Abstract
Background
The characteristic structure of motor neurons (MNs), particularly of the long axons, becomes damaged in the early stages of amyotrophic lateral sclerosis (ALS). However, the molecular pathophysiology of axonal degeneration remains to be fully elucidated.
Method
Two sets of isogenic human-induced pluripotent stem cell (hiPSCs)-derived MNs possessing the single amino acid difference (p.H517D) in the fused in sarcoma (FUS) were constructed. By combining MN reporter lentivirus, MN specific phenotype was analyzed. Moreover, RNA profiling of isolated axons were conducted by applying the microfluidic devices that enable axon bundles to be produced for omics analysis. The relationship between the target gene, which was identified as a pathological candidate in ALS with RNA-sequencing, and the MN phenotype was confirmed by intervention with si-RNA or overexpression to hiPSCs-derived MNs and even in vivo. The commonality was further confirmed with other ALS-causative mutant hiPSCs-derived MNs and human pathology.
Findings
We identified aberrant increasing of axon branchings in FUS-mutant hiPSCs-derived MN axons compared with isogenic controls as a novel phenotype. We identified increased level of Fos-B mRNA, the binding target of FUS, in FUS-mutant MNs. While Fos-B reduction using si-RNA or an inhibitor ameliorated the observed aberrant axon branching, Fos-B overexpression resulted in aberrant axon branching even in vivo. The commonality of those phenotypes was further confirmed with other ALS causative mutation than FUS.
Interpretation
Analyzing the axonal fraction of hiPSC-derived MNs using microfluidic devices revealed that Fos-B is a key regulator of FUS-mutant axon branching.
Fund
Japan Agency for Medical Research and development; Japanese Ministry of Education, Culture, Sports, Science and Technology Clinical Research, Innovation and Education Center, Tohoku University Hospital; Japan Intractable Diseases (Nanbyo) Research Foundation; the Kanae Foundation for the Promotion of Medical Science; and “Inochi-no-Iro” ALS research grant.
Keywords: Amyotrophic lateral sclerosis (ALS), Fused in sarcoma (FUS), Axon branching, Fos-B, Nerve organoid, Human-induced pluripotent stem cell (hiPSC)-derived motor neuron
Research in context.
Evidence before this study
Amyotrophic lateral sclerosis (ALS) is a devastating, adult-onset neurodegenerative disorder characterized by the progressive loss of motor neurons (MNs). Although the axonal mRNA profiles have been extensively investigated in neurodegenerative diseases, studies regarding RNA profiling of MN axons, particularly in terms of ALS pathology, are limited. The disruption of dendritic spine morphology in cortical neurons in FUS knockout or mutant FUS overexpression mice models and the abnormal morphologies of MN axons in transgenic FUS-ALS zebrafish models have previously been reported. However, such processes remain unclear in human MNs without the overexpression of mutant protein.
Added value of this study
The axon pathomechanisms of ALS remain unclear owing to both the inaccessibility of patient-derived primary MNs and the low yields of samples from the axon compartment. Here we present the application of a newly established microfluidic device that facilitates the production of axon bundles for omics analyses. We conducted RNA profiling of isolated axons from human-induced pluripotent stem cell (hiPSCs)-derived MNs. Two sets of isogenic hiPSCs with a single amino acid difference in ALS-causative FUS mutation were compared and we obtained the following results:
-
(i)
complete axon profile using hiPSC-derived MNs
-
(ii)
abnormal increase in axon branching using FUS-ALS MNs
-
(iii)
aberrant axon branching regulated by Fos-B in both cultured MNs and in vivo
-
(iv)
the suppression of aberrant Fos-B rescued the cellular phenotype of FUS-ALS MNs
Implication of all the available evidence
Our study has critical implications based on the high species specificity (human iPSCs) and RNA extraction procedure (large amount of axon RNA were purified) as well as the identification of Fos-B as a key molecule that regulates axon morphology. Our results provide the complete axon profile of human MNs, which has not yet been reported, as well as provides a novel strategy for the analysis of the axon pathology of neurodegenerative diseases including ALS. Fos-B pathway could be a potential therapeutic target in future ALS research.
Alt-text: Unlabelled Box
1. Materials and methods
1.1. Establishment of human-induced pluripotent stem cell (hiPSCs)
All protocols were approved by the ethics committees of Tohoku University School of Medicine (No. 2010–306) and the Keio University School of Medicine (No. 20080016). hiPSCs from a healthy control named 409B2 (Control) and 201B7 (Control-2) were provided by the Center for iPS Cell Research and Application, Kyoto University [1,2]. To generate hiPSCs from a patient with amyotrophic lateral sclerosis (ALS) exhibiting the fused in sarcoma (FUS) mutation on p.H517D (FUS-ALS), a skin-punch biopsy was performed on a 43-year-old Japanese patient with familial ALS [3]. Control and FUS-ALS hiPSCs were generated using episomal vectors transfected into dermal fibroblasts (Supplementary Fig. 1a) [1,4]. Control-2 hiPSCs were generated by infecting dermal fibroblasts with lentivirus (Supplementary Fig. 7f) [2]. Each hiPSC line was confirmed to all the exons of FUS for genotyping. Moreover, each line was confirmed to not contain other ALS-causative mutations, including copper/zinc superoxide dismutase 1 (SOD1), TAR-DNA binding protein coding TDP-43 (TARDBP), and chromosome 9 open reading frame 72 (C9orf72). The hiPSC lines from familial ALS with the SOD1 mutation on p.H46R (SOD1-ALS) [5] and the TARDBP mutation on p.M337 V (TARDBP-ALS) [6] have previously been established using episomal vectors transfected into peripheral blood mononuclear cells (PBMCs) and dermal fibroblasts, respectively (Supplementary Fig. 7f).
1.2. Culture of undifferentiated hiPSCs
The hiPSCs lines were cultured using mitomycin C-treated SNL murine fibroblast feeder cells in hiPSC medium containing DMEM/F12 (Fujifilm), 20% KnockOut serum replacement (KOSR) (Thermo Fisher Scientific), 0.1 mM nonessential amino acids (Fujifilm), 0.1 mM 2-mercaptoethanol (Sigma-Aldrich), 50 units/mL penicillin G, 50 μg/mL streptomycin, and 4 ng/mL fibroblast growth factor 2 (FGF-2) (PeproTech) at 37 °C and 3% CO2. The medium was changed every day. When the hiPSCs reached confluency, hiPSC colonies were detached from the feeder layers using a dissociation solution [0.25% trypsin, 100 μg/mL collagenase IV (Thermo Fisher Scientific), 1 mM CaCl2, and 20% KOSR] and passaged. The culture medium was regularly checked using a Mycoalert kit (Lonza).
1.3. Construction of isogenic hiPSCs possessing FUSH517D mutation using TALEN
TALEN genome editing was performed as described previously [4]. Briefly, to generate isogenic mutant lines, the control hiPSC line 409B2 was cultured under feeder-free conditions using StemFit AK03 (Ajinomoto) according to the manufacturer's protocol. Following dissociation into single cells, the left and right TALEN-expressing plasmids and the targeting donor plasmid were transfected using a NEPA21 electroporator (Nepagene). After puromycin-resistant hiPSCs colonies were obtained, PCR genotyping and Sanger sequencing were performed to identify the knock-in clone. Subsequently, PGK-PurTK cassette-free cells were obtained using AdEFNCre-4FVF (kindly provided by Dr. Yumi Kanegae, University of Tokyo [7]) infection and ganciclovir selection. Following each selection, single ganciclovir-resistant hiPSCs colonies were genotyped using Sanger sequencing. As a result, the FUS-mutant isogenic line (FUSH517D/H517D) was generated. To generate isogenic control lines, the mutation of FUS-ALS was rectified using the same protocol by modifying the targeting donor plasmid thereby resulting in the development of the mutation-corrected line (FUSRescued). Isogenic hiPSCs were transferred to on-feeder cultures from feeder-free conditions, and each line was validated for pluripotency markers, normal karyotypes, genomic integrities, and developmental potency.
1.4. Construction of isogenic hiPSCs possessing FUSP525L mutation by CRISPR/Cas9
For generating an isogenic FUSP525L/+ mutant line, the control hiPSC line 201B7 was cultured under feeder-free conditions using StemFit AK03 as the manufactured protocol. Human codon optimized spCas9 cDNA was sub-cloned into pCAGGS vector [8], and human U6 promoter, CRISPR target sequence and sgRNA scaffold sequence were sub-cloned into pBlueScript II vector (Agilent Technologies). As donor DNA, 90 bp of FUSP525L phosphorothioate modified single stranded OligoDNA (PS-ssODN) and FUSWT PS-ssODN with silent mutations were synthesized. pCAGGS-spCas9 vector, pBlueScript II sgRNA expression vector and PS-ssODN were co-electroporated with Puromycin N-acetyltransferase expression plasmid using Neon Transfection System (Thermo Fisher Schientific). After puromycin-resistant single hiPSCs colonies were picked up, PCR genotyping and Sanger sequencing were performed to identify the knock-in clone. Thus, the FUS-mutant isogenic line (FUSP525L/+) were established. The FUSP525L/+ line was validated for pluripotency markers, normal karyotypes, genomic integrities, and developmental potency (data not shown).
1.5. Fish strains and maintenance
hb9:Venus zebrafish were obtained from National BioResource Project, Zebrafish Core Institution in Japan [9] and were maintained in Tokai University under approved protocols (approval number: 173021). Zebrafish embryos were raised at 28 °C according to standard procedures [10].
1.6. Validation of hiPSCs
To validate pluripotency markers, immunocytochemical (ICC) analysis was performed as described below using appropriate antibodies. The karyotypes were examined at Nihon-Gene Research Laboratories Inc. and the developmental potency was confirmed as previously described [4]. Briefly, hiPSCs were passaged to FGF-2-free hiPSC medium and cultured under floating conditions for 14 days. Subsequently, the cells were plated on PO/fibronectin-coated dishes and further cultured for 14 days until fixation (also refer to Supplementary Fig. 1f).
1.7. Motor neuron (MN) differentiation
MN differentiation was performed as previously described (also refer to Supplementary Fig. 2a) [5]. Typically, 2nd MN precursor cells (MPCs) were dissociated to adhere to poly-L-ornithine/Matrigel (Corning) (PO/M-gel)-coated dishes with 2 × 105 cells/mL for immunostaining. For single molecular fluorescence in situ hybridization (smFISH), 10 μL 2nd MPCs at a concentration of 3 × 106 cells/mL were placed in the microfluidic device (SND450, Xona microfluidics). To examine the axon ends and culture on the nerve organoid microfluidic device (Jiksak Bioengineering), 2nd MPCs were cultured at a concentration of 1 × 104 cells/well in 96-well v-bottom plates (Sumitomo Bakelite), which generated 2nd MPC spheres. The 2nd MPC spheres were plated onto each well of PO/M-gel coated 96-well thin-bottom plates (Thermo Fisher Scientific), glass bottom dishes (Iwaki), and the nerve organoid microfluidic device, which contains a chamber receiving MPCs, a microfluidic channel for axon bundle formation, and a chamber targeted by axon terminals (Fig. 2a–c; also refer to Kawada et al [11]). After plating, the medium was changed every 2–3 days. Moreover, the culture medium was checked using a Mycoalert kit (Lonza) before retrieving RNA samples.
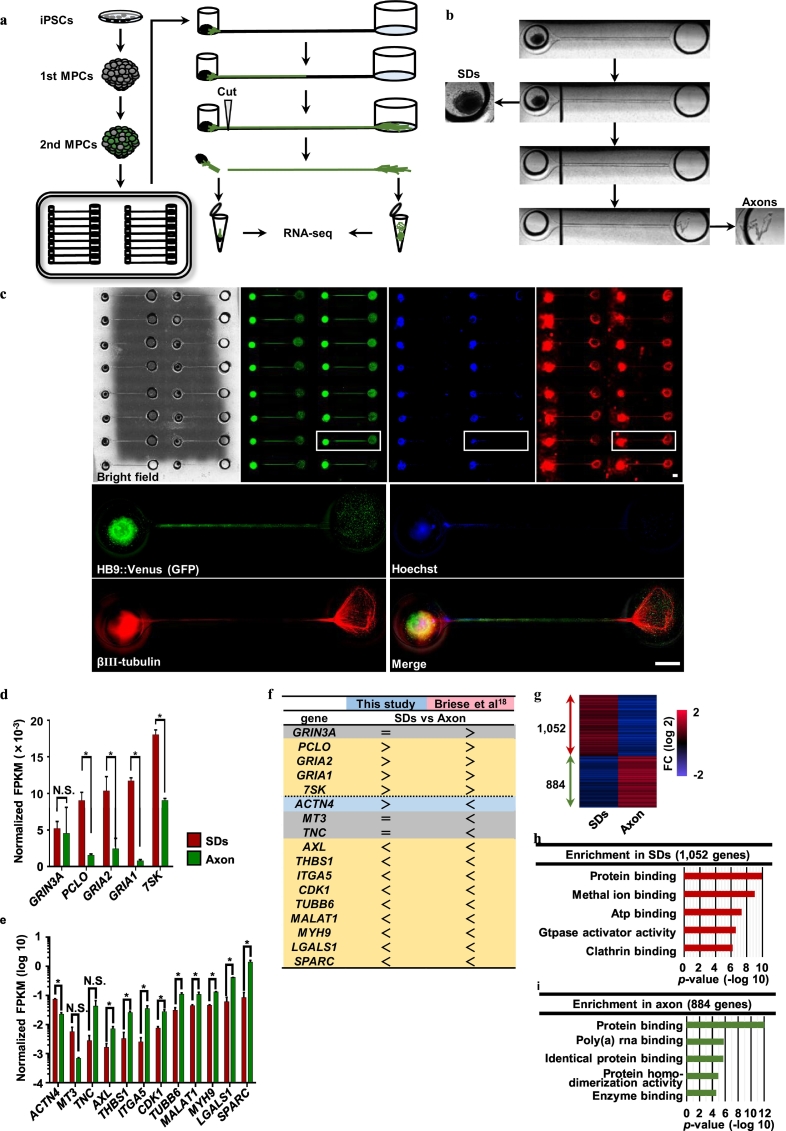
Fig. 2.
MN axons were extracted with a microfluidic device.
(a) The experimental scheme of MN culture using the microfluidic device. The 2nd MPCs with (for ICC) or without (for RNA-seq) HB9 reporter lentivirus infection were plated onto the device. The axons elongated in the microfluidics to the next well. After 20 DPP, the axons were divided from the SDs for RNA extraction. (b) Representative images of axon dividing. Axons were divided from the SDs by cutting the axon bundle at 450 μm away from the sphere to avoid contaminating the cell body and pushing out due to hydraulic pressure. (c) Representative ICC images of MNs on the microfluidic device. The lower four figures represent the enlarged images of white squares of the upper figures. Bars: 1 mm. (d,e) The RNA profiles of the SDs and axonal fractions were compared to check the fractional population. FPKM values were normalized with GAPDH FPKM values of each data set. The presented gene sets were extracted from the reference [19]. The 7SK in the present study indicated the mean FPKM of 7SK genes includes CCNT1, CCNT2, CDK9, DDX21, HEXIM1, and HEXIM2. Student's t-test was used for analysis. See also Supplementary Fig. 4, and Supplementary Table 1,2. (f) Inventory of RNA profile indicated in (d) and (e). Signs of inequality represent the statistically different genes with regard to SDs and axons. Gene profiles that matched with those previously reported are shown with orange highlights. Only ACTN4 exhibited a different profile between that previously reported and that reported in the present study. The other genes are indicated with gray highlights. The RNA profiles matched 84% in the gene sets. (g) Box plot of the SDs or axon-enriched genes. DEGs were extracted with fold changes (FC) |log 2| > 1, Q value <0.05 (Welch's T-test) by Subio platform (ver 1.21). (h,i) The global profiles of the SDs and axon-enriched genes in the present study were analyzed by GO term analysis using Subio platform. The top 5 terms are listed with a p-value (Fisher's exact test). See also Supplementary Table 3.
1.8. Virus production
Lentiviruses were produced in HEK293T cells via the transient transfection of three plasmids: the packaging construct pCAG-HIVgp, the VSV-G and Rev-expressing construct pCMV-VSV-G-RSV-Rev, and the self-inactivating (SIN) lentiviral vector construct. To generate HB9e438::Venus, the pSIN2- HB9e438-bGlo-Venus [12] was used, whereas to induce Venus expression under EF-1α promotor lentiviruses, the CSII-EF-Venus vector was used [13]. To generate Fos-B expressing EF-1α promotor lentiviruses, the human Fos-B mRNA sequence (NM_006732) was amplified using the appropriate primers (Supplementary Table 7). Thereafter, PCR products were subcloned into the pENTR™-D-TOPO vector (Life technologies) using the TOPO reaction. Subsequently, the Fos-B sequence was subcloned into the CSII-EF-RfA and CSII-EF-RfA-IRES2-Venus vectors via the LR reaction using LR clonase II (Life technologies). In this process, the CSII-EF-Fos-B and CSII-EF-Fos-B-IRES2-Venus vectors were constructed. At Day 0, HEK293T cells were passaged in PO-coated dishes in DMEM containing 10% FBS and cultured at 37 °C and 5% CO2 for 24 h. The transfection of each gene as well as that of packaging and coat proteins were performed using polyethylenimine (PEI). For each dish, a cocktail of SIN (pSIN2- HB9e438-bGlo-Venus, CSII-EF-Venus, CSII-EF-Fos-B, or CSII-EF-Fos-B-IRES2-Venus), pCAG-HIVgp, pCMV-VSV-G-RSV-Rev, and PEI was suspended in HBSS (Sigma). Subsequently, the DNA/PEI mixture was added to the cells and after 12 h, the supernatant was changed to DMEM with 10% FBS containing 10 μM forskolin to enhance virus production. After 48 h, the supernatant was collected and filtered through a 0.45 μm filter (Millipore). Therefore, in this process, we produced HB9e438::Venus, EF-1α::Venus, EF-1α::Fos-B, and EF-1α::Fos-B/Venus. Lentiviruses were concentrated using Lenti-X concentrator (Takara) according to the manufacturer's protocol, resuspended in 1/100 volume of MN medium, and frozen at −80 °C until further analysis.
1.9. Viral titration
Viral titration was performed using the two-way method. To determine an infectious unit (IFU) value by independent means using FACS (BD FACSVerse, BD Biosciences), HEK293T cells were infected with viral supernatant (dilution, 1/200 to 1/2000). At 3 days after infection, fluorescent-positive cells were counted and the titration was calculated. For viruses with the HB9 promoter or those without fluorescent expression, a titration was performed by examining the RNA copy number. Using an RNeasy mini kit (Qiagen) with DNase I treatment, genomic viral RNA was purified from a 5 μL aliquot of concentrated viral supernatant. To determine the RNA copy number, cDNA was synthesized using a QuantiTect reverse transcription kit (Qiagen) and the quantitative real-time polymerase chain reaction (qRT–PCR) titration value was estimated by comparing the copy numbers with a reference, which determined the titer using FACS. Each virus was infected at multiplicity of infection (MOI) = 1.0.
1.10. ICC analysis
Cells were fixed with 4% paraformaldehyde (PFA) in PBS, permeabilized, and subsequently blocked for 60 min in 5% serum in PBS. Cells were stained with the indicated primary and secondary antibodies. Fluorescent images were obtained using a Nikon C2si (Nikon) or BZX-900 (Keyence). The antibodies used in the present study are listed in Supplementary Table 6.
1.11. Image analysis using Cellomics
For differentiation efficiency confirmation (Supplementary Fig. 2c), ICC-processed cells were obtained from 5 × 5 fields/well using Cellomics (Thermo Fisher Scientific), resulting in >4000 cells being scored per well, and each image was analyzed using Neuronal Profiling Ver 4. Imaging conditions were as follows: the nuclei, ChAT, and βIII-tubulin were labeled using Hoechst, Alexa Fluor 488, and Alexa Fluor 647 antibodies, respectively. Imaging was performed using the following filter set (excitation/emission): nuclei, broad blues (365/535); ChAT, greens (475/535); βIII tubulin, far reds (630/695). For counting, βIII tubulin positive cell with or without ChAT staining, analysis (Neuronal Profiling Ver 4) began by identifying intact nuclei stained by Hoechst, which were defined as traced nuclei with typical intensity levels lower than the threshold brightness of pyknotic cells. Thereafter, each traced nucleus region was expanded by 50% and cross-referenced with βIII-tubulin. The number of nuclei was analyzed with or without ChAT staining at the βIII-tubulin positive neurites. For stress granule (SG) counting (Supplementary Fig. 2f,g), ICC-processed cells were obtained from 7 × 7 fields/well using Cellomics (Thermo Fisher Scientific), resulting in >10,000 cells being scored per well, and each image was analyzed using co-localization Ver 4. Imaging conditions were as follows: the nuclei, FUS, βIII-tubulin, and Ras GTPase-activating protein-bindingprotein (G3BP) were labeled using Hoechst, Alexa Fluor 488, Alexa Fluor 555, and Alexa Fluor 647 antibodies, respectively. Imaging was performed using the following filter set (excitation/emission): nuclei, broad blues (365/535); FUS, greens (475/535); βIII tubulin, orange red (535/590); and G3BP, far reds (630/695). For FUS mislocalization and SG analysis, analysis (co-localization Ver 4) was initiated by identifying intact nuclei stained by Hoechst, which were defined as traced nuclei with typical intensity levels lower than the threshold brightness of pyknotic cells. Thereafter, each traced nucleus region was expanded by 50% and cross-referenced with βIII-tubulin. The area (pixel) of G3BP granules with or without FUS granules at the βIII-tubulin positive cytosol was analyzed.
1.12. Axon branching counting
For visualizing MNs, a reporter lentivirus (HB9e438::Venus) [12] was infected at the 2nd MPC stage (Fig. 1b), and for Fos-B overexpression, EF-1α::Fos-B was co-infected with HB9e438::Venus. For intervening with si-RNA (Ambicon silencer select s5343 and s223612 for si-Fos-B and 4390846 for si-Scramble), lipofectamine RNA iMax was used for transfection, according to the manufacturer's protocol. T5224 was prepared as a 20 mM stock solution in dimethyl sulfoxide (DMSO) and administered at a final concentration of 100 μM. si-RNA and T5224 were administered at 3 days post plating (DPP) and cultured for an additional 7 days. All figures were acquired at 10 DPP as stated below.
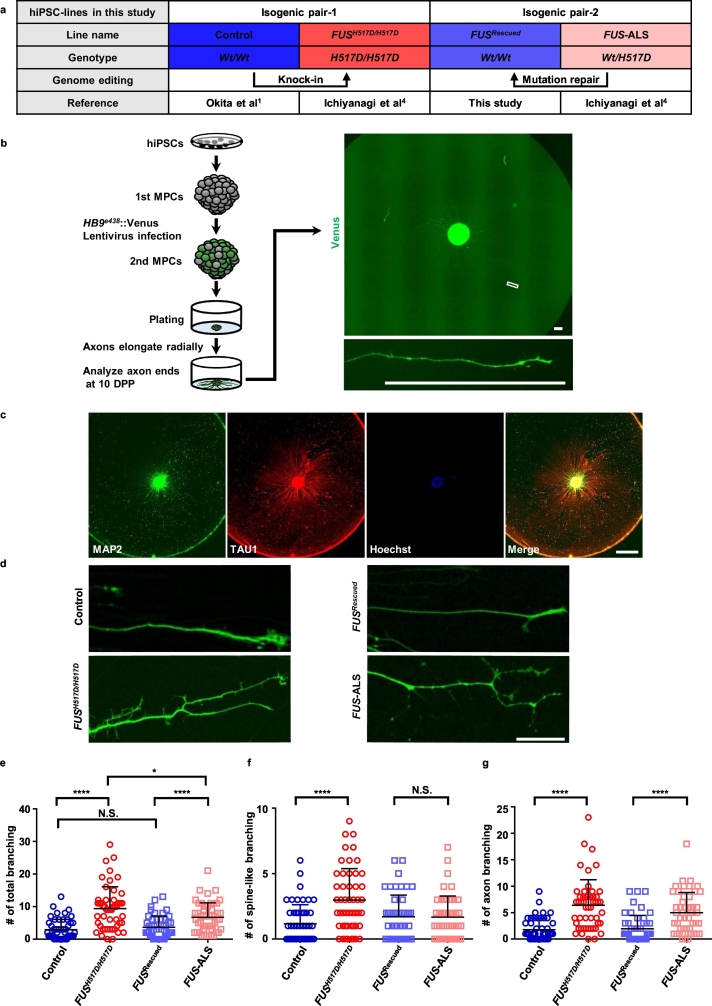
Fig. 1.
The FUS mutation significantly increased axon branching.
(a) Details of hiPSCs derived from healthy donor (Control), a patient with familial ALS (FUS-ALS), and isogenic hiPSCs. FUSH517D/H517D and FUSRescued were established by modifying FUS gene using TALEN genome editing technology from Control and FUS-ALS each other. Therefore, we use two sets of isogenic pairs in the present study. See Supplementary Fig. 1 and Materials and methods (section 1.1. and 1.3.) for more detailed information. (b) The experimental schema for observing MN axon ends. HB9 reporter lentivirus-infected 2nd MPCs were plated onto the center of PO/M-gel coated culture dishes. The enlarged image of the white square in the upper figure presents an example of the axon end that is far from SDs (by at least 1 mm) for counting. Bars: 200 μm. (c) Representative ICC images at 40 DPP (refer to Supplementary Fig. 2). At 40 DPP, nuclear staining was detected only at the center of the well. Bar: 1 mm. (d) Representative images of the axon ends of each cell line at 10 DPP. Bar: 50 μm. (e–g) Quantification of axon branching [see Supplementary Fig. 3 and Materials and methods (section 1.12) for the counting and analysis methods].
Because it was difficult to determine axon branching following fixation because of the aggregation of Venus (data not shown), branching was counted using live cell monitoring. Neurites >450 μm away from the somato dendrites (SDs) were considered axons [14]; therefore, we selected the axons based on distance (>1 mm away from SDs were selected) (Supplementary Fig. 3e). To normalize the evaluation area, we defined branches as those within 200 μm of the distal end of axons. However, because growth cones adopt various formations, we excluded 50 μm of the axon end as a slight modification from the previous report [15]. Accordingly, branching numbers were counted along 150 μm of the axon terminal. Moreover, branches assume various formations; therefore, we classified the branches as two types—the spine-like branching defined as a branching length of 0.5–2 μm with reference to dendritic spine [16] and the axon branching defined as that >2 μm. In the present study, we counted branches from the sole axon (Supplementary Fig. 3g,h). Spines or axon branching originating from other branches were excluded from the counting (Supplementary Fig. 3i,j). Acquired figures were randomly shuffled and structures were counted in a blinded manner. N = 50 based on three independent experiments (16–17 axons were captured from one batch) analyzed using one-way ANOVA (for the following figures; Fig. 1e–g, 5c–e, and Supplementary Fig. 7g–i) or Student's t-test (for the following figures; Fig. 5g–i and Supplementary Fig. 7c–e).
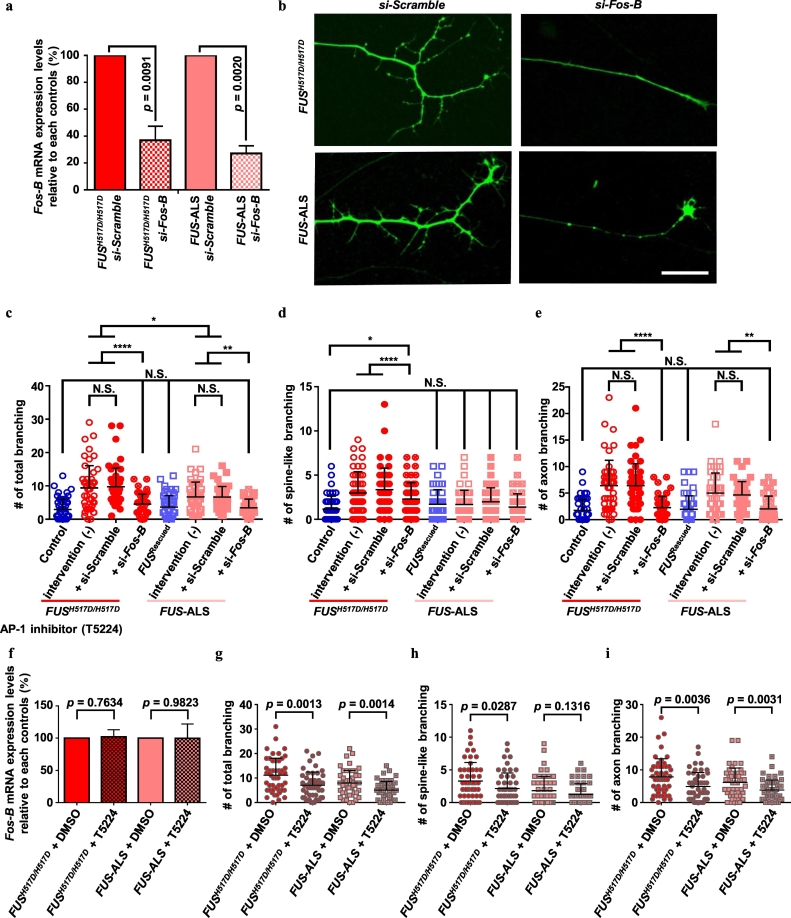
Fig. 5.
Intervention of aberrant Fos-B expression rescues the cellular phenotype of FUS-mutant MNs.
(a–e) Abnormal increases in axon branching was rescued with si-Fos-B. a: Fos-B mRNA was suppressed by si-RNA. The relative expression of si-scrambled control (with ΔΔCt method) is presented. N = 3 independent experiments analyzed by Student's t-test. b: Representative images of the axon ends with or without si-Fos-B. c–e: Quantification of axon branching. To compare the normal condition, untreated data (same data from Fig. 1e–g) were presented and analyzed by one-way ANOVA. (f–i) The number of axon branches decreased with the AP-1 inhibitor, T5224, at 0.1 mM for 7 days. f; RNA expression levels did not differ with T5224 administration. The relative expression of DMSO administrated samples (with the ΔΔCt method) is presented. N = 3 independent experiments analyzed by Student's t-test. g–i; Quantification of axon branching.
1.13. RNA extraction and sequencing analysis
Axons and SDs were retrieved separately by physically cleaving an axon bundle of MNs using a disposable scalpel. A total of 12 axon bundles or SDs were collected to extract the total RNA, which was performed using an RNeasy micro kit (Qiagen) according to the manufacturer's protocol. RNA-sequencing (RNA-seq) libraries were prepared from approximately 12 ng of axonal total RNA and 1 μg of SD total RNA using a SMART-Seq v4 Ultra Low Input RNA Kit (Clontech) and a TruSeq RNA stranded mRNA Sample Prep Kit (Illumina), respectively. These libraries were clonally amplified on a flow cell and sequenced on a HiSeq2500 (HiSeq Control Software v2.2.58, Illumina) with a 51-mer single-end sequence. Image analysis and base calling were performed using Real-Time Analysis Software (v1.18.64, Illumina). For data analysis, UCSC hg19 and RefSeq were used as the reference human genome and gene model, respectively. For gene expression analysis, single-end reads were mapped to the human genome using TopHat (ver. 2.1.0) [17]. Cufflinks (ver. 2.2.1) was used for estimating the gene expression levels based on fragments per kilobase of the exon model per million mapped fragments (FPKM) [18]. The FPKM value was normalized with the GAPDH FPKM for comparing axon- and SDs-dominant RNA profiles as previously described (Fig. 2d–f) [19]. Because of insufficient evidence for the use of GAPDH as a suitable reference gene, we only compared FPKM for Fig. 2g–i and Supplementary Fig. 4b–d. Gene expression levels were compared between Control cells and FUSH517D/H517D cells using Cuffdiff (ver. 2.2.1). The data was uploaded to the DNA Data Bank of Japan (ESUB000323) and GEA (E-GEAD-287).
1.14. Microarray
RNA was prepared using an RNeasy micro kit (Qiagen) and DNase I treatment from Control hiPSC-derived MNs with EF-1α::Venus or EF-1α::Fos-B/Venus-expression lentivirus infection at 10 DPP (Fig. 4i). The microarray was performed using the GeneChip® WT PLUS Reagent Kit. The data was uploaded to Gene Expression Omnibus (accession number: GSE122010).
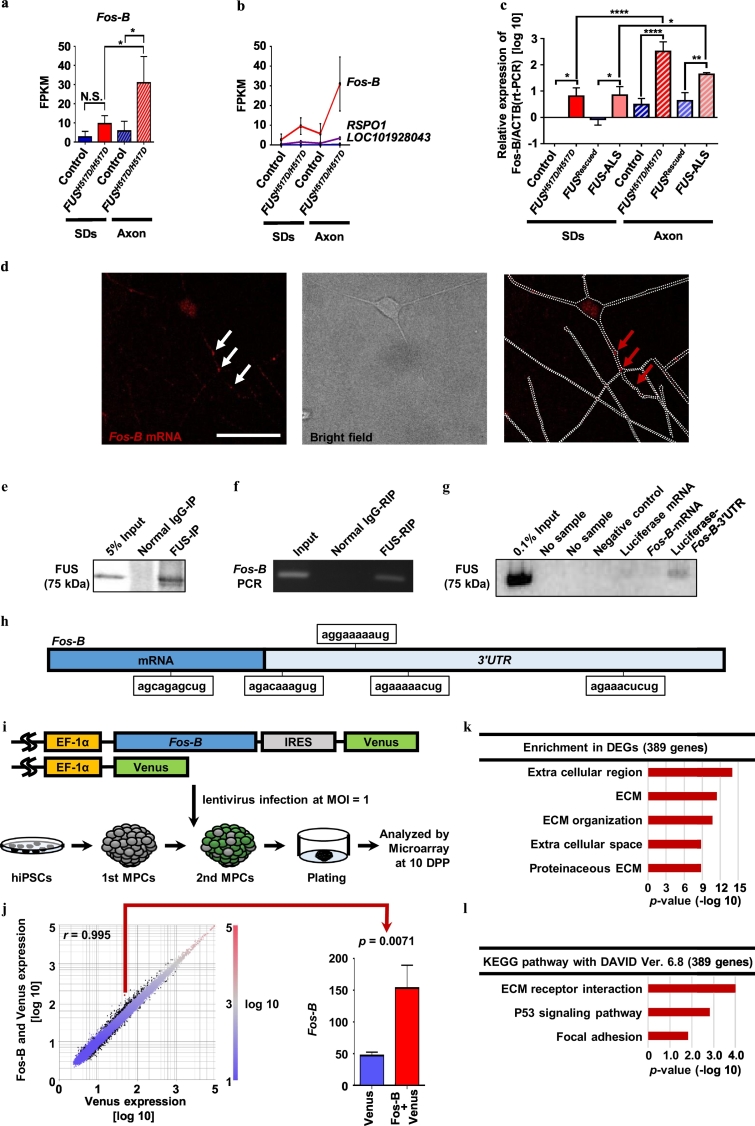
Fig. 4.
Fos-B mRNA binds FUS and affects ECM in hiPSC-derived MNs.
(a) RNA-seq results of Fos-B. Compared with Control, Fos-B was found to be upregulated in FUSH517D/H517D, particularly in the axon fraction (one-way ANOVA). (b) The top 3 gene sets that were observed to be increased in the axon fraction of FUSH517D/H517D. The gene sets were extracted with the findsimilar method of the cummeRbund package of the R (version 3.3.1) platform. (c) Confirming the expression level of Fos-B using qRT–PCR from two sets of isogenic MNs. The relative expression of Control SDs samples (with the ΔΔCt method) are presented; N = 3 independent experiments; one-way ANOVA. (d) Representative image of smFISH for detecting Fos-B mRNA in a FUS-mutant hiPSC-derived MNs. smFISH probes (red) for Fos-B mRNA were detected in the nucleus as well as at the neurites [white (left panel) and red (right panel) arrows]. White dotted lines represent the outline of neurites (see also Supplementary Fig. 6). (e) FUS purified using IP. (f) PCR results with RIP samples. Fos-B mRNA precipitated with the FUS protein. (g) The FUS protein was precipitated with the biotinylated Fos-B 3′UTR sequence. (h) The schema shows the site of the FUS binding sequence [49] on Fos-B. Compared with Fos-B mRNA that includes one binding sequence, the 3′UTR sequence contains four binding sites. (i) The schema of the microarray experiment for Fos-B overexpression. (j) Scatterplots of the transcripts of control MNs with EF-1α::Venus or EF-1α::Fos-B/Venus lentivirus infection. DEGs (FC difference of |log 2| > ± 0.58, Student's t-test <0.05) are represented by black dots (listed in Supplementary Table 5). Fos-B is highlighted with a red point and the low reads are illustrated in a column graph. N = 3 from three independent experiments were analyzed using Student's t-test. (k) The top 5 GO enriched terms and the p-values (Fisher's exact test with Subio platform) of the DEGs shown in (j). (l) Top 3 KEGG pathway analysis results and the p-values of DEGs shown in (j) with DAVID (https://david.ncifcrf.gov/) [66,67]. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
1.15. Immunoblot
Samples were homogenized using IP buffer containing 25 mM HEPES-NaOH (pH 7.4), 150 mM NaCl, 5 mM MgCl2, 1 mM EDTA, and 1% NP-40, along with the supplementation of protease inhibitors (Roche) and RNase inhibitors (Takara). Homogenates were incubated on ice for 20 min and centrifuged at 13,000 rpm for 10 min at 4 °C. Protein concentrations were measured using the BCA protein kit (Thermo Fisher Scientific). Lysates were mixed with Laemmli buffer, heated at 95 °C for 5 min, and separated using SDS–PAGE under 100 V for 40 min. Protein bands were transferred to polyvinylidine difluoride (PVDF) membranes (Millipore; 250 mA for 60 min), which were incubated first with a primary antibody followed by horseradish peroxidase-conjugated secondary antibodies. Chemiluminescence was detected with an enhanced chemiluminescent prime solution (GE healthcare) using a LAS-3000 imaging reader (Fujifilm).
1.16. RNA immunoprecipitation (RIP)
Total cell lysates from Control hiPSC-derived MNs at 10 DPP were prepared as described above, following which they were mixed with the antibodies and rotated at 4 °C for 1 h. Thereafter, protein G Dynabeads (Invitrogen) were added to the mixture and further incubated at 4 °C for 2 h. After washing the beads with IP buffer thrice, bound proteins and RNA were analyzed as described below. To examine protein–protein interactions, beads were directly mixed with 2 × Laemmli buffer, heated at 95 °C for 5 min, and analyzed using Western blotting. To analyze the bound RNA, beads were directly mixed with Trizol reagent (Thermo Fisher Scientific) for purifying the RNA, according to the manufacturer's protocol. RNA was resuspended in RNase-free water in conjunction with reverse transcription using a QuantiTect reverse transcription kit (Qiagen) and analyzed by PCR using appropriate primers (Supplementary Table 7).
1.17. RNA pulldown assay
For Fos-B mRNA synthesis, the RG207004 (Origene) was used by inserting the stop codon using the PrimeStar Mutagenesis kit (Takara) and appropriate primers (Supplementary Table 7), according to the manufacturer's protocol. For Fos-B 3′UTR construction, the HmiT006205-MT05 vector (Genecopoeia) was used. As a control, a luciferase sequence (CmiT00001-MT05) (Genecopoeia) was used. All vectors were enzymatically linearized with XhoI. Subsequently, T7 RNA polymerase (Takara) with a biotin RNA labeling mix (Roche) was used for in vitro transcription. Biotinylated RNA was purified using an RNeasy micro kit (Qiagen), according to the manufacturer's instructions. Lysates from Control and FUSH517D/H517D hiPSC-derived MNs at 10 PDD containing 300 μg of proteins were prepared as described above and incubated with 3 μg of biotinylated RNA for 1 h at RT, following which streptavidin Dynabeads (Invitrogen) were added. After 1 h of incubation at 4 °C, the beads were washed thrice with IP buffer containing RNase inhibitor (Takara), boiled with 2 × Laemmli buffer for 5 min at 95 °C, and analyzed using Western blotting.
1.18. qRT–PCR
RNA was isolated using an RNeasy mini/micro kit (Qiagen) in conjunction with reverse transcription using a QuantiTect reverse transcription kit (Qiagen). qRT–PCR was performed using the SsoFast EvaGreen Supermix (Bio-Rad) and appropriate primers (Supplementary Table 7) and analyzed using a CFX96 Real-Time PCR detection system (Bio-Rad).
1.19. smFISH
Custom Stellaris® FISH Probes were designed against human Fos-B (NM_006732) using the Stellaris® FISH Probe Designer (LGC Biosearch Technologies, Inc., Petaluma, CA), which is available online (www.biosearchtech.com/stellarisdesigner). Control and FUSH517D/H517D hiPSC-derived MNs were hybridized with the Fos-B Stellaris FISH Probe set labeled with Quasar 570 (LGC Biosearch Technologies, Inc.), at 10 DPP according to the manufacturer's instructions (available online at www.biosearchtech.com/support/resources/stellaris-protocols).
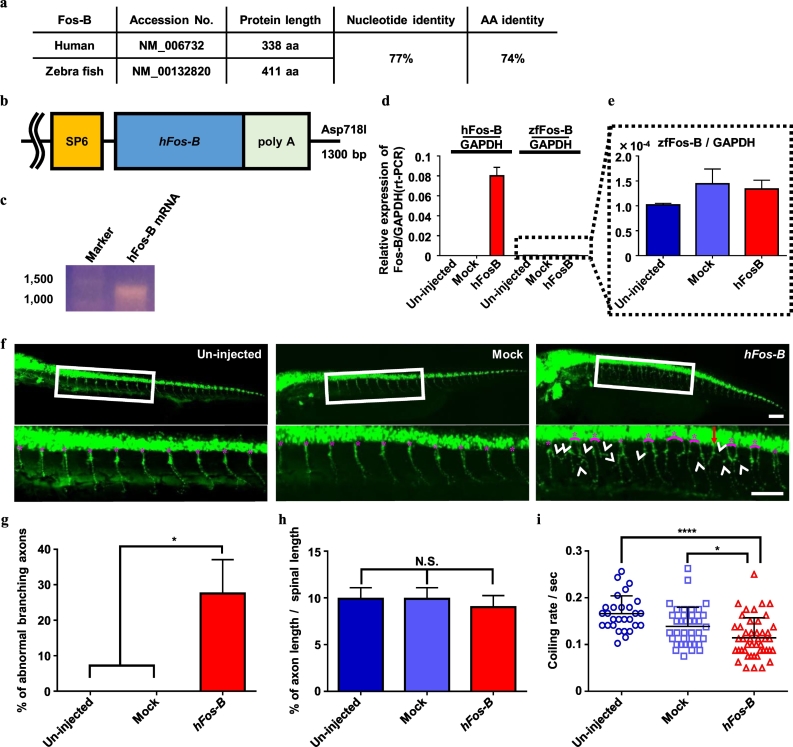
1.20. Administering mRNA injection to zebrafish
For hFos-B mRNA construction, the hFos-B sequence was subcloned into pCS2+ vectors for the administration of the poly-A tail under the hFos-B sequence (Fig. 6b). The constructed vector was digested with Asp718I and hFos-B mRNA-poly-A was synthesized via in vitro transcription using the mMessage mMachine SP6 kit (Ambion). Following RNA purification using the MinElute RNeasy cleanup kit (Qiagen), 800 pg of mRNA was injected into the yolk of fertilized hb9:Venus zebrafish embryos at the one-cell stage using FemtoJet and InjectMan NI2 (Eppendorf). As a mock control, a similar amount of RNase-free water was injected using the same method.
Fig. 6.
hFos-B regulated axon branching in vivo.
(a) Fos-B nucleotide and protein differences between human and zebrafish. Nucleotides correlated at 77%, whereas proteins correlated at 74%. The alignments of human and zebrafish Fos-B cDNA and amino acid (aa) sequences were compared using nucleotide BLAST (https://blast.ncbi.nlm.nih.gov/Blast.cgi) and eBioX, respectively. (b,c) The schema of injected hFos-B mRNA. (d,e) RNA expression levels were confirmed using qRT–PCR. The zf-Fos-B levels were not different among the three lines. (N = 20 embryos were analyzed in one experiment; qRT–PCR was triplicated and analyzEd.) (f) Representative images of the zebrafish axons at 24 hpf. Lower figures represent magnified images of the white squares of the upper figures. Abnormal spinal roots (red arrow) and axon branching (white arrowheads) were confirmed only in the hFos-B mRNA injected line. The pink asterisks represent the one set of spinal roots. Bars: 100 μm. (g) The % of branching axons at 20 axons/line at 24 hpf. Axon branching was significantly increased in the hFos-B mRNA-injected line. No branching was observed at 24 hpf in un-injected or mock control lines. N = 4, one-way ANOVA. (h) Quantification of axonal length at 24 hpf. The axonal length was adjusted by spinal lengths (the length among 20 spinal roots was defined as the spinal length). No differences in spinal length were observed among the three lines (N = 3, one-way ANOVA). (i) Quantification of the coiling rate/s. The coiling rate significantly decreased in the hFos-B-mRNA injected line. N = 28 (un-injected), 38 (mock), and 46 (hFos-B). The 40 s movies (Supplementary video 1, Supplementary video 2) were analyzed with duplication; one-way ANOVA. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
1.21. Axonal counting of zebrafish
At 24–72 hpf, zebrafish embryos were manually deyolked, fixed overnight with 4% PFA, and washed thrice with PBS. Subsequently, the caudal end was cut off and embedded on a glass slide. The axon was visualized by the fluorescence of Venus and was captured using a BZX-900 (Keyence) and counted.
1.22. Motor functional assay of zebrafish
At 24 hpf, zebrafish movement was captured using a digital camera and analyzed for spontaneous coiling rates by observing the coiling number in a 40-s period (also see Supplementary video 1, Supplementary video 2).
1.23. RNA extraction from zebrafish
At 24 hpf, RNA was extracted from 20 embryos using an RNeasy mini kit (Qiagen) according to the manufacturer's protocol.
1.24. Human postmortem samples
Patients were diagnosed with ALS using the revised El Escorial criteria [20], and the diagnosis was further pathologically confirmed by postmortem examination. The spinal cords were removed, and the blocks of each level were immediately placed in 10%-buffered formalin, embedded in paraffin, and subjected to neuropathological examination, or immediately frozen for biochemical examination. For immunohistochemistry (IHC), formalin-fixed, paraffin-embedded 4-μm-thick sections obtained from Controls and patients with ALS were deparaffinized. Following antigen retrieval by heat/autoclaving (5 min at 121 °C in 10 mM sodium citrate buffer, pH 6.0), the sections were incubated with primary antibodies overnight at 4 °C, followed by incubation with secondary antibodies for 30 min and then with biotinylated streptavidin (Histofine Kit; Nichirei Biosciences) for 30 min at RT. For sporadic ALS (sALS) autopsied sample analysis, cases with phosphorylated TDP-43 (pTDP-43) deposits were selected for Fos-B staining and analysis. Samples were captured with a TissueFAXS (Novel science) and analyzed using ImageJ as described below. MNs were defined as neurons with a major radius of >20 μm. For counting Fos-B-positive cells, DAB-stained images were divided into each stained image using the Colour Deconvolution v1.8 plug-in. Fos-B-positive cells were defined as those cells displaying Fos-B staining that was higher than background (>2 fold).
1.25. Quantification and statistical analysis
Graphpad Prism 6 (MDF) was used for statistical analysis. The number of biological replicates, animals, and cells were specified in the figure legends. For axon counting, 50 axons from three independent experiments (16–17 axons were captured from one batch) were analyzed for the following figures: Fig. 1e–g, 5c–e and g–i, and Supplementary Fig. 7c–e and g–i. Data were shown as the mean ± standard error, and P < 0.05 was considered significant. Data with no significant differences are described as “N.S.” From the ANOVA analysis, asterisks are used to denote the level of significance, with *P < 0.05, **P < 0.01, ***P < 0.001, and ****P < 0.0001.
2. Introduction
ALS is a devastating adult-onset neurodegenerative disorder characterized by the progressive loss of upper and lower MNs [21]. Since the identification of SOD1 in 1993, >25 genes, including TARDBP, FUS, and C9orf72 have been reported as the causative genes of familial ALS. Among the candidate genes, RNA-binding protein coding genes, including FUS, have emerged as critical determinants of ALS, and FUS is one of the most frequently mutated genes in familial ALS (~5%) [21]. The C-terminal nuclear localization signal (NLS) site of FUS regulates the localization of this protein and has been found to be a mutation hotspot in familial ALS [22,23]. NLS mutation impairs the nuclear import of FUS, and the importance of mis-localized cytoplasmic FUS has been emphasized in ALS pathology [24]. However, this mislocalization of FUS was identified in various patient-derived cell types other than MNs, such as hiPSCs [4]; therefore, the further investigation of MN-specific phenotypes is warranted.
The long axon is a characteristic structure of MNs and connects the soma of MNs to skeletal muscle, with lengths reaching up to 100 cm in humans. Axonal degeneration occurs prior to the motor phenotype in ALS [25,26]. Moreover, axonal degenerative changes preceding MN death have been reported in the transgenic ALS models and iPSCs-derived MNs [[27], [28], [29]]. Accordingly, the distal axon and synapse have received increasing attention as early targets of ALS pathology. Reportedly, the toxic gain of function from mutant FUS is crucial for neurodegeneration [24,30,31], and FUS is believed to play multiple roles in the nucleus and in the axonal compartments involved in axonal transport as well as in the synaptic function regulation and neuromuscular junction (NMJ) maintenance [30,32]. It has previously been reported that a disruption of survival motor neuron protein distribution in mutant FUS expresses primary cortical axon [33]; in addition, the abnormal morphologies of axons in transgenic FUS-ALS zebrafish model have been reported [34]. Moreover, progressive axonal transport defect was reported with hiPSC-derived MNs [35]. Although the axonal mRNA regulation has been reported for elucidating axon maintenance as well as in neurodegenerative diseases [36], studies regarding RNA profiling of motor axons [19,37], particularly the profiling associated with ALS axon pathology [38,39], are limited. Therefore, the axonal RNA profile remains unclear in ALS-linked human MNs without the mutant protein overexpression. The analysis of axon remains a challenge because of both the inaccessibility of patient-derived primary MNs and the low yield of samples from the axon compartment, even under cell culture conditions. Among all the causative genes of ALS, FUS was confirmed to locate at axon ends [40]. Therefore, in the present study, we focused on the effect of FUS mutation on MN axons.
The establishment of hiPSCs technology represents a major breakthrough for ALS research [41]. However, differences in the genetic backgrounds, other than the target gene (FUS), of patients and control participants lead to a reduced specificity/sensitivity of detection regarding mutation-induced phenotypes. Therefore, to address this issue, we constructed two sets of isogenic hiPSCs possessing the single amino acid difference in the FUS gene using TALEN genome editing technology [42]. By comparing the two sets of isogenic MNs, we identified increased branching in FUS-mutant MN axons compared with isogenic controls. Moreover, in conjunction with our innovative microfluidic device [11], we further revealed the entire in vitro RNA profile of the human MN axon and identified Fos-B mRNA as a target of FUS as well as a causative factor of aberrant axon morphology even in vivo. The improvement in axon morphology with the suppression of the abnormally upregulated Fos-B with FUS-mutants suggests a promising target to which ALS-linked mutations cause axonal retraction and degeneration, which are one of the earliest events in the disease.
3. Results
3.1. Established hiPSC-lines reproduce FUS mislocalization pathology
For analyzing the specific pathology of the FUS mutation, we established two sets of isogenic hiPSCs using TALEN genome editing technology. One of the isogenic pairs included a control hiPSC line that has previously been established and reported as 409B2 (described as the “Control” below) [1] and the isogenic FUS-mutant line, “FUSH517D/H517D,” previously reported as FUSH517D/H517D-1, generated by the knock-in of the p.H517D mutation in the homozygote [4]. The other isogenic pair was a hiPSC line obtained from a patient with FUS-mutated ALS (previously reported as 2e3 [4]; described as “FUS-ALS” hereafter) and the isogenic control line, “FUSRescued,” established in the present study (Fig. 1a and Supplementary Fig. 1a). All lines were validated for substituted codons, pluripotency markers, normal karyotypes, genomic integrity, and developmental potency (Supplementary Fig. 1b–g). We induced spinal MNs that were differentiated from hiPSCs using a previously described protocol based on the chemically transitional-embryoid-body-like state [43] (Supplementary Fig. 2a). Various neuronal and MN markers were further confirmed using ICC (Supplementary Fig. 2b). Constant and high MN induction efficiency (approximately 70% cells were ChAT positive at 20 DPP) was confirmed, which was consistent with the findings of our previous report (Supplementary Fig. 2c) [5].
The mutant FUS protein reportedly mis-localizes from the nucleus to the cytoplasm and co-localizes with SG markers under various stressors [44,45]. Under unstressed conditions, FUS staining was occasionally observed in the nucleus as well as in the cytoplasm (Supplementary Fig. 2d). Following the administration of NaAsO2 (Supplementary Fig. 2e), SGs stained with G3BP showed no difference between the controls and FUS-mutants (Supplementary Fig. 2f). However, the co-staining of FUS with G3BP was found to be increased in FUS-mutants in the cytoplasm (Supplementary Fig. 2g). This abnormal accumulation of cytoplasmic FUS was further confirmed in hiPSCs and MPCs (Supplementary Fig. 2h).
3.2. FUS-mutant MNs demonstrate aberrant axon branching
When MNs were plated into single cells following the dissociation of MPCs, we were unable to discriminate the axon structure from that of the dendrites. Therefore, we plated single 2nd MPC spheres onto the center of single PO/M-gel-coated-wells without dissociation into single cells (Fig. 1b). After an MPC sphere was plated, the neurites radially elongated (Fig. 1b) and the MNs remained attached up to 40 DPP (Fig. 1c). MNs could be visualized using lentivirus carrying reporter gene to express the Venus fluorescence (an altered yellow fluorescent protein [13]) under HB9 gene enhancer (HB9e438::Venus) [12] (Fig. 1b–d), which enabled us to monitor MN axon elongation. At plating, a significant portion of MPCs were observed to be Venus-positive (>30%), with no significant differences between the cell lines (Supplementary Fig. 3a–c). The TAU1 and MAP2 staining patterns changed approximately 450 μm away from SDs (Supplementary Fig. 3d,e), which is consistent with a previous report [14]. MAP2 labels dendrites, whereas TAU1 labels the axons of mature neurons. Therefore, we defined axons with the distance away from SDs and analyzed Venus-positive axons at least 1 mm from the SDs.
We observed an abnormal axon morphology in FUS-mutant MNs compared with that in each of the isogenic MNs at 10 DPP (Fig. 1d) without differences in axon length (Supplementary Fig. 3f). We confirmed that this branching originated from one axon because parallel extended axons typically overlap only up to a maximum of 80 μm (Supplementary Fig. 3g,h). We defined this process as “spine-like branching” and “axon branching” and further quantified these observations (Supplementary Fig. 3i,j). Compared with control MNs, the degree of axon branching was significantly increased in FUS-mutant MNs (Fig. 1e–g), with no correlation with axon length (Supplementary Fig. 3k–m). Our results indicated that the FUS mutation disrupts the normal axon morphology of MNs.
3.3. Global expression profiling in human MN axons using a “nerve organoid” microfluidic device
To further investigate the molecular pathology of FUS-mutant axons, we hypothesized that mutant FUS mislocalization would lead to abnormal amounts of RNA in the axon via the RNA binding function. Therefore, we extracted axon-dominant RNA to analyze the global RNA profile using RNA-seq in conjunction with a recently established device, which we named “nerve organoid” microfluidic device [11]. This device enabled us to culture organoid-mimicking motor nerve tissue and harvest macroscopically observable axon bundles (Fig. 2a). Compared with previously reported axon compartment analyses [19,[37], [38], [39]], our system facilitated the production of a large number of axons (Supplementary Fig. 4a). Macroscopically observable axon bundles were observed to extend from MPCs into the microfluidics at 20 DPP (Fig. 2b). Simultaneously, HB9 reporter-positive MNs extended up to the other well (Fig. 2c). By physically disconnecting the axon bundle (450 μm away from the spheres), the axons were effectively divided (Fig. 2a,b). We subsequently performed RNA-seq using the SDs and individual axons with Control MNs.
Most of the selected genes that have reportedly been expressed in either SDs or the axon compartments of primary mouse spinal MNs were reproducibly expressed in our system (Fig. 2d–f) [19]. Differentially expressed genes (DEGs) were extracted from SDs (n = 1052) and the axon compartment (n = 884) (Fig. 2g, Supplementary Fig. 4b–d, and Supplementary Table 1,2). The enriched genes categorized by gene ontology (GO) revealed protein binding (GO: 0005515), poly (A) RNA binding (GO: 0044822), and enzyme binding (GO: 0019899) genes in the axon compartment, consistent with those observed in the previous report (Fig. 2h,i, and Supplementary Table 3) [38].
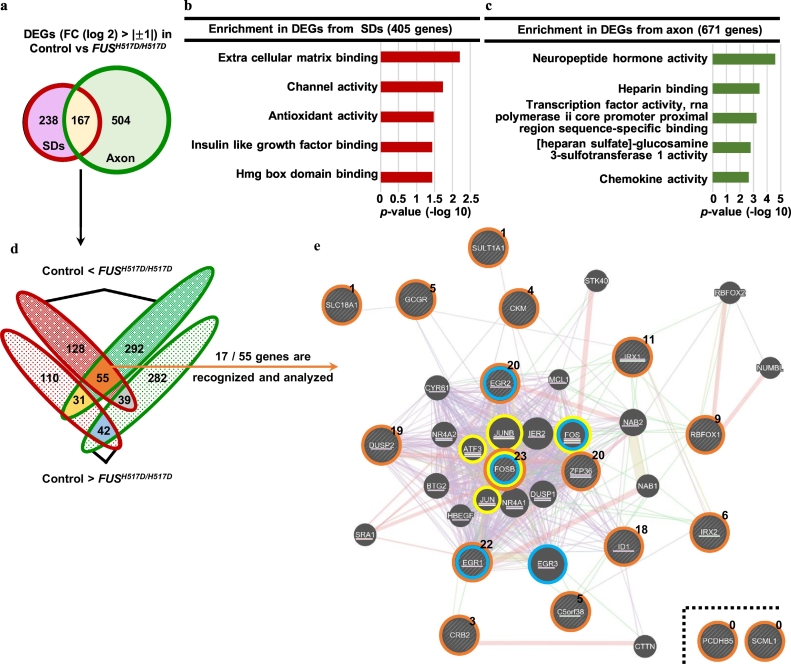
3.4. Aberrant expression of Fos-B mRNA, the binding target of FUS, in FUS-mutant ALS MNs
To understand the pathological target of the aberrant axon morphology observed with the FUS mutation, we compared the RNA profiles of MNs with Control and FUSH517D/H517D hiPSC-derived MNs. The RNA profiles from the SDs indicated that the extracted MNs represented well-matured upper cervical spinal MNs [46,47], with no differences observed among the cell lines (Supplementary Fig. 5a,b). To observe the differences in the global gene expression profile with the FUS mutation on MNs, we performed GO term analysis for the transcripts using the DEGs between Control and its isogenic cell FUSH517D/H517D SDs and axons (Fig. 3a and Supplementary Table 4). Transcripts related to the extracellular matrix (ECM) were observed to be particularly enriched in the SDs. In contrast, we observed that the DEGs with neuropeptide hormone activity were enriched on the axonal side (Fig. 3b,c). We attempted to focus on the genes that specifically changed in FUS-mutant axon because of our hypothesis that axonal transport of mutant FUS transport leads to the abnormal amounts of RNA in the axon via the RNA binding function. We attempted to identify the transcript that is upregulated in the axon and increase the toxic gain of function in the present study. Extracting axonal fraction of MNs using newly developed microfluidic device enabled us to identify novel target genes involved in the FUS-mutant pathology. Among these, we focused on the axon-dominant gene and not the axon-specific gene because of the technical reason that small amount of sample of axonal fraction is unsuitable for the deeper analysis of molecular pathomechanism in protein level (i.e., RIP). To identify the upregulated genes with FUSH517D/H517D compared with Control in both fractions, we performed a network analysis with 55 genes [17 of 55 genes (highlighted with orange in Fig. 3d) were recognized using the GeneMANIA online tool (https://genemania.org/)] [48]. Consequently, we observed that the zinc-finger nucleases activator protein 1 (AP-1) (comprising the Jun-, ATF-, and Fos-families, including Fos-B) and the immediate early gene (IEG)-related genes of the EGR- and Fos-families were accumulated in FUS-mutant MNs (Fig. 3d,e). Because Fos-B contains the largest number of related genes (Fig. 3e) among the aforementioned genes, we considered Fos-B to play a key role in FUS-mutant MNs. Among the 42 genes that were downregulated in FUS-mutant MNs, we were unable to limit the focus although we adapted the network analysis using GeneMANIA (data not shown). In addition to confirming the increase in Fos-B at the protein level, we confirmed the increase in variations in Fos-B associated with FUS mutation (Supplementary Fig. 5c,d). There were no differences in FUS expression among the four cell lines (Supplementary Fig. 5c,d).
Fig. 3.
Global analysis of the FUS mutation in MNs revealed the accumulation of AP-1 related genes.
(a) DEGs (FC difference of |log 2| > 1) of Control and FUSH517D/H517D MNs of the SDs and axon fractions were compared. (b,c) GO enrichment analysis with DEGs in (a). The top 5 terms are listed with a p-value (Fisher's exact test). (d) The detailed results of DEGs in (a). The genes are listed in Supplementary Table 4. (e) Visualization of the gene network using the set of 55 genes of upregulated DEGs in FUS-mutant MNs shown in (d). Using the GeneMANIA online tool (https://genemania.org/) [48], 17 of 55 genes were analyzed (represented by orange circles). The numbers of related genes (connected with lines described below) are denoted using superscript letters. Particularly, Fos-B-related genes are denoted using underlines. Purple lines represent co-expression connections. Red lines represent physical interactions. Yellow lines represent shared protein domains. Blue lines represent co-localization. Green lines represent genetic interactions. The accumulation of AP-1- (yellow circles) and IEG- (blue circles) related genes was observed. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
Therefore, we opted to focus on Fos-B, which represented the top ranked upregulated gene in the FUS-mutant axon fraction (Fig. 4a,b), although some variation was confirmed among AP-1 related genes in RNA-seq results (Supplementary Fig. 5e). Abnormal Fos-B expression was further confirmed using qRT–PCR, and Fos-B upregulation was confirmed in another set of isogenic pairs (Fig. 4c). The presence of Fos-B mRNA in neurites was demonstrated using smFISH (Fig. 4d) in Control and FUSH517D/H517D, and Fos-B mRNA was particularly prevalent in FUSH517D/H517D MNs (Supplementary Fig. 6), which supports our RNA-seq results (Fig. 4a).
Further, we investigated whether Fos-B induces abnormal branching in MNs. Accordingly, we first examined the binding between Fos-B mRNA and the FUS protein. RIP experiments revealed that the FUS protein binds Fos-B mRNA (Fig. 4e,f). Next, RNA pulldown assays using the biotinylated RNA in conjunction with streptavidin beads revealed that the FUS protein bound to Fos-B 3′UTR sequences more efficiently than to the control or the Fos-B mRNA probe (Fig. 4g). The Fos-B 3′UTR contained the FUS binding sequence (Fig. 4h) [49]. To demonstrate the differences in RIP sample from the Control and FUSH517D/H517D, RIP and RNA-pulldown assay was examined for each of the cell lines. As anticipated, Fos-B mRNA was upregulated in RIP sample from FUSH517D/H517D (Supplementary Fig. 5f,g). In addition, the RNA-pulldown assay using the Control and FUSH517D/H517D samples revealed no differences between these samples (Supplementary Fig. 5h,i). Therefore, the affinity of FUS for Fos-B (3’UTR) was estimated to be equivalent. Because FUS contains an RNA recognition motif, which is far from the ALS mutation hotspot (NLS), it is considered as a valid result.
We further investigated the functional pathway that contributes to the morphological abnormality of MNs by Fos-B. For these experiments, we constructed a Venus-expression lentivirus (EF-1α::Venus) and a Fos-B/Venus-expression lentivirus under the EF-1α promotor (EF-1α::Fos-B/Venus). To focus simply on the effect of Fos-B overexpression, because the Fos-B was up-regulated in whole fraction in FUS-mutant MNs, the EF-1α::Fos-B/Venus did not include Fos-B 3’UTR. We introduced these lentiviruses into Control hiPSC-derived 2nd MPCs at the same MOI (MOI = 1) (Fig. 4i) and compared the RNA profiles at 10 DPP using microarray analyses (Supplementary Table 5). GO term and KEGG pathway analyses revealed that the ECM-related genes were affected by Fos-B overexpression (Fig. 4j–l).
3.5. Suppression of aberrant Fos-B expression rescued the cellular phenotype of FUS-mutant MNs
Further, we intervened in the Fos-B pathway using several techniques, including overexpression as well as the use of si-RNA and a chemical compound. Fos-B suppression in FUS-mutant MNs rescued the abnormal increase in axon branching to normal levels (Fig. 5a,b). The quantification of the axonal branching revealed significantly reduced aberrant morphology in the FUS-mutants (Fig. 5c–e). Moreover, T5224, an inhibitor of AP-1 [50], partially normalized the aberrant morphology without affecting Fos-B mRNA expression levels (Fig. 5f–i). On the other hand, Fos-B overexpression using EF-1α::Fos-B lentivirus infection of control MNs showed deteriorated axon morphologies (Supplementary Fig. 7a–e) The EF-1α::Fos-B construct was sufficient for the production of multiple Fos-B variations (Supplementary Fig. 7b) as detected in FUSH517D/H517D-derived MNs (Supplementary Fig. 5c). Moreover, we attempted to examine whether these observed abnormal phenotypes could be reconfirmed with other ALS-causative mutations. To examine other FUS mutations other than p.H517D, we constructed new isogenic FUS-mutant hiPSCs (FUSP525L/+) that possesses a p.P525L mutation—one of most common mutation in FUS-ALS—on FUS of 201B7 healthy control hiPSCs (Control-2; established previously [2]) using CRISPR genome editing (Supplementary Fig. 7f–m). Moreover, SOD1- and TARDBP-mutant hiPSCs were used for examining other ALS-causative mutations than FUS. As a result, abnormal axon branching was detected in FUSP525L/+-, SOD1-, and TARDBP-mutant hiPSC-derived MNs (Supplementary Fig. 7f–i). FUSP525L/+- and TARDBP-mutant hiPSC-derived MNs demonstrated Fos-B dysregulation similar to FUSH517D-mutant MNs, whereas SOD1-mutant hiPSC-derived MNs did not exhibit this dysregulation (Supplementary Fig. 7j–m).
3.6. Fos-B overexpression affects MN axons in zebrafish
Our in vitro results indicated that Fos-B regulates the abnormal axonal morphology in FUS-mutant MNs. To confirm the effect of Fos-B using an in vivo model, we used hb9:Venus zebrafish [9], in which MNs were visualized by Venus. The predicted nucleotide and amino acid sequence of zebrafish Fos-B (zf-Fos-B) demonstrated approximately 77% and 74% similarity with human Fos-B (hFos-B), respectively (Fig. 6a). We injected hFos-B mRNA into hb9:Venus embryos and examined them at 24–72 h post fertilization (hpf). qRT–PCR analysis confirmed that this mRNA injection resulted in a higher expression levels of hFos-B without affecting the endogenous expression of zf-Fos-B (Fig. 6b–e). In addition, we observed significantly increased abnormal branching in Venus-positive MNs in hFos-B mRNA-injected zebrafish (Fig. 6f,g). Moreover, abnormal spinal rout was confirmed only in hFos-B mRNA-injected zebrafish (Fig. 6f). There were no significant differences in axonal length (normalized with spinal length) following hFos-B mRNA injection (Fig. 6h). These phenotypes could be identified with 48 hpf, however, at 72 hpf, it was challenging to count the axon branches because a considerable number of branches were detected at this time-point (Supplementary Fig. 8). Furthermore, hFos-B mRNA-injected zebrafish showed aberrant motor function at 24 hpf. The spontaneous coiling rate was significantly decreased in the hFos-B mRNA-injected line (Fig. 6i, Supplementary video 1 and Supplementary video 2). The findings indicate the aberrant expression of hFos-B mRNA results in MN axon abnormality and motor dysfunction in vivo.
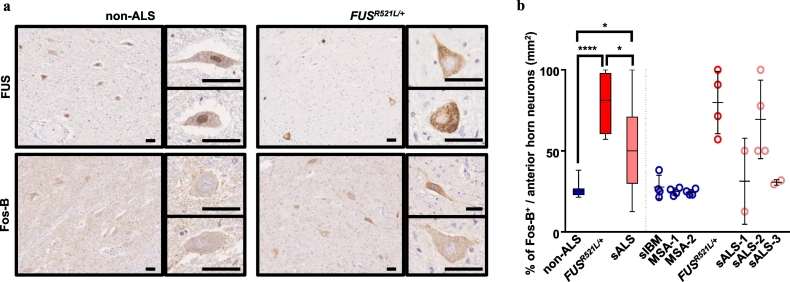
3.7. Abnormal Fos-B upregulation observed in ALS autopsy samples
Finally, to observe the Fos-B expression in human ALS, we used IHC to examine the spinal cords of a patient with familial ALS who exhibited the mutation on FUSR521L/+. Using autopsy samples, we observed FUS mislocalization from the nuclei to the cytoplasm in the patient with FUSR521L/+ (upper panel in Fig. 7a), which was consistent with the findings of a previous study [51]. Fos-B was upregulated in the cytoplasm of the ventral horn neurons in the spinal cord of the patient with FUSR521L/+ (lower panel in Fig. 7a). Moreover, this abnormal upregulation of Fos-B in the ventral horn neurons was detected in sALS autopsy samples with some diversity (Supplementary Fig. 9a,e). A significantly higher number of Fos-B-upregulated ventral horn neurons were observed in ALS autopsy samples (Fig. 7b), without correlation with survival neuron numbers (Supplementary Fig. 9b,c) or the age at death (Supplementary Fig. 9d).
Fig. 7.
Abnormal accumulation of Fos-B was confirmed in the ALS spinal cord.
(a) Representative IHC images of autopsy samples from a patient with sporadic inclusion body myositis (sIBM; non-ALS) and one with FUSR521L/+ mutation. FUS was mislocalized in the FUSR521L/+ sample, whereas it was nuclear dominant in the non-ALS sample. Compared with non-ALS samples that did not stain with Fos-B, the cytoplasmic dominant staining of Fos-B was confirmed in FUSR521L/+ samples. Bars: 50 μm. (b) The quantification of cytoplasmic Fos-B-positive cells with three non-ALS [four sections each from one sIBM and two multiple system atrophy (MSA)], one FUSR521L/+, and three sporadic ALS (sALS) (four sections from one case and two sections from two cases) was analyzed using one-way ANOVA. See also Supplementary Fig. 9.
4. Discussion
The use of patient-derived hiPSCs enabled us to overcome several obstacles typical in animal models, including species-specific differences between humans and rodents [52]. To exclude the genetic variation-derived phenotypes in hiPSCs, we utilized two pairs of FUS isogenic lines using TALEN genome editing technology. The reproducibility of the FUS mislocalization that we observed indicated the usefulness of the two pairs of isogenic lines with physiological levels of mutant FUS (Fig. 1d–g) [24]. On the other hand, we confirmed that FUS mislocalization occurs in MNs as well as in hiPSCs (Supplementary Fig. 2 h), which is consistent with the findings of a previous report [4]. Such mutant FUS mis-localizations have been reported in non-MN cells as well [53], suggesting that FUS mislocalization is insufficient for inducing MN-specific cell death in ALS.
During the examination of the MN phenomenon, we observed an abnormal increase in axon branching in FUS-mutant hiPSC-derived MNs (Fig. 1d). The prevalence of this phenotype in multiple FUS (p.H517D and p.P525L)-, SOD1-, and TARDBP-mutant hiPSC-derived MNs (Supplementary Fig. 7f–m) indicated the importance of the finding. Moreover, abnormal neural branching has previously been detected in zebrafish with mutant FUS overexpression [24] and SOD1G37R mice [54]. We observed the axonal phenotype at 10 DPP, which is a rather early time-point compared with the timing of apoptosis that we have previously reported (approximately 20 DPP is equivalent to 40 days in vitro [5]). This suggests that aberrant axon branching is a surrogate marker for screening drugs for FUS-mutant MNs in the cellular setting. ALS is known as an adult-onset neurodegenerative disorder, and this insight proposes the pathological hallmark progress in the developing stage. Further, we observed abnormal axon morphology and mobility in Fos-B-overexpressed zebrafish, indicating the importance of this overlooked phenotype of axon morphology (Fig. 6). However, abnormal axon branching regulated by Fos-B could be promising in addressing a key question that has not been uncovered in the field: “How do ALS-causative FUS mutations cause axonal retraction and degeneration?” which is one of the earliest events in the disease [28]. The phenomenon might only affect developing neurons; therefore, additional experiments using adult ALS models are warranted.
A nerve organoid microfluidic device with a large canal enabled us to collect adequate samples from the isolated MN axons for RNA-seq [11]. Such a device is useful for the observation of the global profiling of the compartment of the cell (i.e., the axon). Although there are other types of microfluidic devices commercially available, they typically are restricted in their use owing to the limited specimen amounts (Supplementary Fig. 4a). Because only low specimen amounts can be analyzed, various conditions, such as cell purity and neuron status, as well as variations in procedures, could easily influence the results. The microfluidic device we chose enabled us to perform our analysis, with only a few technical biases, by collecting large amounts of macroscopically observable axon bundles adequate for the immunochemical confirmation of the axonal protein TAU1. In addition, nuclear staining signals were absent in the axon fascicle [11]. Although our culture method enabled the production of pure MNs (approximately 70% cells were ChAT positive), RNA profiles from the axon samples reproduced the previously reported MN axon profiles (Fig. 2d–f) [19], justifying the methodology of our new device. Other than the gene sets above, axon-enriched genes did not appear to adequately match with those of previous reports (Axon; 64/884 with Briese et al. and 88/884 with Rotem et al. Supplementary Fig. 4b,c); this finding was consistent with recently reported hiPSC-derived neuronal axon existence genes (Supplementary Fig. 4d). Such differences may be attributed to differences between species (mouse vs. human), cell type, and cell purity as well as the various methodologies applied (Supplementary Fig. 4a). However, our data could be an important resource for the subcellular analysis of human MN axons. Moreover, our concept of the nerve organoid device could be applied to other neuron types or for proteomic analysis using human pluripotent cells.
The comparison of control and FUS homo-mutant axons revealed both cell type and cell fraction effects associated with FUS mutation that have not previously been revealed. From our results of RNA-seq, we opted to focus on Fos-B for several reasons: 1) Because mutant FUS mis-localizes from the nucleus into the cytoplasm, the RNA that increased in the axon together with FUS should be important; 2) Fos-B was the key gene in the upregulated genes observed in FUS-mutant MNs (Fig. 3e); 3) Fos-B was significantly increased, particularly in the FUS-mutated axon fraction (Fig. 4c); 4) Fos-B mRNA was revealed to be the binding target of FUS (Fig. 4e–g); 5) the intervention of the Fos-B pathway regulated the axon morphology in hiPSC-derived MNs (Fig. 5 and Supplementary Fig. 7a–e) and in vivo (Fig. 6f). Although Fos-B was upregulated by kainic acid induced-neural excitation [55], no spontaneous excitation was observed with our hiPSC-derived MNs at 10 DPP with Ca2+ imaging (data not shown). Therefore, Fos-B upregulation was independent from the excitability at the time-point we observed. Previous studies have reported Fos-B mRNA upregulation with an increase in the number of spines [56,57] and number of growth cones [58]. In addition, delta-Fos-B has been observed to modulate immature spines in nucleus accumbens in a drug addiction model [59]. Although the mechanism is yet to be resolved, Fos-B increase in FUS-mutant MNs, even in protein levels (Supplementary Fig. 5c,d) suggested the importance of Fos-B in axon morphogenesis.
T5224 is an AP-1 inhibitor that was developed as a therapy for rheumatoid arthritis [50]; however, it can be used as a drug-repositioning candidate for ALS. Indeed, a recent report regarding AP-1 on MN degeneration in a SOD-1 mutant ALS model has attracted attention [60]. In addition, the suppression of dual leucine zipper kinase, which is the upstream signaling molecule of c-Jun (member of AP-1), is reportedly a therapeutic target for neurodegenerative diseases, including ALS [61]. Although SOD1-ALS MNs did not reveal Fos-B mRNA upregulation in the present study (Supplementary Fig. 7j,m), differences in genetic background or other AP-1 related factors may have influenced the results [60]. In fact, some variations in AP-1-related genes were observed in the present study as well (Supplementary Fig. 5e). Although we did not examine the effects of other AP-1-related genes, it is possible that these genes are important for axon morphogenesis. Such substantial differences of SOD1-ALS from FUS- or TARDBP-ALS have been reported [5]. The common characteristics observed among SOD1-, TARDBP-, and FUS-mutant MNs suggest the importance of AP-1 in ALS-associated neurodegeneration. As reported in the present study, the pathological abnormal accumulation of the Fos-B protein in patients with ALS, including sporadic cases (Fig. 7b), indicates the importance of Fos-B in ALS beyond FUS-ALS. Although FUS mislocalization remains unclarified in sALS pathology (Supplementary Fig. 9a,e), other mechanisms that regulate Fos-B expression could be at play rather than FUS mislocalization, which were observed in FUS-ALS. Reconciling developmental defects and degeneration in the aging human is a major concern in the field of stem cell biology. The axon branching is associated with motor function in zebrafish; however, the link between these two events remains unclear. In addition, the abnormal branching phenomenon might not be a MN-specific event in the FUS-mutant model. To resolve such questions, a FUS-mutant mouse neurodegeneration model would be a useful tool for confirming the therapeutic potential of AP-1 inhibition in a future study. Although the reason for the increased axon branching with Fos-B upregulation and the manner in which the increased axon branching induced ALS pathology remain unclear, we observed that the expression of ECM and focal adhesion family genes fluctuated with FUS mutation and Fos-B expression in hiPSC-derived MNs (Fig. 4k,l). Moreover, the ECM pathway fluctuated with the SDs fraction of FUS-mutant MNs (Fig. 3b) and the previously reported FUS-mutant mouse spinal neurons [62]. ECM and focal adhesion may affect the neuronal growth cone and filopodia morphology [63]. Some previous reports have suggested the important roles of IEGs, including Fos-B in the synaptic modulation of neurons [58]; however, no definitive molecules that were involved in modifying the axon branching were observed. In addition, p53 was the second common pathway regulated by Fos-B expression (Fig. 4l). Although p53 has been reported to be increasingly expressed in FUS-mutant MNs [64], the inhibition of p53 results in a modest improvement in survival in rodent ALS models [65]. In addition, prior to axonal degeneration, the disruption of axonal branches, which results in asynchronous NMJ destruction [54], was identified in SOD1G37R mice. Such findings highlight the significance of the relationship between Fos-B and ALS pathology, including the phenomenon of abnormal axon branching.
In summary, analyzing the axonal fraction of FUS-mutant MNs using an innovative microfluidic device revealed that Fos-B is a key regulator of FUS-mutant axon branching, thereby promising an early pathological hallmark of ALS axon degeneration and providing insights on axogenesis in neurodevelopment.
The following are the supplementary data related to this article.
Coiling movement of mock control zebrafish
Coiling movement of hFos-B injected zebrafish
Supplementary Tables
Supplementary Figs and legends
Author contributions
T.A., N.S., and M.A. developed hypothesis and designed the experiments. T.A. performed all experiments using hiPSCs. M.I., K.F., T.So., S.Mo., T.K., M.Ni., M.No., K.O., and H.Ok. contributed toward hiPSC culture and established isogenic lines. J.K., S.K., Y.I., and T.F. developed and provided microfluidic devices. S.Mi., K.I., H.On., T.Sh., and H.W. contributed toward hiPSC culture. Y.O. contributed toward reporter lentivirus construction. R.F., M.S., T.N., and K.N. contributed toward RNA-seq. S.O., Y.K., A.N., and R.I. assisted with the genetic analysis. H.M. contributed toward the zebrafish experiment. F.F. performed the examination of autopsy samples. T.A., N.S., and M.A. prepared the manuscript with revision and input from all the authors. All authors read and approved the final manuscript.
Sources of funding
This work was supported by Grants-in-Aid for research on rare and intractable diseases (M.A.); Research Committee on Establishment of Novel Treatments for Amyotrophic Lateral Sclerosis (M.A.); the Research Center Network for Realization Research Centers/Projects of Regenerative Medicine (the Program for Intractable Disease Research Utilizing Disease-specific iPS Cells and the Acceleration Program for Intractable Diseases Research Utilizing Disease-specific iPS Cells) and Practical Research Project for Rare/Intractable Diseases (M.A., H.Ok.) (16ek0109013h0003, 16ek0109158h0002, 16bm0609003h0005, 17bm0804003h0001, 18bm0804003h0002, 19bm0804003h0003) from Japan Agency for Medical Research and development, AMED; Grant-in-Aid for Young Scientists (A) (15H05667 to N.S.), Grant-in-Aid for Scientific Research (C) (18 K07519 to N.S.), Grant-in-Aid for Scientific Research (B) (25,293,199 and 16H05318 to M.A.) from Japanese Ministry of Education, Culture, Sports, Science, and Technology, Clinical Research, Innovation and Education Center, Tohoku University Hospital (CRIETO); Japan Intractable Diseases (Nanbyo) Research Foundation (to N.S.), the Kanae Foundation for the Promotion of Medical Science (to N.S.), and “Inochi-no-Iro” ALS research grant (to N.S.).
Declaration of interests
H.Ok. is a Scientific Advisor at SanBio Co.Ltd. and K Pharma Inc. Y.O. is a Scientific Advisor at Kohjin Bio Co., Ltd. Other authors declare no competing financial interests.
Acknowledgments
The authors thank S. Maeda, S. Ishii, N. Ichiyanagi, S. Yoshida, J. Kobayashi, N. Sugeno, T. Hasegawa, H. Aoyama, S. Shibata, A. Intoh, and K. Eggan for their excellent advice; N. Shimakura and A. Machii for their technical assistance; M. Shikama for data acquisition; K. Kuroda, M. Kikuchi, M. Nakagawa, and M Nakagawa for RNA-seq; Y. Aoyama for staining human spinal cords; and A. Miyawaki and H. Miyoshi for donating lentivirus vectors. We would also like to thank Professors Haruhisa Inoue, Shinya Yamanaka, and Masato Nakagawa (Kyoto University) for donating hiPSC clones (409B2, 201B7, and CiRA00026). The hiPSCs were maintained at biomedical research unit of Tohoku University Hospital. Zebrafish line Tg [hb9:Venus] was obtained from National BioResource Project, Zebrafish Core Institution, Japan. This work was supported by Research and Study Project of Tokai University Educational System General Research Organization [to H.M]. Sanger sequencing of DNA constructs was performed by the Support Center for Medical Research and Education, Tokai University.
References
- 1.Okita K., Matsumura Y., Sato Y., Okada A., Morizane A., Okamoto S. A more efficient method to generate integration-free human iPS cells. Nat Methods. 2011 May;8(5):409–412. doi: 10.1038/nmeth.1591. [DOI] [PubMed] [Google Scholar]
- 2.Takahashi K., Tanabe K., Ohnuki M., Narita M., Ichisaka T., Tomoda K. Induction of pluripotent stem cells from adult human fibroblasts by defined factors. Cell. 2007 Nov 30;131(5):861–872. doi: 10.1016/j.cell.2007.11.019. [DOI] [PubMed] [Google Scholar]
- 3.Akiyama T., Warita H., Kato M., Nishiyama A., Izumi R., Ikeda C. Genotype-phenotype relationships in familial amyotrophic lateral sclerosis with FUS/TLS mutations in Japan. Muscle Nerve. 2016 Sep;54(3):398–404. doi: 10.1002/mus.25061. [DOI] [PubMed] [Google Scholar]
- 4.Ichiyanagi N., Fujimori K., Yano M., Ishihara-Fujisaki C., Sone T., Akiyama T. Establishment of in vitro FUS-associated familial amyotrophic lateral sclerosis model using human induced pluripotent stem cells. Stem Cell Rep. 2016 Apr 12;6(4):496–510. doi: 10.1016/j.stemcr.2016.02.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Fujimori K., Ishikawa M., Otomo A., Atsuta N., Nakamura R., Akiyama T. Modeling sporadic ALS in iPSC-derived motor neurons identifies a potential therapeutic agent. Nat Med. 2018 Oct;24(10):1579–1589. doi: 10.1038/s41591-018-0140-5. [DOI] [PubMed] [Google Scholar]
- 6.Egawa N., Kitaoka S., Tsukita K., Naitoh M., Takahashi K., Yamamoto T. Drug screening for ALS using patient-specific induced pluripotent stem cells. Sci Transl Med. 2012 Aug 1;4(145):145ra04. doi: 10.1126/scitranslmed.3004052. [DOI] [PubMed] [Google Scholar]
- 7.Kanegae Y., Lee G., Sato Y., Tanaka M., Nakai M., Sakaki T. Efficient gene activation in mammalian cells by using recombinant adenovirus expressing site-specific Cre recombinase. Nucleic Acids Res. 1995 Oct 11;23(19):3816–3821. doi: 10.1093/nar/23.19.3816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Niwa H., Yamamura K., Miyazaki J. Efficient selection for high-expression transfectants with a novel eukaryotic vector. Gene. 1991 Dec 15;108(2):193–199. doi: 10.1016/0378-1119(91)90434-d. [DOI] [PubMed] [Google Scholar]
- 9.Nakano Y., Fujita M., Ogino K., Saint-Amant L., Kinoshita T., Oda Y. Biogenesis of GPI-anchored proteins is essential for surface expression of sodium channels in zebrafish Rohon-Beard neurons to respond to mechanosensory stimulation. Development. 2010 May;137(10):1689–1698. doi: 10.1242/dev.047464. [DOI] [PubMed] [Google Scholar]
- 10.Nusslein-Volhard CaD . Oxford University Press; New York: 2002. R. Zebrafish: A Practical Approach. 303p (Book) [Google Scholar]
- 11.Kawada J., Kaneda S., Kirihara T., Maroof A., Levi T., Eggan K. Generation of a motor nerve Organoid with human stem cell-derived neurons. Stem cell Rep. 2017 Nov 14;9(5):1441–1449. doi: 10.1016/j.stemcr.2017.09.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Shimojo D., Onodera K., Doi-Torii Y., Ishihara Y., Hattori C., Miwa Y. Rapid, efficient, and simple motor neuron differentiation from human pluripotent stem cells. Mol Brain. 2015 Dec 01;8(1):79. doi: 10.1186/s13041-015-0172-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Nagai T., Ibata K., Park E.S., Kubota M., Mikoshiba K., Miyawaki A. A variant of yellow fluorescent protein with fast and efficient maturation for cell-biological applications. Nat Biotechnol. 2002 Jan;20(1):87–90. doi: 10.1038/nbt0102-87. [DOI] [PubMed] [Google Scholar]
- 14.Taylor A.M., Blurton-Jones M., Rhee S.W., Cribbs D.H., Cotman C.W., Jeon N.L. A microfluidic culture platform for CNS axonal injury, regeneration and transport. Nat Methods. 2005 Aug;2(8):599–605. doi: 10.1038/nmeth777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Spillane M., Gallo G. Involvement of Rho-family GTPases in axon branching. Small GTPases. 2014;5:e27974. doi: 10.4161/sgtp.27974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bourne J.N., Harris K.M. Balancing structure and function at hippocampal dendritic spines. Annu Rev Neurosci. 2008;31:47–67. doi: 10.1146/annurev.neuro.31.060407.125646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Trapnell C., Pachter L., Salzberg S.L. TopHat: discovering splice junctions with RNA-Seq. Bioinformatics (Oxford, England) 2009 May 1;25(9):1105–1111. doi: 10.1093/bioinformatics/btp120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Trapnell C., Hendrickson D.G., Sauvageau M., Goff L., Rinn J.L., Pachter L. Differential analysis of gene regulation at transcript resolution with RNA-seq. Nat Biotechnol. 2013 Jan;31(1):46–53. doi: 10.1038/nbt.2450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Briese M., Saal L., Appenzeller S., Moradi M., Baluapuri A., Sendtner M. Whole transcriptome profiling reveals the RNA content of motor axons. Nucleic Acids Res. 2016 Feb 29;44(4):e33. doi: 10.1093/nar/gkv1027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Brooks B.R., Miller R.G., Swash M., Munsat T.L. El Escorial revisited: revised criteria for the diagnosis of amyotrophic lateral sclerosis. Amyotrophic Lateral Sclerosis other Motor Neuron Disorders. 2000 Dec;1(5):293–299. doi: 10.1080/146608200300079536. [DOI] [PubMed] [Google Scholar]
- 21.Taylor J.P., Brown R.H., Cleveland D.W. Decoding ALS: from genes to mechanism. Nature. 2016 Nov 10;539(7628):197–206. doi: 10.1038/nature20413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Suzuki N., Aoki M., Warita H., Kato M., Mizuno H., Shimakura N. FALS with FUS mutation in Japan, with early onset, rapid progress and basophilic inclusion. J Hum Genet. 2010 Apr;55(4):252–254. doi: 10.1038/jhg.2010.16. [DOI] [PubMed] [Google Scholar]
- 23.Nishiyama A., Niihori T., Warita H., Izumi R., Akiyama T., Kato M. Comprehensive targeted next-generation sequencing in Japanese familial amyotrophic lateral sclerosis. Neurobiol Aging. 2017 Jan 10;53(194):e191–e198. doi: 10.1016/j.neurobiolaging.2017.01.004. [DOI] [PubMed] [Google Scholar]
- 24.Shiihashi G., Ito D., Yagi T., Nihei Y., Ebine T., Suzuki N. Mislocated FUS is sufficient for gain-of-toxic-function amyotrophic lateral sclerosis phenotypes in mice. Brain. 2016 Sep;139(Pt 9):2380–2394. doi: 10.1093/brain/aww161. [DOI] [PubMed] [Google Scholar]
- 25.Fischer L.R., Culver D.G., Tennant P., Davis A.A., Wang M., Castellano-Sanchez A. Amyotrophic lateral sclerosis is a distal axonopathy: evidence in mice and man. Exp Neurol. 2004 Feb;185(2):232–240. doi: 10.1016/j.expneurol.2003.10.004. [DOI] [PubMed] [Google Scholar]
- 26.Roy S., Zhang B., Lee V.M., Trojanowski J.Q. Axonal transport defects: a common theme in neurodegenerative diseases. Acta Neuropathol. 2005 Jan;109(1):5–13. doi: 10.1007/s00401-004-0952-x. [DOI] [PubMed] [Google Scholar]
- 27.Armstrong G.A., Drapeau P. Loss and gain of FUS function impair neuromuscular synaptic transmission in a genetic model of ALS. Hum Mol Genet. 2013 Nov 1;22(21):4282–4292. doi: 10.1093/hmg/ddt278. [DOI] [PubMed] [Google Scholar]
- 28.Naumann M., Pal A., Goswami A., Lojewski X., Japtok J., Vehlow A. Impaired DNA damage response signaling by FUS-NLS mutations leads to neurodegeneration and FUS aggregate formation. Nat Commun. 2018 Jan 23;9(1):335. doi: 10.1038/s41467-017-02299-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tian F., Yang W., Mordes D.A., Wang J.Y., Salameh J.S., Mok J. Monitoring peripheral nerve degeneration in ALS by label-free stimulated Raman scattering imaging. Nat Commun. 2016 Oct 31;7 doi: 10.1038/ncomms13283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lopez-Erauskin J., Tadokoro T., Baughn M.W., Myers B., McAlonis-Downes M., Chillon-Marinas C. ALS/FTD-linked mutation in FUS suppresses intra-axonal protein synthesis and drives disease without nuclear loss-of-function of FUS. Neuron. 2018 Oct 17;100:816–830. doi: 10.1016/j.neuron.2018.09.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sharma A., Lyashchenko A.K., Lu L., Nasrabady S.E., Elmaleh M., Mendelsohn M. ALS-associated mutant FUS induces selective motor neuron degeneration through toxic gain of function. Nat Commun. 2016 Feb 4;7 doi: 10.1038/ncomms10465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.So E., Mitchell J.C., Memmi C., Chennell G., Vizcay-Barrena G., Allison L. Mitochondrial abnormalities and disruption of the neuromuscular junction precede the clinical phenotype and motor neuron loss in hFUSWT transgenic mice. Hum Mol Genet. 2017 Nov 28;27:463–474. doi: 10.1093/hmg/ddx415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Groen E.J., Fumoto K., Blokhuis A.M., Engelen-Lee J., Zhou Y., van den Heuvel D.M. ALS-associated mutations in FUS disrupt the axonal distribution and function of SMN. Hum Mol Genet. 2013 Sep 15;22(18):3690–3704. doi: 10.1093/hmg/ddt222. [DOI] [PubMed] [Google Scholar]
- 34.Kabashi E., Bercier V., Lissouba A., Liao M., Brustein E., Rouleau G.A. FUS and TARDBP but not SOD1 interact in genetic models of amyotrophic lateral sclerosis. PLoS Genet. 2011 Aug;7(8):e1002214. doi: 10.1371/journal.pgen.1002214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Guo W., Naujock M., Fumagalli L., Vandoorne T., Baatsen P., Boon R. HDAC6 inhibition reverses axonal transport defects in motor neurons derived from FUS-ALS patients. Nat Commun. 2017 Oct 11;8(1):861. doi: 10.1038/s41467-017-00911-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Jung H., Yoon B.C., Holt C.E. Axonal mRNA localization and local protein synthesis in nervous system assembly, maintenance and repair. Nat. Rev. Neurosci. 2012 Apr 13;13(5):308–324. doi: 10.1038/nrn3210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Maciel R., Bis D.M., Rebelo A.P., Saghira C., Zuchner S., Saporta M.A. The human motor neuron axonal transcriptome is enriched for transcripts related to mitochondrial function and microtubule-based axonal transport. Exp Neurol. 2018 Sep;307:155–163. doi: 10.1016/j.expneurol.2018.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Rotem N., Magen I., Ionescu A., Gershoni-Emek N., Altman T., Costa C.J. ALS along the axons - expression of coding and noncoding RNA differs in axons of ALS models. Sci. Rep. 2017 March 16;7:44500. doi: 10.1038/srep44500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Nijssen J., Aguila J., Hoogstraaten R., Kee N., Hedlund E. Axon-Seq decodes the motor axon transcriptome and its modulation in response to ALS. Stem Cell Rep. 2018 Dec 11;11(6):1565–1578. doi: 10.1016/j.stemcr.2018.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Schoen M., Reichel J.M., Demestre M., Putz S., Deshpande D., Proepper C. Super-resolution microscopy reveals presynaptic localization of the ALS/FTD related protein FUS in hippocampal neurons. Front Cell Neurosci. 2015;9:496. doi: 10.3389/fncel.2015.00496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Takahashi K., Yamanaka S. Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell. 2006 Aug 25;126(4):663–676. doi: 10.1016/j.cell.2006.07.024. [DOI] [PubMed] [Google Scholar]
- 42.Joung J.K., Sander J.D. TALENs: a widely applicable technology for targeted genome editing. Nat Rev Mol Cell Biol. 2013 Jan;14(1):49–55. doi: 10.1038/nrm3486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Fujimori K., Matsumoto T., Kisa F., Hattori N., Okano H., Akamatsu W. Escape from Pluripotency via inhibition of TGF-beta/BMP and activation of Wnt Signaling accelerates differentiation and aging in hPSC progeny cells. Stem Cell Rep. 2017 Nov 14;9(5):1675–1691. doi: 10.1016/j.stemcr.2017.09.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Dormann D., Rodde R., Edbauer D., Bentmann E., Fischer I., Hruscha A. ALS-associated fused in sarcoma (FUS) mutations disrupt Transportin-mediated nuclear import. EMBO J. 2010 Aug 18;29(16):2841–2857. doi: 10.1038/emboj.2010.143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Bentmann E., Neumann M., Tahirovic S., Rodde R., Dormann D., Haass C. Requirements for stress granule recruitment of fused in sarcoma (FUS) and TAR DNA-binding protein of 43 kDa (TDP-43) J Biol Chem. 2012 Jun 29;287(27):23079–23094. doi: 10.1074/jbc.M111.328757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Davis-Dusenbery B.N., Williams L.A., Klim J.R., Eggan K. How to make spinal motor neurons. Development. 2014 Feb;141(3):491–501. doi: 10.1242/dev.097410. [DOI] [PubMed] [Google Scholar]
- 47.Patani R. Generating diverse spinal motor neuron subtypes from human pluripotent stem cells. Stem Cells Int. 2016;2016 doi: 10.1155/2016/1036974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Warde-Farley D., Donaldson S.L., Comes O., Zuberi K., Badrawi R., Chao P. The GeneMANIA prediction server: biological network integration for gene prioritization and predicting gene function. Nucl. Acids Res. 2010 Jul;38(Web Server issue):W214–W220. doi: 10.1093/nar/gkq537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Choi S., Park C., Kim K.E., Kim K.K. An in vitro technique to identify the RNA binding-site sequences for RNA-binding proteins. BioTechniques. 2017 Jul 1;63(1):28–33. doi: 10.2144/000114567. [DOI] [PubMed] [Google Scholar]
- 50.Aikawa Y., Morimoto K., Yamamoto T., Chaki H., Hashiramoto A., Narita H. Treatment of arthritis with a selective inhibitor of c-Fos/activator protein-1. Nat Biotechnol. 2008 Jul;26(7):817–823. doi: 10.1038/nbt1412. [DOI] [PubMed] [Google Scholar]
- 51.Suzuki N., Kato S., Kato M., Warita H., Mizuno H., Kato M. FUS/TLS-immunoreactive neuronal and glial cell inclusions increase with disease duration in familial amyotrophic lateral sclerosis with an R521C FUS/TLS mutation. J Neuropathol Exp Neurol. 2012 Sep;71(9):779–788. doi: 10.1097/NEN.0b013e318264f164. [DOI] [PubMed] [Google Scholar]
- 52.Okano H., Yamanaka S. Vol. 7. 2014 Mar 31. iPS cell technologies: Significance and applications to CNS regeneration and disease. Molecular brain; p. 22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Lo Bello M., Di Fini F., Notaro A., Spataro R., Conforti F.L., La Bella V. ALS-related mutant FUS protein is Mislocalized to cytoplasm and is recruited into stress granules of fibroblasts from asymptomatic FUS P525L mutation carriers. Neurodegener Dis. 2017;17(6):292–303. doi: 10.1159/000480085. [DOI] [PubMed] [Google Scholar]
- 54.Martineau E., Di Polo A., Vande Velde C., Robitaille R. Dynamic neuromuscular remodeling precedes motor-unit loss in a mouse model of ALS. eLife. 2018 Oct 15;7 doi: 10.7554/eLife.41973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Pereno G.L., Balaszczuk V., Beltramino C.A. Kainic acid-induced early genes activation and neuronal death in the medial extended amygdala of rats. Exp. Toxicol. Pathol. 2011 Mar;63(3):291–299. doi: 10.1016/j.etp.2010.02.001. [DOI] [PubMed] [Google Scholar]
- 56.Cahill M.E., Walker D.M., Gancarz A.M., Wang Z.J., Lardner C.K., Bagot R.C. The dendritic spine morphogenic effects of repeated cocaine use occur through the regulation of serum response factor signaling. Mol Psychiatry. 2018 Jun;23(6):1474–1486. doi: 10.1038/mp.2017.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Lafragette A., Bardo M.T., Lardeux V., Solinas M., Thiriet N. Reduction of cocaine-induced Locomotor effects by enriched environment is associated with cell-specific accumulation of DeltaFosB in striatal and cortical subregions. Int J Neuropsychopharmacol. 2017 Mar 1;20(3):237–246. doi: 10.1093/ijnp/pyw097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Anastasiadou S., Knoll B. The multiple sclerosis drug fingolimod (FTY720) stimulates neuronal gene expression, axonal growth and regeneration. Exp Neurol. 2016 May;279:243–260. doi: 10.1016/j.expneurol.2016.03.012. [DOI] [PubMed] [Google Scholar]
- 59.Grueter B.A., Robison A.J., Neve R.L., Nestler E.J., Malenka R.C. FosB differentially modulates nucleus accumbens direct and indirect pathway function. Proc Natl Acad Sci U S A. 2013 Jan 29;110(5):1923–1928. doi: 10.1073/pnas.1221742110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Bhinge A., Namboori S.C., Zhang X., VanDongen A.M.J., Stanton L.W. Genetic correction of SOD1 mutant iPSCs reveals ERK and JNK activated AP1 as a driver of neurodegeneration in amyotrophic lateral sclerosis. Stem Cell Rep. 2017 Apr 11;8(4):856–869. doi: 10.1016/j.stemcr.2017.02.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Le Pichon C.E., Meilandt W.J., Dominguez S., Solanoy H., Lin H., Ngu H. Loss of dual leucine zipper kinase signaling is protective in animal models of neurodegenerative disease. Sci Transl Med. 2017 Aug 16;9(403) doi: 10.1126/scitranslmed.aag0394. [DOI] [PubMed] [Google Scholar]
- 62.Qiu H., Lee S., Shang Y., Wang W.Y., Au K.F., Kamiya S. ALS-associated mutation FUS-R521C causes DNA damage and RNA splicing defects. J Clin Invest. 2014 Mar;124(3):981–999. doi: 10.1172/JCI72723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Kim M.C., Silberberg Y.R., Abeyaratne R., Kamm R.D., Asada H.H. Computational modeling of three-dimensional ECM-rigidity sensing to guide directed cell migration. Proc Natl Acad Sci U S A. 2018 Jan 16;115(3):E390–E399. doi: 10.1073/pnas.1717230115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.De Santis R., Santini L., Colantoni A., Peruzzi G., de Turris V., Alfano V. FUS mutant human motoneurons display altered transcriptome and microRNA pathways with implications for ALS pathogenesis. Stem cell Rep. 2017 Nov 14;9(5):1450–1462. doi: 10.1016/j.stemcr.2017.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Kuntz Ct, Kinoshita Y., Beal M.F., Donehower L.A., Morrison R.S. Absence of p53: no effect in a transgenic mouse model of familial amyotrophic lateral sclerosis. Exp Neurol. 2000 Sep;165(1):184–190. doi: 10.1006/exnr.2000.7464. [DOI] [PubMed] [Google Scholar]
- 66.Huang da W., Sherman B.T., Lempicki R.A. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc. 2009;4(1):44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- 67.Huang da W., Sherman B.T., Lempicki R.A. Bioinformatics enrichment tools: paths toward the comprehensive functional analysis of large gene lists. Nucleic Acids Res. 2009 Jan;37(1):1–13. doi: 10.1093/nar/gkn923. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Coiling movement of mock control zebrafish
Coiling movement of hFos-B injected zebrafish
Supplementary Tables
Supplementary Figs and legends