Abstract
Brain tumours are the most common cause of cancer death in children. Molecular studies have greatly improved our understanding of these tumours but tumour metabolism is underexplored. Metabolites measured in vivo have been reported as prognostic biomarkers of these tumours but analysis of surgically resected tumour tissue allows a more extensive set of metabolites to be measured aiding biomarker discovery and providing validation of in vivo findings. In this study, metabolites were quantified across a range of paediatric brain tumours using 1H-High-Resolution Magic Angle Spinning nuclear magnetic resonance spectroscopy (HR-MAS) and their prognostic potential investigated. HR-MAS was performed on pre-treatment frozen tumour tissue from a single centre. Univariate and multivariate Cox regression was used to examine the ability of metabolites to predict survival. The models were cross validated using C-indices and further validated by splitting the cohort into two. Higher concentrations of glutamine were predictive of a longer overall survival, whilst higher concentrations of lipids were predictive of a shorter overall survival. These metabolites were predictive independent of diagnosis, as demonstrated in multivariate Cox regression models. Whilst accurate quantification of metabolites such as glutamine in vivo is challenging, metabolites show promise as prognostic markers due to development of optimised detection methods and increasing use of 3 T clinical scanners.
Subject terms: Paediatric cancer, Metabolomics, CNS cancer, Prognostic markers
Introduction
Cancer is a major cause of death from disease in childhood, and brain tumours are the most common cause of cancer-related death in this age group1. Whilst some brain tumours now have a very good prognosis, others have continued to present a challenge, highlighting the need for new techniques for investigation and management. Identifying novel biomarkers of prognosis would allow more accurate treatment stratification to improve survival rates and reduce long-term morbidity. A key strategy in optimizing the clinical management of children with brain tumours is to identify subgroups that have prognostic significance, and this has been particularly successful in medulloblastoma where molecular subgroups have already been incorporated into the accepted diagnostic classification2. Whilst most molecular markers have been defined by tumour genetics, there is an increasing interest in tumour metabolism as both a biomarker of prognosis and potential therapeutic target. Mutations in the metabolic enzyme isocitrate dehydrogenase (IDH) act as a marker of good prognosis in gliomas3 and has led to novel therapeutic targets being identified4. The mutated enzyme produces the metabolite 2-hydroxyglutarate which can be detected both in tissue and in vivo providing a non-invasive test for this subgroup illustrating a key advantage for metabolite biomarkers of prognosis.
Whilst the identification of specific prognostic subgroups has advantages, markers that are applicable across multiple tumour types also give clinical value. Histological markers, such as Ki67 proliferation index, are used regardless of diagnosis to assess tumour aggressiveness, and MYC status is a marker of poor prognosis used clinically in many different tumour types5. Specific metabolites have been proposed as markers of prognosis over a range of different children’s brain tumours using in vivo Magnetic Resonance Spectroscopy (MRS). Specifically, glutamine and N-acetylaspartate (NAA) were found to be markers of good prognosis whilst scyllo-inositol and mobile lipids markers of poor prognosis6.
However, performing MRS on typical clinical scanners is associated with a number of limitations, in particular, the relatively small number of metabolites that can be measured accurately and the requirement for sampling of relatively large tumour volumes. Ex-vivo 1H-High-Resolution Magic Angle Spinning (HR-MAS) can analyse small brain tumour tissue samples to provide quantitative information on a larger number of metabolites7. HR-MAS has identified prognostic metabolic markers in a number of tumour types including prostate8, colorectal9, breast10, neuroblastoma11 and pancreatic adenocarcinomas12. A good agreement has been demonstrated between in vivo and ex vivo methods13,14, providing an indication that HR-MAS-visible metabolites accurately reflect the values present in situ and are potentially observable in vivo. The primary aim of this study was to identify and measure the concentrations of metabolites in a range of paediatric brain tumours using HR-MAS and test the ability of metabolites to predict survival.
Methods
Patients
All patients with brain tumours presenting at the Birmingham Children’s Hospital were eligible to be entered into the study. Approval was obtained from the research ethics committee (NRES East Midlands-Derby, 04/MRE04/41) and informed consent was given by parents/guardians. All experiments were performed in accordance with relevant guidelines and regulations. Patients were accrued between January 1998 and May 2016 and followed up until September 2017. A consensus diagnosis was obtained for each patient by a multidisciplinary team of clinicians that included histopathological diagnosis according to Louis et al.15. Brain tumour tissue, frozen as soon as possible after resection, was requested from the histopathology tissue bank at Birmingham Children’s Hospital. In total, 133 brain tumour samples were released for HR-MAS. Dates of death and clinical information were obtained from the West Midlands Regional Children’s Tumour Registry. Patients were treated with the protocol appropriate for their respective diagnosis. Broadly, this involved maximal safe surgical resection when the tumour was in a favourable location, with adjuvant radiotherapy and/or chemotherapy if necessary.
From the 133 tumour samples available, 19 were excluded from the analysis due to quality reasons or cause of death not being from the tumour diagnosis. The final cohort consisted of 114 primary pre-treatment brain tumour samples including 36 pilocytic astrocytomas (PA), 32 medulloblastomas (MB), 15 ependymomas (EP), 7 glioblastoma multiforme (GBM), 6 atypical teratoid rhabdoid tumours (ATRT), 4 anaplastic astrocytomas (AA), 4 choroid plexus papilloma (CPP), 3 gangliogliomas (GG), 2 atypical choroid plexus papillomas (ACPP), 2 CNS primitive neuroectodermal tumours (PNET), 1 choroid plexus carcinoma (CPC), 1 astroblastoma (AB) and 1 unclassified low grade astrocytic glial tumour. As the tumour tissue was requested before the release of the WHO CNS classification 2016, diagnoses are those specified by the WHO CNS classification 200715. 35 patients died and 79 were alive at the end of the study. The median follow-up time for the whole cohort was 5.31 years (Table 1).
Table 1.
Clinical information for patients included in this study, organised by tumour diagnosis.
| Pilocytic astrocytoma | Medullo-blastoma | Ependy-moma | Gliobla-stoma | Atypical teratoid rhabdoid tumour | Anaplastic astrocytoma | Choroid plexus papilloma | Ganglio-glioma | CNS primitive neuroectodermal tumour | Atypical choroid plexus papilloma | Choroid plexus carcinoma | Astrobla-stoma | Astrocytic glial tumour | Total | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| N | 36 | 32 | 15 | 7 | 6 | 4 | 4 | 3 | 2 | 2 | 1 | 1 | 1 | 114 |
| Number of events (n) | 3 | 13 | 3 | 6 | 4 | 3 | 0 | 0 | 1 | 1 | 1 | 0 | 0 | 35 |
| Gender (male:female) | 18:18 | 26:6 | 11:4 | 5:2 | 4:2 | 1:3 | 4:0 | 1:2 | 1:1 | 1:1 | 1:0 | 0:1 | 1:0 | 74:40 |
| Mean age at diagnosis (years) | 8.30 | 7.11 | 5.53 | 5.68 | 1.10 | 10.49 | 3.40 | 11.07 | 7.24 | 2.95 | 5.09 | 10.18 | 11.58 | 5.93 |
| Age range (years) | 1.2–15.9 | 1.5–14.6 | 0.3–16.3 | 0.03–11.5 | 0.02–4.6 | 4.1–15.5 | 0.4–10.1 | 8.7–14.9 | 2.2–12.3 | 1.6–4.3 | N/A | N/A | N/A | 0.03–16.3 |
| Median survival (years) | 4.71 | 1.82 | 1.68 | 0.94 | 0.31 | 1.54 | N/A | N/A | 0.94 | 1.93 | 1.39 | N/A | N/A | 1.50 |
| Anatomical location (n)[supratentorial:infratentorial:spinal] | 7:29:0 | 0:32:0 | 5:9:1 | 6:1:0 | 1:5:0 | 3:1:0 | 2:2:0 | 2:1:0 | 2:0:0 | 1:1:0 | 1:0:0 | 1:0:0 | 0:1:0 | 31:82:1 |
Sample preparation and HRMAS
The frozen samples were stored at −80 °C until used for HR-MAS. Samples were dissected on dry ice and inserted into either a 12 μl or 50 μl HR-MAS rotor with TMSP (Cambridge Biosciences, Cambridge, UK) as a quantification reference and D2O (Sigma-Aldrich, Dorset, UK) to fill the allocated rotor volume. Sample weights ranged from 3.4 to 35.8 mg. HR-MAS experiments were performed using a Bruker Avance 500 MHz (11.74 T) NMR spectrometer and a 4 mm 3 channel HR-MAS z-PFG band probe (Bruker, Coventry, UK). The samples were maintained at a temperature of 4 °C, a spin rate of 4800 Hz set for each experiment with 256 or 512 scans acquired depending on the signal-to-noise ratio. A standard 1D NOESY pulse sequence preceded by 2 s of water presaturation was acquired, with a repetition time of 4 s. In addition, a 285 ms Carr-Purcell-Meiboom-Gill (CPMG) pulse sequence was also acquired for each sample to aid in metabolite identification by minimizing broad signal contributions from lipids and macromolecules.
Processing of HR-MAS data
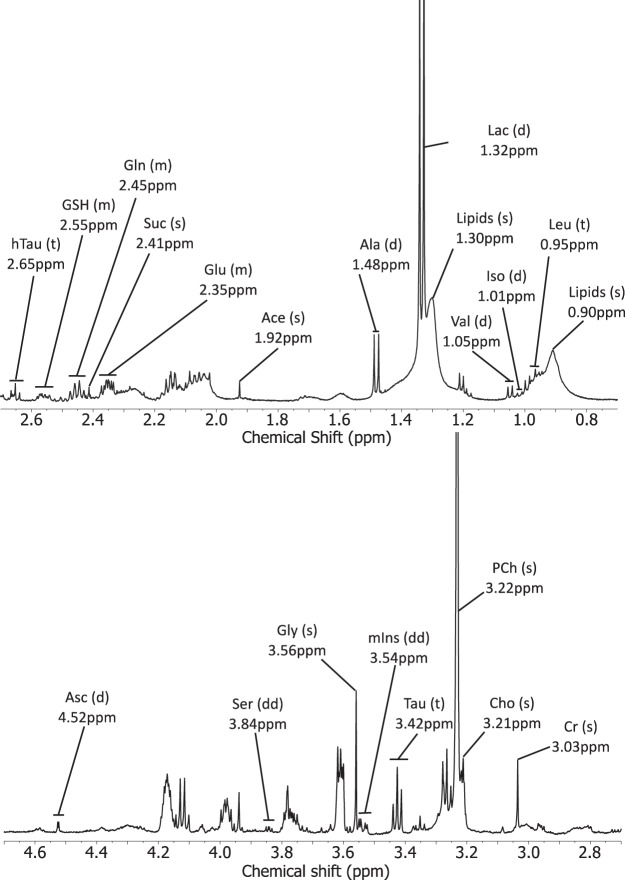
Each free induction decay was Fourier transformed using TOPSPIN (Bruker, Coventry, UK). Phase and baseline correction (Whittaker Smoother) were performed using Mnova NMR 9.0 software suite (2014; Mestrelab Research, Spain). Creatine was used as a chemical shift reference at 3.03 ppm. Spectral fitting was then performed between 0.5 and 4.7 ppm. All spectral peaks were deconvoluted and peak integrals measured followed by normalization to TMSP. The following metabolites were quantified and included in the analyses: acetate (Ace), alanine (Ala), ascorbate (Asc), choline (Cho), creatine (Cr), gamma-aminobutyric acid (GABA), glucose (Glc), glutamine (Gln), glutamate (Glu), glutathione (GSH), glycine (Gly), glycerophosphocholine (GPC), hypotaurine (hTau), isoleucine (Iso), lactate (Lac), leucine (Leu), myo-inositol (mIns), N-acetylaspartate (NAA), phosphocholine (PCh), serine (Ser), scyllo-inositol (sIns), succinate (Suc), taurine (Tau), valine (Val), lipids at 0.9 and lipids at 1.3 ppm (Fig. 1, Supplementary Table 1). Lipids were highly correlated, and so the lipid resonances were summed and termed total lipids. Metabolites were assigned according to Govindaraju et al.16 whilst lipids were assigned according to Moestue et al.17. Metabolite concentrations were normalised to the total sum of non-lipid metabolite concentrations.
Figure 1.
Example spectrum with annotated metabolite resonances and splitting patterns. Not visible in this spectrum are NAA(s) at 2.01ppm, GABA(t) at 2.30ppm, GPC(s) at 3.23ppm, sIns(s) at 3.34ppm and Glc(d) at 4.65ppm. Abbreviations: s, singlet; d, doublet; t, triplet; dd, doublet of doublets; m, multiplet; ppm, parts per million.
Data and statistical analysis
Death caused by the tumour was the endpoint for the study, with time to event calculated from the date of first surgical resection to death. Univariate and multivariate Cox regression were performed on normalised metabolite concentrations and clinical parameters to determine the association of metabolites with survival. Likelihood ratio tests were used to determine significance of Cox models. Clinical parameters found to significantly predict survival in the univariate analyses were included as variables in the multivariate analyses. Univariate P-values were corrected using Benjamini-Hochberg correction to control the false discovery rate.
Validation of the models was performed using leave-one-out cross-validation (LOOCV) of C-indices, as described in Braadland et al.8. For each individual i, a Cox proportional hazards model is created leaving i out. The coefficients, βi, generated are applied to i’s covariates, xi, to obtain a linear predictor ηi = βixi. This is repeated for all individuals in the analysis creating the vector η = η1, η2, η2…ηn, where n is the number of individuals. The resultant vector of linear predictors is used as a single covariate in a Cox proportional hazards model to calculate the C-index. Normalised metabolite concentrations were scaled by dividing each metabolite by its standard deviation prior to univariate and multivariate analysis.
Kaplan-Meier analyses were used to test the difference in survival of patients with brain tumours separated into groups above and below 25%, 50% or 75% quantiles for significant metabolites in the whole cohort. To further test the robustness of the biomarkers, the whole cohort was divided based on the date of tissue collection into an initial cohort (n = 23), comprised of tissue collected during or before June 2006, and a validation cohort (n = 91), comprised of tissue collected after June 2006. The quantile showing the most significant result was recalculated in the initial cohort and the obtained value was applied to the validation cohort. Kaplan-Meier analysis was performed using logrank tests. All survival analyses were performed using the survival library written for the R software package18.
Results
Univariate cox regression
A total of 25 metabolite features were assigned as quantified using HR-MAS (Fig. 1, Supplementary Table 1). The mean metabolite concentrations for the different tumour groups are given in Supplementary Table 2. Univariate Cox regression identified 2 metabolites with prognostic potential (Table 2, Supplementary Table 3); higher Gln was associated with decreased risk of death, whilst a higher concentration of total lipids was associated with an increased risk of death. Whilst Val, Suc and hTau were significant in the univariate analysis, they were no longer significant upon P-value correction. Of the clinical features tested, diagnosis was shown to be a significant predictor of survival; patients diagnosed with AA, ATRT, CPC, GBM, MB and PNET had a significantly higher risk of death than PA patients (Table 2). Similar to Val, Suc and hTau, age at diagnosis was significant in the univariate analysis but was no longer significant after P-value correction. Gender and tumour location were not significant predictors of survival in this analysis (Supplementary Table 4).
Table 2.
Univariate and multivariate Cox models for metabolite concentrations demonstrating their ability to predict overall survival of paediatric brain tumour patients.
| Variable | Univariate analysis | Multivariate analyses | ||||
|---|---|---|---|---|---|---|
| HR (95% CI) | Corrected P-value | Glutamine model | Total lipids model | |||
| HR (95% CI) | P-value | HR (95% CI) | P-value | |||
| Gln | 0.52 (0.31, 0.87) | 0.041 | 0.42 (0.21–0.85) | 0.015 | — | — |
| Total lipids | 2.02 (1.53, 2.66) | 2.48 × 10−4 | — | — | 1.91 (1.35, 2.68) | 2.15 × 10−4 |
| Diagnosis | 4.58 × 10−4 | |||||
| PA | Reference | — | Ref | — | Ref | — |
| AA | 13.03 (2.62, 64.65) | 0.0017 | 9.93(2.99, 49.59) | 0.005 | 15.80 (3.16, 79.16) | 7.86 × 10−4 |
| ACPP | 9.27 (0.95, 90.57) | 0.056 | 3.41 (0.32, 36.58) | 0.31 | 8.08 (0.82, 79.10) | 0.073 |
| ATRT | 17.11 (3.80, 76.95) | 2.1 × 10−4 | 8.63 (1.75, 42.61) | 0.0081 | 11.69 (2.38, 57.53) | 0.0025 |
| CPC | 35.46 (3.52, 357.31) | 0.0025 | 9.48 (0.79, 113.46) | 0.076 | 10.39 (0.96, 112.40) | 0.054 |
| EP | 2.55 (0.51, 12.63) | 0.25 | 1.34 (0.25, 7.09) | 0.73 | 2.61 (0.53, 12.98) | 0.24 |
| GBM | 31.72 (7.77, 129.45) | 1.5 × 10−6 | 32.73 (7.86, 136.23) | 1.64 × 10−6 | 22.29 (5.32, 93.31) | 2.14 × 10−5 |
| MB | 5.91 (1.68, 20.76) | 0.0056 | 2.45 (0.61, 9.89) | 0.21 | 5.71 (1.62, 20.11) | 0.0066 |
| PNET | 14.12 (1.44, 138.02) | 0.023 | 6.03 (0.58, 63.08) | 0.13 | 21.69 (2.15, 218.62) | 0.009 |
As no deaths were recorded for patients diagnosed with GG, AB, CPP or the unclassified glial tumour, these entries have been excluded from this table.
Multivariate cox regression
The two metabolites with prognostic potential in the univariate analyses were further evaluated in a multivariate analysis. Each metabolite was assessed with diagnosis as a second covariate. Gln and total lipids were found to be predictors of survival independent of diagnosis, with higher concentrations of Gln predicting better survival, whilst higher concentrations of total lipids were associated with a higher risk of death (Table 2).
Validation of prognostic markers
Upon validation of the univariate analysis using leave one out cross validation of C-indices, total lipids alone demonstrated a marginally greater predictive accuracy than diagnosis alone. Validation of the multivariate models showed a greater predictive accuracy when either Gln or total lipids were combined with diagnosis when compared with diagnosis alone using the same validation method (Table 3).
Table 3.
Cross validation of the univariate and multivariate models demonstrate improved predictive accuracy after inclusion of either Gln or total lipid concentrations in addition to diagnosis when compared to diagnosis alone.
| Variable | Univariate analysis | Multivariate analysis |
|---|---|---|
| LOOCV C-index | LOOCV C-index | |
| Gln | 0.641 | 0.686 |
| Total lipids | 0.670 | 0.727 |
| Diagnosis | 0.666 | — |
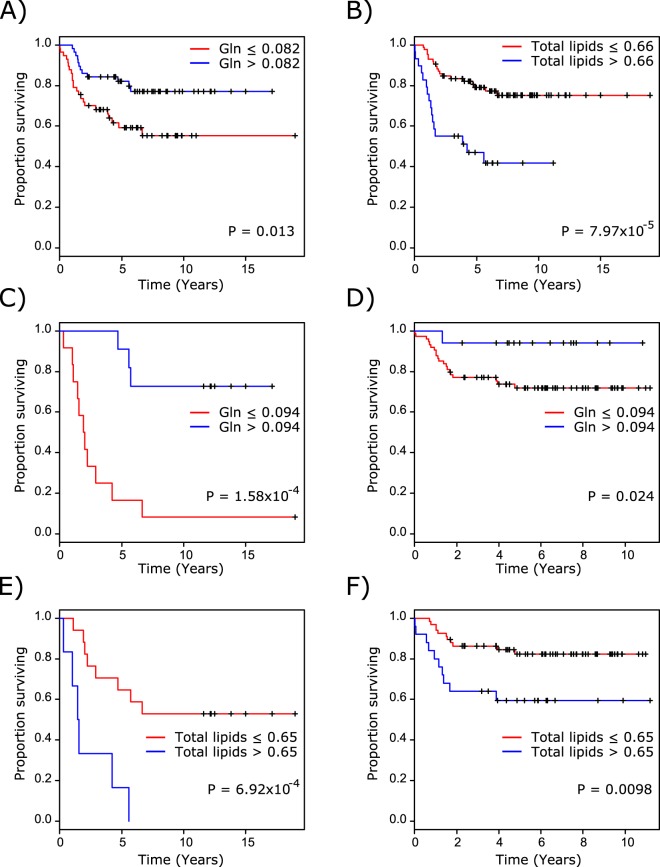
The relationship between survival and metabolic predictors remained when the whole cohort was stratified into high and low concentrations groups using 25%, 50% and 75% quantiles. The difference in survival between patients with high and low Gln was greatest when cases were stratified by the 50% quantile Gln, whilst the difference between high and low lipids patients was greatest upon stratification using the 75% quantile (Fig. 2A,B). Separation of the whole cohort into an initial cohort and validation cohort further demonstrated the prognostic ability of Gln and total lipids. The difference in survival using the 50% quantile Gln concentration in the initial cohort reached prognostic significance, which was maintained when the cut-off was applied to the validation cohort (Fig. 2C,D). The difference in survival using the 75% quantile total lipids in the initial cohort demonstrated prognostic significance, which remained following application to the validation cohort (Fig. 2E,F).
Figure 2.
Stratification of patients in the whole cohort into high and low metabolite concentration groups using (A) 50% quantile Gln and (B) 75% quantile total lipids demonstrates significant differences in survival. Stratification of patients in (C) the initial cohort using 50% quantile Gln demonstrates a significant difference in survival which remains upon application of the determined value to (D) the validation cohort. Stratification of patients in (E) the initial cohort using 75% quantile total lipids demonstrates a significant difference in survival which remains upon application of the determined value to (F) the validation cohort.
Discussion
This study has shown that the concentration of metabolites measured in fresh, frozen diagnostic biopsy tissue from a variety of paediatric brain tumours using HR-MAS varies with disease aggressiveness. Through univariate and multivariate Cox models, it was shown that both Gln and total lipids have prognostic significance independent of tumour diagnosis and that addition of metabolite concentrations can aid in predicting overall survival. Interestingly, through cross-validation, total lipid concentration alone outperformed diagnosis in predicting survival, although the difference was small.
We have previously reported in vivo Gln concentration as a predictor of good prognosis in paediatric brain tumours independent of diagnosis6, and it is encouraging to have this confirmed by ex vivo spectroscopy. Gln is poorly resolved at an MR scanner field strength of 1.5 T due to the overlapping of signals from Glu, NAA and macromolecules. Techniques have been developed to improve the accuracy of Gln quantification; 1D MRS techniques include averaging spectra from different echo times (TE)19, the use of optimal TE’s20-22, very short TE’s23,24 and spectral-selective refocusing25. 2D techniques have also been developed but are not favoured due to the high level of expertise required to ensure reliable results and the time taken to acquire the data26. There have been recent advancements in ultrafast 2D NMR27, although this is still in its infancy. Overall, there is a realistic prospect that it will be possible to routinely measure tumour Gln levels in vivo in the near future.
Gln is the most abundant amino acid in blood plasma, and its metabolism has long been known to be important in tumour biology28; uptake has been shown to increase in brain tumours in order to support cellular bioenergetics and biosynthetic processes needed for proliferation29. Metabolism is under the direct control of major transcription factors such as TP53, Myc and Ras30. Mutations or changes in the regulation of these genes are common in cancers, including brain tumours, and switch the cells’ metabolism from oxidative phosphorylation of glucose to energy inefficient glucose fermentation and glutaminolysis31. This switch to Gln metabolism has been linked to poor prognosis32, therefore Gln concentration may aid stratification of patients into risk groups and inform treatment decisions. Due to the reliance of some cancers on Gln, the metabolism of this amino acid has been identified as a potential target for treatment33.
Lipids have also previously been identified as biomarkers of survival through in vivo MRS of paediatric brain tumours. The lipid signals can be generated by mobile lipids stored in cytoplasmic lipid droplets34,35 or by lipids in areas of tissue necrosis36,37, although it is unlikely that highly necrotic areas of tumour would be sampled. The role of lipids droplets in brain tumours is not well understood; however, there is evidence suggesting the number of lipid droplets increases in response to cellular stress and are associated with growth arrest38 and necrosis34. Furthermore, the number of cells positively stained for adipophilin, a lipid droplet associated protein, was shown to significantly increase with tumour grade39.
Risk stratification is not the only potential benefit of enhanced metabolite quantification. Progression of disease and response to treatment can be monitored using MRS. Increases in Gln concentration in contralateral white matter is linked to glioblastoma migration40, whilst reductions in the concentration of phosphocholine and lactate in glioblastoma cell lines have been observed after treatment with mTor and PI3K inhibitors41, reflecting the observation of reduced total Cho in orthotopic glioblastoma mouse models42. Increases in lipid concentrations have been correlated with responses to treatment in cell lines exposed to chemotherapy agents38,43.
Suc, hTau and Val, which were significant in the univariate analysis before correction for multiple comparisons, warrant further investigation considering the limited understanding of these metabolites in the context of paediatric brain tumours. Increases in both Suc and hTau were associated with a longer survival time, whilst increases in Val were associated with a shorter survival time.
Despite in vivo studies identifying NAA and sIns as prognostic markers, neither metabolite reached prognostic significance in the current study of tumour tissue. In addition to the limited sample size which could explain this, it is worth noting that the population of tumours included is somewhat different to that of in vivo studies and this could also contribute to the finding. There is a lack of available frozen tissue from surgically intractable tumours such as optic pathway gliomas and brain stem tumours unlike in vivo studies. Optic pathway gliomas have been shown to have a high concentration of NAA and a good prognosis whilst brain stem tumours have a higher concentration of sIns and a poor prognosis6.
In conclusion, this study has confirmed that the concentration of Gln and lipids in resected tissue are predictors of survival independent of diagnosis. The importance of Gln and lipids as predictors of prognosis in children’s tumours should lead to their measurement in tumour tissue, the implementation of MRS techniques to accurately measure their concentration non-invasively and the exploration of novel agents which can alter their metabolism for therapeutic effect.
Supplementary information
Acknowledgements
The authors wish to thank all staff in the Histopathology Department at Birmingham Children’s Hospital for storing and making the tissue available. We also thank the staff at Henry Wellcome Building for Biomolecular NMR at the University of Birmingham, and in particular Dr. Sara Whittaker, for all their support with this work. We also wish to thank the West Midlands Tumour Registry for providing the clinical information for this study. Action Medical Research and the Brain Tumour Charity; grant number GN2181. National Institute of Health Research; NIHR-RP-02-12-019. Birmingham Children’s Hospital Research Foundation; BCHRF 353. Children with Cancer; 15/188.
Author Contributions
Conception and design: C.D.B., S.K.G., A.C.P. Collection and organisation of samples and clinical information: C.D.B., S.K.G., S.E.K. Raw data collection and design of 1H-MRS protocols: C.D.B., S.E.K., S.K.G., M.W. Data processing and analysis of metabolite concentrations: C.D.B., S.E.K., S.K.G. Statistical and survival analysis: C.D.B. Writing of draft manuscript: C.D.B. Revision and preparation of final manuscript: C.D.B., S.K.G., D.A.T., S.E.K., A.C.P. Financial support: N.P.D., T.N.A., A.C.P. Overall supervision: A.C.P. All authors read and approved the final manuscript.
Data Availability
The datasets generated and analysed during the current study are available from the corresponding author on reasonable request.
Competing Interests
The authors declare no competing interests.
Footnotes
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Supplementary information accompanies this paper at 10.1038/s41598-019-45900-x.
References
- 1.Ostrom QT, et al. Alex’s Lemonade Stand Foundation Infant and Childhood Primary Brain and Central Nervous System Tumors Diagnosed in the United States in 2007–2011. Neuro Oncol. 2015;16:x1–x36. doi: 10.1093/neuonc/nou327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Louis DN, et al. The 2016 World Health Organization Classification of Tumors of the Central Nervous System: a summary. Acta neuropathologica. 2016;131:803–820. doi: 10.1007/s00401-016-1545-1. [DOI] [PubMed] [Google Scholar]
- 3.Combs SE, et al. Prognostic significance of IDH-1 and MGMT in patients with glioblastoma: One step forward, and one step back? Radiation Oncology (London, England) 2011;6:115–119. doi: 10.1186/1748-717x-6-115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Rohle D, et al. An Inhibitor of Mutant IDH1 Delays Growth and Promotes Differentiation of Glioma Cells. Science. 2013;340:626–630. doi: 10.1126/science.1236062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hutter S, Bolin S, Weishaupt H, Swartling FJ. Modeling and Targeting MYC Genes in Childhood Brain Tumors. Genes. 2017;8:107. doi: 10.3390/genes8040107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wilson M, et al. Magnetic resonance spectroscopy metabolite profiles predict survival in paediatric brain tumours. European journal of cancer (Oxford, England: 1990) 2013;49:457–464. doi: 10.1016/j.ejca.2012.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Griffin JL, Shockcor JP. Metabolic profiles of cancer cells. Nature reviews. Cancer. 2004;4:551–561. doi: 10.1038/nrc1390. [DOI] [PubMed] [Google Scholar]
- 8.Braadland PR, et al. Ex vivo metabolic fingerprinting identifies biomarkers predictive of prostate cancer recurrence following radical prostatectomy. British journal of cancer. 2017;117:1656–1664. doi: 10.1038/bjc.2017.346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Pacholczyk-Sienicka B, Fabianska A, Pasz-Walczak G, Kordek R, Jankowski S. Prediction of survival for patients with advanced colorectal cancer using (1) H High-resolution magic angle spinning nuclear MR spectroscopy. Journal of magnetic resonance imaging: JMRI. 2015;41:1669–1674. doi: 10.1002/jmri.24734. [DOI] [PubMed] [Google Scholar]
- 10.Giskeodegard GF, et al. Lactate and glycine-potential MR biomarkers of prognosis in estrogen receptor-positive breast cancers. NMR in biomedicine. 2012;25:1271–1279. doi: 10.1002/nbm.2798. [DOI] [PubMed] [Google Scholar]
- 11.Imperiale A, et al. Metabolomic pattern of childhood neuroblastoma obtained by (1)H-high-resolution magic angle spinning (HRMAS) NMR spectroscopy. Pediatric blood & cancer. 2011;56:24–34. doi: 10.1002/pbc.22668. [DOI] [PubMed] [Google Scholar]
- 12.Battini S, et al. Metabolomics approaches in pancreatic adenocarcinoma: tumor metabolism profiling predicts clinical outcome of patients. BMC medicine. 2017;15:56. doi: 10.1186/s12916-017-0810-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tzika A, et al. Biochemical characterization of pediatric brain tumors by using in vivo and ex vivo magnetic resonance spectroscopy. Journal of neurosurgery. 2002;96:1023–1031. doi: 10.3171/jns.2002.96.6.1023. [DOI] [PubMed] [Google Scholar]
- 14.Wilson M, Davies NP, Grundy RG, Peet AC. A quantitative comparison of metabolite signals as detected by in vivo MRS with ex vivo 1H HR-MAS for childhood brain tumours. NMR in biomedicine. 2009;22:213–219. doi: 10.1002/nbm.1306. [DOI] [PubMed] [Google Scholar]
- 15.Louis DN, et al. The 2007 WHO Classification of Tumours of the Central Nervous System. Acta neuropathologica. 2007;114:97–109. doi: 10.1007/s00401-007-0243-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Govindaraju V, Young K, Maudsley AA. Proton NMR chemical shifts and coupling constants for brain metabolites. NMR in biomedicine. 2000;13:129–153. doi: 10.1002/1009-1492(200005)13:3<129::AID-NBM619>3.0.CO;2-V. [DOI] [PubMed] [Google Scholar]
- 17.Moestue S, Sitter B, Bathen TF, Tessem MB, Gribbestad IS. HR MAS MR spectroscopy in metabolic characterization of cancer. Current topics in medicinal chemistry. 2011;11:2–26. doi: 10.2174/156802611793611869. [DOI] [PubMed] [Google Scholar]
- 18.Team, R. C. R: A Language and Environment for Statistical Computing. R Foundation for Statistical Computing, Vienna, Austria (2015).
- 19.Hurd R, et al. Measurement of brain glutamate using TE-averaged PRESS at 3T. Magnetic resonance in medicine. 2004;51:435–440. doi: 10.1002/mrm.20007. [DOI] [PubMed] [Google Scholar]
- 20.Schubert F, Gallinat J, Seifert F, Rinneberg H. Glutamate concentrations in human brain using single voxel proton magnetic resonance spectroscopy at 3 Tesla. NeuroImage. 2004;21:1762–1771. doi: 10.1016/j.neuroimage.2003.11.014. [DOI] [PubMed] [Google Scholar]
- 21.Mullins PG, Chen H, Xu J, Caprihan A, Gasparovic C. Comparative reliability of proton spectroscopy techniques designed to improve detection of J-coupled metabolites. Magnetic resonance in medicine. 2008;60:964–969. doi: 10.1002/mrm.21696. [DOI] [PubMed] [Google Scholar]
- 22.Yang S, Hu J, Kou Z, Yang Y. Spectral simplification for resolved glutamate and glutamine measurement using a standard STEAM sequence with optimized timing parameters at 3, 4, 4.7, 7, and 9.4T. Magnetic resonance in medicine. 2008;59:236–244. doi: 10.1002/mrm.21463. [DOI] [PubMed] [Google Scholar]
- 23.Wijtenburg SA, Knight-Scott J. Very short echo time improves the precision of glutamate detection at 3T in 1H magnetic resonance spectroscopy. Journal of magnetic resonance imaging: JMRI. 2011;34:645–652. doi: 10.1002/jmri.22638. [DOI] [PubMed] [Google Scholar]
- 24.Mekle R, et al. MR spectroscopy of the human brain with enhanced signal intensity at ultrashort echo times on a clinical platform at 3T and 7T. Magnetic resonance in medicine. 2009;61:1279–1285. doi: 10.1002/mrm.21961. [DOI] [PubMed] [Google Scholar]
- 25.Choi C, et al. Measurement of brain glutamate and glutamine by spectrally-selective refocusing at 3 Tesla. Magnetic resonance in medicine. 2006;55:997–1005. doi: 10.1002/mrm.20875. [DOI] [PubMed] [Google Scholar]
- 26.Ramadan Saadallah, Lin Alexander, Stanwell Peter. Glutamate and glutamine: a review ofin vivoMRS in the human brain. NMR in Biomedicine. 2013;26(12):1630–1646. doi: 10.1002/nbm.3045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Akoka S, Giraudeau P. Fast hybrid multi-dimensional NMR methods based on ultrafast 2D NMR. Magnetic resonance in chemistry: MRC. 2015;53:986–994. doi: 10.1002/mrc.4237. [DOI] [PubMed] [Google Scholar]
- 28.DeBerardinis RJ, Lum JJ, Hatzivassiliou G, Thompson CB. The Biology of Cancer: Metabolic Reprogramming Fuels Cell Growth and Proliferation. Cell metabolism. 2008;7:11–20. doi: 10.1016/j.cmet.2007.10.002. [DOI] [PubMed] [Google Scholar]
- 29.DeBerardinis RJ, et al. Beyond aerobic glycolysis: Transformed cells can engage in glutamine metabolism that exceeds the requirement for protein and nucleotide synthesis. Proceedings of the National Academy of Sciences. 2007;104:19345–19350. doi: 10.1073/pnas.0709747104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kim MH, Kim H. Oncogenes and Tumor Suppressors Regulate Glutamine Metabolism in Cancer Cells. Journal of Cancer Prevention. 2013;18:221–226. doi: 10.15430/JCP.2013.18.3.221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Elstrom RL, et al. Akt Stimulates Aerobic Glycolysis in Cancer Cells. Cancer research. 2004;64:3892. doi: 10.1158/0008-5472.CAN-03-2904. [DOI] [PubMed] [Google Scholar]
- 32.Yang L, et al. Metabolic shifts toward glutamine regulate tumor growth, invasion and bioenergetics in ovarian cancer. Molecular Systems Biology. 2014;10:728. doi: 10.1002/msb.20134892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Shelton LM, Huysentruyt LC, Seyfried TN. Glutamine targeting inhibits systemic metastasis in the VM-M3 murine tumor model. International journal of cancer. Journal international du cancer. 2010;127:2478–2485. doi: 10.1002/ijc.25431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Opstad KS, Bell BA, Griffiths JR, Howe FA. An investigation of human brain tumour lipids by high-resolution magic angle spinning 1H MRS and histological analysis. NMR in biomedicine. 2008;21:677–685. doi: 10.1002/nbm.1239. [DOI] [PubMed] [Google Scholar]
- 35.Pan X, et al. The size of cytoplasmic lipid droplets varies between tumour cell lines of the nervous system: a 1H NMR spectroscopy study. Magma (New York, N.Y.) 2012;25:479–485. doi: 10.1007/s10334-012-0315-x. [DOI] [PubMed] [Google Scholar]
- 36.Opstad KS, Bell BA, Griffiths JR, Howe FA. Taurine: a potential marker of apoptosis in gliomas. British journal of cancer. 2009;100:789–794. doi: 10.1038/sj.bjc.6604933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Cheng LL, et al. Quantification of microheterogeneity in glioblastoma multiforme with ex vivo high-resolution magic-angle spinning (HRMAS) proton magnetic resonance spectroscopy. Neuro Oncol. 2000;2:87–95. doi: 10.1093/neuonc/2.2.87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mirbahai L, et al. Lipid biomarkers of glioma cell growth arrest and cell death detected by 1 H magic angle spinning MRS. NMR in biomedicine. 2012;25:1253–1262. doi: 10.1002/nbm.2796. [DOI] [PubMed] [Google Scholar]
- 39.Kohe S, Colmenero I, McConville C, Peet A. Immunohistochemical staining of lipid droplets with adipophilin in paraffin-embedded glioma tissue identifies an association between lipid droplets and tumour grade. Journal of Histology and Histopathology. 2017;4:4. doi: 10.7243/2055-091X-4-4. [DOI] [Google Scholar]
- 40.Kallenberg K, et al. Untreated glioblastoma multiforme: increased myo-inositol and glutamine levels in the contralateral cerebral hemisphere at proton MR spectroscopy. Radiology. 2009;253:805–812. doi: 10.1148/radiol.2533071654. [DOI] [PubMed] [Google Scholar]
- 41.Venkatesh HS, et al. Reduced phosphocholine and hyperpolarized lactate provide magnetic resonance biomarkers of PI3K/Akt/mTOR inhibition in glioblastoma. Neuro Oncol. 2012;14:315–325. doi: 10.1093/neuonc/nor209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Koul D, et al. Cellular and in vivo activity of a novel PI3K inhibitor, PX-866, against human glioblastoma. Neuro Oncol. 2010;12:559–569. doi: 10.1093/neuonc/nop058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Pan X, et al. Increased unsaturation of lipids in cytoplasmic lipid droplets in DAOY cancer cells in response to cisplatin treatment. Metabolomics: Official journal of the Metabolomic. Society. 2013;9:722–729. doi: 10.1007/s11306-012-0483-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets generated and analysed during the current study are available from the corresponding author on reasonable request.