Abstract
Background
Peripheral neuropathies are often caused by disruption of genes responsible for myelination or axonal transport. In particular, impairment in mitochondrial fission and fusion are known causes of peripheral neuropathies. However, the causal mechanisms for peripheral neuropathy gene mutations are not always known. While loss of function mutations in MYH14 typically cause non-syndromic hearing loss, the recently described R941L mutation in MYH14, encoding the non-muscle myosin protein isoform NMIIC, leads to a complex clinical presentation with an unexplained peripheral neuropathy phenotype.
Methods
Confocal microscopy was used to examine mitochondrial dynamics in MYH14 patient fibroblast cells, as well as U2OS and M17 cells overexpressing NMIIC. The consequence of the R941L mutation on myosin activity was modeled in C. elegans.
Findings
We describe the third family carrying the R941L mutation in MYH14, and demonstrate that the R941L mutation impairs non-muscle myosin protein function. To better understand the molecular basis of the peripheral neuropathy phenotype associated with the R941L mutation, which has been hindered by the fact that NMIIC is largely uncharacterized, we have established a previously unrecognized biological role for NMIIC in mediating mitochondrial fission in human cells. Notably, the R941L mutation acts in a dominant-negative fashion to inhibit mitochondrial fission, especially in the cell periphery. In addition, we observed alterations to the organization of the mitochondrial genome.
Interpretation
As impairments in mitochondrial fission cause peripheral neuropathy, this insight into the function of NMIIC likely explains the peripheral neuropathy phenotype associated with the R941L mutation.
Fund
This study was supported by the Alberta Children's Hospital Research Institute, the Canadian Institutes of Health Research and the Care4Rare Canada Consortium.
Keywords: Mitochondria, Mitochondrial fission, Peripheral neuropathy, Non-muscle myosin, mtDNA, Caenorhabditis elegans
Research in context.
Evidence before this study
While most pathogenic variants in MYH14, encoding non-muscle myosin IIC (NMIIC), are associated with non-syndromic hearing loss, the R941L pathogenic variant was reported to segregate with a peripheral neuropathy phenotype in two separate families. Although many types of cellular dysfunction can cause peripheral neuropathies, the molecular mechanism underlying this phenotype in patients with the R941L variant in MYH14 was completely unknown. While, other non-muscle myosin proteins related to MYH14 have been implicated in mediating fission, a process that when impaired can cause peripheral neuropathy, a role for NMIIC in fission had not been established previously, nor had pathogenic variants in non-muscle-myosin proteins been associated with a peripheral neuropathy phenotype.
Added value of this study
The current study characterized a third family where the R941L mutation segregates with a peripheral neuropathy phenotype. A completely novel role in mitochondrial fission is established for the NMIIC protein encoded by MYH14, with the R941L mutation having a dominant negative effect that inhibits mitochondrial fission, especially at the cell periphery. Notably, the impaired fission in MYH14 patient fibroblasts also affects distribution of the mitochondrial genome within the mitochondrial network.
Implications of all the available evidence
With three unrelated families showing the R941L mutation segregates with peripheral neuropathy, there is increased confidence that this variant causes the phenotype. The novel role of MYH14 in mitochondrial fission, which is impaired by the R941L variant, provides insight into the underlying biological mechanism likely causing peripheral neuropathy in these patients.
Alt-text: Unlabelled Box
1. Introduction
Peripheral neuropathies are generally caused by defects affecting myelination and/or axonal transport. Mutations in genes regulating mitochondrial fission or fusion are a known cause of Charcot-Marie-Tooth (CMT) disease, a heterogeneous group of inherited peripheral neuropathies [1]. It is hypothesized that peripheral neuropathy is a consequence of mitochondria that are too fragmented or too fused, such that mitochondria are not efficiently transported down the long axons of peripheral neurons [2]. A key regulator of mitochondrial fission is the dynamin-related protein DRP1 (encoded by DNM1L), which is recruited from the cytosol to mitochondrial tubules, where it forms a ring to constrict mitochondria. While Drp1 is generally regarded as the major protein mediating mitochondrial fission, recent studies have shown that additional factors, such as the endoplasmic reticulum (ER) and Dynamin2 (DNM2), also regulate key steps in fission [[3], [4], [5]].
We now appreciate that a pre-constriction step, mediated by the ER, is required to reduce the mitochondrial circumference such that DRP1 rings can encircle mitochondria [3]. Non-muscle myosin family II proteins A and B (NMIIA/B) are ATP-dependent molecular motors that interact with actin and provide the necessary mechanical force for this pre-constriction, and promote DRP1 recruitment to mitochondria [[6], [7], [8]]. In contrast to muscle myosins, which form the contractile filaments in smooth, skeletal, and cardiac muscle, NMII proteins are present in all cells, where they regulate intracellular processes. The human genome contains three NMII isoforms: NMIIA, NMIIB and NMIIC, encoded by the genes MYH9, MYH10, and MYH14, respectively. These NMII isoforms are thought to be partially redundant. However, while NMIIA/B are known to regulate cellular processes such as cytokinesis, cell motility, cell polarity, mtDNA regulation, and mitochondrial fission, the cellular functions of NMIIC have not been thoroughly investigated [[9], [10], [11]]. Additionally, the interplay and redundancy of NMII proteins regarding their role in mitochondrial fission is unknown.
Recently, the R941L mutation in NMIIC has been linked to a complex phenotype including peripheral neuropathy and hearing loss, and has been reported in two distinct families from different geographic regions, and in a single individual from a larger study [[12], [13], [14]]. The role of NMIIA/B in mediating mitochondrial fission [10] led us to hypothesize that NMIIC is also involved in mitochondrial fission, and that the pathogenic R941L mutation would impair fission. While several other pathogenic variants in MYH14 are known to cause autosomal dominant hearing loss via haploinsufficiency [[15], [16], [17], [18], [19], [20]], the peripheral neuropathy phenotype is unique to the R941L mutation. Moreover, MYH14 null mice are reported to have increased susceptibility to hearing loss [21], but no peripheral neuropathy phenotype has been reported.
Prior to this study, the cellular functions of NMIIC, and the molecular mechanism by which the R941L mutation might cause peripheral neuropathy were unknown. Here, in addition to describing a novel family pedigree harbouring the R941L mutation, we address two unanswered questions, namely how the R941L mutation causes peripheral neuropathy, and how this mutation has appeared independently in multiple families. Mechanistically, we describe a previously uncharacaterized role for the NMIIC as a mediator of mitochondrial fission, which is impaired by the pathogenic R941L mutation.
2. Materials and methods
2.1. Ethics statement
Informed consent for enrollment in the Care4Rare project and for publication was obtained by the examined members of this 3 generation pedigree. This study was approved by the conjoint ethics board of the University of Calgary.
2.2. Genetic analysis
Whole exome sequencing was performed at the McGill Genome Centre as part of the Care4Rare Canada initiative. Details of this project including case selection and analytic pipeline are described elsewhere [22,23]. Whole Exome Sequencing (WES) using a SureSelect (Agilent, Santa Clara, California, USA) exome capture kit was performed on the Illumina sequencing platform.
2.3. CpG methylation detection
CpG methylation was assayed using the FauI enzyme (New England Biosystems), which does not cut methylated DNA. Briefly, 50 ng of genomic DNA was digested at 55 °C for 60 min, followed by heat inactivation at 80 °C for 20 min. Next, DNA was amplified using primers specific to MYH14 (MYH14_CpG_For GCGACGCGGGGGAGGC, and MYH14_CpG_Rev CTGTCCAGGGTGGCCTGG), and 10 μL of the PCR product was visualized via agarose gel electrophoresis.
2.4. In silico protein modeling
The amino acid sequence of human MYH14 isoform 1 (NP_001070654·1) was submitted to the Swiss Model in silico protein modeling tool [24]. Though a crystal structure for the head domain of the human NMIIC protein has been solved (RCSB:2YCU), no structure has been published for the tail domain containing the R941L mutation. Thus, a predicted protein structure was created based on the structure of Gallus gallus meromyosin (RCSB:1I84) [25], as this homologous structure contains the protein tail domain, which is not present in the crystal structure of human NMIIC protein. The resulting structure prediction and mutations were visualized using UCSF Chimera [26].
2.5. Genetic modeling in C. elegans
C. elegans were cultured under standard conditions [27] at the indicated temperatures. Complete broods of at least four hermaphrodites were determined, with a total of 450–1300 progeny scored. The following mutations were used: the temperature-sensitive allele mel-11(it26), the null allele nmy-1(sb113) [28] and the R915L CRISPR allele nmy-1(sb139) described here. To introduce the R915L mutation into nmy-1, eft-3::Cas9, pJA58[dpy-10(cn64) gRNA] and a dpy-10(cn64) repair single-stranded oligonucleotide (University of Calgary Core DNA Services) were used as described for “Co-CRISPR” [29]. CRISPR targeting sequence and primers used in the generation of the mutant nmy-1(sb113) allele are provided in the additional methods (Supplemental Table 1). Two nmy-1 gRNA PCR products were created using PCR “stitching”. PCR's on pJW1285 (pha-1 gRNA(F + E), a gift from Jordan Ward, Addgene) using oJW1787 + nmy1sgRNA1rev and oJW1790 + nmy1sgRNA1fwd were stitched together, substituting nmy-1 sequences for those of pha-1 and gel-purified to make nmy-1 gRNA-1 (oligonucleotides and nmy-1 gRNA target sequence are listed below). Similarly, PCR's using oJW1787 + nmy1sgRNA2rev and oJW1790 + nmy1sgRNA2fwd were stitched together and gel purified to make nmy-1 gRNA-2. The single-stranded repair oligonucleotide nmy-1-R915L-rescueoligo (IDT), was designed to have 60 base pair homology arms, a (GC–TT) mutation resulting in R915L and two silent mutations in R911 (G–A) and N916 (C–T) to disrupt nmy-1 gRNA targeting of the new allele and to introduce SalI/MseI restriction enzyme sites respectively. Gravid wild-type hermaphrodites were injected with 50 ng/μL eft-3::Cas9, 25 ng/μL pJA58(dpy-10(cn64) gRNA), 500 nM dpy-10(cn64) oligonucleotide (Arribere et al. 2014), 25 ng/μL nmy-1 gRNA-1, 25 ng/μL nmy-1 gRNA-2, and 500 nM nmy-1-R915L-rescueoligo. Dpy and Rol F1 progeny were screened by PCR and restriction enzyme digestion for insertions. One allele was identified, nmy-1(sb139) and confirmed by sequencing.
2.6. Cloning and plasmids
The wild-type NMIIC-EGFP plasmid was a gift from Dr. John Hammer at the National Institutes of Health, which was originally created in the lab of Dr. Adelstein of the National Institutes of Health. The NMIIC R941L mutant was generated by overlapping PCR mutagenesis and verified by Sanger sequencing. The wild-type mCh-Drp-1 plasmid was purchased from Addgene (plasmid# 49152), which was originally created by Dr. Gia Voeltz.
2.7. Cell culture conditions
Fibroblast cells were generated from skin biopsies and cultured in DMEM containing 10% fetal bovine serum (FBS), supplemented with Penicillin/Streptomycin (100 IU/mL/100 μL/mL). U2OS human osteosarcoma cells were grown in McCoy's 5A (modified) media containing 10% FBS and supplemented with Penicillin/Streptomycin. For transfection, 8 × 104 U2OS cells were initially seeded on glass bottom dishes (Mattek, P35G-1.5-14-C). The following day, cells were transfected with 0·5 μg of plasmid DNA using Lipofectamine 3000 according to the manufacturer's recommended protocol. Finally, U2OS cells were imaged 24 h following transfection. BE(2)-M17 neuroblastoma cells (hereafter M17) were cultured in a 1:1 mixture of Eagle's Minimum Essential Medium and F12 medium with 10% FBS, and supplemented with non-essential amino acids and Penicillin/Streptomycin. For M17 transfection, cells were first differentiated as previously described [30]. Briefly, 1 × 105 M17 cells were seeded on glass bottom dishes pre-coated with poly-D- Lysine for 24 h. Retinoic acid at concentration of 15 μM was added and the cells incubated for 72 h. Following 72 h, M17 cells were transfected with 2 μg of plasmid DNA (wild-type or R941L mutant NMIIC- EGFP, or empty pcDNA vector as a control) using Lipofectamine 3000. Twelve hours after transfection, the media was changed and M17 cells were imaged the following day.
2.8. Microscopy
Cells were imaged as indicated with either of the following two microscope setups. A Zeiss LSM 710 confocal microscope (plan-Apochromat 63×/1·4 oil objective) with image capture and processing performed with Zen Blue and Black software (Carl Zeiss, Jena, Germany). For live cell imaging with the Zeiss microscope, cells were supplemented with 50 mM HEPES and incubated at 37 °C. Alternatively an Olympus spinning disc confocal system (Olympus SD-OSR) (UAPON 100XOTIRF/1·49 oil objective) operated by Metamorph software was utilized. The SD-OSR was equipped with a cellVivo incubation module to maintain cells at 37 °C and 5% CO2 during live cell imaging.
2.9. Live cell imaging
Approximately 24 h prior to imaging, cells were seeded at 8 × 104 cells/mL onto glass bottom dishes. Immediately prior to imaging, cells were treated with the indicated dyes as follows. To label mitochondria, cells were stained for 30 min with 50 nM MitoTracker Red (Thermo Fisher Scientific, M7512), or MitoTracker Deep Red (Thermo Fisher Scientific, M22426), as indicated. To label mDNA nucleoids, cells were co-stained with the DNA dye PicoGreen (Thermo Fisher Scientific, P11495) (3 μL/mL), as previously described [31], in addition to MitoTacker Red. After staining, cells were washed 3 times with pre-warmed 1× phosphate buffered saline (PBS), and normal growth media was added prior to imaging. For the phototoxic-dependent mitochondrial fission assay using the Olympus SD-OSR microscope, MitoTrackerRed-labelled fibroblasts were repeatedly exposed (100 ms at 1 frame per second intervals) to higher levels of the 561 nm excitation laser (100 mW at 4% power), for a total of 5 min.
2.10. Immunofluorescence
Cells were grown on glass coverslips (no. 1·5), then fixed in prewarmed 4% formaldehyde for 10 min at 37 °C, permeabilized in 0·25% Triton X-100, and quenched with 50 mM ammonium chloride. After blocking with 5% FBS in PBS, cells were incubated for 1 h at 37οC with primary antibodies to TOMM20 (rabbit anti-TOMM20, Santa Cruz Biotechnology, FL-145) diluted 1:1000 in 5% FBS in PBS, and subsequently incubated for 1 h at room temperature with appropriate Alexafluor secondary antibodies (Themo Fisher Scientific) diluted 1:1000 in 5% FBS in PBS. Coverslips were then mounted on glass slides with Dako mounting media (Agilent, S3023) and imaged.
2.11. Image analysis
Mitochondrial length was quantified from 30 cells from each patient as described previously [32], using the Tubeness plugin from FIJI for mitochondrial segmentation. For nucleoid analysis, images were cropped, globally adjusted for contrast and brightness, and median filtered using ImageJ (https://imagej.nih.gov/ij/index.html). Mitochondrial DNA nucleoid size and number were analyzed in 10 fibroblast cells from each group using the particle analysis tool on ImageJ FIJI [33]. Briefly, images were scaled and binarized. A region of interest engulfing the entire cell was selected and the particle analysis tool was used to generate surface area and total nucleoid count measurements. Nuclear and nonspecific signal outside the mitochondrial network were manually excluded from the analysis.
2.12. Mitochondrial membrane potential and mitochondrial mass analyses
Mitochondrial membrane potential and mitochondrial mass were assessed in control and patient fibroblasts by flow cytometry, as described previously [34]. Briefly, fibroblasts were seeded at 1 × 105 cells in 6 well plates and allowed to grow for 2 days. Prior to analysis, cells were stained with TMRE (Tetramethylrhodamine, ethyl ester) (50 nM, 20 min) (T669; Life Technologies) or MitoTracker Green (50 nM, 20 min) (M7514; Life Technologies). Cells were subsequently washed with 1× PBS, trypsinized and harvested. Cell pellets were resuspended in media and analyzed using the BD LSR II flow cytometer (BD Bioscience) and the BD FACSDiva software. Signal intensity was recorded from at least 20,000 events and data is presented as percent control. Analyses were performed on triplicates for each fibroblast line.
2.13. mtDNA copy number analysis
Genomic DNA (gDNA) from control and patient fibroblasts was extracted using PureLink Genomic DNA Mini Kit (Thermo Fisher Scientific, K182001) according to manufacturer's instructions. Relative mtDNA copy number was assessed by real-time quantitative PCR (qPCR) using the QuantStudio 6 Flex Real-Time PCR system (Thermo Fisher Scientific). Primer sequences to amplify mtDNA, the nuclear-encoded housekeeping gene 18S, and thermocycling conditions were exactly as described previously [35]. The 20 μL reactions contained 10 μL PowerUp SYBR Green Master Mix (Thermo Fisher Scientific, A25742), 100 ng gDNA and a final concentration of 500 nM forward and 500 nM reverse primers.
2.14. Long-range PCR
To examine mtDNA deletions, the following primers were used to amplify nearly full length mtDNA (16.3 kb), (1482–1516 F: ACCGCCCGTCACCCTCCTCAAGTATACTTCAAAGG; 1180–1146 R: ACCGCCAGGTCCTTTGAGTTTTAAGCTGTGGCTCG) as reported previously [36]. Long range PCR reactions were performed using the Takara LA Taq polymerase (Takara Bio, RR002M), with 250 ng genomic DNA, 200 nM forward and reverse primers. The PCR cycling conditions were as follows: 94 °C for 1 min; 98 °C for 10 s and 68 °C for 11 min (30 cycles); and a final extension cycle at 72 °C for 10 min. PCR products were visualized by electrophoresis on a 0·6% agarose gel, run for approximately 12 h at 20 V.
2.15. Statistical analysis
Mitochondrial morphology was assessed in a blinded fashion by qualitatively assigning morphology into three or four categories. Quantification was performed in at least 50 cells from multiple technical and experimental replicates, as indicated. In order to quantify the peripheral hyper-connectivity in fibroblasts cells, the presence/absence of hyperconnected mitochondrial networks at the cell periphery were scored from three independent experiments. At least 70 cells were assessed per replica. MtDNA nucleoid sizes are presented as the average size of all nucleoids per cell ± SD. Nucleoid counts are presented as mean ± SD. MtDNA copy number relative to 18S was analyzed using the ΔΔCt method and expressed as percent control [37] from at least three independent biological replicates. All results are presented as mean ± SD. p-values were based on unpaired, 2-tailed Student's t-tests comparing data from each patient or experiment to the control, in order to assess statistical significance.
3. Results
3.1. Clinical evaluation and modeling of the R941L pathogenicity
3.1.1. A novel family carrying the R941L mutation
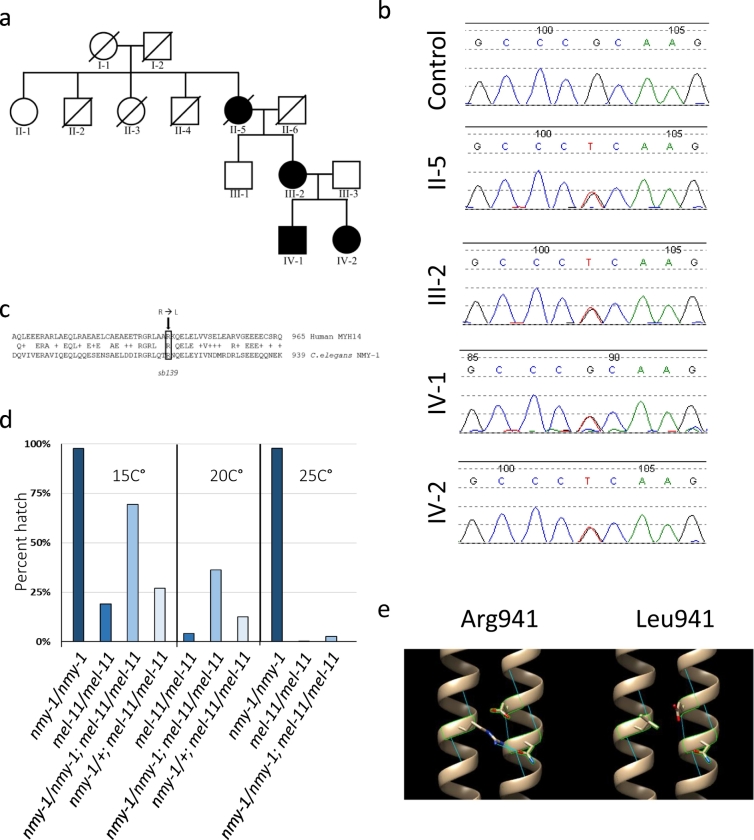
We report a novel family carrying the R941L mutation in MYH14, which has been followed in the Neurogenetics clinic at the Alberta Children's Hospital in Calgary for many years. Across three generations, there are four affected individuals showing a pattern of autosomal dominant inheritance (Fig. 1a). Details regarding available clinical features are summarized in the table (Supplemental Table 2). Individual II-5 was not directly examined in this study, but was diagnosed ‘late-in-life’ with CMT, with a long standing history of high arches, hearing loss, and mild progression of her disease. All individuals directly examined in this study (III-2, IV-1, IV-2) have symptoms and clinical signs consistent with peripheral neuropathy manifesting in 2nd and 3rd decades of life, and sensorineural hearing loss manifesting in early childhood (this was diagnosed in infancy in subject IV-1 and resulted in language delay). Nerve conduction studies in patient III-2 revealed a borderline reduction in conduction velocity and amplitude in the tibial nerve, which was considered to be consistent with axonal polyneuropathy rather than demyelinating polyneuropathy. Limited electromyography study in tibialis anterior identified polyphasic motor units, but was unremarkable in vastus lateralis. Neurophysiology was not obtained in patients IV-1 and IV-2, but the motor neuropathy diagnosis was based upon the length dependent weakness/atrophy pattern, pes cavus, and family history in the mother who had confirmed neuropathy on nerve conduction studies. Echocardiogram and muscle MRI in the upper and lower legs did not show evidence for cardiac involvement or a coexistent myopathy.
Fig. 1.
Genetic analysis of patients and modeling of the R941L mutation. (a) Pedigree of a three generation family with peripheral neuropathy and hearing loss. Shaded sections indicate the presence of both CMT and hearing loss, which were always seen together in the pedigree. Exome sequencing was performed on individuals II-5, III-2 and IV-2. The c.2822G>T p.Arg941Leu mutation was identified in individuals II-5, III-2, IV-1 and IV-2. Note that for subsequent fibroblast studies individuals III-2, IV-1, and IV-2 are patients 1–3, respectively. (b) Sanger sequencing confirms the presence of the c.2822G>T p.Arg941Leu mutation in individuals II-5, III-2, IV-1 and IV-2. (c) Alignment of the human and worm genes showing the conservation in the region surrounding MYH14 R941, which corresponds to R915 of the worm gene. (d) The R915L mutation in C. elegans NMY-1 decreases nonmuscle myosin function. nmy-1(sb139) acts semidominantly at all three temperatures to increase the hatching rate of mel-11 mutations, indicating that sb139/R195L compromises NMY-1 function (the 2.6% hatching at 25° was significant as it represented 25/936 vs. 2/938 for the control). The 27% viability of nmy-1(sb139)/+; mel-11/mel-11 embryos at 15 °C was inferred from the hatching rates of the progeny of mothers of that genotype. These animals segregate a mixture of nmy-1(sb139) homozygotes and heterozygotes, as well as nmy-1(+) homozygotes in a Mendelian 1:2:1 ratio. The hatching rates of homozygous nmy-1(sb139) and nmy-1(+) embryos in the mel-11/mel-11 background are 69% and 19%, respectively. We observed 36% hatching from nmy-1(sb139)/+; mel-11/mel-11 mothers and this corresponds to 27% hatching of their heterozygous offspring, an increase above the 19% hatching nmy-1(+)/nmy-1(+); mel-11/mel-11. Similar calculations demonstrated the semidominance at 20 °C. (e) In silico protein modeling suggests that the wild-type arginine residue at position 941 (left) has hydrophilic interactions with residues on an adjacent alpha-helix during dimerization. The mutant leucine residue (right) is unable to form the same hydrophilic interactions with the opposite myosin chain.
After filtering through established peripheral neuropathy genes, exome sequencing in three individuals (II-5, III-2, and IV-2) identified the c.2822G>T (p.Arg941Leu) (NM_001077186.1) mutation (referred to hereafter as R941L). Sanger sequencing confirmed heterozygosity for the pathogenic variant in all four affected individuals: II-5, III-2, IV-1, IV-2 (Fig. 1b).
3.1.2. Recurrence of the R941L mutation
As the R941L variant has been reported in several geographically discrete regions, we hypothesized that it could be the result of a recurrent mutation rather than identity by descent. Genome-wide methylation studies have identified the recurrently mutated guanine nucleotide (c.2822G) as part a CpG methylation site [38,39]. Notably, we were able to confirm that this site is indeed methylated (Supplemental Fig. 1).
3.1.3. Genetic modeling of the R941L mutation
As there are no established assays to study the function of NMIIC [16], we modeled the R941L mutation in C. elegans, where there is a sensitive assay for non-muscle myosin function [28]. The worm homolog of MYH14, NMY-1, is 48% identical to MYH14, and the R941 residue is conserved (R915 in C. elegans, Fig. 1c). We used CRISPR technology to create an R915L allele, designated sb139. To measure NMY-1 function, we examined the effects of nmy-1(sb139) on C. elegans embryonic morphogenesis. NMY-1 participates in the contraction of the C. elegans epidermal cells, which squeezes the embryo from an ovoid into a long, thin larva [28]. C. elegans MEL-11 represents the myosin phosphatase targeting subunit/MYPT that moderates this contraction. Loss of MEL-11 thus results in death due to hypercontraction, and embryos fail to hatch. Mutations in genes such as nmy-1, which lessen the contractile force, rescue mel-11 lethality. Thus, we can use this rescue as a sensitive readout of NMII protein function. At 15 °C, 19% of the homozygous embryos of the temperature-sensitive allele mel-11(it26) hatched (Fig. 1d). This value increased to 69% with the addition of nmy-1(sb139). Rescue of mel-11 lethality was also seen at 20 °C and 25 °C where mel-11(it26) function is even more compromised (Fig. 1d). This rescue of mel-11 lethality shows that R915L results in decreased nonmuscle myosin function and nmy-1(sb139) appears to act semi-dominantly (Fig. 1d). We previously reported that an nmy-1 null allele [28] also shows semi-dominant rescue of mel-11. However, while null alleles of NMY-1 have a characteristic lumpy body shape [28], sb139 animals are wild-type, indicating retention of at least some myosin function.
3.1.4. Structural modeling of the R941L mutation
In order to begin to understand the consequences of R941L mutation on the tertiary or quaternary structure of the NMIC protein, we employed in silico protein modeling. In general, myosin proteins are comprised of a catalytic head domain, and a coiled-coil tail domain that is required for multimerization. The wild-type arginine residue is predicted to engage in hydrophilic interactions that could potentially influence myosin dimerization (Fig. 1e). Our in silico modeling suggests that the introduction of a leucine residue at position 941 eliminates a predicted hydrophilic interaction with residues on an opposite alpha helix. We thus speculate that this substitution could alter the formation of a functional dimer, which would be consistent with a possible dominant-negative effect, where a defective subunit poisons the complex.
3.2. A role for NMIIC in mitochondrial fission
3.2.1. Characterization of R941L patient fibroblasts
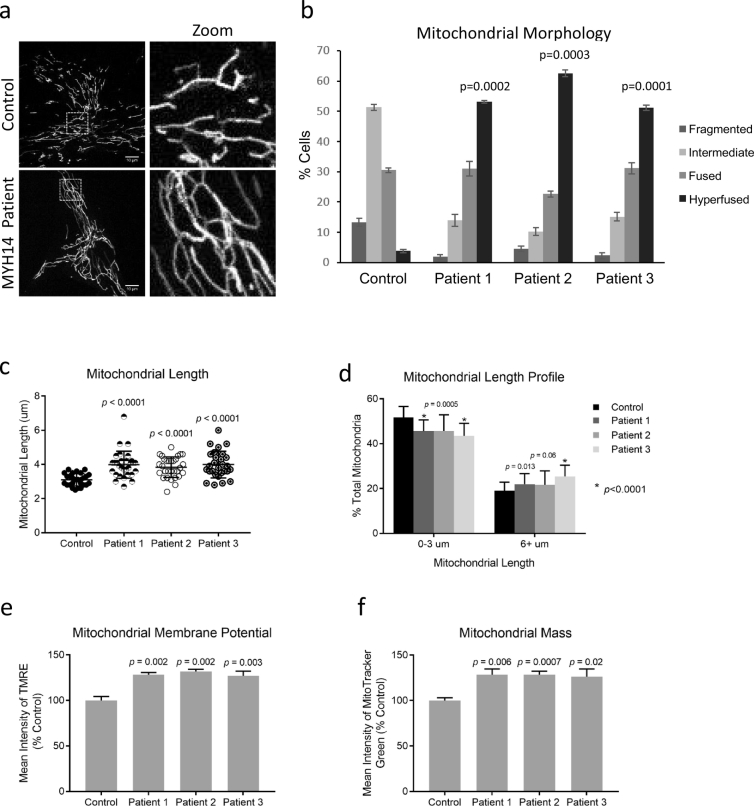
Given the established link between NMIIA/B isoforms and mitochondrial fission, and the fact that impaired mitochondrial fission can cause peripheral neuropathies, we hypothesized that the R941L mutation might impair mitochondrial fission. Thus, we examined mitochondrial morphology in fibroblast cells obtained from our R941L patients. We observed a significant shift towards a more fused mitochondrial network in all three patient fibroblast lines, consistent with impaired mitochondrial fission (Fig. 2a, b). Notably, the mitochondrial morphology in the control fibroblast line we used is similar to other control fibroblast lines (Supplemental Fig. 2). We confirmed these changes in mitochondrial morphology by quantifying the length of individual mitochondria, which were also longer in patient fibroblasts (Fig. 2c, d).
Fig. 2.
R941L Patient Fibroblasts have altered mitochondrial morphology. (a) Representative confocal images of mitochondrial networks taken with an Olympus SD-OSR microscope. Mitochondria in fixed control and patient cells were stained via immunofluorescence using a TOMM20 antibody. Scale bars indicate 10 μm. (b) Quantification of mitochondrial morphology in control fibroblast and R941L patient fibroblast cells. One hundred cells were quantified in three technical replicates for two independent experimental replicates. Error bars indicate standard deviations, while p-values (Student's t-test) were determined by comparison to the number of cells with hyperfused mitochondria in control. (c) Quantification of average mitochondrial length. Data represents automated analyses from at least 30 cells per fibroblast line. (d) Mitochondrial length profile from cells in (c), binned into 0–3 and 6+ μm bins and presented as % of total mitochondria. (e) Mitochondrial membrane potential in control and patient fibroblasts measured with flow cytometry analysis of TMRE-stained cells. (f) Mitochondrial mass measured in MitoTracker Green stained samples by flow cytometry. Data for (e) and (f) are presented as % control. Error bars indicate standard deviations, and p-values (Student's t-test) determined by comparison to the control. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
As changes in mitochondrial morphology are sometimes linked to alterations in mitochondrial function (e.g. more fragmented mitochondrial tend to be less active), we looked at whether there were any functional consequences of reduced fission in MYH14 patient fibroblasts. To this end, we measured mitochondrial membrane potential with the potentiometric dye TMRE, and mitochondrial mass with the Mitotracker Green dye. Although we observed a slight ~20% increase in TMRE signal in MYH14 fibroblasts (Fig. 2e), we also observed a corresponding ~20% increase in Mitotracker Green signal (Fig. 2f), suggesting that the increased TMRE signal is due to increased mitochondrial mass rather than membrane potential.
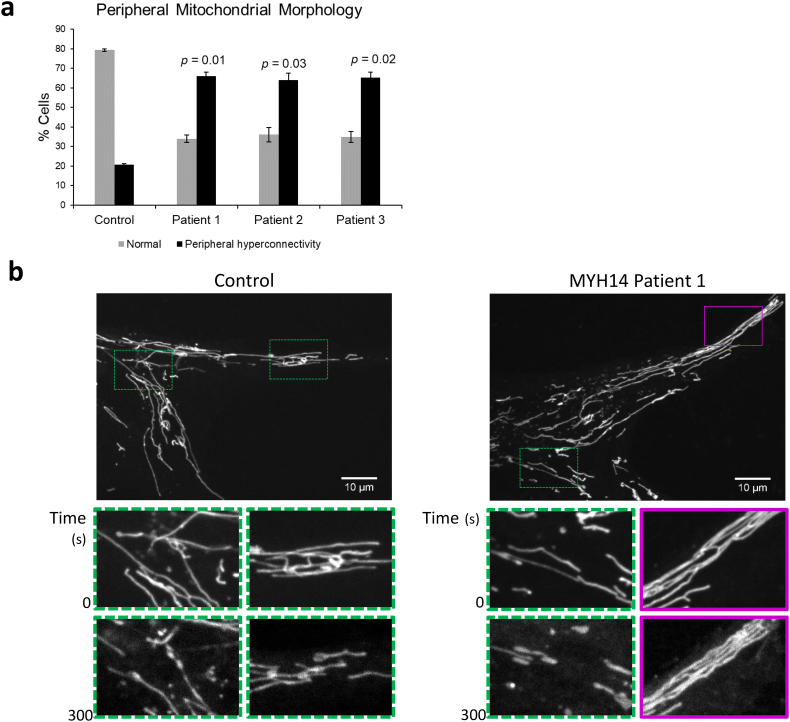
Given that there were no major global changes in mitochondrial function, combined with the notion that peripheral neuropathy linked to impaired mitochondrial fission and fusion are thought to be due to impaired mitochondrial transport, rather than decreased function per se [2], we decided to look at the distribution of mitochondria in MYH14 patient fibroblasts. Surprisingly, a close examination of mitochondrial morphology in patient fibroblasts also revealed an enrichment of hyper-connected mitochondrial networks in the periphery of greater than 60% of cells, compared approximately 20% of control fibroblasts (Fig. 3a). This observation led us to hypothesize that fission may be more impaired at the cell periphery.
Fig. 3.
Hyperconnected mitochondrial networks at the cell periphery in R941L patient fibroblasts are resistant to phototoxicity-induced fission. (a) Quantification of control and patient cells containing hyperconnected mitochondrial networks at the cell periphery. At least 70 cells were quantified from two independent replicates. Error bars indicate standard deviations, and p-values (Student's t-test) were determined by comparison to the number of control cells with hyperfused mitochondria. (b) Representative confocal images of mitochondrial networks taken with an Olympus SD-OSR microscope. Control and patient cells stained with MitoTracker Red were imaged continuously over 5 min with high laser power to induce fission. Inset zoomed boxes represent regions with fragmented mitochondria (green hashed boxes) or resistant to fission (magenta hashed boxes) when imaging commenced, and at the end of 5 min (full video available as Supplemental Video 1). Signal intensity was enhanced for later frames to adjust for photobleaching. Scale bars indicate 10 μm. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
Hyperconnected mitochondrial networks at the cell periphery in R941L patient fibroblasts are resistant to phototoxicity-induced fission. (a) Quantification of control and patient cells containing hyperconnected mitochondrial networks at the cell periphery. At least 70 cells were quantified from two independent replicates. Error bars indicate standard deviations, and p-values (Student's t-test) were determined by comparison to the number of control cells with hyperfused mitochondria. (b) Representative confocal images of mitochondrial networks taken with an Olympus SD-OSR microscope. Control and patient cells stained with MitoTracker Red were imaged continuously over 5 min with high laser power to induce fission. Inset zoomed boxes represent regions with fragmented mitochondria (green hashed boxes) or resistant to fission (magenta hashed boxes) when imaging commenced, and at the end of 5 min (full video available as Supplemental Video 1). Signal intensity was enhanced for later frames to adjust for photobleaching. Scale bars indicate 10 μm. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
To test this notion, we devised a fission assay that allowed us to induce mitochondrial fission and monitor the mitochondrial network throughout the cell. When mitochondrial fission was induced with a phototoxic stress, we found that these peripheral hyper-connected regions remained intact in R941L patient fibroblasts, even though central mitochondrial networks in the same cell were able to fragment (Fig. 3b, Supplemental Video 1). In comparison, mitochondrial networks in control fibroblasts were able to undergo fission throughout the entire cell, even in the few hyper-connected peripheral regions that could be found. Thus, this peripheral mitochondrial fission defect phenotype is unique to R941L patient fibroblasts.
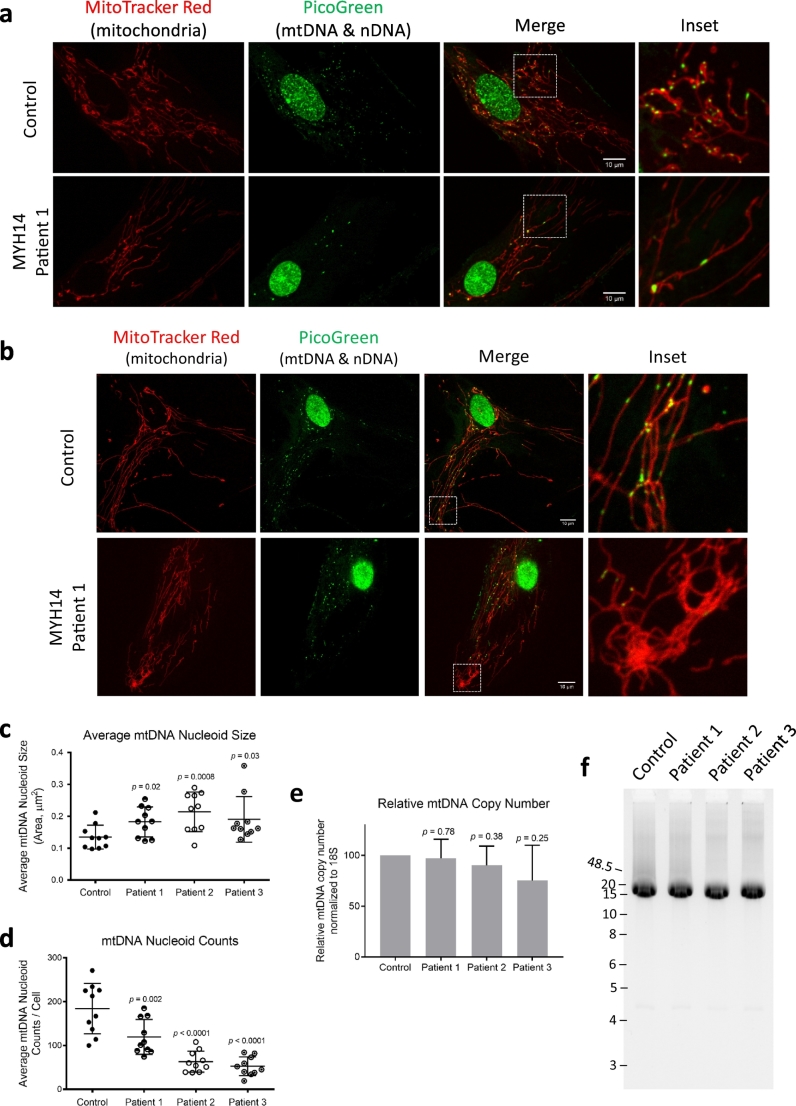
As an independent approach to look at whether mitochondrial fission was impaired in R941L patient fibroblasts cells, we also examined mitochondrial genomes, as mitochondrial fission is required for mtDNA maintenance [[40], [41], [42]]. Specifically, loss of fission leads to enlarged mtDNA nucleoid structures [43]. Moreover, NMIIA and B isoforms have also been implicated in regulating mtDNA [11]. Using live-cell imaging to visualize nucleoid size and distribution, we observed that patient fibroblast cells had fewer, but larger nucleoids than controls cells (Fig. 4a). Moreover, the distribution of mtDNA nucleoids throughout the mitochondrial network was different in MYH14 fibroblasts. In control fibroblasts, mtDNA nucleoids are distributed evenly throughout the network, even in hyper-connected peripheral regions. In contrast, when we looked at the nucleoid distribution in the hyper-connected mitochondria in the cell periphery of R941L patient fibroblasts, we observed a marked lack of nucleoids within these regions (Fig. 4b).
Fig. 4.
Altered mtDNA nucleoids in R941L patient fibroblasts. (a) Representative confocal images of control and patient fibroblast cells taken with an Olympus SD-OSR microscope. Live cells were stained with MitoTracker Red (Red, mitochondria) and PicoGreen (Green, nuclear and mitochondrial DNA). Scale bars indicate 10 μm.. (b) Representative confocal images of peripheral mitochondria and nucleoids as in (a). Quantification of nucleoid size (c) and number (d) from 10 cells for each line. (e) Copy number of mtDNA as determined by quantitative PCR. Error bars indicate standard deviations, and p-values (Student's t-test) were determined by comparison to control fibroblasts. (f) Long range PCR of mtDNA in control and patient fibroblasts showing 16.3 kb amplicons and no mtDNA deletions. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
Next, we quantified the average size and number of mtDNA nucleoids and found that MYH14 fibroblasts has fewer, but larger mtDNA nucleoids (Fig. 4c, d). As decreases in the total number of nucleoids can be due to either loss of mtDNA or clumping of several smaller nucleoids, we also looked at the total amount of mtDNA in these cells. Quantification of the mtDNA copy number by qPCR showed that there were no significant changes in R941L patient fibroblast cells compared to control (Fig. 4e). Finally, we performed long-range PCR to rule out the possibility of mtDNA deletions in MYH14 fibroblasts (Fig. 4f). Altogether, these observations are consistent with impaired mitochondrial fission leading to mtDNA nucleoid alterations.
3.2.2. Wild-type NMIIC induces mitochondrial fission while R941L mutation exerts a dominant-negative effect
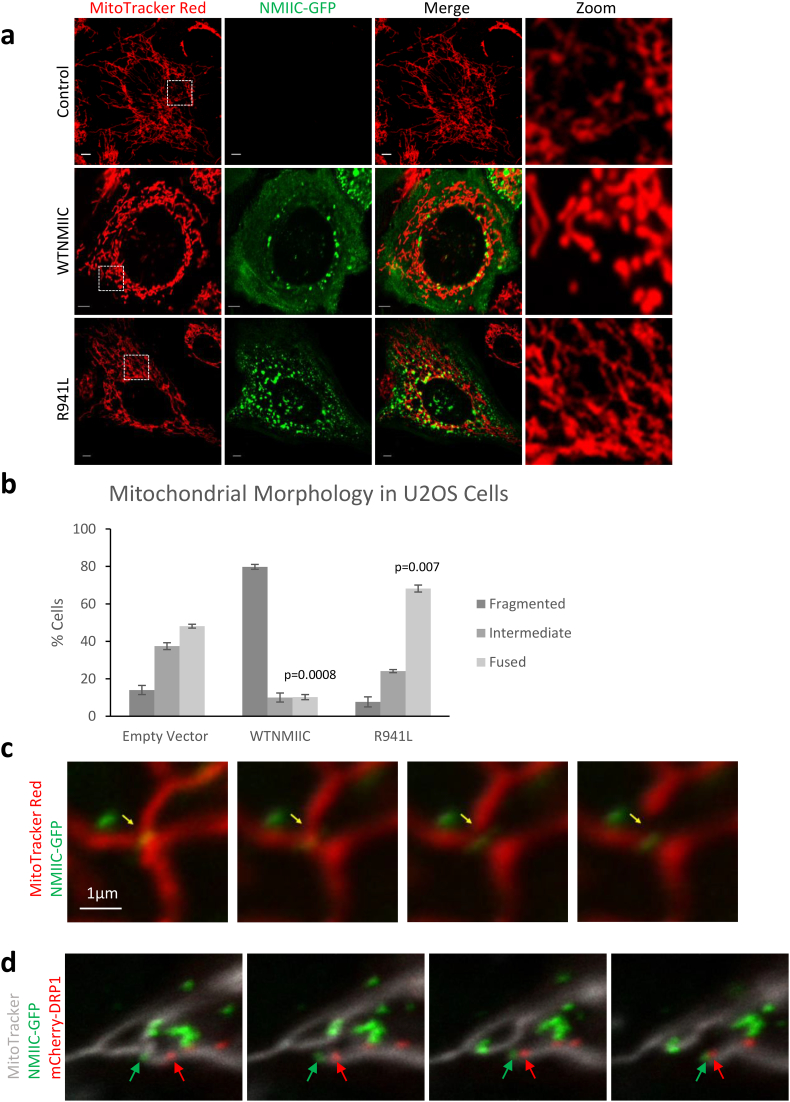
Given the altered mitochondrial networks in patient fibroblast cells, which were both longer and more connected, we wanted to test whether NMIIC was directly involved in regulating mitochondrial structure. We expressed an EGFP-tagged NMIIC protein that has been used previously to study the role of NMIIC in U2OS cells [44]. Consistent with a role for NMIIC in regulating mitochondrial fission, we saw that overexpression of wild-type NMIIC-EGFP in U2OS cells increased fragmentation of the mitochondrial network (Fig. 5a, b). In contrast, when we overexpressed NMIIC containing the R941L mutation, we saw a marked difference, as overexpression of R941L-NMIIC did not promote mitochondrial fragmentation. In fact, we observed a statistically significant increase in the number of cells with hyperfused mitochondrial networks. This finding shows that the R941L mutation not only abrogates the ability of NMIIC to promote mitochondrial fragmentation, but also demonstrates a dominant negative effect on the ability of endogenous wild-type protein to mediate fission.
Fig. 5.
NMIIC puncta localize to mitochondria at sites of mitochondrial fission. (a) Representative confocal images from live U2OS cells transfected with empty vector (control) or NMIIC-EGFP constructs (WTNMIIC = wild-type, R941L = mutant). Mitochondria were stained with Mitotracker Red, and the EGFP signal represents wild-type or R941L mutant NMIIC. Images were captured with a Zeiss LSM microscope. Scale bars indicate 5 μm. (b) Quantification of mitochondrial morphology in U2OS cells transfected as in (a). One hundred cells were quantified in three technical replicates for each of three independent biological replicates. Error bars indicate standard deviations, and p-values (Student's t-test) were determined by comparison to the number of cells with hyperfused mitochondria in the empty vector control. (c) A subset of NMIIC-EGFP puncta co-localize with mitochondria at sites of fission. Single frames from live cell imaging of wild-type NMIIC as described in (a), with yellow arrow indicating fission site (full video available as Supplemental Video 2). (d) NMIIC puncta at fission sites precede Drp1 recruitment and fission. Single frames from live cell imaging of U2OS cell transfected with wild-type NMIIC-EGFP (green), mCherry-DRP1 (red), and mitochondria stained with Mitotracker DeepRed (grey) (full video available as Supplemental Video 3). Green arrow denotes the NMIIC puncta at the site of fusion. Red arrow indicates the DRP1 recruited to the site of fission. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
NMIIC puncta localize to mitochondria at sites of mitochondrial fission. (a) Representative confocal images from live U2OS cells transfected with empty vector (control) or NMIIC-EGFP constructs (WTNMIIC = wild-type, R941L = mutant). Mitochondria were stained with Mitotracker Red, and the EGFP signal represents wild-type or R941L mutant NMIIC. Images were captured with a Zeiss LSM microscope. Scale bars indicate 5 μm. (b) Quantification of mitochondrial morphology in U2OS cells transfected as in (a). One hundred cells were quantified in three technical replicates for each of three independent biological replicates. Error bars indicate standard deviations, and p-values (Student's t-test) were determined by comparison to the number of cells with hyperfused mitochondria in the empty vector control. (c) A subset of NMIIC-EGFP puncta co-localize with mitochondria at sites of fission. Single frames from live cell imaging of wild-type NMIIC as described in (a), with yellow arrow indicating fission site (full video available as Supplemental Video 2). (d) NMIIC puncta at fission sites precede Drp1 recruitment and fission. Single frames from live cell imaging of U2OS cell transfected with wild-type NMIIC-EGFP (green), mCherry-DRP1 (red), and mitochondria stained with Mitotracker DeepRed (grey) (full video available as Supplemental Video 3). Green arrow denotes the NMIIC puncta at the site of fusion. Red arrow indicates the DRP1 recruited to the site of fission. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
3.2.3. NMIIC localizes to future sites of mitochondrial fission
To further characterize the role of NMIIC in mediating mitochondrial fission, we examined the cellular distribution of the NMIIC-EGFP protein via microscopy. In addition to observing NMIIC-EGFP at the periphery of cells, in line with a role in cell migration, we also observed several NMIIC-EGFP puncta throughout the cell, several of which co-localized with mitochondria (Fig. 5a).
Live-cell imaging allowed us to see that a subset of NMIIC-EGFP mitochondrial puncta were located at sites of mitochondrial fission, indicating that NMIIC-EGFP participates in mitochondrial fission (Fig. 5c, Supplemental Video 2). To quantify how often NMIIC-EGFP was located at fission sites, we monitored fission events from 14 cells transfected with NMIIC-EGFP. We found that NMIIC-EGFP was located at fission sites in 62 of 216 total fission events that were observed (29%), over a total of 42 min of video. In addition, we were able to show that NMIIC-EGFP puncta at sites of fission precedes recruitment of the mitochondrial fission protein Drp1 tagged with an mCherry fluorescent protein (mCh-Drp1) (Fig. 5d, Supplemental Video 3, Supplemental Video 4). These findings demonstrate that NMIIC regulates mitochondrial fission in a similar manner as NMIIA and B, which mediate ER-mediated pre-constriction of mitochondrial tubules.
3.2.4. The R941L mutation disrupts neuronal mitochondria
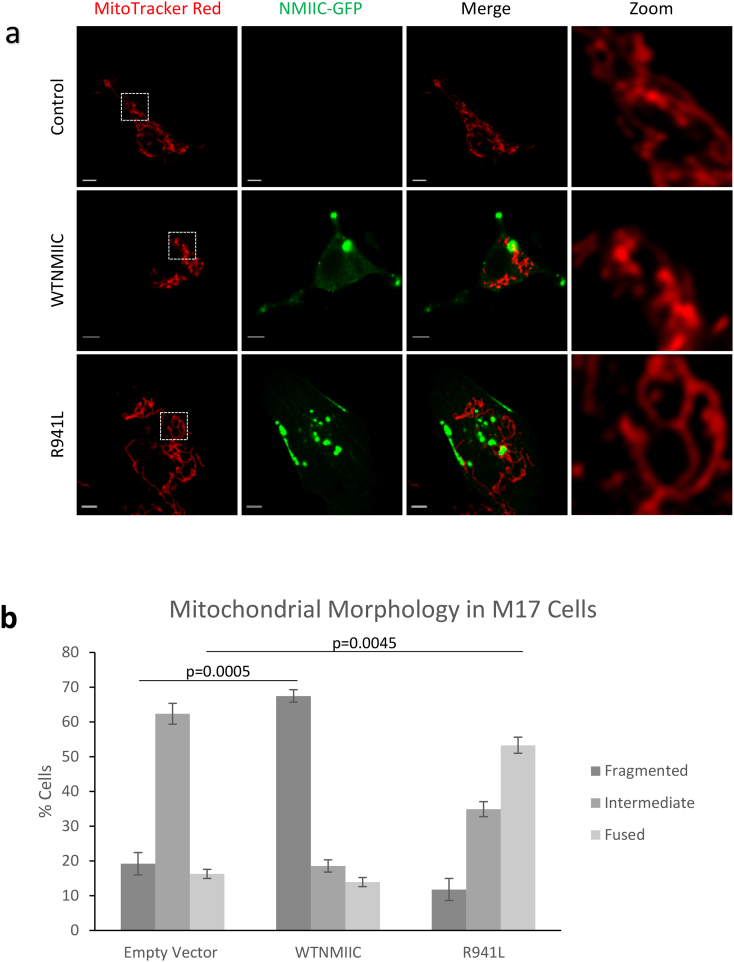
Given the peripheral neuropathy phenotype in patients, which can be caused by impaired axonal mitochondrial transport in other peripheral neuropathies due to alterations to mitochondrial fission, we decided to look at the effects of R941L overexpression in a neuronal cell type. Thus, we turned to M17 neuroblastoma cells, which are amenable to transfection, and which can be differentiated into neuronal-like cells [30,45]. Similar to U2OS cell, and consistent with a role for NMIIC in fission, overexpression of WT-NMIIC promoted mitochondrial fragmentation. Meanwhile, the R941L led to more hyperfused mitochondrial networks, again indicating a dominant-negative function (Fig. 6a, b).
Fig. 6.
The R941L NMIIC does not promote mitochondrial fission in differentiated M17 neuronal cells. (a) Representative confocal images of live M17 cells transfected with empty vector (control) or NMIIC-EGFP constructs (WTNMIIC = wild-type, R941L = mutant). Mitochondria were stained with MitoTracker Red, and the EGFP signal represents wild-type or R941L mutant NMIIC. Images were captured with a Zeiss LSM microscope. Scale bars indicate 10 μm. (b) Quantification of mitochondrial morphology in differentiated M17 cells as transfected as in (a). At least 40 cells were quantified in two independent experimental replicates. Error bars indicate standard deviations, and p-values (Student's t-test) were determined by comparison to the number of cells with fragmented or fused mitochondrial networks (as indicated) in the empty vector control. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
4. Discussion
Prior to this study, although it was shown that the R941L mutation in NMIIC was associated with a peripheral neuropathy phenotype, there was little known about the molecular function of the NMIIC protein, and no insight into the underlying mechanism of the pathology. Here, we describe a third family where the R941L mutation segregates with a peripheral neuropathy phenotype, further strengthening the causal link between the mutation and the phenotype. Identifying a causal gene ends a diagnostic odyssey for patients and can provide families answers, counselling and prognosis. Moreover, diagnosis is also the first step towards therapy, with the next step involving mechanistic studies to understand the molecular basis of the disease. In this context, we also demonstrate a previously unrecognized role for NMIIC in regulating mitochondrial fission, which is impaired by the R941L mutation, and which may explain the peripheral neuropathy phenotype.
Two previous reports of the R941L mutation occurring de novo in independent families exhibiting deafness and peripheral neuropathy have been described in the literature [12,14]. In addition, this mutation has also been reported as part of a larger study of peripheral neuropathies [13]. However, the reason for reoccurrence of the same base pair change was unexplained. In the current report, we describe a new family pedigree harbouring the c.2822G>T mutation resulting in the R941L amino acid substitution in the protein sequence. Thus, the c.2822G>T mutation in MYH14 appears to have occurred de novo four times. Normally, it would be exceedingly rare for the exact same mutation to arise independently. However, our data support existing evidence that the locus in question is a methylated CpG site, increasing the chance of a mutation, and explaining the recurrence of this de novo mutation. Consistent with this notion, there are multiple different nucleotide substitutions at this position in The Exome Aggregation Consortium (ExAC) (http://exac.broadinstitute.org/) [46] (Supplemental Fig. 3), suggesting an increased sensitivity to mutations. Though of note, the R941L variant itself does not appear in the ExAC database.
While several autosomal dominant loss of function mutations in MYH14 have been linked to non-syndromic hearing loss [[15], [16], [17], [18], [19], [20]], the additional peripheral neuropathy phenotype in patients with the R941L mutation suggests that this neuronal phenotype is not likely to be due simply to haploinsufficiency. Nonetheless, it is possible that peripheral neuropathy was not closely examined in these other MYH14 affected individuals with hearing loss alone, or that the disease had not progressed sufficiently to exhibit peripheral neuropathy when hearing loss was first diagnosed. However, the most likely explanation is that the non-syndromal hearing loss associated with other MYH14 mutations is due to haploinsufficiency from loss of function in these alleles. The fact that MYH14-null mice are also susceptible to hearing loss [21], but have no reported peripheral neuropathy phenotype supports this conclusion.
In contrast, our data suggest that the peripheral neuropathy phenotype associated with the R941L mutation is not simply due to loss of function. In this regard, in silico modeling of the R941L mutation suggests that the replacement of the arginine residue with a leucine could impair protein interactions required for dimer formation that could influence the functional activity of the NMIIC protein. Such structural changes that alter dimerization are consistent with a dominant-negative effect where a dimer of mutant with wild-type protein would be inactive. Meanwhile, our modeling of the R941L mutation in C. elegans, where there is a highly sensitive functional assay of NMII protein function, shows that the correlating R915L mutation acts semidominantly, but retains some myosin activity. However, our data does not allow us to conclude if the R915L mutation also has dominant-negative properties in C. elegans. Nonetheless, as it pertains to mitochondrial fission in human cell culture models, we see that overexpression of the R941L mutant protein has the opposite effect on mitochondrial morphology as the wild-type NMIIC protein. Taken together, these findings are consistent with a distinct dominant-negative mechanism underlying the peripheral neuropathy of the R941L mutation.
Critically, we demonstrate a role for NMIIC as a mediator of mitochondrial fission, a cellular function that is known to cause peripheral neuropathy when impaired [1]. To characterize the role of the R941L mutation at endogenous levels, we examined mitochondria in patient fibroblasts. We found that patient fibroblast cells exhibit several phenotypes that are consistent with impaired mitochondrial fission, most notably hyperfused mitochondrial networks. Although such hyperfused mitochondrial networks could be caused by either increased fusion or decreased fission, our data clearly show that NMIIC plays a previously unrecognized role in fission, which is impaired in patient fibroblasts harbouring the R941L mutation. Notably, impaired fission is even more prominent at the cell periphery, as central mitochondria in the same cell could still undergo fragmentation. This intriguing finding demonstrates a definitive defect in mitochondrial fission in the fibroblasts cells, and suggests that NMIIC may play a more important role in mediating mitochondrial fission at the cell periphery. Such a role would be especially relevant in the context of the peripheral neuropathy phenotype in patients with the R941L mutation in MYH14.
A peripheral fission phenotype is also intriguing as it pertains to our understanding of how impaired fission leads to mitochondrial dysfunction and peripheral neuropathies. Rather than the proposed model where impaired mitochondrial fission in the cell body prevents the anterograde transport of large mitochondria down axons [2], impaired fission at the tips of axons could impact local mitochondrial quality control and function in a different fashion. Specifically, the fused state at the ends of axons could prevent the retrograde transport of mitochondria. Additionally, as fission is required for the autophagic removal of mitochondria [47], which has been reported to occur in axons [48,49], impaired peripheral mitochondrial fission could also inhibit mitochondrial autophagy and thus impair neuronal function. The slight increase in mitochondrial mass that we observe in MYH14 fibroblasts would also be consistent with decreased turnover of mitochondria due to reduced mitochondrial autophagy.
In addition to mitochondrial network changes, we observed fewer and larger mtDNA nucleoids in patient fibroblasts, alterations that are also consistent with impaired fission [43]. While the larger mtDNA nucleoids that we visualized could be due to clumping of multiple mtDNA genomes, it is notable that signal from the DNA intercalating dye picogreen can also reflect changes to the supercoiling or packaging state of the mtDNA, not just mtDNA abundance [50,51]. Meanwhile, the decrease in the number of nucleoids could also be due to clumping, or alternatively to loss of total mtDNA. As our quantification of the mtDNA copy number in patient fibroblasts shows there is no significant mtDNA loss, we conclude that the larger nucleoids are most likely due to clumping of multiple nucleoids. This result is consistent with a previous report where inhibition of fission via acute knockdown of DRP1 leads to fewer, but larger nucleoids composed of multiple genomes, without any change in copy number [43]. The notion that fission is playing a key role in the distribution of mtDNA throughout the network is also supported by the lack of mtDNA nucleoids that we observe in the hyper-connected mitochondrial networks in the cell periphery of R941L patient fibroblasts. The mtDNA nucleoid impairments we observed in R941L patient fibroblasts are also notable, as regulation of mtDNA is involved in peripheral neuropathies. For example, peripheral neuropathies can be due to mutations in the mtDNA or mutations in proteins that regulate mtDNA replication [52]. In addition, a peripheral neuropathy is a side-effect of the chemotherapeutic agent cisplatin, which preferentially damages mtDNA [53] and also impairs mitochondrial dynamics in peripheral nerves [54].
Supporting the previously unrecognized role for NMIIC in mediating mitochondrial fission, we found that overexpression of a GFP-tagged NMIIC shifted mitochondrial networks to a more fragmented state. While there are always potential concerns about protein overexpression artifacts, we note that the R941L mutation is not competent to promote mitochondrial fission, arguing against the increased mitochondrial fragmentation resulting from an accumulation of NMIIC protein. In fact, overexpression of the R941L mutation shifted cells towards a more hyperfused network, consistent with a dominant-negative effect of the mutation. Moreover, we could directly visualize NMIIC-GFP puncta at mitochondrial fission sites preceding Drp1 recruitment, demonstrating that NMIIC mediates fission in the same fashion as NMIIA and NMIIB. Although we observed NMIIC-GFP at only 29% of total fission sites, it is important to remember that the U2OS cells express unlabeled NMII proteins [44], which likely mediate ER-mediated constriction at the remaining mitochondrial fission events.
The functional interplay and redundancy of the three NMII isoforms (A, B, and C) remains unknown in the context of regulating mitochondrial fission, but has important implications to the peripheral neuropathy phenotype in patients. In this regard, the situation of NMII isoforms is reminiscent to that of the fusion proteins MFN1 and MFN2. Though MFN1 and MFN2 are functionally redundant for fusion, mutations in MFN2 are thought to lead to a peripheral neuropathy phenotype because MFN1 is not highly expressed in neurons [55]. Thus, it is notable that neurons predominantly express NMIIB and NMIIC [56]. As such, it might be expected that neurons are more sensitive to mutations affecting NMIIC function than other cell types. It is intriguing that the phenotype associated with the R941L mutation is primarily a distal motor neuropathy phenotype, and few reported patients have any sensory symptoms or signs (Patient III-2 from this report and Patient III-8 from Choi et al. [12]). One possible explanation could be that the various NMII isoforms are differentially expressed in nerve fibers and sensory neurons.
In addition to expression differences, it is also possible that the cellular distribution and precise roles of the different NMII protein isoforms varies. Although little is known about the relative cellular functions of the NMII isoforms, based on our findings we would predict that NMIIC is more important for mediating fission at the cell extremities. Finally, it is notable that all three NMII isoforms can form homo or heterodimers with each other [44]. Though our structural modeling suggests that the R941L mutation could affect dimerization, we do not know how this might affect homo versus heterodimers, let alone the resulting functional outcomes on fission.
Due to the fact that we do not know all of the cellular functions of NMIIC, a limitation of this study is that we cannot say with certainty that the peripheral neuropathy is caused specifically by impaired mitochondrial fission. Future work using a peripheral neuron model expressing the R941L mutant would allow us to strengthen this connection. Nonetheless, the role of NMIIC in regulating mitochondrial fission, particularly at the cell periphery is likely to be relevant to the peripheral neuropathy phenotype in R941L patients. Notably, there is a growing list of peripheral neuropathy genes encoding proteins that regulate mitochondrial dynamics via fission (e.g. GDAP1, DNM2, INF2, and MFF), fusion (e.g. MFN2, and OPA1), or transport (e.g. KIF1A, KIF1B, and KIF5A) [1,57,58]. Of particular relevance to the role of the NMIIC protein in fission, the INF2 protein regulates actin, which interacts with NMII proteins during the ER-mediated mitochondrial constriction that initiates fission [59]. Thus, the R941L is likely a second example of how impairing the actin/myosin contraction required for fission can lead to a peripheral neuropathy phenotype. Collectively, our findings provide evidence that NMIIC mediates mitochondrial fission. Moreover, the fact that the R941L mutation is deficient in the ability to promote mitochondrial fission provides additional evidence that impaired mitochondrial fission underlies the peripheral neuropathy in these patients. Thus, MYH14 can be added to the list of genes regulating mitochondrial dynamics, mutations in which lead to peripheral neuropathy.
The following are the supplementary data related to this article.
DNA oligos used for CRISPR editing of nmy-1 gene in C. elegans. gRNA targeting sequences are underlined and mutations highlighted.
Detailed clinical features of patients with the R941L mutation.
Peripheral hyper-connected mitochondria are resistant to phototoxicity –induced fission in R941L patient fibroblasts. Video corresponding to static images in Fig. 3b. Imaged using Olympus SD-OSR microscope.
NMIIC-GFP puncta colocalize to mitochondria at sites of fission. Video corresponding to static images in Fig. 5c. Imaged using a Zeiss LSM microscope.
NMIIC-GFP puncta at fission sites precede Drp1 recruitment. Video corresponding to static images in Fig. 5d. Imaged using a Zeiss LSM microscope.
NMIIC-GFP puncta at fission sites precede Drp1 recruitment. Imaged using Olympus SD-OSR microscope.
Supplemental Figure 1 Evidence for CpG methylation at c.2822G of the MYH14 gene. To verify the presence of a methylated CpG site experimentally, we devised an assay based on the methylation selectivity of the restriction enzyme FauI. a Schematic depicting the experimental strategy, where unmethylated genomic DNA is cleaved by the enzyme, such that the region cannot be PCR amplified. b Digestion with methyl sensitive enzyme shows methylation in whole blood extracted genomic DNA. As a control, DNA lacking methylation marks (i.e., PCR amplified prior to treatment) was digested by FauI, indicating that the enzyme was actively able to cleave the DNA. Meanwhile, the genomic DNA samples extracted from whole blood were readily amplified following incubation with FauI prior to PCR, indicating their resistance to digestion by the enzyme. This result was observed across a broad cross section of genomic DNA samples (data not shown), and is consistent with previous reports that the c.2822G>T mutation occurs at a methylated CpG site.
Supplemental Figure 2 Mitochondrial Morphology in Control Fibroblasts. Quantification of mitochondrial morphology from three different control fibroblast lines. At least 100 cells were quantified in three independent experimental replicates. Error bars indicate standard deviations. No significant differences were observed between the three lines, as determined by Student’s T-test compared to the number of cells in control. Control 1 was used for subsequent fibroblast cell analysis.
Supplemental Figure 3 MYH14 variants reported in the The Exome Aggregation Consortium (ExAC). Alignment of various alleles that correspond to the methylated CpG site examined in this study, as reported in ExAC, an aggregation of exome sequencing data from a variety of large-scale sequencing projects. Notably, the c.2822G>T change leading to the R941L mutation is not in ExAC.
Acknowledgments
Acknowledgements
The authors would like to thank the study participants and their family. We would like to acknowledge Linda MacLaren for her role in clinical support. This work was performed under the Care4Rare Canada Consortium funded by Genome Canada, the Canadian Institutes of Health Research, the Ontario Genomics Institute, Ontario Research Fund, Genome Alberta, Genome BC, Genome Quebec, and Children's Hospital of Eastern Ontario Foundation.
Funding sources
This work was supported by funds provided by the Alberta Children's Hospital Research Institute (T.S.), the UQTR Foundation (M.G.), and the Canadian Institutes of Health Research (P.E.M). W.A. was supported by the Saudi Cultural Bureau. R.S. was supported by a QEII Graduate Scholarship. H.S.I. was supported by a FRQ-NT scholarship. This work was performed under the Care4Rare Canada Consortium funded by Genome Canada, the Canadian Institutes of Health Research, the Ontario Genomics Institute, Ontario Research Fund, Genome Alberta, Genome BC, Genome Quebec, and Children's Hospital of Eastern Ontario Foundation. The funders had no role in study design, data collection and interpretation, or the decision to submit the work for publication.
Declarations of interest
Dr. Shutt received research funds from the Alberta Children's Hospital Research Institute, which supported the research conducted in this manuscript; Ms. Almutawaa reports other from Saudi Arabia Cultural Bureau, during the conduct of the study; Ms. Sabouny reports grants from University of Calgary, during the conduct of the study. The remaining authors declare that they have no conflicts of interests.
Author contributions
Designed and/or performed and analyzed mitochondrial experiments, W.A., C.S., R.S., T.Z., and T.E.S.; Analyzed mitochondrial data, W.A., C.S., R.S., T.Z., H.S.I., J.D.G., M.G., T.E.S, R.W., and L.L.-G.; Designed, performed and analyzed C. elegans studies, R.B.S., and P.E.M.; Clinical evaluation of patients, O.S., G.P., and A.M.I.; Analyzed exome data, C.S., and J.S.P.; Performed skin biopsy from mitochondrial patients, G.P.; Drafted the manuscript or figures, W.A., C.S., R.S., G.P., A.M.I., T.E.S. All authors discussed the results and commented on the manuscript. W.A. and C.S. are joint first authors. T.E.S. and A.M.I. are co-corresponding authors.
Contributor Information
A. Micheil Innes, Email: micheil.innes@ahs.ca.
Timothy E. Shutt, Email: timothy.shutt@ucalgary.ca.
References
- 1.Pareyson D., Saveri P., Sagnelli A., Piscosquito G. Mitochondrial dynamics and inherited peripheral nerve diseases. Neurosci Lett. 2015;596:66–77. doi: 10.1016/j.neulet.2015.04.001. [DOI] [PubMed] [Google Scholar]
- 2.Chen H., Chan D.C. Mitochondrial dynamics–fusion, fission, movement, and mitophagy–in neurodegenerative diseases. Hum Mol Genet. 2009;18(R2):R169–R176. doi: 10.1093/hmg/ddp326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Friedman J.R., Lackner L.L., West M., DiBenedetto J.R., Nunnari J., Voeltz G.K. ER tubules mark sites of mitochondrial division. Science. 2011;334(6054):358–362. doi: 10.1126/science.1207385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kraus F., Ryan M.T. The constriction and scission machineries involved in mitochondrial fission. J Cell Sci. 2017;130(18):2953–2960. doi: 10.1242/jcs.199562. [DOI] [PubMed] [Google Scholar]
- 5.Lee J.E., Westrate L.M., Wu H., Page C., Voeltz G.K. Multiple dynamin family members collaborate to drive mitochondrial division. Nature. 2016;540(7631):139–143. doi: 10.1038/nature20555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hatch A.L., Ji W.K., Merrill R.A., Strack S., Higgs H.N. Actin filaments as dynamic reservoirs for Drp1 recruitment. Mol Biol Cell. 2016;27(20):3109–3121. doi: 10.1091/mbc.E16-03-0193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ji W.K., Hatch A.L., Merrill R.A., Strack S., Higgs H.N. Actin filaments target the oligomeric maturation of the dynamin GTPase Drp1 to mitochondrial fission sites. Elife. 2015;4 doi: 10.7554/eLife.11553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Prudent J., McBride H.M. Mitochondrial dynamics: ER actin tightens the Drp1 noose. Curr Biol. 2016;26(5):R207–R209. doi: 10.1016/j.cub.2016.01.009. [DOI] [PubMed] [Google Scholar]
- 9.Golomb E., Ma X., Jana S.S., Preston Y.A., Kawamoto S., Shoham N.G. Identification and characterization of nonmuscle myosin II-C, a new member of the myosin II family. J Biol Chem. 2004;279(4):2800–2808. doi: 10.1074/jbc.M309981200. [DOI] [PubMed] [Google Scholar]
- 10.Korobova F., Gauvin T.J., Higgs H.N. A role for myosin II in mammalian mitochondrial fission. Curr Biol. 2014;24(4):409–414. doi: 10.1016/j.cub.2013.12.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Reyes A., He J., Mao C.C., Bailey L.J., Di Re M., Sembongi H. Actin and myosin contribute to mammalian mitochondrial DNA maintenance. Nucleic Acids Res. 2011;39(12):5098–5108. doi: 10.1093/nar/gkr052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Choi B.O., Kang S.H., Hyun Y.S., Kanwal S., Park S.W., Koo H. A complex phenotype of peripheral neuropathy, myopathy, hoarseness, and hearing loss is linked to an autosomal dominant mutation in MYH14. Hum Mutat. 2011;32(6):669–677. doi: 10.1002/humu.21488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gonzaga-Jauregui C., Harel T., Gambin T., Kousi M., Griffin L.B., Francescatto L. Exome sequence analysis suggests that genetic burden contributes to phenotypic variability and complex neuropathy. Cell Rep. 2015;12(7):1169–1183. doi: 10.1016/j.celrep.2015.07.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Iyadurai S., Arnold W.D., Kissel J.T., Ruhno C., McGovern V.L., Snyder P.J. Variable phenotypic expression and onset in MYH14 distal hereditary motor neuropathy phenotype in a large, multigenerational North American family. Muscle Nerve. 2017;56(2):341–345. doi: 10.1002/mus.25491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Donaudy F., Snoeckx R., Pfister M., Zenner H.P., Blin N., Di Stazio M. Nonmuscle myosin heavy-chain gene MYH14 is expressed in cochlea and mutated in patients affected by autosomal dominant hearing impairment (DFNA4) Am J Hum Genet. 2004;74(4):770–776. doi: 10.1086/383285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kim B.J., Kim A.R., Han J.H., Lee C., Oh D.Y., Choi B.Y. Discovery of MYH14 as an important and unique deafness gene causing prelingually severe autosomal dominant nonsyndromic hearing loss. J Gene Med. 2017;19(4) doi: 10.1002/jgm.2950. [DOI] [PubMed] [Google Scholar]
- 17.Kim K.Y., Kovacs M., Kawamoto S., Sellers J.R., Adelstein R.S. Disease-associated mutations and alternative splicing alter the enzymatic and motile activity of nonmuscle myosins II-B and II-C. J Biol Chem. 2005;280(24):22769–22775. doi: 10.1074/jbc.M503488200. [DOI] [PubMed] [Google Scholar]
- 18.Kim S.J., Lee S., Park H.J., Kang T.H., Sagong B., Baek J.I. Genetic association of MYH genes with hereditary hearing loss in Korea. Gene. 2016;591(1):177–182. doi: 10.1016/j.gene.2016.07.011. [DOI] [PubMed] [Google Scholar]
- 19.Yang R., Li H., Zhan C.X., Mao H.Y., Zhan T.L., Zhu Z.F. c.359T>C mutation of the MYH14 gene in two autosomal dominant non-syndromic hearing impairment families with common ancestor. Zhonghua Yi Xue Yi Chuan Xue Za Zhi. 2010;27(3):259–262. doi: 10.3760/cma.j.issn.1003-9406.2010.0.005. [DOI] [PubMed] [Google Scholar]
- 20.Yang T., Pfister M., Blin N., Zenner H.P., Pusch C.M., Smith R.J. Genetic heterogeneity of deafness phenotypes linked to DFNA4. Am J Med Genet A. 2005;139(1):9–12. doi: 10.1002/ajmg.a.30989. [DOI] [PubMed] [Google Scholar]
- 21.Fu X., Zhang L., Jin Y., Sun X., Zhang A., Wen Z. Loss of Myh14 increases susceptibility to noise-induced hearing loss in CBA/CaJ mice. Neural Plast. 2016;2016 doi: 10.1155/2016/6720420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Beaulieu C.L., Majewski J., Schwartzentruber J., Samuels M.E., Fernandez B.A., Bernier F.P. FORGE Canada consortium: outcomes of a 2-year national rare-disease gene-discovery project. Am J Hum Genet. 2014;94(6):809–817. doi: 10.1016/j.ajhg.2014.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hartley T., Wagner J.D., Warman-Chardon J., Tetreault M., Brady L., Baker S. Whole-exome sequencing is a valuable diagnostic tool for inherited peripheral neuropathies: outcomes from a cohort of 50 families. Clin Genet. 2018;93(2):301–309. doi: 10.1111/cge.13101. [DOI] [PubMed] [Google Scholar]
- 24.Biasini M., Bienert S., Waterhouse A., Arnold K., Studer G., Schmidt T. SWISS-MODEL: modelling protein tertiary and quaternary structure using evolutionary information. Nucleic Acids Res. 2014;42:W252–W258. doi: 10.1093/nar/gku340. Web Server issue. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wendt T., Taylor D., Trybus K.M., Taylor K. Three-dimensional image reconstruction of dephosphorylated smooth muscle heavy meromyosin reveals asymmetry in the interaction between myosin heads and placement of subfragment 2. Proc Natl Acad Sci U S A. 2001;98(8):4361–4366. doi: 10.1073/pnas.071051098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pettersen E.F., Goddard T.D., Huang C.C., Couch G.S., Greenblatt D.M., Meng E.C. UCSF chimera – a visualization system for exploratory research and analysis. J Comput Chem. 2004;25(13):1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- 27.Brenner S. The genetics of Caenorhabditis elegans. Genetics. 1974;77(1):71–94. doi: 10.1093/genetics/77.1.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Piekny A.J., Johnson J.L., Cham G.D., Mains P.E. The Caenorhabditis elegans nonmuscle myosin genes nmy-1 and nmy-2 function as redundant components of the let-502/Rho-binding kinase and mel-11/myosin phosphatase pathway during embryonic morphogenesis. Development. 2003;130(23):5695–5704. doi: 10.1242/dev.00807. [DOI] [PubMed] [Google Scholar]
- 29.Arribere J.A., Bell R.T., Fu B.X., Artiles K.L., Hartman P.S., Fire A.Z. Efficient marker-free recovery of custom genetic modifications with CRISPR/Cas9 in Caenorhabditis elegans. Genetics. 2014;198(3):837–846. doi: 10.1534/genetics.114.169730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Andres D., Keyser B.M., Petrali J., Benton B., Hubbard K.S., McNutt P.M. Morphological and functional differentiation in BE(2)-M17 human neuroblastoma cells by treatment with trans-retinoic acid. BMC Neurosci. 2013;14:49. doi: 10.1186/1471-2202-14-49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ashley N., Harris D., Poulton J. Detection of mitochondrial DNA depletion in living human cells using PicoGreen staining. Exp Cell Res. 2005;303(2):432–446. doi: 10.1016/j.yexcr.2004.10.013. [DOI] [PubMed] [Google Scholar]
- 32.Ouellet M., Guillebaud G., Gervais V., Lupien St-Pierre D., Germain M. A novel algorithm identifies stress-induced alterations in mitochondrial connectivity and inner membrane structure from confocal images. PLoS Comput Biol. 2017;13(6) doi: 10.1371/journal.pcbi.1005612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Schindelin J., Arganda-Carreras I., Frise E., Kaynig V., Longair M., Pietzsch T. Fiji: an open-source platform for biological-image analysis. Nat Methods. 2012;9(7):676–682. doi: 10.1038/nmeth.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zhao T., Goedhart C.M., Sam P.N., Sabouny R., Lingrell S., Cornish A.J. PISD is a mitochondrial disease gene causing skeletal dysplasia, cataracts, and white matter changes. Life Sci Alliance. 2019;2(2) doi: 10.26508/lsa.201900353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Eaton J.S., Lin Z.P., Sartorelli A.C., Bonawitz N.D., Shadel G.S. Ataxia-telangiectasia mutated kinase regulates ribonucleotide reductase and mitochondrial homeostasis. J Clin Invest. 2007;117(9):2723–2734. doi: 10.1172/JCI31604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Nishigaki Y., Marti R., Hirano M. ND5 is a hot-spot for multiple atypical mitochondrial DNA deletions in mitochondrial neurogastrointestinal encephalomyopathy. Hum Mol Genet. 2004;13(1):91–101. doi: 10.1093/hmg/ddh010. [DOI] [PubMed] [Google Scholar]
- 37.Livak K.J., Schmittgen T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2(−Delta Delta C(T)) method. Methods. 2001;25(4):402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 38.Kent W.J., Sugnet C.W., Furey T.S., Roskin K.M., Pringle T.H., Zahler A.M. The human genome browser at UCSC. Genome Res. 2002;12(6):996–1006. doi: 10.1101/gr.229102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Maunakea A.K., Nagarajan R.P., Bilenky M., Ballinger T.J., D'Souza C., Fouse S.D. Conserved role of intragenic DNA methylation in regulating alternative promoters. Nature. 2010;466(7303):253–257. doi: 10.1038/nature09165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Garrido N., Griparic L., Jokitalo E., Wartiovaara J., van der Bliek A.M., Spelbrink J.N. Composition and dynamics of human mitochondrial nucleoids. Mol Biol Cell. 2003;14(4):1583–1596. doi: 10.1091/mbc.E02-07-0399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Iborra F.J., Kimura H., Cook P.R. The functional organization of mitochondrial genomes in human cells. BMC Biol. 2004;2:9. doi: 10.1186/1741-7007-2-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lewis S.C., Uchiyama L.F., Nunnari J. ER-mitochondria contacts couple mtDNA synthesis with mitochondrial division in human cells. Science. 2016;353(6296) doi: 10.1126/science.aaf5549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ban-Ishihara R., Ishihara T., Sasaki N., Mihara K., Ishihara N. Dynamics of nucleoid structure regulated by mitochondrial fission contributes to cristae reformation and release of cytochrome c. Proc Natl Acad Sci U S A. 2013;110(29):11863–11868. doi: 10.1073/pnas.1301951110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Beach J.R., Shao L., Remmert K., Li D., Betzig E., Hammer J.A., 3rd Nonmuscle myosin II isoforms coassemble in living cells. Curr Biol. 2014;24(10):1160–1166. doi: 10.1016/j.cub.2014.03.071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Macias M.P., Gonzales A.M., Siniard A.L., Walker A.W., Corneveaux J.J., Huentelman M.J. A cellular model of amyloid precursor protein processing and amyloid-beta peptide production. J Neurosci Methods. 2014;223:114–122. doi: 10.1016/j.jneumeth.2013.11.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lek M., Karczewski K.J., Minikel E.V., Samocha K.E., Banks E., Fennell T. Analysis of protein-coding genetic variation in 60,706 humans. Nature. 2016;536(7616):285–291. doi: 10.1038/nature19057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Twig G., Elorza A., Molina A.J., Mohamed H., Wikstrom J.D., Walzer G. Fission and selective fusion govern mitochondrial segregation and elimination by autophagy. EMBO J. 2008;27(2):433–446. doi: 10.1038/sj.emboj.7601963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ashrafi G., Schlehe J.S., LaVoie M.J., Schwarz T.L. Mitophagy of damaged mitochondria occurs locally in distal neuronal axons and requires PINK1 and Parkin. J Cell Biol. 2014;206(5):655–670. doi: 10.1083/jcb.201401070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Maday S., Wallace K.E., Holzbaur E.L. Autophagosomes initiate distally and mature during transport toward the cell soma in primary neurons. J Cell Biol. 2012;196(4):407–417. doi: 10.1083/jcb.201106120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.He J., Mao C.C., Reyes A., Sembongi H., Di Re M., Granycome C. The AAA+ protein ATAD3 has displacement loop binding properties and is involved in mitochondrial nucleoid organization. J Cell Biol. 2007;176(2):141–146. doi: 10.1083/jcb.200609158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Fukuoh A., Cannino G., Gerards M., Buckley S., Kazancioglu S., Scialo F. Screen for mitochondrial DNA copy number maintenance genes reveals essential role for ATP synthase. Mol Syst Biol. 2014;10:734. doi: 10.15252/msb.20145117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Cassereau J., Codron P., Funalot B. Inherited peripheral neuropathies due to mitochondrial disorders. Rev Neurol (Paris) 2014;170(5):366–374. doi: 10.1016/j.neurol.2013.11.005. [DOI] [PubMed] [Google Scholar]
- 53.Canta A., Pozzi E., Carozzi V.A. Mitochondrial dysfunction in chemotherapy-induced peripheral neuropathy (CIPN) Toxics. 2015;3(2):198–223. doi: 10.3390/toxics3020198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Bobylev I., Joshi A.R., Barham M., Neiss W.F., Lehmann H.C. Depletion of Mitofusin-2 causes mitochondrial damage in cisplatin-induced neuropathy. Mol Neurobiol. 2018;55(2):1227–1235. doi: 10.1007/s12035-016-0364-7. [DOI] [PubMed] [Google Scholar]
- 55.Detmer S.A., Chan D.C. Complementation between mouse Mfn1 and Mfn2 protects mitochondrial fusion defects caused by CMT2A disease mutations. J Cell Biol. 2007;176(4):405–414. doi: 10.1083/jcb.200611080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Kneussel M., Wagner W. Myosin motors at neuronal synapses: drivers of membrane transport and actin dynamics. Nat Rev Neurosci. 2013;14(4):233–247. doi: 10.1038/nrn3445. [DOI] [PubMed] [Google Scholar]
- 57.Boyer O., Nevo F., Plaisier E., Funalot B., Gribouval O., Benoit G. INF2 mutations in Charcot-Marie-Tooth disease with glomerulopathy. N Engl J Med. 2011;365(25):2377–2388. doi: 10.1056/NEJMoa1109122. [DOI] [PubMed] [Google Scholar]
- 58.Koch J., Feichtinger R.G., Freisinger P., Pies M., Schrodl F., Iuso A. Disturbed mitochondrial and peroxisomal dynamics due to loss of MFF causes Leigh-like encephalopathy, optic atrophy and peripheral neuropathy. J Med Genet. 2016;53(4):270–278. doi: 10.1136/jmedgenet-2015-103500. [DOI] [PubMed] [Google Scholar]
- 59.Korobova F., Ramabhadran V., Higgs H.N. An actin-dependent step in mitochondrial fission mediated by the ER-associated formin INF2. Science. 2013;339(6118):464–467. doi: 10.1126/science.1228360. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
DNA oligos used for CRISPR editing of nmy-1 gene in C. elegans. gRNA targeting sequences are underlined and mutations highlighted.
Detailed clinical features of patients with the R941L mutation.
Peripheral hyper-connected mitochondria are resistant to phototoxicity –induced fission in R941L patient fibroblasts. Video corresponding to static images in Fig. 3b. Imaged using Olympus SD-OSR microscope.
NMIIC-GFP puncta colocalize to mitochondria at sites of fission. Video corresponding to static images in Fig. 5c. Imaged using a Zeiss LSM microscope.
NMIIC-GFP puncta at fission sites precede Drp1 recruitment. Video corresponding to static images in Fig. 5d. Imaged using a Zeiss LSM microscope.
NMIIC-GFP puncta at fission sites precede Drp1 recruitment. Imaged using Olympus SD-OSR microscope.
Supplemental Figure 1 Evidence for CpG methylation at c.2822G of the MYH14 gene. To verify the presence of a methylated CpG site experimentally, we devised an assay based on the methylation selectivity of the restriction enzyme FauI. a Schematic depicting the experimental strategy, where unmethylated genomic DNA is cleaved by the enzyme, such that the region cannot be PCR amplified. b Digestion with methyl sensitive enzyme shows methylation in whole blood extracted genomic DNA. As a control, DNA lacking methylation marks (i.e., PCR amplified prior to treatment) was digested by FauI, indicating that the enzyme was actively able to cleave the DNA. Meanwhile, the genomic DNA samples extracted from whole blood were readily amplified following incubation with FauI prior to PCR, indicating their resistance to digestion by the enzyme. This result was observed across a broad cross section of genomic DNA samples (data not shown), and is consistent with previous reports that the c.2822G>T mutation occurs at a methylated CpG site.
Supplemental Figure 2 Mitochondrial Morphology in Control Fibroblasts. Quantification of mitochondrial morphology from three different control fibroblast lines. At least 100 cells were quantified in three independent experimental replicates. Error bars indicate standard deviations. No significant differences were observed between the three lines, as determined by Student’s T-test compared to the number of cells in control. Control 1 was used for subsequent fibroblast cell analysis.
Supplemental Figure 3 MYH14 variants reported in the The Exome Aggregation Consortium (ExAC). Alignment of various alleles that correspond to the methylated CpG site examined in this study, as reported in ExAC, an aggregation of exome sequencing data from a variety of large-scale sequencing projects. Notably, the c.2822G>T change leading to the R941L mutation is not in ExAC.