proinflammatory properties of transitional B cells in systemic lupus erythematosus patients, Cell. Mol. Immunol (2018). doi: 10.1038/s41423-018-0010-6. [DOI] [PMC free article] [PubMed]
- [15].Hamilton JA, Wu Q, Yang P, Luo B, Liu S, Li J, Mattheyses AL, Sanz I, Chatham WW, Hsu H-C, Mountz JD, Cutting Edge: Intracellular IFN-β and Distinct Type I IFN Expression Patterns in Circulating Systemic Lupus Erythematosus B Cells, J. Immunol (2018) 10.4049/jimmunol.1800791. [DOI] [PMC free article] [PubMed]
- [16].Sims GP, Ettinger R, Shirota Y, Yarboro CH, Illei GG, Lipsky PE, Identification and characterization of circulating human transitional B cells, Blood 105 (2005) 4390–4398. doi: 10.1182/blood-2004-11-4284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Simon Q, Pers J-O, Cornec D, Pottier LL, Mageed RA, Hillion S, In-depth characterization of CD24highCD38high transitional human B cells reveals different regulatory profiles, J. Allergy Clin. Immunol 137 (2016) 1577–1584. 10.1016/j.jaci.2015.09.014. [DOI] [PubMed] [Google Scholar]
- [18].Tan EM, Cohen AS, Fries JF, Masi AT, McShane DJ, Rothfield NF, Schaller JG, Talal N, Winchester RJ, The 1982 revised criteria for the classification of systemic lupus erythematosus, Arthritis Rheum 25 (1982) 1271–1277. [DOI] [PubMed] [Google Scholar]
- [19].Gladman DD, Ibañez D, Urowitz MB, Systemic lupus erythematosus disease activity index 2000, J. Rheumatol 29 (2002) 288–291. [PubMed] [Google Scholar]
- [20].Franklyn K, Lau CS, Navarra SV, Louthrenoo W, Lateef A, Hamijoyo L, Wahono CS, Chen SL, Jin O, Morton S, Hoi A, Huq M, Nikpour M, Morand EF, Asia-Pacific Lupus Collaboration, Definition and initial validation of a Lupus Low Disease Activity State (LLDAS), Ann. Rheum. Dis 75 (2016) 1615–1621. doi: 10.1136/annrheumdis-2015-207726. [DOI] [PubMed] [Google Scholar]
- [21].Tung JW, Heydari K, Tirouvanziam R, Sahaf B, Parks DR, Herzenberg LA, Herzenberg LA, Modern flow cytometry: a practical approach, Clin. Lab. Med 27 (2007) 453–468, 10.1016/j.cll.2007.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Rice GI, Melki I, Frémond M-L, Briggs TA, Rodero MP, Kitabayashi N, Oojageer A, Bader-Meunier B, Belot A, Bodemer C, Quartier P, Crow YJ, Assessment of Type I Interferon Signaling in Pediatric Inflammatory Disease, J. Clin. Immunol 37 (2017) 123–132. doi: 10.1007/s10875-016-0359-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Dobin A, Davis CA, Schlesinger F, Drenkow J, Zaleski C, Jha S, Batut P, Chaisson M, Gingeras TR, STAR: ultrafast universal RNA-seq aligner, Bioinforma. Oxf. Engl 29 (2013) 15–21. doi: 10.1093/bioinformatics/bts635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Anders S, Pyl PT, Huber W, HTSeq--a Python framework to work with high-throughput sequencing data, Bioinforma. Oxf. Engl 31 (2015) 166–169. doi: 10.1093/bioinformatics/btu638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Love MI, Huber W, Anders S, Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2, Genome Biol 15 (2014) 550. doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Benjamini Y, Hochberg Y, Controlling the False Discovery Rate: A Practical and Powerful Approach to Multiple Testing, J. R. Stat. Soc. Ser. B Methodol 57 (1995) 289–300. [Google Scholar]
- [27].Chaussabel D, Baldwin N, Democratizing systems immunology with modular transcriptional repertoire analyses, Nat. Rev. Immunol 14 (2014) 271–280. doi: 10.1038/nri3642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].McKenna A, Hanna M, Banks E, Sivachenko A, Cibulskis K, Kernytsky A, Garimella K, Altshuler D, Gabriel S, Daly M, DePristo MA, The Genome Analysis Toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data, Genome Res 20 (2010) 1297–1303. doi: 10.1101/gr.107524.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Cingolani P, Patel VM, Coon M, Nguyen T, Land SJ, Ruden DM, Lu X, Using Drosophila melanogaster as a Model for Genotoxic Chemical Mutational Studies with a New Program, SnpSift, Front. Genet 3 (2012) 35. doi: 10.3389/fgene.2012.00035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Cingolani P, Platts A, Wang LL, Coon M, Nguyen T, Wang L, Land SJ, Lu X, Ruden DM, A program for annotating and predicting the effects of single nucleotide polymorphisms, SnpEff: SNPs in the genome of Drosophila melanogaster strain w1118; iso-2; iso-3, Fly (Austin) 6 (2012) 80–92. doi: 10.4161/fly.19695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Taher TE, Ong VH, Bystrom J, Hillion S, Simon Q, Denton CP, Pers J-O, Abraham DJ, Mageed RA, Association of Defective Regulation of Autoreactive Interleukin-6–Producing Transitional B Lymphocytes With Disease in Patients With Systemic Sclerosis, Arthritis Rheumatol 70 (2018) 450–461. doi: 10.1002/art.40390. [DOI] [PubMed] [Google Scholar]
- [32].Palanichamy A, Barnard J, Zheng B, Owen T, Quach T, Wei C, Looney RJ, Sanz I, Anolik JH, Novel human transitional B cell populations revealed by B cell depletion therapy, J. Immunol. Baltim. Md 1950 182 (2009) 5982–5993. doi: 10.4049/jimmunol.0801859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Rodero MP, Crow YJ, Type I interferon-mediated monogenic autoinflammation: The type I interferonopathies, a conceptual overview, J. Exp. Med 213 (2016) 2527–2538. doi: 10.1084/jem.20161596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Schneider WM, Chevillotte MD, Rice CM, Interferon-stimulated genes: a complex web of host defenses, Annu. Rev. Immunol 32 (2014) 513–545. doi: 10.1146/annurev-immunol-032713-120231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Palanichamy A, Bauer JW, Yalavarthi S, Meednu N, Barnard J, Owen T, Cistrone C, Bird A, Rabinovich A, Nevarez S, Knight JS, Dedrick R, Rosenberg A, Wei C, Rangel-Moreno J, Liesveld J, Sanz I, Baechler E, Kaplan MJ, Anolik JH, Neutrophil-mediated IFN activation in the bone marrow alters B cell development in human and murine systemic lupus erythematosus, J. Immunol. Baltim. Md 1950 192 (2014) 906–918. doi: 10.4049/jimmunol.1302112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Goodnow CC, Cyster JG, Hartley SB, Bell SE, Cooke MP, Healy JI, Akkaraju S, Rathmell JC, Pogue SL, Shokat KP, Self-tolerance checkpoints in B lymphocyte development, Adv. Immunol 59 (1995) 279–368. [DOI] [PubMed] [Google Scholar]
- [37].Shlomchik MJ, Sites and stages of autoreactive B cell activation and regulation, Immunity 28 (2008) 18–28. doi: 10.1016/j.immuni.2007.12.004. [DOI] [PubMed] [Google Scholar]
- [38].Ait-Azzouzene D, Verkoczy L, Peters J, Gavin A, Skog P, Vela JL, Nemazee D, An immunoglobulin Cκ-reactive single chain antibody fusion protein induces tolerance through receptor editing in a normal polyclonal immune system, J. Exp. Med 201 (2005) 817–828. doi: 10.1084/jem.20041854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Lang J, Ota T, Kelly M, Strauch P, Freed BM, Torres RM, Nemazee D, Pelanda R, Receptor editing and genetic variability in human autoreactive B cells, J. Exp. Med 213 (2016) 93–108. doi: 10.1084/jem.20151039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Goodnow CC, Vinuesa CG, Randall KL, Mackay F, Brink R, Control systems and decision making for antibody production, Nat. Immunol 11 (2010) 681–688. doi: 10.1038/ni.1900. [DOI] [PubMed] [Google Scholar]
- [41].van Zelm MC, Bartol SJW, Driessen GJ, Mascart F, Reisli I, Franco JL, Wolska-Kusnierz B, Kanegane H, Boon L, van Dongen JJM, van der Burg M, Human CD19 and CD40L deficiencies impair antibody selection and differentially affect somatic hypermutation, J. Allergy Clin. Immunol 134 (2014) 135–144. 10.1016/j.jaci.2013.11.015. [DOI] [PubMed] [Google Scholar]
- [42].von Muenchow L, Engdahl C, Karjalainen K, Rolink AG, The selection of mature B cells is critically dependent on the expression level of the co-receptor CD19, Immunol. Lett 160 (2014) 113–119. doi: 10.1016/j.imlet.2014.01.011. [DOI] [PubMed] [Google Scholar]
- [43].Shivtiel S, Leider N, Sadeh O, Kraiem Z, Melamed D, Impaired light chain allelic exclusion and lack of positive selection in immature B cells expressing incompetent receptor deficient of CD19, J. Immunol. Baltim. Md 1950 168 (2002) 5596–5604. [DOI] [PubMed] [Google Scholar]
- [44].Domeier PP, Chodisetti SB, Schell SL, Kawasawa YI, Fasnacht MJ, Soni C, Rahman ZSM, B-Cell-Intrinsic Type 1 Interferon Signaling Is Crucial for Loss of Tolerance and the Development of Autoreactive B Cells, Cell Rep 24 (2018) 406–418. doi: 10.1016/j.celrep.2018.06.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Munroe ME, Lu R, Zhao YD, Fife DA, Robertson JM, Guthridge JM, Niewold TB, Tsokos GC, Keith MP, Harley JB, James JA, Altered type II interferon precedes autoantibody accrual and elevated type I interferon activity prior to systemic lupus erythematosus classification, Ann. Rheum. Dis 75 (2016) 2014–2021. doi: 10.1136/annrheumdis-2015-208140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].Kato Y, Park J, Takamatsu H, Konaka H, Aoki W, Aburaya S, Ueda M, Nishide M, Koyama S, Hayama Y, Kinehara Y, Hirano T, Shima Y, Narazaki M, Kumanogoh A, Apoptosis-derived membrane vesicles drive the cGAS-STING pathway and enhance type I IFN production in systemic lupus erythematosus, Ann. Rheum. Dis 77 (2018) 1507–1515. doi: 10.1136/annrheumdis-2018-212988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].Giltiay NV, Chappell CP, Sun X, Kolhatkar N, Teal TH, Wiedeman AE, Kim J, Tanaka L, Buechler MB, Hamerman JA, Imanishi-Kari T, Clark EA, Elkon KB, Overexpression of TLR7 promotes cell-intrinsic expansion and autoantibody production by transitional T1 B cells, J. Exp. Med 210 (2013) 2773–2789. doi: 10.1084/jem.20122798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [48].Souyris M, Cenac C, Azar P, Daviaud D, Canivet A, Grunenwald S, Pienkowski C, Chaumeil J, Mejía JE, Guéry J-C, TLR7 escapes X chromosome inactivation in immune cells, Sci. Immunol 3 (2018) 10.1126/sciimmunol.aap8855. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.