Abstract
Duchenne muscular dystrophy (DMD) is an X-linked disorder characterized by progressive muscle degeneration, caused by the absence of dystrophin. Exon skipping by antisense oligonucleotides (ASOs) has recently gained recognition as therapeutic approach in DMD. Conjugation of a peptide to the phosphorodiamidate morpholino backbone (PMO) of ASOs generated the peptide-conjugated PMOs (PPMOs) that exhibit a dramatically improved pharmacokinetic profile. When tested in animal models, PPMOs demonstrate effective exon skipping in target muscles and prolonged duration of dystrophin restoration after a treatment regime. Herein we summarize the main pathophysiological features of DMD and the emergence of PPMOs as promising exon skipping agents aiming to rescue defective gene expression in DMD and other neuromuscular diseases. The listed PPMO laboratory findings correspond to latest trends in the field and highlight the obstacles that must be overcome prior to translating the animal-based research into clinical trials tailored to the needs of patients suffering from neuromuscular diseases.
Abbreviations: ASO, antisense oligonucleotides; CNS, central nervous system; CPP, cell penetrating peptide; DGC, dystrophin glyco-protein complex; DMD, Duchenne muscular dystrophy; FDA, US food and drug administration; PMO, phosphorodiamidate morpholino; PPMO, peptide-conjugated PMOs; PS, phosphorothioate; SMA, Spinal muscular atrophy; 2ʹ-OMe, 2ʹ-O-methyl; 2ʹ-MOE, 2ʹ-O-methoxyethyl; 6MWT, 6-minute walk test
1. Introduction - Duchenne muscular dystrophy
1.1. Clinical presentation
Duchenne muscular dystrophy (DMD) is an X chromosome-linked, progressive, fatal degenerative muscle disorder affecting approximately 1/3,500-5,000 male births worldwide [[1], [2], [3]]. Signs of disease are visible early in childhood (2-5 years) and comprise a delayed ability to walk or waddling gait, impairment in movements, difficulty in running and frequent falls [4,5]. Progressive muscle weakness and joint contractures lead to loss of ambulation and wheelchair dependency around the age of 9-12 [6]. Scoliosis is a frequent complication that starts developing as soon as the loss of autonomous ambulation occurs and causes a significant negative impact on the respiratory system [7,8]. Restriction of diaphragmatic movements and pulmonary expansion further compromises respiratory function and around the age of 20, mechanical ventilation may be necessary to sustain life [9,10]. Myocardial impairment originates in the inferolateral wall and progressively leads to left ventricular fibrosis and dysfunction [11,12]. A cognitive impairment component of DMD with deficits in short term memory, multitasking, procedural learning and problem-solving is attributed to dysfunctions in cerebro-cerebellar pathway and has recently become the topic of thorough investigation [[13], [14], [15]]. Affected individuals succumb due to repeated pulmonary infections arising from mechanical support and/or cardiac muscle impairment [16].
While most DMD patients did not reach adolescence in the 1970s, improvement in current pharmaceutical regimes means that many of them may live up to their fourth decade [17]. However an extension of life expectancy is achieved through adequate and timely management of cardiopulmonary and respiratory complications and not due to a halt in the natural progression of the disease [18]. Therefore, this expanding patient population currently represents a challenge for the medical community, as patients will gradually require more elaborate and multidisciplinary approaches to treatment. A surveillance and management plan adhering to international standards of care should be carefully implemented and closely monitored to maximize the patient’s quality of life [19].
1.2. Genetics and pathophysiology
DMD is caused by mutations in the largest known human gene called dystrophin (DMD), which spans 24 kbs of genomic DNA with its 79 exons [20,21]. DMD encodes dystrophin, a 427 kDa protein localized on the cytoplasmic side of the sarcolemma of skeletal and cardiac muscle fibers as well as cortical/cerebellar synapses [[22], [23], [24]]. Most common mutations are non-randomly distributed deletions (approximately 68%) that may span one or more exons [[25], [26], [27]]. Surprisingly, the extent of the gene deletion does not correlate with the onset or severity of the clinical manifestations [28]. Exonic duplications, missense, frameshift, point or intronic mutations account for the rest of DMD cases [[29], [30], [31], [32]]. Most mutations result in a shift of the open reading frame and generation of premature termination codons leading to exclusion of one or more exons [33]. These aberrant mRNA transcripts undergo nonsense-mediated decay and therefore almost no dystrophin is produced [34]. Because dystrophin normally functions as an anchor between the actin cytoskeleton and the connective tissue via a sarcoplasmic complex, called the dystrophin glyco-protein complex (DGC) (Fig. 1), muscle fibers that lack dystrophin are subjected to increased mechanical stress and are more susceptible to damage upon contraction [27,35]. Disruption of sarcolemmal integrity causes abnormal influx of Ca2+ ions into the cytosol and aberrant activation of calcium binding elements, calcium-dependent proteases and pro-inflammatory cytokines [36]. The above effects further promote skeletal muscle regeneration, as a compensatory action to counterbalance the loss of function [37,38]. The altered myogenic signalling causes impaired proliferative capacity exhaustion of the satellite cell pool and replacement of muscle with fibrotic tissue, eventually resulting in necrosis and muscle wasting [39,40]. This effect is further exacerbated by functional ischemia in the affected muscles, due to the detachment of neuronal nitric oxide synthase from the sarcolemma, where it normally regulates vasoconstriction during muscle exercise [[41], [42], [43]].
Fig. 1.
Extracellular, membrane and cytoplasmatic components of the DGC. The muscle-specific laminin located in extracellular matrix is composed of α2, β1, and γ1 chains. The α2 subunit directly interacts with glycosylated α-dystroglycan (α-DG), which in turn interacts with the transmembrane β-dystroglycan (β-DG). Dystrophin binds to β-DG through cysteine-rich domain (Cys). The transmembrane protein family sarcoglycans (SG) (alpha, beta, gamma and delta) connect the cytoskeleton to the extracellular matrix, conferring structural stability to the sarcolemma. The four subunits of the SG complex interact with each other and with the transmembrane protein sarcospan. The small leucine-rich repeat proteoglycan biglycan (BGN) in the extracellular binds to α- and γ-SG and α-DG. The N- terminal of dystrophin protein (actin binding domain: ABD) binds to F-actin of the cytoskeleton and the C-terminal domain binds to alpha dystrobrevin (α-DB) and syntrophins (Syn). α1 and β1 in dark pink denote α1- and β1-syntrophin, respectively. Aquaporin 4 (AQP4) water channel protein along with syntrophin alpha regulates the efficiency of water transport in myofibers. The cytolinker protein plectin binds β-DG and dystrophin and connects desmin with the DGC. Syntrophins bind directly to α-DB and dystrophin and caveolin 3 (CAV3) through neuronal nitric oxide synthase (nNOS) whereas α1 syntrophin binds to the splice variant of nNOS in skeletal muscle termed nNOSμ.
2. Exon skipping as a therapeutic strategy for DMD
In the less severe allylic form of DMD, called Becker muscular dystrophy, the generated dystrophin reading frame is maintained in approximately 92% of cases and the generated dystrophin protein product is shorter and less abundant but still partially functional [25]. In Becker patients DMD mutations consist of 70% large deletions, 15% duplications and 15% point mutations and dystrophin detection ranging from 3-10% (severe cases) to even 20% (mild cases) [25,44]. Becker patients may remain ambulatory past 15 years of age and have a milder disease progression attributed to the existing functional dystrophin detected in their muscle fibers. This notion has led to the hypothesis that, if correction of the reading frame in DMD patients is successful, it would lead to the production of a truncated albeit semi-functional dystrophin transcript coding for a protein isoform that will remain resistant to proteolytic degradation and could be properly localized to the sarcolemma [45,46]. In this way, the connection between extracellular matrix and cytoskeletal muscle fibers (costamere) could be restored [47] re-establishing the link to the contractile apparatus [48]. Therefore modification of dystrophin pre-mRNA processing that results to the production of an internally deleted protein while preserving the N- and C- terminal domains which link to the cytoskeleton and extracellular matrix respectively might be the key to DMD future gene therapy [49].
The restoration of the disrupted open reading frame for DMD transcripts that generates the BMD phenotype was originally attempted in vitro by targeting exon 19 in the Kobe DMD phenotype [[50], [51], [52]], has now become the basis of the exon skipping therapeutic approach [53]. The strategy uses synthetic single-stranded DNA-like molecules called antisense oligonucleotides (ASOs) that have the potential to hybridize to RNA sequence motifs and to prevent assembly of the spliceosome, restoring the translatable mRNA transcript [54,55]. ASOs are commonly designed to bind to 5’ or 3’ splice junction and to sterically block access of splicing factors to the target site, altering pre-mRNA splicing [56]. ASOs may also bind to an exonic splicing enhancer or silencer to either promote or block the splicing effect [57].
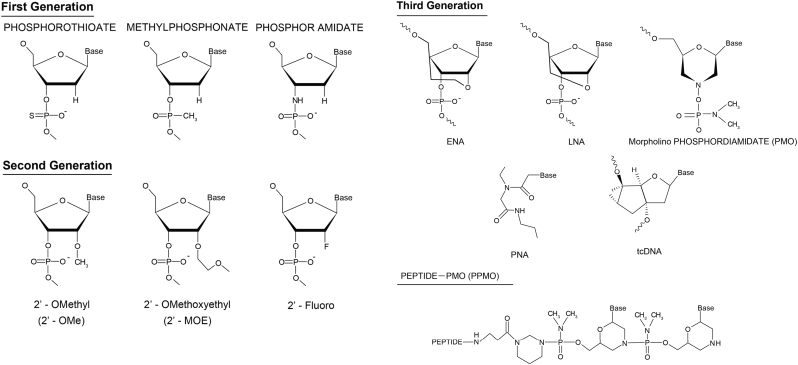
The unmodified ASOs used originally were subject to degradation by endonucleases and/or exonucleases, therefore, their therapeutic potential was de facto very limited [58]. Various chemical modifications of the phosphoribose backbone were performed in order to improve stability, efficacy and pharmacokinetics of ASOs. Substitution of the non-bridging phosphate oxygen with a sulphur atom within the phosphodiester linkage generates the phosphorothioate backbone (PS), which confers increased binding to plasma proteins and resistance to nuclease activity, prolonging the half-life of the ASO [59,60]. In addition, in order to allow for a high-affinity interaction with the target mRNA during splicing to occur rather than mediating mRNA destruction, ASOs should not support ribonuclease H activity [59,61]. Several ASOs fulfilling the above criteria have been tested in cells such as 2ʹ-O-methyl (2ʹ-OMe) and 2ʹ-O-methoxyethyl (2ʹ-MOE) with PS modifications (second generation ASOs) and Locked Nucleic Acids (LNA), ethylene-bridged nucleic acids (ENA), peptide nucleic acids (PNA), tricyclo-DNAs, phosphorodiamidate morpholinos (PMO) (third generation ASOs) [62] (Fig. 2). The most promising ASOs first tested in clinical trials are the 2’-OMe, that consist of methyl modifications to the 2′ position of the sugar moiety and the PMOs [63]. 2ʹ-OMe and 2ʹ-MOE contain a PS backbone and 2ʹ-O-substituted oligoribonucleotide segments. Both 2ʹ-OMe and 2ʹ-MOE ASOs exhibit high nuclease resistance, reduced immune stimulation due to their PS backbone, and are less toxic however they have lower affinity for their target compared to all other modified ASOs [64]. In PMOs, a third generation ASOs, the deoxyribose moiety is substituted by a morpholine ring while the charged phosphodiester inter-subunit linkage is replaced by a non-ionic phosphorodiamidate linkage [65]. This non-ribose based modification renders PMOs immune to nuclease activity but their non-ionic nature minimizes their nuclear uptake [66].
Fig. 2.
Chemical structures of first, second and third generation ASOs in comparison to PPMO. Modifications to the phosphodiester backbone of ASOs yielded several analogues such as phosphothioate, methylphosphonate and phosphoramidate that comprise the 1st generation of ASOs. Modifications to the deoxyribose sugar in ASOs yielded compounds such as 2’-OMe, 2’-MOE and 2’-fluoro that belong to the 2nd generation of ASOs. Third generation ASO modifications in ENA, LNA, PNA, tcDNA and PMO confer resistance to nuclease degradation as well as improve binding affinity of compounds. Peptide conjugated PMOs derive from peptide conjugation at the 5’ (as shown here) or 3’ end of a PMO.
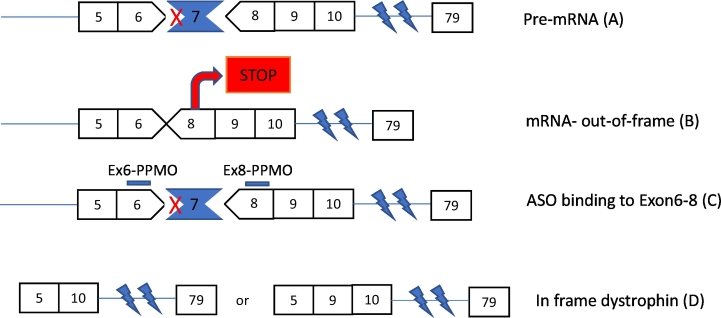
Numerous research groups have shown successful restoration of dystrophin using 2ʹ-OMe and PMO based ASOs in DMD animal models [67]. The most commonly used mdx mouse harbours a spontaneous nonsense mutation in exon 23 which results in early termination codon and mild to moderate DMD histopathological features. On the contrary, the more humanized mdx52 model was generated via targeted deletion of exon 52 which corresponds to the so-called hot spot exon 45-55 region where most DMD mutations are mapped in patients [68,69]. In the mdx mouse, exon 23 removal does not disrupt the DMD reading frame, allowing for mRNA induction and a little shorter dystrophin production that simulates the Becker phenotype [70]. Intramuscular injection of 1 μg of 2ʹ-OMe ASO administered weekly over a 4 week period or even as a single dose (5 μg) in mdx muscle induced dystrophin synthesis and improved functionality of treated muscle [71,72]. Repeated systemic administration of a 2ʹ-OMe ASO elevated dystrophin up to 5% of normal wild type levels in gastrocnemius, intercostal, and abdominal muscles and 1% in quadriceps [73]. Intramuscular or intravenous injections of PMO in mdx mice induced body-wide distribution of dystrophin with meaningful therapeutic levels compared to the 2ʹ-OMe ASO studies, albeit with high variability among samples [74,75]. In the mdx52 mouse model, exon 51 skipping was predicted to generate the Becker phenotype similar to humans. Systemic delivery of a PMO cocktail in the mdx52 mice seven times at weekly intervals induced 20-30% of wild-type dystrophin expression in muscle, a treatment that could theoretically apply to a high percentage of DMD patients [76,77]. Multi-exon skipping performed on the mdx52 mutation hot spot (exons 45-55) was achieved by systemic injections of ten PMOs restoring dystrophin levels up to 15%, accounting for over 60% of patients that harbour deletion mutations [78]. Because the mdx and mdx52 dystrophic phenotype is less severe than the one observed in humans, with mildly impaired muscle function and normal lifespan due to the utrophin compensation, there is a need to find alternative animal models to test efficacy, pharmacodynamics and safety of the drug [79,80]. For this purpose, multi exon skipping (exon 6-8 or exon 6-9) using PMO cocktail was successfully applied in the golden retriever canine dog model in vitro [81] and in canine X linked muscular dystrophy model CXMDJ in vivo [82] where it ameliorated pathological phenotype without triggering any serious side effect. Collaborative projects using the CXMDJ model bred in our facilities have thoroughly investigated the systemic efficacy and safety of 6-9 multi-exon skipping using combinations of PMO cocktails [[82], [83], [84]] and a similar approach has been adopted to our current studies that assess the potency and safety of novel PPMO cocktails (Fig. 3).
Fig. 3.
Exon skipping strategy in CXMDJ dog model using PPMOs. A point mutation in exon 6 is responsible for the loss of exon 7 in dystrophic CXMDJ dog (A) ultimately resulting to out of frame mRNA (B) and disruption of dystrophin protein production. PPMOs sequences manufactured in such a way to bind in exon 6 and 8 (C) cause effective splicing of either exon 6-7-8 or 6-7-8-9, restoring the dystrophin reading frame.
3. ASOs in exon skipping clinical trials targeting DMD
The majority of DMD mutations in humans cluster between exons 45-55 the so-called ‘hot spot’ and mutations in exon 51 represent 13% of the DMD patient population, making this subgroup the most attractive target for clinical trials using the single exon skipping approach [85]. The first experimental clinical trial used 2’-OMe-PS based drug called drisapersen (PRO051/GSK2402968/Kyndrisa) targeting exon 51 was developed by Prosensa, GlaxoSmithKline (GSK) and lately BioMarin. Preliminary data from phase I clinical trial was very encouraging, indicating dystrophin build up in a dose-dependent manner reaching up to 15.6% of that of healthy muscle [86]. Two randomized placebo-controlled phase 2 clinical trials demonstrated improvement of the 6-minute walk test (6MWT) in children treated with 6 mg/kg/week drisapersen administered subcutaneously for a period of 24 weeks the effect which was maintained, albeit with reduced significance, 48 weeks after treatment [87]. However results were not reproducible in the phase III placebo-controlled trial that followed and no dystrophin production was detectable by western blotting in treated patient's muscle obtained through biopsy [88,89] although an increase in sarcolemmal dystrophin myofiber was observed by immunohistochemistry [[90], [91], [92]]. Adverse effects such as skin fragility at the site of injection, proteinuria and presence of alpha microalbumin in the urine occurred and became more prominent when dose was scaled up to 9 mg/kg/week, therefore, the US Food and Drug Administration (FDA) declined approval on the basis that ‘the standards of effectiveness have not been met’ [90]. However, intensified efforts to overcome such toxicity problems have led to the development of optimized stereopure ASOs. A characteristic example is WVE-210201, designed to skip exon 51 in the DMD gene, which has yielded very promising in vitro and mouse-based exon skipping data and its efficacy and safety are currently assessed in a phase 1 clinical trial [93,94].
The FDA recently approved eteplirsen (Exondys 51, AVI4658; Sarepta Therapeutics), targeting exon 51 in DMD patients, which achieved around 42% positive dystrophic fibers when administered intramuscularly to the extensor digitorum brevis in DMD patients in a single-blind placebo-controlled trial [[90], [91], [92]]. Therefore eteplirsen, like drisapersen could successfully mediate 51 exon skipping in dystrophic patients [86,92,95,96]. Subsequently, eteplirsen was tested systemically in a non-randomized (phase I/II) [92] and randomized (phase II) clinical trial [91] showing skipping of exon 51 of variable nature in all patients and substantial increase in dystrophin-positive fibers in the high dose cohort groups or longer treated cohort groups with minimal side effects. Further functional assessments using the 6MWT demonstrated a decreased rate of ambulation loss in eteplirsen treated patients [97]. However more elaborate assessments indicated that variability in exon skipping efficiency occurred among patients and dystrophin was restored non- homogenously between different muscles, limiting the drug’s therapeutic potential [98]. Currently, the success in exon skipping therapy is determined by the percentage efficiency of exon skipping and the resulting expression level of the restored protein but these two parameters are not always well correlated and may not account for the observed phenotype or strongly deviate from earlier laboratory findings [99]. For example, in eteplirsen-treated patients, 30-50 mg/kg doses administered weekly 48 weeks resulted in a mean of 47.3% of dystrophin-positive fibres in original clinical trials, but when the percentage of dystrophin protein for the same patients was re-evaluated by FDA using the western blot technique which is considered a more reliable quantification method, it was as low as 0.93% compared to 0.08% in untreated controls [85,91]. Furthermore, systemic administration of eteplirsen appeared to increase dystrophin positive fibres up to 23% in phase I trials, but the latest data show that eteplirsen may only restore dystrophin up to 0.28%, yielding similar findings with the non-approved drisapersen [97,100,101]. Taking into account that mild dystrophinopathy is observed in patients that possess 10-25% of dystrophin we can make the assumption that achieving at least 10% of DMD mRNA knock up is essential for ameliorating the DMD phenotype [102,103] whereas 4% of dystrophin may be enough to significantly increase survival in severely diseased patients [104]. Both percentage targets mentioned above are far from the ones highlighted by the statistics obtained in the eteplirsen’s trials [105]. Furthermore, whether an elevation in the percentage of dystrophin-positive fibres is significant and can be deemed responsible for the amelioration of the clinical performance of a patient as measured by 6MWT is debatable, especially after pooling statistics in eteplirsen’s case [106].
While approval of eteplirsen was non unanimous and controversial, sparking a debate among FDA members and scientists and it is still not conclusive whether eteplirsen can successfully halt disease progression in diagnosed DMD patients [89,107,108], this decision will hopefully pave the way for more elaborate and robust PMO clinical trial designs. Recently, in our institute we concluded a phase I open-label study in collaboration with Nippon Shinyaku Co. Ltd., dose-escalation clinical trial to evaluate exon 53 skipping efficacy of NS-065/NCNP-01 PMO which could be targeting 8% of DMD patient population [53]. NS-065/NCNP-01 PMO demonstrated dose-dependent exon skipping efficacy and successful dystrophin expression with minimal side effects [109]. Recently released data by Nippon Shinyaku indicate that the level of dystrophin expression in 8 patients participating in Phase I/II clinical trial averaged 5.21%, well-surpassing efficacy data reported with the approved drug eteplirsen. Sarepta Therapeutics has recently announced its plan to submit a new drug application for obtaining accelerated approval of the exon 53 targeting PMO based drug called golodirsen (SRP-4053) after generating promising data in a phase I/II clinical trials [110]. The efficacy and safety of golodirsen and of another PMO drug, called casimersen (SRP-4503), which targets exon 45 of the DMD gene (approximately corresponding to 8% of patient population) are currently evaluated in the ongoing ESSENCE phase III double-blinded assessment (NCT02500381). This new era of ASO based drugs should lead to novel treatment regimens that will succeed in delivering dystrophin restoration with minimal adverse effects, bringing hope to patients suffering from genetic neuromuscular diseases.
4. PMO limitations and the development of PPMOs
Some of the main limitations of PMOs as a therapeutic agent for DMD underlined by numerous animal-based studies are: poor cellular uptake and permeability of membrane barriers, rapid clearance from systemic circulation, inability to cross blood-brain barriers, variability of dystrophin expression and distribution in various target tissues or within the same tissue, short duration of the exon skipping effect requiring repetitive administration and/or high dosage of the drug [45]. Several of the mouse-based studies conducted aimed to ameliorate the poor PMO uptake in systemic administration and enhance intracellular delivery, but little progress has been made so far [[73], [74], [75],111,112]. In fact, high and repeated systemic doses of PMOs are necessary to achieve upregulation of dystrophin in skeletal muscles in animal models [82,113] without any significant effect in diaphragm or heart. These observations are not unexpected when considering PMO’s chemical nature. Originally PMOs were thought as molecules that have no net electrical charge and thus are unable to form complexes with delivery vectors, a fact that minimizes their off-target effects and renders them suitable for intramuscular administration but unfortunately reduces their overall ability to cross cell membranes and thus their systemic efficacy [114]. Based on research data very recently published by our group, PMO may have negative zeta potential which enables them to form complexes [115]. Because PMO uptake can occur by passive diffusion, the dystrophin-deficient leaky muscle fibers can more readily internalize intramuscularly administered PMOs [77,116]. In fact, eteplirsen has the potential to penetrate leaky muscle cells to exert its therapeutic effect however once the treated muscle starts building up dystrophin, it automatically becomes less leaky and thus less penetrable preventing additional entry of PMO and hampering an homogeneous dystrophin build up [98]. This notion is in agreement with interpretations of data derived from PMO trials using the mdx mouse model. It was observed that cycles of muscle regeneration/degeneration had taken place during an intermittent PMO systemic high dose delivery scheme, a fact which can possibly account for the dystrophin fluctuations observed among treated mice and the inability of the delivered dosage to protect dystrophic muscle from the eccentric contraction-induced damage [117].
On the other hand, PMOs are almost always successfully endocytosed but due to the hydrophobicity of the plasma membrane, only traces of internalized PMOs can escape endosomes and reach their target [118]. Therefore, delivery of PMOs in tissue culture is aided by the use of endo-porter which is converted to its poly cationic form inside the acidified endosomal compartment, rendering the endosomal membrane permeable [119]. Covalent linkage of an octaguanidinium dendrimer scaffold on the 3’ end of a PMO ring leads to the generation of a modified morpholino called vivo morpholino (vPMO) [120]. vPMOs have been tested in the mdx52 mouse model as well as dystrophic dog models, and in both cases, they have shown optimized efficiency in splicing modulation and skeletal dystrophin production [[121], [122], [123]].
An effective way to enhance PMO penetration in cell membrane is to conjugate them to short cell penetrating peptides (CPPs). CPPs, also known as protein transduction domains are short peptides of cationic, amphipathic or hydrophobic nature that have the ability to form a complex with cargo molecules and successfully transport active biological conjugates inside the cell [124,125]. Experiments using such CPPs including HIV-1 Tat protein [126,127], Drosophila antennapedia protein and oligoarginine peptides as crosslinkers enhanced PMO uptake in cells [128]. Internalization of HIV-1 Tat protein conjugates can be impaired possibly due to the strong electrostatic interactions that are formed with cellular heparin sulfates during endocytosis [129]. The third alpha helix domain of antennapedia also known as penetratin, is widely used to maximize the efficiency of internalization, but fails to deliver any significant amounts to the nucleus [120]. Moreover, enzyme degradation throughout delivery and instability of the above conjugates in human serum are factors that pose hurdles towards their use in systemic delivery and thus limit their therapeutic potential. To enhance PMO delivery, CPPs were subsequently enriched with arginines, because as cationic amino acids they could potentially facilitate delivery of the neutrally charged PMOs into cell compartments [130,131]. Conjugation of PMOs to a penetratin which contains six arginines residues near the N-terminus (R6-Penetratin) and a bulky side-chain composed of hydrophobic amino acids generated a new class of PMOs called PMO internalization peptides (Pips) with enhanced cellular uptake capacity and stability against serum proteolysis [63,132,133]. Insertions of 6-aminohexanoic acid residues (X) into an R8 peptide increased the corresponding CPP's serum stability and nuclear delivery but failed to prevent intracellular degradation. Incorporation of non-α-amino acids into the oligoarginine (R8) peptides prevented potential endosomal entrapment and thus greatly enhanced their metabolic stability [134] whereas insertion of β-alanines into this skeleton further increased intracellular stability [[134], [135], [136]].
Originally, CPPs used for conjugation derived from naturally occurring proteins already proven to have excellent translocation properties [137], but further understanding of the structural activity relationship of CPPs led to synthesis of CPPs based on predictive algorithms [138] therefore existing CPPs are heterogeneous in nature. The resulting peptide conjugated morpholinos (PPMOs) are taken up in vitro by proliferating myoblasts or terminally differentiated myotubes via a poorly understood process called gymnosis [139], that does not require any vehicle or transfecting reagent for delivery [[140], [141], [142], [143]]. In fact, arginine-rich CPP conjugation to PMO not only remarkably enhanced cellular uptake of PMOs but also improved their pharmacophore potency as described in detail in later sections. For a summary of in vitro and animal studies undertaken in the field of DMD, please refer to table 1.
Table 1.
List of developed PPMOs and their therapeutic effects in experimental models.
| Compound name | Sequence | System | Route of administration | Age begin treatment | Dosage regime | Dystrophin restoration | Ref |
|---|---|---|---|---|---|---|---|
| B-peptide based | (RXRRBR)2XB | CXMDJ | im | 4–5 mo | 3,600 μg/1,200 μg | skeletal muscle | 166 |
| single ic/iv | 5 mo | 12 mg/kg | skeletal and cardiac muscle | ||||
| systemic iv | 4–5 mo | 12 mg/kg | skeletal and cardiac muscle | ||||
| Pip6a-PMO | RXRRBRRXR YQFLI RXRBRXRB | mdx | im | 1 nmole | 1 nmole | enhances DMD rescue by AAV | 155 |
| pretreatment | |||||||
| B-PMO | (RXRRBR)2XB | mdx | ip | 21 wk | 19 mg/kg dose | diaphragm | 166 |
| iv | 21 wk | 19 mg/kg dose | diaphragm, intercostal, sternomastoid | ||||
| M12 | RRQPPRSISSHP | mdx | iv 3x weekly | 6–8 wk | 25 mg/kg | skeletal muscles | 163 |
| iv single | 6–8 wk | 75 mg/kg | skeletal muscles | ||||
| PMOE23 | (RXRRBR)2XB | DKO | iv biweekly | 20–29, 30–39, | 15 mg/kg | early treatment prevents onset | 158 |
| 40–49, 50+ days | |||||||
| Pip6a | RXRRBRRXR YQFLI RXRBRXRB | H2K- mdx | in vitro | 0.125 – 1 μmol/L | exon skipping observed | 165 | |
| mdx | iv | 4 to 5 mo | 12,5 mg | diaphragm,skeletal and cardiac (high) | |||
| Pip6b | RXRRBRRXR IQFLI RXRBRXRB | H2K- mdx | in vitro | 0.125 – 1 μmol/L | exon skipping observed | 165 | |
| mdx | iv | 4 to 5 mo | 12,5 mg | diaphragm (high),skeletal and cardiac | |||
| Pip6c | RXRRBRRXR QFLI RXRBRXRB | H2K- mdx | in vitro | 0.125 – 1 μmol/L | exon skipping observed | 165 | |
| mdx | iv | 4 to 5 mo | 12,5 mg | diaphragm,skeletal and cardiac | |||
| Pip6d | RXRRBRRXR QFL RXRBRXRB | H2K- mdx | in vitro | 0.125 – 1 μmol/L | exon skipping observed | 165 | |
| mdx | iv | 4 to 5 mo | 12,5 mg | diaphragm,skeletal and cardiac | |||
| Pip6e | RXRRBRRX YRFLI RXRBRXRB | H2K- mdx | in vitro | 0.125 – 1 μmol/L | exon skipping observed | 165 | |
| mdx | iv | 4 to 5 mo | 12,5 mg | diaphragm,skeletal | |||
| Pip6f | RXRRBRRXR FQILY RXRBRXRB | H2K- mdx | in vitro | 0.125 – 1 μmol/L | exon skipping observed | 165 | |
| mdx | iv | 4 to 5 mo | 12,5 mg | diaphragm,skeletal and cardiac | |||
| Pip6g | RXRRBRRX YRFRLI XRBRXRB | H2K- mdx | in vitro | 0.125 – 1 μmol/L | exon skipping observed | 165 | |
| mdx | iv | 4 to 5 mo | 12,5 mg | low | |||
| Pip6h | RXRRBRRX ILFRY RXRBRXRB | H2K- mdx | in vitro | 0.125 – 1 μmol/L | exon skipping observed | 165 | |
| mdx | iv | 4 to 5 mo | 12,5 mg | low | |||
| B-peptide based | (RXRRBR)2XB | mdx | iv | 4 to 5 wk | 30 mg/kg | 20-50% in skeletal muscle | 72 |
| iv biweekly/year | 1.5 mg/kg | low | |||||
| iv biweekly/year | 6 mg/kg | skeletal muscles and heart | |||||
| iv monthly/year | 30 mg/kg | skeletal, diaphragm | |||||
| Pip5e | RXRRBRRXR-ILFQY-RXRBRXRB | H2K- mdx | in vitro | 1, 2 μmol/l | exon skipping observed | 150 | |
| mdx | im | 2/ 6 mo | 5 μg/kg | highest TA restoration | |||
| mdx | iv single | 2/ 6 mo | 25, 18.75, 12.5 mg/kg | diaphragm, heart and skeletal | |||
| Pip5f | RXRRBRRXR-ILFQY-RXRXRXRB | H2K- mdx | in vitro | 1, 2 μmol/l | exon skipping observed | 150 | |
| mdx | im | 2/ 6 mo | 5 μg/kg | dystrophin observed | |||
| mdx | iv single | 2/ 6 mo | 25, 18.75, 12.5 mg/kg | heart and skeletal | |||
| Pip5h | RXRRXR-ILFQY-RXRRXR | H2K- mdx | in vitro | 1, 2 μmol/l | exon skipping observed | 150 | |
| mdx | im | 2/ 6 mo | 5 μg/kg | dystrophin observed | |||
| mdx | iv single | 2/ 6 mo | 25, 18.75, 12.5 mg/kg | heart and skeletal | |||
| Pip5j | RBRRXRRBR-ILFQY-RBRXRBRB | H2K- mdx | in vitro | 1, 2 μmol/l | exon skipping observed | 150 | |
| mdx | im | 2/ 6 mo | 5 μg/kg | highest TA restoration | |||
| mdx | iv single | 2/ 6 mo | 25, 18.75, 12.5 mg/kg | heart and skeletal | |||
| Pip5k | RBRRXRRBR-ILFQY-RXRBRXRB | H2K- mdx | in vitro | 1, 2 μmol/l | exon skipping observed | 150 | |
| mdx | im | 2/ 6 mo | 5 μg/kg | dystrophin observed | |||
| mdx | iv single | 2/ 6 mo | 25, 18.75, 12.5 mg/kg | heart and skeletal | |||
| Pip5l | RBRRXRRBR-ILFQY-RXRRXRB | H2K- mdx | in vitro | 1, 2 μmol/l | exon skipping observed | 150 | |
| mdx | im | 2/ 6 mo | 5 μg/kg | dystrophin observed | |||
| mdx | iv single | 2/ 6 mo | 25, 18.75, 12.5 mg/kg | heart and skeletal | |||
| Pip5m | RBRRXRRBR-ILFQY-RXRBRXB | H2K- mdx | in vitro | 1, 2 μmol/l | exon skipping observed | 150 | |
| mdx | im | 2/ 6 mo | 5 μg/kg | dystrophin observed | |||
| mdx | iv single | 2/ 6 mo | 25, 18.75, 12.5 mg/kg | heart and skeletal | |||
| Pip5n | RXRRBRRXR-ILFQY-RXRRXRB | H2K- mdx | in vitro | 1, 2 μmol/l | exon skipping observed | 150 | |
| mdx | im | 2/ 6 mo | 5 μg/kg | highest TA restoration | |||
| mdx | iv single | 2/ 6 mo | 25, 18.75, 12.5 mg/kg | heart and skeletal | |||
| Pip5o | RXRRBRRXR-ILFQY-RXRBRXB | H2K- mdx | in vitro | 1, 2 μmol/l | exon skipping observed | 150 | |
| mdx | im | 5 ug/kg | dystrophin observed | ||||
| mdx | iv single | 2/ 6 mo | 25, 18.75, 12.5 mg/kg | heart and skeletal | |||
| P007 | (RXR)4XB | DKO | ip weekly x 6 | 10 days | 25 mg/kg/week | skeletal muscle, diaphragm | 175 |
| B peptide | (RXRRBR)2XB | mdx | iv single | 6 mo | 18.75 mg/kg | skeletal muscles | |
| PMO-Pep | (RXR)4XB | DKO | ip x 6 | 10 days | 25 mg/kg/week | skeletal muscles, prevents onset | 157 |
| B-PMO | RXRRBRRXRRBRXB | mdx | iv x 4 dailyx2 wk | 8/16 wk | 12 mg/kg/day | skeletal, cardiac, smooth muscles | 185 |
| B-PMO | RXRRBRRXRRBRXB | mdx | iv | 6-8 wk | 25 mg/kg | skeletal muscles, heart | 161 |
| iv X 6 weekly | 6-8 wk | 3 mg/kg/wk | TA, quadriceps | ||||
| iv X 3 weekly | 6-8 wk | 6 mg/kg/wk | skeletal muscles | ||||
| MSP-PMO | ASSLNIAXB | mdx | iv | 6-8 wk | 25 mg/kg | TA and quadriceps | 161 |
| B-MSP-PMO | RXRRBRRXRRBRXB-ASSLNIAXB | mdx | iv x 6 weekly | 6-8 wk | 3 mg/kg/wk | TA and quadriceps | 161 |
| MSP - B*-PMO | ASSLNIAXB-RXRRBRRXRRBRXB | mdx | iv x 6 weekly | 6-8 wk | 6 mg/kg/wk | TA only | 161 |
| P007 | (RXR)4XB | mdx | iv | 6-8 wk | 25 mg/kg | heart, biceps, diaphragm | 160 |
| iv x 3 | 6 mg/kg/wk | skeletal muscles and heart | |||||
| B peptide | (RXRRBR)2XB | mdx | iv | 6-8 wk | 25 mg/kg | skeletal, lower than P007 | 160 |
| iv x 3 | 6 mg/kg/wk | skeletal muscles, lower efficiency | |||||
| J-PMO | (rXr)4XB | EGFP-654 | ip x 4 | 7–8-wk | 12 mg/kg | quadriceps | 159 |
| M23D-B | RXRRBRRXRRBRXB | mdx | sc x 4 | 7–8-wk | 12 mg/kg | cardiac muscle, diaphragm, quadriceps | 159 |
| iv x 4 | 7–8-wk | 12 mg/kg | |||||
| ip x 4 | 7–8-wk | 12 mg/kg | |||||
| PPMOE23 | RXRRBRRXRRBRXB | mdx | im | 4–5 wk | 2 μg | TA | 162 |
| iv | adult | 30 mg/kg | diaphragm, skeletal, cardiac muscle | ||||
| iv x 6 biweekly | adult | 30 mg/kg | diaphragm, skeletal, cardiac muscle | ||||
| Pip1 | RXRRXRRXR IKILFQN RRMKWKK | H2K mdx | in vitro | 1 or 2 μΜ | efficient exon23 skipping | 132 | |
| Pip2a | RXRRXRRXR IdKILFQNd RRMKWHKB | mdx | im | 6-8 wk | 5 mg | dystrophin restoration in TA | |
| Pip2b | RXRRXRRXR IHILFQNd RRMKWHKB | mdx | im | 6-8 wk | 5 mg | dystrophin restoration in TA | |
| MSP | ASSLNIA | H2K mdx | in vitro | 250 nmol/L | exon skipping observed | 127 | |
| mdx | im | 2 mo | 5, 10, and 20 μg | dystrophin upregulation in TA | |||
| mdx | im | 3 wk, 6 mo | 5 μg | dystrophin upregulation in TA | |||
| TAT | YGRKKRRQRRRP | H2K mdx | in vitro | 250 nmol/L | exon skipping observed | 127 | |
| mdx | im | 2 mo | 5, 10, and 20 μg | dystrophin upregulation in TA | |||
| mdx | im | 3 wk, 6 mo | 5 μg | dystrophin upregulation in TA | |||
| AAV6 | TVAVNLQSSSTDPATGDVHVM | H2K mdx | in vitro | 250 nmol/L | exon skipping observed | 127 | |
| mdx | im | 2 mo | 5, 10, and 20 μg | dystrophin upregulation in TA | |||
| mdx | im | 3 wk, 6 mo | 5 μg | dystrophin upregulation in TA | |||
| AAV8 | IVADNLQQQNTAPQIGTVNSQ | H2K mdx | in vitro | 250 nmol/L | exon skipping observed | 127 | |
| mdx | im | 2 mo | 5, 10, and 20 μg | dystrophin upregulation in TA | |||
| mdx | im | 3 wk, 6 mo | 5 μg | dystrophin upregulation in TA | |||
| PMO-Pep | (RXR)4XB | mdx | ip single | neonatal | 1,2,5,10,25 mg/kg | diaphragm | 173 |
| ip weekly x 6 | neonatal | 1,2,5 mg/kg | skeletal muscle, diaphragm | ||||
| ip weekly x 4 | neonatal | 5 mg/kg | skeletal muscle, diaphragm |
R: arginine, B: beta alanine, X: 6 aminohexanoic acid, wk: week, mo: month, im: intramuscular, iv: intravenously, sc: subcutaneously, ic: intracoronary, ip: intraperitoneal
5. Comparison of PPMO and PMO properties
5.1. Improved internalization into cells
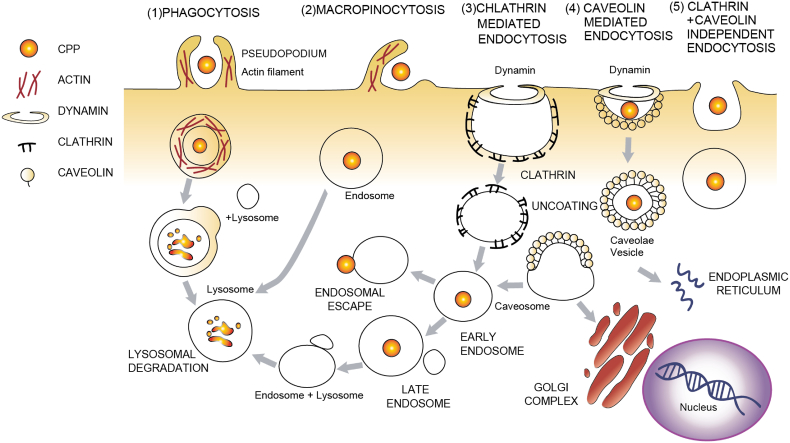
Internalization of PMOs into cells can occur via interchangeable pathways which are dependent upon the nature of the cell (Fig. 4) [144,145]. Cell surface PMO adsorption is mediated by a clathrin and caveolin-dependent or less frequently independent endocytic processes [146] and aided by cell surface receptors such as integrins, G protein- coupled receptors, receptor tyrosine kinase, Toll-like receptors and scavenger receptors [61]. Once ASOs are internalized in cells via endocytosis, they have to effectively be delivered to the nucleus to exert their splicing effect. CPP conjugation is implemented in order to increase cellular uptake of PMOs, which is very poor and requires large dosage and repeated administration to reach their target [147]. Similarly to PMO uptake, the most prevalent theory regarding PPMO uptake is via endocytosis mainly mediated by class A scavenger receptor subtypes (SCARAs) [141,148]. This interaction is greatly enhanced by the propensity of amphipathic PPMOs to self-assemble into nanoparticles [141]. Furthermore, the net charge of PPMOs, called zeta potential, is negative when measured in isotonic media and may influence the interaction of individual PPMOs with the plasma membrane, strengthening the notion that a receptor is necessary for internalization of PPMOs [137]. Addition of penetratin boosted the ability of PPMO to penetrate the cell membrane of differentiated neuronal cultures [149]. Addition of a B peptide has shown to facilitate heparin sulphate proteoglycan binding, thus enhancing internalization of PPMO to the endosomal pathway [114,150]. It was observed that Pip6-PMO was more readily internalized by H2K-mdx52 and C2C12 myotubes rather than myoblasts possibly due to higher endosomal entrapment that limits its availability and this was independent of the leakiness of the membrane [151]. The same group attributed the diminished potency of Pip6-PMO in cardiac cells versus skeletal muscle cells to different endocytic pathway internalization routes.
Fig. 4.
Cellular internalization of CPPs through various endocytotic pathways. ASOs are adsorbed and internalized in the cell via different routes including 1) Phagocytosis 2) Macropinocytosis 3) Clathrin-mediated endocytosis 4) Caveolin- mediated endocytosis and 5) Clathrin/caveolin- independent endocytosis. Once internalized ASOs may traffic from early endosomes to lysosomes and Golgi. To exert their function, ASOs must be able to escape from endosomes and reach the nucleus.
5.2. Enhanced potency at lower doses
A considerable hurdle in PMO endocytosis which contributes to their limited systemic therapeutic effect is their endosomal entrapment [152,153]. Peptides containing polyarginine analogues can induce leakage of endosomes and aid in the release of the conjugated PMOs into the cytosol [154]. PMOs and first-generation PPMOs that had a Tat or penetratin backbone required high concentrations of the compound to induce efficient and targeted exon skipping as well as to escape endosomal entrapment. Addition of R6 penetratin in the Pip compounds allowed efficient splicing at much lower doses [133,136]. In fact, injection of Pip2a or Pip2b conjugated PPMOs in the tibialis anterior of the mdx mouse resulted in efficient exon 23 skipping and significantly higher dystrophin rescue compared to the naked PMOs [132]. Pre-treatment of tibialis anterior muscle of mdx mice with PPMOs allows for a rescue of dystrophin expression at low dosages of AAV administration by prevention of AAV genomic loss, potentiating the microdystrophin based gene therapy [155].
Repeated intraperitoneal administration of a PPMO in the double utrophin/dystrophin KO mouse which shows a more severe phenotype than the mdx mouse and is deemed a more appropriate model to test the therapeutic effect of PMO [156] restored dystrophin expression in most skeletal muscles including diaphragm and prevented the onset of the dystrophic phenotype [157]. However, PPMO administration at a more advanced stage of disease failed to prevent disease progression although significantly delayed the disease progression when applied to mice in an early stage of disease [158].
5.3. Sustained dystrophin production
Evaluation of a series of PPMOs comprising a variable number of 6-aminohexanoic acid (X) and β-alanine (B) residues through intraperitoneal delivery in EGFP-645 mice that use the EGFP-654 pre-mRNA reporter to ascertain PPMO entry to cells. It was shown that the B conjugated peptide was the most effective one in sustaining dystrophic protein expression and targeting heart, diaphragm and quadriceps, key muscles in DMD patients [159]. Independent studies on B-PMO conjugated administered by intravenous injection to mdx mice have confirmed the high efficacy of B-PMO in dystrophin correction in mdx skeletal muscle with no overt hepatic or renal toxicity observed [160]. Direct demonstration that cell penetrating peptides may accentuate in vivo nucleic acid delivery came by the same group one year later. The authors describe how a chimeric fusion peptide generated by conjugation of a muscle-specific heptapeptide and a B peptide can induce effective exon skipping resulting in an efficient restoration of dystrophin in multiple skeletal muscles as well as significant increase in muscle strength in the mdx mouse model [161].
5.4. Improved efficiency of systemic delivery to target tissues
The advantage of PPMOs over PMOs to induce efficient systemic and target specific exon skipping and ameliorate the DMD phenotype was evident from the pioneering studies in mdx mice [121,[159], [160], [161], [162], [163]] and has been confirmed by virtually every study undertaken since then [147,164]. The generation of peptide nucleic acids/ PMO internalization peptides (Pips) series which contain two arginine-rich domains separated by a central short hydrophobic core were designed in order to improve serum stability and drive efficient exon skipping in a variety of target tissues, increasing heart dystrophin production [132,133]. Indeed, both Pip5 and Pip6 PMO series were capable of restoring dystrophin expression body wide following a single intravenous injection [150,165]. Further studies on Pip6 series demonstrated amelioration of DMD pathology and phenotype in exercised mice [151,166]. Dystrophin built up in more tissues aided restoration of DGC integrity with proper localization of beta-dystroglycan and improvement in muscle power and improvement of the phenotype in mice [161] and dogs [167]. Identification of candidate muscle-homing peptides and subsequent conjugation to ASOs, may improve delivery to target tissues, maximizing therapeutic effects [168,169].
5.5. Diaphragm targeting
In DMD patients, diaphragm function is severely compromised, leading to a progressive decline of ventilation and premature death [170]. In the mdx mouse model, diaphragm exhibits a fibrotic pattern with loss of elasticity and increased collagen density, becoming the best muscle to study representative histological changes of dystrophic phenotype in this animal model [171]. Repetitive administration of an (R-X-R)4XB–PMO conjugate (X= aminohexanoic acid and B: β alanine) effectively restored dystrophin expression in the diaphragm of mdx mice, when treatment was applied as early as the neonatal stage, however this effect was discontinuous at longer intervals after the final injection [172]. Dystrophin expression in diaphragm of neonatal mdx mice increased dose-dependently after a single intraperitoneal injection of 1, 2, 5 and 10 mg/kg of an (RXR)4XB peptide-based PPMO, reaching levels comparable to wild-type mice [173]. Systemic administration of the chimeric B-MSP-PMO at 6 mg/kg induced diaphragmatic dystrophin levels similar to gastrocnemius and biceps ones [174]. Intraperitoneal injection of P007-PMO at a dosage of 25 mg/kg/week for six weeks highly restored diaphragmatic levels of dystrophin and ameliorated the severe pathology of dystrophin/utrophin double knock-out mouse [175]. Studies carried out using the same P007-PMO revealed that a single intravenous administration of 25 mg/kg restored up to 25% of diaphragmatic dystrophin but this percentage dropped when a lower dosage of 5 mg/kg was administered systemically in the mdx mice [160]. The Pip5 and Pip6 series improved dystrophin diaphragmatic targeting [150,165]. A single dose of 19 mg/kg of B-PMO administered intraperitoneally markedly restored diaphragmatic dystrophin whereas the same dosage delivered intravenously also elevated dystrophin in intercostal and sternomastoid muscles of mdx mice [176].
5.6. Cardiac muscle targeting
Cardiomyopathy is an unavoidable consequence of DMD and is present in almost all patients over 18 years as a form of dilated cardiomyopathy [177] accounting for 20% of the mortality [178]. In DMD patients, absence of myocardial dystrophin leads to fibrosis, conditions that aggravates cardiac workload and stimulates autonomous system to increase heart rate as a compensation mechanism, further worsening the existent ventricular dysfunction [12].
PMO administration failed to aid cardiac function improvement in animal models tested [75], possibly due to inability of the drug to enter the impermeable cardiomyocytic membrane [179]. Even high doses of PMO (60 mg/kg) administered intravenously at biweekly intervals for one year in the mdx mouse have shown little efficacy in upregulating dystrophin protein levels in myocardium or in improving cardiac output and stress response [180]. Effective exon 23 skipping was observed in cardiac myoblasts obtained from mdx mice in vitro albeit with much higher doses than the ones necessary to produce the same effect in skeletal muscle cells [179,181]. Low levels of skipping efficiency in cardiac muscle (range of 2-3%) upon PMO systemic administration has been observed in almost every PMO study published so far, despite achieving high-efficiency dystrophin expression in most of the skeletal muscles [76]. Notably, intra-cardiac injections of naked morpholino oligos in aged mdx mice yielded very low exon skipping percentages [182].
Up to now, no available treatment is capable of restoring dystrophin protein in the heart of DMD patients. This fact is of pivotal significance, not only because of the high risk of failure of proper cardiac function per se which remains one of the main culprit of premature mortality in DMD patients but also because aggravation of cardiac disease progression occurs as a result of increased work load originating from amelioration of skeletal muscle function and enhanced locomotor activity using the current treatment regime [183,184].
The need for more efficient targeting of in-frame exon skipping particularly in cardiac muscle has led to the exploration of diverse structural PPMOs. Restoration of dystrophin in skeletal muscles and diaphragm in mdx mouse and in the more severely affected dystrophin/utrophin double knock out mouse, in absence of cardiac dystrophin expression, restored cardiac function to wild type levels suggesting that targeting respiratory muscles may prevent cardiomyopathy in DMD patients [175]. However intravenous administration of 19 mg/kg dose of B-PMO delivered intravenously or intraperitoneally restored dystrophin level of respiratory muscles but failed to improve cardiac function in mdx mice [176]. An arginine-rich PPMO with a backbone of (RXRRBR)2XB- targeting exon 23 in the mdx mouse named as PMOE23 successfully restored dystrophin almost to normal levels and at the same time protected heart muscle from damage after dobutamine stress challenge [162]. Systemic administration of the arginine-rich PPMO, AVI-5225 that induces exon 23 skipping, efficiently rescued cardiac dystrophin in the mdx mouse and inhibited onset and progression of cardiomyopathy [159,185]. Up to 20% of dystrophin expression was detected in the heart of mdx mice, three week after administration of a single intravenous injection of P007-PMO which has a (RXR)4XB- backbone and is more effective than the (RXRRBR)2XB- peptide [160]. It was found that a dosage of 6 mg/kg biweekly for a year rather than the higher treatment regime of 30 mg/kg monthly restores dystrophin in cardiac muscle up to 5%, indicating that treatment spacing is equally important to dosage regime [72].
A single intravenous injection of Pip5e-PMO -conjugated peptide induced 50% dystrophin expression in the heart of mdx adult mice, attributed to an increased nuclear delivery of Pip5e-PMO in cardiomyocytes [150]. Generation of the Pip6 series PMO by altering the peptide hydrophobic core sequence of Pip5e-PMO was carried out in order to promote homogeneous dystrophin restoration and particularly to more efficiently target the cardiac muscle. In fact, inversion of the Pip5e-PMO hydrophobic core (Pip6a) yielded cardiac dystrophin recovery score as high as 37% in the mdx mouse model whereas the specific arrangement of hydrophobic residues within the core did not alter the efficacy of the construct’s exon skipping [165]. The same group has shown that administration of Pip6f-PMO (scrambled peptide core) may restore dystrophin protein levels up to 28% in the heart of mdx mice previously subjected to a forced exercise regimen to induce changes that mimic the DMD cardiac phenotype [166]. Moreover, these mice exhibited lower levels of fibrosis, inflammatory and oxidative markers as well as other signs indicative of cardiomyopathy progression.
In the CXMDJ dog model, 4 monthly intravenous injections of the B peptide conjugated PMO cocktail or a single intracoronary or intravenous injection successfully induced 6-9 multi exon skipping and rescued dystrophin expression in most parts of cardiac muscle, as assessed by western blotting [167]. The dystrophin protein expression ameliorated vacuole degeneration in Purkinje fibres and increased Q/R ratio in the treated dogs.
6. The use of PMO and PPMO in neurodegenerative diseases
The term neurodegenerative diseases encompass a range of progressive disorders characterized by the gradual degeneration of the structure and function of the nervous system [186]. Representative examples are Parkinson’s disease, Alzheimer’s disease, Huntington’s disease (HD), Amyotrophic Lateral Sclerosis (ALS), spinal muscular atrophy (SMA) and spinocerebellar ataxias. There is a diverse range in pathophysiology; memory and cognitive functions might be gradually compromised while mobility and speech are unaffected, or the opposite or eventually all functions may be affected [187]. Risk factors and disease severity may be correlated with an advanced age (e.g. Alzheimer’s) or genetic predisposition (e.g. HD) or both (e.g. Parkinson’s, early onset Alzheimer’s) but primary cause for most of the neurodegenerative diseases is yet to be identified. As the percentage of aged population is rapidly expanding worldwide, global efforts to find new cures for neurodegenerative conditions that are linked to changes found in aged brains are intensified [188].
CPPs show improved systemic delivery and cellular uptake and due to their proven transmembrane transporting capacity they have been listed as promising agents in the treatment of central nervous system diseases (CNS). It was assumed that the otherwise impermeable blood-brain barrier (BBB) [189], being a negatively charged membrane formed by endothelial cells, may demonstrate increased affinity for the small size cationic or amphipathic CPPs [190,191] especially if systemic inflammation is present in individuals with abnormal neurological conditions [192]. In cases where CPPs have an arginine core, the high charge density generated may further enhance their BBB influx rates [193]. CPPs can cross the BBB using different transport mechanisms. In adsorptive-mediated transcytosis, the strong electrostatic interactions generated by the negatively charged phospholipids may aid translocation of the CPPs across the hydrophobic core of the membrane [194]. For example such a mechanism was employed in order to deliver the anti-apoptotic protein B-cell lymphoma-extra large (Bcl-xl) fusion protein with Tat to the murine brain as a cure for ischaemic injury [195]. In receptor-mediated transcytosis, interaction of the CPPs with a transporter localized at the endothelial cell surface such as the low-density lipoprotein receptor (LDLR), the low-density lipoprotein receptor-related proteins 1 and 2 (LRP1 and 2), the scavenger receptors class A type I (SR-AI), class B type I (SR-BI), allows CPP passage across the BBB [194].
PPMOs may effectively cross the BBB and reach targets in the CNS if administered via intrathecal injection; however such an invasive strategy, apart from causing discomfort to the patient, can also have multiple side-effects [61,196]. Once PPMOs manage to reach the CNS, vascular barriers may act beneficially to limit their escape towards the periphery, avoiding rapid loss of the drug through peripheral metabolism [64]. However, numerous risk factors associated with intrathecal administration such as infection, spinal headache, neurological injuries, have prompted researchers to further explore intravenous administrative routes in animal models. An arginine rich CPP-conjugated PMO was efficiently delivered to cerebellum and Purkinje cells when administered via tail vein in mice [197] raising hopes that systemic administration of PPMO could become a convenient route to effectively target CNS in the future. Systemic delivery of tricyclic DNA, a conformational constrained oligonucleotide analog, resulted to dystrophin restoration in the brain of mdx mice, although the cellular internalization mechanism of tricyclic DNA and thus the mode of endothelial barrier crossing has not been clarified by the authors [112].
SMA is an autosomal recessive disease caused by progressive loss of spinal motor neuron which leads to muscle atrophy, motor impairment and in its severe Type I manifestation, premature death [198]. It is caused by homozygous deletion or mutations in the survival motor neuron gene (SMN1) whereas phenotypic variations can be attributed to the number of copies of SMN1’s centromeric homolog, called survival motor neuron gene 2 (SMN2) [199]. Most SMN2 transcripts lack exon 7 and interferes with SMN’s ability to oligomerize, so the resulting protein product is rapidly degraded [200,201]. Therefore SMN2 may only partially compensate for the lack of SMN1 [202]. Inclusion of SMN2 exon 7 by targeting its splicing regulatory elements that has successfully increased full-length SMN2 production in vitro [203,204] and in vivo [205], has now become the most prominent ASO therapy-based approach for SMA [206]. ASOs of 2′-OMe chemistry administered by intracerebroventricular bolus injection was successfully taken up by neurons and glial cells in the CNS in SMA model mouse, inducing exon 7 inclusion and improving function and survival of diseased mice [205,207]. Nusinersen (ISIS 396443, Spinranza®), a modified 2'-MOE PS ASO, originally developed by marketed by Biogen is a recently approved FDA drug for the treatment of SMA. Nusinersen’s intrathecal administration resulted in significant improvement of motor milestones and prolonged lifespan of SMA patients with minimal adverse effects [208,209]. To improve systemic delivery and minimize side effects that result from repeated intrathecal injections, PPMO trials have been conducted. Systemic intravenous delivery of Pip6a PMO increased brain and spinal cord SMN2 expression and rescued disease phenotype in the Taiwanese severe SMA mouse model [210]. Recent research has demonstrated that a derivative of an ApoE (141-150) peptide was capable of successfully inducing pre-mRNA exon 7 inclusion of SMN2 in a mouse model of spinal muscular atrophy, ameliorating the phenotype of diseased mice [196].
While in SMA, like DMD, PPMO treatment focuses on increasing the production of functional proteins, in all other neurodegenerative PPMO clinical trials the main scope is to reduce aberrant mRNA transcripts [211]. In HD, expansion of the CAG sequence inside Huntingtin gene results in an elongated glutamine stretch near its amino terminus. The protein product is toxic and leads to neuronal loss mainly in the striatum and cortex in affected individuals [212] that suffer from progressive cognitive and motor impairment and succumb 15–20 years after the clinical onset. ASO technology selectively silenced mouse Huntingtin at identified exonic and intronic single nucleotide polymorphism sites in vitro and in vivo [213]. PMOs designed to target CAG repeat expansions and administered via intracerebroventricular injection significantly decreased Huntingtin’s protein expression and reduced neurotoxicity in a transgenic HD mouse model [214]. Modulation of the epigenetic regulator called repressor element-1 silencing transcription factor through ASO exon skipping in a striatal cell model of HD rescued transcription of neuronal genes, proving that exon skipping may prove beneficial in HD clinical trials in the future [215]. A 2ʹ-MOE chemistry-based drug (IONIS-HTTRx) delivered via intrathecal injection in early stage HD patients achieved a dose-dependent reduction in mutant huntingtin with no adverse effect [216].
In ALS, degeneration of upper and lower motor neurons leads to progressive and irreversible paralysis and ultimately death of the affected individuals [217]. Mutations in the Cu/Zn superoxide dismutase 1 (SOD1) gene have been linked to both sporadic and familial ALS [218,219], thus this gene is one of the main targets in ALS clinical trials [220]. Intrathecal delivery of a 2ʹ-MOE ASO (ISIS 333611) decreased SOD1 mRNA in spinal cord of recruited ALS patients in a phase 1 randomized control trial and demonstrated excellent safety profile [221] albeit concerns regarding toxicity remain [61]. Another 2ʹ-MOE ASO compound targeting SOD1 (IONIS-SOD1Rx, BIIB067) was investigated in a placebo-controlled phase I/II trial [222] in order to establish dosage and address safety issues and is currently in phase III clinical trial (NCT02623699). Use of morpholino oligomers to silence SOD1 after disease onset reduced microgliosis and increased motor neuron survival in a mouse model of ALS [223]. Furthermore, targeted degradation of the hexameric expansion containing RNA foci in the C9orf72 gene using ASOs reduced accumulation of expanded foci and dipeptide repeat proteins without affecting the overall level of C9orf72-encoding mRNAs in patient cells [224].
It is evident that since PMO therapeutic based strategy is already successfully applicable to many neurodegenerative diseases, the most potent PPMOs may hold tremendous therapeutic potential in this field as well as other ASO chemistries.
7. Limitations in PPMO use and future challenges
Targeted exon skipping by PPMOs is a revolutionary treatment that in the future could become applicable to a majority of DMD patients. Used alone or in combination with already approved treatment regimes, exon skipping may aid restoration of a partially functional dystrophin in most human tissues, significantly impacting on the quality and duration of life of dystrophic patients [173]. Recently, Sarepta Therapeutics announced initiation of a Phase I/IIa clinical trial of the novel PPMO SRP-5051, targeting DMD patients amenable to exon 51 skipping (NCT03375255). Pre-clinical studies using five more PPMO drug candidates targeting exons 44, 45, 50, 52 and 53 of DMD are also part of Sarepta’s pipeline now.
Currently, the main limitation in conducting future clinical trials for PPMO based drugs is their toxicity [125,130,162]. While not well understood, toxicity may be dependent on the following factors: species, duration of treatment, a frequency of systemic administration, dosage, exon chosen to be skipped, the peptide’s cationic nature [147]. The toxicity might arise due to immunogenic mechanisms such as complement activation [167,225] and first-generation arginine-rich peptides were found to be more immunogenic than PMOs [164]. Furthermore cell-mediated and humoral responses due to repetitive PPMO treatment or serum circulating antibodies directed against the newly synthesized dystrophin are listed as potential causes of toxicity [226]. Mild manifestations of drug toxicity after low dose systemic administration in rats include lethargy and weight loss [135], but at higher doses, elevated creatinine and BUN were recorded. Similarly, AVI-5038 a PPMO targeting the human dystrophin exon 50, was found to be well tolerated at low doses, however ongoing prolonged intravenous administration caused proximal tubular degeneration in the kidneys of cynomolgus monkeys [227]. Systemic administration of PMO in monkeys has caused tubular injury, with basophilic granulation and tubular vacuolation in the examined kidneys which were deemed to be dose-dependent and reversible upon discontinuation of treatment [228]. A pre-clinical trial using an arginine rich PPMO conducted by Sarepta had to be terminated due to the toxic side effects, that could partly be attributed to the high dosage used [147]. Novel biomarkers of acute kidney injury such as Kim1 and neutrophil gelatinase-associated lipocalinare (N-GAL) are highly specific, sensitive and inexpensive and greatly facilitated monitoring the efficacy of experimental treatments in animal models [229]. In fact, the latest generation of Pip peptides (series 7,8 and 9) purposely designed to contain a reduced number of arginine residues (from 10 to 6) compared to the Pip6 series have shown dramatically improved toxicity profiles whilst maintaining the compound’s splicing potency, rendering their therapeutic index more favourable for clinical development [230]. Further work in a novel PPMO series carried out in our group in collaboration with a new spin out company called Pepgen has resulted in a novel series of PPMO compounds called DPEP with favourable toxicity profiles well suited for future clinical trials. However, it still remains a challenge to estimate any side effect of long-term administration of PPMO in humans prior to we fully understand the pathophysiology of their toxicity.
Dosage and treatment regime is also of pivotal importance in obtaining maximal dystrophin expression while impeding development of severe off-target effects. The off-target effects arise from hybridization-mediated mechanisms and were not an issue for the first generation ASOs due to their limited ability to penetrate cellular membranes [63,231]. Unfortunately, while PPMO conjugation has maximized target delivery [135] it has also facilitated penetration in organs such as the liver, raising concerns about toxicity side effects [232]. Using lower dose or spacing systemic injections at longer intervals during PPMO administration might minimize side effects and potentially make the treatment cost-effective and friendlier to patients. For example, careful selection of a PMO dosing regime in mice allowed significant improvement of DGC expression complex, minimizing the histopathological features of DMD [233]. However the same group showed that intermittent injections with PMO irrespectively of the dosage could not prevent degeneration/regeneration cycles between treatments leading to muscle damage and uneven distribution of dystrophin among tissues, as occurred in the case of the eteplirsen clinical trials [117]. Therefore, prior applying any PPMO treatment regime to DMD patients, the in vivo efficacy of PPMO to prevent degeneration-regeneration must be carefully calculated.
The timing of exon skipping treatment is equally important to the dosage regime. If treatment is applied at the onset of disease or prior manifestation of a more severe phenotype, chances to slow down its progression are maximized, as already demonstrated in the mdx mouse models [157,158,173]. Indeed, delayed onset of treatment in the eteplirsen clinical trials failed to prevent loss of ambulation [91]. It is now established that destabilization of the interactions of satellite cells with the surrounding environment leads to a exacerbation of inflammation and fibrosis, further complicating DMD pathogenesis [234]. Understandably, even if a successful exon skipping restores dystrophin expression in several muscle groups in advanced diseased patients, the newly produced dystrophin won’t be able to reverse those pathological processes that have already taken place due to the destabilization of the DGC complex. More importantly, the beneficial effect of any genetic correction of the DMD defect may be obscured or hampered by the quality of muscle such as advanced fibrosis, exhaustion of the satellite cell pool and reduced myofiber production that are common pathological changes observed in dystrophic muscle. It is therefore not surprising that PPMO treatment is not effective in advanced disease patients where a prolonged absence of dystrophin has led to advanced fibro-fatty degeneration, conditions which do not create an ideal environment for dystrophin restoration. This vicious cycle can only be broken if combination of existing therapies with PPMO skipping are applied, to improve the quality of life for patients.
Systemic delivery of ASOs has been majorly improved with peptide conjugation however tissues like heart and diaphragm do not demonstrate high efficient exon skipping and sustained expression of dystrophin remains challenging. Although most studies attribute this to the impermeable nature of cardiomyocytes, the cardiac levels of a PPMO injected in mdx mice as measured by ELISA were comparable to those of the other tissues [232], hinting that the poor efficacy of PPMOs in the heart may not necessarily be connected to their poor delivery but to their mode of subcellular uptake. A major drawback in order to optimize delivery in tissues is the lack of prediction of PPMO efficiency from their secondary structure [63]. The ability of PMO to successfully induce exon skipping of the DMD gene transcript depends on many factors such as their length, their affinity of binding, proximity to the acceptor splice site, ability to block an exon splicing enhancer or interference with serine/arginine protein binding [235]. In silico pre-screening models based on measurement of parameters such as the binding energetics of ASO to the RNA, the distance of the target site from a splice acceptor site may give up to 89% accurate prediction of the PMO’s exon-skipping efficacy [236].
The animal PPMO studies undertaken so far have been very promising however major care should be taken prior to translating these findings in human studies. In fact, lack of understanding of optimization of PPMO exon skipping efficiency via structural modification hinders the path for discovery of more potent PPMO cocktails. Current exon skipping therapy can target only one exon and thus is applicable to a limited number of individuals harbouring the specific mutation. Ongoing clinical trials with PMOs targeting exon 53, 45 and 52 are conducted by Sarepta Pharmaceutics. Even so, none of the mentioned regimes can address patients that harbour mutations in other exons. Exons 3-9 [237] and 45-55 [238] are mutational hotspots in the DMD gene accounting for 7% - 47% of patients. Therefore, if a multi-exon skipping treatment regime covering these two hot spot regions will be approved, this treatment may be applicable to almost half of the DMD community. At the same time, a drug that targets multi-exon skipping of exons 45–55 would render patients asymptomatic, circumventing the unknown truncated protein stability/function factor issue, as this type of DMD patient exhibit only mild characteristics of disease [239]. Detection of truncated DMD mRNAs around the mutational hot spot reveals the presence, among others, of lowly expressed exon 44-56 multi-exon skipping product of DMD mRNAs which is an ideal induction target for exon skipping therapy [240]. Studies in myotubes transdifferentiated from DMD patient fibroblast cells recently published demonstrated that PMO oligomers could produce a dose-dependent exon 45-55 skipping, indicating that such a therapeutic effect could be more effectively achieved using PPMOs in the near future [239].
The future goal should be a development of a genetic strategy that can apply to all individuals with DMD [241]. In order to achieve an efficient multiple exon skipping that could be potentially applicable to 90% of patients [242] we need to administer PPMOs as a cocktail rather than single drugs. This will require synthesis and screening of different oligomers as well as validation of their safety and efficacy throughout clinical trials. Such an approach will inevitably increase the cost of treatment as well as the risk of potential immunological complications arising by a combination of different chemical substances.
Finally we have to keep in mind that a rather conservative approach is required when reviewing and validating drug efficacy data that are solidly based on dystrophin quantification; because a clear, dose-response effect on the DMD phenotype positively correlating to the amount of dystrophin protein quantified from muscle biopsies has not been established yet [243]. In vitro exon skipping efficacy and quantification of dystrophin expression is mainly performed using primary cell models but such lines are hard to maintain and alternative screening systems are currently under investigation [244]. A more objective post-therapeutic evaluation of muscle function recovery is pending due to the inadequacy in assessment of muscle function in the clinical situation. Besides, muscle biopsy is a highly invasive technique, particularly difficult to excuse its necessity or intensify its frequency if the patients involved are children. Physical evaluation of a patient’s motor activity is of pivotal importance, however this can only be done when the patient visits the hospital and fails to monitor progress in daily tasks [99] and data obtained from functional measures such as the 6MWT were very debatable at all previous clinical trials, leading to the exclusion of many patients [91]. Identification of novel pharmacological biomarkers will hopefully allow for a more patient-friendly assessment approach of PPMO therapeutic effect. Such an example of biomarkers are miRNAs that are released into the bloodstream of DMD patients as a result of fiber damage, can be detected in a serum sample and their fold change elevation levels coincide with the severity of disease whilst their reduction is associated with positive response to exon skipping treatment and dystrophin restoration [245,246].
So what steps we need to take in order to facilitate translation of these novel PPMO treatments to the clinic? It is clear that a multidisciplinary approach is needed to ensure safety of future clinical trials. Firstly, it has to be noted that although a great number of DMD animal models is currently available none of these models can perfectly assimilate human phenotype, posing significant hurdles in drug testing. The widespread used mdx murine model exhibits the pathological characteristics of disease but has a mild phenotype and normal life span and switching to larger mammals where we can more accurately evaluate key clinical milestones poses ethical and financial issues that are prohibiting factors in maintaining such colonies in many research facilities [247]. Therefore it is a priority to establish more appropriate pre-clinical models of disease. Human induced pluripotent stem cells (iPS) have been successfully programmed to produce skeletal muscle constructs expressing the characteristic markers of maturation such as MyoD and myosin heavy chain. Such iPS cells deriving from dystrophic patients can be used in order to create 3D skeletal muscle platforms that are accurately portraying the cellular hallmarks of disease [248]. Multiple studies have been conducted using patient derives iPS cells in order to evaluate approaches of dystrophin restoration such as exon skipping, exon knock in, CRISPR-Cas9 and thus development and evaluation of novel drugs [[249], [250], [251]]. In the future successful engraftment of corrected patient-derived iPS cells may turn to be a safe ex vivo applied gene therapy.
Variability in age and severity of clinical symptoms amongst the same species are common factors not only in existing DMD animal models but also in human patients. For that reason, there is a need to increase the number of clinical trials in humans in order to establish proof of concept with emphasis should be given to small clinical trials. Adequate preparation of necessary start up registries and follow up documents containing a detailed natural history of each patient is pivotal in order to accurately monitor beneficial and potential side effects of the drugs tested. Since the onset and clinical course of DMD is well described, emphasis by clinicians is currently given towards accurately monitoring disease milestones and progress (ability to walk, stand, climb stairs); therefore 6MWT test and timed function tests represent the current clinical endpoints [252,253]. Biochemical endpoints are correlated with the ability of gene therapy treatments to restore dystrophin and reduce fibrosis and to correlate such dystrophin production with improved muscle strength and physical activity especially in younger patients. The closest monitoring and pairing of both biochemical and clinical end points is essential in order to evaluate drug efficacy in early clinical trials. Improvement of existing clinical endpoints can be achieved by the use of novel technology; e.g. more sophisticated devices to effectively monitor and evaluate physical activity in patient’s natural environment.
8. Conclusions
Novel therapeutic strategies using antisense oligonucleotides have tremendously altered the clinical outcome, life expectancy and prognosis in patients in the field of neuromuscular disease. Over the last decade, two ASO based drugs have obtained approval from the FDA for the treatment of DMD and SMA respectively, paving the way for novel discoveries in molecular therapy. PPMO compounds show increased efficacy as splice correcting agents at lower doses than naked ASOs and effective restoration of dystrophin body wide distribution in skeletal tissues when administered systemically. Unfortunately attempts to utilize PPMOs in the treatment of neurodegenerative diseases have so far been plagued by their lack of delivery to the CNS plus their toxicity. Ongoing research will aid clarification of the pharmacodynamics and mode of action of PPMOs and will unravel their mechanism of beneficial action. This will hopefully enable synthesis of novel PPMO drugs that will minimize off-target effects and maximize efficient uptake in skeletal, respiratory and cardiac tissues and ultimately in the CNS, bringing hope for a better quality of life to patients.
Acknowledgements
The authors gratefully acknowledge fruitful discussions with Eijiro OZAWA. This work was supported by a Japan Society for the Promotion of Science Grant-in-Aid for Scientific Research (C) [grant number 18K07544 to Y.A.], Grants-in-Aid for Research on Nervous and Mental Disorders [grant number 28-6 to Y.A.], and the Japan Agency for Medical Research and Development [grant numbers 18ek0109239h0002, 18lm0203066h0001, and 18lm0203069h0001 to Y.A.].
References
- 1.Dickson G., Brown S.C. Duchenne muscular dystrophy. Mol Cell Biol Hum Dis Ser. 1995;5:261–280. doi: 10.1007/978-94-011-0547-7_14. [DOI] [PubMed] [Google Scholar]
- 2.Emery A.E.H. Population frequencies of inherited neuromuscular diseases-A world survey. Neuromuscul Disord. 1991;1:19–29. doi: 10.1016/0960-8966(91)90039-u. [DOI] [PubMed] [Google Scholar]
- 3.Hoffman E.P., Brown R.H., Kunkel L.M. Dystrophin: The protein product of the duchenne muscular dystrophy locus. Cell. 1987;51:919–928. doi: 10.1016/0092-8674(87)90579-4. [DOI] [PubMed] [Google Scholar]
- 4.Flanigan K.M. Duchenne and Becker Muscular Dystrophies. Neurol Clin. 2014;32:671–688. doi: 10.1016/j.ncl.2014.05.002. [DOI] [PubMed] [Google Scholar]
- 5.Henricson E.K., Abresch R.T., Cnaan A. The cooperative international neuromuscular research group Duchenne natural history study: Glucocorticoid treatment preserves clinically meaningful functional milestones and reduces rate of disease progression as measured by manual muscle testing and othe. Muscle Nerve. 2013;48:55–67. doi: 10.1002/mus.23808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Yiu E.M., Kornberg A.J. Duchenne muscular dystrophy. J Paediatr Child Health. 2015;51:759–764. doi: 10.1111/jpc.12868. [DOI] [PubMed] [Google Scholar]
- 7.Archer J.E., Gardner A.C., Roper H.P., Chikermane A.A., Tatman A.J. Duchenne muscular dystrophy: the management of scoliosis. J Spine Surg. 2016;2:185–194. doi: 10.21037/jss.2016.08.05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Blat Y., Blat S. Drug discovery of therapies for duchenne muscular dystrophy. J Biomol Screen. 2015;20:1189–1203. doi: 10.1177/1087057115586535. [DOI] [PubMed] [Google Scholar]
- 9.Lo Cascio C.M., Latshang T.D., Kohler M., Fehr T., Bloch K.E. Severe metabolic acidosis in adult patients with duchenne muscular dystrophy. Respiration. 2014;87:499–503. doi: 10.1159/000358439. [DOI] [PubMed] [Google Scholar]
- 10.Kohler M., Clarenbach C.F., Böni L., Brack T., Russi E.W., Bloch K.E. Quality of life, physical disability, and respiratory impairment in Duchenne muscular dystrophy. Am J Respir Crit Care Med. 2005;172:1032–1036. doi: 10.1164/rccm.200503-322OC. [DOI] [PubMed] [Google Scholar]
- 11.Mavrogeni S., Markousis-Mavrogenis G., Papavasiliou A., Kolovou G. Cardiac involvement in Duchenne and Becker muscular dystrophy. World J Cardiol. 2015;7:410–414. doi: 10.4330/wjc.v7.i7.410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fayssoil A., Abasse S., Silverston K. Cardiac Involvement Classification and Therapeutic Management in Patients with Duchenne Muscular Dystrophy. J Neuromuscul Dis. 2017;4:17–23. doi: 10.3233/JND-160194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rae M.G., O’Malley D. Cognitive dysfunction in Duchenne muscular dystrophy: a possible role for neuromodulatory immune molecules. J Neurophysiol. 2016;116:1304–1315. doi: 10.1152/jn.00248.2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Battini R., Chieffo D., Bulgheroni S. Cognitive profile in Duchenne muscular dystrophy boys without intellectual disability: The role of executive functions. Neuromuscul Disord. 2018;28:122–128. doi: 10.1016/j.nmd.2017.11.018. [DOI] [PubMed] [Google Scholar]
- 15.Vicari S., Piccini G., Mercuri E. Implicit learning deficit in children with Duchenne muscular dystrophy: Evidence for a cerebellar cognitive impairment? PLoS One. 2018;13 doi: 10.1371/journal.pone.0191164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Eagle M., Baudouin S.V., Chandler C., Giddings D.R., Bullock R., Bushby K. Survival in Duchenne muscular dystrophy: Improvements in life expectancy since 1967 and the impact of home nocturnal ventilation. Neuromuscul Disord. 2002;12:926–929. doi: 10.1016/s0960-8966(02)00140-2. [DOI] [PubMed] [Google Scholar]
- 17.Wagner K.R., Lechtzin N., Judge D.P. Current treatment of adult Duchenne muscular dystrophy. Biochim Biophys Acta. 1772;2007:229–237. doi: 10.1016/j.bbadis.2006.06.009. [DOI] [PubMed] [Google Scholar]
- 18.Passamano L., Taglia A., Palladino A. Improvement of survival in Duchenne Muscular Dystrophy: Retrospective analysis of 835 patients. Acta Myol. 2012;31:121–125. [PMC free article] [PubMed] [Google Scholar]
- 19.Bushby K., Bourke J., Bullock R., Eagle M., Gibson M., Quinby J. The multidisciplinary management of Duchenne muscular dystrophy. Curr Paediatr. 2005;15:292–300. [Google Scholar]
- 20.Ahn A.H., Kunkel L.M. The structural and functional diversity of dystrophin. Nat Genet. 1993;3:283–291. doi: 10.1038/ng0493-283. [DOI] [PubMed] [Google Scholar]
- 21.Roberts R.G., Coffey A.J., Bobrow M., Bentley D.R. Exon Structure of the Human Dystrophin Gene. Genomics. 1993;16:536–538. doi: 10.1006/geno.1993.1225. [DOI] [PubMed] [Google Scholar]
- 22.Hoffman E.P., Knudson C.M., Campbell K.P., Kunkel L.M. Subcellular fractionation of dystrophin to the triads of skeletal muscle. Nature. 1987;330:754–758. doi: 10.1038/330754a0. [DOI] [PubMed] [Google Scholar]
- 23.Kim T.-W., Wu K., Black I.B. Deficiency of brain synaptic dystrophin in human duchenne muscular dystrophy. Ann Neurol. 1995;38:446–449. doi: 10.1002/ana.410380315. [DOI] [PubMed] [Google Scholar]
- 24.Constantin B. Dystrophin complex functions as a scaffold for signalling proteins. Biochim Biophys Acta - Biomembr. 1838;2014:635–642. doi: 10.1016/j.bbamem.2013.08.023. [DOI] [PubMed] [Google Scholar]
- 25.Koenig M., Beggs A.H., Moyer M. The molecular basis for duchenne versus becker muscular dystrophy: correlation of severity with type of deletion. Am J Hum Genet. 1989;45:498–506. [PMC free article] [PubMed] [Google Scholar]
- 26.Beggs A.H., Koenig M., Boyce F.M., Kunkel L.M. Detection of 98% of DMD/BMD gene deletions by polymerase chain reaction. Hum Genet. 1990;86:45–48. doi: 10.1007/BF00205170. [DOI] [PubMed] [Google Scholar]
- 27.Aartsma-Rus A., Ginjaar I.B., Bushby K. The importance of genetic diagnosis for Duchenne muscular dystrophy. J Med Genet. 2016;53:145–151. doi: 10.1136/jmedgenet-2015-103387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Duchenne Matsuo M., Becker Muscular dystrophy: from gene diagnosis to molecular therapy. IUBMB Life (International Union Biochem Mol Biol Life) 2002;53:147–152. doi: 10.1080/15216540212333. [DOI] [PubMed] [Google Scholar]
- 29.Gao Q.Q., McNally E.M. The dystrophin complex: Structure, function, and implications for therapy. Compr Physiol. 2015;5:1223–1239. doi: 10.1002/cphy.c140048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Muntoni F., Torelli S., Ferlini A. Dystrophin and mutations: one gene, several proteins, multiple phenotypes. Lancet Neurol. 2003;2:731–740. doi: 10.1016/s1474-4422(03)00585-4. [DOI] [PubMed] [Google Scholar]
- 31.Monaco A.P., Bertelson C.J., Liechti-Gallati S., Moser H., Kunkel L.M. An explanation for the phenotypic differences between patients bearing partial deletions of the DMD locus. Genomics. 1988;2:90–95. doi: 10.1016/0888-7543(88)90113-9. [DOI] [PubMed] [Google Scholar]
- 32.Roberts R.G., Bobrow M., Bentley D.R. Point mutations in the dystrophin gene. Proc Natl Acad Sci U S A. 1992;89:2331–2335. doi: 10.1073/pnas.89.6.2331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Meregalli M., Maciotta S., Angeloni V., Torrente Y. Duchenne muscular dystrophy caused by a frame-shift mutation in the acceptor splice site of intron 26. BMC Med Genet. 2016;17:55. doi: 10.1186/s12881-016-0318-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kerr T.P., Sewry C.A., Robb S.A., Roberts R.G. Long mutant dystrophins and variable phenotypes: Evasion of nonsense-mediated decay? Hum Genet. 2001;109:402–407. doi: 10.1007/s004390100598. [DOI] [PubMed] [Google Scholar]
- 35.Petrof B.J., Shrager J.B., Stedman H.H., Kelly A.M., Sweeney H.L. Dystrophin protects the sarcolemma from stresses developed during muscle contraction. Proc Natl Acad Sci. 1993;90:3710–3714. doi: 10.1073/pnas.90.8.3710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Allen D.G., Whitehead N.P., Froehner S.C. Absence of dystrophin disrupts Skeletal muscle signaling: roles of Ca 2+, reactive oxygen species, and nitric oxide in the development of muscular dystrophy. Physiol Rev. 2016;96:253–305. doi: 10.1152/physrev.00007.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ohlendieck K. The pathophysiological role of impaired calcium handling in muscular dystrophy. Bookshelf. 2000:1–11. [Google Scholar]
- 38.Mozzetta C., Minetti G., Puri P.L. Regenerative pharmacology in the treatment of genetic diseases: The paradigm of muscular dystrophy. Int J Biochem Cell Biol. 2009;41:701–710. doi: 10.1016/j.biocel.2008.08.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Collins C.A., Olsen I., Zammit P.S. Stem cell function, self-renewal, and behavioral heterogeneity of cells from the adult muscle satellite cell niche. Cell. 2005;122:289–301. doi: 10.1016/j.cell.2005.05.010. [DOI] [PubMed] [Google Scholar]
- 40.Klingler W., Jurkat-Rott K., Lehmann-Horn F., Schleip R. The role of fibrosis in Duchenne muscular dystrophy. Acta Myol. 2012;31:184–195. [PMC free article] [PubMed] [Google Scholar]
- 41.Fairclough R.J., Bareja A., Davies K.E. Progress in therapy for Duchenne muscular dystrophy. Exp Physiol. 2011;96:1101–1113. doi: 10.1113/expphysiol.2010.053025. [DOI] [PubMed] [Google Scholar]
- 42.Chang W.J., Iannaccone S.T., Lau K.S. Neuronal nitric oxide synthase and dystrophin-deficient muscular dystrophy. Proc Natl Acad Sci U S A. 1996;93:9142–9147. doi: 10.1073/pnas.93.17.9142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Brenman J.E., Chao D.S., Xia H., Aldape K., Bredt D.S. Nitric oxide synthase complexed with dystrophin and absent from skeletal muscle sarcolemma in Duchenne muscular dystrophy. Cell. 1995;82:743–752. doi: 10.1016/0092-8674(95)90471-9. [DOI] [PubMed] [Google Scholar]
- 44.Hoffman E.P., Kunkel L.M., Angelini C., Clarke A., Johnson M., Harris J.B. Improved diagnosis of Becker muscular dystrophy by dystrophin testing. Neurology. 1989;39:1011–1017. doi: 10.1212/wnl.39.8.1011. [DOI] [PubMed] [Google Scholar]
- 45.Lu Q.-L., Yokota T., Takeda S., Garcia L., Muntoni F., Partridge T. The status of exon skipping as a therapeutic approach to duchenne muscular dystrophy. Mol Ther. 2011;19:9–15. doi: 10.1038/mt.2010.219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Cirak S., Arechavala-Gomeza V., Guglieri M. Exon skipping and dystrophin restoration in patients with Duchenne muscular dystrophy after systemic phosphorodiamidate morpholino oligomer treatment: An open-label, phase 2, dose-escalation study. Lancet. 2011;378:595–605. doi: 10.1016/S0140-6736(11)60756-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Matsumura K., Tomé F.M.S., Collin H. Expression of dystrophin-associated proteins in dystrophin-positive muscle fibers (revertants) in Duchenne muscular dystrophy. Neuromuscul Disord. 1994;4:115–120. doi: 10.1016/0960-8966(94)90002-7. [DOI] [PubMed] [Google Scholar]
- 48.García-Pelagio K.P., Bloch R.J., Ortega A., González-Serratos H. Biomechanics of the sarcolemma and costameres in single skeletal muscle fibers from normal and dystrophin-null mice. J Muscle Res Cell Motil. 2011;31:323–336. doi: 10.1007/s10974-011-9238-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.England S.B., Nicholson L.V.B.B., Johnson M.A. Very mild muscular dystrophy associated with the deletion of 46% of dystrophin. Nature. 1990;343:180–182. doi: 10.1038/343180a0. [DOI] [PubMed] [Google Scholar]
- 50.Matsuo M., Masumura T., Nishio H. Exon skipping during splicing of dystrophin mRNA precursor due to an intraexon deletion in the dystrophin gene of Duchenne muscular dystrophy Kobe. J Clin Invest. 1991;87:2127–2131. doi: 10.1172/JCI115244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Matsuo M., Masumura T., Nakajima T. A very small frame-shifting deletion within exon 19 of the Duchenne muscular dystrophy gene. Biochem Biophys Res Commun. 1990;170:963–967. doi: 10.1016/0006-291x(90)92185-3. [DOI] [PubMed] [Google Scholar]
- 52.Matsuo M. Duchenne/Becker muscular dystrophy: from molecular diagnosis to gene therapy. Brain Dev. 1996;18:167–172. doi: 10.1016/0387-7604(96)00007-1. [DOI] [PubMed] [Google Scholar]
- 53.Aartsma-Rus A., Fokkema I., Verschuuren J. Theoretic applicability of antisense-mediated exon skipping for Duchenne muscular dystrophy mutations. Hum Mutat. 2009;30:293–299. doi: 10.1002/humu.20918. [DOI] [PubMed] [Google Scholar]
- 54.Shin J., Tajrishi M.M., Ogura Y., Kumar A. Wasting mechanisms in muscular dystrophy. Int J Biochem Cell Biol. 2013;45:2266–2279. doi: 10.1016/j.biocel.2013.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Lee J.J.A., Yokota T. Translational Research in Muscular Dystrophy. Springer Japan; Tokyo: 2016. Translational research in nucleic acid therapies for muscular dystrophies; pp. 87–102. [Google Scholar]
- 56.Bennett C.F., Swayze E.E. RNA targeting therapeutics: molecular mechanisms of antisense oligonucleotides as a therapeutic platform. Annu Rev Pharmacol Toxicol. 2010;50:259–293. doi: 10.1146/annurev.pharmtox.010909.105654. [DOI] [PubMed] [Google Scholar]
- 57.Havens M.A., Duelli D.M., Hastings M.L. Targeting RNA splicing for disease therapy. Wiley Interdiscip Rev RNA. 2013;4:247–266. doi: 10.1002/wrna.1158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Eder P.S., Devine R.J., Dagle J.M., Walder J.A. Substrate specificity and kinetics of degradation of antisense oligonucleotides by a 3′ exonuclease in plasma. Antisense Res Dev. 1991;1:141–151. doi: 10.1089/ard.1991.1.141. [DOI] [PubMed] [Google Scholar]
- 59.Aoki Y., Yokota T., Wood M.J.A. Development of multiexon skipping antisense oligonucleotide therapy for duchenne muscular dystrophy. Biomed Res Int. 2013 doi: 10.1155/2013/402369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Freier S.M., Altmann K.H. The ups and downs of nucleic acid duplex stability: structure-stability studies on chemically-modified DNA:RNA duplexes. Nucleic Acids Res. 1997;25(22):4429–4443. doi: 10.1093/nar/25.22.4429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Juliano R.L. The delivery of therapeutic oligonucleotides. Nucleic Acids Res. 2016;44:6518–6548. doi: 10.1093/nar/gkw236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Nakamura A., Takeda S. Exon-skipping therapy for Duchenne muscular dystrophy. Neuropathology. 2009;29:494–501. doi: 10.1111/j.1440-1789.2009.01028.x. [DOI] [PubMed] [Google Scholar]
- 63.Rinaldi C., Wood M.J.A.A. Antisense oligonucleotides: the next frontier for treatment of neurological disorders. Nat Rev Neurol. 2017 doi: 10.1038/nrneurol.2017.148. [DOI] [PubMed] [Google Scholar]
- 64.Evers M.M., Toonen L.J.A., van Roon-Mom W.M.C. Antisense oligonucleotides in therapy for neurodegenerative disorders. Adv Drug Deliv Rev. 2015;87:90–103. doi: 10.1016/j.addr.2015.03.008. [DOI] [PubMed] [Google Scholar]
- 65.Summerton J., Weller D. Morpholino antissense oligomers: design, preparation, and properties. Antisense Nucleic Acid Drug Dev. 1997;7:187–195. doi: 10.1089/oli.1.1997.7.187. [DOI] [PubMed] [Google Scholar]
- 66.Warfield K.L., Panchal R.G., Aman M.J., Bavari S. Antisense treatments for biothreat agents. Curr Opin Mol Ther. 2006;8:93–103. [PubMed] [Google Scholar]
- 67.van Deutekom J.C., de Kimpe S.J., Campion G.V. Antisense oligonucleotides as personalized medicine for Duchenne muscular dystrophy. Drug Discov Today Ther Strateg. 2013;10:e149–e156. [Google Scholar]
- 68.Araki E., Nakamura K., Nakao K. Targeted disruption of exon 52 in the mouse dystrophin gene induced muscle degeneration similar to that observed in duchenne muscular dystrophy. Biochem Biophys Res Commun. 1997;238:492–497. doi: 10.1006/bbrc.1997.7328. [DOI] [PubMed] [Google Scholar]
- 69.Yucel N., Chang A.C., Day J.W., Rosenthal N., Blau H.M. Humanizing the mdx mouse model of DMD: the long and the short of it. npj Regen Med. 2018 doi: 10.1038/s41536-018-0045-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Graham I.R., Hill V.J., Manoharan M., Inamati G.B., Dickson G. Towards a therapeutic inhibition of dystrophin exon 23 splicing in mdx mouse muscle induced by antisense oligoribonucleotides (splicomers): Target sequence optimisation using oligonucleotide arrays. J Gene Med. 2004;6:1149–1158. doi: 10.1002/jgm.603. [DOI] [PubMed] [Google Scholar]
- 71.Mann C.J., Honeyman K., Cheng A.J. Antisense-induced exon skipping and synthesis of dystrophin in the mdx mouse. Proc Natl Acad Sci U S A. 2001;98:42–47. doi: 10.1073/pnas.011408598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Wu B., Lu P., Cloer C. Long-term rescue of dystrophin expression and improvement in muscle pathology and function in dystrophic mdx mice by peptide-conjugated morpholino. Am J Pathol. 2012;181:392–400. doi: 10.1016/j.ajpath.2012.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Lu Q.L., Rabinowitz A., Chen Y.C. Systemic delivery of antisense oligoribonucleotide restores dystrophin expression in body-wide skeletal muscles. Proc Natl Acad Sci. 2005;102:198–203. doi: 10.1073/pnas.0406700102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Gebski B.L., Mann C.J., Fletcher S., Wilton S.D. Morpholino antisense oligonucleotide induced dystrophin exon 23 skipping in mdx mouse muscle. Hum Mol Genet. 2003;12:1801–1811. doi: 10.1093/hmg/ddg196. [DOI] [PubMed] [Google Scholar]
- 75.Alter J., Lou F., Rabinowitz A. Systemic delivery of morpholino oligonucleotide restores dystrophin expression bodywide and improves dystrophic pathology. Nat Med. 2006;12:175–177. doi: 10.1038/nm1345. [DOI] [PubMed] [Google Scholar]
- 76.Aoki Y., Nakamura A., Yokota T. In-frame dystrophin following exon 51-skipping improves muscle pathology and function in the exon 52-deficient mdx mouse. Mol Ther. 2010;18:1995–2005. doi: 10.1038/mt.2010.186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Aoki Y., Nagata T., Yokota T. Highly efficient in vivo delivery of PMO into regenerating myotubes and rescue in laminin-α2 chain-null congenital muscular dystrophy mice. Hum Mol Genet. 2013;22:4914–4928. doi: 10.1093/hmg/ddt341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Aoki Y., Yokota T., Nagata T. Bodywide skipping of exons 45-55 in dystrophic mdx52 mice by systemic antisense delivery. Proc Natl Acad Sci. 2012;109:13763–13768. doi: 10.1073/pnas.1204638109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Chamberlain J.S., Metzger J., Reyes M., Townsend D., Faulkner J.A. Dystrophin-deficient mdx mice display a reduced life span and are susceptible to spontaneous rhabdomyosarcoma. FASEB J. 2007;21:2195–2204. doi: 10.1096/fj.06-7353com. [DOI] [PubMed] [Google Scholar]
- 80.Tanganyika-De Winter C.L., Heemskerk H., Karnaoukh T.G. Long-term exon skipping studies with 2'-o-methyl phosphorothioate antisense oligonucleotides in dystrophic mouse models. Mol Ther - Nucleic Acids. 2012;1 doi: 10.1038/mtna.2012.38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.McClorey G., Moulton H.M., Iversen P.L., Fletcher S., Wilton S.D. Antisense oligonucleotide-induced exon skipping restores dystrophin expression in vitro in a canine model of DMD. Gene Ther. 2006;13:1373–1381. doi: 10.1038/sj.gt.3302800. [DOI] [PubMed] [Google Scholar]
- 82.Yokota T., Lu Q.L., Partridge T. Efficacy of systemic morpholino exon-skipping in duchenne dystrophy dogs. Ann Neurol. 2009;65:667–676. doi: 10.1002/ana.21627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Lim K.R.Q., Echigoya Y., Nagata T. Efficacy of multi-exon skipping treatment in duchenne muscular dystrophy dog model neonates. Mol Ther. 2019 doi: 10.1016/j.ymthe.2018.10.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Miskew Nichols B., Aoki Y., Kuraoka M., Lee J.J.A., Takeda S., Yokota T. Multi-exon skipping using cocktail antisense oligonucleotides in the canine X-linked muscular dystrophy. J Vis Exp. 2016 doi: 10.3791/53776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Niks E.H., Aartsma-Rus A. Exon skipping: a first in class strategy for Duchenne muscular dystrophy. Expert Opin Biol Ther. 2017;17:225–236. doi: 10.1080/14712598.2017.1271872. [DOI] [PubMed] [Google Scholar]
- 86.Goemans N.M., Tulinius M., van den Akker J.T. Systemic administration of PRO051 in Duchenne's muscular dystrophy. N Engl J Med. 2011;364:1513–1522. doi: 10.1056/NEJMoa1011367. [DOI] [PubMed] [Google Scholar]
- 87.Voit T., Topaloglu H., Straub V. Safety and efficacy of drisapersen for the treatment of Duchenne muscular dystrophy (DEMAND II): An exploratory, randomised, placebo-controlled phase 2 study. Lancet Neurol. 2014;13:987–996. doi: 10.1016/S1474-4422(14)70195-4. [DOI] [PubMed] [Google Scholar]
- 88.Goemans N., Mercuri E., Belousova E. A randomized placebo-controlled phase 3 trial of an antisense oligonucleotide, drisapersen, in Duchenne muscular dystrophy. Neuromuscul Disord. 2018;28:4–15. doi: 10.1016/j.nmd.2017.10.004. [DOI] [PubMed] [Google Scholar]
- 89.Kesselheim A.S., Avorn J. Approving a problematic muscular dystrophy drug. JAMA. 2016;316:2357. doi: 10.1001/jama.2016.16437. [DOI] [PubMed] [Google Scholar]
- 90.Mendell J.R., Sahenk Z., Rodino-Klapac L.R. Clinical trials of exon skipping in Duchenne muscular dystrophy. Expert Opin Orphan Drugs. 2017;5:683–690. [Google Scholar]
- 91.Mendell J.R., Rodino-Klapac L.R., Sahenk Z. Eteplirsen for the treatment of Duchenne muscular dystrophy. Ann Neurol. 2013;74:637–647. doi: 10.1002/ana.23982. [DOI] [PubMed] [Google Scholar]
- 92.Kinali M., Arechavala-Gomeza V., Feng L. Local restoration of dystrophin expression with the morpholino oligomer AVI-4658 in Duchenne muscular dystrophy: a single-blind, placebo-controlled, dose-escalation, proof-of-concept study. Lancet Neurol. 2009;8:918–928. doi: 10.1016/S1474-4422(09)70211-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Wood M., Zhang J., Bowman K. WVE-210201, an investigational stereopure oligonucleotide therapy for Duchenne muscular dystrophy, induces Exon 51 skipping and dystrophin protein restoration. Neuromuscul Disord. 2017;27:S217. [Google Scholar]
- 94.Panzara M., Zhang J., Rinaldi C. Preclinical studies of WVE-210201, an investigational stereopure antisense oligonucleotide in development for the treatment of patients with duchenne muscular dystrophy (DMD) J Neurol Sci. 2017;381:277–278. [Google Scholar]
- 95.van Deutekom J.C., Janson A.A., Ginjaar I.B. Local dystrophin restoration with antisense oligonucleotide PRO051. N Engl J Med. 2007;357:2677–2686. doi: 10.1056/NEJMoa073108. [DOI] [PubMed] [Google Scholar]
- 96.Charleston J.S., Schnell F.J., Dworzak J. Eteplirsen for the treatment of duchenne muscular dystrophy (DMD) Neurology. 2017;88 [Google Scholar]
- 97.Mendell J.R., Goemans N., Lowes L.P. Longitudinal effect of eteplirsen versus historical control on ambulation in Duchenne muscular dystrophy. Ann Neurol. 2016;79:257–271. doi: 10.1002/ana.24555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Yong-fu L. In vivo delivery of morpholino oligos as therapeutics: what barriers still exist ? J Drug Discov Dev Deliv. 2016;3:1–2. [Google Scholar]
- 99.Nakamura A. Moving towards successful exon-skipping therapy for Duchenne muscular dystrophy. J Hum Genet. 2017;62:871–876. doi: 10.1038/jhg.2017.57. [DOI] [PubMed] [Google Scholar]
- 100.Lim K.R., Maruyama R., Yokota T. Eteplirsen in the treatment of Duchenne muscular dystrophy. Drug Des Devel Ther. 2017;11:533–545. doi: 10.2147/DDDT.S97635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Aslesh T., Maruyama R., Yokota T. Skipping multiple exons to treat DMD—promises and challenges. Biomedicines. 2018;6:1. doi: 10.3390/biomedicines6010001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Hoffman E.P., Bronson A., Levin A.A. Restoring dystrophin expression in duchenne muscular dystrophy muscle. Am J Pathol. 2011;179:12–22. doi: 10.1016/j.ajpath.2011.03.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Lu Q.L., Cirak S., Partridge T. What can we learn from clinical trials of exon skipping for DMD? Mol Ther - Nucleic Acids. 2014;3 doi: 10.1038/mtna.2014.6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Aartsma-Rus A., Krieg A.M. FDA approves eteplirsen for duchenne muscular dystrophy: the next chapter in the Eteplirsen Saga. Nucleic Acid Ther. 2017;27:1–3. doi: 10.1089/nat.2016.0657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Young C.S., Pyle A.D. Exon skipping therapy. Cell. 2016;167:1144. doi: 10.1016/j.cell.2016.10.050. [DOI] [PubMed] [Google Scholar]
- 106.Randeree L., Eslick G.D. Eteplirsen for paediatric patients with Duchenne muscular dystrophy: A pooled-analysis. J Clin Neurosci. 2018;49:1–6. doi: 10.1016/j.jocn.2017.10.082. [DOI] [PubMed] [Google Scholar]
- 107.Stein C.A. Eteplirsen approved for duchenne muscular dystrophy: the FDA faces a difficult choice. Mol Ther. 2016;24:1884–1885. doi: 10.1038/mt.2016.188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.González Torre J.A., Cruz-Gómez Á.J., Belenguer A., Sanchis-Segura C., Ávila C., Forn C. Hippocampal dysfunction is associated with memory impairment in multiple sclerosis: A volumetric and functional connectivity study. Mult Scler. 2017;23:1854–1863. doi: 10.1177/1352458516688349. [DOI] [PubMed] [Google Scholar]
- 109.Komaki H., Nagata T., Saito T. Systemic administration of the antisense oligonucleotide NS-065/NCNP-01 for skipping of exon 53 in patients with Duchenne muscular dystrophy. Sci Transl Med. 2018;10:eaan0713. doi: 10.1126/scitranslmed.aan0713. [DOI] [PubMed] [Google Scholar]
- 110.Muntoni F., Frank D., Sardone V. Golodirsen induces exon skipping leading to sarcolemmal dystrophin expression in duchenne muscular dystrophy patients with mutations amenable to Exon 53 skipping (S22.001) Neurology. 2018;90 http://n.neurology.org/content/90/15_Supplement/S22.001.abstract [Google Scholar]
- 111.Wood M.J.A. Toward an oligonucleotide therapy for duchenne muscular dystrophy: a complex development challenge. Sci Transl Med. 2010;2:25ps15. doi: 10.1126/scitranslmed.3000512. [DOI] [PubMed] [Google Scholar]
- 112.Goyenvalle A., Griffith G., Babbs A. Functional correction in mouse models of muscular dystrophy using exon-skipping tricyclo-DNA oligomers. Nat Med. 2015;21:270–275. doi: 10.1038/nm.3765. [DOI] [PubMed] [Google Scholar]
- 113.Moulton H.M., Wu B., Jearawiriyapaisarn N., Sazani P., Lu Q.L., Kole R. Peptide-morpholino conjugate: a promising therapeutic for duchenne muscular dystrophy. Ann N Y Acad Sci. 2009;1175:55–60. doi: 10.1111/j.1749-6632.2009.04976.x. [DOI] [PubMed] [Google Scholar]
- 114.Douglas A.G.L., Wood M.J.A. Splicing therapy for neuromuscular disease. Mol Cell Neurosci. 2013;56:169–185. doi: 10.1016/j.mcn.2013.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Miyatake S., Mizobe Y., Tsoumpra M.K. Scavenger receptor Class A1 mediates uptake of morpholino antisense oligonucleotide into dystrophic skeletal muscle. Mol Ther - Nucleic Acids. 2019 doi: 10.1016/j.omtn.2019.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Mokri B., Engel A.G. Duchenne dystrophy: electron microscopic findings pointing to a basic or early abnormality in the plasma membrane of the muscle fiber. Neurology. 1975;25:1111–1120. doi: 10.1212/wnl.25.12.1111. [DOI] [PubMed] [Google Scholar]
- 117.Malerba A., Sharp P.S., Graham I.R. Chronic systemic therapy with low-dose morpholino oligomers ameliorates the pathology and normalizes locomotor behavior in mdx mice. Mol Ther. 2011;19:345–354. doi: 10.1038/mt.2010.261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Moulton J.D., Jiang S. Gene knockdowns in adult animals: PPMOs and vivo-morpholinos. Molecules. 2009;14:1304–1323. doi: 10.3390/molecules14031304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Summerton J.E. Endo-Porter: a novel reagent for safe, effective delivery of substances into cells. Ann N Y Acad Sci. 2005;1058:62–75. doi: 10.1196/annals.1359.012. [DOI] [PubMed] [Google Scholar]
- 120.Morcos P.A., Li Y., Jiang S. Vivo-morpholinos: a non-peptide transporter delivers morpholinos into a wide array of mouse tissues. Biotechniques. 2008;45:613–623. doi: 10.2144/000113005. [DOI] [PubMed] [Google Scholar]
- 121.Wu B., Li Y., Morcos P.A., Doran T.J., Lu P., Lu Q.L. Octa-guanidine morpholino restores dystrophin expression in cardiac and skeletal muscles and ameliorates pathology in dystrophic mdx mice. Mol Ther. 2009;17:864–871. doi: 10.1038/mt.2009.38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Yokota T., Nakamura A., Nagata T. Extensive and prolonged restoration of dystrophin expression with vivo-morpholino-mediated multiple exon skipping in dystrophic dogs. Nucleic Acid Ther. 2012;22:306–315. doi: 10.1089/nat.2012.0368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Echigoya Y., Aoki Y., Miskew B. Long-term efficacy of systemic multiexon skipping targeting Dystrophin exons 45-55 with a cocktail of vivo-morpholinos in Mdx52 mice. Mol Ther - Nucleic Acids. 2015;4 doi: 10.1038/mtna.2014.76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Guidotti G., Brambilla L., Rossi D. Cell-penetrating peptides: from basic research to clinics. Trends Pharmacol Sci. 2017;38:406–424. doi: 10.1016/j.tips.2017.01.003. [DOI] [PubMed] [Google Scholar]
- 125.Said Hassane F., Saleh A.F., Abes R., Gait M.J., Lebleu B. Cell penetrating peptides: overview and applications to the delivery of oligonucleotides. Cell Mol Life Sci. 2010;67:715–726. doi: 10.1007/s00018-009-0186-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Moulton H.M., Hase M.C., Smith K.M., Iversen P.L. HIV Tat peptide enhances cellular delivery of antisense morpholino oligomers. Antisense Nucleic Acid Drug Dev. 2003;13:31–43. doi: 10.1089/108729003764097322. [DOI] [PubMed] [Google Scholar]
- 127.Yin H., Lu Q., Wood M. Effective exon skipping and restoration of dystrophin expression by peptide nucleic acid antisense oligonucleotides in mdx mice. Mol Ther. 2008;16:38–45. doi: 10.1038/sj.mt.6300329. [DOI] [PubMed] [Google Scholar]
- 128.Moulton H.M., Nelson M.H., Hatlevig S.A., Reddy M.T., Iversen P.L. Cellular uptake of antisense morpholino oligomers conjugated to arginine-rich peptides. Bioconjug Chem. 2004;15:290–299. doi: 10.1021/bc034221g. [DOI] [PubMed] [Google Scholar]
- 129.Richard J.P., Melikov K., Vives E. Cell-penetrating peptides: a reevaluation of the mechanism of cellular uptake. J Biol Chem. 2003;278:585–590. doi: 10.1074/jbc.M209548200. [DOI] [PubMed] [Google Scholar]
- 130.Abes R., Moulton H.M., Clair P. Delivery of steric block morpholino oligomers by (R-X-R)4peptides: structure-activity studies. Nucleic Acids Res. 2008;36:6343–6354. doi: 10.1093/nar/gkn541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Mitchell D.J., Steinman L., Kim D.T., Fathman C.G., Rothbard J.B. Polyarginine enters cells more efficiently than other polycationic homopolymers. J Pept Res. 2000;56:318–325. doi: 10.1034/j.1399-3011.2000.00723.x. [DOI] [PubMed] [Google Scholar]
- 132.Ivanova G.D., Arzumanov A., Abes R. Improved cell-penetrating peptide-PNA conjugates for splicing redirection in HeLa cells and exon skipping in mdx mouse muscle. Nucleic Acids Res. 2008;36:6418–6428. doi: 10.1093/nar/gkn671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Abes S., Turner J.J., Ivanova G.D. Efficient splicing correction by PNA conjugation to an R6-Penetratin delivery peptide. Nucleic Acids Res. 2007;35:4495–4502. doi: 10.1093/nar/gkm418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Youngblood D.S., Hatlevig S.A., Hassinger J.N., Iversen P.L., Moulton H.M. Stability of cell-penetrating peptide-morpholino oligomer conjugates in human serum and in cells. Bioconjug Chem. 2007;18:50–60. doi: 10.1021/bc060138s. [DOI] [PubMed] [Google Scholar]
- 135.Amantana A., Moulton H.M., Cate M.L. Pharmacokinetics, biodistribution, stability and toxicity of a cell-penetrating peptide - Morpholino oligomer conjugate. Bioconjug Chem. 2007;18:1325–1331. doi: 10.1021/bc070060v. [DOI] [PubMed] [Google Scholar]
- 136.Abes S., Moulton H.M., Clair P. Vectorization of morpholino oligomers by the (R-Ahx-R)4peptide allows efficient splicing correction in the absence of endosomolytic agents. J Control Release. 2006;116:304–313. doi: 10.1016/j.jconrel.2006.09.011. [DOI] [PubMed] [Google Scholar]
- 137.Lehto T., Ezzat K., Wood M.J.A., EL Andaloussi S. Peptides for nucleic acid delivery. Adv Drug Deliv Rev. 2016;106:172–182. doi: 10.1016/j.addr.2016.06.008. [DOI] [PubMed] [Google Scholar]
- 138.Hansen M., Kilk K., Langel Ü. Predicting cell-penetrating peptides. Adv Drug Deliv Rev. 2008;60:572–579. doi: 10.1016/j.addr.2007.09.003. [DOI] [PubMed] [Google Scholar]
- 139.Stein C.A., Hansen J.B., Lai J. Efficient gene silencing by delivery of locked nucleic acid antisense oligonucleotides, unassisted by transfection reagents. Nucleic Acids Res. 2009;38 doi: 10.1093/nar/gkp841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Soifer H.S., Koch T., Lai J. Silencing of gene expression by gymnotic delivery of antisense oligonucleotides. Methods Mol Biol. 2012;815:333–346. doi: 10.1007/978-1-61779-424-7_25. [DOI] [PubMed] [Google Scholar]
- 141.Ezzat K., Aoki Y., Koo T. Self-assembly into nanoparticles is essential for receptor mediated uptake of therapeutic antisense oligonucleotides. Nano Lett. 2015;15:4364–4373. doi: 10.1021/acs.nanolett.5b00490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Juliano R.L., Carver K. Cellular uptake and intracellular trafficking of oligonucleotides. Adv Drug Deliv Rev. 2015;87:35–45. doi: 10.1016/j.addr.2015.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.González-Barriga A., Nillessen B., Kranzen J. Intracellular distribution and nuclear activity of antisense oligonucleotides after unassisted uptake in myoblasts and differentiated myotubes in vitro. Nucleic Acid Ther. 2017;27:144–158. doi: 10.1089/nat.2016.0641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Crooke S.T., Wang S., Vickers T.A., Shen W., Liang X. Cellular uptake and trafficking of antisense oligonucleotides. Nat Biotechnol. 2017;35:230–237. doi: 10.1038/nbt.3779. [DOI] [PubMed] [Google Scholar]
- 145.Cleal K., He L., Watson P.D., Jones A.T. Endocytosis, intracellular traffic and fate of cell penetrating peptide based conjugates and nanoparticles. Curr Pharm Des. 2013;19:2878–2894. doi: 10.2174/13816128113199990297. [DOI] [PubMed] [Google Scholar]
- 146.Koller E., Vincent T.M., Chappell A., De S., Manoharan M., Bennett C.F. Mechanisms of single-stranded phosphorothioate modified antisense oligonucleotide accumulation in hepatocytes. Nucleic Acids Res. 2011;39:4795–4807. doi: 10.1093/nar/gkr089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Moulton H.M., Moulton J.D. Morpholinos and their peptide conjugates: Therapeutic promise and challenge for Duchenne muscular dystrophy. Biochim Biophys Acta - Biomembr. 1798;2010:2296–2303. doi: 10.1016/j.bbamem.2010.02.012. [DOI] [PubMed] [Google Scholar]
- 148.Ezzat K., Helmfors H., Tudoran O. Scavenger receptor-mediated uptake of cell-penetrating peptide nanocomplexes with oligonucleotides. FASEB J. 2012;26:1172–1180. doi: 10.1096/fj.11-191536. [DOI] [PubMed] [Google Scholar]
- 149.Joliot A., Pernelle C., Deagostini-Bazin H., Prochiantz A. Antennapedia homeobox peptide regulates neural morphogenesis. Proc Natl Acad Sci. 1991;88:1864–1868. doi: 10.1073/pnas.88.5.1864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Yin H., Saleh A.F., Betts C. Pip5 transduction peptides direct high efficiency oligonucleotide-mediated dystrophin exon skipping in heart and phenotypic correction in mdx mice. Mol Ther. 2011;19:1295–1303. doi: 10.1038/mt.2011.79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Lehto T., Alvarez A.C., Gauck S. Cellular trafficking determines the exon skipping activity of Pip6a-PMO in mdx skeletal and cardiac muscle cells. Nucleic Acids Res. 2014;42 doi: 10.1093/nar/gkt1220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Godfrey C., Desviat L.R., Smedsrød B. Delivery is key: lessons learnt from developing splice-switching antisense therapies. EMBO Mol Med. 2017;9:545–557. doi: 10.15252/emmm.201607199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Abes S., Williams D., Prevot P., Thierry A., Gait M.J., Lebleu B. Endosome trapping limits the efficiency of splicing correction by PNA-oligolysine conjugates. J Control Release. 2006;110:595–604. doi: 10.1016/j.jconrel.2005.10.026. [DOI] [PubMed] [Google Scholar]
- 154.Najjar K., Erazo-Oliveras A., Mosior J.W. Unlocking Endosomal Entrapment with Supercharged Arginine-Rich Peptides. Bioconjug Chem. 2017;28:2932–2941. doi: 10.1021/acs.bioconjchem.7b00560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Peccate C., Mollard A., Le Hir M. Antisense pre-treatment increases gene therapy efficacy in dystrophic muscles. Hum Mol Genet. 2015;25:3555–3563. doi: 10.1093/hmg/ddw201. [DOI] [PubMed] [Google Scholar]
- 156.Deconinck A.E., Rafael J.A., Skinner J.A. Utrophin-dystrophin-deficient mice as a model for Duchenne muscular dystrophy. Cell. 1997;90:717–727. doi: 10.1016/s0092-8674(00)80532-2. [DOI] [PubMed] [Google Scholar]
- 157.Goyenvalle A., Babbs A., Powell D. Prevention of dystrophic pathology in severely affected dystrophin/utrophin-deficient mice by morpholino-oligomer-mediated exon-skipping. Mol Ther. 2010;18:198–205. doi: 10.1038/mt.2009.248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Wu B., Cloer C., Lu P. Exon skipping restores dystrophin expression, but fails to prevent disease progression in later stage dystrophic dko mice. Gene Ther. 2014;21:785–793. doi: 10.1038/gt.2014.53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Jearawiriyapaisarn N., Moulton H.M., Buckley B. Sustained dystrophin expression induced by peptide-conjugated morpholino oligomers in the muscles of mdx mice. Mol Ther. 2008;16:1624–1629. doi: 10.1038/mt.2008.120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Yin H., Moulton H.M., Seow Y. Cell-penetrating peptide-conjugated antisense oligonucleotides restore systemic muscle and cardiac dystrophin expression and function. Hum Mol Genet. 2008;17:3909–3918. doi: 10.1093/hmg/ddn293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Yin H., Moulton H.M., Betts C. A fusion peptide directs enhanced systemic dystrophin exon skipping and functional restoration in dystrophin-deficient mdx mice. Hum Mol Genet. 2009;18:4405–4414. doi: 10.1093/hmg/ddp395. [DOI] [PubMed] [Google Scholar]
- 162.Wu B., Moulton H.M., Iversen P.L. Effective rescue of dystrophin improves cardiac function in dystrophin-deficient mice by a modified morpholino oligomer. Proc Natl Acad Sci. 2008;105:14814–14819. doi: 10.1073/pnas.0805676105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Gao X., Zhao J., Han G. Effective dystrophin restoration by a novel muscle-homing peptide-morpholino conjugate in dystrophin-deficient mdx mice. Mol Ther. 2014;22:1333–1341. doi: 10.1038/mt.2014.63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Muntoni F., Wood M.J.A. Targeting RNA to treat neuromuscular disease. Nat Rev Drug Discov. 2011;10:621–637. doi: 10.1038/nrd3459. [DOI] [PubMed] [Google Scholar]
- 165.Betts C., Saleh A.F., Arzumanov A.A. Pip6-PMO, a new generation of peptide-oligonucleotide conjugates with improved cardiac exon skipping activity for DMD treatment. Mol Ther - Nucleic Acids. 2012;1 doi: 10.1038/mtna.2012.30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Betts C.A., Saleh A.F., Carr C.A. Prevention of exercised induced cardiomyopathy following Pip-PMO treatment in dystrophic mdx mice. Sci Rep. 2015;5 doi: 10.1038/srep08986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Echigoya Y., Nakamura A., Nagata T. Effects of systemic multiexon skipping with peptide-conjugated morpholinos in the heart of a dog model of Duchenne muscular dystrophy. Proc Natl Acad Sci. 2017;114:4213–4218. doi: 10.1073/pnas.1613203114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Jirka S.M.G., Heemskerk H., Tanganyika-de Winter C.L. Peptide conjugation of 2′- O -methyl phosphorothioate antisense oligonucleotides enhances cardiac uptake and exon skipping in mdx mice. Nucleic Acid Ther. 2014;24:25–36. doi: 10.1089/nat.2013.0448. [DOI] [PubMed] [Google Scholar]
- 169.Jirka S.M.G., ’t Hoen PAC, Diaz Parillas V, et al. Cyclic peptides to improve delivery and exon skipping of antisense oligonucleotides in a mouse model for duchenne muscular dystrophy. Mol Ther. 2017;26:1–16. doi: 10.1016/j.ymthe.2017.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Mead A.F., Petrov M., Malik A.S. Diaphragm remodeling and compensatory respiratory mechanics in a canine model of Duchenne muscular dystrophy. J Appl Physiol. 2014;116 doi: 10.1152/japplphysiol.00833.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Stedman H.H., Sweeney H.L., Shrager J.B. The mdx mouse diaphragm reproduces the degenerative changes of Duchenne muscular dystrophy. Nature. 1991;352:536–539. doi: 10.1038/352536a0. [DOI] [PubMed] [Google Scholar]
- 172.Moulton H.M., Fletcher S., Neuman B.W. Cell-penetrating peptide–morpholino conjugates alter pre-mRNA splicing of DMD (Duchenne muscular dystrophy) and inhibit murine coronavirus replication in vivo. Biochem Soc Trans. 2007;35:826–828. doi: 10.1042/BST0350826. [DOI] [PubMed] [Google Scholar]
- 173.Fletcher S., Honeyman K., Fall A.M. Morpholino oligomer-mediated exon skipping averts the onset of dystrophic pathology in the mdx mouse. Mol Ther. 2007;15:1587–1592. doi: 10.1038/sj.mt.6300245. [DOI] [PubMed] [Google Scholar]
- 174.Yin H., Moulton H.M., Betts C. Functional rescue of dystrophin-deficient mdx mice by a chimeric peptide-PMO. Mol Ther. 2010 doi: 10.1038/mt.2010.151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Crisp A., Yin H.F., Goyenvalle A. Diaphragm rescue alone prevents heart dysfunction in dystrophic mice. Hum Mol Genet. 2011;20:413–421. doi: 10.1093/hmg/ddq477. [DOI] [PubMed] [Google Scholar]
- 176.Betts C.A., Saleh A.F., Carr C.A. Implications for cardiac function following rescue of the dystrophic diaphragm in a mouse model of duchenne muscular dystrophy. Sci Rep. 2015;5:1–13. doi: 10.1038/srep11632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Nigro G., Comi L.I., Politano L., Bain R.J.I. The incidence and evolution of cardiomyopathy in Duchenne muscular dystrophy. Int J Cardiol. 1990;26:271–277. doi: 10.1016/0167-5273(90)90082-g. [DOI] [PubMed] [Google Scholar]
- 178.Finsterer J., Stöllberger C. The heart in human dystrophinopathies. Cardiology. 2003;99:1–19. doi: 10.1159/000068446. [DOI] [PubMed] [Google Scholar]
- 179.Wu B., Lu P., Benrashid E. Dose-dependent restoration of dystrophin expression in cardiac muscle of dystrophic mice by systemically delivered morpholino. Gene Ther. 2010;17:132–140. doi: 10.1038/gt.2009.120. [DOI] [PubMed] [Google Scholar]
- 180.Wu B., Xiao B., Cloer C. One-year treatment of morpholino antisense oligomer improves skeletal and cardiac muscle functions in dystrophic mdx mice. Mol Ther. 2011;19:576–583. doi: 10.1038/mt.2010.288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Heemskerk H., De Winter C., Van Kuik P. Preclinical PK and PD studies on 2′-O-methyl-phosphorothioate RNA antisense oligonucleotides in the mdx mouse model. Mol Ther. 2010;18:1210–1217. doi: 10.1038/mt.2010.72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Vitiello L., Bassi N., Campagnolo P. In vivo delivery of naked antisense oligos in aged mdx mice: analysis of dystrophin restoration in skeletal and cardiac muscle. Neuromuscul Disord. 2008;18:597–605. doi: 10.1016/j.nmd.2008.05.011. [DOI] [PubMed] [Google Scholar]
- 183.Malerba A., Boldrin L., Dickson G. Long-term systemic administration of unconjugated morpholino oligomers for therapeutic expression of dystrophin by exon skipping in skeletal muscle: implications for cardiac muscle integrity. Nucleic Acid Ther. 2011;21:293–298. doi: 10.1089/nat.2011.0306. [DOI] [PubMed] [Google Scholar]
- 184.Townsend D.W., Yasuda S., Li S., Chamberlain J.S., Metzger J.M. Emergent dilated cardiomyopathy caused by targeted repair of dystrophic skeletal muscle. Mol Ther. 2008;16:832–835. doi: 10.1038/mt.2008.52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Jearawiriyapaisarn N., Moulton H.M., Sazani P., Kole R., Willis M.S. Long-term improvement in mdx cardiomyopathy after therapy with peptide-conjugated morpholino oligomers. Cardiovasc Res. 2010;85:444–453. doi: 10.1093/cvr/cvp335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Gao H.M., Hong J.S. Why neurodegenerative diseases are progressive: uncontrolled inflammation drives disease progression. Trends Immunol. 2008;29:357–365. doi: 10.1016/j.it.2008.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Gitler A.D., Dhillon P., Shorter J. Neurodegenerative disease: models, mechanisms, and a new hope. Dis Model Mech. 2017;10:499–502. doi: 10.1242/dmm.030205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.Wyss-Coray T. Ageing, neurodegeneration and brain rejuvenation. Nature. 2016;539:180–186. doi: 10.1038/nature20411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.Zhang D., Wang J., Xu D. Cell-penetrating peptides as noninvasive transmembrane vectors for the development of novel multifunctional drug-delivery systems. J Control Release. 2016;229:130–139. doi: 10.1016/j.jconrel.2016.03.020. [DOI] [PubMed] [Google Scholar]
- 190.Gao H. Progress and perspectives on targeting nanoparticles for brain drug delivery. Acta Pharm Sin B. 2016;6:268–286. doi: 10.1016/j.apsb.2016.05.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191.Mäe M., Langel U. Cell-penetrating peptides as vectors for peptide, protein and oligonucleotide delivery. Curr Opin Pharmacol. 2006;6:509–514. doi: 10.1016/j.coph.2006.04.004. [DOI] [PubMed] [Google Scholar]
- 192.Varatharaj A., Galea I. The blood-brain barrier in systemic inflammation. Brain Behav Immun. 2017;60:1–12. doi: 10.1016/j.bbi.2016.03.010. [DOI] [PubMed] [Google Scholar]
- 193.Stalmans S., Bracke N., Wynendaele E. Cell-penetrating peptides selectively cross the blood-brain barrier in vivo. PLoS One. 2015;10 doi: 10.1371/journal.pone.0139652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194.Zou L.-L., Ma J.-L., Wang T., Yang T.-B., Liu C.-B. Cell-penetrating peptide-mediated therapeutic molecule delivery into the central nervous system. Curr Neuropharmacol. 2013;11:197–208. doi: 10.2174/1570159X11311020006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 195.Cao G., Pei W., Ge H. In vivo delivery of a Bcl-xL fusion protein containing the TAT protein transduction domain protects against ischemic brain injury andneuronal apoptosis. J Neurosci. 2002;22:5423–5431. doi: 10.1523/JNEUROSCI.22-13-05423.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196.Shabanpoor F., Hammond S.M., Abendroth F., Hazell G., Wood M.J.A., Gait M.J. Identification of a peptide for systemic brain delivery of a morpholino oligonucleotide in mouse models of spinal muscular atrophy. Nucleic Acid Ther. 2017;27:130–143. doi: 10.1089/nat.2016.0652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 197.Du L., Kayali R., Bertoni C. Arginine-rich cell-penetrating peptide dramatically enhances AMO-mediated ATM aberrant splicing correction and enables delivery to brain and cerebellum. Hum Mol Genet. 2011;20:3151–3160. doi: 10.1093/hmg/ddr217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 198.Faravelli I., Nizzardo M., Comi G.P., Corti S. Spinal muscular atrophy—recent therapeutic advances for an old challenge. Nat Rev Neurol. 2015;11:351–359. doi: 10.1038/nrneurol.2015.77. [DOI] [PubMed] [Google Scholar]
- 199.Feldkötter M., Schwarzer V., Wirth R., Wienker T.F., Wirth B. Quantitative analyses of SMN1 and SMN2 based on real-time lightCycler PCR: fast and highly reliable carrier testing and prediction of severity of spinal muscular atrophy. Am J Hum Genet. 2002;70:358–368. doi: 10.1086/338627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 200.Lorson C.L., Strasswimmer J., Yao J.M. SMN oligomerization defect correlates with spinal muscular atrophy severity. Nat Genet. 1998 doi: 10.1038/ng0598-63. [DOI] [PubMed] [Google Scholar]
- 201.Burnett B.G., Munoz E., Tandon A., Kwon D.Y., Sumner C.J., Fischbeck K.H. Regulation of SMN protein stability. Mol Cell Biol. 2009 doi: 10.1128/MCB.01262-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 202.Hamilton G., Gillingwater T.H. Spinal muscular atrophy: going beyond the motor neuron. Trends Mol Med. 2013;19:40–50. doi: 10.1016/j.molmed.2012.11.002. [DOI] [PubMed] [Google Scholar]
- 203.Lim S.R., Hertel K.J. Modulation of survival motor neuron Pre-mRNA splicing by inhibition of alternative 3′ splice site pairing. J Biol Chem. 2001 doi: 10.1074/jbc.M107632200. [DOI] [PubMed] [Google Scholar]
- 204.Hua Y., Sahashi K., Hung G. Antisense correction of SMN2 splicing in the CNS rescues necrosis in a type III SMA mouse model. Genes Dev. 2010;24:1634–1644. doi: 10.1101/gad.1941310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 205.Passini M.A., Bu J., Richards A.M. Antisense oligonucleotides delivered to the mouse CNS ameliorate symptoms of severe spinal muscular atrophy. Sci Transl Med. 2011;3 doi: 10.1126/scitranslmed.3001777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 206.Sardone V., Zhou H., Muntoni F., Ferlini A., Falzarano M. Antisense oligonucleotide-based therapy for neuromuscular disease. Molecules. 2017;22:563. doi: 10.3390/molecules22040563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 207.Rigo F., Chun S.J., Norris D.A. Pharmacology of a central nervous system delivered 2'-O-methoxyethyl-modified survival of motor neuron splicing oligonucleotide in mice and nonhuman primates. J Pharmacol Exp Ther. 2014;350:46–55. doi: 10.1124/jpet.113.212407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 208.Finkel R.S., Mercuri E., Darras B.T. Nusinersen versus sham control in infantile-onset spinal muscular atrophy. N Engl J Med. 2017;377:1723–1732. doi: 10.1056/NEJMoa1702752. [DOI] [PubMed] [Google Scholar]
- 209.Parente V., Corti S. Advances in spinal muscular atrophy therapeutics. Ther Adv Neurol Disord. 2018;11 doi: 10.1177/1756285618754501. (175628561875450) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 210.Hammond S.M., Hazell G., Shabanpoor F. Systemic peptide-mediated oligonucleotide therapy improves long-term survival in spinal muscular atrophy. Proc Natl Acad Sci. 2016;113:10962–10967. doi: 10.1073/pnas.1605731113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 211.van der Bent M.L., Paulino da Silva Filho O., van Luijk J., Brock R., Wansink D.G. Assisted delivery of antisense therapeutics in animal models of heritable neurodegenerative and neuromuscular disorders: a systematic review and meta-analysis. Sci Rep. 2018;8:4181. doi: 10.1038/s41598-018-22316-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 212.Raymond L.A., André V.M., Cepeda C., Gladding C.M., Milnerwood A.J., Levine M.S. Pathophysiology of Huntington’s disease: time-dependent alterations in synaptic and receptor function. Neuroscience. 2011;198:252–273. doi: 10.1016/j.neuroscience.2011.08.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 213.Carroll J.B., Warby S.C., Southwell A.L. Potent and selective antisense oligonucleotides targeting single-nucleotide polymorphisms in the huntington disease gene/allele-specific silencing of mutant huntingtin. Mol Ther. 2011;19:2178–2185. doi: 10.1038/mt.2011.201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 214.Sun X., Marque L.O., Cordner Z. Phosphorodiamidate morpholino oligomers suppress mutant huntingtin expression and attenuate neurotoxicity. Hum Mol Genet. 2014;23:6302–6317. doi: 10.1093/hmg/ddu349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 215.Chen G.L., Ma Q., Goswami D., Shang J., Miller G.M. Modulation of nuclear REST by alternative splicing: a potential therapeutic target for Huntington’s disease. J Cell Mol Med. 2017;21:2974–2984. doi: 10.1111/jcmm.13209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 216.Tabrizi S., Leavitt B., Kordasiewicz H. Effects of IONIS-HTTRx in patients with early Huntington's disease, results of the first HTT-lowering drug trial (CT.002) Neurology. 2018;90 [Google Scholar]
- 217.Brown R.H., Al-Chalabi A. Amyotrophic lateral sclerosis. N Engl J Med. 2017;2:162–172. doi: 10.1056/NEJMra1603471. [DOI] [PubMed] [Google Scholar]
- 218.Rotunno M.S., Bosco D.A. An emerging role for misfolded wild-type SOD1 in sporadic ALS pathogenesis. Front Cell Neurosci. 2013;7 doi: 10.3389/fncel.2013.00253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 219.Rosen D.R., Siddique T., Patterson D. Mutations in Cu/Zn superoxide dismutase gene are associated with familial amyotrophic lateral sclerosis. Nature. 1993;362:59–62. doi: 10.1038/362059a0. [DOI] [PubMed] [Google Scholar]
- 220.Bali T., Self W., Liu J. Defining SOD1 ALS natural history to guide therapeutic clinical trial design. J Neurol Neurosurg Psychiatry. 2017;88:99–105. doi: 10.1136/jnnp-2016-313521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 221.Miller T.M., Pestronk A., David W. An antisense oligonucleotide against SOD1 delivered intrathecally for patients with SOD1 familial amyotrophic lateral sclerosis: A phase 1, randomised, first-in-man study. Lancet Neurol. 2013;12:435–442. doi: 10.1016/S1474-4422(13)70061-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 222.Schoch K.M., Miller T.M. Antisense oligonucleotides: translation from mouse models to human neurodegenerative diseases. Neuron. 2017;94:1056–1070. doi: 10.1016/j.neuron.2017.04.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 223.Nizzardo M., Simone C., Rizzo F. Morpholino-mediated SOD1 reduction ameliorates an amyotrophic lateral sclerosis disease phenotype. Sci Rep. 2016;6 doi: 10.1038/srep21301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 224.Lagier-Tourenne C., Baughn M., Rigo F. Targeted degradation of sense and antisense C9orf72 RNA foci as therapy for ALS and frontotemporal degeneration. Proc Natl Acad Sci. 2013;110:E4530–E4539. doi: 10.1073/pnas.1318835110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 225.Henry S.P., Beattie G., Yeh G. Complement activation is responsible for acute toxicities in rhesus monkeys treated with a phosphorothioate oligodeoxynucleotide. Int Immunopharmacol. 2002;2:1657–1666. doi: 10.1016/s1567-5769(02)00142-x. [DOI] [PubMed] [Google Scholar]
- 226.Nagaraju K., Vila M., Novak J. Humoral and cell mediated immune response to new dystrophin after morpholino-induced exon skipping therapy in dystrophin-deficient mdx mice. Neuromuscul Disord. 2018 [Google Scholar]
- 227.Sazani P., Gemignani F., Kang S.H. Systemically delivered antisense oligomers upregulate gene expression in mouse tissues. Nat Biotechnol. 2002;20:1228–1233. doi: 10.1038/nbt759. [DOI] [PubMed] [Google Scholar]
- 228.Sazani P., Ness K.P.V., Weller D.L., Poage D.W., Palyada K., Shrewsbury S.B. Repeat-dose toxicology evaluation in cynomolgus monkeys of AVI-4658, a phosphorodiamidate morpholino oligomer (PMO) drug for the treatment of duchenne muscular dystrophy. Int J Toxicol. 2011;30:313–321. doi: 10.1177/1091581811403505. [DOI] [PubMed] [Google Scholar]
- 229.Zhang A., Uaesoontrachoon K., Shaughnessy C. The use of urinary and kidney SILAM proteomics to monitor kidney response to high dose morpholino oligonucleotides in the mdx mouse. Toxicol Rep. 2015 doi: 10.1016/j.toxrep.2015.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 230.Gait M.J., Arzumanov A.A., McClorey G. Cell-penetrating peptide conjugates of steric blocking oligonucleotides as therapeutics for neuromuscular diseases from a historical perspective to current prospects of treatment. Nucleic Acid Ther. 2018 doi: 10.1089/nat.2018.0747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 231.Henry S.P., Bolte H., Auletta C., Kornbrust D.J. Evaluation of the toxicity of ISIS 2302, a phosphorothioate oligonucleotide, in a four-week study in cynomolgus monkeys. Toxicology. 1997;120:145–155. doi: 10.1016/s0300-483x(97)03661-5. [DOI] [PubMed] [Google Scholar]
- 232.Burki U., Keane J., Blain A. Development and application of an ultrasensitive hybridization-based ELISA method for the determination of peptide-conjugated phosphorodiamidate morpholino oligonucleotides. Nucleic Acid Ther. 2015;25:275–284. doi: 10.1089/nat.2014.0528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 233.Malerba A., Thorogood F.C., Dickson G., Graham I.R. Dosing regimen has a significant impact on the efficiency of morpholino oligomer-induced exon Skipping in mdx mice. Hum Gene Ther. 2009;20:955–965. doi: 10.1089/hum.2008.157. [DOI] [PubMed] [Google Scholar]
- 234.Kharraz Y., Guerra J., Pessina P., Serrano A.L., Muñoz-Cánoves P. Understanding the process of fibrosis in duchenne muscular dystrophy. Biomed Res Int. 2014;2014:1–11. doi: 10.1155/2014/965631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 235.Popplewell L.J., Trollet C., Dickson G., Graham I.R. Design of phosphorodiamidate morpholino oligomers (PMOs) for the induction of exon skipping of the human DMD gene. Mol Ther. 2009;17:554–561. doi: 10.1038/mt.2008.287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 236.Echigoya Y., Mouly V., Garcia L., Yokota T., Duddy W. In silico screening based on predictive algorithms as a design tool for exon skipping oligonucleotides in duchenne muscular dystrophy. PLoS One. 2015;10 doi: 10.1371/journal.pone.0120058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 237.Nakamura A., Fueki N., Shiba N. Deletion of exons 3-9 encompassing a mutational hot spot in the DMD gene presents an asymptomatic phenotype, indicating a target region for multiexon skipping therapy. J Hum Genet. 2016;61:663–667. doi: 10.1038/jhg.2016.28. [DOI] [PubMed] [Google Scholar]
- 238.Nakamura A., Shiba N., Miyazaki D. Comparison of the phenotypes of patients harboring in-frame deletions starting at exon 45 in the Duchenne muscular dystrophy gene indicates potential for the development of exon skipping therapy. J Hum Genet. 2017;62:459–463. doi: 10.1038/jhg.2016.152. [DOI] [PubMed] [Google Scholar]
- 239.Lee J., Echigoya Y., Duddy W. Antisense PMO cocktails effectively skip dystrophin exons 45-55 in myotubes transdifferentiated from DMD patient fibroblasts. PLoS One. 2018;13 doi: 10.1371/journal.pone.0197084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 240.Suzuki H., Aoki Y., Kameyama T. Endogenous multiple exon skipping and back-splicing at the DMD mutation hotspot. Int J Mol Sci. 2016;17 doi: 10.3390/ijms17101722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 241.Abdul-Razak H., Malerba A., Dickson G. Advances in gene therapy for muscular dystrophies. F1000Research. 2016;5:2030. doi: 10.12688/f1000research.8735.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 242.Yokota T., Duddy W., Partridge T. Optimizing exon skipping therapies for DMD. Acta Myol Myopathies Cardiomyopathies. 2007;26:179–184. [PMC free article] [PubMed] [Google Scholar]
- 243.Aartsma-Rus A., Straub V., Hemmings R. Development of exon skipping therapies for duchenne muscular dystrophy: a critical review and a perspective on the outstanding issues. Nucleic Acid Ther. 2017;27(5):251–259. doi: 10.1089/nat.2017.0682. (nat.2017.0682) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 244.Nguyen Q., Yokota T. Immortalized muscle cell model to test the exon skipping efficacy for duchenne muscular dystrophy. J Pers Med. 2017;7:13. doi: 10.3390/jpm7040013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 245.Cacchiarelli D., Legnini I., Martone J. miRNAs as serum biomarkers for Duchenne muscular dystrophy. EMBO Mol Med. 2011 doi: 10.1002/emmm.201100133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 246.Giordani L., Sandoná M., Rotini A., Puri P.L., Consalvi S., Saccone V. Muscle-specific microRNAs as biomarkers of Duchenne Muscular Dystrophy progression and response to therapies. Rare Dis (Austin, Tex) 2014 doi: 10.4161/21675511.2014.974969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 247.Wells D.J. Tracking progress: an update on animal models for Duchenne muscular dystrophy. Dis Model Mech. 2018 doi: 10.1242/dmm.035774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 248.Maffioletti S.M., Sarcar S., Henderson A.B.H. Three-dimensional human iPSC-derived artificial skeletal muscles model muscular dystrophies and enable multilineage tissue engineering. Cell Rep. 2018 doi: 10.1016/j.celrep.2018.03.091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 249.Li H.L., Fujimoto N., Sasakawa N. Precise correction of the dystrophin gene in duchenne muscular dystrophy patient induced pluripotent stem cells by TALEN and CRISPR-Cas9. Stem Cell Reports. 2015 doi: 10.1016/j.stemcr.2014.10.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 250.Ifuku M., Iwabuchi K.A., Tanaka M., Siu M., Lung Y., Hotta A. Exon Skipping and Inclusion Therapies: Methods and Protocols. 2018. Restoration of dystrophin protein expression by exon skipping utilizing CRISPR-Cas9 in myoblasts derived from DMD patient iPS cells. [DOI] [PubMed] [Google Scholar]
- 251.Shoji E., Sakurai H., Nishino T. Early pathogenesis of Duchenne muscular dystrophy modelled in patient-derived human induced pluripotent stem cells. Sci Rep. 2015 doi: 10.1038/srep12831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 252.Merlini L., Sabatelli P. Improving clinical trial design for Duchenne muscular dystrophy. BMC Neurol. 2015 doi: 10.1186/s12883-015-0408-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 253.McDonald C.M., Henricson E.K., Han J.J. The 6-minute walk test as a new outcome measure in duchenne muscular dystrophy. Muscle Nerve. 2010 doi: 10.1002/mus.21544. [DOI] [PubMed] [Google Scholar]