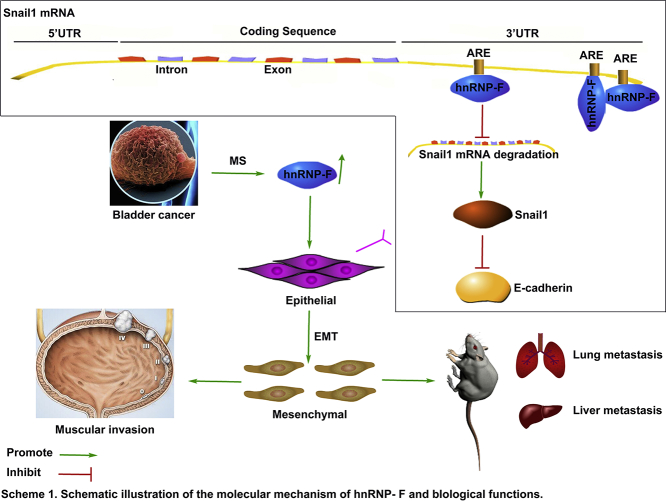
Abstract
Background
Heterogeneous nuclear ribonucleoprotein F (hnRNP-F) has been implicated in multiple cancers, suggesting its role in tumourigenesis, but the potential oncogenic role and mechanism of hnRNP-F in bladder cancer (BC) remain incompletely understood.
Methods
HnRNP-F was identified by proteomic methods. A correlation of hnRNP-F expression with prognosis was analysed in 103 BC patients. Then, we applied in vitro and in vivo methods to reveal the behaviours of hnRNP-F in BC tumourigenesis. Furthermore, the interaction between hnRNP-F and Snail1 mRNA was examined by RNA immunoprecipitation (RIP), and Snail1 mRNA stability was measured after treatment with actinomycin D. Finally, the binding domain between hnRNP-F and Snail1 mRNA was verified by constructing Snail1 mRNA truncations and mutants.
Finding
HnRNP-F is significantly upregulated in BC tissue, and its increased expression is associated with a poor prognosis in BC patients. HnRNP-F is necessary for tumour growth, inducing epithelial-mesenchymal transition (EMT) and metastasis in BC. The changes in Snail1 expression were positively correlated with hnRNP-F at both the mRNA and protein levels when hnRNP-F was silenced or enhanced, suggesting that Snail1 is likely a downstream target of hnRNP-F that mediates its effects on enhancing invasion, metastasis and EMT in BC. The overexpression of hnRNP-F caused an increase in the stability of Snail1 mRNA. Our RNA chip analysis revealed that hnRNP-F could combine with Snail1 mRNA, and we further demonstrated that hnRNP-F could directly bind to the 3′ untranslated region (3′ UTR) of Snail1 mRNA to enhance its stability.
Interpretation
Our findings suggest that hnRNP-F mediates the stabilization of Snail1 mRNA by binding to its 3′ UTR, subsequently regulating EMT.
Keywords: Bladder cancer, Heterogeneous nuclear ribonucleoprotein F, Snail1, Stabilization, Epithelial-mesenchymal transition
Graphical abstract
Research in context.
Evidence before this study
HnRNP-F, an RNA-binding protein, has previously been implicated in multiple steps of RNA metabolism and gene expression regulation such as mRNA stabilization, transcriptional regulation and post-transcriptional modification. Recently, hnRNP-F has been demonstrated in a number of cancers, suggesting its roles in tumorigenesis, but the potential oncogenic role and mechanism of hnRNP-F in BC remains incompletely understood.
Added value of this study
We first found that hnRNP-F was significantly up-regulated in BC tissues via the 2D-DIGE method. Furthermore, its increased expression was associated with poor prognosis in 103 BC patients. We applied in vitro and in vivo methods to reveal the functions of hnRNP-F in BC tumorigenesis, which showed that hnRNP-F was necessary for tumour growth, EMT and metastasis in BC, and suggested that Snail is likely to be the target of hnRNP-F. We further demonstrated that hnRNP-F could directly bind to the 3′ untranslated region (3′ UTR) of Snail1 mRNA to enhance its stability.
Implications of all the available evidence
The current study may allow us to conclude that Snail1 is a mediator of hnRNP-F affecting EMT, which could lead to the development of a new approach for BC therapy in the future.
Alt-text: Unlabelled Box
1. Introduction
Bladder cancer (BC) is the second most prevalent malignant urinary disease [1]. At diagnosis, ~70% of new BC cases are superficial non-muscle-invasive tumours. Moreover, up to 50–70% of those cases will recur, and 10–20% of recurrent tumours will progress into deep tissue layers. The extent of tumour invasion is closely correlated with the clinical treatment of BC, and muscle-invasive BC can progress to life-threatening metastasis and result in BC-related mortality [2]. Therefore, elucidating the pathological mechanisms underlying BC invasion and metastasis may facilitate the prevention of the development of BC and improve its prognosis.
Epithelial-mesenchymal transition (EMT) plays key roles in cancer cells with respect to promoting migration, invasion, and subsequent dissemination [3]. Recent studies have demonstrated that multiple factors, including EMT transcription factors (EMT-TFs), may regulate the EMT process. EMT-TFs, which are activated and therefore promote the EMT programme, are necessary for the metastatic cascade in cancer [4]. Snail1, one of the principal EMT-TFs, has been demonstrated to induce EMT in various epithelial cancer cell lines and primary mammary tumour cells [[5], [6], [7]]. Notably, Snail1 is a highly unstable protein with a short half-life (t1/2) of ~25 min. Snail1 mRNA bears multiple AU-rich elements (AREs) in the 3′ untranslated region (3′ UTR) that are responsible for the stability of Snail1 mRNA [8].
Heterogeneous nuclear ribonucleoprotein F (hnRNP-F) is an RNA-binding protein belonging to the hnRNP family that has been implicated in multiple steps of RNA metabolism and gene expression regulation, such as alternative splicing, mRNA stabilization, transcriptional regulation and posttranscriptional modification [9]. HnRNP-F was found to be a cofactor in a subset of tristetraprolin proteins, which regulate the stabilities of mRNAs containing AREs [10]. Increased hnRNP-F levels have been observed in a number of cancers [5,11,12], suggesting its role in tumourigenesis. However, the potential oncogenic role and mechanism of hnRNP-F in BC remain poorly understood.
In this study, hnRNP-F was first determined to exhibit enhanced expression in BC patients compared with controls using the proteomic technique of two-dimensional fluorescent differential gel electrophoresis (2D-DIGE). Then, we examined the effects of aberrant hnRNP-F expression on the cellular biological behaviour of BC cells. Through our investigation of the contribution of hnRNP-F overexpression to EMT by regulating Snail1 expression, we present evidence of a role for hnRNP-F in regulating Snail1 mRNA stability in BC by binding to its 3′ UTR. Our findings suggest important new roles for hnRNP-F in BC pathogenesis and a potential therapeutic target for BC.
2. Materials and methods
2.1. Specimen collection and preparation
For the proteomic analysis, a total of eight BC specimens and their adjacent normal tissues were obtained at Nanfang Hospital (Guangzhou, China) between January and April 2015. There were four patients with noninvasive papillary urothelial carcinoma and infiltrating urothelial carcinoma. All tissues were pathologically diagnosed after surgical operations. Tissue specimens were prepared for further analysis as described in our previous studies [13]. All patients provided written informed consent, and all procedures were approved by the bioethics committee of our hospital.
2.2. Proteomic analysis
An equal amount of protein from eight BC patients and eight controls was pooled for 2D-DIGE analysis as described in our previous studies [13]. Briefly, 50 μg of protein samples from BC patients and controls were minimally labelled with 400 pmol of Cy3 and Cy5 fluorescent dyes, and an internal standard pool was labelled with Cy2. All three labelled samples were mixed and resolved in one gel. After isoelectric focusing and second-dimension electrophoresis, the gel was stained with Coomassie brilliant blue. Then, the gel images were analysed with DeCyder 5.0 software (GE Healthcare). The spots whose ratios of Cy5/Cy2 and Cy3/Cy2 were changed >2-fold were considered for identification by matrix-assisted laser desorption time-of-flight mass spectrometry (MALDI-TOF/TOF MS). All raw files and search results have been deposited in ProteomeXchange via iProX (ww.iprox.org) with the identification no.:IPX0001621000 or PXD014078.
2.3. Real-time quantitative polymerase chain reaction (RT-qPCR)
RT-qPCR was performed with a standard SYBR Green PCR Kit (TAKARA, Japan) on an Applied Biosystems 7500 Real-Time PCR system (Foster City, CA). Glyceraldehyde 3-phosphate dehydrogenase (GAPDH) was used for normalization. The primer sequences used are summarized in Table S2.
2.4. Western blot analysis
Protein lysates were separated using 10% SDS-PAGE and transferred onto a PVDF membrane (Roche, Switzerland). Then, membranes were incubated with antibodies against hnRNP-F (1:1000, Abcam, England), E-cadherin, N-cadherin, vimentin, Snail1, Twist1 and GADPH purchased from Cell Signaling Technology (USA) (1:1000–5000), followed by incubation with secondary antibodies. Finally, the proteins were detected using an enhanced chemiluminescence (ECL, Pierce, Rockford, IL, USA) detection system.
2.5. Cell culture
Human BC cell lines (T24, EJ, UMUC-3, BIU-87 and 5637) were used in this study. The cells were cultured in RPMI-1640 medium (Gibco, USA) supplemented with 10% foetal bovine serum (Bioind, Israel) and incubated at 37 °C in 5% CO2. The cell lines were authenticated by short tandem repeat (STR) profiling before receipt and were propagated for <6 months after resuscitation.
2.6. Construction of the lentivirus and transfection
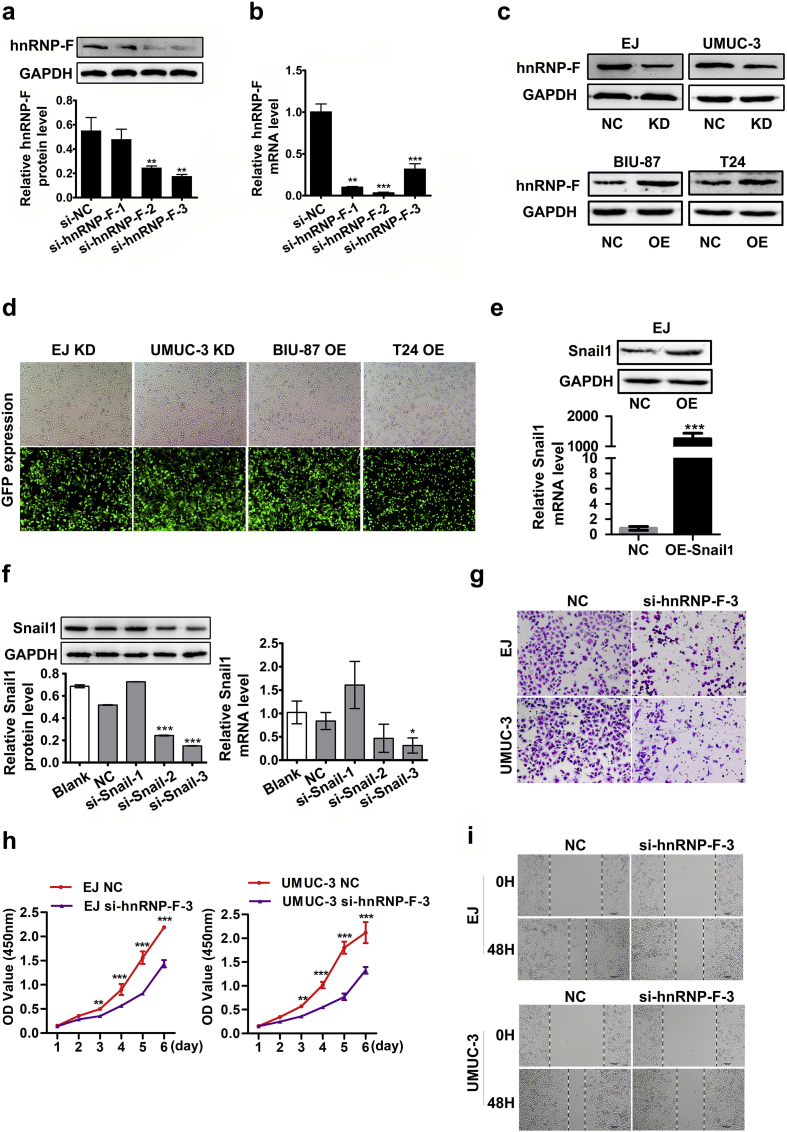
Lentiviral constructs repressing hnRNP-F purchased from GenePharma (Suzhou, China) were used to establish stable cell lines. The fragment with the highest efficiency verified by RT-qPCR and western blot was selected from three interference fragments of hnRNP-F and Snail1 (Fig. S1a–b, f). si-hnRNP-F-2 was used to construct Lentiviral vectors, si-Snail1-3 was used in the rescue assays. The target sequence of the hnRNP-F short hairpin RNA (shRNA) in Lentiviral was 5′-GGTACATTGAGGTGTTCAA-3′. Empty vectors were used as controls. The target sequence of the Snail small interfering RNA (siRNA) was 5′-ACUCAGAUGUCAAGAAGUA-3′ (Table S3). The pENTER-Snail1 plasmid and the empty vector were purchased from Vigene Bioscience (Shandong, China). In the rescue experiments, transfection was performed using Lipofectamine 2000 (Invitrogen, USA) according to the manufacturer's instructions. The transfection efficiency of the Snail1 plasmid was monitored by RT-qPCR and western blot (Fig. S1e).
2.7. Immunohistochemistry (IHC)
The IHC analysis comprised 103 BC tissues and 46 adjacent nontumour tissues, with follow-up durations ranging from 11 to 82 months. A comprehensive set of clinicopathological data was collected, and the median survival duration was 46 months. According to the recommendations of the S-P kit (Zhongshan Biotechnology Co. Ltd., Beijing, China), tissue sections were retrieved with citrate buffer and incubated with an anti-hnRNP-F primary antibody (1:100, Abcam, England). The degree of staining in the sections was observed and scored independently by two pathologists according to the following criteria: (1) staining degree: 0, none; 1, weak staining; 2, moderate staining; and 3, strong staining; and (2) staining scale: <30%: 1; 31%~60%: 2; and >60%: 3. The positive intensity of staining in each case was calculated as follows: positive intensity = (1) × (2); a score of 0–3 was regarded as negative expression, and a score > 3 was regarded as positive expression [14].
2.8. Cell proliferation assay in vitro
Cell proliferation was evaluated using a Cell Counting Kit-8 (CCK-8, Dojindo, Japan) according to the manufacturer's instructions. Briefly, 10 μl of CCK-8 solution was added to the culture medium and incubated for 2 h in 5% CO2 at 37 °C. The absorbance was determined at a wavelength of 450 nm, with a reference wavelength of 570 nm. The cell proliferation assay was performed on days 1, 2, 3, 4, 5 and 6.
2.9. Wound scratch healing and invasion assays
Cells were seeded onto 6-well plates. The monolayer was scratched with a 10 μl pipette tip and then incubated in serum-free RPMI-1640 medium. Images of the wells were taken at different time points (0 h and 48 h) on an inverted microscope (Axioskop 2 Plus; Zeiss, Germany). The length of the open area was calculated with software.
Cell invasion was evaluated using transwell inserts with 8 μm pores (BD Biosciences, San Jose, CA, USA). Briefly, cells were released using trypsin-EDTA and sequentially rinsed with RPMI-1640 medium containing 10% FBS. The rinsed cells were resuspended in serum-free RPMI-1640 medium, and 200 μl of the cell suspension (1 × 105 cells) was added to the transwell insert chamber with a filter that was coated with Matrigel. In the lower chamber, 500 μl of RPMI-1640 medium containing 10% FBS was added as a chemoattractant. After 24 h of incubation, the cells in the lower chamber were fixed, stained with haematoxylin and counted under a microscope.
2.10. Animal models
All experimental procedures were approved by the ethical committee of our hospital. To evaluate tumour growth in vivo, xenograft tumours were generated by subcutaneously injecting 1 × 107 cells. The volume of tumours were measured every 5 days until day 30 after injection with a caliper as follows: (length × width2)/2. To examine metastasis, a total of 5 × 106 cells were injected into the tail veins of BALB/c nude mice, and the animals were sacrificed six weeks after injection. Serial sections of lung and liver tissues were performed every 5 mm for histological examination. The area ratios of tumours in lung and liver were calculated by Image J software.
2.11. RNA immunoprecipitation (RIP)
RIP was performed according to the instructions provided by the Magna RIP RNA-Binding Protein Immunoprecipitation Kit (Millipore, Billerica, MA, USA). Briefly, cells were collected and suspended in 200 μl of lysis buffer containing 1 μl of protease inhibitor cocktail and 0.5 μl of RNase inhibitor after centrifugation. Magnetic beads were pretreated with an anti-rabbit IgG or anti-rabbit hnRNP-F antibody for 1 h at room temperature, and lysates were then immunoprecipitated with the beads-antibody complexes at 4 °C overnight. RNA was purified from the RNA-protein complexes that bound to the beads and analysed by RT-qPCR and agarose gel electrophoresis.
2.12. Half-life of Snail1 mRNA
For mRNA stability measurements, cells were treated with actinomycin D (2 μg/ml). Total RNA was extracted at the indicated time points (0, 15, 30, 45, and 60 min). Snail1 mRNA expression levels were normalized to those of 18S RNA.
2.13. Construction of truncations and mutants
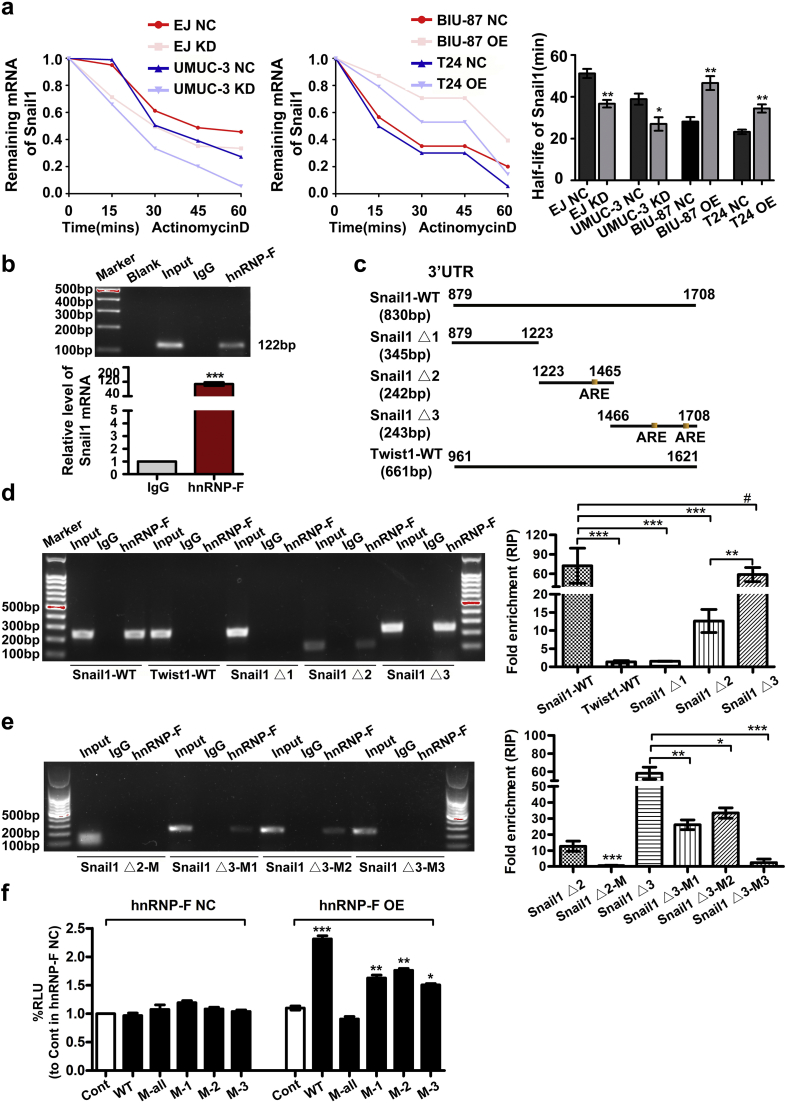
The Snail1 mRNA 3′ UTR was divided into three fragments that were cloned into the eukaryotic expression vector pcDNA3.1 and defined as Snail1 Δ1, Snail1 Δ2 and Snail1 Δ3, respectively (Fig. 6c). The full-length sequence of the Snail1 3′ UTR was used as a positive control (Snail1 wild-type (WT)). The Twist1 3′ UTR was also examined.
Fig. 6.
HnRNP-F regulates the stability of Snail1 mRNA by binding to its 3′ UTR. a. The half-life of Snail1 mRNA was determined by RT-qPCR after treatment with actinomycin D (2 μg/ml) for the indicated times following hnRNP-F KD and OE; bars show the half-lives of different groups; b. A RIP experiment was performed in EJ cells using an hnRNP-F antibody and a nonspecific IgG antibody to determine whether hnRNP-F binds to Snail1 mRNA. RNA enrichment was determined by RT-PCR; c. Schematic view of the constructs containing the 3′ UTR of Snail1, WT Twist1, three 3′ UTR Snail1 mRNA truncations and three AREs; d. The relative RNA abundance of three truncations obtained from RIP assays was assessed by agarose gel electrophoresis and RT-qPCR in EJ cells; e. The relative RNA abundance of the corresponding ARE mutants obtained from RIP assays was assessed by agarose gel electrophoresis and RT-qPCR in EJ cells (*p < .05, **p < .01, ***p < .001, #p ≥ .05); f. Luciferase reporter vectors containing mutated AREs in the 3′ UTR of Snail1 were tested for their responsiveness to hnRNP-F OE and controls (NC) in 293 T cells. (*p < .05, **p < .01, ***p < .001, #p ≥ .05). Student's t-test was performed to analyze statistical significance. Error bars denote standard deviation.
Notably, Snail1 Δ1 does not contain any AREs. However, Snail1 Δ2 contains one ARE, and Snail1 Δ3 contains two AREs. The AREs (ATTTA) of Snail1△2 and Snail1△3 truncations were mutated to CGCGC respectively, marked as Snail △2-M, Snail1△3-M1(one ARE was mutated), Snail1△3-M2 (the other one ARE was mutated) and Snail1△3-M3(two AREs were mutated). All of the above corresponding plasmids were transfected into EJ cells for further RIP assays. The sequences of these truncations and mutants are listed in Table S4, and the RT-qPCR primers are shown in Table S2.
2.14. Luciferase reporter gene assay
Plasmid transfections for luciferase assays in 293 T cells were performed with 0.2 μg Renilla reniformis and 0.8 μg of the reporter construct in conjunction with a variety of mutations in the Snail1 3′ UTR. Each ARE in Snail1-WT was mutated and marked as Snail1-M1, Snail1-M2 and Snail1-M3, Snail1-M-all (all ARE were mutated) respectively. Snail1-WT represents the wild-type Snail1 3′ UTR, luciferase plasmid without Snail1 UTR were as negative control. Then, 0.8 μg of the hnRNP-F plasmid was added to each well of a 24-well plate and transfected with Hieff Trans TM Liposomal Transfection Reagent (Yeasen) according to the manufacturer's instructions. Luciferase activity was measured 48 h posttransfection with the Dual Luciferase Reporter Assay System (Promega) according to the manufacturer's instructions. The corresponding sequences of these mutant plasmids are shown in the Table S5.
2.15. Statistical analysis
Data are expressed as the mean ± standard deviation (SD), and the experiments were repeated three times. Student's t-test or one-way analysis of variance (ANOVA) was applied where appropriate. The correlation of hnRNP-F with various clinicopathologic characteristics was tested by the χ2 test. Survival curves were plotted by the Kaplan-Meier method and compared by the log-rank test. The significance of variables for survival was analysed by the Cox proportional hazards model for multivariate analyses. All statistical analyses were performed with SPSS 22.0 software (SPSS, Inc., Chicago, USA). P < .05 was regarded as significant.
3. Results
3.1. HnRNP-F is upregulated in BC, as determined by proteomics
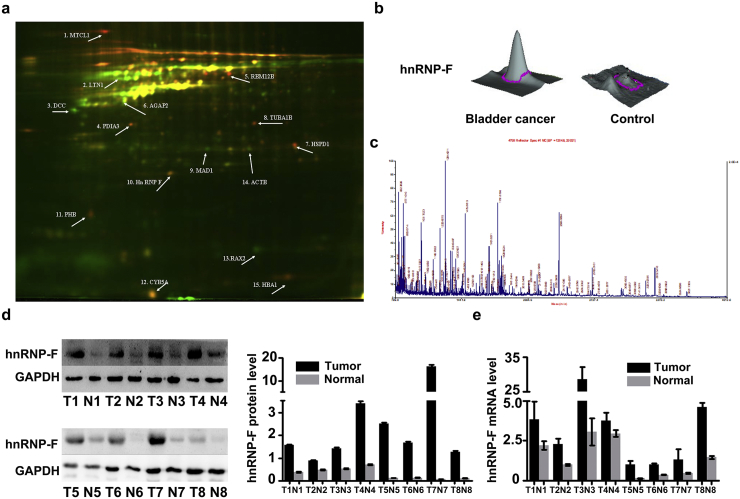
After screening differentially expressed proteins in BC tissues by the 2D-DIGE proteomic method, a total of 15 differential protein spots whose volumes changed by at least 2-fold were selected for further identification by MALDI-TOF/TOF MS (Fig. 1a). In BC tissues, nine proteins were significantly upregulated, and six proteins were downregulated. The upregulated proteins included MTCL1, PDIA3, RBM12B, HSPD1, TUBA1B, hnRNP-F, PHB, CYB5A and HBA1. The downregulated proteins included LTN1, DCC, AGAP2, MAD1, RAX2 and ACTB (Fig. 1a, Table 1).
Fig. 1.
HnRNP-F protein is upregulated in BC. a. An image of all 15 differentially expressed protein spots between BC patients and controls by DIGE presented by DeCyder 5.0 software (GE Healthcare); b. A 3-D view of the hnRNP-F protein was enlarged to show the expression of the hnRNP-F protein in tissues from BC patients and controls presented by DeCyder 5.0 software (GE Healthcare); c. MS analysis of the in-gel trypsin digests of the protein and analysis of the depicted peptide spectrum resulted in the identification of hnRNP-F (identified by MALDI-TOF/TOF MS); d–e. Relative expression of hnRNP-F in eight BC tissues and their adjacent normal tissues was examined by western blot (d) and RT-qPCR (e) (*p < .05, **p < .01, ***p < .001, #p ≥ .05) Paired t-test was performed to analyze statistical significance. Error bars denote standard deviation.
Table 1.
The association of hnRNP-F with clinicalpathologic characteristics in 103 patients with BC.
| Clinical-pathologic characteristics | n | HnRNP-F protein expression |
P valueb | |
|---|---|---|---|---|
| Negative (%) | Positive (%) | |||
| Age (years) | ||||
| ≤65a | 51 | 17 (32.7) | 34 (67.3) | 0.588 |
| >65 | 52 | 20 (39.2) | 32 (60.8) | |
| Gender | ||||
| Male | 82 | 32 (39.0) | 50 (61.0) | 0.195 |
| Female | 21 | 5 (23.8) | 16 (76.2) | |
| Pathology grade | ||||
| I–II | 38 | 17 (44.7) | 21 (55.3) | 0.154 |
| III | 65 | 20 (35.9) | 45 (64.1) | |
| Clinical stage | ||||
| 0a, 0is, I | 38 | 22 (57.9) | 16 (42.1) | |
| II | 31 | 6 (19.4) | 25 (80.6) | 0.002 |
| III–IV | 34 | 9 (26.5) | 25 (73.5) | |
Median age.
p value is from χ2-test.
To further verify the expression of hnRNP-F identified by the 2D-DIGE proteomic method, we quantified the expression levels of hnRNP-F by western blotting and RT-qPCR assays in human BC tissues. The mRNA and protein expression levels of hnRNP-F were significantly upregulated in BC patients compared with those in the controls, consistent with our 2D-DIGE results (Fig. 1b–e, p < .001).
3.2. Elevated hnRNP-F is associated with a poor prognosis in BC patients
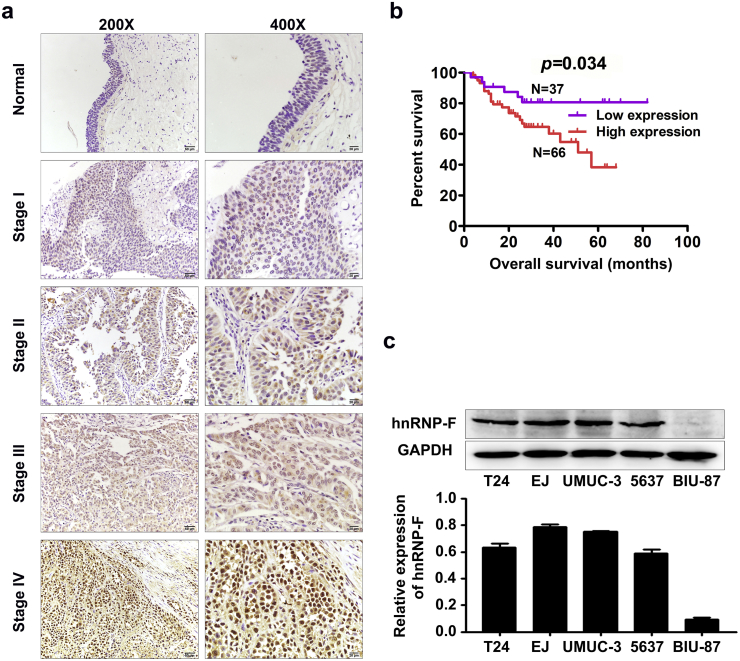
HnRNP-F protein was detected in the cytoplasm and nuclei of normal bladder transitional cells and cancerous cells by IHC. The staining intensity was stronger in the BC group than in the corresponding adjacent normal mucosa (Fig. 2a).
Fig. 2.
HnRNP-F expression was examined in BC patient tissues and BC cell lines. a. Expression of hnRNP-F in tissues from BC patients (classified by clinical stage) and controls by IHC; b. The Kaplan-Meier overall survival curve of BC patients (n = 103) according to hnRNP-F protein expression (p = .034), Kaplan-Meier test was performed to analyze statistical significance; c. The expression of hnRNP-F protein in five human BC cell lines.
The relationship between hnRNP-F levels and the clinical features of BC is presented in Table 1. High hnRNP-F expression was positively associated with an advanced clinical stage (p = .002, Fig. 2a). Notably, Kaplan-Meier analysis indicated that BC patients with high hnRNP-F protein levels had poor overall survival (Fig. 2b, log-rank, p = .034). Furthermore, the multivariate analysis showed that increased hnRNP-F expression may be a risk factor for poor overall survival in BC patients (Table 2, p = .016). These results indicate that hnRNP-F might play a key role in BC progression.
Table 2.
Univariate and multivariate analysis of different prognostic parameters in BC patients by Cox regression analysis.
| Covariates | Univariate analysis |
Multivariate analysis |
||
|---|---|---|---|---|
| p | HR (95% CI) | p | HR (95% CI) | |
| Age | 0.330 | 1.018 (0.982–1.054) | 0.396 | 1.019 (0.976–1.064) |
| Gender | 0.935 | 1.038 (0.418–2.580) | 0.879 | 0.921 (0.316–2.679) |
| HnRNP-F | 0.021 | 3.130 (1.192–8.221) | 0.016 | 6.205 (1.397–27.567) |
| Pathology grade | 0.160 | 1.798 (0.794–4.074) | 0.595 | 1.314 (0.481–3.591) |
| Clinical stage | 0.002 | 1.987 (1.293–3.053) | 0.027 | 1.677 (1.062–2.650) |
Abbreviations: HR: Hazard Ratio; CI: confidence interval.
3.3. HnRNP-F promotes the proliferation, migration and invasion of BC in vitro
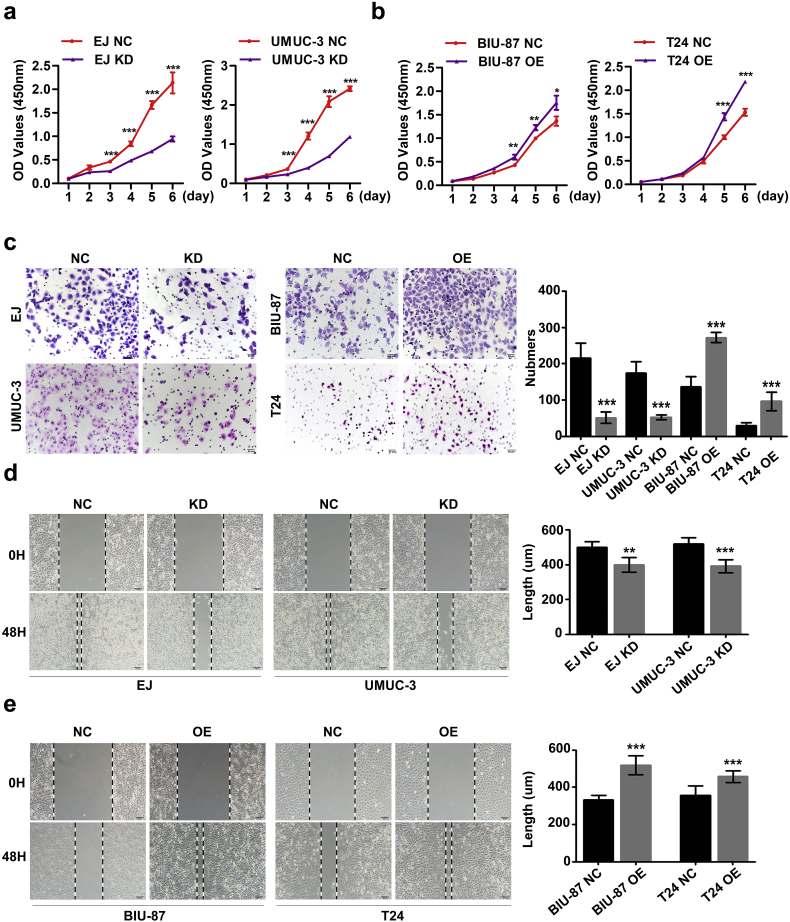
To explore the oncogenic functions of hnRNP-F in cell proliferation and invasion, we investigated the effect of hnRNP-F knockdown or overexpression on BC cell behaviours. Following analysis of the endogenous hnRNP-F expression levels in five human BC cell lines by western blot analysis (Fig. 2c), EJ and UMUC-3 cells were transfected with a lentivirus containing the shRNA sequence of hnRNP-F to generate stable transfectants, whereas T24 and BIU-87 cells were selected and transfected with LV-hnRNP-F. Transfection efficiency was examined by western blot and GFP expression (Fig. S1c–d).
The effect of hnRNP-F on proliferation in BC cell lines was determined by the CCK-8 assay. The CCK-8 assay revealed that proliferative ability was significantly increased by hnRNP-F overexpression but inhibited by hnRNP-F depletion (Fig. 3a–b, Fig. S1h).
Fig. 3.
HnRNP-F enhances the proliferation, invasion, and migration of BC in vitro. a–b. Effects of hnRNP-F OE and KD on BC cell proliferation in vitro by the CCK-8 assay. A two-way analysis of variance (ANOVA) test was performed to determine the statistically significant differences; c. Effects of hnRNP-F OE and KD on BC cell invasion in vitro by the Boyden chamber assay. Scale bars represent 50 μm; d–e. Effects of hnRNP-F OE and KD on BC cell migration in vitro by the wound scratch healing assay. (*p < .05, **p < .01, ***p < .001, #p ≥ .05). Student's t-test was performed to analyze statistical significance. Error bars denote standard deviation. Scale bars represent 100 μm.
EMT is considered a key step in promoting migration, invasion, and subsequent dissemination. Therefore, migratory and invasive abilities were examined by wound scratch healing and transwell assays in vitro. The results indicated that hnRNP-F overexpression significantly promoted the migratory and invasive abilities of T24 and BIU-87 cells, and hnRNP-F knockdown significantly inhibited cell migration and invasion in vitro (Fig. 3c–e, Fig. S1g, i).
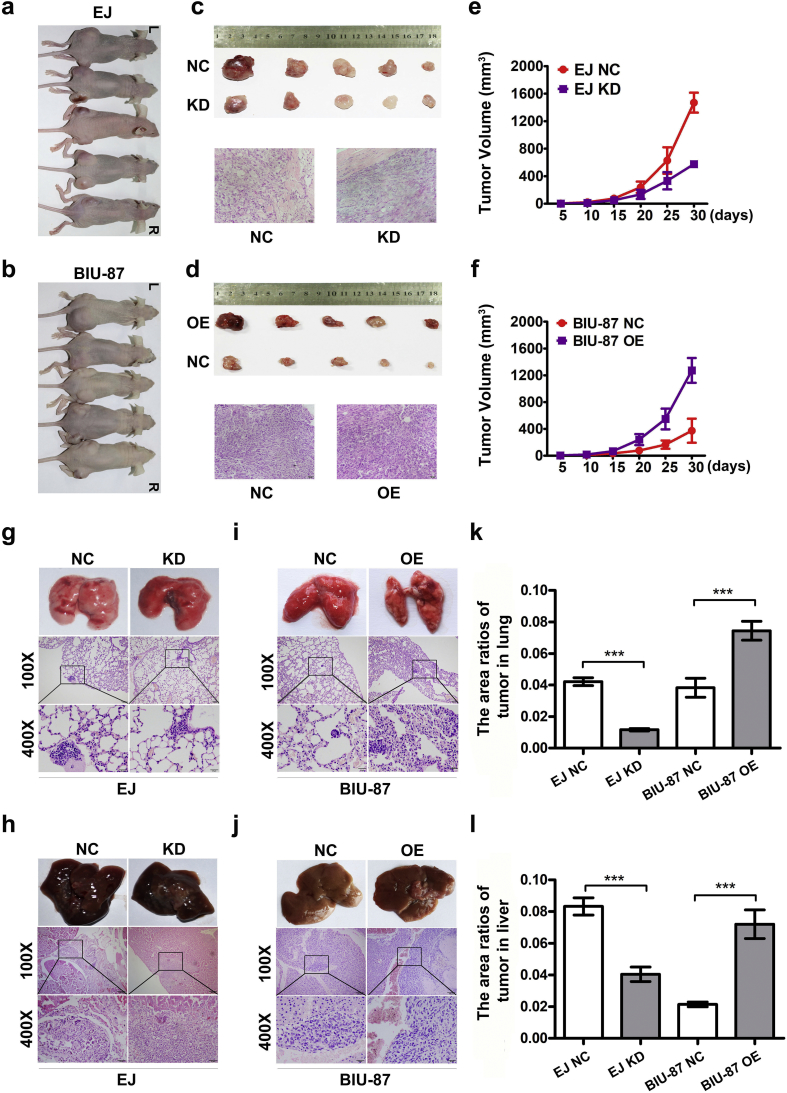
3.4. HnRNP-F accelerates tumour growth and metastasis in vivo
To test the effect of hnRNP-F on tumour growth and migration in vivo, an orthotropic metastasis nude mouse model was established. The results indicated that hnRNP-F KD led to smaller tumours in EJ KD cells than in control cells (Fig. 4a, c, e) and OE promoted proliferation in BIU-87 OE cells relative to control (Fig. 4b, d, f). Furthermore, the area ratios of metastasis tumour in lung in the EJ negative control and EJ knockdown groups were about 4.1% and 1.0%, respectively, whereas the area ratios of tumour in lung in the BIU-87 negative control and overexpression group were about 4.0% and 7.1%, respectively (Fig. 4g, i, k). The area ratios of metastasis tumour in liver were about 8.1%, 4.0%, 2.1% and 7.4%, respectively in the EJ negative control, EJ knockdown, BIU-87 negative control and BIU-87 overexpression groups (Fig. 4h, j, l). These data suggest that hnRNP-F accelerates tumour growth and metastasis in vivo.
Fig. 4.
HnRNP-F promotes tumour growth and metastasis in vivo. a–b. Images of mice injected on right side (R) with KD cells (a) or OE cells (b) and left side (L) with Control cells (n = 5); c–d. Images of HE staining of tumours excised from the mice following hnRNP-F KD (c) and hnRNP-F OE (d), Scale bars represent 100 μm; e–f. Growth curves of the tumour xenografts following hnRNP-F KD (e) and hnRNP-F OE (f). The tumour size was monitored every five days with a Vernier caliper; g–h. Gross and microscopic observations of lung metastases (g) and liver metastases (h) in mice after injection with EJ-hnRNP-F KD cells into tail vein; (n = 5); i–j. Gross and microscopic observations of lung metastases (i) and liver metastases (j) in mice after injection with BIU-87-hnRNP-F OE cells into tail vein; (n = 5). k–l. The bars present the area ratios of tumour in lung (k) and liver (l) after injection with BC cells into tail vein. (*p < .05, **p < .01, ***p < .001, #p ≥ .05). Student's t-test was performed to analyze statistical significance. Error bars denote standard deviation. Scale bars of 100× represent 100 μm, Scale bars of 400× represent 20 μm.
3.5. HnRNP-F induces EMT in BC
E-cadherin is one of the most commonly used markers for the epithelial trait, and N-cadherin and vimentin are the most commonly used markers for the mesenchymal trait [[15], [16], [17]]. The results indicated that hnRNP-F overexpression inhibited the expression of E-cadherin protein but increased the expression levels of vimentin and N-cadherin (Fig. 5b). By contrast, the protein levels of the mesenchymal markers vimentin and N-cadherin were significantly reduced, and the epithelial marker E-cadherin was increased in the hnRNP-F knockdown group compared to that in the control group (Fig. 5a). To summarize these results, hnRNP-F induced EMT in BC cell lines.
Fig. 5.
Snail1 is required for the hnRNP-F-enhanced EMT in BC. a–b. E-cadherin (E-cad), N-cadherin (N-cad), vimentin, Snail1 and Twist1 expression was assessed by western blot following hnRNP-F KD (a) and OE (b); c–d. Relative mRNA expression of hnRNP-F, Snail1 and Twist1 was examined by RT-qPCR following hnRNP-F KD (c) and OE (d); e. E-cad, N-cad, vimentin, Snail1 and Twist1 expression was assessed by western blot in EJ cells with Snail1 OE and/or hnRNP-F KD; f. E-cad, N-cad, vimentin, Snail1 and Twist1 expression was assessed by western blot in BIU-87 cells with Snail1 KD and/or hnRNP-F OE (*p < .05, **p < .01, ***p < .001, #p ≥ .05). Student's t-test was performed to analyze statistical significance. Error bars denote standard deviation.
3.6. Snail1 is required for the hnRNP-F-enhanced invasion, metastasis and EMT
Interestingly, the expression levels of the transcription factor Snail1 were increased in the hnRNP-F overexpression group and decreased in the hnRNP-F knockdown group according to western blot analysis (Fig. 5a–b). Surprisingly, the protein expression level of the other transcription factor Twist1 did not change appreciably. In addition, the alterations in Twist1 mRNA were not significant when hnRNP-F was knocked down (Fig. 5c) or overexpressed (Fig. 5d), as assessed by RT-qPCR, whereas the mRNA level of Snail1 was positively associated with hnRNP-F mRNA (Fig. 5c–d). These results suggest that Snail1 is a likely target of hnRNP-F that mediates its effect on invasion, metastasis and EMT regulation in BC.
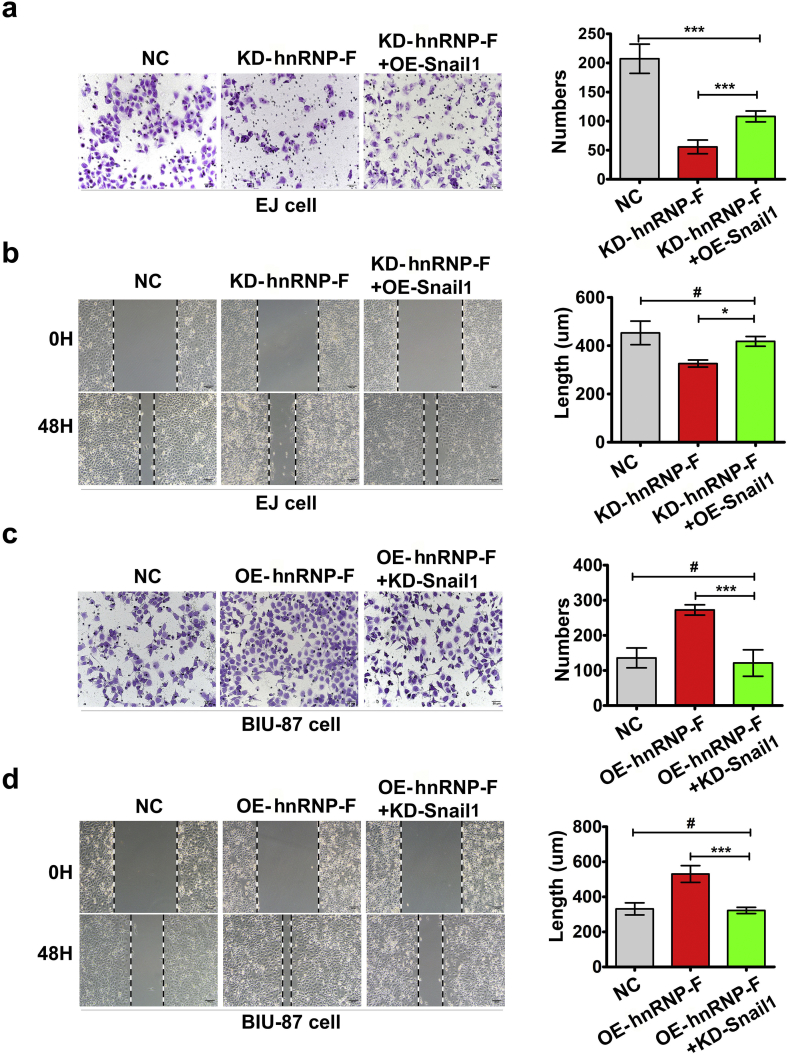
To verify whether hnRNP-F mediated BC progression through Snail1, we examined whether Snail1 overexpression and knockdown would reverse or amplify the effects of hnRNP-F knockdown and overexpression in EJ and BIU-87 cells. When we silenced Snail1 in hnRNP-F overexpression cells, western blot analysis showed that the expression patterns of the EMT markers were reversed in BIU-87 cells (Fig. 5f). As anticipated, increased invasion and migration were completely blocked when Snail1 was silenced in hnRNP-F overexpression cells (Fig. S2c–d). Conversely, Snail1 overexpression in hnRNP-F-depleted cells had the opposite effect on EMT markers and impaired the invasion and migration of EJ cells (Fig. 5e, Fig. S2a–b). The above findings indicate that Snail1 is required for the effect of hnRNP-F-enhanced invasion, metastasis and EMT.
3.7. HnRNP-F regulates the half-life of Snail1 mRNA
Considering the function of hnRNPs in the regulation of mRNA stability, we examined the effect of hnRNP-F protein on Snail1 mRNA stability. Actinomycin D (2 μg/ml) was used to block genetic transcription, and the remaining Snail1 mRNA and its rate of decay were analysed by RT-qPCR. As shown in Fig. 6a, the half-life of Snail1 mRNA in EJ cells significantly declined from 51.13 ± 3.78 min to 36.70 ± 3.14 min when hnRNP-F was silenced. Similarly, the half-life of Snail1 mRNA decreased from 38.86 ± 4.57 min to 26.97 ± 5.54 min upon hnRNP-F depletion in UMUC-3 cells (p < .05). Not surprisingly, the half-life of Snail1 mRNA increased from 23.20 ± 1.9 min to 34.38 ± 3.44 min and from 28.12 ± 3.73 min to 46.55 ± 5.78 min in T24 and BIU-87 cell lines, respectively, when hnRNP-F was overexpressed (p < .05). Moreover, the half-life of Twist1 mRNA showed no significant difference when the levels of hnRNP-F protein were changed (p > .05) (Fig. S3a–c).
3.8. HnRNP-F binds to the 3′ UTR of Snail1 mRNA
The Snail1 mRNA 3′ UTR contains three AREs that are responsible for the stability of Snail1 mRNA [[18], [19], [20]]. The aforementioned finding that Snail1 mRNA stability was regulated by hnRNP-F prompted us to explore whether hnRNP-F binds to the 3′ UTR of Snail1 mRNA. We first performed a RIP assay and found significant Snail1 RNA enrichment by using the hnRNP-F antibody compared with the results obtained with a nonspecific antibody (IgG control) (Fig. 6b, p < .001). This result showed that hnRNP-F protein was physically associated with Snail1 mRNA in vitro. Furthermore, we generated constructs with the 3′ UTR of Snail1 mRNA, three 3′ UTR truncations of Snail1 mRNA and the 3′ UTR of Twist1 mRNA (Fig. 6c). RNA agarose gel electrophoresis and RT-qPCR were conducted to examine the qualitative and quantitative results of RNA enrichment after incubation with the hnRNP-F antibody. Null-specific sequences were used to eliminate interference from endogenous sequences. As shown in Fig. 6d, the quantitative results demonstrated that the transcribed Snail1 3′ UTR was more enriched in the hnRNP-F antibody group than in the Twist1 mRNA 3′ UTR group. Moreover, of the three 3′ UTR truncation fragments of Snail1 mRNA, only Snail1 Δ2 and Snail1 Δ3, which contained AREs, bound to the hnRNP-F antibody. Furthermore, the enrichment in Snail1 Δ3 was approximately 4.65 times that in Snail1 Δ2. By contrast, no apparent enrichment was observed for the Twist1 3′ UTR or Snail1 Δ1.
To further determine whether hnRNP-F could bind to the AREs in the 3′ UTR of Snail1 mRNA, we mutated AREs in the Snail1 Δ2 and Snail1 Δ3 truncations. The results demonstrated that the binding strength between hnRNP-F and Snail1 Δ3 was reduced to ~58% and 47% (for Snail1 Δ3-M1 and Snail1 Δ3-M2, respectively). In addition, the binding between hnRNP-F and Snail1 Δ2 and Snail1 Δ3 almost disappeared (in Snail1 Δ2-M and Snail1 Δ3-M3) (Fig. 6e).
To further verify above results, luciferase reporter gene assays were applied in our study. As shown in Fig. 6f, the luciferase reporter analysis demonstrated that Snail1-WT but not Snail1-M-all had a significant effect on luciferase reporter activity compared to the hnRNP-F NC group, while Snail1-M1, Snail1-M2 and Snail1-M3 were downregulated relative to Snail1-WT in hnRNP-F overexpression group. This result indicates that the interaction between hnRNP-F and the Snail1 3′ UTR is impaired after mutating one ARE and is completely inhibited in the Snail1-M-all group. Briefly, the above results may indicate there is a dose-dependent relationship between AREs and hnRNP-F.
4. Discussion
There is growing interest in identifying pathological mechanisms to aid medical professionals in improving the prognosis of BC patients. In the past few years, proteomic approaches have emerged as a strategy for discovering diagnostic and prognostic protein biomarkers as well as novel therapeutic targets [21,22]. In this study, we examined protein expression profiles in tissue samples from BC patients and controls by 2D-DIGE and MS. Fifteen differentially expressed proteins in BC patients were identified. Of these identified proteins, hnRNP-F, which belongs to the hnRNP family and is responsible for diverse cellular processes, such as modulating splicing, miRNA maturation, mRNA transport and mRNA stabilization, was upregulated in the BC group [9,23].
A growing body of evidence has shown that the expression levels of hnRNPs are altered in many types of cancer and play important roles in tumourigenesis [9]. HnRNP A2/B1 is involved in MDA-MB-231 cells, wherein the knockdown of its expression inhibits cell invasion and thereby the proliferation of tumour cells [24]. Similarly, hnRNP C may act as a key regulator that controls the metastatic potential of breast cancer [25]. HnRNP-H was reported to be associated with poor differentiation in oesophageal squamous cell carcinoma [26] and colorectal cancer node metastasis [27]. It was also reported to be an oncogene that promotes prostate cancer cell growth [28]. HnRNP-F was demonstrated to have an oncogenic role in breast cancer [29], colorectal cancer [5] and cervical cancer [30], but its expression in liver cancer is significantly decreased [30]. However, the roles and underlying mechanisms of hnRNP-F in the development of BC remain unclear. Therefore, of those differentially expressed proteins in BC identified by the proteomic method, the hnRNP-F protein was chosen for further study.
In our study, we first found that hnRNP-F was significantly upregulated in BC tissues via 2D-DIGE. Western blot and RT-qPCR analyses further confirmed our results. Furthermore, the findings of our IHC analysis, which was based on a cohort of 103 BC patients, demonstrated that high expression levels of hnRNP-F were significantly associated with an advanced clinical stage. The multivariate analysis showed that elevated hnRNP-F expression was associated with poor overall survival in BC patients, indicating that hnRNP-F expression may be a potential prognostic biomarker for BC patients. Moreover, we applied in vitro and in vivo methods to reveal the functions of hnRNP-F in BC tumourigenesis, which showed that hnRNP-F was necessary for tumour growth and metastasis in BC. Briefly, these data suggest that hnRNP-F plays a vital role in carcinogenesis, particularly in BC.
Tissue invasion and metastasis are major problems in treating BC. EMT is a vital process that causes stationary epithelial cells to gain the ability to migrate and invade as single cells, which has a critical role in various human diseases, especially cancer [31,32]. EMT also plays important roles in BC, enhancing cancer cells with invasive and metastatic properties that are closely related to the development and prognosis of BC [33]. Our results showed that hnRNP-F had the ability to downregulate epithelial marker expression, upregulate mesenchymal marker expression, promote cell invasion and induce EMT in human BC cells. The downstream targets of hnRNP-F are not fully known, and this issue was the main focus of our present investigation. In our investigation of the potential downstream regulator of hnRNP-F, Snail1, but not Twist1, expression was decreased at the mRNA and protein levels when hnRNP-F was silenced by a siRNA. In contrast, hnRNP-F overexpression led to an increase in both the RNA and protein levels of Snail1, suggesting that Snail1 is likely a target of hnRNP-F that mediates its effects on enhancing invasion, metastasis and EMT in BC.
Furthermore, the effect of hnRNP-F on inducing EMT was blocked when Snail1 was silenced. In this context, the regulation of Snail1 by hnRNP-F is a novel mechanism identified in this report by which hnRNP-F may induce EMT in BC. Snail1 binds to the E-box sequences in the promoter of E-cadherin to repress transcription and trigger EMT, which could elicit associated pathological characteristics such as invasion, metastasis and stemness in cancer cells [34]. Roth et al [35] reported that silencing Snail1 completely blocked circulating tumour cell production and regional/distant metastasis in a BC xenograft model. The stability and activity of Snail1 protein expression are mainly regulated by various posttranslational modification factors. The regulation of mRNA stability is a vital element of gene expression, especially for a gene with an unstable mRNA, such as Snail1, with its short t1/2 of ~25 min. As previously reported, the AREs in the 3′ UTR are responsible for the stability of Snail1 mRNA [17,20].
Many previous studies have implicated hnRNPs in the regulation of mRNA stability. It has also been demonstrated that hnRNP-F can interact with the tristetraprolin family of zinc finger proteins, leading to the stability of ARE-mRNAs, including cytokines, transcription factors and protooncogenes. Notably, the interaction of hnRNP-F with tristetraprolin proteins is not dependent on the extent of hnRNP-F binding to the mRNA but is mediated by a nuclear interaction [10]. Surprisingly, our RNA chip analysis revealed that hnRNP-F was able to bind to Snail1 mRNA. Interestingly, our findings also showed that the decay of Snail1 mRNA was significantly slower after treatment with actinomycin D in the hnRNP-F overexpression group than in the control group. This finding indicated that the stability of Snail1 mRNA was enhanced in the hnRNP-F overexpression group. Therefore, we speculate that hnRNP-F binds to the 3′ UTR of Snail1 mRNA, resulting in distinct consequences for mRNA stability and gene/protein expression. In support of this result, our experiments further revealed that hnRNP-F could directly bind to the 3′ UTR of Snail1 mRNA in the regions containing the AREs of Snail1 Δ2 and Snail1 Δ3, which stabilized Snail1 and promoted invasion, metastasis and EMT in BC. Undoubtedly, the 3′ UTR truncations into pcDNA.3 could reflect the specific binding relationship between AREs and hnRNP-F. Whereas the reporter transcripts with the mutated UTR could reflect binding relationship from the original and the whole angle compared with 3′ UTR truncations into pcDNA.3, which investigated a positive relationship between binding and AREs and would reinforce the conclusions of this part.
Taken together, the results of our study show that hnRNP-F, identified by proteomic methods, is significantly upregulated in BC tissues. Its increased expression is a potentially powerful predictor of a poor prognosis for BC patients. HnRNP-F is critical for promoting BC cell invasion and metastasis and mediates the stabilization of Snail1 mRNA by binding to its 3′ UTR, subsequently regulating EMT. These findings may allow us to conclude that Snail1 is a mediator of hnRNP-F affecting EMT, which could lead to the development of a new approach for BC therapy in the future.
The following are the supplementary data related to this article.
Fig. S1.
a–b. HnRNP-F mRNA and protein expression was examined in EJ cells following transfection with a negative control siRNA (NC), si-hnRNP-F-1, si-hnRNP-F-2 and si-hnRNP-F-3; c. HnRNP-F protein expression levels were examined in four BC cell lines by western blot analysis following hnRNP-F KD and OE; d. GFP expression was assessed in four BC cells after transfection with the corresponding lentivirus vectors to examine transfection efficiency, Scale bars represent 100 μm; e. The protein expression of Snail1 was examined by western blot analysis following Snail1 OE in EJ cells; f. Snail1 mRNA and protein expression was examined in EJ cells following transfection with the NC, si-Snail1-1, si-Snail1-2 and si-Snail1-3; g. The effect of hnRNP-F on BC cell invasion was assessed in vitro by the Boyden chamber assay after transfection with si-hnRNP-F-3. Scale bars represent 50 μm; h. The effect of hnRNP-F on BC cell proliferation was assessed in vitro by the CCK-8 assay after transfection with si-hnRNP-F-3, A two-way analysis of variance (ANOVA) test was performed to determine the statistically significant differences; i. The effect of hnRNP-F on BC cell migration was assessed in vitro by the wound scratch healing assay after transfection with si-hnRNP-F-3. Scale bars represent 100 μm (*p < .05, **p < .01, ***p < .001, #p ≥ .05). Student's t-test was performed to analyze statistical significance. Error bars denote standard deviation.
Fig. S2.
a–b. Constitutive OE of Snail1 rescues invasion (a) and migration (b) in EJ cells following hnRNP-F KD; c–d. Blocking Snail1 expression counteracted the enhanced invasion (c) and migration (d) in BIU-87 cells following hnRNP-F OE; Student's t-test was performed to analyze statistical significance. Error bars denote standard deviation. Scale bars (a and c) represent 50 μm, Scale bars (b and d) represent 100 μm;
Fig. S3.
a–c. The half-life of Twist1 mRNA was determined by RT-qPCR after treatment with actinomycin D (2 μg/ml) following hnRNP-F KD and OE. Bars (c) show the half-lives of different groups (*p < .05, **p < .01, ***p < .001, #p ≥ .05). Student's t-test was performed to analyze statistical significance. Error bars denote standard deviation; a. The expression of hnRNP-F mRNA in four human BC cell lines and 293 T cell line.
Identification of proteins differentially expressed between BC and control.
All qRT-PCR Primer.
Sequences of interference fragment and overexpression transcript.
Sequences of truncations and mutants.
Sequences of plasmids in Luciferase reporter gene assay.
Acknowledgments
Acknowledgments
We thank Pro. Shuang Wang (The Department of Pathology, Nanfang Hospital of Southern Medical University) for assistance in our study.
Funding sources
This study was supported by Natural Science Foundation Committee of China (NSFC 81802941) (F.L.), and the Natural Science Foundation of Guangdong Province of China (2018A0303130287) (F.L.). These funding sources had no role in the study design, data collection, data analysis, interpretation, or writing of this manuscript.
Declarations of interests
The authors declare no conflicts of interest.
Author contributions
FL, HFZ, MQS and ZY performed and conceived the experiments. FL, LNH and WLT wrote the manuscript. MQS, ZYF and WWX provided clinical information. HFZ, ZY and MQS provided the paraffin-embedded tumour specimens and analysed the IHC results. YJD coordinated the project. FL and WLT assisted in improving the quality of language. All authors read and approved the final manuscript.
Contributor Information
Zhe Yu, Email: yuzhe@smu.edu.cn.
Lina Hou, Email: linahou@smu.edu.cn.
Wanlong Tan, Email: twl@smu.edu.cn.
References
- 1.Torre L.A., Bray F., Siegel R.L., Ferlay J., Lortet-Tieulent J., Jemal A. Global cancer statistics, 2012. CA Cancer J Clin. 2015;65:87–108. doi: 10.3322/caac.21262. [DOI] [PubMed] [Google Scholar]
- 2.Li F., Hong X., Hou L., Lin F., Chen P., Pang S. A greater number of dissected lymph nodes is associated with more favorable outcomes in bladder cancer treated by radical cystectomy: a meta-analysis. Oncotarget. 2016;7:61284–61294. doi: 10.18632/oncotarget.11343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Batlle E., Sancho E., Francí C., Domínguez D., Monfar M., Baulida J. The transcription factor Snail1 is a repressor of E-cadherin gene expression in epithelial tumour cells. Nat Cell Biol. 2000;2(2):84–89. doi: 10.1038/35000034. [DOI] [PubMed] [Google Scholar]
- 4.Khan F.M., Marquardt S., Gupta S.K., Knoll S., Schmitz U., Spitschak A. Unraveling a tumor type-specific regulatory core underlying E2F1-mediated epithelial-mesenchymal transition to predict receptor protein signatures. Nat Commun. 2017;8(1):198. doi: 10.1038/s41467-017-00268-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Balasubramani M., Day B.W., Schoen R.E., Getzenberg R.H. Altered expression and localization of creatine kinase B, heterogeneous nuclear ribonucleoprotein F, and high mobility group box 1 protein in the nuclear matrix associated with colon cancer. Cancer Res. 2006;66(2):763–769. doi: 10.1158/0008-5472.CAN-05-3771. [DOI] [PubMed] [Google Scholar]
- 6.Zhou X., Liu S., Cai G., Kong L., Zhang T., Ren Y. Long non coding RNA MALAT1 promotes tumor growth and metastasis by inducing epithelial-mesenchymal transition in oral squamous cell carcinoma. Sci Rep. 2015;5 doi: 10.1038/srep15972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yeung K.T., Yang J. Epithelial-mesenchymal transition in tumor metastasis. Mol Oncol. 2017;11(1):28–39. doi: 10.1002/1878-0261.12017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chen C.Y., Shyu A.B. AU-rich elements, characterization and importance in mRNA degradation. Trends Biochem Sci. 1995;20(11):465–470. doi: 10.1016/s0968-0004(00)89102-1. [DOI] [PubMed] [Google Scholar]
- 9.Geuens T., Bouhy D., Timmerman V. The hnRNP family: insights into their role in health and disease. Hum Genet. 2016;135(8):851–867. doi: 10.1007/s00439-016-1683-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Reznik B., Clement S.L., Lykke-Andersen J. hnRNP F complexes with tristetraprolin and stimulates ARE-mRNA decay. PLoS One. 2014;9(6) doi: 10.1371/journal.pone.0100992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Goh E.T., Pardo O.E., Michael N., Niewiarowski A., Totty N., Volkova D. Involvement of heterogeneous ribonucleoprotein F in the regulation of cell proliferation via the mammalian target of rapamycin/S6 kinase 2 pathway. J Biol Chem. 2010;285(22):17065–17076. doi: 10.1074/jbc.M109.078782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Garneau D., Revil T., Fisette J.F., Chabot B. Heterogeneous nuclear ribonucleoprotein F/H proteins modulate the alternative splicing of the apoptotic mediator Bcl-x. J Biol Chem. 2005;280(24):22641–22650. doi: 10.1074/jbc.M501070200. [DOI] [PubMed] [Google Scholar]
- 13.Li F., Chen D.N., He C.W., Zhou Y., Olkkonen V.M., He N. Identification of urinary Gc-globulin as a novel biomarker for bladder cancer by two-dimensional fluorescent differential gel electrophoresis (2D-DIGE) J Proteomics. 2012;77:225–236. doi: 10.1016/j.jprot.2012.09.002. [DOI] [PubMed] [Google Scholar]
- 14.Maréchal R., Demetter P., Nagy N., Berton A., Decaestecker C., Polus M. High expression of CXCR4 may predict poor survival in resected pancreatic adenocarcinoma. Br J Cancer. 2009;100:1444–1451. doi: 10.1038/sj.bjc.6605020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Nieto M.A., Huang R.Y., Jackson R.A., Thiery J.P. EMT: 2016. Cell. 2016;166(1):21–45. doi: 10.1016/j.cell.2016.06.028. [DOI] [PubMed] [Google Scholar]
- 16.Xu M.Y., Chen R., Yu J.X., Liu T., Qu Y., Lu L.G. AZGP1 suppresses epithelial-to-mesenchymal transition and hepatic carcinogenesis by blocking TGFβ1-ERK2 pathways. Cancer Lett. 2016;374(2):241–249. doi: 10.1016/j.canlet.2016.02.025. [DOI] [PubMed] [Google Scholar]
- 17.Barbera M.J., Puig I., Dominguez D., Julien-Grille S., Guaita-Esteruelas S., Peiró S., García de Herreros A. Regulation of Snail1 transcription during epithelial to mesenchymal transition of tumor cells. Oncogene. 2004;23:7345–7354. doi: 10.1038/sj.onc.1207990. [DOI] [PubMed] [Google Scholar]
- 18.Zhou B.P., Deng J., Xia W., Xu J., Li Y.M., Gunduz M. Dual regulation of Snail1 by GSK-3beta-mediated phosphorylation in control of epithelial-mesenchymal transition. Nat Cell Biol. 2004;6(10):931–940. doi: 10.1038/ncb1173. [DOI] [PubMed] [Google Scholar]
- 19.Liu Z.C., Wang H.S., Zhang G., Liu H., Chen X.H., Zhang F. AKT/GSK-3β regulates stability and transcription of Snail1 which is crucial for bFGF-induced epithelial-mesenchymal transition of prostate cancer cells. Biochim Biophys Acta. 2014;1840(10):3096–3105. doi: 10.1016/j.bbagen.2014.07.018. [DOI] [PubMed] [Google Scholar]
- 20.Yoo J.O., Kwak S.Y., An H.J., Bae I.H., Park M.J., Han Y.H. miR-181b-3p promotes epithelial-mesenchymal transition in breast cancer cells through Snail1 stabilization by directly targeting YWHAG. Biochim Biophys Acta. 2016;1863(7):1601–1611. doi: 10.1016/j.bbamcr.2016.04.016. [DOI] [PubMed] [Google Scholar]
- 21.Li J., Guo W., Li F., He J., Yu Q., Wu X. HnRNP L as a key factor in spermatogenesis: lesson from functional proteomic studies of azoospermia patients with sertoli cell only syndrome. J Proteomics. 2012;75(10):2879–2891. doi: 10.1016/j.jprot.2011.12.040. [DOI] [PubMed] [Google Scholar]
- 22.Shao Q., Byrum S.D., Moreland L.E., Mackintosh S.G., Kannan A., Lin Z. A proteomic study of human Merkel cell carcinoma. J Proteomics Bioinform. 2013;6:275–282. doi: 10.4172/jpb.1000291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Du J., Wang Q., Ziegler S.F., Zhou B. FOXP3 interacts with hnRNPF to modulate pre-mRNA alternative splicing. J Biol Chem. 2018;293(26):10235–10244. doi: 10.1074/jbc.RA117.001349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Loh T.J., Moon H., Cho S., Jang H., Liu Y.C., Tai H. CD44 alternative splicing and hnRNP A1 expression are associated with the metastasis of breast cancer. Oncol Rep. 2015;34(3):1231–1238. doi: 10.3892/or.2015.4110. [DOI] [PubMed] [Google Scholar]
- 25.Anantha R.W., Alcivar A.L., Ma J., Cai H., Simhadri S., Ule J. Requirement of heterogeneous nuclear ribonucleoprotein C for BRCA gene expression and homologous recombination. PLoS One. 2013;8(4) doi: 10.1371/journal.pone.0061368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sun Y.L., Liu F., Liu F., Zhao X.H. Protein and gene expression characteristics of heterogeneous nuclear ribonucleoprotein H1 in esophageal squamous cell carcinoma. World J Gastroenterol. 2016;22(32):7322–7331. doi: 10.3748/wjg.v22.i32.7322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hope N.R., Murray G.I. The expression profile of RNA-binding proteins in primary and metastatic colorectal cancer: relationship of heterogeneous nuclear ribonucleoproteins with prognosis. Hum Pathol. 2011;42(3):393–402. doi: 10.1016/j.humpath.2010.08.006. [DOI] [PubMed] [Google Scholar]
- 28.Yang Y., Jia D., Kim H., Abd Elmageed Z.Y., Datta A., Davis R. Dysregulation of miR-212 promotes castration resistance through hnRNPH1-mediated regulation of AR and AR-V7: implications for racial disparity of prostate cancer. Clin Cancer Res. 2016;22(7):1744–1756. doi: 10.1158/1078-0432.CCR-15-1606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Dong X., Yang M., Sun H., Lü J., Zheng Z., Li Z. Combined measurement of CA 15-3 with novel autoantibodies improves diagnostic accuracy for breast cancer. Onco Targets Ther. 2013;6:273–279. doi: 10.2147/OTT.S43122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Honoré B., Vorum H., Baandrup U. hnRNPs H, H' and F behave differently with respect to posttranslational cleavage and subcellular localization. FEBS Lett. 1999;456(2):274–280. doi: 10.1016/s0014-5793(99)00911-4. [DOI] [PubMed] [Google Scholar]
- 31.Hensley P.J., Zetter D., Horbinski C.M., Strup S.E., Kyprianou N. Association of epithelial-mesenchymal transition and nuclear cofilin with advanced urothelial cancer. Hum Pathol. 2016;57:68–77. doi: 10.1016/j.humpath.2016.06.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Singh R., Ansari J.A., Maurya N., Mandhani A., Agrawal V., Garg M. Epithelial-to-mesenchymal transition and its correlation with clinicopathologic features in patients with urothelial carcinoma of the bladder. Clin Genitourin Cancer. 2017;15(2):e187–e197. doi: 10.1016/j.clgc.2016.07.021. [DOI] [PubMed] [Google Scholar]
- 33.Zhu B., Qi L., Liu S., Liu W., Ou Z., Chen M. CLASP2 is involved in the EMT and early progression after transurethral resection of the bladder tumor. BMC Cancer. 2017;17(1):105. doi: 10.1186/s12885-017-3101-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Comijn J., Berx G., Vermassen P., Verschueren K., van Grunsven L., Bruyneel E. The two-handed E box binding zinc finger protein SIP1 downregulates E-cadherin and induces invasion. Mol Cell. 2001;7(6):1267–1278. doi: 10.1016/s1097-2765(01)00260-x. [DOI] [PubMed] [Google Scholar]
- 35.Roth B., Jayaratna I., Sundi D., Cheng T., Melquist J., Choi W. Employing an orthotopic model to study the role of epithelial-mesenchymal transition in BC metastasis. Oncotarget. 2017;8(21):34205–34222. doi: 10.18632/oncotarget.11009. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Identification of proteins differentially expressed between BC and control.
All qRT-PCR Primer.
Sequences of interference fragment and overexpression transcript.
Sequences of truncations and mutants.
Sequences of plasmids in Luciferase reporter gene assay.