Abstract
Background
Perihilar cholangiocarcinoma (PHCC) is the most common type of cholangiocarcinoma with the worst prognosis. Radical resection of PHCC is difficult; thus, few effective biomarkers or useful molecular profiles for PHCC have been reported in recent years. Therefore, in this study, we aimed to assess biomarkers for PHCC.
Methods
We screened potential biomarkers for PHCC using exome and transcriptome sequencing with PHCC tissues and paired normal tissues. Transcription factor 7 (TCF7) expression was evaluated using quantitative reverse transcription polymerase chain reaction, western blotting, and immunohistochemistry. The correlations between TCF7 and clinicopathological factors were analyzed with Chi-square test, and the prognostic significance of TCF7 was evaluated with univariate and multivariate analyses. The functions of TCF7 and its main effectors in PHCC cells were investigated in vitro and in vivo.
Findings
TCF7 expression was upregulated in PHCC and was an unfavorable prognostic biomarker. c-Myc was a main effector of TCF7 in PHCC cells and modulated TCF7-induced proliferation, invasion, and migration. FOS-like antigen 1 (FOSL1) was identified as a downstream target of TCF7 and was required in TCF7-induced PHCC proliferation. Triple-positive expression of TCF7, c-Myc, and FOSL1 predicted a much worse prognosis in patients with PHCC than TCF7 expression alone.
Interpretation
Postoperative detection of TCF7, c-Myc, and FOSL1 may be useful for stratifying patients with a high risk of unfavorable prognosis, and suppressing TCF7 or its downstream effectors may be a promising strategy for the treatment of PHCC.
Keywords: Transcription factor 7, Perihilar cholangiocarcinoma, C-Myc, FOS-like antigen 1, Prognosis, Progression
Abbreviations: CCA, Cholangiocarcinoma; PHCC, Perihilar cholangiocarcinoma; IHCC, Intrahepatic cholangiocarcinoma; DCC, Distal cholangiocarcinoma; AJCC/UICC, American joint committee on cancer/Union for International Cancer Control; TCF7, Transcription factor 7; FOSL1, FOS-like antigen 1; CCND1, Cyclin D1; MMP7, Matrix metalloproteinase 7; PPAR, Peroxisome proliferating activation receptor; CD44, Cluster of differentiation 44; AXIN2, Axis inhibition protein 2; MMP26, Matrix metalloproteinase 26; CDX1, Caudal type homeobox 1; CDX4, Caudal type homeobox 4; GJA1, Gap junction alpha-1; GJB6, Gap junction beta-6; BIRC5, Baculoviral IAP repeat containing 5; SFRP1, Secreted frizzled related protein 1; SOX2, SRY-box 2; COX2, Cyclooxygenase 2; CCN1, Cysteine rich 61; ID2, Inhibitor of DNA binding 2; HATH1, Human atonal homolog 1; TWIST1, TWIST family BHLH transcription factor 1; FOXN1, Forkhead box N1; SALL4, Spalt like transcription factor 4; TCF4, Transcription factor 4; IRX3, Iroquois Homeobox 3; SOX9, SRY-box 9; PTTG1, Pituitary tumor-transforming 1; RUNX2, Runt related transcription factor 2; FGF3, Fibroblast growth factor 3; FGF4, Fibroblast growth factor 4; FGF9, Fibroblast growth factor 9; CDH1, Cadherin 1; AP-1, Activating protein-1; TMA, Tissue microarray; OS, Overall survival rate; FBS, Fetal bovine serum; PBS, Phosphate buffer saline; IHC, Immunohistochemistry; SDS-PAGE, Sodium dodecyl sulfate polyacrylamide gel electrophoresis; PVDF, Polyvinylidene fluoride; qRT-PCR, Quantitative real-time PCR; CCK-8, Cell counting kit-8; KRAS, Kirsten rat sarcoma viral oncogene; MAPK, Mitogen-activated protein kinase; ERK, Extracellular regulated protein kinases
Research in context.
Evidence before this study
Perihilar cholangiocarcinoma (PHCC) is the most common type of cholangiocarcinoma with the worst prognosis. Most patients with PHCC are diagnosed with unresectable tumors because of the silent symptoms. Even for the patients who can undergo radical resection, the 5 year overall survival rate of PHCC is only approximately 30%, owing in part to a lack of effective adjuvant therapies. Indeed, the effects of chemotherapy and radiotherapy to PHCC are limited, and there are no targeted drugs currently available. Thus, new biomarkers or target therapies are urgently needed to improve outcomes in patients with PHCC.
Added value of this study
We found TCF7 expression was upregulated in PHCC with exome and transcriptome sequencing, and identified TCF7 as an unfavorable prognostic biomarker in two independent retrospective cohorts of PHCC. With in vitro and in vivo experiments, we showed that c-Myc was a main effector of TCF7 in PHCC cells and responsible for TCF7-induced proliferation, invasion, and migration of PHCC cells. With qRT-PCR screening, FOSL1 was also identified as another downstream target gene of TCF7 and was required in TCF7-induced PHCC proliferation. In addition, patients with triple-positive expression of TCF7, c-Myc, and FOSL1 were demonstrated to have a much worse prognosis than patients with TCF7 overexpression alone.
Implications of all the available evidence
TCF7 promoted progression and led to poor prognosis in PHCC by elevating expression of c-Myc and FOSL1. Postoperative detection of TCF7, c-Myc, and FOSL1 may be useful for stratifying patients with a high risk of unfavorable prognosis and that suppressing TCF7 or its downstream effectors may be a promising strategy for the treatment of PHCC.
Alt-text: Unlabelled Box
1. Introduction
Cholangiocarcinoma (CCA) is a type of epithelial cancer arising from the biliary tree [1]. In the eighth edition of American Joint Committee on Cancer/Union for International Cancer Control TNM staging classification, CCA is classified as intrahepatic cholangiocarcinoma (IHCC), perihilar cholangiocarcinoma (PHCC), and distal cholangiocarcinoma (DCC) according to the anatomical location. These subtypes of CCA have distinct risk factors, molecular pathogenesis, therapeutic options, and prognoses [2]. PHCC accounts for >50% of all CCA [3] and is characterized by late diagnosis and dismal poor outcomes. Most patients with PHCC are diagnosed with unresectable tumors because of the silent symptoms. Even for patients who can undergo radical resection, the 5-year overall survival rate of PHCC is only approximately 30% [4], owing in part to a lack of effective adjuvant therapies. Indeed, the effects of chemotherapy and radiotherapy on PHCC are limited, and there are no targeted drugs currently available. Thus, new biomarkers or target therapies are urgently needed to improve the outcomes in patients with PHCC.
The Wnt signaling pathway is a highly conserved molecular mechanism involved in many fundamental cellular processes, including cell development, proliferation, and differentiation [5]. Ectopic activation of Wnt signaling is observed in many types of cancers [6,7], and a recent study showed that Wnt signaling promotes the growth of IHCC [8]. In canonical Wnt signaling, the transcription factor (TCF)/β-catenin complex is the key effector determining the target genes downstream of Wnt [9]. TCF7, one of four members in the TCF family [10], binds to nuclear β-catenin to activate target gene transcription in response to Wnt signaling [11]. The expression and function of TCFs are tissue- and context-specific, constituting a precise and complex network downstream of the Wnt signal. However, the expressions and functions of TCFs in PHCC have not been elucidated.
In 2007, PHCC was first defined as an independent subtype of CCA, and emerging evidence has demonstrated that PHCC exhibits biological features different from those of IHCC and DCC, suggesting that IHCC, PHCC, and DCC should be investigated and treated separately [12]. However, fewer studies have evaluated the progression and prognosis of PHCC compared with that of IHCC, primarily because radical surgery and specimen collection are more difficult for PHCC [13]. Recently, high-throughput sequencing has revealed the molecular profiles of IHCC in several independent lines, and this approach has been shown to be useful for stratifying patients more precisely and guiding individual treatment of IHCC [14]. However, such results have not been reported for PHCC.
Accordingly, in this study, we performed exome and transcriptome sequencing with three pairs of PHCC tissues and their corresponding tumor adjacent tissues. We then evaluated the expression of TCF7 in two independent retrospective cohorts of PHCC. In addition, the function of TCF7 in PHCC progression was investigated using in vitro and in vivo experiments. We further screened and identified target genes of TCF7 responsible for TCF7-induced PHCC progression.
2. Materials and methods
2.1. Cells and reagents
The human PHCC cell lines QBC-939 and FRH-0201, IHCC cell line RBE, biliary epithelial cell line HIBEpiC, gallbladder carcinoma cell lines GBC-SD and NOZ, and hepatic carcinoma cell line HepG2 were purchased from the Cell Bank of the Chinese Academy of Sciences (Shanghai, China). The IHCC cell line HCCC-9810 was purchased from American Type Culture Collection (Manassas, VA, USA). All cell lines were maintained in Dulbecco's modified Eagle's medium (Gibco, Grand Island, NY, USA), except HIBEpiC cells, which were cultured in RPMI-1640 (Gibco), supplemented with 10% fetal bovine serum (Lonsera, Uruguay) and penicillin/streptomycin (HyClone, Logan, UT, USA). All cell lines were authenticated using short tandem repeat analysis and the databases of the Chinese Academy of Sciences or American Type Culture Collection as references. Anti-TCF7 antibodies were obtained from Cell Signal Technology (Danvers, MA, USA; cat. no. 2203S). Anti-c-Myc antibodies (cat. no. ab32072), anti-FOS-like antigen 1 (FOSL1) antibodies (cat. no. ab232745), anti-cyclin D1 (CCND1) antibodies (cat. no. ab16663), anti-matrix metalloproteinase (MMP) 7 antibodies (cat. no. ab5706), and anti-LaminB1 antibodies (cat. no. ab16048) were purchased from Abcam (Cambridge, UK). The Wnt signal agonist Wnt-1 was purchased from Selleck Chemicals (Houston, TX, USA; cat. no. S8178). All other agents were from Sigma-Aldrich (St. Louis, MO, USA).
2.2. Retrospective cohorts and follow-up
In total, 338 patients were diagnosed with PHCC at Qilu Hospital of Shandong University and underwent surgical resection from 2010 to 2017, constituting primary cohort 1. Validation cohort 1 consisted of 103 patients who were selected from the primary cohort based on the following criteria: (i) patients underwent radical resection with clear surgical margins; (ii) available formalin-fixed tumor tissues, follow-ups, and complete medical records; (iii) postoperational survival time >1 month; and (iv) no history of other malignancies. Validation cohort 2 comprised 57 patients with PHCC who were selected from 223 patients who underwent PHCC surgery at Taizhou Hospital of Zhejiang Province from 2011 to 2016 according to the same criteria. All samples were obtained with prior consent of patients, and the study was approved by the Ethics Committee of Shandong University.
2.3. Tissue microarray (TMA)
Representative paraffin-embedded sections of PHCC were used for TMA construction and immunohistochemical analysis [15]. Before immunohistochemical analysis, hematoxylin and eosin staining was performed to confirm the histological features of all samples. For TMA construction, core biopsies measuring 1.5 mm in diameter were taken from each sample and arranged into TMA slides.
2.4. Immunohistochemistry (IHC) and scoring
IHC was performed with TMA slides according to previous studies [15,16]. Stained TMAs were screened using a TMA scanner (Pannoramic MIDI; 3D HISTECH, Budapest, Hungary), and the IHC results were quantified with Quant center software. The tumor area was selected by a senior pathologist and evaluated by Quant Center software to stratify the staining intensity as weak, moderate, or strong and to calculate the area of each staining. The IHC score was also generated by Quant Center software and defined as follows: IHC score = (percentage of cells of weak intensity × 1) + (percentage of cells of moderate intensity × 2) + (percentage of cells of strong intensity × 3), according to previous studies [17,18]. Cut-offs were used to divide the cohort into different groups according to the IHC score [19]. The cut-offs were identified as the point with the highest sum of specificity and sensitivity in receiver operating characteristic curves.
2.5. Tumor xenograft model
All animal experiments were approved by the Medical Ethics Committee of Shandong University. Female BALB/c nude mice (5–6 weeks old, 16–18 g) were purchased from GemPharmatech Company (Nanjing, China). Mice were randomly divided into three groups (n = 6/group). Stable clones of QBC939 cells (5 × 106 cells), transfected with shcontrol, shTCF7–1, or shTCF7–2, were subcutaneously injected into the right flanks of nude mice. Tumor diameters were measured with an external caliper every 3 days. Tumor volume was calculated according to the formula V = (L × W2)/2, where V is the volume (mm3), L is the length (mm), and W is the width (mm). Similar methods were used to establish xenografts with FOSL1 knockdown. Wnt agonist 1 (3.5 mg/kg) was injected daily via the tail vein from day 3 after tumor cell injection.
2.6. Statistical analysis
SPSS 17.0 and GraphPad Prism 5.0 software were used to perform statistical analyses. The correlations between TCF7 and clinicopathological factors were assessed by χ2 test. Survival curves were plotted using the Kaplan-Meier method and compared using the log-rank test. The independent prognostic significance of clinicopathological factors was analyzed in multivariate analysis with the Cox proportional hazards regression model. Student's t-tests were used for comparisons of two independent groups. One- or two-way analysis of variance was applied to compare statistical differences between groups. Results with P values of <0.05 were considered significant.
3. Results
3.1. Expression of TCF7 in PHCC and normal common bile duct tissues
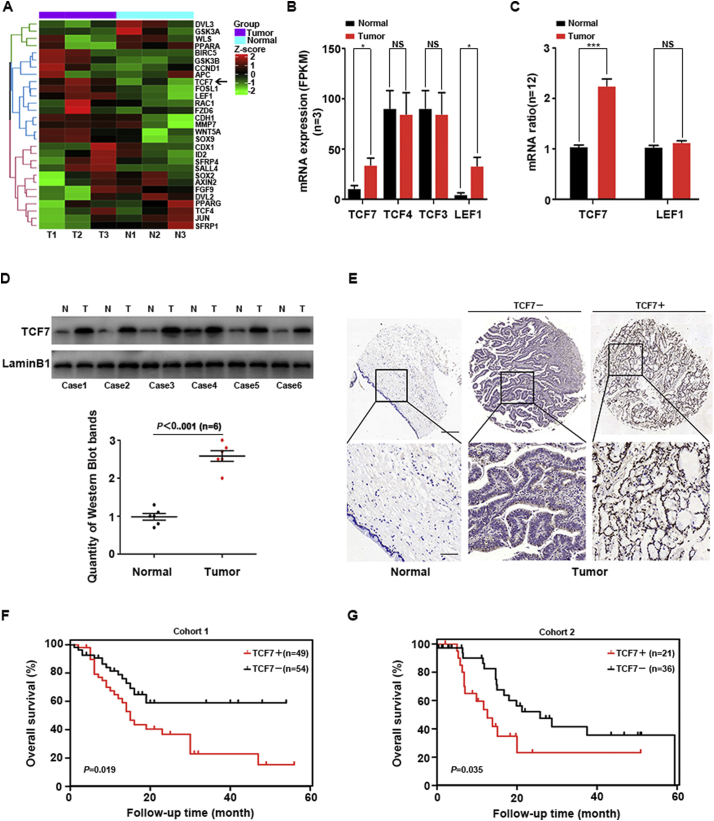
Three pairs of PHCC tissues and patient-matched normal common bile duct tissues were used for exome and transcriptome sequencing. The results were uploaded in NCBI SRA database (accession no. PRJNA517030), and SNP data of these PHCC tissues were detailed in Supplemental Table 1. Wnt signaling related genes in transcriptome sequencing were displayed in Fig. 1A. The mRNA level in transcriptome sequencing of TCF family(TCF7, TCF3, TCF4 and LEF1) were evaluated with FPKM (Fig. 1B), showing that TCF7 and LEF1 expression was substantially upregulated in PHCC tissues. Quantitative reverse transcription polymerase chain reaction (qRT-PCR) of TCF7 and LEF1 in 12 pairs of PHCC and corresponding normal bile duct tissues further confirmed the up-regulation of TCF7 but not LEF1 in PHCC (Fig. 1C). In addition, the expression of TCF7 in another six PHCC samples were detected with western blotting (Fig. 1D). All above results suggested a possible oncogenic role of TCF7 in PHCC.
Fig. 1.
Expression and prognostic significance of TCF7 in PHCC. (A) Heatmap and hierarchical clustering analysis revealed the different Wnt-related gene expression profiles in three pairs of PHCC tissues (T1, T2, and T3) and adjacent normal tissues (N1, N2, and N3). High and low expression levels are indicated in red and blue, respectively. (B) mRNA levels of TCF family were determined by mRNA sequencing in FPKM method. (C) qRT-PCR was performed to detect TCF7 and LEF1 mRNA levels in PHCC tissues and adjacent normal tissues. (D) Western blotting was performed to compare TCF7 expression in PHCC tissues and adjacent normal tissues. (E) Representative images of immunohistochemical staining for TCF7 in PHCC and normal bile duct specimens in the tissue microarray (top, 100× magnification; bottom, 400× magnification). (F) IHC scores for TCF7 in 39 pairs of PHCCs and adjacent normal tissues. (G and H) Overall survival curves for patients with PHCC in cohort 1 (G) and cohort 2 (H) were stratified according to TCF7 expression (Kaplan-Meier method). Patients with high TCF7 expression had significantly poorer overall survival rates than those with low TCF7 expression. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
To evaluate the prognostic value of TCF7, we detected the expression of TCF7 with IHC in two retrospective cohorts of PHCC patients who underwent radical resection, consisted of 103 and 57 patients respectively(Fig. 1E). By evaluating the IHC scores, and these cohorts were divided into subgroups with low or high expression of TCF7. In both cohorts, patients with high expression of TCF7 had significantly lower survival rates than patients with low TCF7 expression, suggesting that TCF7 was a prognostic biomarker of PHCC (Fig. 1G and H).
3.2. Clinical significance of TCF7
The correlations between TCF7 and clinicopathological factors in these two cohorts were then analyzed. TCF7 expression was significantly associated with tumor differentiation in cohort 1 (Supplemental Table 2) and correlated with tumor size and lymphatic invasion (N stage) in cohort 2 (Supplemental Table 3). Patients with high TCF7 expression tended to have poor differentiation, larger tumor size, and positive lymphatic invasion, indicating that TCF7 may be involved in the progression of PHCC.
The prognostic value of TCF7 and other clinicopathological factors was evaluated with univariate and multivariate analyses (Table 1). Univariate analysis demonstrated that high TCF7 expression, positive lymphatic invasion, and advanced TNM stage were both correlated with unfavorable prognosis in cohort 1. In cohort 2, larger tumor size, advanced T stage, and TNM stage were associated with poor prognosis in addition to TCF7 expression (Table 1). Prognostic factors with P values of <0.030 in univariate analysis were subjected to Cox-regression models for multivariate analysis, except for TNM stage. TCF7 was identified as an independent prognostic biomarker in cohort 1 (P = 0.024). In cohort 2, the prognostic significance of TCF7 as an independent prognostic biomarker was not statistically significant (P = 0.087).
Table 1.
The prognostic significance of TCF7 and clinicopathological factors in PHCC.
| Clinicopathologic factors | Cohort 1 |
Cohort 2 |
||||||
|---|---|---|---|---|---|---|---|---|
| Univariate analysis |
Multivariate analysis |
Univariate analysis |
Multivariate analysis |
|||||
| 3-year OS | Pa | HR | Pb | 3-year OS | Pa | HR | Pb | |
| Age (years) | ||||||||
| <65 | 44.3 | 0.055 | 1 | 38.8 | 0.22 | 1 | ||
| ≥65 | 25.0 | 1.84 | 0.070 | 22.5 | 1.46 | 0.510 | ||
| Gender | ||||||||
| Male | 37.4 | 0.638 | 44.4 | 0.338 | ||||
| Female | 36.0 | 13.9 | ||||||
| Tumor size | ||||||||
| <3 cm | 42.2 | 0.237 | 1 | 50.3 | 0.072 | 1 | ||
| ≥3 cm | 29.6 | 1.16 | 0.632 | 14.2 | 1.69 | 0.250 | ||
| Differentiation | ||||||||
| Well | 38.9 | 0.421 | 66.7 | 0.354 | ||||
| Moderate/Poor | 35.4 | 31.2 | ||||||
| T stage | ||||||||
| T1 + T2 | 37.1 | 0.244 | 1 | 45.0 | 0.01 | 1 | ||
| T3 + T4 | 26.3 | 0.56 | 0.310 | 16.2 | 3.05 | 0.163 | ||
| N stage | ||||||||
| N0 | 46.2 | 0.031 | 1 | 36.0 | 0.357 | |||
| N1/N2 | 21.6 | 1.56 | 0.188 | 32.4 | ||||
| M stage | ||||||||
| M0 | 34.7 | 0.951 | 36.2 | <0.001 | 1 | |||
| M1 | 50.0 | 0 | 4.11 | 0.069 | ||||
| TNM stage | ||||||||
| I + II | 36.4 | 0.014 | 48.5 | 0.014 | ||||
| III + IV | 30.1 | 18.1 | ||||||
| Neural invasion | ||||||||
| N | 41.1 | 0.147 | 1 | 41.3 | 0.109 | 1 | ||
| P | 30.8 | 1.72 | 0.146 | 23.1 | 0.52 | 0.421 | ||
| Hepatolith | ||||||||
| N | 40 | 0.105 | 1 | 35.4 | 0.641 | |||
| P | 0 | 2.65 | 0.083 | 25.0 | ||||
| TCF7 | ||||||||
| Low | 59.0 | 0.019 | 1 | 41.6 | 0.035 | 1 | ||
| High | 23.0 | 2.06 | 0.024 | 23.2 | 1.70 | 0.087 | ||
Abbreviations:OS = overall survival; HR = hazard ratio;95%CI = 95% confidence interval;TCF7 = Transcription Factor 7;PHCC = perihilar cholangiocarcinoma.
Calculated by log-rank test.
Calculated by Cox-regression Hazard model.
3.3. TCF7 promoted the proliferation and invasion of PHCC cells
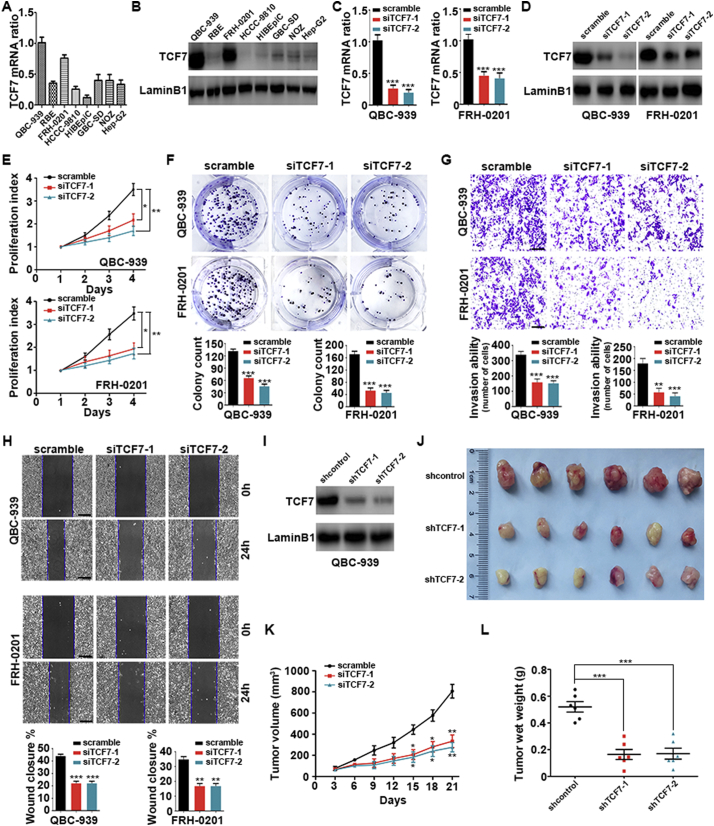
Next, we aimed to further verify the roles of TCF7 in PHCC progression in vitro. The expression of TCF7 was detected in a series of cell lines, including the PHCC cell lines QBC939 and FRH0201, IHCC cell lines RBE and HCCC-9810, gallbladder carcinoma cell lines GBC-SD and NOZ, normal biliary epithelium cell line HIBEpiC, and hepatocellular carcinoma cell line HepG2 using qRT-PCR and western blotting (Fig. 2A and B). Intriguingly, TCF7 expression was upregulated in PHCC cell lines compared with that in IHCC or gallbladder carcinoma cell lines. TCF7 expression was silenced with two independent siRNAs in QBC939 and FRH0201 cells, and successful knockdown was verified with qRT-PCR and western blotting (Fig. 2C and D). Both Cell Counting Kit-8 (CCK8) assays and soft agar colony formation assays indicated that TCF7 knockdown markedly impaired the proliferation of PHCC cells, suggesting an essential role of TCF7 in PHCC proliferation (Fig. 2E and F). In addition, matrigel transwell assays and wound healing assays demonstrated that TCF7 was also required for the invasion and migration of PHCC cells (Fig. 2G and H).
Fig. 2.
TCF7 knockdown inhibited the proliferation, migration, and invasion of PHCC cells. (A and B) TCF7 expression in different human cholangiocarcinoma cell lines, gallbladder carcinoma cell lines, HIBEpiC cells, and HepG2 cells were as detected with qRT-PCR (A) and western blotting (B). (C and D) Successful knockdown of TCF7 by two independent siRNAs was verified with qRT-PCR (C) and western blotting (D) in QBC-939 and FRH-0201 PHCC cells. (E and F) CCK8 assays (E) and colony formation assays (F) to determine the effects of TCF7 silencing on the proliferation of QBC-939 and FRH-0201 cells. (G) Transwell invasion assays in QBC-939 and FRH-0201 cells with TCF7 knockdown. Scale bar: 200 μm. (H) Wound healing assays in QBC939 and FRH-0201 cells with TCF7 knockdown. Data in (E–H) were from at least three independent experiments and are shown as means ± SEMs. Statistical significance was analyzed using Student's t-tests; *, **, and *** indicate P < 0.05, P < 0.01, and P < 0.001, respectively, compared with the control group transfected with scrambled siRNA. (I) Western blotting showing the successful establishment of QBC939 cell lines with stable TCF7 knockdown using the two indicated shRNAs. (J) Stable QBC939-TCF7i cells and control cells (transfected with empty vector) were injected subcutaneously into the right flanks of BALB/c nude mice (n = 6/group). Three weeks after implantation, xenograft tumors were observed in the three groups. (K) Tumor volumes of xenografts were measured every 3 days. (L) Weights of xenograft tumors after 21 days. *, **, and *** indicate P < 0.05, P < 0.01, and P < 0.001, respectively, compared with the control group transfected with empty vector. Statistical significance was analyzed with Student's t-tests.
Experiments in vivo with a xenograft model were then performed. QBC939 cells showing stable knockdown of TCF7 were generated by the infection with a lentivirus carrying two independent TCF7 shRNAs and the selection with puromycin (Fig. 2I). Cells infected with control shRNA or shRNAs targeting TCF7 were injected subcutaneously into BALB/C nude mice (Fig. 2J). Notably, the volumes and weights of xenograft tumors with TCF7 knockdown were smaller and lower, respectively, than those in the control group (Fig. 2K and L). Furthermore, we confirmed the knockdown of TCF7 in xenografts from cell stably expressing TCF7 shRNA (Supplemental Fig. 2A and 2B).
3.4. c-Myc was upregulated by TCF7 and modulated TCF7-induced PHCC progression
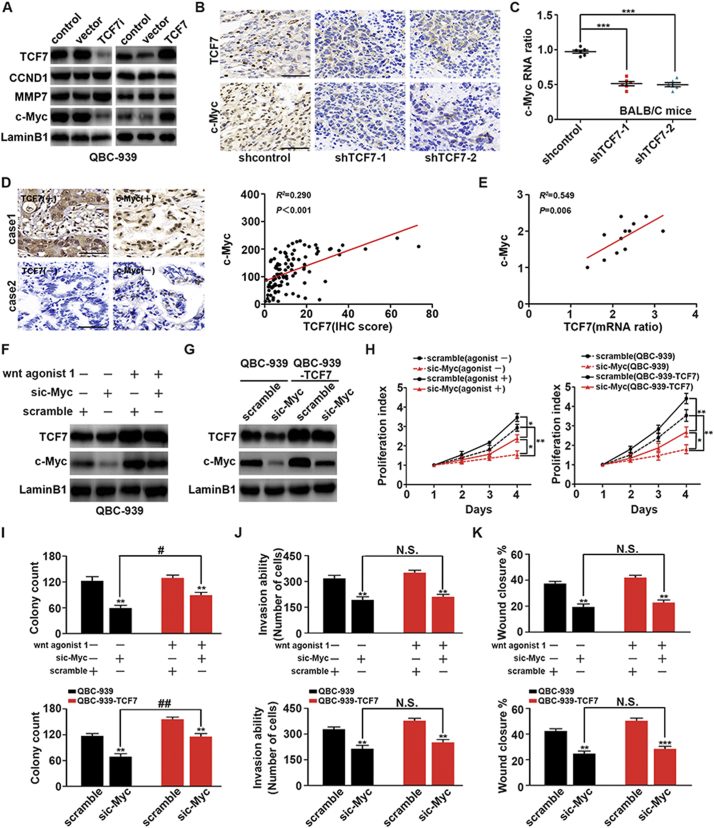
c-Myc, CCND1, and MMP7 are all well-known targets of TCF7 [20]. To explore the underlying mechanisms of TCF7-induced PHCC progression, we evaluated the changes in the expression of c-Myc, CCND1, and MMP7 after silencing or overexpressing TCF7. In QBC939 cells, c-Myc expression was upregulated along with TCF7 overexpression and downregulated when TCF7 was knocked down; in contrast, no significant changes in CCND1 or MMP7 were observed (Fig. 3A). This result indicated that c-Myc may be a main effector of TCF7 in QBC939 cells. Moreover, we analyzed the correlations between TCF7 and c-Myc expression in xenografts using qRT-PCR and IHC. The mRNA levels and IHC staining of c-Myc in xenografts with TCF7 knockdown were markedly lower than those in the control group (Fig. 3B and C). The correlations between c-Myc and TCF7 were further detected in human PHCC tissues. Consequently, IHC revealed a strong correlation between c-Myc and TCF7 expression in cohort 1 (Fig. 3D). In fresh PHCC tissues, c-Myc mRNA was also positively correlated with TCF7 mRNA (Fig. 3E). Taken together, these results indicated that c-Myc was regulated by TCF7 and that c-Myc was a main effector of TCF7 in PHCC.
Fig. 3.
c-Myc was required for TCF7-induced proliferation, migration, and invasion. (A) After silencing or overexpressing TCF7 in QBC939 cells, expression levels of c-Myc, CCND1, and MMP7 were detected by western blotting. (B) Representative c-Myc IHC staining in xenograft tumors with transfection of empty vector or shTCF7. Scale bar: 50 μm. (C) mRNA level of c-Myc in control xenografts and xenografts with TCF7 knockdown. (D) Correlation of IHC scores for TCF7 and c-Myc in PHCC tissues. (E) Association of c-Myc mRNA levels with TCF7 levels in eight fresh human PHCCs. (F) Effects of Wnt agonist 1 (20 μM, 8 h) on TCF7 and c-Myc expression in QBC939 cells. (G) Expression of c-Myc was knocked down in normal QBC939 cells or QBC939 cells overexpressing TCF7. (H) CCK8 assays showing the effects of c-Myc silencing and Wnt agonist 1 on proliferation in QBC939 cells. * and ** represent P < 0.05 and P < 0.01, respectively, between the indicated subgroups. (I) Effects of TCF7 knockdown, TCF7 overexpression, and c-Myc knockdown on colony formation ability in QBC939 cells. (J and K) Effects of c-Myc knockdown with TCF7 overexpression or Wnt agonist 1 stimulation on soft agar transwell assays (J) and wound healing assays (K) in QBC939 cells. *, **, and *** indicate P < 0.05, P < 0.01, and P < 0.001, respectively, compared with the corresponding control groups. ## represents P < 0.01 between the indicated groups. Analyzed data were from at least three independent experiments.
Wnt agonist 1 is a well-known agonist of Wnt signaling [21]. In PHCC cells, stimulation of Wnt signaling with Wnt agonist 1 promoted the expression of TCF7 and c-Myc (Fig. 3F). Moreover, TCF7-overexpressing and c-Myc-knockdown QBC939 cells were then established for functional studies (Fig. 3G). After c-Myc knockdown, PHCC cell proliferation was significantly attenuated, although cells with Wnt agonist or TCF7 overexpression still showed higher proliferation rates than the cells with only c-Myc knockdown (Fig. 3H). Similar results were observed in colony formation assays (Fig. 3I), suggesting that c-Myc was required for TCF7-induced proliferation of PHCC. Additionally, matrigel transwell assays (Fig. 3J) and wound healing assays (Fig. 3K) indicated that c-Myc was also essential for TCF7-induced invasion and migration. However, it was interesting to note that c-Myc knockdown almost completely abolished the increases in invasion and migration induced by TCF7 overexpression or Wnt agonist treatment, but the proliferation of TCF7-overexpressing or Wnt-treated cells was still increased after c-Myc knockdown. Thus, these findings suggested that another target protein of TCF7 was responsible for the TCF7-induced proliferation but not invasion in PHCC.
3.5. TCF7 promoted the transcription and expression of FOSL1
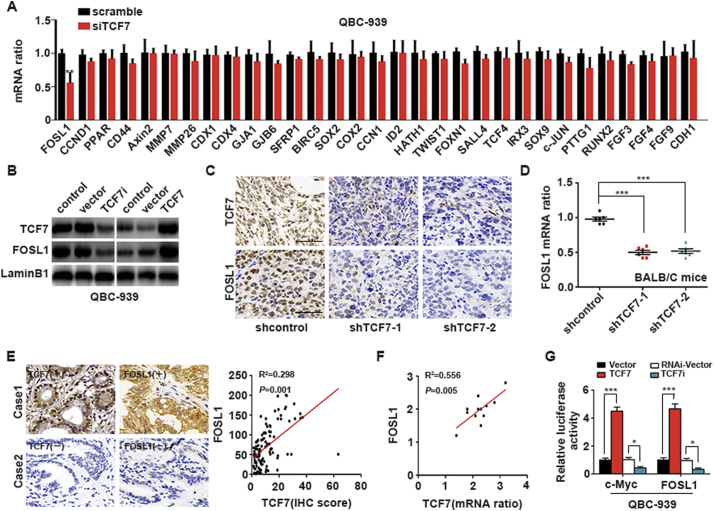
Genome-based screening has revealed that approximate 110 candidate Wnt targets are involved in various cellular processes, including the cell cycle, cell adhesion, and endocrine signaling [22]. By literature reviewing, we selected 31 proliferation-involved genes from these candidate target genes and detected their mRNA levels after TCF7 knockdown. The results showed that FOSL1 mRNA was substantially decreased when TCF7 was silenced (Fig. 4A), indicating that FOSL1 may be a potential target of TCF7 in PHCC. To further verify these findings, FOSL1 expression was evaluated in cells with TCF7 overexpression or knockdown; the results showed that FOSL1 expression was regulated by TCF7 in QBC939 cells (Fig. 4B). In addition, we further analyzed the correlations between FOSL1 and TCF7 in PHCC xenografts and patient samples with IHC and qRT-PCR and demonstrated that FOSL1 expression was decreased in xenografts when TCF7 was knocked down (Fig. 4C and D). In human formalin-fixed PHCC tissues and fresh PHCC tissues, FOSL1 expression was significantly correlated with TCF7 expression (Fig. 4E and F). These results suggested that TCF7 could regulate the expression of FOSL1. Moreover, luciferase assays demonstrated that TCF7 enhanced the expression of c-Myc and FOSL1 by promoting their transcription (Fig. 4G), suggesting that c-Myc and FOSL1 were direct target genes of TCF7 in PHCC cells.
Fig. 4.
FOSL 1 transcription was promoted by TCF7 in PHCC. (A) FOSL1 mRNA was decreased in TCF7-silenced QBC939 cells among the 31 candidate genes screened with qRT-PCR. (B) Effects of TCF7 knockdown or overexpression on FOSL1 expression. (C) Representative FOSL1 IHC staining of xenograft tumors derived from cells transfected with empty vector or shTCF7. Scale bar: 50 μm. (D) FOSL1 mRNA in xenografts transfected with shTCF7 or empty vector. (E) Association of IHC scores for FOSL1 and TCF7 in human PHCC tissues. (F) Correlations of mRNA levels of TCF7 and FOSL1 in fresh PHCC tissues. (G) Luciferase assays for analysis of the effects of TCF7 on the transcription of c-Myc and FOSL1. *, **, and *** represent P < 0.05, P < 0.01, and P < 0.001, respectively, between the indicated subgroups. Statistical significance was analyzed using Student's t-tests.
3.6. FOSL1 was required in TCF7-induced proliferation of PHCC
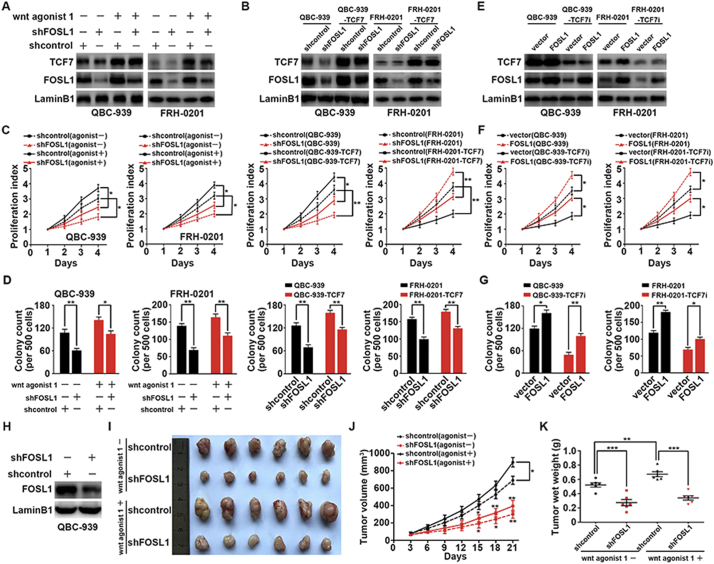
In functional assays with c-Myc knockdown, we suspected that another protein was responsible for PHCC proliferation. Accordingly, we further evaluated the influence of FOSL1 on PHCC proliferation in vitro and in vivo. In QBC939 and FRH0201 cells, FOSL1 was knocked down by shRNA, and Wnt/TCF7 signaling was activated by Wnt agonist 1 or TCF7 overexpression (Fig. 5A and B). Both CCK8 assays and colony formation assays showed that FOSL1 knockdown markedly attenuated the proliferation of PHCC cells (Fig. 5C and D). To confirm the role of FOSL1 in TCF7-induced proliferation, we performed a FOLS1 rescue assay by overexpressing FOSL1 in QBC939 and FRH0201 cells after knocking down TCF7 expression (Fig. 5E). As a result, both CCK8 and soft agar assays revealed that FOSL1 overexpression could rescue the reduced proliferation induced by TCF7 knockdown in QBC939 and FRH0201 cells to some extent (Fig. 5F and G). Stable FOSL1-silenced QBC939 cells were established by infection with lentivirus and verified with western blotting (Fig. 5H). Xenografts were established in nude mice using stable FOSL1-silenced cells or stable cells transfected with control shRNA, in the presence or absence of Wnt agonist 1 (Fig. 5I). The volumes and weights of xenografts were decreased when FOSL1 was knocked down (Fig. 5J and K), revealing that FOSL1 was required for PHCC cell proliferation in vivo.
Fig. 5.
FOSL1 was required for TCF7-induced proliferation of PHCC. (A) Effects of Wnt agonist 1 on the expression of TCF7 and FOSL1 in QBC939 and FRH0201 cells. (B) FOSL1 knockdown in QBC939/FRH0201 cells or stable QBC939/FRH0201 cell overexpressing TCF7. (C) CCK8 assays showing the effects of FOSL1, Wnt agonist 1, and TCF7 on the proliferation of QBC939 and FRH0201 cells. (D) Effects of FOSL1 knockdown on the proliferation of QBC939 and FRH0201 cells in soft agar colony assays. (E) FOSL1 was overexpressed by lentivirus infection of FOSL1 plasmid after silencing TCF7 in QBC939 and FRH0201 cells. (F and G) CCK8 (F) and soft agar (G) assays revealed that FOLS1 overexpression rescued the reduced proliferation induced by TCF7 knockdown. (H) Establishment of a stable QBC939 cell line with FOSL1 knockdown using a lentivirus carrying FOSL1 shRNA. (I) Xenografts from nude mice were generated with or without FOSL1 knockdown. Mice were intravenously injected in presence or absence of Wnt agonist 1 at 3.5 mg/kg. (J and K) FOSL1 knockdown decreased the tumor volume and weight of xenografts.
3.7. Triple upregulation of TCF7/FOSL1/c-Myc was a sensitive marker of poor prognosis
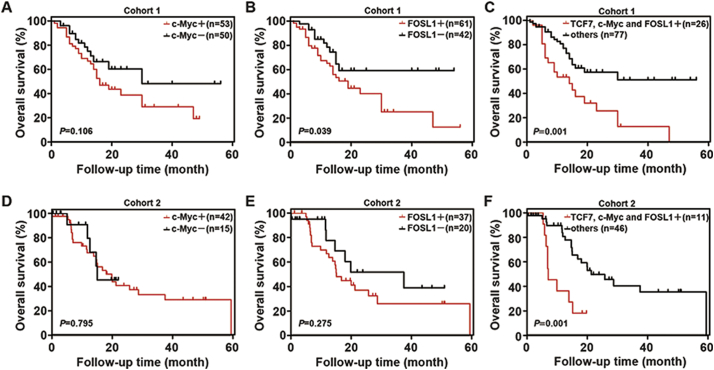
Our above results showed that TCF7 was a prognostic biomarker of PHCC and that high expression of TCF7 predicted poor prognosis (Fig. 1F and G). However, high expression of c-Myc or FOSL1 was not significantly associated with low survival rates in cohort 1 (Fig. 6A and B). Because TCF7 promoted the progression of PHCC by inducing expression of c-Myc or FOSL1 independently, we compared survival rates in patients with triple upregulation of TCF7/FOSL1/c-Myc and other patients. The results showed that patients with triple upregulation of TCF7/c-Myc/FOSL1 had much worse prognoses than other patients (Fig. 6C). Similar results were also confirmed in cohort 2 (Fig. 6D–F). These results suggested that triple overexpression of TCF7/c-Myc/FOSL1 was a more sensitive prognostic marker and was more effective for predicting prognosis than detection of TCF7 alone in patients with PHCC.
Fig. 6.
Triple upregulation of TCF7/c-Myc/FOSL1 was a sensitive marker predicting unfavorable prognosis. (A–C) In cohort 1, the overall survival curves were stratified according to c-Myc (A) and FOSL1 (B) expression or triple overexpression of TCF7/c-Myc/FOSL1 (C). (D–F) In cohort 2, the overall survival curves were stratified according to c-Myc (D) and FOSL1 (E) expression or triple overexpression of TCF7/c-Myc/FOSL1 (F). Patients with high expression of TCF7 had poor prognoses, while patients with triple upregulation of TCF7/c-Myc/FOSL1 had much worse prognoses than other patients in both cohort 1 and cohort 2 (P < 0.001 and P = 0.001).
4. Discussion
Patients with PHCC usually have poor prognoses partially owing to the lack of effective adjuvant therapy for PHCC. The benefits of traditional chemotherapy or radiotherapy in patients with PHCC are limited, and there are no targeted drugs approved for the treatment of PHCC to date, even though high-throughput sequencing has revealed new therapeutic targets for CCA and has led to the development of several small-molecular inhibitors [23]. The development of new drug targets and drug therapies is typically based on the identification of novel biomarkers, which largely relies on cohort studies including large numbers of patients. Unfortunately, most patients with PHCC are not surgical candidates and only undergo palliative resection owing to a lack of detection of early symptoms and the surgical complexity of PHCC, increasing the difficulty of obtaining specimens and establishing the cohorts [24]. In our study, we enrolled patients with PHCC who underwent radical resection at two medical centers and confirmed TCF7 as a prognostic biomarker in these two independent cohorts. To the best of our knowledge, this is the first verification of prognostic biomarkers in two independent PHCC cohorts with radical resection. Moreover, we further demonstrated that patients with triple-positive expression of TCF7, c-Myc, and FOSL1 had a significantly worse prognosis than other patients in these two cohorts, suggesting that triple-positive expression of TCF7/c-Myc/FOSL1 was a much more sensitive factor for predicting unfavorable prognosis than TCF7 alone. Thus, postoperative detection of TCF7, c-Myc, and FOSL1 may be a helpful approach to stratify patients with a high risk of poor prognosis, which is important for post-operation surveillance and precise treatment.
Aberrant activation of Wnt signaling, which is essential for cell proliferation and differentiation, is observed in many types of tumors, including IHCC [8,25]; however, the function of Wnt signaling in PHCC has not been investigated before partially because of difficulties in obtaining PHCC specimens. In our study, we demonstrated that TCF7 promoted PHCC progression and identified target genes responsible for TCF7-dependent progression. This result indicated that inhibitors blocking Wnt signaling, TCF7, or their downstream molecules may have applications as potential targeted drugs for the treatment of PHCC. Notably, several small-molecule inhibitors of Wnt signaling have been developed recently. For example, ICG001 and C59 were found to inhibit Wnt signaling in animal models without severe adverse effects [8,26]. However, further studies are needed to determine whether inhibitors of Wnt signaling can suppress PHCC progression because the Wnt signal pathway is a complex network, and other cellular compensation mechanisms may balance out the inhibitory effects of these compounds. Direct inhibition of TCF7 may be a more precise way to treat PHCC; however, no specific inhibitors of TCF7 have been reported to date.
Compared with well-known Wnt signaling effectors, such as c-Myc and CCND1, FOSL1 has not been extensively studied since it was first identified as a target of Wnt/β-catenin signal in 2007 [27]. In our study, we demonstrated for the first time that FOSL1 transcription was increased by TCF7 and that FOSL1 promoted PHCC proliferation. These findings expanded our understanding of TCF7 downstream target genes and the importance of Wnt signaling in tumor progression. In tumorigenesis, FOSL1 mainly functions as a constitutor of activating protein-1 (AP-1) transcription factors and induces the expression of potentially tumor-relevant events, thereby strongly influencing the aggressive phenotype after receiving activation signals, e.g., mitogen-activated protein kinase (MAPK)/extracellular signal-regulated kinase (ERK) [28,29]. The expression and function of FOSL1/AP-1 is context-dependent and tissue-specific [30]. Mutations in KRAS lead to constitutive activation of MAPK/ERK, an important upstream activator of FOSL1/AP-1, and FOSL1 has been reported to facilitate the progression of cancers with mutant KRAS [31]. This correlation between FOSL1 and mutant KRAS is interesting because KRAS mutations are the most frequent mutations and most extensively reported fingerprints of CCA [32]. In PHCC, Wnt signaling may show crosstalk with KRAS/ERK signaling, and FOSL1 may be a key molecule regulating that process.
In summary, we identified TCF7 as an independent prognostic biomarker of PHCC in two independent cohorts and demonstrated that TCF7 could promote the proliferation and invasion of PHCC cells by inducing c-Myc expression. FOSL1 was a target effector of Wnt/TCF7 signaling and was responsible for TCF7-induced proliferation of PHCC. Patients with triple-positive expression of TCF7, c-Myc, and FOSL1 had significantly worse prognoses than other patients. These results suggested that postoperative detection of TCF7, c-Myc, and FOSL1 may be a helpful approach for stratifying patients with high-risk of unfavorable prognosis and that suppressing TCF7 or its downstream effectors may be a promising therapy for treatment of PHCC.
Conflicts of interest
We declare no conflicts of interest.
Funding
Our study was supported by National Natural Science Foundation of China (grant no. 81601668), Shandong Province Major Research and Design Program (grant no. 2018GSF118169), Jinan City Science and Technology Development Program (grant nos. 201805017, 201805013), and Hengrui Hepatobiliary and Pancreatic Foundation (grant no. Y-2017-144). The funders did not play a role in manuscript design, data collection, data analysis, data interpretation or writing of the manuscript.
Availability of data and materials
The dataset used and/or analyzed during the current study are available from the corresponding author on reasonable request.
Authors' contributions
LZL, SRQ, ZXM, QB, and LZP contributed to the experimental procedures. ZZL and CTL collected the specimens. ZZL and XYF collected the funding. XYF designed the research and analyzed the data. ZZL supervised all the work. All authors read and approved the final manuscript.
Ethics approval and consent to participate
This study was approved by the Qilu Hospital of Shandong University. The Laboratory Animal Care and Use Committees of Qilu Hospital approved all experimental procedures. Written informed consent was obtained from all patients.
Acknowledgments
Not applicable.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.ebiom.2019.06.023.
Contributor Information
Yunfei Xu, Email: xuyunfei1988@126.com.
Zongli Zhang, Email: zzlzzl1900@163.com.
Appendix A. Supplementary data
Supplementary material 1
Supplementary material 2
References
- 1.Razumilava N., Gores G.J. Cholangiocarcinoma. Lancet. 2014;383(9935):2168–2179. doi: 10.1016/S0140-6736(13)61903-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Rizvi S., Gores G.J. Emerging molecular therapeutic targets for cholangiocarcinoma. J Hepatol. 2017;67(3):632–644. doi: 10.1016/j.jhep.2017.03.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Siriwardena A.K. Klatskin Tumor. J Clin Oncol. 2017;35(36):4091–4092. doi: 10.1200/JCO.2017.75.1586. [DOI] [PubMed] [Google Scholar]
- 4.Groot Koerkamp B., Wiggers J.K., Gonen M., Doussot A., Allen P.J., Besselink M.G. Survival after resection of perihilar cholangiocarcinoma-development and external validation of a prognostic nomogram. Ann Oncol Off J Eur Soc Med Oncol/ESMO. 2016;27(4):753. doi: 10.1093/annonc/mdw063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Clevers H., Nusse R. Wnt/beta-catenin signaling and disease. Cell. 2012;149(6):1192–1205. doi: 10.1016/j.cell.2012.05.012. [DOI] [PubMed] [Google Scholar]
- 6.Monga S.P. Beta-catenin signaling and roles in liver homeostasis, injury, and tumorigenesis. Gastroenterology. 2015;148(7):1294–1310. doi: 10.1053/j.gastro.2015.02.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Liu W., Dong X., Mai M., Seelan R.S., Taniguchi K., Krishnadath K.K. Mutations in AXIN2 cause colorectal cancer with defective mismatch repair by activating beta-catenin/TCF signalling. Nat Genet. 2000;26(2):146–147. doi: 10.1038/79859. [DOI] [PubMed] [Google Scholar]
- 8.Boulter L., Guest R.V., Kendall T.J., Wilson D.H., Wojtacha D., Robson A.J. WNT signaling drives cholangiocarcinoma growth and can be pharmacologically inhibited. J Clin Invest. 2015;125(3):1269–1285. doi: 10.1172/JCI76452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Behrens J., von Kries J.P., Kuhl M., Bruhn L., Wedlich D., Grosschedl R. Functional interaction of beta-catenin with the transcription factor LEF-1. Nature. 1996;382(6592):638–642. doi: 10.1038/382638a0. [DOI] [PubMed] [Google Scholar]
- 10.Arce L., Yokoyama N.N., Waterman M.L. Diversity of LEF/TCF action in development and disease. Oncogene. 2006;25(57):7492–7504. doi: 10.1038/sj.onc.1210056. [DOI] [PubMed] [Google Scholar]
- 11.Cadigan K.M., Waterman M.L. TCF/LEFs and Wnt signaling in the nucleus. Cold Spring Harb Perspect Biol. 2012;4(11) doi: 10.1101/cshperspect.a007906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Farges O., Fuks D., Le Treut Y.P., Azoulay D., Laurent A., Bachellier P. AJCC 7th edition of TNM staging accurately discriminates outcomes of patients with resectable intrahepatic cholangiocarcinoma: by the AFC-IHCC-2009 study group. Cancer. 2011;117(10):2170–2177. doi: 10.1002/cncr.25712. [DOI] [PubMed] [Google Scholar]
- 13.Xu Y.F., Liu Z.L., Pan C., Yang X.Q., Ning S.L., Liu H.D. HMGB1 correlates with angiogenesis and poor prognosis of perihilar cholangiocarcinoma via elevating VEGFR2 of vessel endothelium. Oncogene. 2019 Feb;38(6):868–880. doi: 10.1038/s41388-018-0485-8. [DOI] [PubMed] [Google Scholar]
- 14.Sia D., Hoshida Y., Villanueva A., Roayaie S., Ferrer J., Tabak B. Integrative molecular analysis of intrahepatic cholangiocarcinoma reveals 2 classes that have different outcomes. Gastroenterology. 2013;144(4):829–840. doi: 10.1053/j.gastro.2013.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Xu Y.F., Yang X.Q., Lu X.F., Guo S., Liu Y., Iqbal M. Fibroblast growth factor receptor 4 promotes progression and correlates to poor prognosis in cholangiocarcinoma. Biochem Biophys Res Commun. 2014;446(1):54–60. doi: 10.1016/j.bbrc.2014.02.050. [DOI] [PubMed] [Google Scholar]
- 16.Xu Y.F., Liu H.D., Liu Z.L., Pan C., Yang X.Q., Ning S.L. Sprouty2 suppresses progression and correlates to favourable prognosis of intrahepatic cholangiocarcinoma via antagonizing FGFR2 signalling. J Cell Mol Med. 2018;22(11):5596–5606. doi: 10.1111/jcmm.13833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Azim H.A., Jr., Peccatori F.A., Brohee S., Branstetter D., Loi S., Viale G. RANK-ligand (RANKL) expression in young breast cancer patients and during pregnancy. Breast Cancer Res: BCR. 2015;17:24. doi: 10.1186/s13058-015-0538-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yeo W., Chan S.L., Mo F.K., Chu C.M., Hui J.W., Tong J.H. Phase I/II study of temsirolimus for patients with unresectable hepatocellular carcinoma (HCC)- a correlative study to explore potential biomarkers for response. BMC Cancer. 2015;15:395. doi: 10.1186/s12885-015-1334-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Liu H., Xu Y., Zhang Q., Yang H., Shi W., Liu Z. Prognostic significance of TBL1XR1 in predicting liver metastasis for early stage colorectal cancer. Surg Oncol. 2017;26(1):13–20. doi: 10.1016/j.suronc.2016.12.003. [DOI] [PubMed] [Google Scholar]
- 20.Hovanes K., Li T.W., Munguia J.E., Truong T., Milovanovic T., Lawrence Marsh J. Beta-catenin-sensitive isoforms of lymphoid enhancer factor-1 are selectively expressed in colon cancer. Nat Genet. 2001;28(1):53–57. doi: 10.1038/ng0501-53. [DOI] [PubMed] [Google Scholar]
- 21.Pang Y., Liu J., Li X., Zhang Y., Zhang B., Zhang J. Nano Let7b sensitization of eliminating esophageal cancer stemlike cells is dependent on blockade of Wnt activation of symmetric division. Int J Oncol. 2017;51(4):1077–1088. doi: 10.3892/ijo.2017.4104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Vlad A., Rohrs S., Klein-Hitpass L., Muller O. The first five years of the Wnt targetome. Cell Signal. 2008;20(5):795–802. doi: 10.1016/j.cellsig.2007.10.031. [DOI] [PubMed] [Google Scholar]
- 23.Hezel A.F., Deshpande V., Zhu A.X. Genetics of biliary tract cancers and emerging targeted therapies. J Clin Oncol. 2010;28(21):3531–3540. doi: 10.1200/JCO.2009.27.4787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Xu Y.F., Liu Z.L., Pan C., Yang X.Q., Ning S.L., Liu H.D. HMGB1 correlates with angiogenesis and poor prognosis of perihilar cholangiocarcinoma via elevating VEGFR2 of vessel endothelium. Oncogene. 2019;38(6):868–880. doi: 10.1038/s41388-018-0485-8. [DOI] [PubMed] [Google Scholar]
- 25.Perugorria M.J., Olaizola P., Labiano I., Esparza-Baquer A., Marzioni M., Marin J.J.G. Wnt-beta-catenin signalling in liver development, health and disease. Nat Rev Gastroenterol Hepatol. 2019;16(2):121–136. doi: 10.1038/s41575-018-0075-9. [DOI] [PubMed] [Google Scholar]
- 26.Virshup D.M. Moving upstream in the war on WNTs. J Clin Invest. 2015;125(3):975–977. doi: 10.1172/JCI80819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Abe K., Takeichi M. NMDA-receptor activation induces calpain-mediated beta-catenin cleavages for triggering gene expression. Neuron. 2007;53(3):387–397. doi: 10.1016/j.neuron.2007.01.016. [DOI] [PubMed] [Google Scholar]
- 28.Maurus K., Hufnagel A., Geiger F., Graf S., Berking C., Heinemann A. The AP-1 transcription factor FOSL1 causes melanocyte reprogramming and transformation. Oncogene. 2017;36(36):5110–5121. doi: 10.1038/onc.2017.135. [DOI] [PubMed] [Google Scholar]
- 29.Vallejo A., Valencia K., Vicent S. All for one and FOSL1 for all: FOSL1 at the crossroads of lung and pancreatic cancer driven by mutant KRAS. Mol Cell Oncol. 2017;4(3) doi: 10.1080/23723556.2017.1314239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhang K., Myllymaki S.M., Gao P., Devarajan R., Kytola V., Nykter M. Oncogenic K-Ras upregulates ITGA6 expression via FOSL1 to induce anoikis resistance and synergizes with alphaV-class integrins to promote EMT. Oncogene. 2017;36(41):5681–5694. doi: 10.1038/onc.2017.177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Vallejo A., Perurena N., Guruceaga E., Mazur P.K., Martinez-Canarias S., Zandueta C. An integrative approach unveils FOSL1 as an oncogene vulnerability in KRAS-driven lung and pancreatic cancer. Nat Commun. 2017;8 doi: 10.1038/ncomms14294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rizvi S., Gores G.J. Pathogenesis, diagnosis, and management of cholangiocarcinoma. Gastroenterology. 2013;145(6):1215–1229. doi: 10.1053/j.gastro.2013.10.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material 1
Supplementary material 2
Data Availability Statement
The dataset used and/or analyzed during the current study are available from the corresponding author on reasonable request.