Abstract
Purpose
To construct and validate an efficient artificial neural network (ANN) based on parameters with statistical correlation to live birth, to be used as a comprehensive tool for the prediction of the clinical outcome for patients undergoing ART.
Methods
Data from 257 infertile couples that underwent a total of 426 IVF/ICSI cycles from 2010 to 2017 was collected on an ensemble of 118 parameters for each cycle. Statistical correlation of the parameters with the outcome of live birth was performed, using either t test or χ2 test, and the parameters that demonstrated statistical significance were used to construct the ANN. Cross-validation was performed by random separation of data and repeating the training-testing procedure by 10 times.
Results
12 statistically significant parameters out of the initial ensemble were used for the ANN construction, which exhibited a cumulative sensitivity and specificity of 76.7% and 73.4%, respectively. During cross-validation, the system exhibited the following: sensitivity 69.2% ± 2.36%, specificity 69.19% ± 2.8% (OR 5.21 ± 1.27), PPV 36.96 ± 3.44, NPV 89.61 ± 1.09, and OA 69.19% ± 2.69%. A rather small standard deviation in the performance indices between the training and test sets throughout the validation process indicated a stable performance of the constructed ANN.
Conclusions
The constructed ANN is based on statistically significant variables with the outcome of live birth and represents a stable and efficient system with increased performance indices. Validation of the system allowed an insight of its clinical value as a supportive tool in medical decisions, and overall provides a reliable approach in the routine practice of IVF units in a user-friendly environment.
Keywords: Artificial neural network, Artificial intelligence, Prediction model, Assisted reproduction, Live birth, Personalized treatment
Introduction
An estimated 8 to 12% of couples in reproductive age face challenges in achieving pregnancy within a year of regular, timed, and unprotected intercourse [1–3], with global surveys reporting as many as 186 million individuals suffering from infertility [4]. For the past 40 years, assisted reproduction technologies (ART) utilize scientific knowledge and sophisticated technology for infertility management, even though compared with the degree of intervention, the success rates remain readily low, with only 30% of the embryos produced in vitro, being ultimately transferred to the uterus and only 10–30% of transferred embryos progressing to live birth [5–9].
This realization calls for a fresh view on infertility management, along with the new perspectives on modern lifestyle and social structure that delays family planning. In this context, the utilization of some rapidly evolving fields, such as computer-based prediction models and artificial intelligence (AI) systems, conforms to the tendency towards automation of the procedures performed in a modern in vitro fertilization (IVF) Unit. Such systems are already implemented in other medical fields with numerous examples, including cancer research [10, 11], neurology [12, 13], cardiology [14, 15], drug design [16], stem cell, cell transplantation, and immune therapies [17].
In the field of ART, prediction models have been engaged in embryo selection as a complementary tool for decision-making and for the intrinsic assessment of various factors in relation to their contribution to the clinical outcome [18]. The so far applied models have demonstrated usefulness in performing correlations between the analyzed factor/s and treatment outcome or causative source, in this case, infertility factor. However, there are varying levels of accuracy and restrictions limiting their effectiveness that withhold routine implementation in IVF procedures [18]. In this notion, more sophisticated AI systems were introduced, such as artificial neural networks (ANNs). These appear advantageous due to their remarkable information-processing characteristics pertinent mainly to nonlinearity, high level of parallelism, noise and fault tolerance, and learning, generalization, and self-adapting capabilities [19]. The first ANN constructed with application on assisted reproduction was proposed by Kauffman et al. in 1997 [20] with onwards reporting of similar systems developed with AI targeting on various aspects of ART, in an attempt to predict clinical outcomes and especially live birth, but with varying input variables and predictive power [21–32].
In 2011, our team underlined the need to employ AI in IVF [33] and, in 2016, we proposed the construction and implementation of a supervised ANN with a flexible architecture that can assist clinicians to offer subfertile couples a novel approach in personalized treatment and act as an accessible software platform for routine use in the IVF unit [34]. By acquiring experience from previous attempts in ANN employment in ART, our aim was to construct and validate a functional ANN by including parameters that exert a meaningful effect on live birth following assisted reproduction.
Population and methods
Patient population
Data was retrospectively collected on a previously diagnosed infertile population of 257 infertile couples who underwent 426 IVF/ICSI cycles, at the Assisted Reproduction Unit of the Third Department of Obstetrics and Gynecology, “Attikon” University Hospital in Greece, from July 2010 to February 2017. For the included population, conclusive data was available on demographics, medical/reproductive history for both partners, and previous IVF cycle parameters and outcomes. Consent to use anonymous data for research purposes and for the construction of ANN was obtained from all participants and the research protocol was approved by the Scientific Council and the Bioethics Committee of the Hospital, prior to study initiation (EVD 1172/26-11-15). The population of the cohort had been previously diagnosed with infertility (tubal factor, male factor, combined tubal/male, and unexplained infertility) and met the criteria for undergoing ovarian stimulation protocols with GnRH agonists/antagonists followed by IVF/ICSI and fresh embryo transfer (ET) or frozen-thawed embryo transfer. No other interventions were applied during cycles, apart from the routine protocols described in the regular practice of the IVF unit, that conform to International trends and European standards for appropriate medical management. Overall, success rates and laboratory performance demonstrate stability in the clinical setting of the current study throughout the years.
Criteria for exclusion were the advanced age of the female partner (> 43 years), increased follicle-stimulating hormone (FSH) serum levels on the third day of the menstrual cycle (> 15 IU/L), other factors of infertility than the reported, previous poor ovarian response according to the 2011 Bologna criteria [35] or over-response (polycystic ovaries or ovarian syndrome (PCO/PCOS), ovarian hyperstimulation syndrome (OHSS)) [36], known genetic factors contributing to infertility, ovarian cystectomy or oophorectomy, endometriosis, or pathology affecting the endometrial cavity. For all cycles performed, the clinical outcome was available, whereas it was possible to monitor each successful cycle for live birth, or adverse outcomes.
Statistical methods and construction-validation of the ANN
For the total of 426 IVF cycles, a dynamic database was generated with Microsoft Excel and used for data storage and pre-processing. In total, 118 parameters were available, as categorically reported here: cycle characteristics of previous ART cycles and their outcome; infertility duration and factor, medical and reproductive history, selective lifestyle information and demographics linked to fertility dynamics, and pathology linked to infertility factors for both partners; age at cycle, parity, and hormonal profile along with selected information on menstrual cycle characteristics for the female partner; and sperm analysis for the male partner.
The core of the AI system was an ANN with a classical multilayer feedforward architecture with one hidden layer and training through back propagation of the error algorithm (Levenberg-Marquardt variant), according to the workflow in Fig. 1. As described previously [33, 34], data collection and processing was performed and the outcome of the system was live birth, as defined in the latest published International Glossary [37].
Fig. 1.
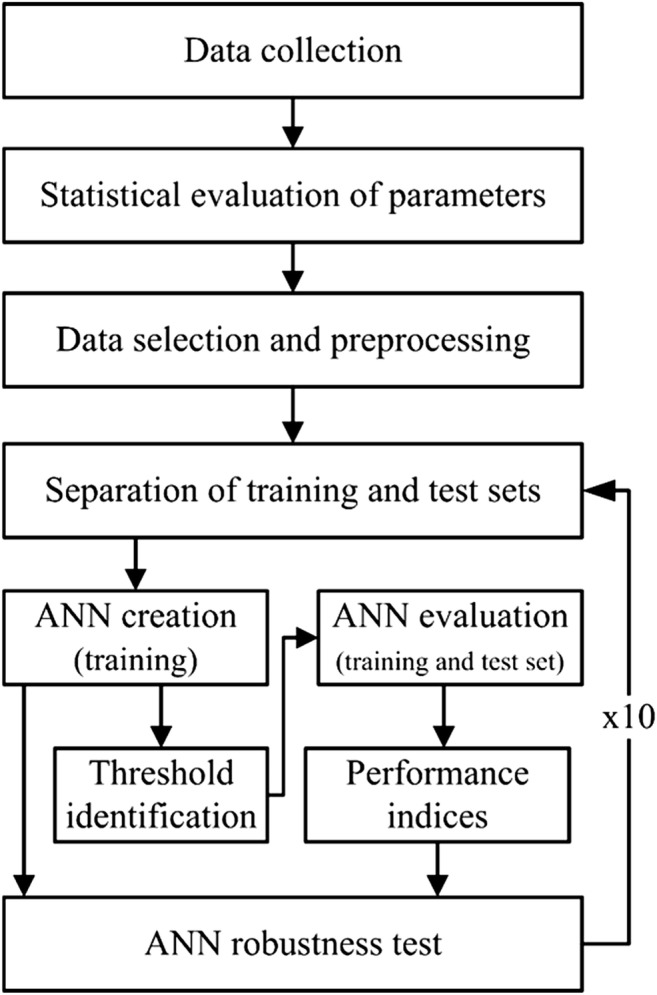
Work flow with basic steps on the ANN system construction
Initially, the performance of ANN was assessed by the input of the parameter ensemble and statistical correlation was thereafter performed for each one of the 118 available parameters separately, in order to signify those with a direct effect on live birth. Analysis was performed with the SAS 9.4 platform (SAS Institute Inc., NC, USA) [38], by employing either Student’s t test for numeric parameters or the χ2 test for categorical parameters. The latter group of categorical values was subsequently mapped to represent numerical values, which were scaled in the range between + 0.1 and + 0.9, in order to correspond with the range of the produced outputs of the neurons of the ANN. Data cohorts attributing to study participants were then separated into two sets; the training set, used for the training of the ANN, and the test set, used for the evaluation of the ANN performance on “unknown data” [39, 40]. The separation was performed through stratified random sampling: 70% of cycles’ data were assigned to the training set and 30% to the test set.
A threshold was required to be determined since the ANN output was numeric, in order to identify the output for which the inserted data would expect a positive clinical outcome, in this case, live birth. By using the ANN outputs obtained for the training set at various thresholds from zero to one using a step of 0.001, the sensitivity and specificity for each individual threshold value was calculated and the optimal threshold was determined at the minimum difference between sensitivity and specificity. This threshold was applied throughout the processes and during the validation of the system. The algorithms for the threshold calculation were implemented in a MATLAB programming environment (MathWorks Inc.).
Cross-validation of the system was performed by random data allocation into training and test sets and by repeating the training-testing procedure by 10 times. The performance indices for the construction and validation of the ANN were sensitivity, specificity, positive predictive value (PPV), negative predictive value (NPV), false positive rate (FPR), false negative rate (FNR), overall accuracy (OA), and odds ratios (ORs). These indices were also calculated for the sum of the 10 trained systems during the validation process and separately for the training and test sets. In addition, mean values and standard deviations of the replicated ANN structures and the differences between the performance in the training and test sets were calculated.
Results
An ensemble of 118 parameters and variables was available for 426 IVF cycles, sourcing from a total of 257 infertile couples, with overall successful clinical outcome with live birth in 21.6% of the cycles included. Statistical analysis attributed a correlation with the clinical outcome of live birth to only a fraction of the available parameters, as expected: age of the female partner, age group (≤ 35 years, 36–40 years, > 40 years), age at menarche, age at menarche at a threshold of 12 years of age, history of dyspareunia, total dose of gonadotrophins administered during ovarian stimulation, endometrial thickness prior to oocyte retrieval, number of top-quality embryos (TQE) on day three, ratio of the number of TQE D3 to the number of fertilized oocytes (2PN), number of embryos transferred to the uterus to the total number of 2PN, embryo transfer in fresh cycle (ET) or following embryo cryopreservation (FET), and difficulty during ET (Table 1). The statistically significant parameters constituted the final input nodes for the ANN construction and throughout the validation of the system.
Table 1.
Quantitative and categorical parameters that were statistically different for the cycles resulting in live birth and for the cycles with negative outcome (t test was performed for the quantitative parameters and χ2 test for the categorical parameters)
| Live birth | No live birth | |||||
|---|---|---|---|---|---|---|
| Quantitative variables | Mean | SD | Mean | SD | p value | Difference characterization |
| Age (female) | 34.64 | 3.9989 | 36.01 | 4.0968 | 0.0044 | Somewhat |
| Age at menarche | 13.2 | 1.9834 | 12.28 | 1.7064 | < 0.0001 | Highly |
| Difficulty during ET | 1.78 | 0.4147 | 1.93 | 0.6125 | 0.0062 | Weak |
| Endometrium thickness prior to OR | 10.26 | 1.8045 | 9.63 | 1.9172 | 0.0101 | Weak |
| ET/2PN | 0.48 | 0.2614 | 0.6 | 0.3104 | 0.0035 | Somewhat |
| TQE D3 | 1.51 | 1.4577 | 0.85 | 1.2947 | 0.0002 | Somewhat |
| TQE D3/2PN | 0.29 | 0.2608 | 0.18 | 0.2571 | 0.0016 | Somewhat |
| Total gonadotropins | 2620.7 | 996.5 | 2906.3 | 1138.1 | 0.0472 | Weak |
| Categorical variables | Chi-square | Odds ratio | 95% lower CI | 95% upper CI | p value | Difference characterization |
| Age group | 8.8028 | NA | NA | NA | 0.012 | Weak |
| Dyspareunia | 9.9988 | 3.9512 | 1.5914 | 9.8101 | 0.002 | Somewhat |
| Fresh or frozen cycle | 7.5309 | 0.4201 | 0.2230 | 0.7914 | 0.006 | Weak |
| Menarche > 12 years | 19.7164 | 0.2292 | 0.1145 | 0.4588 | < 0.0001 | Highly |
ET, embryo transfer; OR, oocyte retrieval; ET/2PN, ratio of number of embryos transferred to the uterus to oocytes fertilized (2PN) following OR; TQE D3, top-quality embryos at day 3 of development; TQE D3/2PN, ratio of top-quality embryos at day 3 of development to oocytes fertilized (2PN); total gonadotropins, total dose of gonadotropins administered (IU) during the stimulation protocol in a single cycle
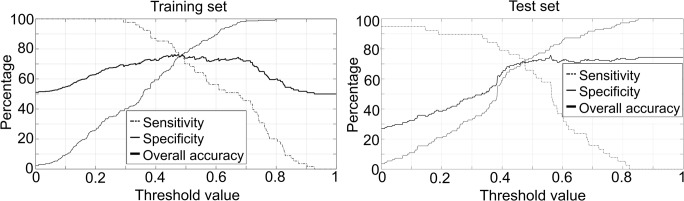
The constructed ANN was restricted to < 1000 iterations and the memory factor (μ) initiated from 0.001, while μ increase and decrease factors were 10 and 0.1, respectively. Notably, the ANN converged in just seven iterations. Two hidden neurons and one single output neuron were used, as the selection of a higher number of neurons would not contribute to the robustness of the system, while a single output was decided to depict a positive or negative outcome according to the threshold. The performance indices of the constructed ANN are presented in Fig. 2 for both the training and the test set at variable thresholds, during the step-up procedure to determine the optimal threshold. Minimum difference between sensitivity and specificity was determined at a threshold of 0.48, where the performance of the ANN in the training and test demonstrated a cumulative sensitivity and specificity of 76.7% and 73.4%, respectively (Table 2).
Fig. 2.
Sensitivity, specificity, and overall accuracy of the ANN, trained with statistically important parameters. Horizontal axis represents various thresholds of the ANN output. a Training set (left). b Test set (right)
Table 2.
Performance indicators of the ANN trained by the statistically significant parameters on reproductive outcome
| Performance indicator | Training set | Test set | All data |
|---|---|---|---|
| Sensitivity | 77.7 | 71.1 | 76.7 |
| Specificity | 74.6 | 70.1 | 73.4 |
| Positive predictive value | 75.3 | 45.7 | 69.3 |
| Negative predictive value | 77.0 | 87.6 | 80.1 |
| False positive rate | 25.4 | 29.1 | 26.6 |
| False negative rate | 22.3 | 28.9 | 23.3 |
| Overall accuracy | 76.1 | 70.9 | 74.8 |
| Odds ratio | 10.2 | 6.0 | 9.1 |
Possible study biases were avoided by testing the stability of the system, a procedure that was repeated 10 times with random data allocation to training and test sets (Table 3). ANN architecture was maintained (10-2-1 neurons) and the number of iterations required for convergence was in the range of seven to nine(7.6 ± 0.8). There were no significant differences in the performance indices between training and test sets: mean sensitivity (2.03% (69.20% vs. 67.17%)), mean specificity (2.22% (69.19% vs. 66.97%)), and overall accuracy (2.19% (69.19% vs. 67.01%)) (p values > 0.05). These results indicate that the performance of the ANNs, during the validation procedure, was stable with no statistical difference between the training and test sets.
Table 3.
Performance indicators for the training and test sets of the 10 ANNs trained by ten different random allocations of the dataset
| Dataset name | Sensitivity | Specificity | PPV | NPV | FPR | NPR | OA | OR | |
|---|---|---|---|---|---|---|---|---|---|
| Training set | ANNDataSet0 | 66.67 | 64.76 | 32.73 | 88.31 | 35.24 | 33.33 | 65.15 | 3.68 |
| ANNDataSet1 | 71.43 | 71.22 | 40.40 | 90.12 | 28.78 | 28.57 | 71.26 | 6.19 | |
| ANNDataSet2 | 66.67 | 67.86 | 33.33 | 89.41 | 32.14 | 33.33 | 67.63 | 4.22 | |
| ANNDataSet3 | 72.00 | 73.53 | 40.00 | 91.46 | 26.47 | 28.00 | 73.23 | 7.14 | |
| ANNDataSet4 | 69.09 | 68.64 | 33.93 | 90.50 | 31.36 | 30.91 | 68.73 | 4.89 | |
| ANNDataSet5 | 70.91 | 71.01 | 39.39 | 90.18 | 28.99 | 29.09 | 70.99 | 5.97 | |
| ANNDataSet6 | 72.58 | 72.52 | 42.45 | 90.45 | 27.48 | 27.42 | 72.54 | 6.99 | |
| ANNDataSet7 | 66.67 | 66.51 | 34.55 | 88.27 | 33.49 | 33.33 | 66.54 | 3.97 | |
| ANNDataSet8 | 67.27 | 67.15 | 35.24 | 88.54 | 32.85 | 32.73 | 67.18 | 4.20 | |
| ANNDataSet9 | 68.75 | 68.67 | 37.61 | 88.89 | 31.33 | 31.25 | 68.69 | 4.82 | |
| Mean value ± SD | 69.20 ± 2.36 | 69.19 ± 2.80 | 36.96 ± 3.44 | 89.61 ± 1.09 | 30.81 ± 2.80 | 30.80 ± 2.36 | 69.19 ± 2.69 | 5.21 ± 1.27 | |
| Test set | ANNDataSet0 | 69.23 | 68.56 | 38.46 | 88.70 | 31.44 | 30.77 | 68.71 | 4.91 |
| ANNDataSet1 | 63.89 | 63.57 | 32.86 | 86.32 | 36.43 | 36.11 | 63.64 | 3.09 | |
| ANNDataSet2 | 63.16 | 65.46 | 38.71 | 83.72 | 34.55 | 36.84 | 64.87 | 3.25 | |
| ANNDataSet3 | 64.18 | 63.79 | 33.86 | 86.05 | 36.21 | 35.82 | 63.88 | 3.16 | |
| ANNDataSet4 | 70.27 | 70.41 | 47.27 | 86.25 | 29.59 | 29.73 | 70.37 | 5.62 | |
| ANNDataSet5 | 67.57 | 66.14 | 36.77 | 87.50 | 33.86 | 32.43 | 66.46 | 4.07 | |
| ANNDataSet6 | 63.33 | 62.50 | 31.15 | 86.42 | 37.50 | 36.67 | 62.68 | 2.88 | |
| ANNDataSet7 | 65.71 | 65.55 | 35.94 | 86.67 | 34.45 | 34.29 | 65.58 | 3.65 | |
| ANNDataSet8 | 72.97 | 72.44 | 43.55 | 90.20 | 27.56 | 27.03 | 72.56 | 7.10 | |
| ANNDataSet9 | 71.43 | 71.29 | 40.82 | 90.00 | 28.71 | 28.57 | 71.32 | 6.21 | |
| Mean value ± SD | 67.17 ± 3.63 | 66.97 ± 3.49 | 37.94 ± 4.96 | 87.18 ± 1.98 | 33.03 ± 3.49 | 32.83 ± 3.63 | 67.01 ± 3.50 | 4.39 ± 1.49 |
In addition, multiple ANNs were initially constructed by employing the total of 118 available parameters, during the initial phase: these ANNs exhibited different numbers of neurons in the hidden layer (varying from 1 to 50), while their performance was inferior to the presented and validated ANN based on the 12 statistically significant parameters. In more detail, specificity and sensitivity indices in the training set ranged from 60.9 to 70.3 and from 55.5 to 67.3, respectively, while in the test set the respective ranges were 46.7–60.0 and 48.2–59.5. These differences between the training and test sets indicated an unstable behavior of the initial system, contrary to the increased stability obtained by the validated ANN presented here.
Discussion
The initial aim of this study was to construct and validate an ANN, as an attempt to utilize AI in routine medical practice and to offer the “opinion” of an objective intelligent system trained to overview data from multiple cycles. By including 426 ART cycles with an initial recording of 118 parameters and signifying the statistically relevant parameters to live birth, a concise system with high relevance to its application was constructed that furthermore exhibited significant and balanced sensitivity and specificity of almost 70% in both training and test sets, as furthermore favorable characteristics during its validation. A rather “shallow” ANN architecture was selected with 12 input neurons, two neurons in the hidden layer and one output neuron, as this design was efficient, straightforward, and suitable for the creation of a stable system. The mean number of the training cycles required during the construction of the ANN was 7.6 ± 0.8, indicating that the ANN self-converged quickly and consequently overtraining was avoided.
The present constructed and validated system differs from previously published approaches on ANNs and prediction models, in terms of methodological design, outcomes, completeness, efficiency, and stability of the system. The first reported ANN in ART [20] demonstrated an overall accuracy of 59% in predicting clinical pregnancy with PPV of 39% and NPV of 82%. An increased predictive accuracy of 82% was obtained by Wald et al. [21] that focused entirely in predicting the outcome of ART cycles with male factor infertility, where sperm was surgically retrieved for fertilization through IVF/ICSI. Based on embryo selection and implantation as an outcome, Uyar et al. [22] offered another methodological view by utilizing a mixed original IVF dataset for classification of embryos to obtain a PPV of 65.6% and NPV of 67.5%. Banerjee et al. [23] utilized the characteristics of previous failed ART cycles to predict live birth for a boosted-tree model that demonstrated improved characteristics compared with age-based prediction and reported an area under the curve (AUC) for receiver-operator characteristics curve of 0.8. Uyar et al. [24] applied the Naïve Bayes classifier to an original IVF dataset in order to discriminate embryos according to the implantation potentials and determined an optimum threshold decision of 0.3 with a true positive rate of 64.4% and a false positive rate of 30.6%, while in a later approach [31], they proposed a model for implantation outcome of individual embryos in an IVF cycle that resulted in an overall accuracy of 75.7% compared with expert judgment alone, without population characteristics and age groups or other factors that could affect the outcome. Ballester et al. [25] developed a nomogram in a small population sample of patients with endometriosis, concentrating solely on the ART outcome following this pathology and determined a sensitivity and specificity of 72%. A model named PreIVF-D built by Choi et al. [26] involved data obtained from different IVF units, employing a boosted regression tree approach with over 20 characteristics involved in the system development with a personalized success rate of > 45% with PPV of 59.4% and NPV 94.9% as summarized in the review by Simopoulou et al. [18]. Durairaj et al. [27] developed an ANN with an accuracy of 73%, with basic variables in a smaller mixed population sample with potential pathological infertility factors. Manna et al. [28] used AI systems for embryo or oocyte scoring/selection in ART programs that demonstrated interesting classification performance, although these results are reported as preliminary by the same authors. Milewski et al. [29] compared ANN with logistic regression models only to establish that ANNs were superior with reported sensitivity of 69.0% and 60.3% specificity and a chance of about 70% in correctly predicting the establishment of early biochemical pregnancy. Durairaj and Nandhakumar [30] suggested an integrated methodology of ANN with data mining techniques and the experimental model exhibited an overall accuracy of 90%, thus proposed the applied techniques for finding the minimum set of influential parameters in order to predict a success rate of IVF. In another approach on AI utilization, Milewski et al. [32] revisited ANNs by combining embryo morphokinetic data to determine embryo implantation potential and the model presented was able to correctly predict approximately 70% of pregnancies, although no other variables were utilized.
In a previous theoretical evaluation of the usefulness of AI systems for personalized management in ART, we proposed an ANN with the respective parameters to be included in the analysis and later construction of the system [34]. From the previously proposed qualitative and quantitative inputs, in the present cohort of data, we defined that 12 variables were significantly associated with live birth. Some of these parameters have been previously recognized as significant prognostic factors of the success in ART outcome, such as endometrial thickness [41, 42], dyspareunia when linked to endometriosis [43], maternal age [44, 45], total gonadotrophins administered, favoring lowered doses [46, 47], embryo quality [48], embryo transfer technique [49], fresh ET or FET [50], and age at menarche [51].
In the present study, a threshold was used to obtain a balanced sensitivity and specificity; however, it is possible to employ different thresholds according to clinical requirements and applications of the system. For example, in case higher accuracy is required for the prediction of live birth, thus increased sensitivity, the threshold may be adjusted accordingly, although this is accompanied by a reduction in specificity. Furthermore, the clinically relevant variables that were ultimately employed in the present ANN do differ from the approaches employed by other workgroups, as does the clinical outcome. This difference possibly reflects inter-laboratory variations, represents differences of the local population characteristics, and highlights the impact of patient recruitment, data recording, and processing methods and overall the variability in the approach of IVF unit in the management of individual patients.
Apart from the noted discrepancies, the reporting of the associations from different research groups could enable a multicenter collaboration, based mainly on ethnic group assortment, as an attempt to maintain at least a single set of variables stable after acknowledging the usefulness of ANNs. Looking at the limitations of this study, we could suggest certain improvements: incorporation of a higher number of cycles irrespectively of outcome and infertility factor, as well as cycles that failed to reach the stage of embryo transfer. In addition, although a general proportion of the infertile population was covered, more work on a targeted ANN needs to be adjusted, specifically for poor or high responders and patients with a history of endometriosis, with an adequate sample size and additional recorded parameters, since these cases are complex by nature. External validation and multicenter collaboration is encouraged and sought, to reaffirm the usefulness of this ANN and contribute to its advancement by learning from new cases. Another limitation could be the selection of training parameters in a univariate manner (χ2 and t test): this approach could mask multivariate effects, i.e., exclude parameters that might have impact when used combinatorial. However, in our study, the initial ANN trained with the sum of 118 parameters did not exhibit better performance; thus, we anticipate that parameters were not masked in this study; of note, in larger settings, these parameters may be influential. Finally, another approach to reduce dimensionality could be the application of principal components analysis (PCA); nevertheless, this approach does not highlight the statistical importance of each of the parameters that were included in the study.
As a closing remark, the ANN utilized here represents a stable system that has employed clinically meaningful variables, associated with live birth. It can be easily replicated and validated by the incorporation of data from other IVF units, in a user-friendly interface [34, 52]. Further expansion of the system by incorporating data from research collaborations to include cycles with an adverse outcome or other more complex pathological states of infertility could prove beneficial for the research community or at routine IVF implementation.
Acknowledgments
The authors wish to thank the Medical, Paramedical and Laboratory Team of the Assisted Reproduction Unit of “Attikon” University Hospital, Greece.
Compliance with ethical standards
Conflict of interest
The authors declare that they have no conflict of interest.
Ethical approval
All procedures performed in studies involving human participants were in accordance with the ethical standards of the Scientific Council and the Bioethics Committee of “Attikon” University Hospital (EVD 1172/26-11-15) and with the 1964 Helsinki Declaration and its later amendments or comparable ethical standards.
Footnotes
Capsule Summary: An Artificial Neural Network was constructed and cross-validated to predict live birth for patients undergoing assisted reproduction technologies, with demonstrated increased efficiency and stability.
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Vander Borght M, Wyns C. Fertility and infertility: definition and epidemiology. Clin Biochem. 2018;62:2–10. doi: 10.1016/j.clinbiochem.2018.03.012. [DOI] [PubMed] [Google Scholar]
- 2.Mascarenhas MN, Flaxman SR, Boerma T, Vanderpoel S, Stevens GA. National, regional, and global trends in infertility prevalence since 1990: a systematic analysis of 277 health surveys. PLoS Med. 2012;9:e1001356. doi: 10.1371/journal.pmed.1001356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ombelet W, Cooke I, Dyer S, Serour G, Devroey P. Infertility and the provision of infertility medical services in developing countries. Hum Reprod Update. 2008;14:605–621. doi: 10.1093/humupd/dmn042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Inhorn MC, Patrizio P. Infertility around the globe: new thinking on gender, reproductive technologies and global movements in the 21st century. Hum Reprod Update. 2015;21:411–426. doi: 10.1093/humupd/dmv016. [DOI] [PubMed] [Google Scholar]
- 5.De Geyter C, Calhaz-Jorge C, Kupka MS, Wyns C, Mocanu E, Motrenko T, et al. The European IVF-monitoring Consortium (EIM) for the European Society of Human Reproduction and Embryology (ESHRE); ART in Europe, 2014: results generated from European registries by ESHRE: the European IVF-monitoring Consortium (EIM) for the European Society of Human Reproduction and Embryology (ESHRE) Hum Reprod. 2018;33:1586–1601. doi: 10.1093/humrep/dey242. [DOI] [PubMed] [Google Scholar]
- 6.Baker VL, Luke B, Brown MB, Alvero R, Frattarelli JL, Usadi R, Grainger DA, Armstrong AY. Multivariate analysis of factors affecting probability of pregnancy and live birth with in vitro fertilization: an analysis of the Society for Assisted Reproductive Technology Clinic Outcomes Reporting System. Fertil Steril. 2010;94:1410–1416. doi: 10.1016/j.fertnstert.2009.07.986. [DOI] [PubMed] [Google Scholar]
- 7.Lieberman BA, Falconer D, Brison DR. Presentation of in-vitro fertilisation results. Lancet. 2001;357(357):397. doi: 10.1016/S0140-6736(05)71534-8. [DOI] [PubMed] [Google Scholar]
- 8.De Mouzon J, Goossens V, Bhattacharya S, Castilla JA, Ferraretti AP, Korsak V, et al. Assisted reproductive technology in Europe, 2006: results generated from European registers by ESHRE. Hum Reprod. 2010;25:1851–1862. doi: 10.1093/humrep/deq124. [DOI] [PubMed] [Google Scholar]
- 9.Kovalevsky G, Patrizio P. High rates of embryo wastage with use of assisted reproductive technology: a look at the trends between 1995 and 2001 in the United States. Fertil Steril. 2005;84:325–330. doi: 10.1016/j.fertnstert.2005.04.020. [DOI] [PubMed] [Google Scholar]
- 10.Somashekhar SP, Kumarc R, Rauthan A, Arun KR, Patil P, Ramya YE. Double blinded validation study to assess performance of IBM artificial intelligence platform, Watson for oncology in comparison with Manipal multidisciplinary tumour board – first study of 638 breast cancer cases. Cancer Res. 2017;77:S6–07. [Google Scholar]
- 11.Esteva A, Kuprel B, Novoa RA, Ko J, Swetter SM, Blau HM, Thrun S. Dermatologist-level classification of skin cancer with deep neural networks. Nature. 2017;542:115–118. doi: 10.1038/nature21056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bouton CE, Shaikhouni A, Annetta NV, Bockbrader MA, Friedenberg DA, Nielson DM, Sharma G, Sederberg PB, Glenn BC, Mysiw WJ, Morgan AG, Deogaonkar M, Rezai AR. Restoring cortical control of functional movement in a human with quadriplegia. Nature. 2016;533:247–250. doi: 10.1038/nature17435. [DOI] [PubMed] [Google Scholar]
- 13.Farina D, Vujaklija I, Sartori M, Kapelner T, Negro F, Jiang N, Bergmeister K, Andalib A, Principe J, Aszmann OC. Man/machine interface based on the discharge timings of spinal motor neurons after targeted muscle reinnervation. Nat Biomed Eng. 2017;1:0025. doi: 10.1038/s41551-016-0025. [DOI] [Google Scholar]
- 14.Dilsizian SE, Siegel EL. Artificial intelligence in medicine and cardiac imaging: harnessing big data and advanced computing to provide personalized medical diagnosis and treatment. Curr Cardiol Rep. 2014;16:441. doi: 10.1007/s11886-013-0441-8. [DOI] [PubMed] [Google Scholar]
- 15.Johnson KW, Torres Soto J, Glicksberg BS, Shameer K, Miotto R, Ali M, Ashley E, Dudley JT. Artificial intelligence in cardiology. J Am Coll Cardiol. 2018;71(23):2668–2679. doi: 10.1016/j.jacc.2018.03.521. [DOI] [PubMed] [Google Scholar]
- 16.Jing Y, Bian Y, Hu Z, Wang L, Xie X. Deep learning for drug design: an artificial intelligence paradigm for drug discovery in the big data era. AAPS J. 2018;20:58. doi: 10.1208/s12248-018-0210-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sniecinski I, Seghatchian J. Artificial intelligence: a joint narrative on potential use in pediatric stem and immune cell therapies and regenerative medicine. Transfus Apher Sci. 2018;57:422–424. doi: 10.1016/j.transci.2018.05.004. [DOI] [PubMed] [Google Scholar]
- 18.Simopoulou M, Sfakianoudis K, Maziotis E, Antoniou N, Rapani A, Anifandis G, Bakas P, Bolaris S, Pantou A, Pantos K, Koutsilieris M. Are computational applications the “crystal ball” in the IVF laboratory? The evolution from mathematics to artificial intelligence. J Assist Reprod Genet. 2018;35:1545–1557. doi: 10.1007/s10815-018-1266-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Basheer IA, Hajmeer M. Artificial neural networks: fundamentals, computing, design, and application. J Microbiol Methods. 2000;43:3–31. doi: 10.1016/S0167-7012(00)00201-3. [DOI] [PubMed] [Google Scholar]
- 20.Kaufmann SJ, Eastaugh JL, Snowden S, Smye SW, Sharma V. The application of neural networks in predicting the outcome of in-vitro fertilization. Hum Reprod. 1997;12:1454–1457. doi: 10.1093/humrep/12.7.1454. [DOI] [PubMed] [Google Scholar]
- 21.Wald M, Sparks A, Sandlow J, Van-Voorhis B, Syrop CH, Niederberger CS. Computational models for prediction of IVF/ICSI outcomes with surgically retrieved spermatozoa. Reprod BioMed Online. 2005;11:325–331. doi: 10.1016/S1472-6483(10)60840-1. [DOI] [PubMed] [Google Scholar]
- 22.Uyar A, Bener A, Ciray H, Bahceci M. A frequency based encoding technique for transformation of categorical variables in mixed IVF dataset. Conf Proc IEEE Eng Med Biol Soc. 2009;2009:6214–6217. doi: 10.1109/IEMBS.2009.5334548. [DOI] [PubMed] [Google Scholar]
- 23.Banerjee P, Choi B, Shahine LK, Jun SH, O’Leary K, Lathi RB, et al. Deep phenotyping to predict live birth outcomes in in vitro fertilization. Proc Natl Acad Sci U S A. 2010;107:13570–13575. doi: 10.1073/pnas.1002296107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Uyar A, Bener A, Nadir Ciray H, Bahceci M. Handling the imbalance problem of IVF implantation prediction. IAENG Int J Comput Sci. 2010;37:2. [Google Scholar]
- 25.Ballester M, Oppenheimer A, D’Argent EM, Touboul C, Antoine JM, Coutant C, et al. Nomogram to predict pregnancy rate after ICSI-IVF cycle in patients with endometriosis. Hum Reprod. 2012;27:451–456. doi: 10.1093/humrep/der392. [DOI] [PubMed] [Google Scholar]
- 26.Choi B, Bosch E, Lannon BM, Leveille MC, Wong WH, Leader A, Pellicer A, Penzias AS, Yao MWM. Personalized prediction of first-cycle in vitro fertilization success. Fertil Steril. 2013;99:1905–1911. doi: 10.1016/j.fertnstert.2013.02.016. [DOI] [PubMed] [Google Scholar]
- 27.Durairaj M, Thamilselvan P. Applications of artificial neural network for IVF data analysis and prediction. JEC AS. 2013;2(9):11–5.
- 28.Manna C, Nanni L, Lumini A, Pappalardo S. Artificial intelligence techniques for embryo and oocyte classification. Reprod BioMed Online. 2013;26:42–49. doi: 10.1016/j.rbmo.2012.09.015. [DOI] [PubMed] [Google Scholar]
- 29.Milewski R, Milewska AJ, Więsak T, Morgan A. Comparison of artificial neural networks and logistic regression analysis in pregnancy prediction using the in vitro fertilization treatment. SLGR. 2013;35:39–48. [Google Scholar]
- 30.Durairaj M, Nandhakumar R. An integrated methodology of artificial neural network and rough set theory for analyzing IVF data. 2014 International Conference on Intelligent Computing Applications, Coimbatore, 2014, pp. 126–129
- 31.Uyar A, Bener A, Ciray HN. Predictive modeling of implantation outcome in an in vitro fertilization setting: an application of machine learning methods. Med Decis Mak. 2015;35:714–725. doi: 10.1177/0272989X14535984. [DOI] [PubMed] [Google Scholar]
- 32.Milewski R, Kuczyńska A, Stankiewicz B, Kuczyński W. How much information about embryo implantation potential is included in morphokinetic data? A prediction model based on artificial neural networks and principal component analysis. Adv Med Sci. 2017;62:202–206. doi: 10.1016/j.advms.2017.02.001. [DOI] [PubMed] [Google Scholar]
- 33.Siristatidis C, Pouliakis A, Chrelias C, Kassanos D. Artificial intelligence in IVF: a need. Syst Biol Reprod Med. 2011;57:179–185. doi: 10.3109/19396368.2011.558607. [DOI] [PubMed] [Google Scholar]
- 34.Siristatidis C, Vogiatzi P, Pouliakis A, Trivella M, Papantoniou N, Bettocchi S. Predicting IVF outcome: a proposed web-based system using artificial intelligence. In Vivo. 2016;30:507–512. doi: 10.21873/invivo.11018. [DOI] [PubMed] [Google Scholar]
- 35.Ferraretti AP, La Marca A, Fauser BC, Tarlatzis B, Nargund G, Gianaroli L, ESHRE working group on Poor Ovarian Response Definition ESHRE consensus on the definition of ‘poor response’ to ovarian stimulation for in vitro fertilization: the Bologna criteria. Hum Reprod. 2011;26:1616–1624. doi: 10.1093/humrep/der092. [DOI] [PubMed] [Google Scholar]
- 36.Mascarenhas M, Balen AH. The high responder: a review of pathophysiology and outcomes during IVF treatment. Hum Fertil (Camb) 2017;20:155–167. doi: 10.1080/14647273.2017.1293851. [DOI] [PubMed] [Google Scholar]
- 37.Zegers-Hochschild F, Adamson GD, Dyer S, Racowsky C, de Mouzon J, Sokol R, Rienzi L, Sunde A, Schmidt L, Cooke ID, Simpson JL, van der Poel S. The international glossary on infertility and fertility care, 2017. Fertil Steril. 2017;108:393–406. doi: 10.1016/j.fertnstert.2017.06.005. [DOI] [PubMed] [Google Scholar]
- 38.DiMaggio C. SAS for epidemiologists: applications and methods. New York: Springer; 2013. [Google Scholar]
- 39.Haykin SS. Neural networks: a comprehensive foundation. 2. New York: Prentice Hall; 1998. [Google Scholar]
- 40.Swingler K. Applying neural networks: a practical guide, vol. 109. 3rd ed: Academic Press; 2001. p. 2001.
- 41.Holden EC, Dodge LE, Sneeringer R, Moragianni VA, Penzias AS, Hacker MR. Thicker endometrial linings are associated with better IVF outcomes: a cohort of 6331 women. Hum Fertil (Camb) 2017;18:1–6. doi: 10.1080/14647273.2017.1334130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zhao J, Zhang Q, Wang Y, Li Y. Endometrial pattern, thickness and growth in predicting pregnancy outcome following 3319 IVF cycle. Reprod BioMed Online. 2014;29:291–298. doi: 10.1016/j.rbmo.2014.05.011. [DOI] [PubMed] [Google Scholar]
- 43.Harb HM, Gallos ID, Chu J, Harb M, Coomarasamy A. The effect of endometriosis on in vitro fertilisation outcome: a systematic review and meta-analysis. BJOG. 2013;120:1308–1320. doi: 10.1111/1471-0528.12366. [DOI] [PubMed] [Google Scholar]
- 44.Cimadomo D, Fabozzi G, Vaiarelli A, Ubaldi N, Ubaldi FM, Rienzi L. Impact of maternal age on oocyte and embryo competence. Front Endocrinol (Lausanne) 2018;9:327. doi: 10.3389/fendo.2018.00327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Yan J, Wu K, Tang R, Ding L, Chen ZJ. Effect of maternal age on the outcomes of in vitro fertilization and embryo transfer (IVF-ET) Sci China Life Sci. 2012;55:694–698. doi: 10.1007/s11427-012-4357-0. [DOI] [PubMed] [Google Scholar]
- 46.Alper MM, Fauser BC. Ovarian stimulation protocols for IVF: is more better than less? Reprod BioMed Online. 2017;34:345–343. doi: 10.1016/j.rbmo.2017.01.010. [DOI] [PubMed] [Google Scholar]
- 47.Yilmaz N, Yilmaz S, Inal H, Gorkem U, Seckin B, Turkkani A, Gulerman C. Is there a detrimental effect of higher gonadotrophin dose on clinical pregnancy rate in normo-responders undergoing ART with long protocol? Arch Gynecol Obstet. 2013;287:1039–1044. doi: 10.1007/s00404-012-2673-z. [DOI] [PubMed] [Google Scholar]
- 48.Berger DS, Zapantis A, Merhi Z, Younger J, Polotsky AJ, Jindal SK. Embryo quality but not pronuclear score is associated with clinical pregnancy following IVF. J Assist Reprod Genet. 2014;31:279–283. doi: 10.1007/s10815-013-0162-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Bortoletto P, Bakkensen J, Anchan RM. Embryo transfer: timing and techniques. Minerva Endocrinol. 2018;43(1):57–68. doi: 10.23736/S0391-1977.17.02649-9. [DOI] [PubMed] [Google Scholar]
- 50.Wong KM, van Wely M, Mol F, Repping S, Mastenbroek S. Fresh versus frozen embryo transfers in assisted reproduction. Cochrane Database Syst Rev. 2017;3:CD011184. doi: 10.1002/14651858.CD011184.pub2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Vogiatzi P, Pouliakis A, Bettocchi S, Daskalakis G, Vrantza T, Siristatidis C. Age at menarche and clinical outcomes following medically assisted reproduction (MAR): a cohort study. Gynecol Endocrinol. 2019;18:1–5. doi: 10.1080/09513590.2018.1538344. [DOI] [PubMed] [Google Scholar]
- 52.Perroti R, Pouliakis A, Margari N, Panopoulou E, Karakitsou E, Iliopoulou D, et al. CytoNet, a versatile web-based system for accessing advisory cytology services: application of artificial intelligence. IJRQEH. 2018;7:37–56. [Google Scholar]