Abstract
Background
With the development of RNA-seq technology, tens of thousands of circular RNAs (circRNAs), a novel class of RNAs, have been identified. However, little is known about circRNA formation and biogenesis in hepatocellular carcinoma (HCC).
Methods
We performed ribosomal-depleted RNA-seq profiling of HCC and para-carcinoma tissues and analyzed the expression of a hotspot circRNA derived from the 3’UTR of the stearoyl-CoA desaturase (SCD) gene, termed SCD-circRNA 2.
Findings
It was significantly upregulated in HCC and correlated with poor patient prognosis. Moreover, we observed that the production of SCD-circRNA 2 was dynamically regulated by RNA-binding protein 3 (RBM3). RBM3 overexpression was indicative of a short recurrence-free survival and poor overall survival for HCC patients. Furthermore, by modulating the RBM3 or SCD-circRNA 2 levels, we found that RBM3 promoted the HCC cell proliferation in a SCD-circRNA 2 dependent manner.
Interpretation
Herein, we report that RBM3 is crucial for the SCD-circRNA 2 formation in HCC cells, which not only provides mechanistic insights into cancer-related circRNA dysregulation but also establishes RBM3 as an oncogene with both therapeutic potential and prognostic value.
Fund
This work was supported by the National Key Research and Development Program of China (2016YFC1302303), the National Natural Science Foundation of China (Grant No. 81672345 and 81,402,269). The funders did not have any roles in study design, data collection, data analysis, interpretation, writing of the report.
Keywords: Circular RNA, Hepatocellular carcinoma, Oncogene, RBM3, Stearoyl-CoA desaturase
Abbreviations: circRNA, Circular RNA; HCC, hepatocelluar carcinoma; RBM3, RNA-binding protein 3; RFS, recurrence-free survival; OS, overall survival; SCD, Stearoyl-CoA Desaturase; RIP, RNA immunoprecipitation
Research in context.
Evidence before this study
The published literature on circular RNA (circRNA) has grown exponentially over the past few years. These studies indicate that circRNAs widely exist in human normal tissues, cancerous tissues, serum and plasma. Furthermore, they have been reported to play important roles in the development of cancers, such as bladder, gastric and colon cancer. However, the role of circRNAs in hepatocellular carcinoma (HCC) was unclear.
Added value of this study
The data presented in this study show that a circRNA derived from the 3’UTR of the stearoyl-CoA desaturase (SCD) gene, termed SCD-circRNA 2, is up-regulated in human HCC and high SCD-circRNA 2 expression is positively associated with incomplete encapsulation and AFP. SCD-circRNA 2 is an independent risk factor for predicting prognosis of HCC patients. We also found that the biogenesis of SCD-circRNA 2 was promoted by RNA-binding protein 3 (RBM3). Strikingly, RBM3 expression is increased dramatically in HCC tissues, and the RBM3 level correlates positively with poor survival. In addition, the protein level of RBM3 was positively associated with the expression of SCD-circRNA 2 in human HCC tissues.
Implications of all the available evidence
Our findings demonstrate that circRNAs in HCC are regulated by cell type-specific mechanisms, suggesting they play specific biological roles in HCC. SCD-circRNA 2 can potentially serve as an indicator of disease progression and prognosis of patients with HCC.
Alt-text: Unlabelled Box
1. Introduction
In the last decade, hepatocellular carcinoma (HCC) has been one of the most common cancers, ranking sixth in cancer mortality worldwide [1]. With the progress of medical technology, the early diagnosis and surgical resection of primary HCC have significantly increased. However, according to the current diagnostic criteria, many patients are not diagnosed during early stages of the disease and are thus missing the opportunity for early resection. To improve the therapeutic efficacy and prognostic accuracy, it is important to identify more effective biomarkers and therapeutic targets for HCC to increase the early diagnosis rate and improve patient prognosis.
Circular RNA (circRNA), an emerging endogenous noncoding RNA (ncRNA), has received a lot of attention in recent years. The 3′ end of circRNA is connected with the 5′ end to yield a fully annular structure by exon cyclization and intron cyclization. Compared with microRNAs and long noncoding RNAs, circRNAs exhibit higher stability and sequence conservation among mammalian cells. Studies on circRNA have revealed that they are widely expressed in a tissue- and developmental-stage-specific patterns, and a subset displays conservation across species. Identification of circRNAs and exploration of their roles in regulating other genes have increased in recent years [[2], [3], [4], [5]]. Several possible functions of circRNA have been proposed, such as the regulation of gene transcription and protein translation through the binding of circRNA to microRNA [[6], [7], [8]] or protein [9,10]. Therefore, circRNAs may act as emerging biomarkers for diagnosing cancer and other diseases [4,11,12]. As studies on tumor circRNA have increased, two issues have aroused investigators' attention: What regulates circRNA in cancer development and progression? What is the oncogenic function of circRNA?
RNA-binding proteins (RBPs) participate in each step of RNA metabolism, including RNA splicing, polyadenylation, sequence-editing, RNA transfer, maintenance of RNA stabilization and degradation, intracellular localization and translational control. These processes are also closely intertwined with the occurrence and progression of cancer. For example, Hu Antigen R is compatible with mRNA. It promotes the deadenylation and degradation of mRNA by directly or indirectly regulating the cis-regulatory element [13]. Recent studies have shown that many types of RBPs could be connected to circRNA to form RNA-protein complexes (RPCs), which could regulate the formation of circRNA. Simom and others have found that many circRNAs are differentially expressed in the epithelial-mesenchymal transition process, and more than one-third of circRNAs are regulated by quaking (QKI) [14]. RNA-Binding Motif Protein 20 is crucial for the formation of a subset of circRNAs that originate from the I-band of the titin gene [15]. Therefore, RBPs might play a critical role in regulating cancer through the regulation of circRNA.
The present study revealed that circRNAs were differentially expressed in HCC compared to adjacent non-tumor tissues. In the search for a circRNA dysregulation checkpoint in HCC tissues, we examined the effect of eight selected RNA-binding proteins on circRNA formation in HCC cells and identified RNA-binding protein 3 (RBM3) as a potential crucial player in regulating circRNA formation.
2. Materials and methods
2.1. Patients, samples and follow-up data
In total, 173 pairs of HCC (Tumor) and adjacent non-tumor tissue (Peri-tumor, < 2 cm apart from the tumor tissues) obtained from surgical resections of patients without preoperative treatment at Eastern Hepatobiliary Surgery Hospital (Shanghai, China). Among them, two pairs were used for circRNA sequencing to compare expression of circRNA between cancerous (Tumor) and adjacent non-tumor tissues (Peri-tumor), and differential expression of circRNA was validated using paired carcinoma (Tumor) and para-carcinoma tissues (Peri-tumor, <2 cm apart from the tumor tissues) from 20 patients (cohort 1) by quantitative real-time polymerase chain reaction (qRT-PCR). The other 151 pairs (cohort 2) were used for quantification of SCD-circRNA 2 and RBM3, then analysis of the correlation between their expression and outcome of patients with HCC after hepatectomy. Clinical characteristics of the HCC patients used in this study are presented in Supplementary Table 1. All recurrent HCCs were excluded. Surgically removed tissues were quickly frozen in liquid nitrogen and then stored at the −80 °C until analysis. All the resected samples were independently identified by two pathologists.
All of used tumor and adjacent non-tumor tissue samples were collected from HCC patients between April 2012 and December 2014. The follow-up of cohort 2 were ended on May 31st, 2017. The recurrence-free survival (RFS) was defined as the time from tumor resection until detection of tumor recurrence, death, or the last observation. The overall survival (OS) was defined as the time from tumor resection to either the death of the patient or the last follow-up visit. We defined tumor differentiation according to Edmondson's grading system and micrometastases as tumors adjacent to the border of the main tumor that were only observed under the microscope. Tumor staging was defined according to the sixth edition of the tumor node metastasis (TNM) classification system and Barcelona Clinic Liver Cancer (BCLC) staging system.
This research study was approved by the ethics committee of the Second Military Medical University, and all patients signed the informed consent provided by the committee. Patients whose consents were unavailable were excluded from this study.
2.2. RNA preparation and sequencing
Total RNA from paired samples (Patient 1 T, Patient 1 N, Patient 2 T and Patient 2 N) was extracted and purified using the RNeasy mini kit (Cat#74106; QIAGEN, GmbH, Germany) following the manufacturer's instructions, followed by an assessment for RIN to inspect RNA integration using the Agilent Bioanalyzer 2100 (Agilent Technologies, Santa Clara, CA, USA). Detection of circRNA was performed as described previously [4,16]. Briefly, total RNA samples were treated using the RiboMinus™ Eukaryote Kit (A10837–08; Qiagen, GmbH, Germany) to remove ribosomal RNA. Next, ribosome-depleted RNA was treated with RNase R (RNR07250; Epicentre, Wisconsin, USA). Strand-specific RNA-seq libraries were prepared using the NEBNext Ultra Directional RNA Library Prep Kit for Illumina (E7530L, NEB, Ipswich, MA, USA) following the manufacturer's instructions. Library quality was evaluated using the Qubit® 2.0 Fluorometer (Q32866; Thermo Fisher Scientific Inc., USA) and Bioanalyzer 2100 (Agilent). Strand-specific RNA-seq libraries were sequenced using the Illumina HiSeq 2500 platform.
2.3. Detection and annotation of HCC circRNAs
Raw reads were cleaned using FASTX (version: 0.0.13), and cleaned reads were subsequently mapped to the human reference genome GRCh37/hg19 using Bowtie 2 BWT [17]. Reads that aligned contiguously to the genomes were discarded. From the remaining reads, we used the find_circ [18] pipeline to identify potential circRNAs. Briefly, unmapped reads were extracted 20mers from both ends and aligned independently to identify unique anchoring positions within spliced exons. Anchors that aligned in the reversed orientation (head-to-tail) indicated circRNA splicing. Next, anchor alignments were extended such that the complete read aligned and the breakpoints were flanked by GU/AG splice sites. Only circRNAs with 2 or more supporting reads within single samples were kept. The total number of reads supporting a particular head-to-tail junction was used as an absolute measure of circRNA abundance. To estimate its relative expression, we normalized the counts of read mapping across an identified back-splice by the read length and number of read mappings (spliced reads per billion mapping (SRPBM)) [4]. Predicted circRNAs by this method are exactly the same as the location information found in circBase [19], which is expressed as recorded circRNA and newly identified circRNAs. All circRNAs identified from the sequencing data of HCC and paired normal liver samples were intersected with known gene models. Depending on the type of intersection with a known transcript, the circRNAs were classified, and the exon-intron structure within a circRNA was inferred from the known exons as described previously [18]. Fold-changes were also estimated according to SRPBM in each sample. The p-value significance threshold in multiple tests was set by the false discovery rate (FDR). Differentially expressed circRNAs were selected using the following filter criteria: Tumor vs Peri-tumor fold changes >5.0 or smaller than 0.2; P < .01; FDR < 0.01.
2.4. Cell lines and cell culture
Huh7, HepG2, HCT-15 and NCI-N87 cells were purchased between 2014 and 2018 from the biological cell bank of the Chinese Academy of Science (Shanghai, China). Good cell fidelity was ensured throughout the use of cells by monitoring changes in morphological and cell growth rate, neither of which changed throughout the duration of our experiments. Cells were cultured in high-glucose DMEM culture solution containing 10% fetal calf serum (HyClone, USA) at 37 °C and 5% CO2 with the humidity maintained at a certain level. Short tandem repeat (STR) profiling (Genetic Testing Biotechnology Corporation, Suzhou, China) was used to confirm that all cell lines were not contaminated by Hela or other cells.
2.5. Total RNA extraction, cDNA synthesis and quantitative real-time PCR
The method of extraction for total RNA of cells and tissue was carried out in strict accordance with the experimental procedures for Trizol reagent (Takara, Dalian, China). Concentration and purity of total RNA were measured by Nanodrop 2000 and agarose gel electrophoresis to further confirm RNA structural integrity. Only those samples with value of OD260 / OD280 between 1.8 and 2.0 and without obvious smearing or degradation were used for further analysis. The method of cDNA synthesis was in accordance with the experimental procedure of the RR047A reagent kit (Takara, Dalian, China), a 20 μl system containing 1 μg of total RNA after the genomic DNA was removed. Quantitative real-time PCR (qRT-PCR) was performed according to the GoTaq® qPCR Master Mix (Promega, USA) protocol using the ABI7500 system (StepOnePlus system) (Applied Biosystems, USA) using 18S as an endogenous control. Relative expression of circRNA was based on Ct, calculated by subtracting the expression of 18S. See Supplementary Table 2 for the specific primer sequence.
2.6. Vector construction
shRNA targeting sequences for the RBM3, SCD cirRNA-2 and scrambled RNA sequence (negative-shRNA) are shown in Supplementary Table 3. The GV248 vector encoding RBM3 or SCD cirRNA-2 specific shRNA and helper vectors (GeneChem, Shanghai, China) were co-transfected into the 293 T cells using Lipofectamine 3000 (Invitrogen, Carlsbad, USA) to generate shRNA lentiviruses. After 48 h, supernatants were collected and filtered using 0.22 μm filter membranes. Next, shRNA lentiviruses were harvested by ultracentrifugation at 4 °C, 17,000 rpm, for 60 min using Beckman SW32 rotors, followed by storage at −80 °C until further use. Recombinant lentivirus for the expression of RBM3 and negative control lentivirus (NC) were purchased from Genechem (Shanghai, China) and were transduced into Huh7 or HepG2 cells following the manufacturer's instructions. To obtain cell lines stably expressing RBM3, Huh7 or HepG2 cells were infected with the RBM3 overexpression lentivirus or NC and selected with puromycin (2 μg/ml) for three weeks. For functional studies, we selected two Huh7 clones (Huh7 Over-1 and Over-2) and two HepG2 (HepG2 Over-1 and Over-2), all of which showed approximately 4-fold overexpression to resemble a more physiological situation.
To generate a vector suitable for the overexpression of SCD-circRNA 2, we amplified the genomic region of SCD-circRNA 2 with its back-splicing site flanking 50-bp, 200-bp or 500-bp sequence (Fig. 6a) using Ex Taq HS (Takara, Dalian, China). The PCR products were inserted into the pZW1 vector [16,20] using the Hieff Clone™ Plus One Step Cloning Kit (10911ES25, Yeasen Biotechnology, Shanghai).
Fig. 6.
RBM3 increases expression of SCD-circRNA 2 depending on the sequence of the flanking back-splicing site. Genomic fragments corresponding to SCD-circRNA 2 with different sequence of flanking back-splicing site were cloned into pZW1 plasmid (a). These plasmids were transfected into Huh7 or HepG2 cells for 48 h, and then expression of SCD-circRNA 2 was measured using qRT-PCR (b). RNA immunoprecipitation (RIP) experiments were performed using the RBM3 antibody to immunoprecipitate Huh7 (c) or HepG2 (d) cell extracts. Then, SCD 3’UTR and 18S (negative control) were detected using specific primers (Supplementary Table 2). RIP enrichment was determined as RNA associated with RBM3 immunoprecipitation relative to an input control. Data are presented as means ± SD; n = 3. Student's t-test was used. * P < .05.
To generate a vector suitable for Sanger sequencing of SCD-circRNA 2, we amplified the full-length of SCD-circRNA 2 (Supplementary Fig. 2) using Ex Taq HS (Takara, Dalian, China). The PCR products were inserted into T vector.
Supplementary Fig. 2.
The characteristics of the circular RNA SCD-circRNA 2. (a) Scheme illustrating the circular RNA SCD-circRNA 2. PCR primers used to specifically clone full-length of SCD-circRNA 2 are indicated by arrows. Northern blot probes targeting SCD-circRNA 2 are indicated by blue bars. (b) Northern blot showed the expression of SCD-circRNA 2 in 4 paired HCC (Tumor) and adjacent normal liver tissues (Peri-tumor). (c) Random hexamer or oligo (dT)18 primers were used in the reverse transcription experiments. The relative RNA levels in Huh7 cells were analyzed by qRT-PCR and normalized to the value using random hexamer primers. (d) The relative RNA levels in Huh7 cells were analyzed by qRT-PCR and normalized to the value detected in the mock group. (e) Sanger sequencing showing the back-spliced events (red arrow) and full-length of SCD-circRNA 2. PCR primers used to specifically clone full-length of SCD-circRNA 2 are indicated by black arrows.
2.7. Animal experiment
All of animal experiments were approved by the Institutional Animal Care and Use Committee of the Second Military Medical University (Shanghai, China) and performed in accordance with the National Institutes of Health Guide for the Care and Use of Laboratory Animals. We chose 5-week-old female mice in this study, fed with certified standard diet and tap water ad libitum and established a light / dark cycle of 12 h on / 12 h off. The temperature was controlled between 21 °C and 23 °C, and the humidity was controlled between 30% and 60%. After one week of acclimatization, the experiment began. We injected the cells of the control group (Huh7 Mock), vector control group (Huh7 over-NC, HepG2 over-NC), RBM3 overexpression group (Huh7 over-1 and Huh7 over-2), and RBM3 overexpression with SCD-circRNA 2 downexpression group (HepG2 Over-1 / SCD-circRNA 2 shRNA) into the oxter of the left anterior limb of naked BALB/C mice. The tumor size was measured once a week. All of the naked mice were killed 4 weeks after the subcutaneous injection of tumor cells to take the subcutaneous tumor, and each group had their tumor mass weighed.
2.8. RNA immunoprecipitation
We performed RNA immunoprecipitation (RIP) experiments to detected the connection between RBM3 and SCD 3’UTR in HepG2 and Huh7 cells using the Magna RIP™ RNA-Binding Protein Immunoprecipitation Kit (#17–700, Millipore, HONG KONG). The RBM3 antibodies used for RIP were bought from Proteintech Group (14363–1-AP). We detected the coprecipitated RNAs using reverse-transcription polymerase chain reaction. Total RNAs (input controls) and isotype controls were assayed simultaneously to demonstrate that the detected signals were from RNAs specifically binding to RBM3 (n = 3). The gene-specific primers used for detecting SCD 3’UTR and 18S are presented in Supplementary Table 2.
2.9. Northern blot analysis
Four pairs of HCC tumor and peri-tumor tissues' total RNA was isolated with an RNA extract kit (Takara, Dalian, China). Concentration and purity of total RNA were measured by Nanodrop 2000 and agarose gel electrophoresis to confirm RNA structural integrity. Northern blot analysis was performed using a Northern blot kit (#AM1940, Ambion, USA) following the manufacturer's instructions. Briefly, 30 μg total RNAs were denatured in formaldehyde and then electrophoresed in a 1% agarose–formaldehyde gel. The RNAs were then transferred onto a Hybond-N+ nylon membrane (Amersham, USA) and hybridized with digoxigenin-labelled DNA probes (Supplementary Table 2). The Digoxin Chromogenic Detection kit (Thermo Scientific, USA) was used to develop the bound RNAs.
2.10. Protein extraction and western blotting
Total proteins were harvested as indicated in figure legends, separated using sodium dodecyl sulfate polyacrylamide gel electrophoresis. Proteins were transferred to nitrocellulose membrane and incubated with antibodies against RBM3 (ab134946, abcam), p-ERK, ERK, p-p38, p38, p-JNK, JNK (Cell Signaling Tecknology, USA) and GAPDH (sigma-Aldrich, Germany). Then, the blots were probed with Rdye800-conjugated goat anti-rabbit IgG and IRdye700-conjugated goat anti-mouse IgG and detected by an Odyssey infrared scanner (Li-Cor; Lincoln, NE).
2.11. Cell proliferation assay
The control group (Huh7 Mock and HepG2 Mock), vector control group (Huh7 over-NC, HepG2 over-NC), RBM3 overexpression group (Huh7 over-1, Huh7 over-2, HepG2 over-1 and HepG2 over-2), and RBM3 overexpression with SCD-circRNA 2 downexpression group (HepG2 Over-1 / SCD-circRNA 2 siRNA, and Huh7 Over-1 / SCD-circRNA 2 siRNA) cells were used for cell proliferation assay as indicated in figure legends. Generally, 2 × 103 cells were grown in 96-well plates. At the time of planning, we added 10 μL of CCK8 reagent (Dojindo, Japan) per well, and cells were incubated at 37 °C and 5% CO2 for another 2 h. The OD450 value for each well was obtained with Synergy 2 (BioTek, USA), and each group was assayed in triplicate at 12 h intervals after consecutive seeding for up to 2 days. We also measure cell proliferation using a 5-Ethynyl-2′-deoxyuridine (EdU) (sigma-Aldrich, Germany) according to the instruction provided by the manufacturer. The proportion of cells that incorporated EdU was then determined using fluorescence microscopy (Zeiss, Germany).
2.12. Cell cycle experiments
The control group (Huh7 Mock and HepG2 Mock), vector control group (Huh7 over-NC, HepG2 over-NC), RBM3 overexpression group (Huh7 over-1, Huh7 over-2, HepG2 over-1 and HepG2 over-2), and RBM3 overexpression with SCD-circRNA 2 downexpression group (HepG2 Over-1/ SCD-circRNA 2 siRNA, and Huh7 Over-1/ SCD-circRNA 2 siRNA) cells were used for cell cycle experiments as indicated in figure legends. The cell cycle reagent kit (Beyotime, China) was used for these experiments as recommended by the manufacturer. Briefly, the cells in each group were inoculated into a 12-well plate for 24 h. Each group comprised three repeat wells, and EDTA-free trypsinized cells were removed. After cells were washed twice with pre-cooled PBS, then were incubated with pre-cooled 70% ethanol for fixation overnight. Prepared PI dye was added to the samples in each group after the ethanol was washed away, and samples were incubated in the dark for 30 min at 37 °C and stored at 4 °C. Testing was carried out within 24 h.
2.13. Statistical analysis
All the data were statistically analyzed using SPSS software version 17.0, and the qualitative variables were statistically analyzed by χ2 test. Comparison of continuous variables was performed by Student's t-test or the Wilcoxon test. Correlations were measured by Pearson correlation analysis. The optimal cut-off value of the relative expression of SCD-circRNA 2 or RBM3 in HCC was determined by a ROC curve (Euclidean distance) analysis in Cutoff Finder (http://molpath.charite.de/cutoff/). The Kaplan–Meier method was used to calculate the survival curves. The log-rank test was used to compare the survival curves to determine whether there was a statistically significant difference. We considered results statistically significant when P < .05.
3. Results
3.1. Expression profile of circRNAs in HCC
To compare the expression levels of circular RNA between HCCs and paired tumor-adjacent normal liver tissues, we characterized circRNA transcripts using RNA-seq analysis of ribosomal RNA-depleted and RNase R-treated total RNA from paired HCC (Patients 1 T and 2 T) and adjacent non-tumor tissues (Patients 1 N and 2 N) (Supplementary Table 1). First, we generated an average of 129,704,837 (range: 96,787,102-142,857,320) reads and 16,213,104,563 (range: 12,098,387,750-17,857,165,000) bases of sequenced nucleotides per sample (Supplementary Table 4). Second, we identified circRNAs using a computational pipeline based on the anchor alignment of unmapped reads (average of 218,203 unmapped reads per sample (range: 103634–302,308), Supplementary Table 4). Next, we normalized the counts of read mapping across an identified back-splice by SRPBM [4]. Using this method, we identified 48,737 unique back-splice events throughout the genome. Among these, 13,914 back-splice events were recorded in the circBase. The other 71.5% (34,823/48737) of the back-splice events were newly identified.
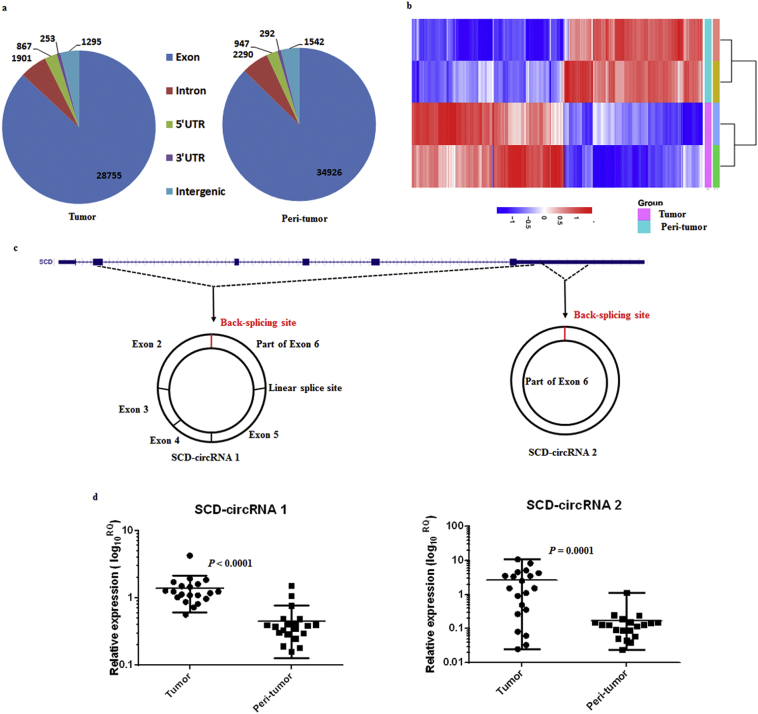
We classified circRNA back-splice sites that were sequenced in the tumors (Tumor) and adjacent non-tumor tissues (Peri-tumor). We found that an overwhelming majority of circRNAs were located in exonic regions, regardless of the tissues examined (Fig. 1a). We performed hierarchical clustering analysis of 1811 circRNAs that were differentially expressed between HCC samples (Tumor) and paired adjacent non-tumor tissues (Teri-tumor) (Tumor vs Peri-tumor fold changes >5.0 or smaller than 0.2; P < .01; FDR < 0.01) (Fig. 1b). Interestingly, we noted that there were 15 circRNAs spliced from stearoyl-CoA desaturase (SCD) pre-mRNA, which were all markedly upregulated in HCC (Supplementary Table 5). SCD encodes an enzyme involved in monounsaturated fatty acids biosynthesis and has been associated with the development of cancer [21], metabolic syndrome [22] and stem cell characteristics [23]. Furthermore, SCD is involved in the development [24] and progression [25,26] of HCC. However, all those studies were focused on the protein production of SCD. The role of SCD pre-mRNA as formed circRNA in HCC remains unknown. We analyzed its circRNA expression using a qRT-PCR in 20 pairs of HCC and adjacent non-tumor tissues. Using a melting curve, electrophoresis and production of PCR sequencing data, we confirmed that the primers for SCD-circRNA 1 (circ_novel_4976) and SCD-circRNA 2 (circ_novel_4982) were correct. Further results indicated that SCD-circRNA 1 and SCD-circRNA 2 were upregulated in tumor tissues compared with their expression in adjacent non-tumor tissues, consistent with the results obtained by direct sequencing (n = 20, Fig. 1d).
Fig. 1.
Differential expression of circRNAs in paired HCC and adjacent non-tumor tissues. (a) CircRNAs detected in HCC (Tumor) and paired non-tumor (Peri-tumor) samples are mostly derived from the protein coding sequence. (b) Hierarchical clustering analysis of 1811 circRNAs that were differentially expressed between HCC samples (Tumor) and paired non-tumor samples (Peri-tumor) (P < .01, FDR < 0.01 and fold change >5 or < 0.2). Expression values are represented in shades of red and blue, indicating expression above and below the median expression value across all samples, respectively. (c) Scheme illustrating the production of SCD-circRNA 1 and SCD-circRNA 2. (d) The results of SCD-circRNA 1 and SCD-circRNA 2 upregulation in tumor tissues (Tumor) compared with those in adjacent non-tumor tissues (Peri-tumor) verify the RNA-seq results. Cohort 1, n = 20, Wilcoxon matched-pairs signed-rank test. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
3.2. High SCD-circRNA 2 expression predicts poor prognosis of HCC patients
To further define the expression patterns of SCD-circRNA 1 and SCD-circRNA 2, we examined their expression in tumor tissues and paired adjacent non-tumor tissues from an additional 151 HCC patients (cohort 2) independent from the 20 HCC patients in cohort 1 by qRT-PCR. As expected, we observed a clear and significant increase of SCD-circRNA 1 and SCD-circRNA 2 in HCC tissues (Supplementary Fig. 1). Next, we sought to determine whether SCD-circRNA 1 or SCD-circRNA 2 expression levels in HCC were associated with specific clinicopathological characteristics. Using ROC curve (Euclidean distance) analysis in Cutoff Finder (http://molpath.charite.de/cutoff/), we found that the optimal cut-off value of the relative expression of SCD-circRNA 2 in HCC was 2.927 for death, which divided the 151 HCC patients into a high SCD-circRNA 2-expression group (n = 82) and a low SCD-circRNA 2-expression group (n = 69). High SCD-circRNA 2 expression levels were observed in HCC tissues with incomplete encapsulation (P = .046), with high serum AFP levels (> 20 μg/L, P = .008), ALT (> 40 U/L, P = .027) and AST (> 40 U/L, P = .000) (Supplementary Table 6). In contrast, there was no correlation between SCD-circRNA 1 and any clinicopathological characteristic in cohort 2 patients. Next, we examined whether the SCD-circRNA 1 or SCD-circRNA 2 expression level of HCC tissues correlated with clinical outcome of HCC patients after hepatectomy in cohort 2. Using Kaplan-Meier survival curves, we found that patients with a higher SCD-circRNA 2 expression levels exhibited both worse recurrence-free survival (RFS; P = .001; Fig. 2a) and overall survival (OS; P = .000; Fig. 2b). Significantly, the multivariate analysis revealed that high SCD-circRNA 2 expression level in HCC tissues was an independent predictor for reduced RFS and OS in HCC patients (Fig. 2c-d). SCD-circRNA 1 expression levels were not associated with HCC patient outcome after hepatectomy.
Supplementary Fig. 1.
circRNA expression in HCC. The results of SCD-circRNA 1 and SCD-circRNA 2 upregulation in tumor tissues (Tumor) compared with those in adjacent normal liver tissues (Peri-tumor) verified the RNA-seq results. n = 151, Wilcoxon matched-pairs signed-rank test.
Fig. 2.
SCD-circRNA 2 may be an independent prognostic factor for predicting recurrence-free survival (RFS) and overall survival (OS). (a and b) Kaplan-Meier analyses of correlations between SCD-circRNA 2 expression level in HCC and RFS or OS in 151 HCC patients (cohort 2) is shown. Log-rank test was used. Multivariate analysis of hazard ratios (HRs) for RFS (c) and OS (d). HRs are presented as the means (95% CI). The Cox proportional hazards model was used.
3.3. Characterization of SCD-circRNA 2 in HCC
We chose the circRNA, SCD-circRNA 2, derived from 3’UTR of the SCD gene for further study (Fig. 1c). The reasons were as follows: (i) SCD-circRNA 2 was one of the most abundant circRNAs of those that were differentially expressed according to its RPM in our RNA-seq data (Supplementary Table 5). (ii) Northern blot (Supplementary Fig. 2B) and qRT-PCR (Supplementary Fig. 2b and Fig. 1d right) showed that SCD-circRNA 2 was unregulated in HCC tissues comparing to corresponding adjacent non-tumor tissue. (iii) High SCD-circRNA 2 expression levels in HCC were associated with incomplete encapsulation, high serum AFP, ALT and AST levels (Supplementary Table 6). More important, higher SCD-circRNA 2 expression levels exhibited both worse RFS (P = .001; Fig. 2a) and OS (P = .000; Fig. 2b).
To further confirm the circular characteristics of SCD-circRNA 2, we used random hexamer or oligo (dT)18 primers in reverse transcription experiments using total RNA from HepG2 cells. It was clear that the relative level of SCD-circRNA 2 was significantly downregulated, when the oligo (dT)18 primers were used (Supplementary Fig. 2c). This finding indicated that SCD-circRNA 2 had no poly-A tail. SCD-circRNA 2 was resistant to RNase R, a highly processive 3′ to 5′ exoribonuclease that digests linear RNAs, indicating that SCD-circRNA 2 is circular (Supplementary Fig. 2d). Moreover, we successfully validated the full-length of SCD-circRNA 2 using circRNA specific divergent primers (Supplementary Fig. 2a) and Sanger sequencing (Supplementary Fig. 2e).
Altogether, these findings demonstrated that SCD-circRNA2 is an abundant and circular transcript, significantly upregulated in HCC.
3.4. RMB3 is required for SCD-circRNA 2 up-regulation in HCC cells
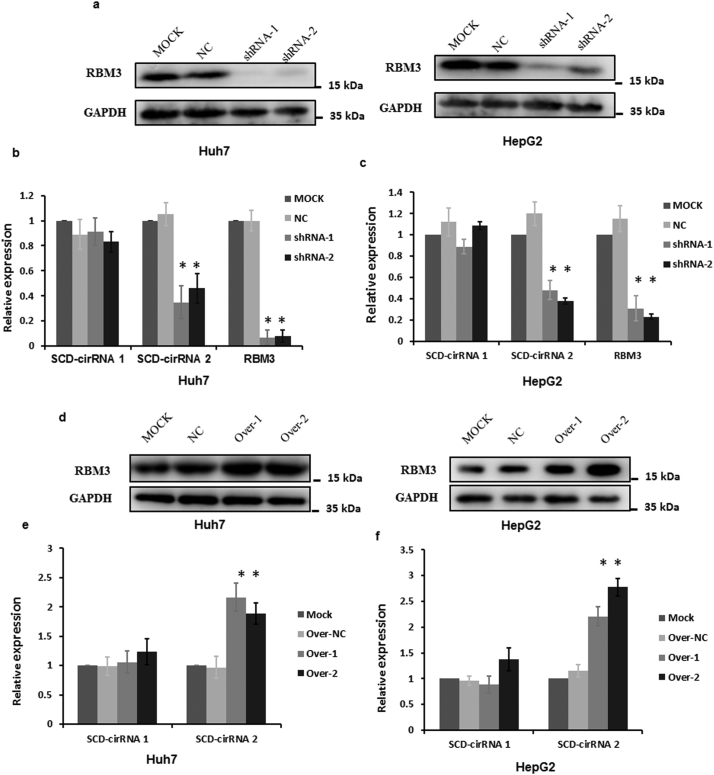
Recent studies have indicated that RNA-binding proteins interact with circRNA by forming an RNA-protein complex, which regulate the formation of circRNA [14,15]. By analyzing and integrating the data sets for the expression profiles between HCC tissue and normal liver tissues (GSE55092, GSE45267, GSE41804, and GSE14520) in GCBI (https://www.gcbi.com.cn/gclib/html/index) and reviewing the related literature [14,27], we focused on the following 8 RNA-binding proteins that were most likely to be differentially expressed in HCC: RBMS3 (NM_001003792), IGF2BP3 (NM_006547), IGF2BP2 (NM_001007225), RBM3 (NM_001017430), TARBP1 (NM_005646), NOVA1 (NM_002515), NOVA2 (NM_002516) and QKI (NM_006775) (Supplementary Table 7). To investigate the possible roles of these RBPs in the regulation of SCD-circRNA 1 and SCD-circRNA 2 formation, we examined the intracellular circRNA expression levels in response to decreased expression of these RBPs using lentiviral vectors harboring the shRNA sequence (Supplementary Table 3). We found that when RBM3 expression was inhibited at two different sites (Fig. 3a), expression of SCD-circRNA 2 was significantly downregulated in both Huh7 and HepG2 cell lines, which was not the case for SCD-circRNA 1 (Fig. 3b-c). However, no similar phenomena were observed when the other seven proteins were inhibited (Supplementary Fig. 3). To further determine whether RBM3 was a key protein for the upregulation of SCD-circRNA 2 in tumors, we overexpressed RBM3 in Huh7 cells and HepG2 cells. Expression of RBM3 was visualized by western blotting analysis (Fig. 3d). The qRT-PCR results showed that, compared with mock and negative control (NC) groups, SCD-circRNA 2 in the RBM3 overexpression groups (Over-1 and Over-2) was upregulated, while SCD-circRNA 1 was not (Fig. 3e-f).
Fig. 3.
Role of RBM3 in controlling circRNA formation. RBM3 depletion by shRNA lentivirus was verified by western blotting (a). qRT-PCR results indicated that downexpression of RBM3 could result in downregulated expression of SCD-circRNA 2 but not SCD-circRNA 1 in the Huh7 (b) and HepG2 (c) cell lines. RBM3 overexpression was verified by western blotting (d), resulting in the upregulated expression of SCD-circRNA 2 but not SCD-circRNA 1 in the Huh7 (e) and HepG2 (f) cell lines. Student's t-test was used. * P < .05.
Supplementary Fig. 3.
Expression of SCD-circRNA 2 or SCD-circRNA 1 could not be downregulated in RBMS3 (a), IGF2BP3 (b), IGF2BP2 (c), TARBP1 (d), NOVA1 (e), NOVA2 (f), QKI (g) depleted Huh7 cells.
3.5. RBM3 promotes HCC growth in vitro and in vivo
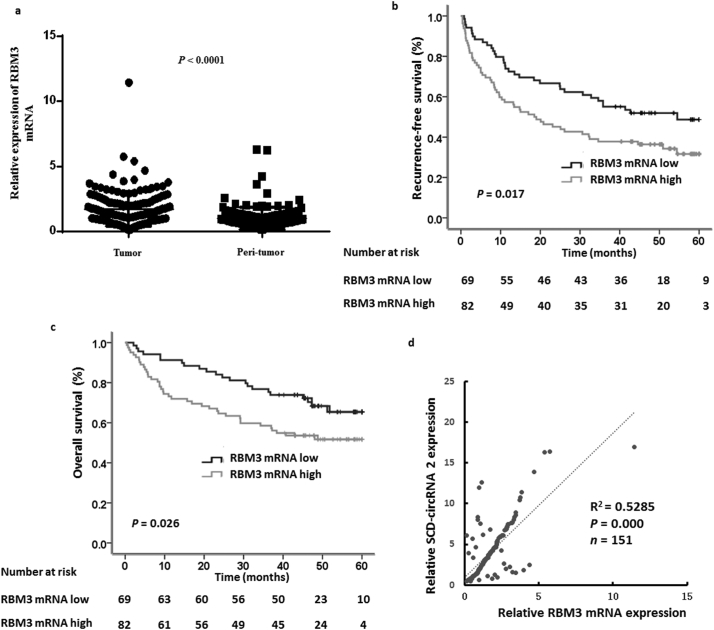
RBM3 is an important cold stress protein in mammalian cells, and its expression level is increased during cold stress. Numerous studies have shown that RBM3 expression could be an important marker in various tumors. We analyzed the relative mRNA expression level of RBM3 by qRT-PCR in HCC and adjacent non-tumor tissues tissues from cohort 2 patients. As shown in Fig. 4a, the qRT-PCR analysis demonstrated higher expression of RBM3 in patient HCC compared to paired adjacent non-tumor tissues. Next, we studied whether there was a correlation between RMB3 expression and patient prognosis. Using ROC curve (Euclidean distance) analysis in Cutoff Finder (http://molpath.charite.de/cutoff/), we found that the optimal cut-off value of the relative expression of RBM3 in HCC was 1.336 for death, which divided the 151 HCC patients into a high RBM3-expression group (n = 82) and a low RBM3-expression group (n = 69). High RBM3 expression levels were observed in HCC tissues with high serum AFP levels (> 20 μg/L, P = .021), AST (> 40 U/L, P = .001) and high SCD-circRNA 2 expression (P = .000) (Supplementary Table 8). Kaplan-Meier survival analysis indicated that the RBM3-high group had significantly worse RFS (P = .017, Fig. 4b) and OS (P = .026 Fig. 4c) than the RBM3-low group. We then analyzed the relationship between RBM3 and SCD-circRNA 2 expression levels in 151 patient HCC tissues (cohort 2). Our results showed a correlation between RBM3 mRNA and SCD-circRNA 2 expression levels in HCC tissues (Fig. 4d, Spearman-test, r2 = 0.5285, P = .000, n = 151), supporting the notion that RBM3 upregulates SCD-circRNA 2 expression in HCC tissues.
Fig. 4.
RBM3 is upregulated in hepatocellular carcinoma. We performed qRT-PCR test of RBM3 mRNA that included 151 HCC specimens (cohort 2) to explore the clinical significance of RBM3. (a) RBM3 mRNA is upregulated in HCC tissues (Tumor) compared with those in adjacent non-tumor tissues (Peri-tumor). n = 151. Wilcoxon matched-pairs signed-rank test. Kaplan–Meier analysis was used to compare the subgroups with RBM3 high expression (n = 82) to those with RBM3 low expression (n = 69). Remarkably, patients showing high expression of RBM3 exhibited worse recurrence-free survival (b) and overall survival (c) (P = .017 and P = .026, respectively). Log-rank test was used. (d) Relative expression of SCD-circRNA 2 was plotted against RBM3 RNA expression level in cohort 2 HCC tissues. Pearson correlation analysis was used.
To verify the biological role of RBM3 in HCC, we established Huh7 and HepG2 cell lines overexpressing RBM3. As shown in Fig. 5a, CCK8 assay demonstrated that cell viability of RBM3-overexpressing Huh7 Over-1 and Huh7 Over-2 cells was much higher than that of Huh7-Mock and Huh7-Over-NC cells. Next, we measured the periodic variation of these four cell types by flow cytometry. As shown in Fig. 5b, the proportion of G1 / G0 phase cells in Huh7 Over-1 and Huh7 Over-2 cells was significantly lower than that in Huh7-Mock cells. Furthermore, the proportion of S phase cells in Huh7 Over-1 and Huh7 Over-2 cells was much higher than that in Huh7-Mock cells. Consistent results were achieved in RBM3-overexpressing HepG2 cells (Fig. 5c and d), suggesting that RBM3 promotes HCC cell proliferation. To confirm the biological function of RBM3 in vivo, we established tumor models with Huh7-Mock, Huh7 Over-NC, Huh7 Over-1, and Huh7 Over-2 cells subcutaneously implanted tumor models in nude mice. We monitored tumor growth by measuring the tumor volumes for four weeks. Our results showed that the xenografted HCCs derived from Huh7 Over-1 or Huh7 Over-2 cells grew much faster (Fig. 5e and f) than those derived from control groups. Four weeks post inoculation, the xenografts were resected, and the Huh7 Over-1 and Huh7 Over-2-derived tumors were significantly heavier than those in the Mock or Over-NC group (Fig. 5g). Considering that several reports have shown high expression of RBM3 as a marker of better prognosis of intestinal-type gastric cancer [28] and metastatic colorectal cancer [29], we further confirmed the roles of RBM3 in colorectal cancer cells HCT-15 and gastric cancer cells NCI-N87. We found that when RBM3 expression was upregulated or downregulated in HCT-15 or NCI-N87 cell lines, the expression of SCD-circRNA 2 was significantly up- or downregulated, respectively (Supplementary Fig. 4a). Even so, the proliferation of HCT-15 or NCI-N87 cell lines was not related to the expression of RBM3 (Supplementary Fig. 4b and 4c). These results indicate that the roles of RBM3-regulated SCD-circRNA 2 in HCT-15 or NCI-N87 cells need further investigation.
Fig. 5.
RBM3 promotes tumor growth in vitro and in vivo. Results of CCK8 indicated that cell counts for RBM3-overexpressing Huh7 (a) and HepG2 (c) cells were much higher than for of mock and negative control cells. The G1/G0 phase of RBM3-overexpressing Huh7 (b) and HepG2 (d) cells was much lower than in mock and negative control cells. For (a-d), data are presented as means ± SD; n = 3. Student's t-test was used. In vivo xenograft-transplanted nude mouse tumor models of human HCC growth were established with RBM3-overexpressing Huh7 cells. Tumor images are presented in (e). (f) In vivo subcutaneous tumor growth curves for RBM3-overexpressing Huh7 cells (Over-1 and Over-2) and control cells (Mock and Over-NC). (g) Weights of subcutaneous tumors derived from RBM3-overexpressing Huh7 cells (Over-1 and Over-2) and control cells (Mock and Over-NC) at four weeks after neoplasm seeding. For (f-g), data were presented as means ± SEM. Mann-Whitney U test was used. n = 5. * P < .05.
3.6. The flanking sequence of the back-splicing site is crucial for RBM3-regulated SCD-circRNA 2 formation
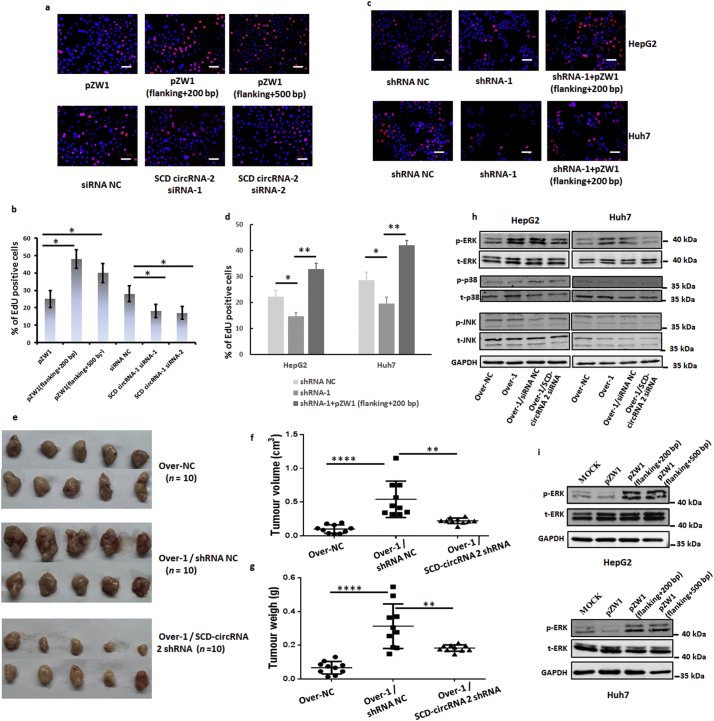
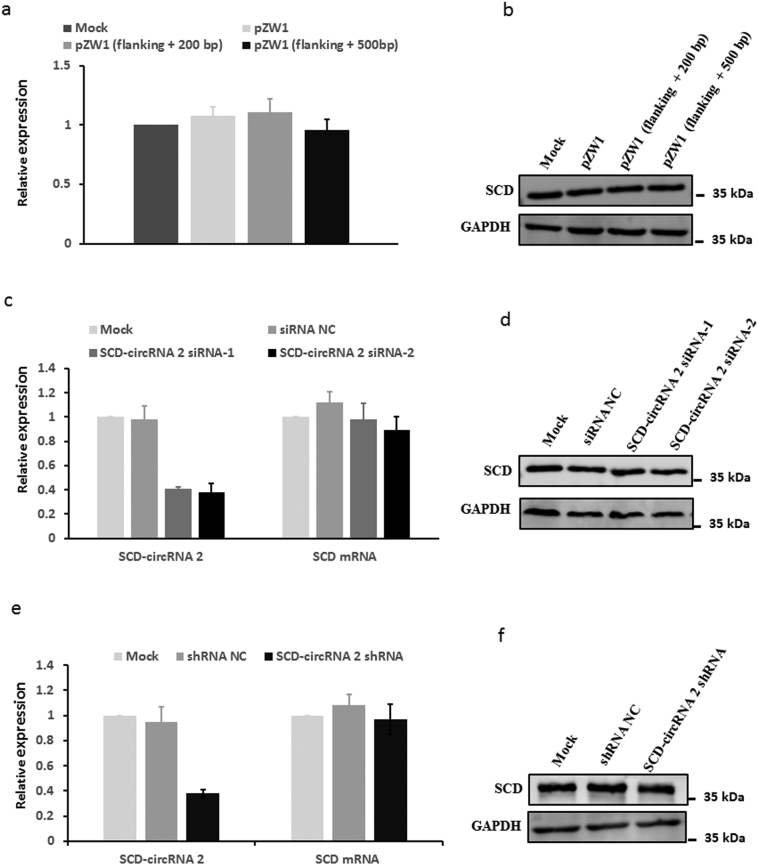
To generate an SCD-circRNA 2 overexpression vector, we amplified the genomic region for SCD-circRNA 2 with its segmental flanking sequence (Fig. 6a). We then cloned the PCR products into the pZW1 vector, a suitable system for circRNA expression in mammalian cells [16]. As shown in Fig. 6b, transfection of the pZW1 vector carrying SCD-circRNA 2 with the 200- or 500-bp flanking sequence of its back-splicing site resulted in SCD-circRNA 2 overexpression in both Huh7 and HepG2 cells. SCD mRNA and protein levels did not show significant changes after overexpressing SCD-circRNA 2 (Supplementary Fig. 5a and 5b). To further verify the role of RBM3 in SCD-circRNA 2 generation, we performed RNA immunoprecipitation (RIP) with an antibody against RBM3 from Huh7 and HepG2 cell extracts. We observed significant enrichment for SCD 3’UTR with the RBM3 antibody in both Huh7 and HepG2 cells (Fig. 6c and d), but there was no enrichment for 18S relative to the nonspecific IgG control antibody. All these results indicated that RBM3 might bind to SCD mRNA 3’ UTR and increase expression of SCD-circRNA 2 depending on the sequence of the flanking back-splicing site.
3.7. RBM3 promotes HCC cells proliferation through SCD-circRNA 2
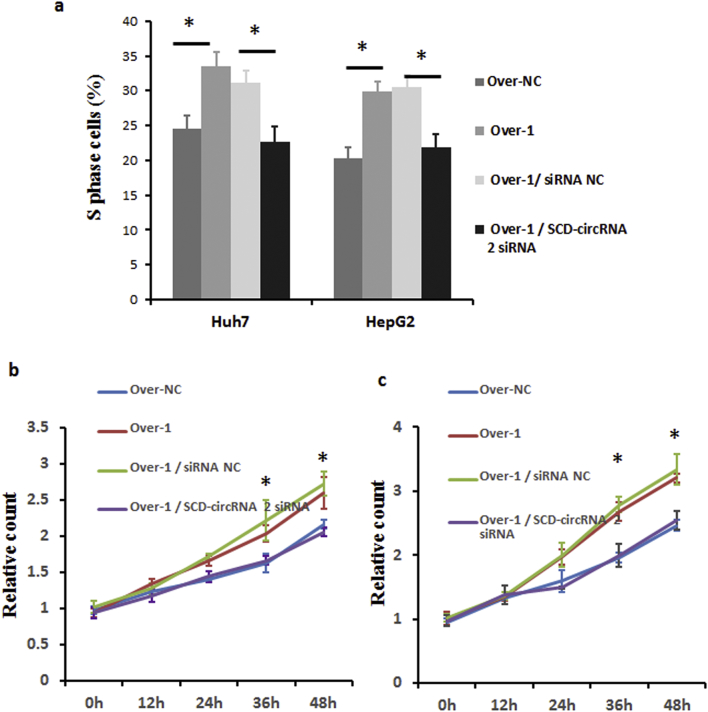
In addition, using small interfering RNAs (siRNAs) or shRNA targeting the back-splice sequence (Supplementary Table 2), we successfully knocked down SCD circRNA-2 in Huh7 and HepG2 cells (Supplementary Fig. 5c and 5e). SCD mRNA and protein levels did not show significant changes after downexpressing SCD-circRNA 2 (Supplementary Fig. 5c-5f). Next, we conducted EdU (5-Ethynyl-2′-deoxyuridine) experiments to verify whether SCD circRNA-2 is associated with the cell cycle. As shown in Fig. 7a-7b, SCD-circRNA 2 overexpression groups exhibited high levels of DNA replication, while low replication levels were seen in siRNA groups, indicating that SCD circRNA-2 levels are positively correlated with the cell cycle. More importantly, the inhibition of cell proliferation induced by RBM3 knockdown could be reversed by introducing SCD circRNA-2 expression (Fig. 7c and d). We further investigated whether SCD-circRNA 2 is involved in the RBM3-promoted cell cycle transition in HCC cells. As shown in Supplementary Fig. 6a, interference of SCD-circRNA 2 completely abrogated the RBM3-enhanced proportion of cells in the S phase, suggesting that SCD-circRNA 2 is required for RBM3-promoted HCC cell proliferation. Also, CCK8 assay demonstrated that cell viability of RBM3-overexpressing Huh7 and HepG2 cells was abrogated by SCD-circRNA 2 siRNA (Supplementary Fig. 6b and 6c). To further confirm the biological function of RBM3 / SCD-circRNA 2 in vivo, we established subcutaneously implanted tumor models with HepG2 Over-NC (HepG2 cells infected with overexpression negative control lentivirus), HepG2 Over-1 / shRNA NC (HepG2 cells infected with RBM3 overexpression lentivirus and shRNA negative control lentivirus), and HepG2 Over-1 / SCD-circRNA 2 shRNA (RBM3 overexpressed HepG2 cells infected with SCD-circRNA 2 shRNA lentivirus) cells in nude mice. Our results showed that the xenografted HCCs derived from HepG2 Over-1 / SCD-circRNA 2 shRNA cells grew much slower (Fig. 7e-7f) than those derived from HepG2 Over-1/shRNA NC groups. Four weeks post inoculation, the xenografts were resected, and the HepG2 Over-1 / SCD-circRNA 2 shRNA derived tumors were significantly lighter than those in the HepG2 Over-1 / shRNA NC groups (Fig. 7g). Together, these results indicate that SCD-circRNA 2 is a downstream target of RBM3. To identify the molecular mechanisms involved in the control of HCC cell proliferation by RBM3, we focused on the role of MAPKs based on data from previous reports [30,31]. Using western blotting, we investigated whether RBM3 induces phosphorylation of MAPKs in HCC cells. Interesting, overexpression of RBM3 increased phosphorylation of ERK, but not phosphorylation of JNK or p38 (Fig. 7h). More importantly, increased ERK phosphorylation induced by RBM3 could be abrogated with SCD-circRNA 2 siRNAs transfection (Fig. 7 h). In addition, the ERK phosphorylation could be increased by overexpression of SCD-circRNA 2 in HepG2 and Huh7 cells (Fig. 7i). Together, the available results indicate that RBM3 promotes HCC cells proliferation through SCD-circRNA 2 / ERK pathway.
Fig. 7.
RBM3 activates cell proliferation through SCD-circRNA 2. EdU experiments were conducted to verify whether SCD circRNA-2 is associated with the cell cycle. SCD-circRNA 2 groups revealed high levels of DNA replication, while low replication levels were observed in siRNA groups relative to those in control (a-b). Data are presented as means ± SD; n = 3. Student's t-test was used. * P < .05. Scale bars, 100 μm. (c) The effect of RBM3 shRNA on the proliferation of HepG2 and Huh7 cell lines is reversed by SCD circRNA-2 overexpression. Scale bars, 100 μm. Statistics for quantitative data are shown in (d). Data are presented as means ± SD; n = 3. Student's t-test was used. To further demonstrated that SCD-circRNA2 was the downstream of RBM3, we established subcutaneously implanted tumor models with HepG2 cells infected with overexpression negative control lentivirus (HepG2 Over-NC), HepG2 cells infected with RBM3 overexpression lentivirus and shRNA negative control lentivirus (HepG2 Over-1 / shRNA NC), and RBM3 overexpressed HepG2 cells infected with SCD-circRNA 2 shRNA lentivirus (HepG2 Over-1 / SCD-circRNA 2 shRNA) in nude mice. Tumor images are presented in (e). Volume (f) and weights (g) of subcutaneous tumors derived from HepG2 Over-NC, HepG2 Over-1 / shRNA NC and HepG2 Over-1 / SCD-circRNA 2 shRNA cells at four weeks after neoplasm seeding. For (f-g), data were presented as means ± SEM. Mann-Whitney U test was used. All P values are defined as: *P < .05, **P < .01 and ***P < .001. (h) RBM3-overexpressing HepG2 and Huh7 cells were transfected with SCD-circRNA 2 siRNA. Forty-eight hours after transfection, cells were harvested. ERK, p38 and JNK phosphorylation was assessed by western blotting analysis with the indicated antibodies. (i) The overexpression of SCD-circRNA 2 was performed as described in Fig. 6a. The relative expression of phosphorylated ERK significantly increased after SCD-circRNA 2 overexpressing in HepG2 (up) and Huh7 (down) cells.
4. Discussion
Despite advances in medical technology that have improved prognosis for HCC patients after surgical treatment, the molecular mechanism underlying hepatocarcinogenesis remains largely unknown, and overall survival time of HCC patients currently remains limited [1,32]. Moreover, functional biomarkers that are extensively involved in HCC development and could play a predictive role in prognosis of patients have not been well established to date [33,34]. Identifying these biomarkers and clarifying their functions in HCC development could facilitate exploration of novel therapeutic targets.
Recent studies have suggested that circRNA could be closely associated with the occurrence and development of multiple tumor types, primarily manifesting as circRNA dysregulation in tumor tissues [7,14,35]. Back-splicing, which is far less efficient than canonical splicing, is also coincidentally the mechanism for the biogenesis of circular RNA [[36], [37], [38]]. Consistent with findings in other tumors, circRNA was abundant in HCC tumor tissues in our experiments. Tissue RNA sequencing of circRNA (including those recorded in CIRCBASE [19] and newly discovered circRNA) extracted from HCC tissue and para-carcinoma tissue showed that expression levels of SCD-circRNA 2 were significantly increased in cancer tissue, which was confirmed by qRT-PCR. However, circRNA abundance did not seem to be due to its long half-life with respect to the rapid growth of solid tumors [36,39,40]. Instead, its abundance was due to the action of RNA-binding proteins that facilitate circRNAs expression in unique patterns [41,42]. Similarly, in our experiments, depleting RBM3, which is highly expressed in liver cancer cells, caused marked downregulation of SCD-circRNA 2, indicating that SCD-circRNA 2 could be a regulatory target for RBM3 in HCC cells. Moreover, the SCD-circRNA 2 expression level is an independent risk factor for overall survival and progression-free survival of HCC patients. However, it should also be pointed out that although this type of studies by us and others provide novel biomarkers for predicting disease progression and prognosis of HCC patients, more studies are needed to confirm the applicability of biomarker testing for accurately predicting outcomes and treatment response of patients.
RBM3 is considered a valuable biomarker that is highly expressed in cells subjected to harsh stressors [43]. Numerous studies indicate that expression of RBM3 is upregulated in various human tumors, including colorectal cancer [44,45], breast cancer [46], and prostate cancer [47,48]. Our study demonstrated for the first time that a high level of RBM3 in HCC tissues correlates with poor survival of HCC patients. Subsequently, our in vitro and in vivo experiments demonstrated that overexpression of RBM3 promotes HCC cell proliferation and xenografted tumor growth. While the exact mechanism underlying the regulation of RBM3 expression in HCC is not well understood, we do know that RBM3 is an evolutionarily conserved RNA-binding protein that is transcriptionally upregulated in response to cold [49] and hypoxia [50]. Furthermore, RBM3 is upregulated by chronic inflammation [51] and NF-κB p65 activity [52]. Hypoxia and chronic inflammation are both key characteristics of HCC [53], which may explain the increased expression of RBM3 in HCC. Nevertheless, the mechanisms of RBM3’ tumorigenic activities are far from being well understood. Therefore, more efforts are still needed to characterize the comprehensive molecular traits of RBM3 before we can target this protein safely and efficaciously in patients.
In the present study, our data demonstrated that overexpression of RMB3 leads to SCD-circRNA 2 accumulation in HCC cells. Consistently, that upregulated SCD-circRNA 2 expression was associated with the high levels of RBM3 in patient HCC tissues. Although various studies have emphasized the important role of circRNA, few have focused on the formation of circRNA with respect to tumor progression. Our results suggested that production of SCD-circRNA 2 could be dynamically regulated by RBM3, providing mechanistic insights into cancer-related circRNA dysregulation. Our functional study revealed that interference with SCD-circRNA 2 expression could abrogate the enhanced cell cycle transition promoted by RBM3, indicating that SCD-circRNA 2 may be a downstream target molecule of RBM3 that plays specific roles in HCC development. Overall, our study demonstrated that circRNAs in HCC are regulated by cell-type specific mechanisms and may represent therapeutic targets for HCC.Our data indicate that hundreds of circRNAs were either overexpressed or underexpressed in HCC when compared to adjacent non-tumor tissue. It is likely that these observed changes may have a biological effect. However, this possibility needs to be further confirmed by specific, hypothesis-driven studies. Secondly, whether RBM3 could promote the formation of other circRNA molecules is an interesting question. Thirdly, this study demonstrated that SCD-circRNA2 was the downstream of RBM3 and the upregulation of p-ERK-mediated by RBM3 could be abolished by SCD-circRNA2 silence. Nevertheless, the exactly mechanism that SCD-circRNA2 silence regulated p-ERK is still unclear. Understanding the precise molecular mechanisms by which circRNAs function in HCC will be critical for exploring these potential new strategies for early diagnosis and therapy of HCC.
The following are the supplementary data related to this article.
Supplementary Fig. 4.
(a) RBM3 overexpression or downregulation could result in the upregulated or decreased expression of SCD-circRNA 2 in the HCT-15 and NCI-N87 cell lines. Data are presented as means ± SD; n = 3. Student’s t test was used. *P < 0.05. RBM3 could not regulate colorextal cancer cells HCT-15 (b) and gastric cancer cells NCI-N87 (c) proliferation. Scale bars, 100 μm.
Supplementary Fig. 5.
The mRNA and protein levels of SCD were not changed in SCD-circRNA 2 upregulated or downregulated HCC cells. The overexpression of SCD-circRNA 2 was performed as described in Fig. 6a. The relative expression of SCD mRNA (a) and protein (b) did not significantly change after overexpressing. (c) The relative expression of SCD-circRNA 2 was downregulated in SCD-circRNA 2 siRNA-1 and SCD-circRNA 2 siRNA-2 transfected Huh7. qRT-PCR and western blot showed that the mRNA (c) and protein (d) level of SCD did not significantly change after silencing SCD-circRNA 2. (e) The expression level of SCD-circRNA 2 was downregulated in SCD-circRNA 2 shRNA lentivirus infected HepG2 cells. qRT-PCR and western blot showed that the mRNA (e) and protein (f) level of SCD did not significantly change after silencing SCD-circRNA 2.
Supplementary Fig. 6.
RBM3 activates cell proliferation through SCD-circRNA 2. (a) S phase was recovered in RBM3-upregulated Huh7 and HepG2 cells by downregulating the expression of SCD-circRNA 2. The results of CCK8 arrays indicated that cell counts was recovered in RBM3-upregulated HepG2 (b) and Huh7(c) cells by downregulating the expression of SCD-circRNA 2. Data are presented as means ± SD; n = 3. Student’s t test was used.
Clinical characteristics of the HCC patients used in this study.
Oligonucleotide sequences used in this study.
Oligonucleotide sequences used in this study.
Summary of RNA-Seq data by sample.
circRNAs spliced from stearoyl-CoA desaturase (SCD) pre-mRNA.
Clinical characteristics of 151 HCC patients according to SCD-circRNA 2 expression level.
HCC-regulated RNA bind proteins.
Clinical characteristics of 151 HCC patients according to RBM3 expression level.
Author contributions
F.Y. and H.L designed the study, analyzed the data and wrote the paper. W.D., ZH. D., CM.G. and FC.L. performed the experiments, analyzed the data, performed the animal studies and statistical analysis. XG. G, H.L, and J.D. provided the HCC patient samples. F.Y., H.L. and J.D. supervised the study. F.Y., H.L. and J.D. edited and reviewed the manuscript. All authors read and approved the final version of the paper.
Funding and acknowledgement
This work was supported by the National Key Research and Development Program of China (2016YFC1302303), the National Natural Science Foundation of China (Grant No. 81672345 and 81402269). The funders did not have any roles in study design, data collection, data analysis, interpretation, writing of the report.
Disclosure statement
There is none of any conflict of interest, financial or otherwise, related to the submitted work.
Contributor Information
Jin Ding, Email: dingjin1103@163.com.
Hui Liu, Email: liuhuigg@hotmail.com.
Fu Yang, Email: yangfusq1997@smmu.edu.cn.
References
- 1.Waller L.P., Deshpande V., Pyrsopoulos N. Hepatocellular carcinoma: A comprehensive review. World J Hepatol. 2015;7:2648–2663. doi: 10.4254/wjh.v7.i26.2648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ebbesen K.K., Kjems J., Hansen T.B. Circular RNAs: identification, biogenesis and function. Biochim Biophys Acta. 2016;1859:163–168. doi: 10.1016/j.bbagrm.2015.07.007. [DOI] [PubMed] [Google Scholar]
- 3.Qu S., Yang X., Li X., Wang J., Gao Y., Shang R. Circular RNA: a new star of noncoding RNAs. Cancer Lett. 2015;365:141–148. doi: 10.1016/j.canlet.2015.06.003. [DOI] [PubMed] [Google Scholar]
- 4.Li Y., Zheng Q., Bao C., Li S., Guo W., Zhao J. Circular RNA is enriched and stable in exosomes: a promising biomarker for cancer diagnosis. Cell Res. 2015;25:981–984. doi: 10.1038/cr.2015.82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Li Z., Huang C., Bao C., Chen L., Lin M., Wang X. Exon-intron circular RNAs regulate transcription in the nucleus. Nat Struct Mol Biol. 2015;22 doi: 10.1038/nsmb.2959. 256–4. [DOI] [PubMed] [Google Scholar]
- 6.Wang K., Long B., Liu F., Wang J.X., Liu C.Y., Zhao B. A circular RNA protects the heart from pathological hypertrophy and heart failure by targeting miR-223. Eur Heart J. 2016;37:2602–2611. doi: 10.1093/eurheartj/ehv713. [DOI] [PubMed] [Google Scholar]
- 7.Zheng Q., Bao C., Guo W., Li S., Chen J., Chen B. Circular RNA profiling reveals an abundant circHIPK3 that regulates cell growth by sponging multiple miRNAs. Nat Commun. 2016;7 doi: 10.1038/ncomms11215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hansen T.B., Jensen T.I., Clausen B.H., Bramsen J.B., Finsen B., Damgaard C.K. Natural RNA circles function as efficient microRNA sponges. Nature. 2013;495:384–388. doi: 10.1038/nature11993. [DOI] [PubMed] [Google Scholar]
- 9.Granados-Riveron J.T., Aquino-Jarquin G. The complexity of the translation ability of circRNAs. Biochim Biophys Acta. 2016;1859:1245–1251. doi: 10.1016/j.bbagrm.2016.07.009. [DOI] [PubMed] [Google Scholar]
- 10.Du W.W., Yang W., Liu E., Yang Z., Dhaliwal P., Yang B.B. Foxo3 circular RNA retards cell cycle progression via forming ternary complexes with p21 and CDK2. Nucleic Acids Res. 2016;44:2846–2858. doi: 10.1093/nar/gkw027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Qu S., Liu Z., Yang X., Zhou J., Yu H., Zhang R. The emerging functions and roles of circular RNAs in cancer. Cancer Lett. 2017;414:301–309. doi: 10.1016/j.canlet.2017.11.022. [DOI] [PubMed] [Google Scholar]
- 12.Huang G., Li S., Yang N., Zou Y., Zheng D., Xiao T. Recent progress in circular RNAs in human cancers. Cancer Lett. 2017;404:8–18. doi: 10.1016/j.canlet.2017.07.002. [DOI] [PubMed] [Google Scholar]
- 13.Blanco F.F., Jimbo M., Wulfkuhle J., Gallagher I., Deng J., Enyenihi L. The mRNA-binding protein HuR promotes hypoxia-induced chemoresistance through posttranscriptional regulation of the proto-oncogene PIM1 in pancreatic cancer cells. Oncogene. 2016;35:2529–2541. doi: 10.1038/onc.2015.325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Conn S.J., Pillman K.A., Toubia J., Conn V.M., Salmanidis M., Phillips C.A. The RNA binding protein quaking regulates formation of circRNAs. Cell. 2015;160:1125–1134. doi: 10.1016/j.cell.2015.02.014. [DOI] [PubMed] [Google Scholar]
- 15.Khan M.A., Reckman Y.J., Aufiero S., van den Hoogenhof M.M., van der Made I., Beqqali A. RBM20 regulates circular RNA production from the titin gene. Circ Res. 2016;119:996–1003. doi: 10.1161/CIRCRESAHA.116.309568. [DOI] [PubMed] [Google Scholar]
- 16.Zhang Y., Zhang X.O., Chen T., Xiang J.F., Yin Q.F., Xing Y.H. Circular intronic long noncoding RNAs. Mol Cell. 2013;51:792–806. doi: 10.1016/j.molcel.2013.08.017. [DOI] [PubMed] [Google Scholar]
- 17.Langmead B., Salzberg S.L. Fast gapped-read alignment with Bowtie 2. Nat Methods. 2012;9:357–359. doi: 10.1038/nmeth.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Memczak S., Jens M., Elefsinioti A., Torti F., Krueger J., Rybak A. Circular RNAs are a large class of animal RNAs with regulatory potency. Nature. 2013;495:333–338. doi: 10.1038/nature11928. [DOI] [PubMed] [Google Scholar]
- 19.Glazar P., Papavasileiou P., Rajewsky N. circBase: a database for circular RNAs. Rna. 2014;20:1666–1670. doi: 10.1261/rna.043687.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wang Z., Rolish M.E., Yeo G., Tung V., Mawson M., Burge C.B. Systematic identification and analysis of exonic splicing silencers. Cell. 2004;119:831–845. doi: 10.1016/j.cell.2004.11.010. [DOI] [PubMed] [Google Scholar]
- 21.Igal R.A. Stearoyl-CoA desaturase-1: a novel key player in the mechanisms of cell proliferation, programmed cell death and transformation to cancer. Carcinogenesis. 2010;31:1509–1515. doi: 10.1093/carcin/bgq131. [DOI] [PubMed] [Google Scholar]
- 22.Walle P., Takkunen M., Mannisto V., Vaittinen M., Lankinen M., Karja V. Fatty acid metabolism is altered in non-alcoholic steatohepatitis independent of obesity. Metabolism. 2016;65:655–666. doi: 10.1016/j.metabol.2016.01.011. [DOI] [PubMed] [Google Scholar]
- 23.Noto A., De Vitis C., Pisanu M.E., Roscilli G., Ricci G., Catizone A. Stearoyl-CoA-desaturase 1 regulates lung cancer stemness via stabilization and nuclear localization of YAP/TAZ. Oncogene. 2017;36:4573–4584. doi: 10.1038/onc.2017.75. [DOI] [PubMed] [Google Scholar]
- 24.Lai K.K.Y., Kweon S.M., Chi F., Hwang E., Kabe Y., Higashiyama R. Stearoyl-CoA desaturase promotes liver fibrosis and tumor development in mice via a Wnt positive-signaling loop by stabilization of low-density lipoprotein-receptor-related proteins 5 and 6. Gastroenterology. 2017;152:1477–1491. doi: 10.1053/j.gastro.2017.01.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Huang G.M., Jiang Q.H., Cai C., Qu M., Shen W. SCD1 negatively regulates autophagy-induced cell death in human hepatocellular carcinoma through inactivation of the AMPK signaling pathway. Cancer Lett. 2015;358:180–190. doi: 10.1016/j.canlet.2014.12.036. [DOI] [PubMed] [Google Scholar]
- 26.Ma M.K.F., Lau E.Y.T., Leung D.H.W., Lo J., Ho N.P.Y., Cheng L.K.W. Stearoyl-CoA desaturase regulates sorafenib resistance via modulation of ER stress-induced differentiation. J Hepatol. 2017;67:979–990. doi: 10.1016/j.jhep.2017.06.015. [DOI] [PubMed] [Google Scholar]
- 27.Zhang Y.A., Zhu J.M., Yin J., Tang W.Q., Guo Y.M., Shen X.Z. High expression of neuro-oncological ventral antigen 1 correlates with poor prognosis in hepatocellular carcinoma. PLoS One. 2014;9 doi: 10.1371/journal.pone.0090955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ye F., Jin P., Cai X., Cai P., Cai H. High RNA-binding motif protein 3 (RBM3) expression is independently associated with prolonged overall survival in intestinal-type gastric Cancer. Med Sci Monit. 2017;23:6033–6041. doi: 10.12659/MSM.905314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Siesing C., Sorbye H., Dragomir A., Pfeiffer P., Qvortrup C., Ponten F. High RBM3 expression is associated with an improved survival and oxaliplatin response in patients with metastatic colorectal cancer. PLoS One. 2017;12 doi: 10.1371/journal.pone.0182512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kim D.Y., Kim K.M., Kim E.J., Jang W.G. Hypothermia-induced RNA-binding motif protein 3 (RBM3) stimulates osteoblast differentiation via the ERK signaling pathway. Biochem Biophys Res Commun. 2018;498:459–465. doi: 10.1016/j.bbrc.2018.02.209. [DOI] [PubMed] [Google Scholar]
- 31.Zhuang R.J., Ma J., Shi X., Ju F., Ma S.P., Wang L. Cold-inducible protein RBM3 protects UV irradiation-induced apoptosis in neuroblastoma cells by affecting p38 and JNK pathways and Bcl2 family proteins. J Mol Neurosci. 2017;63:142–151. doi: 10.1007/s12031-017-0964-3. [DOI] [PubMed] [Google Scholar]
- 32.Llovet J.M., Villanueva A., Lachenmayer A., Finn R.S. Advances in targeted therapies for hepatocellular carcinoma in the genomic era. Nat Rev Clin Oncol. 2015;12:408–424. doi: 10.1038/nrclinonc.2015.103. [DOI] [PubMed] [Google Scholar]
- 33.Sia D., Villanueva A., Friedman S.L., Llovet J.M. Liver Cancer cell of origin, molecular class, and effects on patient prognosis. Gastroenterology. 2017;152:745–761. doi: 10.1053/j.gastro.2016.11.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hardy T., Mann D.A. Epigenetics in liver disease: from biology to therapeutics. Gut. 2016;65:1895–1905. doi: 10.1136/gutjnl-2015-311292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lasda E., Parker R. Circular RNAs: diversity of form and function. Rna. 2014;20:1829–1842. doi: 10.1261/rna.047126.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Westholm J.O., Miura P., Olson S., Shenker S., Joseph B., Sanfilippo P. Genome-wide analysis of drosophila circular RNAs reveals their structural and sequence properties and age-dependent neural accumulation. Cell Rep. 2014;9:1966–1980. doi: 10.1016/j.celrep.2014.10.062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zhang Y., Xue W., Li X., Zhang J., Chen S., Zhang J.L. The biogenesis of nascent circular RNAs. Cell Rep. 2016;15:611–624. doi: 10.1016/j.celrep.2016.03.058. [DOI] [PubMed] [Google Scholar]
- 38.Guo J.U., Agarwal V., Guo H., Bartel D.P. Expanded identification and characterization of mammalian circular RNAs. Genome Biol. 2014;15:409. doi: 10.1186/s13059-014-0409-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Enuka Y., Lauriola M., Feldman M.E., Sas-Chen A., Ulitsky I., Yarden Y. Circular RNAs are long-lived and display only minimal early alterations in response to a growth factor. Nucleic Acids Res. 2016;44:1370–1383. doi: 10.1093/nar/gkv1367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Jeck W.R., Sorrentino J.A., Wang K., Slevin M.K., Burd C.E., Liu J. Circular RNAs are abundant, conserved, and associated with ALU repeats. Rna. 2013;19:141–157. doi: 10.1261/rna.035667.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ivanov A., Memczak S., Wyler E., Torti F., Porath H.T., Orejuela M.R. Analysis of intron sequences reveals hallmarks of circular RNA biogenesis in animals. Cell Rep. 2015;10:170–177. doi: 10.1016/j.celrep.2014.12.019. [DOI] [PubMed] [Google Scholar]
- 42.Wilusz J.E. Circular RNAs: unexpected outputs of many protein-coding genes. RNA Biol. 2016:1–11. doi: 10.1080/15476286.2016.1227905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zhou R.B., Lu X.L., Zhang C.Y., Yin D.C. RNA binding motif protein 3: a potential biomarker in cancer and therapeutic target in neuroprotection. Oncotarget. 2017;8:22235–22250. doi: 10.18632/oncotarget.14755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Melling N., Simon R., Mirlacher M., Izbicki J.R., Stahl P., Terracciano L.M. Loss of RNA-binding motif protein 3 expression is associated with right-sided localization and poor prognosis in colorectal cancer. Histopathology. 2016;68:191–198. doi: 10.1111/his.12726. [DOI] [PubMed] [Google Scholar]
- 45.Hjelm B., Brennan D.J., Zendehrokh N., Eberhard J., Nodin B., Gaber A. High nuclear RBM3 expression is associated with an improved prognosis in colorectal cancer. Proteomics Clin Appl. 2011;5:624–635. doi: 10.1002/prca.201100020. [DOI] [PubMed] [Google Scholar]
- 46.Jogi A., Brennan D.J., Ryden L., Magnusson K., Ferno M., Stal O. Nuclear expression of the RNA-binding protein RBM3 is associated with an improved clinical outcome in breast cancer. Mod Pathol. 2009;22:1564–1574. doi: 10.1038/modpathol.2009.124. [DOI] [PubMed] [Google Scholar]
- 47.Jonsson L., Gaber A., Ulmert D., Uhlen M., Bjartell A., Jirstrom K. High RBM3 expression in prostate cancer independently predicts a reduced risk of biochemical recurrence and disease progression. Diagn Pathol. 2011;6:91. doi: 10.1186/1746-1596-6-91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Grupp K., Wilking J., Prien K., Hube-Magg C., Sirma H., Simon R. High RNA-binding motif protein 3 expression is an independent prognostic marker in operated prostate cancer and tightly linked to ERG activation and PTEN deletions. Eur J Cancer. 2014;50:852–861. doi: 10.1016/j.ejca.2013.12.003. [DOI] [PubMed] [Google Scholar]
- 49.Danno S., Nishiyama H., Higashitsuji H., Yokoi H., Xue J.H., Itoh K. Increased transcript level of RBM3, a member of the glycine-rich RNA-binding protein family, in human cells in response to cold stress. Biochem Biophys Res Commun. 1997;236:804–807. doi: 10.1006/bbrc.1997.7059. [DOI] [PubMed] [Google Scholar]
- 50.Wellmann S., Buhrer C., Moderegger E., Zelmer A., Kirschner R., Koehne P. Oxygen-regulated expression of the RNA-binding proteins RBM3 and CIRP by a HIF-1-independent mechanism. J Cell Sci. 2004;117:1785–1794. doi: 10.1242/jcs.01026. [DOI] [PubMed] [Google Scholar]
- 51.Sakurai T., Kashida H., Komeda Y., Nagai T., Hagiwara S., Watanabe T. Stress response protein RBM3 promotes the development of colitis-associated Cancer. Inflamm Bowel Dis. 2017;23:57–65. doi: 10.1097/MIB.0000000000000968. [DOI] [PubMed] [Google Scholar]
- 52.Ushio A., Eto K. RBM3 expression is upregulated by NF-kappaB p65 activity, protecting cells from apoptosis, during mild hypothermia. J Cell Biochem. 2018;119:5734–5749. doi: 10.1002/jcb.26757. [DOI] [PubMed] [Google Scholar]
- 53.Zhang J., Zhang Q., Lou Y., Fu Q., Chen Q., Wei T. Hypoxia-inducible factor-1alpha/interleukin-1beta signaling enhances hepatoma epithelial-mesenchymal transition through macrophages in a hypoxic-inflammatory microenvironment. Hepatology. 2018;67:1872–1889. doi: 10.1002/hep.29681. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Clinical characteristics of the HCC patients used in this study.
Oligonucleotide sequences used in this study.
Oligonucleotide sequences used in this study.
Summary of RNA-Seq data by sample.
circRNAs spliced from stearoyl-CoA desaturase (SCD) pre-mRNA.
Clinical characteristics of 151 HCC patients according to SCD-circRNA 2 expression level.
HCC-regulated RNA bind proteins.
Clinical characteristics of 151 HCC patients according to RBM3 expression level.