Abstract
Grapevine is one of the most important fruit species in the world. In order to better understand genetic basis of traits variation and facilitate the breeding of new genotypes, we sequenced, assembled, and annotated the genome of the American native Vitis riparia, one of the main species used worldwide for rootstock and scion breeding. A total of 164 Gb raw DNA reads were obtained from Vitis riparia resulting in a 225X depth of coverage. We generated a genome assembly of the V. riparia grape de novo using the PacBio long-reads that was phased with the 10x Genomics Chromium linked-reads. At the chromosome level, a 500 Mb genome was generated with a scaffold N50 size of 1 Mb. More than 34% of the whole genome were identified as repeat sequences, and 37,207 protein-coding genes were predicted. This genome assembly sets the stage for comparative genomic analysis of the diversification and adaptation of grapevine and will provide a solid resource for further genetic analysis and breeding of this economically important species.
Subject terms: Plant domestication, Plant breeding
| Design Type(s) | sequence assembly objective • genetic mapping and linkage analysis objective • sequence annotation objective |
| Measurement Type(s) | whole genome sequencing assay |
| Technology Type(s) | DNA sequencing |
| Factor Type(s) | |
| Sample Characteristic(s) | Vitis riparia • leaf |
Machine-accessible metadata file describing the reported data (ISA-Tab format)
Background & Summary
Since few decades and the development of sequencing technologies, the number of species whose genome has been totally sequenced has increased exponentially. There is a large variability for the quality of all the sequences assemblies. In 2017, 72 plant reference quality genome assemblies were reported in NCBI1. For plant breeding, the availability of a contiguous genome sequence provides a tool to better identify genes underlying traits and how they may be regulated by various environmental parameters in different genetic backgrounds. At the simplest, it allows for association of genetic markers for selection and introgression of traits across germplasm to enable the development of novel products for consumers2,3.
As an important crop, Vitis vinifera was one of the first higher plant species whose genome was sequenced by a French-Italian consortium4. The consortium decided to sequence a near homozygous V. vinifera cultivar related to Pinot Noir (PN40024) in order to facilitate the sequence assembly by limiting sequence variability. To date, this genome still stands as the reference for the grapevine community, but grapevine intra species and interspecies diversity makes using a single reference genome inadequate for studying the function of other genotypes. In order to address the variations in a cultivated V. vinifera variety, the Pinot Noir genome was sequenced using Sanger sequencing providing a high quality draft of the genome with about 10X coverage5. Next Generation Sequencing reads are too short to resolve abundant repeats in particular in plants genome, leading to incomplete or ambiguous assemblies6. Few attempts to produce high quality grapevine genomes were undertaken in grapevine and produced valuable data to study the genetic variations of V. vinifera cv. Tannat7 and cv. Thompson seedless8 through comparison with the reference genomes.
The last few years have seen rapid innovations in sequencing technologies and improvement in assembly algorithms that enabled the creation of highly contiguous genomes. The development of third generation sequencing technologies that deliver long reads from single molecules and carry the necessary information to phase haplotypes over several kilobases have greatly improved the feasibility of de novo assemblies9–11. Sequences of V. vinifera cv. Cabernet Sauvignon were first released12 using PacBio sequencing and FALCON, and FALCON-Unzip pipeline12. This generated a 591 Mbp haplotype genome from a set of 718 primary contigs, and a set of correlated 2,037 haplotigs spanning 367 Mbp. The total p-contig size was larger than the estimated genome size of V. vinifera (~500 Mbp) suggesting that in some cases FALCON-Unzip underestimated the alternative haplotype sequences because of high heterozygosity between homologous regions, which is common in grapevine13,14. Later, the PacBio assembly and annotation of V. vinifera cv Chardonnay variety provided after curation of artefactual contig assignment, 854 p-tigs and 1883 h-tigs, totaling 490 Mb and 378 Mb15. More recently, another version of the Chardonnay genome was proposed with a different level of curation at 605 Mb16.
An evaluation of genetic diversity based on a panel of 783 V. vinifera varieties using 10 K SNPs revealed a high level of diversity (He = 0.32) and confirmed the close pedigree relationship within the cultivated grapevine due to the wide use of the most interesting parents during domestication and early selection by humans17. Considering that grape cultivation currently faces severe pathogen pressures and climate change, we assume that the exploitation of the natural genetic diversity may ensure the long-term sustainability of the grape and wine industries18. Grapes belong to the genus Vitis, which includes over 60 inter-fertile species. The most common grape cultivars derive their entire ancestry from the species V. vinifera, but wild relatives have also been exploited to create hybrid cultivars, often with increased disease resistances19.
To date, no wild Vitis genomes have been released so far and the only whole genome sequences for grape are from V. vinifera varieties and yet there is a clear need for genetic resources20. Here, we report the first de novo assembly and genome annotation of the North American native grape V. riparia. Using the latest sequencing technologies, we show that 10x Genomics Chromium data can be combined with long read PacBio sequencing to effectively determine genome phasing. The phased haplotypes of V. riparia genome will greatly contribute to give more insight into the functional consequences of genetic variants.
Methods
Sample collection, library construction and whole genome sequencing
The Vitis riparia Gloire de Montpellier (RGM) selection was obtained in 1880 by L. Vialla and R. Michel from North American collections and is the only commercially available pure V. riparia stock. RGM clone #1030 and the European native Vitis vinifera Cabernet sauvignon (CS) clone #15 were grown at INRA, Bordeaux (France). A F1 segregating population of 114 individuals named CSxRGM1995-1 was derived from the cross between CS and RGM21. This population was genotyped using the GBS approach22 to create a high resolution genetic map to assist in anchoring and orienting the assembled V. riparia genome scaffolds.
Total DNA was isolated and extracted using QIAGEN Genomic-tips 100/G kit (Cat No./ID: 10243) following the tissue protocol extraction. Briefly, 1 g of young leaf material was ground in liquid nitrogen with mortar and pestle. After 3 h of lysis and one centrifugation step, the DNA was immobilized on the column. After several washing steps, DNA is eluted from the column, then desalted and concentrated by alcohol precipitation. The pellet is resuspended in TE buffer.
Three PacBio libraries with a 20-kb insert size were also constructed and sequenced on RSII platforms (97.71 Gb data; ~118-fold covering), following the standard PacBio protocol of Sequencing Kit 1.2.1 (Pacific Biosciences, USA). Four 10x Chromium Genomics libraries were constructed using the ChromiumTM Genome Solution (10X Genomics, USA), and 2 × 150 bp sequenced on Illumina HiSeq3000, producing ~350 million paired-end linked-reads (~ 107-fold covering). Finally, 2 libraries for 2 × 100 bp sequencing were built with different insert sizes: 500 bp for paired-end (PE) and 6 kb for mate-pair (MP), based on the standard Illumina protocol and sequenced on the Illumina HiSeq2500. The raw reads were trimmed before being used for subsequent genome assembly. For Illumina HiSeq sequencing, the adaptor sequences, the reads containing more than 10% ambiguous nucleotides, as well as the reads containing more than 20% low-quality nucleotides (quality score less than 5), were all removed. After data cleaning and data preprocessing, we obtained a total of 164 Gb of clean data (52 Gb PacBio data, 59 Gb 10X Genomics, 33 Gb PE reads and 20 Gb MP reads,), representing 331X coverage of the V. riparia genome (Table 1).
Table 1.
Data count and library informations for Vitis riparia genome sequencing.
| Sequencing platform | Insert size (bp) | Read length (bp) | Number of sequences (million) | Number of bases (billion) | Sequence depth | Application |
|---|---|---|---|---|---|---|
| PacBio | NA | 7054 | 8.3 | 59 | 118X | Genome assembly |
| 10X Chromium | 400 | 2 × 150 | 350 | 52 | 107X | Genome scaffolding and phasing |
| Illumina | 400 (pair end) | 2 × 100 | 331 | 33 | 66X | Genome survey and genomic base correction |
| 6,000 (mate pair) | 2 × 100 | 200 | 20 | 40X | ||
| Total | 164 | 331X |
Genome size and heterozygosity estimation
Lodhi and Reisch23 estimated the genome size in grape to be approximately 475 Mb based on measurements using flow cytometry for 19 species including wild Vitis species, V. vinifera and V. labrusca cultivars. The measurements showed intraspecific variation in genome size between different varieties of Vitis vinifera ranging from 1C = 415 to 511 Mb, and between different North America Vitis species ranging from 1C = 411 to 541 Mb, with V. riparia around 470 Mb. Genome sequencing of different V. vinifera varieties gave values in the same range or greater depending on the methods of sequencing and assembly. In order to verify these values, we estimated genome size of V. riparia by the k-mer method24,25 using data from pair-end and mate-pair Illumina sequencing. By analyzing the 21-mers depth distribution, a total of ~50 billion k-mers were estimated with a peak frequency of 100, corresponding to a genome size of 494 Mb and the estimated repeat sequencing ratio was 33.74%. In this study, V. riparia heterozygosity was estimated to be 0.46% (mean distance 1 SNP each 217 bp between heterozygous SNPs) from 10x Chromium Genomics data processing.
De novo Genome assembly and scaffolding of the Vitis riparia genome
We employed a hybrid de novo whole-genome assembly strategy, combining both short linked-reads and PacBio long reads data. Genome assembly was first performed on full PacBio cleaned reads using FALCON v0.3.026. Error correction and pre-assembly were carried out with the FALCON/FALCON Unzip pipeline after evaluating the outcomes of using different parameters in FALCON during the pre-assembly process. Based on the contig N50 results, a length_cutoff of 5 kb and a length_cutoff_pr of 8 kb for the assembly step were ultimately chosen. The draft assembly was polished using Quiver27, which mapped the PacBio reads to the assembled genome with the BLASER pipeline28. Haplotypes were separated during assembly using FALCON-Unzip and the preliminary genome assembly was approximately 530 Mb (1,964 primary-contigs) and 317 Mb (3,344 haplotigs). A summary of the assembly statistics can be found in Table 1. Assembly was then processed with Purge Haplotigs13 to investigate the proper assignment of contigs, followed by 2 rounds of polishing to correct residual SNP and INDELs errors with Pilon v1.22 software29 using high-coverage (~106X) Illumina paired-end and mate pair data.
The 10x Chromium Genomics linked-reads were used to produce a separate V. riparia assembly using the Supernova assembler option–style = pseudohap2 and created two parallel pseudohaplotypes30. The mean input DNA molecule length reported by the Supernova assembler was 45 kb and the assembled genome size was 424 Mb with a N50 scaffold of 711 kb.
Subsequently, the PacBio assembly was scaffolded with the 10x Chromium Genomics one using the hybrid assembler LINKS31 with 7 iterations, producing 870 scaffolds spanning 500 Mb with N50 = 964 kb and L50 = 255 (Table 2). Finally, genome phasing was reconstituted using Long Ranger analysis pipeline that processes Chromium sequencing output to align reads and call and phase SNPs, indels, and structural variants on the basis of molecular barcodes information.
Table 2.
Summary of the V. riparia genome assembly and comparison with with V. vinifera varieties.
| Vitis vinifera | Vitis riparia | |||
|---|---|---|---|---|
| PN40024 | Cabernet sauvignon | Chardonnay | Riparia Gloire de Montpellier | |
| Technology | Sanger | PacBio | PacBio | PacBio/10X Chromium |
| Genome coverage | 12X | 140X | 115X | 225X |
| Contig length (Mb) | NA |
591 p-tigs 368 h-tgs |
490 p-tigs 378 h-tgs |
530 p-tigs 317 h-tgs |
| Number of contigs | 14,665 |
718 p-tigs 2,037 h-tgs |
854 p-tigs 1,883 h-tgs |
1,964 p-tigs 3,344 h-tgs |
| Number of scaffolds | 2,065 | NA | NA | 174 |
| N50 (kb) | 103 | 2,170 | 935 | 964 |
| Total length (Mb) | 486 | 591 | 490 | 500 |
| Number of coding genes | 42,414 (Cost.v3) | 36,687 | 29,675 | 37,207 |
| BUSCO | C:95.8% F:1.5% M:2.7% | C:94.0% F:2.0% M:4.0% | C:95.0% F:1.6% M:3.4% | C:95.4% F:1.1% M:3.5% |
Genotyping by Sequencing and genetic mapping
Two 96-plex GBS libraries (Keygene N.V. owns patents and patent applications protecting its Sequence Based Genotyping technologies) were constructed for the two parents (two replicates for each) and the 114 F1 plants of the cross CS × RGM. Raw reads were checked with FastQC32, demultiplexed with a custom script and cleaned with CutAdapt33. Cleaned reads were then mapped to the V. riparia RGM scaffolds previously obtained, the V. vinifera Cabernet Sauvignon contigs12 and V. vinifera PN40024 genome assemblies4 for SNP calling. Aligned on these genomes were performed using BWA34, SAMtools35 and Picard tools36 and SNP genotypes were detected with GATK37 using the hardfilter parameters38. In the variant call format (VCF) output file only sites with less than 20% missing data and a minimum allele frequency (MAF) ≥ 0.2 were retained. The SNP set was parsed into two data sets based on a pseudo-test cross mapping strategy39 using major_minor and get_pseudo_test_cross scripts from Hetmapps40. The segregation ratios of markers in the population were examined by Chi-square analysis. Markers with segregation ratios that differed from expected 1:1 at P < 0.05 were classified as segregation distortion markers and discarded. The RGM and CS sets contain 1591 and 2359 SNPs respectively. Linkage groups (LGs) were determined using software JoinMap® 4.141,42 and Rqtl43. LG were formed with a logarithm of odds (LOD) threshold of 6 and a maximum recombination frequency of 0.45. The 19 LGs that corresponded to the 19 chromosomes of grapevine were reconstructed and leaded to a total genetic map length of 2,268 cM and 2,514 cM for RGM and CS respectively.
Pseudo-molecule construction
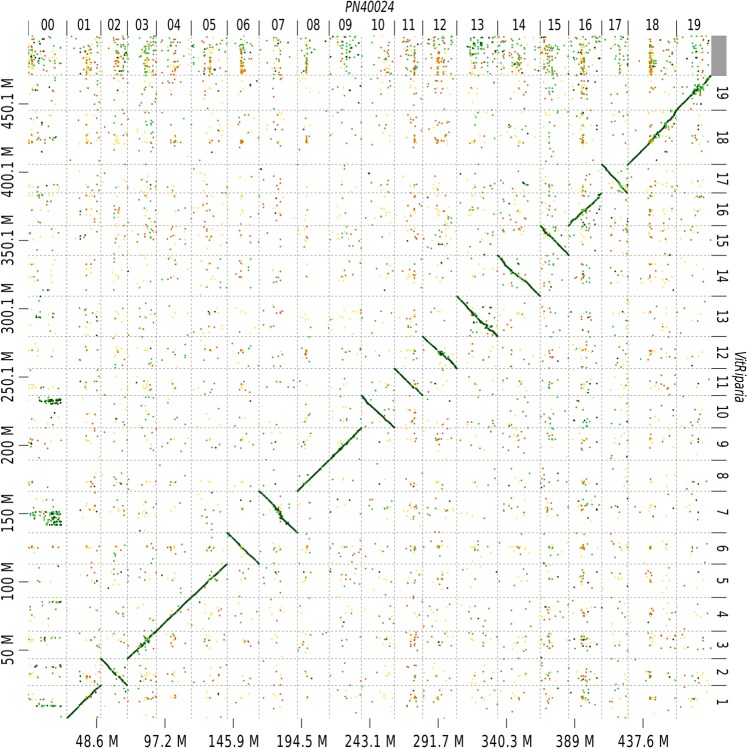
The PacBio/10x Chromium Genomics hybrid scaffolding was organized into pseudo-molecules using GBS markers information from the CS × RGM genetic map. Scaffolds were anchored and oriented SNP using AllMaps44 with the unequal weights2 parameters for a single run for the entire genome. Final pseudo-molecules were named according to Vitis vinifera PN40024 reference genome using SNP identification through SNP calling on this reference. Since PN40024 genome is the only one available who has been scaffolded into pseudo-molecules, collinearity with V. riparia was evaluated using D-GENIES45 and showed extremely high conservation along the 19 chromosomes of the species (Fig. 1) even if the North American and Eurasian Vitis species diverged approximately 46.9 million years ago46.
Fig. 1.
Comparison of Vitis riparia hybrid scaffolds with the reference PN40024 assembly. Hybrid scaffolds (Y-axis) were aligned to all 19 PN40024 chromosomes (X-axis) using D-GENIES and alignments were subsequently filtered for 1-on-1 alignments and rearrangements with a 20 Kbps length cutoff.
Genome annotation and gene prediction
Consistent with observations that long reads sequencing technologies are a better solution for resolving repeat sequences, we found that known repetitive elements accounted for 170 Mb (33.94%) of the genome in V. riparia. This is a lower proportion among grape genomes when comparing published values to date. However, when comparisons are performed with the same analysis workflow and tools47,48, the percentages obtained between the two genotypes were in the same range (Online-only Table 1). Similar to other grape genomes, long terminal repeat (LTR) elements constituted the highest proportion of all repeated elements in V. riparia, (21.44%) with Copia and Gypsy families accounting for 8.33% and 12.66% respectively. The Long Interspersed Nuclear Elements (LINEs) and Miniature Inverted-repeat Transposable Elements (MITEs) represented 3.61% and 6.02% of the whole genome respectively.
Online-only Table 1.
Repeated elements present in the Vitis riparia genome.
| number of elements | length occupied (base pairs) | percentage of sequence | ||
|---|---|---|---|---|
| Retroelements | 95,441 | 125,292,108 | 25.05% | |
| SINEs | 0 | 0 | 0.00% | |
| Penelope | 0 | 0 | 0.00% | |
| LINEs: | 17,557 | 18,048,495 | 3.61% | |
| CRE/SLACS | 0 | 0 | 0.00% | |
| L2/CR1/Rex | 0 | 0 | 0.00% | |
| R1/LOA/Jockey | 0 | 0 | 0.00% | |
| R2/R4/NeSL | 0 | 0 | 0.00% | |
| RTE/Bov-B | 0 | 0 | 0.00% | |
| L1/CIN4 | 17,557 | 180,48,495 | 3.61% | |
| LTR elements: | 77,884 | 107,243,613 | 21.44% | |
| BEL/Pao | 0 | 0 | 0.00% | |
| Ty1/Copia | 27,636 | 41,679,808 | 8.33% | |
| Gypsy/DIRS1 | 48,176 | 63,337,662 | 12.66% | |
| Retroviral | 0 | 0 | 0.00% | |
| DNA transposons | 80,801 | 30,107,834 | 6.02% | |
| hobo-Activator | 14,757 | 6,431,569 | 1.29% | |
| Tc1-IS630-Pogo | 0 | 0 | 0.00% | |
| En-Spm | 436 | 1,086,471 | 0.22% | |
| MuDR-IS905 | 0 | 0 | 0.00% | |
| PiggyBac | 0 | 0 | 0.00% | |
| Tourist/Harbinger | 34,070 | 82,90 ,818 | 1.66% | |
| Other (Mirage, P-element, Transib) | 0 | 0 | 0.00% | |
| Rolling-circles | 0 | 0 | 0.00% | |
| Unclassified | 53 | 67,393 | 0.01% | |
| Total interspersed repeats | 155,467,335 | 31.09% | ||
| Small RNA | 202 | 43,383 | 0.01% | |
| Satellites | 1,282 | 1,975,618 | 0.40% | |
| Simple repeats | 222,322 | 9,483,835 | 1.90% | |
| Low complexity | 54,205 | 2,848,156 | 0.57% | |
After repeat masking, the genome was ab initio annotated using MAKER-P pipeline49,50, SNAP51 and Augustus52 gene finder with 3 rounds of Maker and an Augustus prediction. Structural annotation was then followed with an Interproscan functional annotation and putative gene function assignation using BLAST on UniProtKB. MAKER-P quality metrics with a threshold of AED < 0.5 were chosen to retain the set of predicted genes. We finally generated a gene set of 37,207 protein-coding genes (11,434_AED < 0.1; 8,638_0.1 ≤ AED < 0.2; 5,748_0.2 ≤ AED < 0.3; 5,418_0.3 ≤ AED < 0.4; 5,969_0.4 ≤ AED < 0.5) with 31,240 of them coupled with an evidence of protein function.
To facilitate genomic investigations for the community, a JBrowse Genome Browser53 was set up for V. riparia pseudo-molecules and is available from https://www6.bordeaux-aquitaine.inra.fr/egfv/.
Data Records
The V. riparia genome project was deposited at NCBI under BioProject number PRJNA512170 and BioSample SAMN10662253. The DNA sequencing data from Illumina, PacBio and 10x Genomics have been deposited in the Sequence Read Archive (SRA) database under accession SRP17486654 from SRX5189632 to SRX5189680. This Whole Genome Shotgun project has been deposited at DDBJ/ENA/GenBank under the accession SJAQ0000000055. The versions described in this paper are version SJAQ01000000 and genome ID 13150 (https://www.ncbi.nlm.nih.gov/genome/13150). Genetic mapping data and structural and functional annotation file of the Vitis riparia assembly are available on figshare56.
Technical Validation
To evaluate the accuracy and completeness of the V. riparia assembly, genome features were compared to those of V. vinifera (Table 2). We found that both contig and scaffold N50 lengths of Vitis riparia reached considerable continuity. The Guanine-Cytosine content (GC = 34.32%) was similar to those of V. vinifera Chardonnay (34.43%).
To further assess the accuracy of the V. riparia genome assembly, the NGS-based short reads from whole-genome sequencing data were also aligned against the genome assembly using BWA mem57. We found that 98.4% of the reads were reliably aligned to the genome assembly, and 95.8% of the reads were properly aligned to the genome with their mates. Paired-end reads data were not used during the contig assembly, thus the high alignment ratio demonstrated the high quality of contig assembly.
The assembled genome was also subjected to Benchmarking Universal Single-Copy Orthologs58, which quantitatively assesses genome completeness using evolutionarily informed expectations of gene content from near-universal single-copy orthologs, using the genes in the embryophyta release 9 dataset (embryophyta.odb9). The BUSCO results showed that 96.5% of conserved BUSCO proteins were detected in the V. riparia assembly, including 1.1% of fragment BUSCO proteins (Table 2). Overall, these metrics compare well with other recently published grape genomes, providing a high quality genome sequences for the following functional investigations.
ISA-Tab metadata file
Acknowledgements
We thank Dr. Gregory Gambetta for critical review of the manuscript. We thank the INRA Genotoul bioinformatics platform (http://genotoul.toulouse.inra.fr) for providing computational resources and support and INRA-CNRGV (http://cnrgv.toulouse.inra.fr/) for V. riparia HMW DNA extraction. Sequencing was performed at the GeT core facility, Toulouse, France, supported in part by France Génomique National infrastructure (contract ANR-10-INBS-09). We thank Dr. Timothée Flutre and Amandine Launay for their help in coordinating the FruitSelGen project and in acquiring GBS data. This work was supported by a grant overseen by the French National Research Agency (ANR) as part of the ANR-09-GENM-024 “Vitsec” program and by the University of Bordeaux.
Online-only Table
Author Contributions
Author P.-F.B. and N.O. conceived the project. N.G. and P.-F.B. assembled the genomes, performed the genome annotation and downstream analyses. B.R. performed GBS analysis and genetic mapping. C.L.R. and S.V. performed sequencing. P.-F.B. wrote the paper.
Code Availability
1. GBS demultiplexing
2. Filters FASTQ files with CASAVA 2.20
fastq_illumina_filter –keep N -v -v -o good_reads.fq raw_reads.fastq
3. Cutadapt (regular 3′ adapter)
https://cutadapt.readthedocs.io/en/stable/guide.html
cutadapt -a AGATCGGAAGAGCGGTTCAGCAGGAATGCCGAG
-A AGATCGGAAGAGCGTCGTGTAGGGAAAGAGTGT
-G CTCGGCATTCCTGCTGAACCGCTCTTCCGATCT
-g ACACTCTTTCCCTACACGACGCTCTTCCGATCT -u7 -U7 -m10
4. Burrows-Wheeler Alignment – BWA-MEM
http://bio-bwa.sourceforge.net/bwa.shtml
bwa mem ref.fa read1.fastq.gz read2.fastq.gz > aligned.reads.sam with these options:
-M Mark shorter split hits as secondary (for Picard compatibility)
-R Complete read group header line with ‘\t’ used in STR to be converted to a TEB in the output SAM. An example is ‘@RG\tID:\tSM:\tPL:\tLB:’
5. Picard tools
https://broadinstitute.github.io/picard/
SortSam : java –jar picard.jar SortSam with these options: INPUT (BAM file), OUTPUT (BAM file), SORT_ORDER
MarkDuplicates : java –jar picard.jar MarkDuplicates with these options: INPUT (BAM file), OUTPUT (BAM file), METRIC_FILE (file)
BuildBamIndex : java –jar picard.jar BuildBamIndex with these options: INPUT (BAM file)
6. GATK tools
HaplotypeCaller : java –jar GenomeAnalysisTK.jar –T HaplotypeCaller –R ref.fasta –I file.bam –genotyping_mode DISCOVERY –drf DuplicateRead –emitRefConfidence GVCF –o file.g.vcf
CombineGVCFs : java –jar GenomeAnalysisTK.jar –T CombineGVCFs –R ref.fasta –drf DuplicateRead –G Standard –G AS_Standard –variant sample1 to sample‘n’.g.vcf –o cohort_file.g.vcf
GenotypeGVCFs : java –jar GenomeAnalysisTK.jar –T GenotypeGVCFs –R ref.fasta –drf DuplicateRead –G Standard –G AS_Standard –variant cohort_file.g.vcf –o final_file.vcf
SelectVariants : java –jar GenomeAnalysisTK.jar –T SelectVariants –R ref.fasta –V final_file.vcf –selectType SNP –o file_snps.vcf
VariantFiltration : java –jar GenomeAnalysisTK –T VariantFiltration –R ref.fasta –V file_snps.vcf–filterExpression « QD < 2.0 || FS > 60.0 || MQ < 40.0 || MQRankSum < −12.5 || ReadPosRankSum < −8.0 » –filteredName «FILTER » -o filtered_snps.vcf
7. VCF filtering
vcftools –vcf filtered_snps.vcf –remove-filtered-all –recode –out filteredFinal_snps.vcf
8. Falcon and Falcon_Unzip Assembly for SMRT sequencing
https://github.com/PacificBiosciences/FALCON/wiki
https://github.com/PacificBiosciences/FALCON_unzip/wiki
Main parameters: length_cutoff = 5000, length_cutoff_pr = 5000
pa_HPCdaligner_option = -v -dal128 -e0.70 -M40 -l2500 -k17 -h500 -w7 -s100
ovlp_HPCdaligner_option = -v -dal128 -M40 -k19 -h500 -e.96 -l1500 -s100
pa_DBsplit_option = -a -x500 -s200
ovlp_DBsplit_option = -s200
falcon_sense_option = –output_multi –output_dformat –min_idt 0.80 –min_cov 4 max_n_read 400 –n_core 16
falcon_sense_skip_contained = False
overlap_filtering_setting = –max_diff 120 –max_cov 120 –min_cov 4 –n_core 24
9. Purge Haplotigs
https://bitbucket.org/mroachawri/purge_haplotigs/src/master/
purge_haplotigs readhist -b aligned.bam -g genome.fasta
10. Supernova Assembly for 10x Chromium sequencing
https://support.10xgenomics.com/de-novo-assembly/software/overview/latest/welcome
Option pseudohap2 style output
11. Scaffolding Falcon assembly with LINKS using Supernova outputs Assembly
https://github.com/bcgsc/LINKS
LINKS -f.fa -s fileofname.fofn -b cns1-linked_draft -d 5000 -t 100 -k 19 -l 5 -a 0.3
LINKS -f.fa -s fileofname.fofn -b cns2-linked_draft -d 6000 -t 80 -k 19 -l 15 -a 0.3
LINKS -f.fa -s fileofname.fofn -b cns3-linked_draft -d 7000 -t 60 -k 19 -l 20 -a 0.3
LINKS -f.fa -s fileofname.fofn -b cns4-linked_draft -d 10000 -t 30 -k 19 -l 20 -a 0.3
LINKS -f.fa -s fileofname.fofn -b cns5-linked_draft -d 15000 -t 30 -k 19 -l 20 -a 0.3
LINKS -f.fa -s fileofname.fofn -b cns6-linked_draft -d 50000 -t 30 -k 19 -l 30 -a 0.3
LINKS -f.fa -s fileofname,fofn -b cns7-linked_draft -d 75000 -t 30 -k 19 -l 40 -a 0.3
12. Improving quality with PILON and Illumina sequencing
https://github.com/broadinstitute/pilon/wiki/Requirements-&-Usage
13. Allmaps pseudomolecules scaffolding
https://github.com/tanghaibao/jcvi/wiki/ALLMAPS
14. Assembly evaluation with BUSCO v3
15. Vitis TE(s) Identification using RepeatMasker
16. Annotation with MAKER_P pipeline, SNAP and Augustus gene finder
http://www.yandell-lab.org/publications/pdf/maker_current_protocols.pdf
https://bmcbioinformatics.biomedcentral.com/articles/10.1186/1471-2105-5-59
https://github.com/Gaius-Augustus/Augustus
DB (vitis) AND “Vitis”[porgn] from https://www.ncbi.nlm.nih.gov
EST DB (vitis) AND “Vitis”[porgn] from https://www.ncbi.nlm.nih.gov
- First run : rm_pass = 0, est2genome = 1 and protein2genome = 1
gff3_merge -d master_datastore_index.log
maker2zff -c 0 -e 0 -o 0 -x 0.05 maker1.gff
fathom -categorize 1000 genome.ann genome.dna
fathom -export 1000 -plus uni.ann uni.dna
forge export.ann export.dna
hmm-assembler.pl RGM. >snap1.hmm
- Second run: rm_pass = 1, est2genome = 0, protein2genome = 0, maker_gff = maker1.gff, snaphmm = snap1.hmm leading to a maker2.gff3 and a snap2.hmm files.
gff3_merge -d master_datastore_index.log
maker2zff -c 0 -e 0 -o 0 -x 0.05 maker2.gff
fathom -categorize 1000 genome.ann genome.dna
fathom -export 1000 -plus uni.ann uni.dna
forge export.ann export.dna
hmm-assembler.pl RGM. >snap2.hmm
Run Augustus:
zff2gff3.pl genome.ann | perl -plne ‘s/\t(\S+)$/\t\.\t$1/’ > genome.gff3
autoAug.pl –genome = ../pilon2.fasta –species = RGM18 –cdna = sequence_est_ncbi.fasta –trainingset = genome.gff3 –singleCPU –v –useexisting
- Third run : rm_pass = 1, est2genome = 0, protein2genome = 0, maker_gff = maker2.gff, snaphmm = snap2.hmm, augustus_species = RGM18 leading to a maker3.gff3, maker3.transcripts.fasta and maker3.proteins.fasta structural prediction.
gff3_merge -d master_datastore_index.log
fasta_merge -d master_datastore_index.log
17. Interproscan functional annotation and putative gene function assignation
Download protein DB from http://www.uniprot.org
makeblastdb -in protein_db.fasta -input_type fasta -dbtype prot
blastp -db protein_db.fasta -query maker3.proteins.fasta -out maker3.proteins.blastp -evalue 0.000001 -outfmt 6 -max_hsps 1
maker_functional_gff protein_db.fasta maker3.proteins.blastp maker3.gff3 ≫ maker3.putative.gff3
maker_functional_fasta protein_db.fasta maker3.proteins.blastp maker3.proteins.fasta ≫ maker3.putative.proteins.fa
maker_functional_fasta protein_db.fasta maker3.proteins.blastp maker3.transcripts.fasta ≫ maker3.putative.transcripts.fa
Run Interproscan
interproscan.sh –iprlookup –goterms -f tsv -i maker3.putative.proteins.fa -pa -b RGM.annotated.proteins
18. Assembly validation using WBA mem
bwa mem -M -t 20 VitRiparia.fasta reads_pe.R1.fastq reads_pe.R2.fastq > aln_pe_reads.sam
samtools view -bS aln_pe_reads.sam -o aln_pe_reads.bam #| samtools sort - aln_reads.sorted.bam
samtools sort -o aln_pe_reads.sorted.bam aln_pe_reads.bam
bamtools stats -in aln_pe_reads.sorted.bam > bamstat_pe.reads
Competing Interests
The authors declare no competing interests.
Footnotes
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Change history
10/30/2019
An amendment to this paper has been published and can be accessed via a link at the top of the paper
ISA-Tab metadata
is available for this paper at 10.1038/s41597-019-0133-3.
References
- 1.Peterson, D. G. & Arick, M. Sequencing Plant Genomes. (Progress in Botany. Springer, Berlin, Heidelberg 2018).
- 2.Nguyen KL, Grondin A, Courtois B, Gantet P. Next-generation sequencing accelerates crop gene discovery. Trends Plant Sci. 2018;24:263–274. doi: 10.1016/j.tplants.2018.11.008. [DOI] [PubMed] [Google Scholar]
- 3.Scheben A, Yuan Y, Edwards D. Advances in genomics for adapting crops to climate change. Curr. Plant Biol. 2016;6:2–10. [Google Scholar]
- 4.Jaillon O, et al. The grapevine genome sequence suggests ancestral hexaploidization in major angiosperm phyla. Nature. 2007;449:463–467. doi: 10.1038/nature06148. [DOI] [PubMed] [Google Scholar]
- 5.Velasco R, et al. A High Quality Draft Consensus Sequence of the Genome of a Heterozygous Grapevine Variety. PLoS ONE. 2007;2:e1326. doi: 10.1371/journal.pone.0001326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Alkan C, Sajjadian S, Eichler EE. Limitations of next-generation genome sequence assembly. Nat. Methods. 2010;8:61–65. doi: 10.1038/nmeth.1527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Da Silva C, et al. The high polyphenol content of grapevine cultivar Tannat berries is conferred primarily by genes that are not shared with the reference genome. Plant Cell. 2013;25:4777–4788. doi: 10.1105/tpc.113.118810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Di Genova A, et al. Whole genome comparison between table and wine grapes reveals a comprehensive catalog of structural variants. BMC Plant Biol. 2014;14:7. doi: 10.1186/1471-2229-14-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jiao WB, Schneeberger K. The impact of third generation genomic technologies on plant genome assembly. Curr. Opin. Plant Biol. 2017;36:64–70. doi: 10.1016/j.pbi.2017.02.002. [DOI] [PubMed] [Google Scholar]
- 10.Li C, Lin F, An D, Wang W, Huang R. Genome Sequencing and Assembly by Long Reads in Plants. Genes. 2018;9:6. doi: 10.3390/genes9010006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Treangen TJ, Salzberg SL. Repetitive DNA and next-generation sequencing: computational challenges and solutions. Nat. Rev. Genet. 2012;13:36–46. doi: 10.1038/nrg3117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chin CS, et al. Phased diploid genome assembly with single-molecule real-time sequencing. Nat. Methods. 2016;13:1050–1054. doi: 10.1038/nmeth.4035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Roach MJ, Schmidt SA, Borneman AR. Purge Haplotigs: allelic contig reassignment for third-gen diploid genome assemblies. BMC Bioinformatics. 2018;19:460. doi: 10.1186/s12859-018-2485-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Vinson JP, et al. Assembly of polymorphic genomes: algorithms and application to Ciona savignyi. Genome Research. 2005;15:1127–35. doi: 10.1101/gr.3722605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Roach MJ, et al. Population sequencing reveals clonal diversity and ancestral inbreeding in the grapevine cultivar Chardonnay. PLoS Genet. 2018;14:e1007807. doi: 10.1371/journal.pgen.1007807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhou, Y. S. et al. Structural variants, clonal propagation, and genome evolution in grapevine (Vitis vinifera) Preprint at, 10.1101/508119 (2018).
- 17.Laucou V, et al. Extended diversity analysis of cultivated grapevine Vitis vinifera with 10 K genome-wide SNPs. PLoS One. 2018;13:e0192540. doi: 10.1371/journal.pone.0192540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Myles S, et al. Genetic structure and domestication history of the grape. Proc Natl Acad Sci USA. 2011;108:3530–3535. doi: 10.1073/pnas.1009363108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Migicovsky Z, et al. Genomic ancestry estimation quantifies use of wild species in grape breeding. BMC Genomics. 2016;17:478. doi: 10.1186/s12864-016-2834-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.FAO Commission on genetic resources for food and agriculture assessment. The state of the world’s biodiversity for food and agriculture (2019).
- 21.Marguerit E, et al. Genetic dissection of sex determinism, inflorescence morphology and downy mildew resistance in grapevine. Theor. Appl. Genet. 2009;118:1261–1278. doi: 10.1007/s00122-009-0979-4. [DOI] [PubMed] [Google Scholar]
- 22.Elshire RJ, et al. A robust, simple genotyping-by-sequencing (GBS) approach for high diversity species. PLoS ONE. 2011;6:e19379. doi: 10.1371/journal.pone.0019379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lodhi MA, Reisch BI. Nuclear DNA content of Vitis species, cultivars, and other genera of the Vitaceae. Theor. Appl. Genet. 1995;90:11–16. doi: 10.1007/BF00220990. [DOI] [PubMed] [Google Scholar]
- 24.Liu, B. et al. Estimation of genomic characteristics by analyzing k-mer frequency in de novo genome projects. Preprint at, https://arxiv.org/abs/1308.2012 (2012).
- 25.Marcais G, Kingsford K. A fast, lock-free approach for efficient parallel counting of occurrences of k-mers. Bioinformatics. 2011;27:764–770. doi: 10.1093/bioinformatics/btr011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.PacBio FALCON, https://github.com/PacificBiosciences/FALCON.
- 27.Pacific Biosciences, SMRT tools, https://www.pacb.com/wp-content/uploads/SMRT-Tools-Reference-Guide-v4.0.0.pdf.
- 28.Chaisson MJ, Tesler G. Mapping single molecule sequencing reads using basic local alignment with successive refinement (BLASR): application and theory. BMC Bioinformatics. 2012;13:238. doi: 10.1186/1471-2105-13-238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Walker BJ, et al. Pilon: an integrated tool for comprehensive microbial variant detection and genome assembly improvement. PLoS One. 2014;9:e112963. doi: 10.1371/journal.pone.0112963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Weisenfeld NI, Kumar V, Shah P, Church DM, Jaffe DB. Direct determination of diploid genome sequences. Genome research. 2017;27:757–767. doi: 10.1101/gr.214874.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Warren RL, et al. LINKS: Scalable, alignment-free scaffolding of draft genomes with long reads. Gigascience. 2015;4:35. doi: 10.1186/s13742-015-0076-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Andrews, S. Fastqc: a quality control tool for high throughput sequence data, http://www.bioinformatics.babraham.ac.uk/projects/fastqc (2010).
- 33.Martin M. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet.journal. 2011;17:10–12. [Google Scholar]
- 34.Li H, et al. The Sequence Alignment/Map format and SAMtools. Bioinformatics. 2009;25:2078–2079. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Li H, Durbin R. Fast and accurate long-read alignment with Burrows–Wheeler transform. Bioinformatics. 2010;26:589–595. doi: 10.1093/bioinformatics/btp698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Picard Tools - By Broad Institute. Available from, http://broadinstitute.github.io/picard.
- 37.DePristo MA, et al. A framework for variation discovery and genotyping using next-generation DNA sequencing data. Nat Genet. 2011;43:491–498. doi: 10.1038/ng.806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Van der Auwera GA, et al. From FastQ Data to High‐Confidence Variant Calls: The Genome Analysis Toolkit Best Practices Pipeline. Current Protocols in Bioinformatics. 2013;43:1–11. doi: 10.1002/0471250953.bi1110s43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Grattapaglia D, Bertolucci FLG, Sederoff R. Genetic mapping of QTLs controlling vegetative propagation in Eucalyptus grandis and E. urophylla using a pseudo‐testcross mapping strategy and RAPD markers. Theor. Appl. Genet. 1995;90:933–947. doi: 10.1007/BF00222906. [DOI] [PubMed] [Google Scholar]
- 40.Hyma KE, et al. Heterozygous mapping strategy (HetMappS) for high resolution Genotyping-By-Sequencing markers: a case study in grapevine. PLoS One. 2015;10:e0134880. doi: 10.1371/journal.pone.0134880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Stam, P. & Van Ooijen, J. W. JOINMAP version 2.0: software for the calculation of genetic linkage maps (1995).
- 42.Van Ooijen, J. W. JoinMap® 4.0, Software for the calculation of genetic linkage maps in experimental populations. Kyazma B.V. Wageningen, Netherlands (2006).
- 43.Broman KW, Wu H, Sen S, Churchill GA. R/qtl: QTL mapping in experimental crosses. Bioinformatics. 2003;19:889–890. doi: 10.1093/bioinformatics/btg112. [DOI] [PubMed] [Google Scholar]
- 44.Tang H, et al. ALLMAPS: robust scaffold ordering based on multiple maps. Genome Biol. 2015;16:3–10. doi: 10.1186/s13059-014-0573-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Cabanettes F, Klopp C. D-GENIES: dot plot large genomes in an interactive, efficient and simple way. PeerJ. 2018;6:e4958. doi: 10.7717/peerj.4958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ma ZY, et al. Phylogenomics, biogeography, and adaptive radiation of grapes. Molecular phylogenetics and evolution. 2018;129:258–267. doi: 10.1016/j.ympev.2018.08.021. [DOI] [PubMed] [Google Scholar]
- 47.Tarailo-Graovac M, Chen N. Using RepeatMasker to identify repetitive elements in genomic sequences. Curr Protoc Bioinformatics. 2009;25:4–10. doi: 10.1002/0471250953.bi0410s25. [DOI] [PubMed] [Google Scholar]
- 48.Bao W, Kojima KK, Kohany O. Repbase Update, a database of repetitive elements in eukaryotic genomes. Mobile DNA. 2015;6:11. doi: 10.1186/s13100-015-0041-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Campbell MS, Holt C, Moore B, Yandell M. Genome Annotation and Curation Using MAKER and MAKER-P. Curr Protoc Bioinformatics. 2014;48:1–39. doi: 10.1002/0471250953.bi0411s48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Campbell MS, et al. MAKER-P: a tool kit for the rapid creation, management, and quality control of plant genome annotations. Plant Physiol. 2014;164:513–24. doi: 10.1104/pp.113.230144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Korf I. Gene finding in novel genomes. BMC Bioinformatics. 2004;5:59. doi: 10.1186/1471-2105-5-59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Stanke M, et al. AUGUSTUS: a web server for gene finding in eukaryotes. Nucleic Acids Research. 2004;32:309–312. doi: 10.1093/nar/gkh379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Buels R, et al. JBrowse: a dynamic web platform for genome visualization and analysis. Genome Biology. 2016;12:17–66. doi: 10.1186/s13059-016-0924-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.2018. NCBI Sequence Read Archive. SRP174866
- 55.Bert P-F, Girollet N, Rubio B. 2019. Vitis riparia cultivar Riparia Gloire de Montpellier isolate 1030, whole genome shotgun sequencing project. GenBank. SJAQ00000000
- 56.Girollet N, Rubio B, Bert P-F. 2019. De novo phased assembly of the Vitis riparia grape genome. figshare. [DOI] [PMC free article] [PubMed]
- 57.Li, H. Aligning sequence reads, clone sequences and assembly contigs with BWA-MEM. Preprint at, https://arxiv.org/abs/1303.3997 (2013)
- 58.Simão FA, Waterhouse RM, Ioannidis P, Kriventseva EV, Zdobnov EM. BUSCO: assessing genome assembly and annotation completeness with single-copy orthologs. Bioinformatics. 2015;31:3210–3212. doi: 10.1093/bioinformatics/btv351. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Citations
- 2018. NCBI Sequence Read Archive. SRP174866
- Bert P-F, Girollet N, Rubio B. 2019. Vitis riparia cultivar Riparia Gloire de Montpellier isolate 1030, whole genome shotgun sequencing project. GenBank. SJAQ00000000
- Girollet N, Rubio B, Bert P-F. 2019. De novo phased assembly of the Vitis riparia grape genome. figshare. [DOI] [PMC free article] [PubMed]
Supplementary Materials
Data Availability Statement
1. GBS demultiplexing
2. Filters FASTQ files with CASAVA 2.20
fastq_illumina_filter –keep N -v -v -o good_reads.fq raw_reads.fastq
3. Cutadapt (regular 3′ adapter)
https://cutadapt.readthedocs.io/en/stable/guide.html
cutadapt -a AGATCGGAAGAGCGGTTCAGCAGGAATGCCGAG
-A AGATCGGAAGAGCGTCGTGTAGGGAAAGAGTGT
-G CTCGGCATTCCTGCTGAACCGCTCTTCCGATCT
-g ACACTCTTTCCCTACACGACGCTCTTCCGATCT -u7 -U7 -m10
4. Burrows-Wheeler Alignment – BWA-MEM
http://bio-bwa.sourceforge.net/bwa.shtml
bwa mem ref.fa read1.fastq.gz read2.fastq.gz > aligned.reads.sam with these options:
-M Mark shorter split hits as secondary (for Picard compatibility)
-R Complete read group header line with ‘\t’ used in STR to be converted to a TEB in the output SAM. An example is ‘@RG\tID:\tSM:\tPL:\tLB:’
5. Picard tools
https://broadinstitute.github.io/picard/
SortSam : java –jar picard.jar SortSam with these options: INPUT (BAM file), OUTPUT (BAM file), SORT_ORDER
MarkDuplicates : java –jar picard.jar MarkDuplicates with these options: INPUT (BAM file), OUTPUT (BAM file), METRIC_FILE (file)
BuildBamIndex : java –jar picard.jar BuildBamIndex with these options: INPUT (BAM file)
6. GATK tools
HaplotypeCaller : java –jar GenomeAnalysisTK.jar –T HaplotypeCaller –R ref.fasta –I file.bam –genotyping_mode DISCOVERY –drf DuplicateRead –emitRefConfidence GVCF –o file.g.vcf
CombineGVCFs : java –jar GenomeAnalysisTK.jar –T CombineGVCFs –R ref.fasta –drf DuplicateRead –G Standard –G AS_Standard –variant sample1 to sample‘n’.g.vcf –o cohort_file.g.vcf
GenotypeGVCFs : java –jar GenomeAnalysisTK.jar –T GenotypeGVCFs –R ref.fasta –drf DuplicateRead –G Standard –G AS_Standard –variant cohort_file.g.vcf –o final_file.vcf
SelectVariants : java –jar GenomeAnalysisTK.jar –T SelectVariants –R ref.fasta –V final_file.vcf –selectType SNP –o file_snps.vcf
VariantFiltration : java –jar GenomeAnalysisTK –T VariantFiltration –R ref.fasta –V file_snps.vcf–filterExpression « QD < 2.0 || FS > 60.0 || MQ < 40.0 || MQRankSum < −12.5 || ReadPosRankSum < −8.0 » –filteredName «FILTER » -o filtered_snps.vcf
7. VCF filtering
vcftools –vcf filtered_snps.vcf –remove-filtered-all –recode –out filteredFinal_snps.vcf
8. Falcon and Falcon_Unzip Assembly for SMRT sequencing
https://github.com/PacificBiosciences/FALCON/wiki
https://github.com/PacificBiosciences/FALCON_unzip/wiki
Main parameters: length_cutoff = 5000, length_cutoff_pr = 5000
pa_HPCdaligner_option = -v -dal128 -e0.70 -M40 -l2500 -k17 -h500 -w7 -s100
ovlp_HPCdaligner_option = -v -dal128 -M40 -k19 -h500 -e.96 -l1500 -s100
pa_DBsplit_option = -a -x500 -s200
ovlp_DBsplit_option = -s200
falcon_sense_option = –output_multi –output_dformat –min_idt 0.80 –min_cov 4 max_n_read 400 –n_core 16
falcon_sense_skip_contained = False
overlap_filtering_setting = –max_diff 120 –max_cov 120 –min_cov 4 –n_core 24
9. Purge Haplotigs
https://bitbucket.org/mroachawri/purge_haplotigs/src/master/
purge_haplotigs readhist -b aligned.bam -g genome.fasta
10. Supernova Assembly for 10x Chromium sequencing
https://support.10xgenomics.com/de-novo-assembly/software/overview/latest/welcome
Option pseudohap2 style output
11. Scaffolding Falcon assembly with LINKS using Supernova outputs Assembly
https://github.com/bcgsc/LINKS
LINKS -f.fa -s fileofname.fofn -b cns1-linked_draft -d 5000 -t 100 -k 19 -l 5 -a 0.3
LINKS -f.fa -s fileofname.fofn -b cns2-linked_draft -d 6000 -t 80 -k 19 -l 15 -a 0.3
LINKS -f.fa -s fileofname.fofn -b cns3-linked_draft -d 7000 -t 60 -k 19 -l 20 -a 0.3
LINKS -f.fa -s fileofname.fofn -b cns4-linked_draft -d 10000 -t 30 -k 19 -l 20 -a 0.3
LINKS -f.fa -s fileofname.fofn -b cns5-linked_draft -d 15000 -t 30 -k 19 -l 20 -a 0.3
LINKS -f.fa -s fileofname.fofn -b cns6-linked_draft -d 50000 -t 30 -k 19 -l 30 -a 0.3
LINKS -f.fa -s fileofname,fofn -b cns7-linked_draft -d 75000 -t 30 -k 19 -l 40 -a 0.3
12. Improving quality with PILON and Illumina sequencing
https://github.com/broadinstitute/pilon/wiki/Requirements-&-Usage
13. Allmaps pseudomolecules scaffolding
https://github.com/tanghaibao/jcvi/wiki/ALLMAPS
14. Assembly evaluation with BUSCO v3
15. Vitis TE(s) Identification using RepeatMasker
16. Annotation with MAKER_P pipeline, SNAP and Augustus gene finder
http://www.yandell-lab.org/publications/pdf/maker_current_protocols.pdf
https://bmcbioinformatics.biomedcentral.com/articles/10.1186/1471-2105-5-59
https://github.com/Gaius-Augustus/Augustus
DB (vitis) AND “Vitis”[porgn] from https://www.ncbi.nlm.nih.gov
EST DB (vitis) AND “Vitis”[porgn] from https://www.ncbi.nlm.nih.gov
- First run : rm_pass = 0, est2genome = 1 and protein2genome = 1
gff3_merge -d master_datastore_index.log
maker2zff -c 0 -e 0 -o 0 -x 0.05 maker1.gff
fathom -categorize 1000 genome.ann genome.dna
fathom -export 1000 -plus uni.ann uni.dna
forge export.ann export.dna
hmm-assembler.pl RGM. >snap1.hmm
- Second run: rm_pass = 1, est2genome = 0, protein2genome = 0, maker_gff = maker1.gff, snaphmm = snap1.hmm leading to a maker2.gff3 and a snap2.hmm files.
gff3_merge -d master_datastore_index.log
maker2zff -c 0 -e 0 -o 0 -x 0.05 maker2.gff
fathom -categorize 1000 genome.ann genome.dna
fathom -export 1000 -plus uni.ann uni.dna
forge export.ann export.dna
hmm-assembler.pl RGM. >snap2.hmm
Run Augustus:
zff2gff3.pl genome.ann | perl -plne ‘s/\t(\S+)$/\t\.\t$1/’ > genome.gff3
autoAug.pl –genome = ../pilon2.fasta –species = RGM18 –cdna = sequence_est_ncbi.fasta –trainingset = genome.gff3 –singleCPU –v –useexisting
- Third run : rm_pass = 1, est2genome = 0, protein2genome = 0, maker_gff = maker2.gff, snaphmm = snap2.hmm, augustus_species = RGM18 leading to a maker3.gff3, maker3.transcripts.fasta and maker3.proteins.fasta structural prediction.
gff3_merge -d master_datastore_index.log
fasta_merge -d master_datastore_index.log
17. Interproscan functional annotation and putative gene function assignation
Download protein DB from http://www.uniprot.org
makeblastdb -in protein_db.fasta -input_type fasta -dbtype prot
blastp -db protein_db.fasta -query maker3.proteins.fasta -out maker3.proteins.blastp -evalue 0.000001 -outfmt 6 -max_hsps 1
maker_functional_gff protein_db.fasta maker3.proteins.blastp maker3.gff3 ≫ maker3.putative.gff3
maker_functional_fasta protein_db.fasta maker3.proteins.blastp maker3.proteins.fasta ≫ maker3.putative.proteins.fa
maker_functional_fasta protein_db.fasta maker3.proteins.blastp maker3.transcripts.fasta ≫ maker3.putative.transcripts.fa
Run Interproscan
interproscan.sh –iprlookup –goterms -f tsv -i maker3.putative.proteins.fa -pa -b RGM.annotated.proteins
18. Assembly validation using WBA mem
bwa mem -M -t 20 VitRiparia.fasta reads_pe.R1.fastq reads_pe.R2.fastq > aln_pe_reads.sam
samtools view -bS aln_pe_reads.sam -o aln_pe_reads.bam #| samtools sort - aln_reads.sorted.bam
samtools sort -o aln_pe_reads.sorted.bam aln_pe_reads.bam
bamtools stats -in aln_pe_reads.sorted.bam > bamstat_pe.reads