Abstract
Background
Long noncoding RNAs (lncRNAs) have been identified as regulators of a number of developmental and tumorigenic processes. However, the functions of most lncRNAs in glioma remain unknown and the mechanisms governing the proliferation of tumor cells remain poorly defined.
Methods
Both in vitro and in vivo assays were performed to investigate the roles of lncRNAs in the pathophysiology of gliomas. lncRNA arrays were used to identify differentially expressed lncRNAs. Subcutaneous tumor formation and a brain orthotopic tumor model in nude mice were used to investigate the functions of lncRNAs in vivo. The in vitro functions of lncRNAs were analyzed by fluorescence-activated cell sorting, colony formation, and western blot analyses. RNA fluorescence in situ hybridization and immunoprecipitation were used to explore the underlying mechanisms.
Findings
Here, we describe the newly discovered noncoding RNA RP11-732M18.3, which is highly overexpressed in glioma cells and interacts with 14-3-3β/α to promote glioma growth, acting as an oncogene. Overexpression of lncRNA RP11-732 M18.3 was associated with the proliferation of glioma cells and tumor growth in vitro and in vivo. Remarkably, lncRNA RP11-732M18.3 promoted cell proliferation and G1/S cell cycle transition. lncRNA RP11-732M18.3 is predominately localized in the cytoplasm. Mechanistically, the interaction of lncRNA RP11-732M18.3 with 14-3-3β/α increases the degradation of the p21 protein. lncRNA RP11-732M18.3 promoted the recruitment of ubiquitin-conjugating enzyme E2 E1 to 14-3-3β/α and the binding of 14-3-3β/α with ubiquitin-conjugating enzyme E2 E1 (UBE2E1) promoted the degradation of p21.
Interpretation
Overall these data demonstrated that lncRNA RP11-732M18.3 regulates glioma growth through a newly described lncRNA-protein interaction mechanism. The inhibition of lncRNA RP11-732M18.3 could provide a novel therapeutic target for glioma treatment.
Keywords: lncRNA RP11-732M18.3, 14-3-3β/α, p21, Tumorigenesis, Glioma
Research in context.
Evidence before this study
It has been shown that long noncoding RNAs (lncRNA) accumulates at the transcriptional site and performs its function through cis or trans-regulatory elements, or outputs to the cytoplasm and regulates the activity of interacting proteins. In addition, lncRNAs play an important role in glioma development.
Added value of this study
This research article highlights a new lncRNA modulates the degradation of key cell cycle protein p21. We showed that lncRNA RP11-732 M18.3 is highly overexpressed in glioma tissues. lncRNA RP11-732 M18.3 promotes tumor cell proliferation both in vivo and in vitro. We identified a new degradation mechanism of p21, involved with lncRNA RP11-732M18.3, 14-3-3β/α, and p21. We showed that lncRNA RP11-732M18.3 promotes the recruitment of ubiquitin-conjugating enzyme E2 E1 (UBE2E1) to 14-3-3β/α, which promotes the degradation activity of UBE2E1 on p21. This study deepens the understanding of the mechanism of lncRNAs in glioma and provides a therapeutic target for glioma.
Implications of all the available evidence
Cancer is fundamentally a genetic disease that alters cellular information flow to modify cellular homeostasis and promote growth. Recently, the evidence is emerging of important roles of long noncoding RNAs in the pathogenesis of cancer development. However, few have been identified that regulate glioma proliferation and development. In addition, the mechanism of lncRNAs in glioma is not fully understood. The regulation of lncRNA RP11-732M18.3 in tumorigenesis and the mechanism by which lncRNA RP11-732M18.3 promotes glioma cell proliferation will elucidate the future development of lncRNA-based glioma cancer therapy.
Alt-text: Unlabelled Box
1. Introduction
Long noncoding RNAs (lncRNAs) are transcribed RNAs >200 nucleotides in length, although >90% are not translated into appreciable peptide products [1,2]. Noncoding genes, which outnumber protein-coding genes, are exquisitely regulated, but restricted to specific cell types [3,4]. lncRNAs are unique and are expressed in a spatiotemporal and tissue-specific manner [5,6].
Glioblastoma is the most common and aggressive type of primary brain tumor, thus there is great urgency for the development of useful therapeutic targets of these lesions. Although many lncRNAs have been annotated, few have been functionally characterized in gliomas. Of these, lncRNA CASC2 acts as a suppressor of glioma cell growth by negative regulation of miR-21 [7], while lncRNA CRNDE promotes glioma cell growth and invasion through mTOR signaling [8]. However, the biological roles and molecular functions of the overwhelming majority of lncRNAs in gliomas remain unexplored or elusive.
lncRNAs have been shown to accumulate at sites of transcription and execute their functions via cis- or trans-regulatory elements, or are exported to the cytoplasm and modulate the activity and abundance of interacting proteins [9]. It is now widely understood that the subcellular fate of lncRNAs may provide new insights into the specialized functions of these molecules [10]. The lncRNAs in the nucleus have been shown to regulate gene transcription by organizing subnuclear structures or mediating chromosomal interactions [11]. The lncRNAs in the cytoplasm are known to modulate the activity of interacting proteins or act as miRNA sponges by competitively interacting with miRNAs to reduce availability to target mRNAs [12,13]. The 14-3-3 proteins are a highly conserved family with a subunit mass of approximately 30 kDa that play key roles in various cellular processes, such as signal transduction, cell cycle control, apoptosis, stress responses, and malignant transformation [14]. The14-3-3 proteins alter the activities, modifications, and intracellular localization of target proteins [15]. Therefore, it is of great interest to uncover new functions of the 14-3-3β/α protein mediated by lncRNA in certain biological processes.
The mechanisms of cell division are routinely subjected to endogenous and exogenous stimuli [16]. For example, the cell cycle checkpoint mechanisms are often defective in cancer cells, which very likely contributes to tumorigenesis and progression [17]. The cyclin-dependent kinase (CDK) inhibitor 1A, also known as p21, is a factor that inhibits cell cycle arrest in response to a variety of stimuli. Targeting cell cycle checkpoints, such as p21, may substantially improve cancer therapies. Although there have been immense efforts in the development of drugs targeting key players in the G1/S and G2/M transition checkpoints, efficient and effective treatment modalities are still needed [18]. Therefore, it is of great interest to uncover new therapeutic agents targeting tumorigenesis.
Here, we report the identification of a previously uncharacterized lncRNA, RP11-732M18.3, which is a transcript of about 424 nucleotides that interacts with 14-3-3β/α (also named YWHAB, belonging to the 14-3-3 family, members of which mediate signal transduction by binding to cell components) and promotes the proliferation of glioma cells. In brief, we found that: 1) the expression of lncRNA RP11-732M18.3 was increased in human glioma tissues; 2) lncRNA RP11-732 M18.3 promotes tumor cell proliferation both in vivo and in vitro; 3) lncRNA RP11-732M18.3 promotes cell cycle G1/S transition through the p21/CDK2/CCNE1 pathway; 4) lncRNA RP11-732M18.3 interacts with 14-3-3β/α and increases the degradation of p21; 5) lncRNA RP11-732M18.3 promotes the recruitment of ubiquitin-conjugating enzyme E2 E1 (UBE2E1) to 14-3-3β/α, which promotes the degradation activity of UBE2E1 on p21.
2. Experimental procedures
2.1. Bioinformatics analysis
The coding potentials of lncRNAs were analyzed using the NCBI ORF Finder graphical analysis tool (https://www.ncbi.nlm.nih.gov/orffinder/) and the UCSC genome browser (http://genome.ucsc.edu/) [19].
2.2. Patients and specimens
Frozen and normal glioma tissues were randomly collected with informed consent from patients who initially underwent surgery for a diagnosis of glioma at Nanfang Hospital, Southern Medical University (Guangzhou, China). Ethical consent was granted from the Committee for Ethical Review of Research Involving Human Subjects of Southern Medical University.
2.3. Animals
Female BALB/C nude mice (5 weeks old) were purchased from Vital River Laboratories Co., Ltd. (Beijing, China) and maintained under specific pathogen-free conditions. The animal studies were approved by the Institutional Animal Care and Use Committee of Nanfang Hospital.
2.4. Cell lines
Human U87MG, A172, and U251 glioma cell lines (ATCC, Manassas, VA, USA) were cultured in Dulbecco's modified Eagle's medium (Gibco, Carlsbad, CA, USA) supplemented with 10% fetal bovine serum (Gibco), 100 U/mL of penicillin, and 100 mg/mL of streptomycin (Gibco) in a humidified 5% CO2 incubator at 37 °C.
2.5. RNA isolation and analysis
Quantitative real-time polymerase chain reaction (qRT-PCR), western blot analysis, and cell proliferation, cell migration, immunofluorescence, and immunohistochemical analyses were performed as previously described [20]. The antibodies used in this study are listed in Supplementary Table 1.
2.6. Flow cytometry
Cells (1 × 10 [6]) were trypsinized and resuspended in phosphate-buffered saline. The single cell suspension was fixed in 75% ethanol at 4 °C overnight, then stained with propidium iodide (Nanjing Keygen Biotech, China) and analyzed with an LSRFortessaflow cytometer (BD Biosciences, San Jose, CA, USA) and FlowJo software (Tree Star, Inc., Ashland, OR, USA).
2.7. Lentivirus (LV) construction and cell transfection
LV vectors (overexpression or short hairpin RNA) were prepared as previously described [21]. The human U87MG, U251, and A172 cell lines were cultured in six-well plates for 12 h to 50%–70% confluence before use. The cells were transfected LV vectors with polybrene reagent (Santa Cruz Biotechnology, Inc., Dallas, TX, USA) in OptiMEM according to the manufacturer's instructions at a multiplicity of infection of 1. The stable overexpression or knocked down cell clones were obtained after 2 weeks using puromycin and the lncRNA-RP11-732M18.3 level was evaluated by qRT-PCR.
2.8. Short interfering (si)RNAs
Cells were transfected with siRNAs specific for protein 14-3-3 β/α, UBE2E1, EP300 (Supplementary Table 2), and scrambled siRNA (RiboBio Co., Guangzhou, China) using Lipofectamine reagent (Invitrogen Corporation, Carlsbad, CA, USA) according to the manufacturer's instructions. Transfected cells were grown for 48 h before analysis.
2.9. In vivo tumorigenicity assay
Cells (1 × 107 cells) suspended in 200 μL of phosphate-buffered saline were subcutaneously injected into the underarm area of female BALB/C nude mice. The formation of tumors was monitored using the FX Pro system (Bruker Corporation, Billerica, MA, USA). Tumor growth was examined every 5 days for at least 30 days before the mice were killed and the tumors were recovered. The weight of each tumor was determined and a portion was fixed in 4% paraformaldehyde and embedded in paraffin for staining with hematoxylin-eosin (HE) staining and other agents. All animal experiments were approved by the Animal Experimental Committee of Nanfang Hospital and conducted in accordance with the institutional guidelines for the use of laboratory animals.
2.10. Fluorescence in situ hybridization (FISH)
The FISH analysis was performed as previously described [20], with minor modifications. FISH signals were visualized using an LSM 880 system with an Airyscan microscope (Carl Zeiss AG, Oberkochen, Germany).
2.11. Chromatin isolation by RNA purification (ChIRP) analysis
ChIRP analysis, which exploits the specificity of anti-sense tiling oligonucleotides to allow the enumeration of lncRNA-bound genomic sites [22], was performed as previously described [20].
2.12. Statistical analysis
IBM SPSS Statistics for Windows, version 20.0 (IBM Corporation, Armonk, NY, USA) and GraphPad Prism 6.0 software (GraphPad Software, Inc., La Jolla, CA, USA) were used for data analyses. Data are presented as the mean ± standard error. The χ2 test was used to examine the relationship between lncRNA RP11-732M18.3 expression and clinicopathological characteristics. The two-tailed Student's t-test was used for comparisons of two independent groups. A probability (p) value of <0.05 was considered statistically significant.
3. Results
3.1. LncRNA RP11-732 M18.3 is highly expressed in glioma tissues
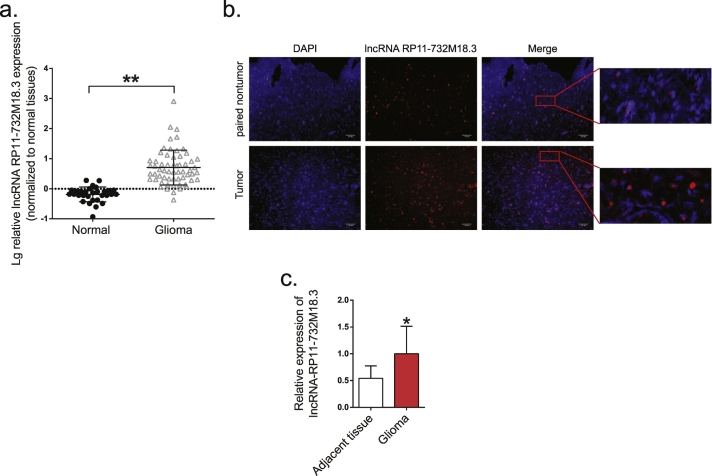
To identify glioma cancer-relevant lncRNAs, the expression profiles of lncRNAs in tumor samples as compared to paired peritumoral samples with determined using the lncRNA Array (v. 2.0) (ArrayStar, Inc., Rockville, MD, USA), as described in a previous study [20]. According to the array analysis results, overexpressed lncRNAs were selected according to the fold change and p-value and validated in small group samples (data not show). Among these aberrantly expressed lncRNAs, lncRNA RP11-732M18.3 was over-expressed in glioma (2.53-fold change, p < .05) and consistently over-expressed in small group glioma tissues (data not show). Thus, lncRNA RP11-732M18.3 was chosen for further research. Because lncRNAs may encode conserved peptides [23], we examined the coding potential of lncRNA RP11-732M18.3 using ORF Finder and PhyloCSF [19,24]. Both analyses confirmed the non-coding potential of lncRNA RP11-732M18.3, which received a low codon substitution frequency (CSF) value similar to that of other well-characterized lncRNAs (Fig. S1a–d). To further study lncRNA RP11-732M18.3 expression in human glioma tissues, we examined a panel of paired tumor and normal tissue specimens that were collected from patients with glioma (n = 60) and normal brain tissues (n = 30). lncRNA RP11-732M18.3 transcripts were expressed at higher levels in the tumor tissues, as compared to the normal tissues, after normalizing to U6 transcript expression by qRT-PCR analyses (Figs. 1a, Student's t-test). The expression levels of lncRNA RP11-732 M18.3 in glioma cell lines were higher than the normal cells (Figs. S1e, Student's t-test). To further confirm this expression pattern, the expression patterns of lncRNA RP11-732M18.3 were analyzed in six paired tumor and normal primary tissue specimens by RNA FISH. As shown in Fig. 1b and c, the lncRNA RP11-732M18.3 transcript was over-expressed in the glioma specimens and the sub-cellular location of this lncRNA in tumors was predominately the cytoplasm. Next, the relationship between lncRNA RP11-732M18.3 expression levels and the clinicopathological characteristics of 58 tumor tissue samples was examined. Correlation regression analysis showed that the overexpression of lncRNA RP11-732M18.3 was significantly correlated with the proliferation marker Ki67 (Table 1) (two specimens without MGMT promoter methylation were excluded). The expression level of lncRNA RP11-732M18.3 between the normal cells and glioma cell lines were detected and compared, found that expression of lncRNA RP11-732M18.3 was higher in glioma cells than normal cells (Figs. 1e). Therefore, the high expression of lncRNA RP11-732M18.3 might be associated with the proliferation of glioma cells and contribute to the accelerated growth and development of gliomas.
Fig. 1.
lncRNA-RP11-732 M18.3 overexpression in glioma tissues.
(a) lncRNA-RP11-732 M18.3 expression in glioma samples and normal tissues was analyzed by qRT-PCR. The log10 transformation was applied to the expression levels, which were normalized to that of U6 (**p < .01, Student's t-test). (b) Representative images of lncRNA-RP11-732 M18.3 expression from paired non-tumor and tumor tissues by RNA FISH. All experiments were performed in triplicate (n = 6). (c) Quantification of immunofluorescence RNA FISH. Data are presented as the mean ± standard deviation (SD) (n = 6, *p < .05, Student's t-test).
Table 1.
Clinical and molecular pathology features of Glioma samples in association with RP11-732 M18.3 expression.
| Low | High | P | |
|---|---|---|---|
| Gender, female/male | 12/10 | 17/19 | 0.588 |
| Age at diagnosis, y | 33.46 ± 16.29 | 49.66 ± 12.94 | 0.469 |
| IDH1 mutation (no mutation/mutation) | 14/8 | 15/21 | 0.104 |
| MGMT promoter methylation (unmethylation/methylation) | 13/9 | 13/23 | 0.088 |
| Ki-67 (low/high) | 15/7 | 12/24 | 0.010⁎ |
P values <.05 were considered statistically significant.
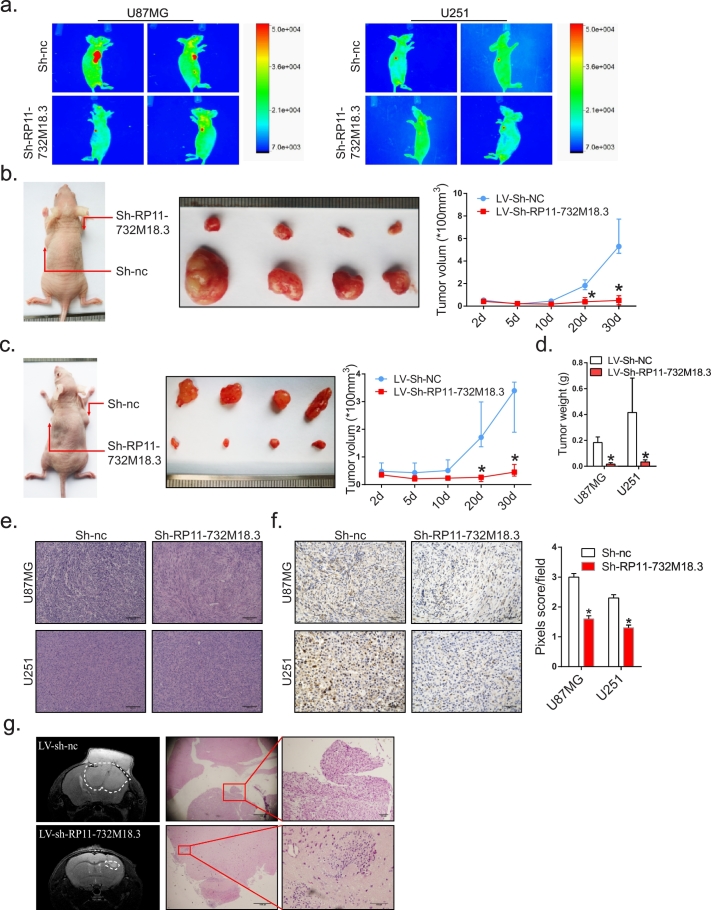
3.2. Knockdown of lncRNA RP11-732M18.3 inhibits glioma growth in vivo
Next, we focused on the correlation between lncRNA RP11-732M18.3 and proliferation. The glioma cell lines U87MG and U251 were infected with a lentivirus coding for the enhanced green fluorescent protein and a short hairpin RNA molecule targeting lncRNA RP11-732M18.3 (Fig. S2a, Student's t-test). The two stable knockout cell lines with the lowest lncRNA RP11-732M18.3 expression levels were subcutaneously injected into the axilla of nude mice. After 4 weeks, the fluorescence signal intensities in the knockout group were significantly decreased as compared with the controls (Fig. 2a). To further confirm the effects of lncRNA RP11-732M18.3 on tumorigenesis in vivo, a mouse xenograft assay was conducted by injecting the same two knockout cell lines into the axilla of nude mice and tumor formation was assessed after 4 weeks. As shown in Fig. 2b, decreased expression of lncRNA RP11-732M18.3 weakened tumor growth in vivo, as compared with the controls. The same results were observed with the U251 cell line group, indicating that the position of cell injected did not affect tumor growth (Fig. 2c, Student's t-test). The tumor weight of the knockout group was less than that of the control group (Fig. 2d, Student's t-test). The expression levels of RP11-732M18.3 in allograft tumors was lower than in controls (Fig. S1f, Student's t-test). In these xenograft tissues, lncRNA RP11-732M18.3 knockdown effectively reduced Ki67 expression (Fig. 2e and f, Student's t-test). In addition, a nude mouse model of orthotopic tumors showed that lncRNA RP11-732M18.3 knockdown decreased tumor volume (Fig. 2g). Thus, lncRNA RP11-732M18.3 has a certain impact on the proliferation of glioma cells.
Fig. 2.
Knockdown of lncRNA-RP11-732 M18.3 inhibits glioma growth in vivo.
(a) Representative fluorescence intensity images of mice over time after axilla injection with the indicated cell clones (n = 4). (b) Effects of lncRNA-RP11-732 M18.3 knockdown on tumor growth in vivo. Left: Representative images of nude mice injected subcutaneously with U87MG cells with knockdown of lncRNA-RP11-732 M18.3. Middle: Representative images of tumors. Right: Tumor growth curves. All experiments were performed in triplicate (n = 4, *p < .05, Student's t-test). (c) Effects of lncRNA-RP11-732 M18.3 knockdown on tumor growth in vivo. Left: Representative images of nude mice injected subcutaneously with U251 cells with knockdown of lncRNA RP11-732 M18.3. Middle: Representative images of tumors. Right: Tumor growth curves. All experiments were performed in triplicate (n = 4, *p < .05, Student's t-test). (d) Quantification of tumor weights from (b) and (c). All experiments were performed in triplicate (n = 4, *p < .05, Student's t-test). (e) Representative images of hematoxylin and eosin staining of xenografts. (f) Left: Representative images of Ki67 staining of xenografts. Right: Quantification of Ki67+ cells (*p < .05, Student's t-test). (g) Nude mice were implanted intracranially with U87MG cells stably knocking down lncRNA RP11-732 M18.3. Left: Representative, contrast-enhanced, T2-weighted images of the mouse brain were obtained using a small animal MRI system (PharmaScan 70/16 US; Bruker, Billerica, MA, USA). Right: An hematoxylin-eosin stained histological section showing tumor cells.
3.3. lncRNA RP11-732M18.3 promotes G1/S transition
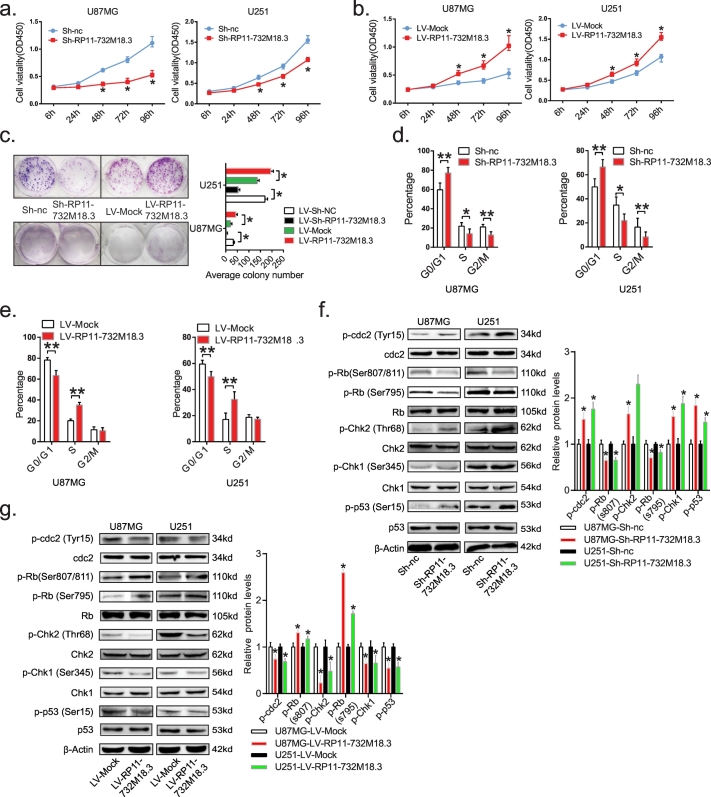
Given the impact of lncRNA RP11-732M18.3 depletion on the proliferation of glioma cells in vivo, several assays were conducted with the three stable knockdown or overexpression cell lines to study this phenotype in vitro (Fig. S2a and S2b, Student's t-test). The results of the cell counting kit-8 and colony formation assays in vitro showed that knockdown of lncRNA RP11-732M18.3 decreased the proliferative capacity of U87MG, U251, and A172 cells, compared with that of parallel stable cell lines containing empty vectors (Fig. 3a, c, and S3a, Student's t-test). In contrast, overexpression of endogenous lncRNA RP11-732M18.3 dramatically increased the proliferative capacity of glioma cells (Fig. 3b, c, and S3a, Student's t-test).
Fig. 3.
lncRNA-RP11-732 M18.3 promotes the G1/S transition.
(a) Cell growth rates were determined with the cell counting kit-8 assay. Knockdown of lncRNA-RP11-732 M18.3 in U87MG and U251 cells significantly inhibited cell proliferation, relative to control cells. All experiments were performed in triplicate (n = 3, *p < .05, Student's t-test). (b) lncRNA-RP11-732 M18.3 overexpression enhanced the proliferation of U87MG and U251 cells. All experiments were performed in triplicate. (n = 3, *p < .05, Student's t-test). (c) lncRNA-RP11-732 M18.3 depletion inhibited the clonal formation, overexpression enhanced. All experiments were performed in triplicate. (n = 3, *p < .05, Student's t-test). (d) FACS analysis showing significant increases or decreases of U87MG and U251 cells with knockdown of lncRNA-RP11-732 M18.3 in the G1 or S phase, respectively. All experiments were performed in triplicate. (n = 3, *p < .05, Student's t-test). (e) U87MG and U251 cells in S phase increased significantly by overexpression of lncRNA-RP11-732 M18.3 (n = 3, *p < .05, Student's t-test). (f and g) Western blot analysis of the phosphorylation level of G1/S checkpoint key proteins in U87MG and U251 cells with lncRNA-RP11-732 M18.3 depletion or overexpression. All experiments were performed in triplicate (n = 3, *p < .05, Student's t-test).
To gain insights into the mechanism by which lncRNA RP11-732M18.3 enhances glioma cell proliferation, differences in cell-cycle distributions were analyzed after lncRNA RP11-732M18.3 silencing or overexpression by fluorescence-activated cell sorting (FACS). As shown in Figs. 3d, and S2c, knockdown of lncRNA RP11-732M18.3 decreased the proportion of cells that entered S phase, as compared with control U87MG and U251 cells. In contrast, overexpression of endogenous lncRNA RP11-732M18.3 increased the proportion of cells that entered S phase (Figs. 3e, and S2d). FACS results of A172 cells also showed that the G1/S checkpoint was indeed compromised with the lowest expression of lncRNA RP11-732M18.3 (Figs. S3b and S3c, Student's t-test).
The cell division cycle must be precise to avoid the accumulation of genetic defects. This process is controlled by molecular circuits called “checkpoints” that are common to all eukaryotic cells [25,26]. Consistent with the FACS data, lncRNA RP11-732M18.3 knockdown increased the phosphorylation levels of cdc2 (Tyr15), Chk2 (Thr68), Chk1 (Ser345), and p53 (Ser15), while the phosphorylation levels of Rb (Ser807/811) and Rb (Ser795) decreased significantly (Fig. 3f, Student's t-test). The total expression levels of these proteins were tested and used for quantification control combined with β-actin. In contrast, overexpression of lncRNA RP11-732 M18.3 reduced the phosphorylation levels of cdc2 (Tyr15), Chk2 (Thr68), Chk1 (Ser345), and p53 (Ser15), and increased the phosphorylation levels of Rb (Ser807/811) and Rb (Ser795) (Fig. 3g, Student's t-test).
3.4. lncRNA-RP11-732M18.3 promotes G1/S transition via p21 regulation
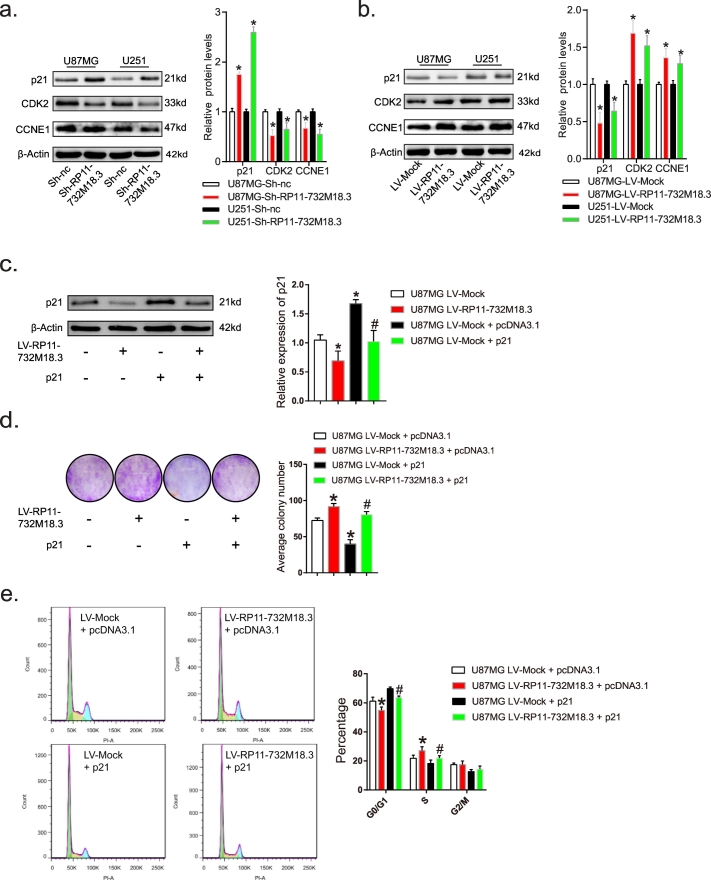
Cdk2 phosphorylates Rb as cells progress through G1 [25]. Previous studies have suggested that the p21 CDK-interacting protein Cip1 is a potent inhibitor of G1 cyclin-dependent kinases [[27], [28], [29]]. In addition, the cyclin E-Cdk2 complex is necessary for the G1/S transition and the expression levels of CDK2 and CCNE1 are closely related to G1/S transition [29,30]. Hence, the expression levels of p21, CDK2, and CCNE1 were examined after overexpression/depletion of lncRNA RP11-732 M18.3. The results showed that knockdown of lncRNA RP11-732M18.3 increased the expression level of p21, while that of CDK2 and CCNE1 was decreased (Fig. 4a, Student's t-test). In contrast, overexpression of lncRNA RP11-732 M18.3 had the opposite effect (Fig. 4b, Student's t-test). To confirm this finding, A172 cells were subjected to western blot analysis, which showed that lncRNA RP11-732 M18.3 regulates the expression of p21 (Figs. S3d, Student's t-test). In addition, inhibition of RP11-732 M18.3 increased the expression of p21 in allograft tumor cells (Figs. S4, Student's t-test).
Fig. 4.
lncRNA-RP11-732 M18.3 promotes G1/S transition via p21 regulation.
(a) lncRNA-RP11-732 M18.3 inhibits the expression of p21, CCNE1, and CDK2 after lncRNA-RP11-732 M18.3 silencing in U87MG and U251 cells. All experiments were performed in triplicate (n = 3, *p < .05, Student's t-test). (b) Overexpression of lncRNA-RP11-732 M18.3 decreased the levels of p21, CCNE1, and CDK2 in U87MG and U251 cells. All experiments were performed in triplicate (n = 3, *p < .05, Student's t-test). (c) Western blot analysis of p21 following the indicated treatments showing successful overexpression. All experiments were performed in triplicate (n = 3, *p < .05, Student's t-test). (d) Enforced expression of the p21 rescue of the proliferation phenotype caused by RP11-732 M18.3. pcDNA3.1 = plasmid control vector. P21 = enforced p21 expression plasmid vector. All experiments were performed in triplicate (n = 3, *p < .05, Student's t-test). (e) Enforced expression of p21 rescued G1/S transition caused by RP11-732 M18.3. Representative images of cell cycle distribution in U87MG and U251 cells following the indicated treatments. Enforced expression of p21 rescued G1/S transition caused by RP11-732 M18.3. The results are expressed as the mean ± SD. All experiments were performed in triplicate (n = 3, *p < .05, Student's t-test) vs. the first group, #p < .05 vs. the second group.
Further research showed that enforced expression of p21 rescued the proliferation phenotype and cell cycle G1/S transition caused by RP11-732 M18.3 (Fig. 4c, d, e, Student's t-test). These results suggest that lncRNA RP11-732 M18.3 regulates the G1/S checkpoint in a p21-dependent manner.
3.5. lncRNA RP11-732M18.3 binding with 14-3-3β/α promotes p21 degradation
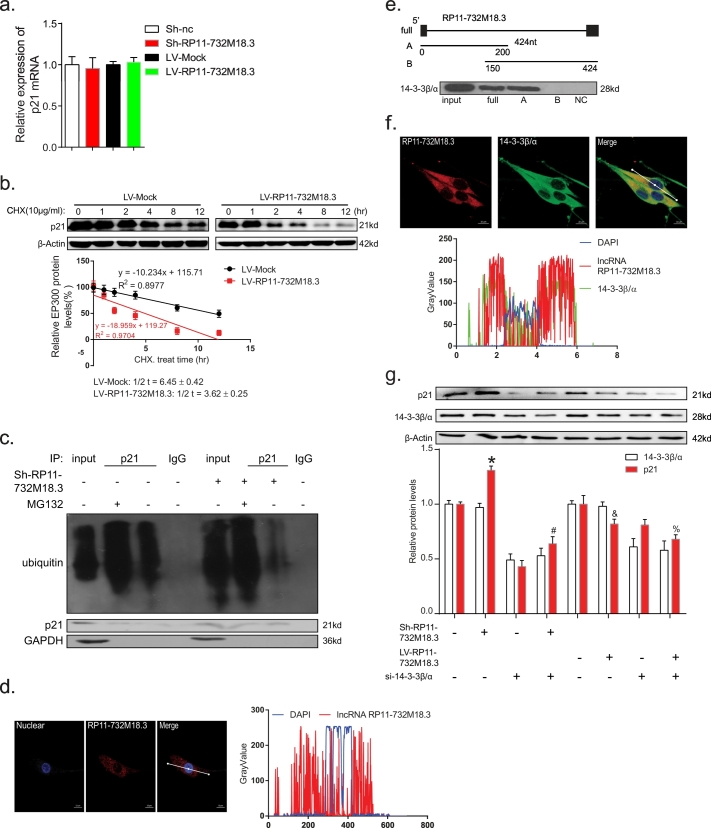
To investigate the molecular mechanism by which lncRNA RP11-732M18.3 is associated with p21 expression, p21 mRNA expression was determined by qRT-PCR. The results showed that lncRNA RP11-732M18.3 had no effect on p21 mRNA levels (Fig. 5a, Student's t-test). However, degradation assay results revealed that lncRNA RP11-732M18.3 promoted p21 degradation (Fig. 5b, Student's t-test). In addition, MG132 decreased the degradation of p21 and knockdown lncRNA RP11-732M18.3 inhibition the ubiquitination of p21 (Fig. 5c). Collectively, these data indicate that lncRNA RP11-732M18.3 promoted glioma cell proliferation by promoting p21 degradation.
Fig. 5.
The interaction of lncRNA-RP11-732 M18.3 with 14–3-3β/α promoted p21 degradation.
(a) lncRNA-RP11-732 M18.3 had no effect on the mRNA level of p21. All experiments were performed in triplicate in U87MG cell lines (n = 3, *p < .05, Student's t-test). (b) Endogenous p21 protein levels in U87MG cells overexpressing lncRNA-RP11-732 M18.3 were monitored at the indicated time points after cycloheximide (CHX) (10 μg/ml) treatment in U87MG cell lines. The upper panels are western blots using the antibodies indicated to the left and the lower graph represents a quantification of p21 normalized to β-actin as a function of time after CHX treatment. The half degradation of lncRNA-RP11-732 M18.3 overexpression, as compared with the control group (3.62 ± 0.25 vs. 6.45 ± 0.42 h, respectively). All experiments were performed in triplicate (n = 3, *p < .05, Student's t-test). (c) Ubiquitin-dependent degradation of p21. Knockdown of lncRNA-RP11-732 M18.3 inhibited the ubiquitin level of p21 in U87MG cell lines. (d) Image showing RNA FISH targeting lncRNA-RP11-732 M18.3 in U87MG cells. The graph below shows the overlap of fluorescence intensity peaks along with profiles spanning the cells. Scale bars, 10 mm. DAPI, 4′,6-diamidino-2-phenylindole. (n = 3). (e) Schematic outline of the pull-down strategy. The 5′-end (0–150 nt) of lncRNA-RP11-732 M18.3 is essential for the association between lncRNA-RP11-732 M18.3 and 14–3-3β/α. (f) Co-localization analysis: RNA FISH assay of lncRNA-RP11-732 M18.3 combined with immunofluorescence detection of 14–3-3β/α in U87MG cells. The graph below shows the overlap of fluorescence intensity peaks along with profiles spanning the cells. Scale bars, 10 mm. DAPI. (n = 3). (g) Western blot analysis of p21 in U87MG cells with the indicated treatments. Sh = lentiviruses encoding lncRNA-RP11-732 M18.3 short hairpin RNA; Lv = lentiviruses encoding lncRNA-RP11-732 M18.3 overexpress RNA; Si = small interfering RNAs. The results are expressed as the mean ± SD. All experiments were performed in triplicate. (n = 3, *p < .05 vs. the first group, #p < .05 vs. the second group, &p < .05 vs. the fifth group, and %p < .05 vs. the sixth group, One-Way ANOVA).
Next, the mechanism by which lncRNA RP11-732M18.3 regulates p21 expression was addressed. Recent studies have revealed that the identification of the subcellular locations of lncRNAs can potentially provide new insights into the functions of lncRNAs [10]. RNA FISH demonstrated that the lncRNA RP11-732M18.3 transcript is predominately localized in the cytoplasm, as the strongest signal was localized in the cytoplasm in U87MG cell lines and about half in the cytoplasm of U251 cells (Figs. 5d, S5a, and S5b), where it can interfere with the post-translational modifications of proteins, leading to abnormal signal transduction [31]. Next, chromatin isolation by RNA purification followed by mass spectrometry (ChIRP-MS) was applied to capture lncRNA RP11-732M18.3 and identify the target proteins, as described in a previous study [20] (Fig. S5c). Biological process analysis revealed that cell growth and/or maintenance were among the most enriched processes (Fig. S5d). Among the proteins associated with cell growth, 14-3-3β/α had higher scores and a good peptide match (Fig. S5e, and S5f). The association between lncRNA RP11-732M18.3 and 14-3-3β/α was further validated with an affinity pull-down assay of 14-3-3β/α using in vitro transcribed biotin-labeled lncRNA RP11-732M18.3. The results of catRAPID express [32] predicted that 14-3-3β/α might interact with the forward part of lncRNA RP11-732M18.3 (Fig. S5 g). Notably, deletion analysis indicated that the 5′-end (0–150 nt) of lncRNA RP11-732M18.3 was essential for this association (Fig. 5e). Moreover, RNA FISH followed by immunofluorescence showed that lncRNA RP11-732M18.3 colocalized with 14-3-3β/α in the cytoplasm (Fig. 5f), indicating that lncRNA RP11-732M18.3 may regulate the activity of cytoplasmic 14-3-3β/α. In addition, 14-3-3β/α silencing reinforced the effect of lncRNA RP11-732M18.3 in inducing p21 expression (Fig. 5g, One-Way ANOVA), indicating that 1433β/α may be involved in the regulation of P21 by lncRNA RP11-732M18.3.
3.6. lncRNA RP11-732M18.3 promoted the recruitment of UBE2E1 to 14-3-3β/α and the association of 14-3-3β/α with UBE2E1 promoted p21 degradation
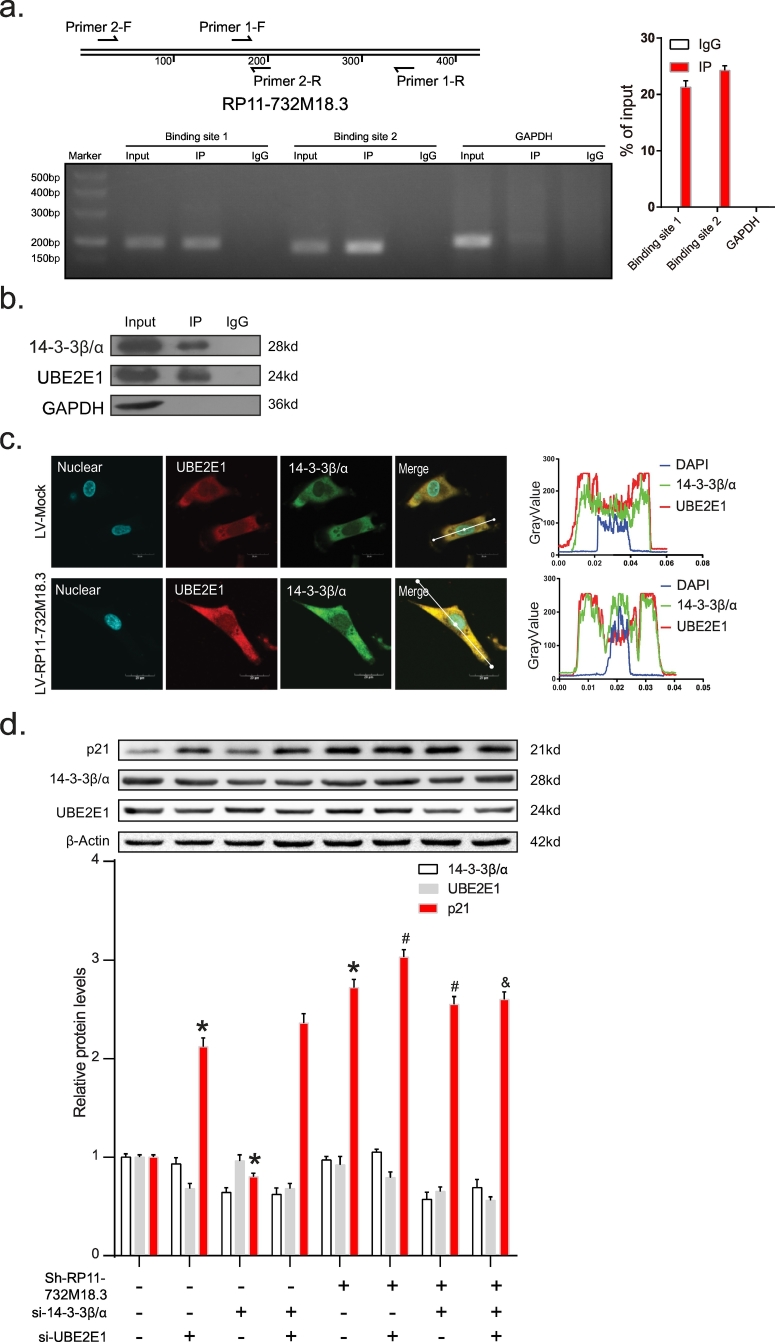
To gain insights into the mechanism by which lncRNA RP11-732M18.3 promotes p21 protein degradation, the target protein 14-3-3β/α was specifically co-immunoprecipitated from the U87MG cell extracts and the immunoprecipitates were validated by tandem mass spectrometry (Fig. S5 h). Next, we focused on the degradation of proteins involved with various biological processes. UBE2E1 was found in the protein complex purified by ChIRP-MS. UBE2E1 has been reported to play an important role in the regulation of protein mono-ubiquitination [33,34]. In addition, the RNA IP results revealed an association between UBE2E1 and lncRNA RP11-732M18.3 (Fig. 6a). Co-IP analysis of total cell lysates with 14-3-3β/α antibody confirmed interactions between 14 and 3-3β/α and UBE2E1 (Fig. 6b). Hence, UBE2E1 was chosen for further analysis. We anticipated that UBE2E1 might affect the degradation of p21. Colocalization immunofluorescence detection of UBE2E1 combined with 14-3-3β/α in U87MG cells found that overexpression of lncRNA RP11-732M18.3 promoted the recruitment of UBE2E1 to 14-3-3β/α (Fig. 6c). In addition, UBE2E1 silencing increased p21 protein levels (Fig. 6d), suggesting that UBE2E1 is required for the degradation of p21. Moreover, UBE2E1 silencing promoted the p21 inducing by lncRNA RP11-732M18.3 knockdown (Fig. 6d, One-Way ANOVA). These data suggest that lncRNA RP11-732M18.3 promoted the recruitment of UBE2E1 to 14-3-3β/α and the association of 14-3-3β/α with UBE2E1 might promote the degradation activity of UBE2E1 on p21 (Fig. 7).
Fig. 6.
lncRNA-RP11-732 M18.3 promoted the recruitment of UBE2E1 to 14–3-3β/α.
(a) U87MG total cell lysates were immunoprecipitated with either UBE2E1 antibody or immunoglobulin (IgG) as a control group. RNAs were detected by qRT-PCR as indicated. (b) Co-IP of endogenous UBE2E1 and 14–3-3β/α in U87MG cell lysates using antibodies specific for UBE2E1. (c) Colocalization analysis: Immunofluorescence assay of UBE2E1 combined with immunofluorescence detection of 14–3-3β/α in U87MG cells. The graph on the right shows the overlap of fluorescence intensity peaks along with profiles spanning the cells. Blue = nuclear; Green = 14–3-3β/α, and Red = UBE2E1. Scale bars, 20 mm. DAPI. (n = 3). (d) Western blot analysis of p21 in U87MG cells following the indicated treatments. Si = small interfering RNAs. The results are expressed as the mean ± SD. All experiments were performed in triplicate (n = 3, *p < .05, One-Way ANOVA) vs. the first group, #p < .05 vs. the fifth group, and &p < .05 vs. the seventh group. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
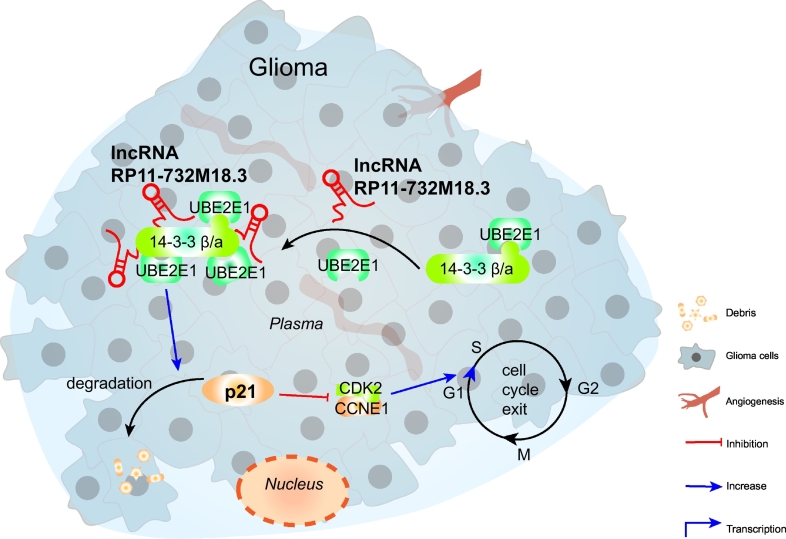
Fig. 7.
Model depicting the roles of lncRNA-RP11-732 M18.3 in the regulation of the cell cycle and tumor growth.
lncRNA-RP11-732 M18.3 promotes the recruitment of UBE2E1 to 14–3-3β/α, which promotes the degradation activity of UBE2E1 on p21, and promotes cell proliferation and glioma development.
4. Discussion
Nearly 95% of the human genome does not encode proteins and it is clear that abnormalities in the “non-coding” genome drive important cancer phenotypes [35,36]. Recently, more and more evidence has confirmed the functional roles of lncRNAs in glioma formation. For example, knockdown of the lncRNA XIST exerted tumor-suppressive effects in human glioblastoma stem cells via up-regulation of miR-152 [37]. Furthermore, lncRNA TUG1 enhanced tumor-induced angiogenesis in human glioblastomas through the inhibition of microRNA-299 [38]. The findings of this study showed that the newly identified noncoding RNA RP11-732M18.3, which is highly expressed in glioma cells, interacts with 14-3-3β/α and promotes glioma cell proliferation and subsequent tumor growth. These higher lncRNA RP11-732M18.3 levels were associated with the proliferation of glioma cells in vivo. Our results demonstrate that lncRNA RP11-732M18.3 promoted G1/S transition via regulation of p21, which resulted in the proliferation of glioma cells. The lncRNA RP11-732M18.3 transcript was found to be associated with 14-3-3β/α to promote p21 degradation in a process that may be mediated by UBE2E1. Thus, our results indicate that lncRNA RP11-732M18.3 plays a critical role in cell proliferation, which may increase the understanding of the roles of lncRNAs in oncogenesis and potentials as targets for glioma therapy.
The mammalian cell cycle is a highly organized and regulated to ensure appropriate gene transcription and cell division. Cancer is characterized by aberrant cell cycle activity [18,39]. The p21 protein, which was the first p53 transcriptional target to be identified, usually acts as a tumor suppressor [40,41]. The stability of p21 is tightly and differentially regulated by ubiquitination and proteasome-mediated degradation during various stages of the cell cycle [42]. In the present work, we identified a novel degradation pathway for p21, which involved the newly discovered lncRNA RP11-732M18.3. lncRNA RP11-732M18.3 primary locates in the plasma in glioma cell lines. The interaction of lncRNA RP11-732M18.3 with 14-3-3β/α increases the degradation of p21. Knockdown of 14-3-3β/α decreased the expression of p21, while silencing of UBE2E1 had the opposite effect, indicating that both 14-3-3β/α and UBE2E1 are involved in the degradation of p21. However, the role of 14-3-3β/α in the regulation of p21 degradation must be further verified. Cancer is characterized by uncontrolled proliferation resulting from the aberrant activity of various cell cycle proteins. This work provides a new regulatory pathway for p21 in glioma. p21 is an important therapeutic target for cancer therapies and iron may be a good therapeutic agent for p21 in cancer [43,44]. The determination of whether lncRNA RP11-732M18.3 is involved in the regulation of iron (Fe) on p21 warrants further research.
There are still several questions posed in this article that have not been addressed. In the present study, overexpression of lncRNA RP11-732M18.3 promoted the recruitment of UBE2E1 to 14-3-3β/α and the subsequent degradation of p21. The 0–150 nucleotides of lncRNA RP11-732M18.3 were essential for the association between lncRNA RP11-732M18.3 and 14-3-3β/α. However, further experiments are needed to confirm the protein-specific binding site of 14-3-3β/α. Interesting, the expression level of p21 was decreased in cells only treated with siRNA of 14-3-3β/α. We Hypothesis that 14-3-3β/α may act as a frame for p21 degradation or involved in other pathways. Liquid phase condensation or biomolecular condensates are involved in a variety of processes including DNA damage response, RNA metabolism, signal transduction, and other cell biological processes [45,46]. Therefore, it is of great interest to uncover whether liquid phase condensation involved in the protective role of 14-3-3β/α for p21. However, it needed further research to confirm. To elucidate the mechanisms of how lncRNA RP11-732M18.3 influences the binding between UBE2E2 and 14-3-3β/α will require improvements in methodologies and the RNA-protein interaction theory. E1s and E3s play important roles in the regulation of protein degradation, thus it would be interesting to investigate whether E1s or E3s is involved in the regulation of lncRNA RP11-732M18.3 and p21 degradation and to explore the function of lncRNA RP11-732M18.3 in the nuclei of U251 or A172 cells.
In summary, our data point to a newly discovered lncRNA, RP11-732M18.3, as a key modulator of tumor progression, which interacts with 14-3-3β/α in the regulation of p21 protein degradation. This work highlights a new lncRNA involved in p21 protein degradation. However, the mechanism underlying the up-regulation of lncRNA RP11-732M18.3 in glioma cells remains unknown. Nonetheless, the regulation of lncRNA RP11-732M18.3 in tumorigenesis and the illustration of the mechanism of lncRNA RP11-732M18.3 to promote the proliferation of glioma cells will shed light on the future development of lncRNA-based glioma cancer therapies.
The following are the supplementary data related to this article.
Biological information analysis of lncRNA-RP11-732M18.3. (a) Prediction of putative proteins encoded by lncRNA-RP11-732M18.3 using ORF Finder. (b) lncRNA-RP11-732M18.3 lacks the potential to encode any recognizable protein domains, based on standard protein BLAST analysis of ORF Finder from (a). (c) The CSF scores of lncRNA-RP11-732M18.3 were <0. (d) The CSF scores of lncRNA-MALAT1 and glyceraldehyde 3-phosphate dehydrogenase (GAPDH). (e) The expression levels of lncRNA RP11-732 M18.3 in glioma cell lines were higher than the normal cells (n = 3, *p < .05, Student's t-test). (f) Relative expression of lncRNA-RP11-732 M18.3 in xenograft tumor cells. All experiments were performed in triplicate (n = 3, *p < .05, Student's t-test).
lncRNA-RP11-732 M18.3 promoted cell cycle transition. (a) lncRNA-RP11-732M18.3 expression was detected in U87MG, U251, and A172 cells by qRT-PCR after transfection of lentiviruses encoding lncRNA-RP11-732M18.3 short hairpin RNA or an empty vector. All experiments were performed in triplicate (n = 3, *p < .05, Student's t-test). (b) lncRNA-RP11-732M18.3 expression was detected in U87MG, U251, and A172 cells by qRT-PCR after transfection of lentivirus harboring the full-length human lncRNA-RP11-732M18.3 sequence or an empty vector. All experiments were performed in triplicate (n = 3, *p < .05, Student's t-test). (c) Representative images of cell cycle distribution in U87MG and U251 cells following the indicated treatments. (d) Representative images of cell cycle distribution in U87MG and U251 cells following the indicated treatments.
lncRNA-RP11-732M18.3 promoted cell proliferation and inhibited p21 expression in A172 cell lines. (a) Knockdown of lncRNA-RP11-732M18.3 inhibited colony formation of A172 cells. All experiments were performed in triplicate (n = 3, *p < .05, Student's t-test). (b and c) Knockdown of lncRNA-RP11-732 M18.3 inhibited G1/S transition but was promoted by lncRNA-RP11-732 M18.3 overexpression. All experiments were performed in triplicate (n = 3, **p < .01, Student's t-test). (d) lncRNA-RP11-732 M18.3 depletion increased p21 expression, which was decreased by overexpression. All experiments were performed in triplicate (n = 3, **p < .01, Student's t-test).
Knockdown of lncRNA-RP11-732M18.3 promoted p21 expression in xenograft tumor cells. Knockdown of lncRNA-RP11-732M18.3 increased p21 levels in xenograft tumor cells. Left: Representative images of p21 staining of xenografts. Right: Quantitative analysis p21 index. All experiments were performed in triplicate (n = 3, *p < .05, Student's t-test).
lncRNA-RP11-732M18.3 interacted with 14-3-3β/α. (a) Positive and negative controls used for RNA FISH analysis of U87MG cells. (b) RNA FISH assays with the lncRNA-RP11-732M18.3 probe in U251 and A172 cells. (c) Silver staining, SDS-PAGE, and ChIRP showing enrichment of human lncRNA-RP11-732M18.3 in U87MG cells. (d) Biological process analysis of proteins from ChIRP/MS. (e) Peptide summary report of 14-3-3β/α. (f) Mass spectra of 14-3-3β/α. (g) catRAPID express predicted that 14-3-3β/α might interact with the forward part of lncRNA-RP11-732M18.3. (h) Silver staining, sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE), and co-immunoprecipitation enrichment of human 14-3-3β/α in U87MG cells. The arrow indicates the position of 14-3-3β/α. (i) Expression levels of the 14-3-3β/α, and UBE2E1 proteins were detected in U87MG cells by qRT-PCR after transfection of the indicated small interfering RNAs. All experiments were performed in triplicate. (n = 3, *p < .05, Student's t-test).
Funding sources
This study was supported by grants from the National Natural Sciences Foundation of China (grant no. 81772244), the Guangdong Natural Science Foundation (2016A030313525), the Science and Technology Program of Guangzhou (201607010015), the President Foundation of Nanfang Hospital, Southern Medical University (2017C044), the Science and Technology Program of Guangzhou (201604020015, 201704020213), and the Natural Science Fund of Guangdong (2017A030313535). The funders did not participate in study design, data collection, data analysis, interpretation, and writing of the report.
Declaration of interests
The authors declare no competing financial interests.
Author contributions
Qian Wang, Yu-Rong Qiu, and Yan-Wei Hu conceived the study; Chun-Min Kang and Yan-Wei Hu designed the experiments; Chun-Min Kang, Huan-Lan Bai, Zhi-Feng Lu, and Lei Zheng performed experiments; Chun-Min Kang performed the in vivo experiments; Jing-Jing Zhao contributed the patient samples; Xue-Heng Li contributed to the construction of the orthotopic tumor model; Huang Rui-Ying contributed to the immunohistochemical analysis; Yuan-Jun Xu performed the pathological analyses; Chun-Min Kang wrote the manuscript; Xiao-Yan Dai contributed to the immunoprecipitation analysis. All authors revised the manuscript.
Contributor Information
Yu-Rong Qiu, Email: qiuyuronggz@126.com.
Yan-Wei Hu, Email: ywhu0618@163.com.
Qian Wang, Email: nfyywangqian@163.com.
References
- 1.Engreitz J.M. Local regulation of gene expression by lncRNA promoters, transcription and splicing. Nature. 2016;539:452–455. doi: 10.1038/nature20149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kato M. An endoplasmic reticulum stress-regulated lncRNA hosting a microRNA megacluster induces early features of diabetic nephropathy. Nat Commun. 2016;7 doi: 10.1038/ncomms12864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Parikshak N.N. Genome-wide changes in lncRNA, splicing, and regional gene expression patterns in autism. Nature. 2016;540 doi: 10.1038/nature20612. 423-+ [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gloss B.S., Dinger M.E. The specificity of long noncoding RNA expression. Biochim Biophys Acta. 2016;1859:16–22. doi: 10.1016/j.bbagrm.2015.08.005. [DOI] [PubMed] [Google Scholar]
- 5.Kato M. An endoplasmic reticulum stress-regulated lncRNA hosting a microRNA megacluster induces early features of diabetic nephropathy. Nat Commun. 2016;7:16. doi: 10.1038/ncomms12864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Liu X. LncRNA NBR2 engages a metabolic checkpoint by regulating AMPK under energy stress. Nat Cell Biol. 2016;18 doi: 10.1038/ncb3328. 431-+ [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wang P. Long non-coding RNA CASC2 suppresses malignancy in human gliomas by miR-21. Cell Signal. 2015;27:275–282. doi: 10.1016/j.cellsig.2014.11.011. [DOI] [PubMed] [Google Scholar]
- 8.Wang Y. CRNDE, a long-noncoding RNA, promotes glioma cell growth and invasion through mTOR signaling. Cancer Lett. 2015;367:122–128. doi: 10.1016/j.canlet.2015.03.027. [DOI] [PubMed] [Google Scholar]
- 9.Quinn J.J., Chang H.Y. Unique features of long non-coding RNA biogenesis and function. Nat Rev Genet. 2016;17:47–62. doi: 10.1038/nrg.2015.10. [DOI] [PubMed] [Google Scholar]
- 10.Chen L.-L. Linking long noncoding RNA localization and function. Trends Biochem Sci. 2016;41:761–772. doi: 10.1016/j.tibs.2016.07.003. [DOI] [PubMed] [Google Scholar]
- 11.Hu W.L. GUARDIN is a p53-responsive long non-coding RNA that is essential for genomic stability. Nat Cell Biol. 2018;20:492–502. doi: 10.1038/s41556-018-0066-7. [DOI] [PubMed] [Google Scholar]
- 12.Lin A. The LINK-A lncRNA activates normoxic HIF1 alpha signalling in triple-negative breast cancer. Nat Cell Biol. 2016;18 doi: 10.1038/ncb3295. 213-+ [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cai H. Long non-coding RNA taurine upregulated 1 enhances tumor-induced angiogenesis through inhibiting microRNA-299 in human glioblastoma. Oncogene. 2017;36:318–331. doi: 10.1038/onc.2016.212. [DOI] [PubMed] [Google Scholar]
- 14.Aghazadeh Y., Papadopoulos V. The role of the 14-3-3 protein family in health, disease, and drug development. Drug Discov Today. 2016;21:278–287. doi: 10.1016/j.drudis.2015.09.012. [DOI] [PubMed] [Google Scholar]
- 15.Toshima J.Y., Toshima J., Watanabe T., Mizuno K. Binding of 14-3-3beta regulates the kinase activity and subcellular localization of testicular protein kinase 1. J. Biol. Chem. 2001;276:43471–43481. doi: 10.1074/jbc.M104620200. [DOI] [PubMed] [Google Scholar]
- 16.Visconti R., Della Monica R., Grieco D. Cell cycle checkpoint in cancer: a therapeutically targetable double-edged sword. J Exp Clin Cancer Res. 2016;35 doi: 10.1186/s13046-016-0433-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kastan M.B., Bartek J. Cell-cycle checkpoints and cancer. Nature. 2004;432:316–323. doi: 10.1038/nature03097. [DOI] [PubMed] [Google Scholar]
- 18.Otto T., Sicinski P. Cell cycle proteins as promising targets in cancer therapy. Nat Rev Cancer. 2017;17:93–115. doi: 10.1038/nrc.2016.138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lin M.F., Jungreis I., Kellis M. PhyloCSF: a comparative genomics method to distinguish protein coding and non-coding regions. Bioinformatics. 2011;27:I275–I282. doi: 10.1093/bioinformatics/btr209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hu Y.-W. LncRNA PLAC2 down-regulates RPL36 expression and blocks cell cycle progression in glioma through a mechanism involving STAT1. J Cell Mol Med. 2018;22:497–510. doi: 10.1111/jcmm.13338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hu Y.W. RP5-833A20.1/miR-382-5p/NFIA-dependent signal transduction pathway contributes to the regulation of cholesterol homeostasis and inflammatory reaction. Arterioscler Thromb Vasc Biol. 2015;35:87–101. doi: 10.1161/ATVBAHA.114.304296. doi:ATVBAHA.114.304296 [pii] 10.1161/ATVBAHA.114.304296 [doi] [DOI] [PubMed] [Google Scholar]
- 22.Chu C., Qu K., Zhong F.L., Artandi S.E., Chang H.Y. Genomic maps of long noncoding RNA occupancy reveal principles of RNA-chromatin interactions. Mol Cell. 2011;44:667–678. doi: 10.1016/j.molcel.2011.08.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Anderson D.M. A micropeptide encoded by a putative long noncoding RNA regulates muscle performance. Cell. 2015;160:595–606. doi: 10.1016/j.cell.2015.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Qu L. Exosome-transmitted lncARSR promotes Sunitinib resistance in renal cancer by acting as a competing endogenous RNA. Cancer Cell. 2016;29:653–668. doi: 10.1016/j.ccell.2016.03.004. [DOI] [PubMed] [Google Scholar]
- 25.Sherr C.J. Cancer cell cycles. Science (New York, NY) 1996;274:1672–1677. doi: 10.1126/science.274.5293.1672. [DOI] [PubMed] [Google Scholar]
- 26.Harbour J.W., Luo R.X., Dei Santi A., Postigo A.A., Dean D.C. Cdk phosphorylation triggers sequential intramolecular interactions that progressively block Rb functions as cells move through G1. Cell. 1999;98:859–869. doi: 10.1016/s0092-8674(00)81519-6. [DOI] [PubMed] [Google Scholar]
- 27.Harper J.W., Adami G.R., Wei N., Keyomarsi K., Elledge S.J. The p21 Cdk-interacting protein Cip1 is a potent inhibitor of G1 cyclin-dependent kinases. Cell. 1993;75:805–816. doi: 10.1016/0092-8674(93)90499-g. [DOI] [PubMed] [Google Scholar]
- 28.Deng C., Zhang P., Harper J.W., Elledge S.J., Leder P. Mice lacking p21CIP1/WAF1 undergo normal development, but are defective in G1 checkpoint control. Cell. 1995;82:675–684. doi: 10.1016/0092-8674(95)90039-x. [DOI] [PubMed] [Google Scholar]
- 29.Du W.W. Foxo3 circular RNA retards cell cycle progression via forming ternary complexes with p21 and CDK2. Nucleic Acids Res. 2016;44:2846–2858. doi: 10.1093/nar/gkw027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ohtsubo M., Theodoras A.M., Schumacher J., Roberts J.M., Pagano M. Human cyclin E, a nuclear protein essential for the G1-to-S phase transition. Mol Cell Biol. 1995;15:2612–2624. doi: 10.1128/mcb.15.5.2612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhou C.-c. Systemic genome screening identifies the outcome associated focal loss of long noncoding RNA PRAL in hepatocellular carcinoma. Hepatology. 2016;63:850–863. doi: 10.1002/hep.28393. [DOI] [PubMed] [Google Scholar]
- 32.Agostini F. catRAPID omics: a web server for large-scale prediction of protein-RNA interactions. Bioinformatics. 2013;29:2928–2930. doi: 10.1093/bioinformatics/btt495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chen M. The nuclear transport receptor Importin-11 is a tumor suppressor that maintains PTEN protein. J Cell Biol. 2017;216:641–656. doi: 10.1083/jcb.201604025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wheaton K. UbE2E1/UBCH6 Is a Critical in Vivo E2 for the PRC1-catalyzed Ubiquitination of H2A at Lys-119. J Biol Chem. 2017;292:2893–2902. doi: 10.1074/jbc.M116.749564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gupta R.A. Long non-coding RNA HOTAIR reprograms chromatin state to promote cancer metastasis. Nature. 2010;464:1071–U1148. doi: 10.1038/nature08975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ulitsky I., Bartel D. P. lincRNAs: genomics, evolution, and mechanisms. Cell. 2013;154:26–46. doi: 10.1016/j.cell.2013.06.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yao Y. Knockdown of long non-coding RNA XIST exerts tumor-suppressive functions in human glioblastoma stem cells by up-regulating miR-152. Cancer Lett. 2015;359:75–86. doi: 10.1016/j.canlet.2014.12.051. [DOI] [PubMed] [Google Scholar]
- 38.Katsushima K. Targeting the notch-regulated non-coding RNA TUG1 for glioma treatment. Nat Commun. 2016;7 doi: 10.1038/ncomms13616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Georgakilas A.G., Martin O.A., Bonner W.M. p21: a two-faced genome Guardian. Trends Mol Med. 2017;23:310–319. doi: 10.1016/j.molmed.2017.02.001. [DOI] [PubMed] [Google Scholar]
- 40.Karimian A., Ahmadi Y., Yousefi B. Multiple functions of p21 in cell cycle, apoptosis and transcriptional regulation after DNA damage. DNA Repair. 2016;42:63–71. doi: 10.1016/j.dnarep.2016.04.008. [DOI] [PubMed] [Google Scholar]
- 41.Fischer M., Quaas M., Steiner L., Engeland K. The p53-p21-DREAM-CDE/CHR pathway regulates G(2)/M cell cycle genes. Nucleic Acids Res. 2016;44:164–174. doi: 10.1093/nar/gkv927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lu, Z. & Hunter, T. Ubiquitylation and proteasomal degradation of the p21(Cip1), p27(Kip1) and p57(Kip2) CDK inhibitors. [DOI] [PMC free article] [PubMed]
- 43.Moussa R.S., Park K.C., Kovacevic Z., Richardson D.R. Ironing out the role of the cyclin-dependent kinase inhibitor, p21 in cancer: novel iron chelating agents to target p21 expression and activity. Free Radical Biol Med. 2019;133:276–294. doi: 10.1016/j.freeradbiomed.2018.03.027. [DOI] [PubMed] [Google Scholar]
- 44.Behan F.M. Prioritization of cancer therapeutic targets using CRISPR-Cas9 screens. Nature. 2019 doi: 10.1038/s41586-019-1103-9. [DOI] [PubMed] [Google Scholar]
- 45.Banani S.F., Lee H.O., Hyman A.A., Rosen M.K. Biomolecular condensates: organizers of cellular biochemistry. Nat Rev Mol Cell Biol. 2017;18:285–298. doi: 10.1038/nrm.2017.7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Shin Y., Brangwynne C.P. Liquid phase condensation in cell physiology and disease. Science. 2017;357 doi: 10.1126/science.aaf4382. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Biological information analysis of lncRNA-RP11-732M18.3. (a) Prediction of putative proteins encoded by lncRNA-RP11-732M18.3 using ORF Finder. (b) lncRNA-RP11-732M18.3 lacks the potential to encode any recognizable protein domains, based on standard protein BLAST analysis of ORF Finder from (a). (c) The CSF scores of lncRNA-RP11-732M18.3 were <0. (d) The CSF scores of lncRNA-MALAT1 and glyceraldehyde 3-phosphate dehydrogenase (GAPDH). (e) The expression levels of lncRNA RP11-732 M18.3 in glioma cell lines were higher than the normal cells (n = 3, *p < .05, Student's t-test). (f) Relative expression of lncRNA-RP11-732 M18.3 in xenograft tumor cells. All experiments were performed in triplicate (n = 3, *p < .05, Student's t-test).
lncRNA-RP11-732 M18.3 promoted cell cycle transition. (a) lncRNA-RP11-732M18.3 expression was detected in U87MG, U251, and A172 cells by qRT-PCR after transfection of lentiviruses encoding lncRNA-RP11-732M18.3 short hairpin RNA or an empty vector. All experiments were performed in triplicate (n = 3, *p < .05, Student's t-test). (b) lncRNA-RP11-732M18.3 expression was detected in U87MG, U251, and A172 cells by qRT-PCR after transfection of lentivirus harboring the full-length human lncRNA-RP11-732M18.3 sequence or an empty vector. All experiments were performed in triplicate (n = 3, *p < .05, Student's t-test). (c) Representative images of cell cycle distribution in U87MG and U251 cells following the indicated treatments. (d) Representative images of cell cycle distribution in U87MG and U251 cells following the indicated treatments.
lncRNA-RP11-732M18.3 promoted cell proliferation and inhibited p21 expression in A172 cell lines. (a) Knockdown of lncRNA-RP11-732M18.3 inhibited colony formation of A172 cells. All experiments were performed in triplicate (n = 3, *p < .05, Student's t-test). (b and c) Knockdown of lncRNA-RP11-732 M18.3 inhibited G1/S transition but was promoted by lncRNA-RP11-732 M18.3 overexpression. All experiments were performed in triplicate (n = 3, **p < .01, Student's t-test). (d) lncRNA-RP11-732 M18.3 depletion increased p21 expression, which was decreased by overexpression. All experiments were performed in triplicate (n = 3, **p < .01, Student's t-test).
Knockdown of lncRNA-RP11-732M18.3 promoted p21 expression in xenograft tumor cells. Knockdown of lncRNA-RP11-732M18.3 increased p21 levels in xenograft tumor cells. Left: Representative images of p21 staining of xenografts. Right: Quantitative analysis p21 index. All experiments were performed in triplicate (n = 3, *p < .05, Student's t-test).
lncRNA-RP11-732M18.3 interacted with 14-3-3β/α. (a) Positive and negative controls used for RNA FISH analysis of U87MG cells. (b) RNA FISH assays with the lncRNA-RP11-732M18.3 probe in U251 and A172 cells. (c) Silver staining, SDS-PAGE, and ChIRP showing enrichment of human lncRNA-RP11-732M18.3 in U87MG cells. (d) Biological process analysis of proteins from ChIRP/MS. (e) Peptide summary report of 14-3-3β/α. (f) Mass spectra of 14-3-3β/α. (g) catRAPID express predicted that 14-3-3β/α might interact with the forward part of lncRNA-RP11-732M18.3. (h) Silver staining, sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE), and co-immunoprecipitation enrichment of human 14-3-3β/α in U87MG cells. The arrow indicates the position of 14-3-3β/α. (i) Expression levels of the 14-3-3β/α, and UBE2E1 proteins were detected in U87MG cells by qRT-PCR after transfection of the indicated small interfering RNAs. All experiments were performed in triplicate. (n = 3, *p < .05, Student's t-test).