Abstract
Background
Severe malarial anaemia (SMA) is a leading cause of childhood mortality in holoendemic Plasmodium falciparum regions.
Methods
To gain an improved understanding of SMA pathogenesis, whole genome and transcriptome profiling was performed in Kenyan children (n = 144, 3–36 months) with discrete non-SMA and SMA phenotypes. Leukocyte associated immunoglobulin like receptor 1 (LAIR1) emerged as a predictor of susceptibility to SMA (P < 1 × 10−2, OR: 0.44–1.37), and was suppressed in severe disease (−1.69-fold, P = 0.004). To extend these findings, the relationship between LAIR1 polymorphisms [rs6509867 (16231C>A); rs2287827 (18835G>A)] and clinical outcomes were investigated in individuals (n = 1512, <5 years) at enrolment and during a 36-month longitudinal follow-up.
Findings
Inheritance of the 16,231 recessive genotype (AA) increased susceptibility to SMA at enrolment (OR = 1.903, 95%CI: 1.252–2.891, P = 0.003), and longitudinally (RR = 1.527, 95%CI: 1.119–2.083, P = 0.008). Carriage of the 18,835 GA genotype protected against SMA cross-sectionally (OR = 0.672, 95%CI: 0.480–0.9439, P = 0.020). Haplotype carriage (C16231A/G18835A) also altered cross-sectional susceptibility to SMA: CG (OR = 0.717, 95%CI: 0.527–0.9675, P = 0.034), CA (OR = 0.745, 95%CI: 0.536–1.036, P = 0.080), and AG (OR = 1.641, 95%CI: 1.160–2.321, P = 0.005). Longitudinally, CA carriage was protective against SMA (RR = 0.715, 95%CI: 0.554–0.923, P = 0.010), while AG carriage had an additive effect on enhanced SMA risk (RR = 1.283, 95%CI: 1.057–1.557, P = 0.011). Variants that protected against SMA had elevated LAIR1 transcripts, while those with enhanced risk had lower expression (P < 0.05). Inheritance of 18,835 GA reduced all-cause mortality by 44.8% (HR = 0.552, 95%CI: 0.329–0.925, P = 0.024), while AG haplotype carriage increased susceptibility by 68% (HR = 1.680, 95%CI: 1.020–2.770, P = 0.040).
Interpretation
These findings suggest LAIR1 is important for modulating susceptibility to SMA and all-cause childhood mortality.
Keywords: Leukocyte associated immunoglobulin like receptor 1, Plasmodium falciparum malaria, Severe malarial anaemia, All-cause mortality
Research in context.
Evidence before this study
Leukocyte associated immunoglobulin like receptor 1 (LAIR1) encodes for a transmembrane inhibitory receptor expressed by peripheral blood mononuclear cells (PBMCs), and is required for regulation of gene pathways involved in leukocyte inflammatory mediator production and cytotoxicity. While a considerable number of the gene pathways subject to LAIR1 regulation could be important in the pathogenesis of paediatric severe malarial anaemia (SMA), the impact of LAIR1 genetic variation on susceptibility to SMA remains to be investigated.
Added value of this study
In this study, we determined the role of LAIR1 polymorphisms and altered expression profiles in modulating susceptibility to SMA in Kenyan children. Two novel LAIR1 SNPs, and their haplotypic constructs, altered susceptibility to SMA at enrolment and longitudinally over a 36-month follow-up period. Carriage of one of the haplotypes that enhanced susceptibility to SMA was also associated with all-cause mortality. Genetic variants associated with increased risk of SMA had reduced LAIR1 transcript expression, while those linked with protection against severe disease had elevated enhanced transcript levels.
Implications of all the available evidence
Since our findings demonstrate that LAIR1 influences susceptibility to severe malaria and all-cause mortality during the critical phases of naturally-acquired immunity, LAIR1 appears to be an important novel gene involved in conditioning the pathogenesis of severe malaria.
Alt-text: Unlabelled Box
1. Introduction
Plasmodium falciparum malaria remains a significant public health problem, as illustrated by an increased incidence of clinical cases from 211 to 216 million between 2015 and 2016 [1]. The burden of malaria is predominantly in sub-Saharan Africa, which accounts for 90% of the global morbidity and 91% of the mortality, with the majority of the deaths occurring in children under 5 years (285,000, 64% of the global malaria deaths) [1]. Due to lack of acquired malarial immunity, children under 5 years in holoendemic transmission regions are susceptible to developing severe malaria [1]. Severe manifestations vary in presentation according to transmission intensity and include cerebral malaria, metabolic acidosis, respiratory distress and severe malarial anaemia (SMA) defined by haemoglobin (Hb) <5.0 g/dL [1]. In developing countries, SMA is a leading cause of illness and death in paediatric populations residing in holoendemic transmission regions such as Siaya County, western Kenya, which has been the focus of our scientific investigations over the last 18 years [[2], [3], [4]].
We have previously shown that polymorphic variation in host immune response genes are associated with altered susceptibility to SMA in Kenyan children [[5], [6], [7], [8], [9]]. Identification of genetic markers that underlie different malaria clinical phenotypes can reveal host genetic pathways and networks that influence the development of SMA, and thereby discover novel therapeutic targets.
To identify gene networks that influence susceptibility to SMA, we performed a pilot study using high-throughput genotyping and whole transcriptome profiling in a subset of parasitemic children with polarized clinical phenotypes from western Kenya (n = 144, aged, 3–36 months). To identify robust signals that could be validated in the entire cohort of children, the exploratory studies included malaria-infected children with high and low Hb concentrations: uncomplicated malaria (Hb = 8.0–10.9 g/dL) and SMA (Hb < 5.0 g/dL), respectively. Leukocyte associated immunoglobulin like receptor 1 (LAIR1, CD305) emerged as a gene strongly associated with SMA.
LAIR1 is an inhibitory transmembrane receptor expressed by peripheral blood mononuclear cells (PBMCs) which maps to a region of chromosome 19q13.4 known as the leukocyte receptor complex (LRC) [[10], [11], [12], [13], [14]]. The LRC comprises a large cluster of cell surface receptors, some of which influence susceptibility to malaria e.g., Killer immunoglobulin like receptors (KIR) [15]. Leukocyte signalling through LAIR1 is an important mechanism required for regulation of inflammatory mediator production and leukocyte cytotoxicity [16].
Previous investigations showed that reduced production of LAIR1 was associated with increased susceptibility to systemic lupus erythematous (SLE) and chronic lymphocytic leukaemia (CLL) [[17], [18], [19], [20]]. To date, there only appears to be one published manuscript describing the impact of LAIR1 genetic variants on disease outcomes [20]. In that study, single nucleotide polymorphisms (SNPs, i.e., rs56802430 and rs11084332) altered the risk of developing pemphigus in a Brazilian population.
Recent studies highlight the potentially important role of LAIR1 in human malaria. For example, antibodies from individuals residing in malaria endemic regions can acquire broad reactivity through a novel mechanism in which insertion of a DNA fragment between the V and DJ segments encodes the entire collagen-binding domain of LAIR1 [21,22]. Although the insert is necessary for binding to specific RIFINs, LAIR1-containing antibodies do not appear to enhance protection against febrile malaria [21,22]. Given the potential clinical significance of LAIR1, the current study examined the relationship between LAIR1 variants and susceptibility to malaria and SMA (upon enrolment and over 36 mos. of longitudinal follow-up). In addition, we determined the association between LAIR1 variants and all-cause mortality throughout the 36 mos. Period.
2. Materials and methods
2.1. Study site and participants
Study participants (n = 1512) were enrolled at the Siaya County Referral Hospital (SCRH), Siaya County, western Kenya. Enrolment for the study participants began 4/2004 and completion of all follow-up visits ended on 9/2015. Siaya is a holoendemic P. falciparum transmission region largely inhabited by the Luo ethnic tribe (>96%), thus providing a homogeneous population for genetic-based studies. Written informed consent in the language of choice (English, Kiswahili, or Dhuoluo) was obtained from the parent/guardian of each child participating in the study. A questionnaire was used to collect demographic and clinical information. Children were excluded from the study if they tested positive for non-P. falciparum species, were previously hospitalized, or were diagnosed with cerebral malaria. The aparasitemic group comprised children with a P. falciparum negative blood smear who presented for childhood vaccinations, and those who presented at hospital for non-malarial diseases. Children with any density P. falciparum malaria were stratified into SMA (Hb < 5.0 g/dL) and non-SMA (Hb ≥ 5.0 g/dL) based on haemoglobin concentrations. The cross-sectional and longitudinal analyses used these clinical definitions. All children were tested for HIV-1 and bacteraemia since these co-infections have been shown to influence malarial anaemia severity [23,24]. Pre- and post-test HIV counselling was provided to the parents/guardians of the study participants. The study was approved by the Scientific Ethics and Research Committee of the Kenya Medical Research Institute (KEMRI), Maseno University Ethics Research Committee (MUERC), and the University of New Mexico Institutional Review Board. Patients were treated according to the Ministry of Health (MOH)-Kenya guidelines.
2.2. Laboratory procedures
Venous blood samples (≤3.0 mL) were collected in EDTA-containing vacutainer tubes, prior to treatment. Trophozoite counts in peripheral blood smears and the reticulocyte production index (RPI) were determined according to our previous methods [25]. Complete haematological parameters were determined using a Beckman Coulter® AcT diff2™ (Beckman–Coulter Corporation). Screening for α-thalassaemia deletional determinant (−α3.7) was performed through nested PCR as previously described [26]. Presence of sickle-cell trait (HbAS) was determined by cellulose acetate electrophoresis per the manufacturer's conditions (Helena Bio-Sciences, Oxford, United Kingdom). Glucose-6-phosphate dehydrogenase (G6PD) deficiency was determined using TaqMan® polymerase chain reaction (PCR) assays as previously published [27]. HIV-1 exposure was determined by serological testing, while HIV infection was assessed by HIV-1 proviral DNA PCR testing according to our published methods [23]. Bacteraemia diagnosis was determined using API biochemical galleries (bioMérieux, Louvres, France) as previously described [24].
2.3. High-throughput genotyping and whole transcriptome profiling
Genomic DNA was extracted from buccal swabs, using the MasterAmp™ Buccal swab DNA extraction kit (Epicentre Biotechnologies, Madison, WI). Individuals (n = 144, aged 3–36 mos.) were selected from a parasitemic cohort (n = 1220), excluding those with HIV-1 (exposed and +ve), bacteraemia, α-thalassaemia (double deletion), HbSS, and G6PD deficiency since these factors can affect Hb levels. Children were stratified into ‘polarized extremes’ of non-SMA [(Hb = 8.0–10.9 g/dL; avg. Hb = 9.8), n = 74] and SMA; [(Hb < 5.0 g/dL; avg. Hb = 4.1), n = 70]. To identify SNPs associated with SMA, we performed a genome-wide association study (GWAS) using the Illumina® Infinium® HD Super Assay in conjunction with Illumina's® Human Omni2.5-8v1 BeadChip (with >2.45 M markers), and high-throughput genotyping with the Human Immunochip (coated with >196 K markers, Illumina®, CA, USA). GWAS and Immunochip® data were analysed using logistic regression analysis with an additive mode of inheritance.
Similarly, for global transcriptome profiling, samples were selected to represent “polarized extremes” of clinical phenotypes as indicated above; non-SMA [(Hb, 8.0–10.9 g/dL; avg. Hb = 9.4 g/dL), n = 35] and SMA [(Hb < 5.0 g/dL; avg. Hb = 3.6 g/dL), n = 13], excluding children with co-infections and haemoglobinopathies (as listed above). Global gene expression profiling was performed using the Illumina® HumanHT-12 v4 beadchip (Illumina®, CA, USA) covering >19,185 transcripts on the Illumina® “iScanSQ” platform. Illumina GenomeStudio® software was used to subtract the background signal from the transcript signal, which generated a list of differentially expressed transcripts and were further analysed using SNP & Variation Suite 8.8.3 Software (Golden Helix, USA). Whole transcriptome expression data were used to identify transcripts that were differentially expressed (P < 0.01) with fold-change expression differences ≥1.5.
2.4. LAIR1 genotyping
Genomic DNA was amplified using GenomiPhi™ (GE Healthcare Life Sciences, Amersham, UK). LAIR1 (16231C>A, rs6509867 and 18835G>A, rs2287827) genotyping was carried out using TaqMan 5′ allelic discrimination Assay-By-Design method according to the manufacturer's instructions; [Assay ID: C_33962823_10 for (16231C>A), C_7798687_10 for (18835G > A); Applied Biosystems, Inc.]. PCR was performed in a total reaction volume of 10 μL with the following amplification cycles: initial denaturation (60 °C for 30s and 95 °C for 10 min) followed by 40 cycles of (95 °C for 15 s and 60 °C for 1 min) and a final extension (60 °C for 30 s). Thereafter, the genotype of each individual was determined using allele-specific fluorescence on the StepOnePlus™ Real-Time PCR Systems (Applied Biosystems, Inc. Foster City, CA, USA). StepOne™ Software Version 2.3 was used for allelic discrimination (Applied Biosystems, Inc. Foster City, CA, USA).
2.5. LAIR1 transcript expression profiling
RNA was isolated from peripheral white blood cells obtained from children with malaria using a combination of Trizol-based and RNEasy® mini kit (Qiagen) techniques. RNA from in vitro PBMC cultures was extracted using the RNEasy® mini kit according to the manufacturer's instructions. From 1.0 μg of total RNA, complementary DNA (cDNA) was prepared with the transcriptor first strand cDNA synthesis kit (Roche). For measurement of LAIR1 expression levels, 0.5 μg of resulting cDNA was used for gene-specific TaqMan® qPCR assays [Assay ID: Hs00253790_m1; Applied Biosystems, Inc. Foster City, CA, USA]. The constitutively expressed housekeeping gene β-actin was used as an endogenous control [Assay ID: Hs01060665_g1; Applied Biosystems, Inc. Foster City, CA, USA], to normalize the target gene expression data in a quantitative gene expression assay on the StepOnePlus™ Real-Time PCR System (Applied Biosystems, Inc. Foster City, CA, USA). LAIR1 Ct values were normalized to β-actin Ct values, and mRNA differences were determined using the delta-delta Ct method.
2.6. Longitudinal follow-up
Upon enrolment of the children into the study (n = 1512, Day 0), parents/guardians were asked to return with their child every 3 mos. throughout a 3-year follow-up period. If the parent/guardian had not returned to hospital by 1:00 pm on the day of the quarterly follow-up visit, our study staff visited the child's residence to check on their health status, including mortality. Since we determined the exact location of each child's residence with our GIS/GPS surveillance system, we could readily locate each child. In addition, since children experience multiple episodes of malaria, and other paediatric infectious diseases in this region, parents/guardians were asked to return to the hospital during their child's febrile episode(s). All laboratory tests required for proper clinical management of the patients were performed at each acute and quarterly visit, including complete haematological indices, malaria parasitaemia measures, and evaluation of bacteraemia (if clinically indicated). In addition, all-cause mortality data were collected throughout the 3-year follow-up. Mortality data, clinical, and laboratory measures were used to evaluate the association between LAIR1 genetic variants and longitudinal outcomes of SMA and mortality.
2.7. Statistical analyses
Cross-sectional (enrolment) data were analysed using SPSS, version 20.0 (SPSS Inc., Chicago, IL). Comparisons of demographic, clinical, and laboratory characteristics, and gene expression data across the groups were explored using the Kruskal-Wallis tests. When significant differences were observed, pairwise comparisons were performed using the Mann-Whitney U tests. Pearson's Chi Square (χ2) and Fisher's exact test were used for comparisons of proportions. Correlations between LAIR1 transcript levels and Hb concentrations were determined using Spearman correlation test. Linkage disequilibrium between the 16231C>A and 18835G>A SNPs was determined using Haploview software (version 4.2) [28].
The relationship between LAIR1 genotypes/haplotypes and susceptibility to malaria and SMA upon enrolment were analysed by performing a logistic regression to predict disease state (malaria or not and SMA or non-SMA) from LAIR1 genotypes/haplotypes, controlling for the effects of age, sex, HIV-1 and bacteraemia (presence/absence), sickle cell trait, and α-thalassaemia and G6PD status. Construction of haplotypes was performed using HPlus (Version 2.5) [29].
The association between LAIR1 genotypes/haplotypes and longitudinal clinical outcomes were analysed using R (version 3.1.3). Clinical outcome definitions of malaria and SMA in the longitudinal analyses were prospective and were independent of the enrolment definition. Bidirectional elimination stepwise loglinear regression (R glm function, family = Poisson) was used to investigate the relationship between LAIR1 genotypic/haplotypic variants and the number of malaria (and SMA) episodes over the 36 mo. follow-up period. Additional covariates (i.e., age at enrolment, sex, HIV-1 and bacteraemia (presence/absence), sickle cell trait, and α-thalassaemia and G6PD status) were entered into the model as potential confounding risk factors. For the loglinear models, we accounted for the varying length of observation, by treating the logarithm of the length of the observational window as an offset. Finally, the relationship between LAIR1 genotypic/haplotypic variants and all-cause mortality was investigated using Cox regression/survival analysis (R survival package [v. 2.38.2] coxph function). Covariates in the model included age at enrolment, sex, HIV-1, bacteraemia, and sickle cell, α-thalassaemia, and G6PD status. In all our statistical analysis, we provide both the standard error of the estimate and its P-value. Statistical significance was set at P ≤ 0.050. Bonferroni correction was used to adjust for multiple comparisons.
3. Results
3.1. Demographic, clinical, and laboratory characteristics upon enrolment
Study participants (n = 1512) were stratified into three categories; aparasitemic (n = 292), non-SMA (Hb ≥ 5.0 g/dL, n = 986) and SMA (Hb < 5.0 g/dL, n = 234). The demographic, clinical, and laboratory characteristics of the study participants are presented in Table 1. The distribution of males and females was comparable across the groups (P = 0.834). Age differed across the groups (P = 0.002) with the SMA group being the youngest (P < 0.001).
Table 1.
Demographic, clinical, and laboratory characteristics upon enrolment.
| Characteristics | Aparasitemic | Non-SMA (Hb ≥ 5.0 g/dL) | SMA (Hb < 5.0 g/dL) | P |
|---|---|---|---|---|
| Demographic indices | ||||
| Sample size (n) | 292 | 986 | 234 | |
| Gender, n (%) | ||||
| Male | 147 (50.34) | 502 (50.91) | 114 (48.72) | 0.834a |
| Female | 145 (49.70) | 484 (49.09) | 120 (51.28) | |
| Age, (months) | 11.28 (13.00) | 12.70 (10.46) | 9.51 (10.73)*** | 0.002b |
| Haematological and parasitological indices | ||||
| Haemoglobin, g/dL | 10.40 (2.40) | 7.70 (2.80) | 4.30 (1.00)*** | <0.001b |
| Hematocrit, (Hct. %) | 32.90 (6.30) | 25.30 (8.90) | 14.20 (3.70) *** | <0.001b |
| RBC, (×1012/μL) | 4.67 (1.03) | 3.78 (1.39) | 1.90 (0.63) *** | <0.001b |
| WBC (×103/μL) | 10.80 (7.00) | 11.70 (6.30) | 14.30 (9.90) *** | <0.001b |
| Lymphocytes (×103/μL) | 5.90 (4.20) | 5.20 (3.60) | 6.70 (5.20) *** | <0.001b |
| Monocytes (×103/μL) | 0.80 (0.70) | 0.90 (0.80) | 1.40 (1.30) *** | <0.001b |
| Granulocytes (×103/μL) | 3.80 (3.10) | 5.12 (4.00) | 5.20 (4.80) | <0.001b |
| Platelet counts (×103/μL) | 350.00 (220.00) | 152.50 (124.00) | 142.50 (100.00)* | 0.003b |
| Parasite density/μL | 0.00 (0.00) | 28,902.75 (78,961) | 23,507.85 (62,646) | 0.004c |
| Genetic variants | ||||
| G6PD gene, n. (%) | ||||
| Normal | 193 (76.60) | 687 (75.40) | 181 (79.00) | |
| Intermediate | 49 (19.40) | 185 (20.30) | 36 (15.70) | 0.589a |
| Deficient | 10 (4.00) | 39 (4.30) | 12 (5.20) | |
| α-thalassaemia, n. (%) | ||||
| αα/αα | 109 (45.00) | 374 (43.40) | 90 (41.90) | |
| -α/αα | 71 (29.30) | 331 (38.40) | 86 (40.00) | 0.031a |
| -α/−α | 62 (25.60) | 157 (18.20) | 39 (18.10) | |
| Sickle cell trait, n. (%) | ||||
| HbAA | 230 (80.40) | 807 (82.50) | 208 (89.70)*** | |
| HbAS | 53 (18.50) | 169 (17.30) | 20 (8.60) | <0.001a |
| HbSS | 3 (1.00) | 2 (0.20) | 4 (1.70) | |
| Co-infections | ||||
| Bacteremia | 27 (9.25) | 59 (5.98) | 20 (8.55) | 0.489a |
| HIV-1 | 12 (4.11) | 25 (2.54) | 13 (5.56)* | 0.039a |
Study participants (n = 1512) were stratified into three groups, aparasitemic, SMA (i.e., Hb < 5.0 g/dL with any density parasitaemia) or non-SMA (Hb ≥ 5.0 g/dL with any density parasitaemia). Data presented are medians (interquartile range, IQR), unless otherwise stated.
Statistical significance determined by the chi-square analysis.
Differences were determined using Kruskal-Wallis tests, and where significant differences were observed, pairwise comparisons between the non-SMA and SMA groups were performed using Mann-Whitney U tests.
Exclusion of the aparasitemic group, differences between the non-SMA and SMA groups determined using Mann-Whitney U tests. Bold indicates P ≤ 0.050. *Represents significant pairwise comparisons between the non-SMA and SMA groups at P < 0.050, **represents P < 0.01, and ***represents P < 0.001. G6PD is presented as absence/presence of G6PD alleles (G202A and A376G), α-thalassaemia is presented as absence/presence of the deletional determinant (−α3.7/−α3.7), and sickle cell trait is presented as the three genotypes. In some samples, we were not able to determine G6PD (n = 120), α-thalassaemia (n = 193), and sickle cell trait (n = 16).
There was a progressive decline in Hb concentrations, hematocrit, and RBC counts across the clinical categories [aparasitaemic → non-SMA → SMA (P < 0.001, respectively)]. The WBC, lymphocyte, and monocyte counts were elevated in children with SMA (P < 0.001 for all). Granulocyte counts progressively increased across the groups (P < 0.001), while platelet counts progressively decreased (P = 0.003), and were lowest in children with SMA. Peripheral parasite density was lower in children with SMA compared to the non-SMA group (P = 0.004).
Carriage of genetic variants that can influence clinical outcomes in children with malaria were also characterized. The frequency of G6PD deficiency was comparable across the clinical groups (P = 0.589), while the α-thalassaemia double deletion (−α3.7/−α3.7) decreased across the groups (P = 0.031). The frequency of HbAS declined across the groups and was lowest in children with SMA (P < 0.001).
Since we have previously shown that bacteraemia and HIV-1 influence the development of SMA [23,24], the status/presence of these co-infections were determined in the study population. The frequency of bacteraemia was comparable across the three groups (P = 0.489). HIV-1 frequency differed across the groups (P = 0.039) and was the highest in children with SMA (P = 0.017). Taken together, the demographic, clinical, and laboratory findings support our previous studies showing that children with SMA have distinct clinical features [24,30].
3.2. Global genomic and transcriptome profiling
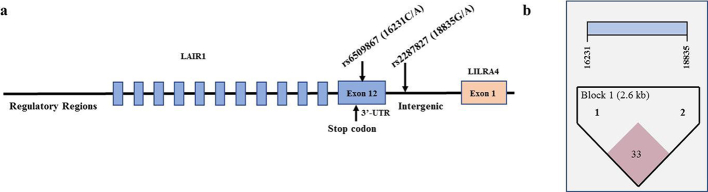
There were a total of 36 SNPs from Human Omni2.5-8v1 BeadChip® and 43 SNPs from the Immunochip® that were located in intra- and intergenic loci of LAIR1. Of those, seven SNPs were significantly associated with susceptibility to SMA (P < 0.01, OR: 0.43–1.37). Since SMA is a polygenic disease, we were interested in significant SNPs with minor allele frequencies (MAF)>10% in Kenyan populations, Luhya (LWK) and Maasai (MKK), from the International HapMap Project. If unavailable for the Kenyan populations, the MAF was determined in populations classified as African (AFR) by the 1000 genomes project. Of the seven significant SNPs, only three had a MAF>10%: -20907A>G (rs11084333), 16231C>A (rs6509867), and 18835G>A (rs2287827). Transcription factor binding analyses (i.e., Alggen-Promo, v3.0.2) revealed that the promoter SNP (−20907A>G) did not impart any (potential) functional changes in the binding site. As such, we investigated 16231C>A located in the 3’-UTR (in the last exon, 12) and 18835G>A located in an intergenic region between LAIR1 and LILRA4 (1902 bp downstream of exon 12 in LAIR1 and 12,553 bp upstream of exon 1 in LILRA4). Both of these genes are in the LRC. The MAF for the 16231C>A variant was 40% in AFR populations, while the 18835G>A variant was 46% in the LWK and 34% in the MKK reference populations, respectively. The structure of the LAIR1 gene is shown in Fig. 1A. The polymorphic loci had a low level of linkage disequilibrium (LD): D': 0.339, LOD: 30.31, r2: 0.079, (Fig. 1B).
Fig. 1.
LAIR1 gene structure and linkage disequilibrium.
(a) The LAIR1 gene is composed of 12 exons located on chromosome 19q13.4 in the leukocyte receptor complex. The SNPs selected for investigation were rs6509867 (16231C>A) and rs2287827 (18835G>A), respectively. The 16231C>A variant is located in the 3’ UTR, while the 18835G>A SNP is located in the intergenic region between LAIR1 and LILRA4 (leukocyte immunoglobulin like receptor A4).
(b) Linkage disequilibrium between the selected LAIR1 SNPs: 16231C>A and 18835G>A (D’: 0.339, LOD: 30.31, r2 squared: 0.079).
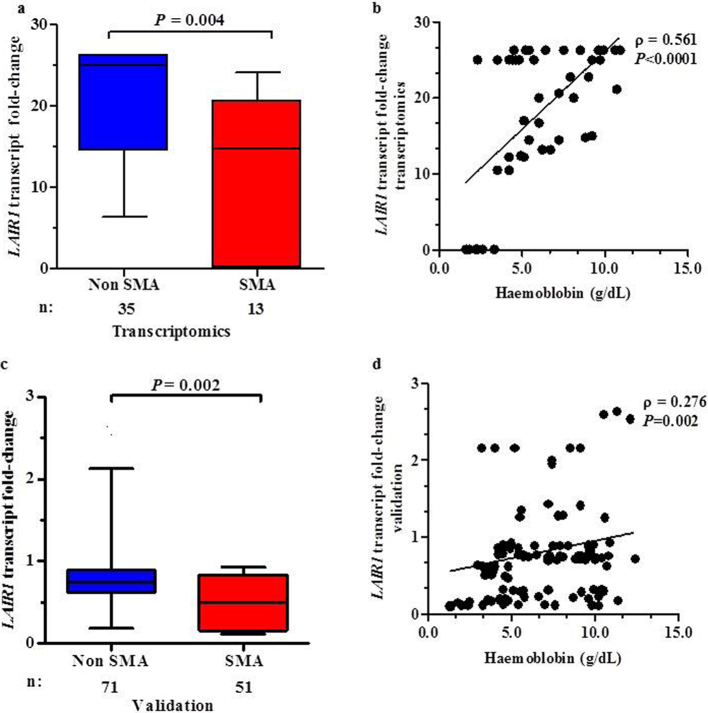
Whole transcriptome expression profiling analysis showed that LAIR1 transcripts were down-regulated in children with SMA relative to the non-SMA group (−1.69 fold-change, P = 0.004, Fig. 2A). In addition, LAIR1 transcript levels were positively correlated with Hb concentrations (ρ = 0.561, P < 0.0001, Fig. 2B). Validation of transcript expression was performed by qRT-PCR using RNA isolates from 122 study participants; non-SMA = 71 and SMA = 51. Consistent with whole transcriptome findings, LAIR1 transcripts were down-regulated in children with SMA relative to non-SMA (−1.50 fold, P < 0.002, Fig. 2C) with a positive relationship between LAIR1 transcript expression and Hb levels (ρ = 0.276, P = 0.002, Fig. 2D). Thus, there were significant markers (SNPs) in LAIR1 that were associated with susceptibility to SMA, and differential expression in LAIR1 between the two groups.
Fig. 2.
LAIR1 transcripts in children with malaria stratified into non-SMA and SMA.
Data are presented as box-plots (fold-change) where the box represents the interquartile range, the line through the box is the median, and whiskers show the 10th and 90th percentiles. Mann-Whitney U test was used to compare transcript levels between the two groups. Correlations between LAIR1 transcript expression and haemoglobin concentrations were determined by Spearman correlation test. Since co-infections and haemoglobinopathies can influence Hb levels, children with HIV-1 (exposed and + ve), bacteraemia, α-thalassaemia (double deletion), HbSS, and G6PD deficiency were excluded from the analyses.
(a) Whole transcriptome expression profiling using Illumina® HumanHT-12 v4 beadchip (Illumina®, CA, USA) covering >19,185 transcripts in total RNA isolated from WBC pellets (n = 48). LAIR1 transcripts were down-regulated in children with SMA (n = 13) relative to the non-SMA group (n = 35).
(b) Correlation between whole transcriptome derived LAIR1 transcript levels and Hb concentrations in parasitemic children (n = 48). LAIR1 transcript levels were positively correlated with Hb concentrations.
(c) TaqMan® gene expression analysis in WBC pellets from children with malaria (n = 122). LAIR1 transcripts were significantly down-regulated in children with SMA (n = 51) compared to non-SMA (n = 71).
(d) Correlation between TaqMan® LAIR1 transcript expression in parasitemic children (n = 122). LAIR1 transcript levels were positively correlated with Hb levels.
3.3. Distribution of LAIR1 genotypes and haplotypes
The distribution of the LAIR1 genotypes (16231C>A and 18835G>A) and haplotypes (16231C>A/18835G>A) is shown in Table 2. The proportion of 16231C>A genotypes differed between the groups (P = 0.037) with the AA (mutant) genotype carried more frequently in children with SMA, suggesting that it potentially enhances susceptibility to SMA. Allele frequencies were C = 0.64 and A = 0.36 in the overall population, C = 0.66 and A = 0.34 in the aparasitemic group, C = 0.65 and A = 0.35 in non-SMA group, and C = 0.58 and A = 0.42 in children with SMA. There was departure from Hardy-Weinberg Equilibrium (HWE) in the overall population (χ2 = 4.63, P = 0.031). However, departure from HWE was not observed within each of the groups: aparasitemic (χ2 = 1.80, P = 0.179), non-SMA (χ2 = 0.82, P = 0.364), and SMA (χ2 = 3.518, P = 0.074).
Table 2.
Distribution of LAIR1 genotypes and haplotypes.
| Genotypes/Haplotypes | Aparasitemic | Non-SMA Hb ≥ 5.0 g/dL | SMA Hb < 5.0 g/dL | P | Total |
|---|---|---|---|---|---|
| 16231C>A | |||||
| Sample size (n) | n = 292 | n = 976 | n = 233 | n = 1501 | |
| CC, n (%) | 134 (45.90) | 417 (42.70) | 86 (36.90) | 0.037 | 637 (42.44) |
| CA, n (%) | 120 (41.10) | 432 (44.30) | 100 (42.90) | 652 (43.44) | |
| AA, n (%) | 38 (13.00) | 127 (13.00) | 47 (20.20) | 212 (14.12) | |
| 18835G>A | |||||
| n = 292 | n = 968 | n = 232 | n = 1492 | ||
| GG, n (%) | 99 (33.90) | 315 (32.50) | 95 (40.90) | 0.007† | 509 (34.12) |
| GA, n (%) | 109 (37.30) | 439 (45.40) | 84 (36.20) | 632 (42.36) | |
| AA, n (%) | 84 (28.80) | 214 (22.10) | 53 (22.80) | 351 (23.53) | |
| 16231C>A/18835G>A | |||||
| Sample size (n) | n = 292 | n = 986 | n = 234 | n = 1512 | |
| Non-CG | 102 (34.90) | 291 (29.50) | 84 (35.90) | 0.157 | 477 (31.55) |
| CG (1 copy) | 127 (43.50) | 489 (49.60) | 101 (43.20) | 717 (47.542) | |
| CG (2 copies) | 63 (21.60) | 206 (20.90) | 49 (20.90) | 318 (21.03) | |
| Non-CA | 186 (63.70) | 662 (67.10) | 172 (73.50) | 0.047 | 1020 (67.46) |
| CA (1 copy) | 77 (26.40) | 263 (26.60) | 49 (20.90) | 389 (25.73) | |
| CA (2 copies) | 29 (9.90) | 61 (6.20) | 13 (5.60) | 103 (6.81) | |
| Non-AG | 245 (83.90) | 820 (83.20) | 174 (74.40) | <0.001† | 1239 (81.94) |
| AG (1 copy) | 40 (13.70) | 144 (14.60) | 41 (17.50) | 225 (14.88) | |
| AG (2 copies) | 7 (2.40) | 22 (2.20) | 19 (8.10) | 48 (3.17) | |
| Non-AA | 170 (58.20) | 543 (55.1) | 135 (57.70) | 0.553 | 848 (56.08) |
| AA (1 copy) | 102 (34.90) | 388 (39.40) | 83 (35.50) | 573 (37.90) | |
| AA (2 copies) | 20 (6.80) | 55 (5.60) | 16 (6.80) | 91 (6.02) | |
Data are presented as proportions [n (%)] of genetic variants within the study groups. Study participants were categorized into three groups, aparasitemic, SMA (i.e., Hb < 5.0 g/dL with any density parasitaemia) or non-SMA (Hb ≥ 5.0 g/dL with any density parasitaemia). Statistical significance was determined by the chi-square analysis. Bold indicates P ≤ 0.050. †Significant after Bonferroni correction for multiple comparisons.
The frequency of 18835G>A genotypes differed between the groups (P = 0.007) and remained significant after testing for multiple comparisons. The lower prevalence of the heterozygous (GA) genotype in the SMA group suggests a potential protective effect against severe malaria. Allele frequencies were G = 0.55 and A = 0.45 in the overall population, G = 0.53 and A = 0.47 in aparasitemic children, G = 0.55 and A = 0.45 in the non-SMA group, and G = 0.59 and A = 0.41 in the SMA group. Significant departure from HWE was observed in the overall population (χ2 = 30.60, P < 0.001), and within the aparasitaemic (χ2 = 18.546, P < 0.001), non-SMA (χ2 = 6.67, P = 0.010), and SMA (χ2 = 14.65, P < 0.001) groups.
Next, we compared the frequency distributions of the constructed haplotypes (carriers vs. non-carriers) for the two SNPs (16231C>A/18835G>A, Table 2). Significant differences were observed between absence and presence (1 and 2 copies) of the CA (P = 0.047) and AG (P < 0.001) haplotypes, but not for the CG (P = 0.157) and AA (P = 0.553) haplotypes. After Bonferroni correction, only the AG haplotypes remained significantly different.
3.4. Impact of LAIR1 genetic variants on susceptibility to malaria upon enrolment
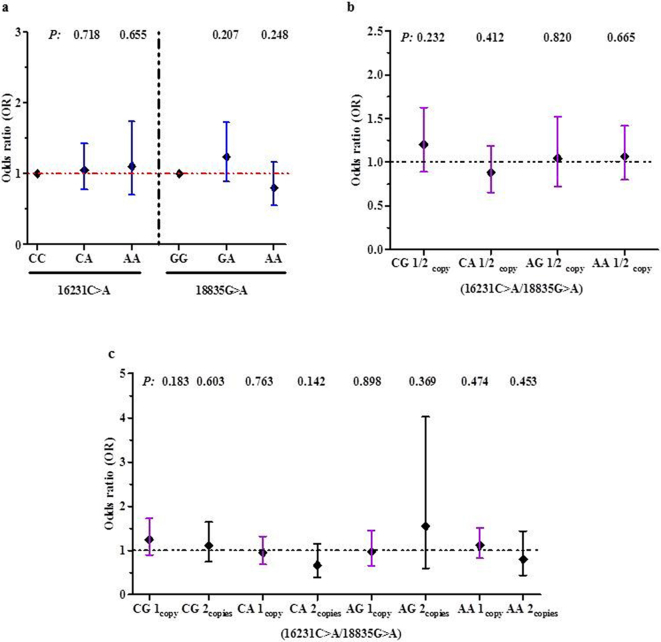
Binomial logistic regression was performed to determine the impact of LAIR1 genotypes on susceptibility to malaria upon enrolment. There were no significant associations between the genotypes and susceptibility to malaria (Fig. 3A).
Fig. 3.
Relationship between LAIR1 genotypes/haplotypes and susceptibility to malaria upon enrolment.
Data presented as Odds Ratios (OR) and 95% Confidence intervals (CI) as determined by binomial logistic regression analyses. Covariates in the models included age, sex, HIV-1 and bacteraemia (presence/absence), sickle cell trait, and α-thalassaemia and G6PD status.
(a) Association between LAIR1 genotypes and susceptibility to malaria. None of the LAIR1 genotypes were significantly associated with susceptibility to malaria.
(b) Association between absence (0 copies) and presence (1 or 2 copies) of the haplotypes and susceptibility to malaria. None of the haplotypes significantly altered susceptibility to malaria.
(c) Additive effect of 0, 1, or 2 copies of the haplotypes on susceptibility to malaria. No significant relationships were found for any of the haplotypes.
To determine the multi-loci impact of the SNPs on susceptibility to malaria, we performed binomial logistic regression using non-carriage (0 copies) vs. carriage (1 or 2 copies) of the haplotypes as the predictor variable (Fig. 3B). None of the haplotypes influenced susceptibility to malaria. Susceptibility to malaria was also investigated in individuals stratified into 0, 1, and 2 copies of the haplotypes to determine if there was an additive impact of haplotypic carriage (Fig. 3C). None of the haplotypes demonstrated an additive effect on susceptibility to malaria.
3.5. Impact of LAIR1 variants on susceptibility to SMA upon enrolment
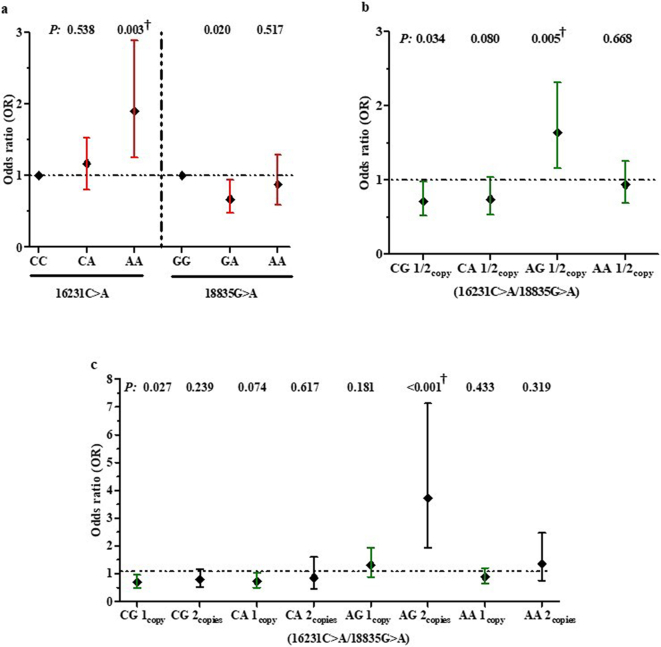
To determine the cross-sectional impact of LAIR1 genotypes on susceptibility to SMA, a (binomial) logistic regression was performed to predict the impact of LAIR1 variants on SMA status at recruitment (Fig. 4A). Inheritance of the 16,231 homozygous recessive genotype (AA) increased susceptibility to SMA (CC vs. AA, OR = 1.903, 95%CI: 1.252–2.891, P = 0.003). The relationship between the AA locus and susceptibility to SMA remained significant after testing for multiple comparisons. In addition, the genotypic model revealed that carriage of the 18,835 heterozygous genotype (GA) conferred protection against SMA (GG vs. GA, OR = 0.672, 95%CI: 0.480–0.9439, P = 0.020). None of the other genotypes for either the 16,231 or 18,835 genotypes were significantly associated with susceptibility to SMA.
Fig. 4.
Relationship between LAIR1 genotypes/haplotypes and susceptibility to SMA upon enrolment.
Data presented as Odds Ratios (OR) and 95% Confidence intervals (CI) as determined by binomial logistic regression analyses. Covariates in the models included age, sex, HIV-1 and bacteraemia (presence/absence), sickle cell trait, and α-thalassaemia and G6PD status. †Significant after Bonferroni correction for multiple comparisons.
(a) Association between LAIR1 genotypes and susceptibility to SMA. Carriage of the 16,231 AA genotype was associated with increased susceptibility to SMA, while inheritance of the 18,835 GA genotype conferred protection against SMA.
(b) Association between absence (0 copies) and presence (1 or 2 copies) of the haplotypes and susceptibility to SMA. Carriers of the CG haplotype had a reduced risk of acquiring SMA, whereas carriers of the AG haplotype had an increased risk of developing SMA.
(c) Additive effect of 0, 1, or 2 copies of the haplotypes on susceptibility to SMA. Inheritance of 1 copy of the CG haplotype was associated with protection against SMA, while inheritance of 2 copies of the AG haplotype was associated with an increased risk of SMA.
The impact of the haplotypes [absence (0 copies) vs. presence (1 or 2 copies)] was also explored with a binomial logistic regression to predict SMA status at recruitment (Fig. 4B). There was a 28.3% reduction in the risk of SMA for carriers of the CG haplotype (OR = 0.717, 95%CI: 0.527–0.9675, P = 0.034), and a 25.0% reduced risk in carriers of the CA haplotype (OR = 0.745, 95%CI: 0.536–1.036, P = 0.080). In contrast, carriers of the AG haplotype were 64.1% more likely to develop SMA (OR = 1.641, 95%CI: 1.160–2.321, P = 0.005 and significant after testing for multiple comparisons). Non-carriage vs. carriage of the AA haplotype did not influence susceptibility to SMA.
The additive effect of the haplotypes (0, 1, or 2 copies) on susceptibility to SMA was also investigated in a logistic regression model (Fig. 4C). Children with 1 copy of the CG haplotype had a 31.1% reduced risk of developing SMA (OR = 0.689, 95%CI: 0.494–0.959, P = 0.027), while carriage of 2 copies of CG did not significantly alter susceptibility to SMA (OR = 0.784, 95%CI: 0.523–1.176, P = 0.239), indicating a non-additive effect of the CG haplotype. Consistent with the carrier vs. non-carrier model, the additive model showed that 1 copy of the CA haplotype was associated with a reduced risk of developing SMA (OR = 0.720, 95%CI: 0.502–1.032, P = 0.0.074), while 2 copies did not enhance the protective effect (OR = 0.851, 95%CI: 0.454–1.598, P = 0.617). Although 1 copy of AG haplotype did not significantly influence susceptibility to SMA (OR = 1.308, 95%CI: 0.883–1.940, P = 0.181), children with 2 copies of the AG haplotype had increased susceptibility to SMA (OR = 3.713, 95%CI: 1.934–7.127, P < 0.001). This relationship remained significant after Bonferroni correction. Carriage of 1 or 2 copies of the AA haplotypes did not significantly alter the risk of developing SMA. Collectively, the cross-sectional results demonstrate that inheritance of particular LAIR1 genetic variants are associated with altered risk profiles for both malaria and SMA.
3.6. Functional associations between LAIR1 variants and gene expression levels upon enrolment
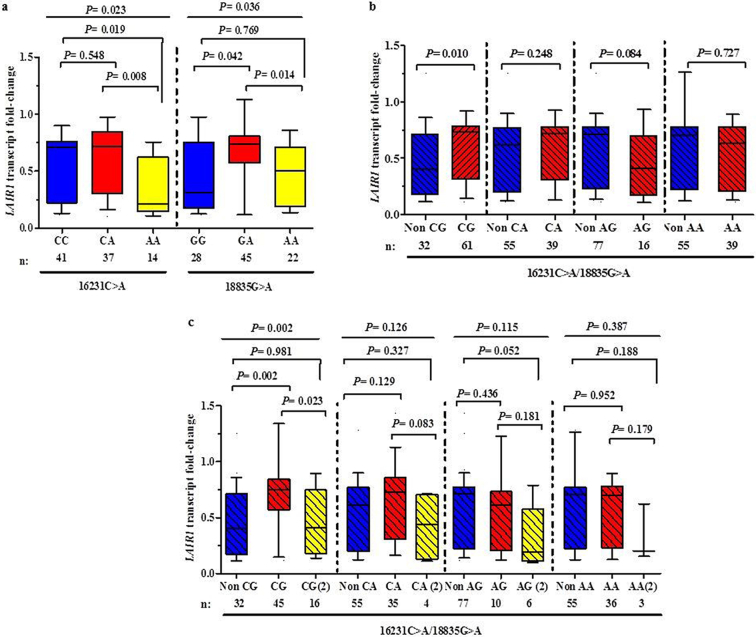
Since LAIR1 variants emerged as important predictors of clinical outcomes, we determined if carriage of the different genotypes/haplotypes were associated with functional changes in LAIR1 transcript levels. LAIR1 median transcript levels differed across the 16231C>A genotypes (P = 0.023, Fig. 5A). Relative to 16,231 wild-type genotype (CC), carriers of the ‘susceptible’ 16,231 homozygous recessive genotype (AA) had lower transcript levels (−3.34 fold, P = 0.019). Carriers of the 16,231 AA genotype also had lower transcript levels than the CA group (+3.35 fold, P = 0.008). LAIR1 expression levels also differed across the 18835G > A genotypic groups (P = 0.036). Relative to 18,835 wild-type genotype (GG), children with the ‘protective’ 18,835 heterozygous genotype (GA) had elevated transcript levels (+2.36 fold, P = 0.042). LAIR1 transcript levels were also higher in GA carriers relative to the homozygous recessive (AA) genotype (+1.47 fold, P = 0.014).
Fig. 5.
LAIR1 transcript levels stratified according to genotypes/haplotypes upon enrolment.
Data are presented as box-plots (fold-change) where the box represents the interquartile range, the line through the box is the median, and whiskers show the 10th and 90th percentiles. Kruskal-Wallis tests were used to compare transcript levels across the clinical groups, and Mann-Whitney U tests were used to compare two groups.
(a) LAIR1 transcript levels were stratified according to genotypes. At 16231, LAIR1 transcript levels differed across the genotypes. Compared to both the CC and CA genotypes, carriers of the AA genotype had lower transcriptional expression. At 18835, LAIR1 transcript levels differed across the genotypes with GA carriers having the highest transcript levels.
(b) LAIR1 transcript levels stratified into non-carriers (0 copies) vs. carriers (1 or 2 copies) of the haplotypes. Inheritance of the ‘protective’ CG haplotype was associated with higher LAIR1 expression. Conversely, carriers of the ‘susceptible’ AG haplotype had marginally lower transcript levels than non-carriers.
(c) LAIR1 transcript levels stratified into 0, 1, or 2 copies of the haplotypes. Carriers of 1 copy of the ‘protective’ CG haplotype had elevated transcript levels relative to non-carriers and carriers of 2 copies. Inheritance of 2 copies of the ‘susceptible’ AG haplotype was associated with marginally lower transcript levels than non-carriers and carriers of 1 copy of the haplotype.
Next, we examined LAIR1 transcript levels in non-carriers (0 copies) vs. carriers (1 or 2 copies, Fig. 5B). Inheritance of the ‘protective’ CG haplotype was associated with higher LAIR1 expression (+1.80 fold, P = 0.010). Conversely, carries of the ‘susceptible’ AG haplotype had marginally lower transcript levels than non-carriers (−1.47 fold, P = 0.084).
Transcript expression of LAIR1 was also compared in haplotypic groups stratified according to 0, 1, or 2 copies of the four haplotypes. LAIR1 transcript levels differed across 0, 1 or 2 copies of the CG haplotypes. (P = 0.002, Fig. 5C). Carriers of 1 copy of the ‘protective’ CG haplotype had higher transcript levels compared to non-carriers (+1.85 fold, P = 0.002). The across group and between group comparison remained significant after testing for multiple comparisons. Children with 1 copy of CG haplotype also had higher LAIR1 transcript levels than those with 2 copies of the haplotype (+1.28 fold, P = 0.023). LAIR1 transcript levels did not differ across absence/presence of CA (P = 0.126), AG (P = 0.115), and AA haplotypes (P = 0.387). However, carriers of 2 copies of the ‘susceptible’ AG haplotype had marginally lower transcript expression than non-carriers (−3.63 fold, P = 0.052). These associations highlight important trends whereby presence of ‘protective’ LAIR1 genetic markers are associated with elevated transcript levels, while those associated with enhanced susceptibility to SMA have lower transcript expression, suggesting that the selected variants may impart functional changes of LAIR1 expression.
3.7. Relationship between LAIR1 variants and frequency of malaria episodes
Since children in holoendemic P. falciparum transmission regions typically suffer from multiple episodes of malaria infections prior to the development of naturally-acquired immunity, we examined the impact of LAIR1 variants on the number of malaria episodes over a 36-month follow-up period (Table 3). To determine the impact of LAIR1 genotypic and haplotypic carriage on the number of malaria and SMA episodes during the 36-month follow-up, loglinear regression analysis was performed with a model that included genotype as well as age at enrolment, sex, co-infections (HIV-1 and bacteraemia), and sickle cell, α-thalassaemia, and G6PD status as covariates. The logarithm of the length of the observational period for each child was taken as an offset. The analysis reveals that genotypes at the 16231C>A locus are not significant risk factors for the number of malaria episodes during the longitudinal follow-up (n = 7326 events). There were 7293 malaria episodes recorded for children in which the 18,835 locus was genotyped. Carriage of the GA genotype increased the risk of having multiple malaria episodes (RR = 1.111, 95%CI: 1.051–1.173, P < 0.001 and significant after Bonferroni correction).
Table 3.
Relationship between LAIR1 genetic variants and longitudinal outcomes.
| Genotypes | Malaria episodes |
SMA episodes |
Mortality |
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Episodes (n) | RR | 95%CI | P | Episodes(n) | RR | 95%CI | P | Episodes(n) | HR | 95%CI | P | |
| 16231C>A | Total = 7326 | Total = 345 | Total = 89 | |||||||||
| CC | 3095 | REF | 140 | REF | 33 | REF | ||||||
| CA | 3214 | 0.998 | 0.948–1.050 | 0.932 | 143 | 1.039 | 0.815–1.324 | 0.758 | 43 | 1.410 | 0.866–2.300 | 0.168 |
| AA | 1017 | 1.014 | 0.942–1.092 | 0.709 | 62 | 1.527 | 1.119–2.083 | 0.008† | 13 | 1.400 | 0.702–2.800 | 0.338 |
| 18835G>A | Total = 7293 | Total = 344 | Total = 88 | |||||||||
| GG | 2393 | REF | 131 | REF | 36 | REF | ||||||
| GA | 3227 | 1.111 | 1.051–1.173 | <0.001† | 138 | 0.848 | 0.661–1.089 | 0.197 | 30 | 0.552 | 0.329–0.925 | 0.024 |
| AA | 1673 | 1.032 | 0.967–1.102 | 0.341 | 75 | 0.774 | 0.576–1.040 | 0.089 | 22 | 0.667 | 0.377–1.180 | 0.164 |
| Haplotypes | ||||||||||||
| Total = 7381 | Total = 346 | Total = 89 | ||||||||||
| Non-CG | 2298 | REF | 114 | REF | 30 | REF | ||||||
| CG | 5083 | 1.192 | 1.024–1.163 | 0.007*a | 232 | 0.829 | 0.652–1.054 | 0.126 | 59 | 0.967 | 0.615–1.520 | 0.886 |
| Non-CA | 4901 | REF | 251 | REF | 64 | REF | ||||||
| CA | 2480 | 1.129 | 1.060–1.203 | <0.001a | 95 | 0.715 | 0.554–0.923 | 0.010 | 25 | 0.714 | 0.439–1.160 | 0.174 |
| Non-AG | 6003 | REF | 271 | REF | 67 | REF | ||||||
| AG | 1378 | 1.130 | 1.052–1.213 | 0.001a | 75 | 1.032 | 0.755–1.411 | 0.843* | 22 | 1.680 | 1.020–2.770 | 0.040 |
| Non-AA | 4148 | REF | 199 | REF | 49 | REF | ||||||
| AA | 3233 | 1.083 | 1.023–1.145 | 0.006a | 147 | 0.861 | 0.667–1.111 | 0.243 | 40 | 0.943 | 0.613–1.450 | 0.792 |
Data are presented as relative risk (RR) and 95% confidence intervals (CI) determined using loglinear regression with the following covariates in the models: age at enrolment, sex, HIV-1 and bacteraemia (presence/absence), sickle cell trait, and α-thalassaemia, and G6PD status. The reference groups in the regression analysis were non-carriers of the respective genotypes/haplotypes. Carriage of the respective haplotypes included presence of 1 or 2 copies. * Represents haplotypes that displayed an additive effect on malaria (RR = 0.969, 95%CI: 0.939–1.001, P = 0.059) and SMA (RR = 1.283, 95%CI: 1.057–1.557, P = 0.011). Bold indicates a P value of ≤0.050.
Significant after Bonferroni correction for multiple comparisons.
There were 7381 malaria episodes recorded for the four LAIR1 haplotypes. Carrier children with 1 or 2 copies of all four LAIR1 haplotypes had on average more malaria episodes over the follow-up period than non-carrier children (0 copies): [CG (RR = 1.192, 95%CI: 1.024–1.163, P = 0.007), CA (RR = 1.129, 95%CI: 1.060–1.203, P < 0.001), AG (RR = 1.130, 95%CI: 1.052–1.213, P = 0.001), and AA (RR = 1.083, 95%CI: 1.023–1.145, P = 0.006)]. Significance was maintained for all four haplotypes after testing for multiple comparisons. Loglinear regression analyses were also used to determine if any of the haplotypes had an additive impact on the number of malaria episodes during the follow-up period. There was a marginally significant additive effect for the CG haplotype which reduced the expected number of malaria episodes (RR = 0.969, 95%CI: 0.939–1.001, P = 0.059). None of the other haplotypes showed additive associations.
3.8. Relationship between LAIR1 variants and frequency of SMA episodes
Next, we examined the relationship between LAIR1 genotypes and the development of SMA episodes over the 36-month follow-up period using loglinear regression analyses (Table 3). Carriers of the 16,231 AA genotype were more likely to develop SMA (RR = 1.527, 95%CI: 1.119–2.083, P = 0.008 and significant after Bonferroni correction). Conversely, a marginal protective effect was associated to the 18,835 AA genotype (RR = 0.774, 95%CI: 0.576–1.040, P = 0.089).
To investigate the impact of LAIR1 haplotypes on predicting the number of SMA events (n = 346 events) a loglinear regression model was used to distinguish between non-carriage (0 copies) vs. carriage (1 or 2 copies) of the haplotypes (Table 3). Consistent with the analyses performed at enrolment, there was a lower risk of developing SMA across the 36-month follow-up in carriers of the CA haplotype (RR = 0.715, 95%CI: 0.554–0.923, P = 0.010, but not significant after Bonferroni correction). None of the other haplotypes significantly influenced the longitudinal risk of SMA. We also determined if any of the haplotypes had an additive association with susceptibility to SMA throughout the 36-month follow-up. Only the AG haplotype showed a significant additive effect with carriage of the AG haplotype associated with increased susceptibility to SMA (RR = 1.283, 95%CI: 1.057–1.557, P = 0.011).
3.9. Relationship between LAIR1 variants and mortality
Since SMA is a significant cause of paediatric mortality in holoendemic malaria transmission regions [4,31], Cox regression survival analysis was used to determine the impact of LAIR1 variants on all-cause mortality (Table 3). Inheritance of the 18,835 GA genotype was associated with 44.8% reduced risk of all-cause mortality (RR = 0.552, 95%CI: 0.329–0.925, P = 0.024). However, after correction for multiple comparisons, the relationship was not significant. None of the other genotypes emerged as significant predictors of mortality.
The impact of LAIR1 haplotypes on mortality were also explored with Cox regression models with identical covariates (Table 3). Consistent with the increased risk of malaria, and enhanced risk of SMA in an additive model, presence of 1 or 2 copies of the AG haplotype was associated with a 68% increase in the risk of childhood mortality (RR = 1.680, 95%CI: 1.020–2.770, P = 0.040). After Bonferroni correction, the impact of the AG haplotype on mortality was not significant.
4. Discussion
The current study determined the impact of selected LAIR1 variants on malaria and SMA in Kenyan children residing in a holendemic P. falciparum transmission region. In addition, since malaria is a primary cause of childhood mortality in such areas [1,32], we also explored the relationship between LAIR1 polymorphic variation and all-cause mortality. Selection of the LAIR1 SNPs was driven by results from our ‘pilot’ global genomic studies showing a significant relationship between LAIR1 and susceptibility to SMA in a phenotypically distinct group of children with mild and severe forms of malarial anaemia. Exploration of the role of LAIR1 in malaria was also based on findings from the transcriptomics investigations in which LAIR1 expression was significantly lower in children with SMA compared to those with mild disease. To our knowledge, this is the first investigation exploring the relationship between host genetic variation in LAIR1 and disease outcomes in human malaria.
The strong selective pressure placed on the human genome from malaria has created an enrichment of genetic variation in the immune response genes of African populations [33,34]. Consistent with this premise, the overall major and minor allele frequencies (p/q) at the 16,231 locus were C = 0.64 and A = 0.36 in our study population vs. C = 0.91 and A = 0.09 in ethnic groups classified as Northern and Western European ancestry (HapMap-CEU) from the international HapMap project [35]. This was also the case for the 18,835 locus in which G = 0.55 and A = 0.45 in Kenyan cohort, and G = 0.90 and A = 0.10 in the Northern and Western European ancestry population (HapMap-CEU) [36].
To better understand the potential impact of LAIR1 variation on malaria disease outcomes, we performed cross-sectional (enrolment) and longitudinal (36-month follow-up) analyses. The cross-sectional investigation did not find any significant relationship between the genotypes/haplotypes and susceptibility to malaria. However, two genotypes and two haplotypes significantly altered SMA risk profiles. The 16,231 homozygous recessive genotype (AA) increased susceptibility to SMA, while the 18,835 heterozygous genotype (GA) conferred protection. After testing for multiple comparisons, only the AA finding remained significant. The relationships with SMA susceptibility are consistent with variant frequencies at the two loci for the non-SMA and SMA groups.
Since haplotypes can identify disease association patterns potentially not revealed by individual SNPs, we examined the impact of LAIR1 haplotypic combinations on malaria disease outcomes. Although none of the haplotypes significantly altered susceptibility to malaria at enrolment, two haplotypic constructs emerged as significant predictors for the cross-sectional risk of acquiring SMA. Carriage of the 16231C/18835G (CG) haplotype protected against SMA, while carriage of the 16231A/18835G (AG) haplotype increased vulnerability to SMA. However, only the AG findings remained significant after testing for multiple comparisons. The relationship between the AG haplotype and enhanced susceptibility to SMA was most pronounced in individuals who inherited 2 copies.
Results from the global transcriptomic experiments revealed that LAIR1 transcripts were significantly lower in children with SMA compared to those with mild disease (P = 0.004) in samples collected upon presentation at hospital, prior to any treatment interventions. These results were confirmed by qRT-PCR analyses in a subset of individuals with SMA (n = 51) and non-SMA (n = 71, P = 0.002). In addition, the relationship between down-regulated LAIR1 expression and enhanced anaemia is supported by the positive correlation between LAIR1 transcripts and Hb concentrations in both the transcriptomic (ρ = 0.561, P < 0.0001) and validation (ρ = 0.276, P = 0.002) experiments. LAIR1 expression was not measured serially due to IRB restrictions on multiple blood draws from the anaemic children. Since LAIR1 expression is important for preventing the overproduction of pro-inflammatory mediators [[37], [38], [39]]., down-regulation of LAIR1 in children with SMA may contribute to enhanced disease severity. For example, we have shown that enhanced production of pro-inflammatory mediators is a significant predictor of SMA [9,40,41]. Consistent with reduced LAIR1 expression being associated with enhanced disease severity, previous investigations observed down-regulation of LAIR1 in severe cases of CLL and SLE [18,19].
To identify LAIR1 variants associated with functional changes in transcript expression, we performed qRT-PCR expression analyses in a subset of individuals in which peripheral white blood cells were available. Transcript levels were compared in groups stratified according to genotypes and haplotypes. LAIR1 genotypes associated with susceptibility to SMA (i.e., 16,231 AA genotype) had reduced transcript levels, whereas those that protected against SMA (i.e., 18,835 GA genotype) had elevated transcript expression. Consistent with this trend, stratification of the haplotypes revealed that the ‘susceptible’ AG haplotype (2 copies) had reduced LAIR1 transcripts, while transcript expression increased in those who inherited the ‘protective’ CG haplotype (1 copy). These findings suggest that LAIR1 variants, which either enhance or reduce susceptibility to SMA, may do so through reduced and enhanced transcriptional regulation, respectively. However, since the underlying mechanism(s) by which the variants functionally alter LAIR1 expression remains undetermined, we are currently exploring this line of investigation. For example, the 16231C>A SNP in the 3’-UTR may serve as a target for microRNAs and thereby affect LAIR1 transcription/translation, whereas the 18835G>A SNP may act as an enhancer/silencer of LAIR1 transcription.
Since children in holoendemic transmission regions typically experience many repeated episodes of malaria prior to developing clinical immunity, cross-sectional genetic analyses may not capture the changing dynamics of the disease burden [42,43]. To address this challenge, we performed longitudinal analyses to determine the influence of the LAIR1 variants on malaria and SMA episodes, and all-cause mortality. Increased susceptibility to malaria over the follow-up period was associated with inheritance of the 18,835 GA genotype, and all four LAIR1 haplotypes (CG, CA, AG, and AA). All of these relationship remained significant after correction for multiple comparisons. Although the reason for detecting a significant relationship between LAIR1 variants and susceptibility to malaria longitudinally, but not cross-sectionally is unknown, it is likely related to the fact that malaria is a dynamic process that changes across time. For example, in holoendemic transmission regions, an individual who presents with an acute infection (case) during a one-time sampling point may present as a non-infected (control) on other occasions, and vice versa. Longitudinal analyses have an enhanced power to discover the impact of genetic variation on polygenic infectious disease outcomes due to repeated measures within an individual. In addition, longitudinal analyses can more accurately catalogue the development of naturally-acquired immunity, a process that is difficult to capture with one-time measures, inherent in cross-sectional models. The fact that all four haplotypes showed an association with longitudinal malaria episodes implies that susceptibility over time is strongly influenced by multi-loci genetic variations.
Since the severe disease manifestation of malaria in holoendemic regions is SMA [4,31], we determined the impact of LAIR1 variation on longitudinal susceptibility to SMA. Consistent with the cross-sectional findings, biallelic carriage of the 16,231 AA genotype increased the risk of developing SMA. Although children who inherited the 18,835 GA genotype had cross-sectional protection against SMA, in the longitudinal analyses, there was a trend towards protection that marginally increased with added carriage of the minor A allele. Longitudinal analyses of SMA outcomes also revealed that carriage of the CA haplotype conferred a 25.8% reduced risk of developing SMA, a finding that parallels the cross-sectional results. This result reflects the combination of carriage of the major allele at 16,231 and minor allele at 18,835, both of which were associated with protection. Inheritance of the AG allele enhanced susceptibility to SMA in the cross-sectional investigations that was most pronounced in carriers with 2 copies. Although the longitudinal model comparing non-carriers with carriers of the AG haplotype failed to detect an increased risk of SMA, significance was observed in the additive model. Collectively, the longitudinal analyses show that inheritance of the CA haplotype confers protection against developing SMA, and that increased copies of the AG haplotype enhance the risk of acquiring SMA. This finding is consistent with the frequency of 1 copy of the “protective” CA haplotype in the overall population at 25.73%, in contrast to the 8-fold lower distribution of 2 copies of the “susceptible” AG haplotype at 3.17%. Thus, the substantially higher minor allele frequencies of the two SNPs in study population, compared to those of European ancestry, appears to offer combinatorial diversity of haplotypes which may favour selective pressure for mitigating the risk of SMA.
In malaria holoendemic regions of western Kenya, such as Siaya, childhood mortality can be as high as 16.7%, of which 48.2% can be attributed to malaria [44]. However, in our study cohort, the all-cause mortality rate was 5.51%. The lower rate of mortality can be attributed to implementation of triage case management, improved and more rapid laboratory diagnostics, antimicrobial susceptibility testing with the availability of 2nd line treatments, health education, and provision of bednets for study participants (and their families) during the study period. Since SMA is a leading cause of childhood mortality in the study population, we carried out longitudinal investigations to determine if LAIR1 genetic variants that predicted SMA also influenced childhood mortality. However, since the number of deaths in the overall cohort was low, and slightly over 50% of the children died at home, we did not have the statistical power to examine malaria-related deaths, and therefore, investigated all-cause mortality. Inheritance of the 16,231 AA genotype that enhanced susceptibility to SMA (both cross-sectionally and longitudinally) also increased the risk of all-cause mortality by 40%. This finding, however, did not reach statistical significance. Inheritance of the 18,835 GA genotype that reduced susceptibility to SMA in the cross-sectional models, and trended towards reduced, but non-significant protection in the longitudinal analyses, reduced all-cause mortality by 44.8%. Carriage of the CA haplotype protected against SMA upon presentation at hospital and longitudinally, and also decreased the risk of mortality by 28.6%, albeit non-significantly. Childhood mortality was also influenced by carriage of the AG haplotype that increased the risk of all-cause mortality by 68%. This finding is consistent with the results showing that inheritance of the AG haplotype enhanced susceptibility to SMA in the cross-sectional analyses, and in an additive longitudinal model. Although carriage of particular LAIR1 variants had a marked impact on mortality, the findings did not reach significance after testing for multiple comparisons, likely due to the reduced statistical power associated with the low mortality rate. Nonetheless, consistency amongst susceptibility to SMA and all-cause mortality points to the important impact of LAIR1 variation on disease outcomes.
To our knowledge, this is the first investigation exploring the role of LAIR1 variation in susceptibility to human malaria. However, the importance of this inhibitory receptor has been reported in relation to binding RIFINs, a family of clonally variant proteins derived from P. falciparum which are expressed on the surface of infected red blood cells [45]. Previous investigations in individuals from a malaria-endemic region identified unique antibodies that contained a mutated LAIR1 insert that bound to specific RIFINs, whereas binding of wild-type LAIR1 to RIFINs was absent [21,22]. Although engagement of LAIR1 with RIFIN did not significantly enhance LAIR1 reporter cells, another inhibitory receptor in the LRC family, i.e., leukocyte immunoglobulin like receptor B1 (LILRB1), interacted with RIFINs and resulted in signalling inhibition of B-cells and an NK cell line [45]. It is postulated that binding of RIFINs to LAIR1 and LILRB1 have evolved as potential immune evasion strategies. Since LAIR1 and LILRB1 are both in the LRC region on chromosome 19, we examined linkage disequilibrium (SNP & Variation Suite 8.8.3 Software) using results from the pilot GWAS. The EM-D' and EM-r2 values revealed very low LD between the regions, including the two LAIR1 SNPs investigated here. The strongest LD was observed between LAIR1 (kgp8992506) and LILRB1 (kgp21512897 - 255 kb apart) with an r2 value of 0.354.
Findings presented here demonstrate that LAIR1 variants at 16231 and 18,835, and their haplotypes, are associated with altered susceptibility to malaria, SMA, and all-cause mortality over a 36-month follow-up period. In addition, ‘protective’ LAIR1 variants were associated with elevated LAIR1 transcript expression, while variants that enhanced susceptibility to SMA had reduced mRNA levels. Based on the important role of LAIR1 in modulating downstream gene pathways known to be important in malaria pathogenesis, we performed as series of in vivo and in vitro experiments to better understand the potential role of LAIR1 in malaria pathogenesis. These investigations are presented in a companion manuscript presented in this issue of EbioMedicine. Findings from this series of experiments revealed that phagocytosis of malarial pigment (haemozoin, PfHz) down-regulates LAIR1 mRNA and protein, and blocks the inhibitory LAIR1 signal through phosphorylation of LAIR1 and SH2-domain containing phosphatase-1 (SHP-1). These molecular events increase NF-κB activation, resulting in enhanced production of IL-6, IL-1β, and TNF-α, inflammatory mediators that can enhance the pathogenesis of SMA. Thus, in addition to LAIR1 playing a role in malarial immunity through its interaction with RIFINs, an additional mechanism is the direct effect of malarial products (i.e., PfHz) on the LAIR1 signalling pathway. The findings presented here, along with the results presented in the companion article, illustrate that LAIR1 represents a novel gene pathway that is important for human malaria pathogenesis. Since severe malaria is holoendemic regions is often accompanied by co-infections, it will be important in future studies to determine the impact of LAIR1 variation on other clinical syndromes.
Declaration of Competing Interests
The authors declare that they have no competing interests.
Authors' contributions
AOA: participated in SNP selection, generated genotyping and Taqman expression data, performed data analyses, and manuscript writing.
NWH: longitudinal data analyses, statistical interpretations, wrote portions of R code, and manuscript review and editing.
ER: data analyses, project supervision, and manuscript review and editing manuscript.
QC: data analyses, technical support, and manuscript review and editing.
SBA: data analyses, project supervision, and manuscript review and editing manuscript.
NL: wrote portions of R code for longitudinal data analyses.
BG: technical advice, and manuscript review and editing.
IFH: data analyses, and manuscript review and editing.
JMO: project supervision, and manuscript review and editing.
BHM: longitudinal data analysis, wrote portions of R code, and manuscript review and editing.
CO: project supervision, and manuscript review and editing.
CGL: genomic and longitudinal analyses, wrote portions of R code, and manuscript review and editing.
DJP designed clinical and experimental studies, data analyses, manuscript writing and editing, provided clinical samples and data, and supplied reagents, materials, and analyses tools.
Acknowledgments and funding
The authors gratefully acknowledge the assistance of the University of New Mexico-Kenya Research Team (Nicholas Otieno Ondiek, Vincent Odhiambo Otieno, Anne A Ong'ondo, Chrispine Wasonga Ochieng, Everlyne A Modi, Joan L A Ochieng, Joseph Oduor, Moses Ebungure, Moses Lokorkeju, Rodney B Mongare, and Vincent Omanje). We are also grateful to all of the parents, guardians and children who participated in the study.
The work was supported by National Institutes of Health (NIH) Research Grants R01AI130473, R01AI51305 and D43TW05884 (DJP), and the University of New Mexico Health Sciences Clinical and Translational Science Center (UL1TR001449). The content is solely the responsibility of the authors and the funders did not have any role in study design, data collection, data analysis, interpretation, or writing of the report.
References
- 1.WHO World Malaria Report. 2017. http://appswhoint/iris/bitstream/handle/10665/259492/9789241565523-engpdf;jsessionid=7C4AC215FEF706839BD95175ADE65ABA?sequence=1
- 2.Breman J.G., Egan A., Keusch G.T. The intolerable burden of malaria: a new look at the numbers. Am J Trop Med Hyg. 2001;64(1–2 Suppl) doi: 10.4269/ajtmh.2001.64.iv. (iv-vii) [DOI] [PubMed] [Google Scholar]
- 3.Ong'echa J.M., Keller C.C., Were T. Parasitaemia, anemia, and malarial anemia in infants and young children in a rural holoendemic Plasmodium falciparum transmission area. Am J Trop Med Hyg. 2006;74(3):376–385. [PubMed] [Google Scholar]
- 4.Obonyo C.O., Vulule J., Akhwale W.S., Grobbee D.E. In-hospital morbidity and mortality due to severe malarial anemia in western Kenya. Am J Trop Med Hyg. 2007;77(6 Suppl):23–28. [PubMed] [Google Scholar]
- 5.Munde E.O., Raballah E., Okeyo W.A., Ong'echa J.M., Perkins D.J., Ouma C. Haplotype of non-synonymous mutations within IL-23R is associated with susceptibility to severe malaria anemia in a P. falciparum holoendemic transmission area of Kenya. BMC Infect Dis. 2017;17(1):291. doi: 10.1186/s12879-017-2404-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ouma C., Davenport G.C., Awandare G.A. Polymorphic variability in the interleukin (IL)-1beta promoter conditions susceptibility to severe malarial anemia and functional changes in IL-1beta production. J Infect Dis. 2008;198(8):1219–1226. doi: 10.1086/592055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ouma C., Davenport G.C., Garcia S. Functional haplotypes of fc gamma (Fcgamma) receptor (FcgammaRIIA and FcgammaRIIIB) predict risk to repeated episodes of severe malarial anemia and mortality in Kenyan children. Hum Genet. 2012;131(2):289–299. doi: 10.1007/s00439-011-1076-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Anyona S.B., Kempaiah P., Raballah E. Functional promoter haplotypes of interleukin-18 condition susceptibility to severe malarial anemia and childhood mortality. Infect Immun. 2011;79(12):4923–4932. doi: 10.1128/IAI.05601-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Awandare G.A., Martinson J.J., Were T. MIF (macrophage migration inhibitory factor) promoter polymorphisms and susceptibility to severe malarial anemia. J Infect Dis. 2009;200(4):629–637. doi: 10.1086/600894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Meyaard L., Adema G.J., Chang C. LAIR-1, a novel inhibitory receptor expressed on human mononuclear leukocytes. Immunity. 1997;7(2):283–290. doi: 10.1016/s1074-7613(00)80530-0. [DOI] [PubMed] [Google Scholar]
- 11.Meyaard L. The inhibitory collagen receptor LAIR-1 (CD305) J Leukoc Biol. 2008;83(4):799–803. doi: 10.1189/jlb.0907609. [DOI] [PubMed] [Google Scholar]
- 12.Poggi A., Tomasello E., Ferrero E., Zocchi M.R., Moretta L. p40/LAIR-1 regulates the differentiation of peripheral blood precursors to dendritic cells induced by granulocyte-monocyte colony-stimulating factor. Eur J Immunol. 1998;28(7):2086–2091. doi: 10.1002/(SICI)1521-4141(199807)28:07<2086::AID-IMMU2086>3.0.CO;2-T. [DOI] [PubMed] [Google Scholar]
- 13.Van der Vuurst de Vries A.R.C.H., Logtenberg T., Meyaard L. Leukocyte-associated immunoglobulin-like receptor-1 (LAIR-1) is differentially expressed during human B cell differentiation and inhibits B cell receptor-mediated signaling. Eur J Immunol. 1999;29(10):3160–3167. doi: 10.1002/(SICI)1521-4141(199910)29:10<3160::AID-IMMU3160>3.0.CO;2-S. [DOI] [PubMed] [Google Scholar]
- 14.Xu M., Zhao R., Zhao Z.J. Identification and characterization of leukocyte-associated Ig-like receptor-1 as a major anchor protein of tyrosine phosphatase SHP-1 in hematopoietic cells. J Biol Chem. 2000;275(23):17440–17446. doi: 10.1074/jbc.M001313200. [DOI] [PubMed] [Google Scholar]
- 15.Yindom L.M., Forbes R., Aka P. Killer-cell immunoglobulin-like receptors and malaria caused by Plasmodium falciparum in the Gambia. Tissue Antigens. 2012;79(2):104–113. doi: 10.1111/j.1399-0039.2011.01818.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Son M., Diamond B. C1q-mediated repression of human monocytes is regulated by leukocyte-associated Ig-like receptor 1 (LAIR-1) Mol Med. 2014;20(1):559–568. doi: 10.2119/molmed.2014.00185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Olde Nordkamp M.J., van Roon J.A., Douwes M., de Ruiter T., Urbanus R.T., Meyaard L. Enhanced secretion of leukocyte-associated immunoglobulin-like receptor 2 (LAIR-2) and soluble LAIR-1 in rheumatoid arthritis: LAIR-2 is a more efficient antagonist of the LAIR-1-collagen inhibitory interaction than is soluble LAIR-1. Arthritis Rheum. 2011;63(12):3749–3757. doi: 10.1002/art.30612. [DOI] [PubMed] [Google Scholar]
- 18.Colombo B.M., Canevali P., Magnani O. Defective expression and function of the leukocyte associated Ig-like receptor 1 in B lymphocytes from systemic lupus erythematosus patients. PLoS One. 2012;7(2) doi: 10.1371/journal.pone.0031903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Perbellini O., Falisi E., Giaretta I. Clinical significance of LAIR1 (CD305) as assessed by flow cytometry in a prospective series of patients with chronic lymphocytic leukemia. Haematologica. 2014;99(5):881–887. doi: 10.3324/haematol.2013.096362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Camargo C.M., Augusto D.G., Petzl-Erler M.L. Differential gene expression levels might explain association of LAIR2 polymorphisms with pemphigus. Hum Genet. 2016;135(2):233–244. doi: 10.1007/s00439-015-1626-6. [DOI] [PubMed] [Google Scholar]
- 21.Tan J., Pieper K., Piccoli L. A LAIR1 insertion generates broadly reactive antibodies against malaria variant antigens. Nature. 2016;529(7584):105. doi: 10.1038/nature16450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pieper K., Tan J., Piccoli L. Public antibodies to malaria antigens generated by two LAIR1 insertion modalities. Nature. 2017;548(7669):597–601. doi: 10.1038/nature23670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Otieno R.O., Ouma C., Ong'echa J.M. Increased severe anemia in HIV-1-exposed and HIV-1-positive infants and children during acute malaria. AIDS. 2006;20(2):275–280. doi: 10.1097/01.aids.0000200533.56490.b7. [DOI] [PubMed] [Google Scholar]
- 24.Were T., Davenport G.C., Hittner J.B. Bacteremia in Kenyan children presenting with malaria. J Clin Microbiol. 2011;49(2):671–676. doi: 10.1128/JCM.01864-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Were T., Hittner J.B., Ouma C. Suppression of RANTES in children with Plasmodium falciparum malaria. Haematologica. 2006;91(10):1396–1399. [PubMed] [Google Scholar]
- 26.Jia X., Liu J., Wang L. A rapid detection for alpha-thalassaemia by PCR combined with dissociation curve analysis. Exp Mol Pathol. 2011;91(2):626–630. doi: 10.1016/j.yexmp.2011.06.013. [DOI] [PubMed] [Google Scholar]
- 27.Hsu J., Fink D., Langer E. PCR-based allelic discrimination for glucose-6-phosphate dehydrogenase (G6PD) deficiency in Ugandan umbilical cord blood. Pediatr Hematol Oncol. 2014;31(1):68–75. doi: 10.3109/08880018.2013.860649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Barrett J.C., Fry B., Maller J., Daly M.J. Haploview: analysis and visualization of LD and haplotype maps. Bioinformatics. 2005;21(2):263–265. doi: 10.1093/bioinformatics/bth457. [DOI] [PubMed] [Google Scholar]
- 29.Li S.S., Khalid N., Carlson C., Zhao L.P. Estimating haplotype frequencies and standard errors for multiple single nucleotide polymorphisms. Biostatistics. 2003;4(4):513–522. doi: 10.1093/biostatistics/4.4.513. [DOI] [PubMed] [Google Scholar]
- 30.Novelli E.M., Hittner J.B., Davenport G.C. Clinical predictors of severe malarial anaemia in a holoendemic Plasmodium falciparum transmission area. Br J Haematol. 2010;149(5):711–721. doi: 10.1111/j.1365-2141.2010.08147.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Olliaro P. Mortality associated with severe Plasmodium falciparum malaria increases with age. Clin Infect Dis. 2008;47(2):158–160. doi: 10.1086/589288. [DOI] [PubMed] [Google Scholar]
- 32.Amek N.O., Van Eijk A., Lindblade K.A. Infant and child mortality in relation to malaria transmission in KEMRI/CDC HDSS, Western Kenya: validation of verbal autopsy. Malar J. 2018;17(1):37. doi: 10.1186/s12936-018-2184-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kwiatkowski D.P. How malaria has affected the human genome and what human genetics can teach us about malaria. Am J Hum Genet. 2005;77(2):171–192. doi: 10.1086/432519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hedrick P.W. Population genetics of malaria resistance in humans. Heredity (Edinb) 2011;107(4):283–304. doi: 10.1038/hdy.2011.16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.NCBI_dbSNP_rs6509867 2019. https://wwwncbinlmnihgov/projects/SNP/snp_refcgi?rs=6509867
- 36.NCBI_dbSNP_rs2287827 2019. https://www.ncbi.nlm.nih.gov/projects/SNP/snp_ref.cgi?rs=2287827
- 37.Merlo A., Tenca C., Fais F. Inhibitory receptors CD85j, LAIR-1, and CD152 down-regulate immunoglobulin and cytokine production by human B lymphocytes. Clin Diagn Lab Immunol. 2005;12(6):705–712. doi: 10.1128/CDLI.12.6.705-712.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bonaccorsi I., Cantoni C., Carrega P. The immune inhibitory receptor LAIR-1 is highly expressed by plasmacytoid dendritic cells and acts complementary with NKp44 to control IFNα production. PLoS One. 2010;5(11) doi: 10.1371/journal.pone.0015080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kim Y.K., Chu S.H., Hsieh J.Y. Incorporation of a ligand peptide for immune inhibitory receptor LAIR-1 on biomaterial surfaces inhibits macrophage inflammatory responses. Adv Healthc Mater. 2017;6(24) doi: 10.1002/adhm.201700707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Perkins D.J., Were T., Davenport G.C., Kempaiah P., Hittner J.B., Ong'echa J.M. Severe malarial anemia: innate immunity and pathogenesis. Int J Biol Sci Spec Issue. 2011;7(9):1427–1442. doi: 10.7150/ijbs.7.1427. [Special Issue on Innate Immunity] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ong'echa J.M., Davenport G.C., Vulule J.M., Hittner J.B., Perkins D.J. Identification of inflammatory biomarkers for pediatric malarial anemia severity using novel statistical methods. Infect Immun. 2011;79(11):4674–4680. doi: 10.1128/IAI.05161-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Mwangi T.W., Fegan G., Williams T.N., Kinyanjui S.M., Snow R.W., Marsh K. Evidence for over-dispersion in the distribution of clinical malaria episodes in children. PLoS One. 2008;3(5) doi: 10.1371/journal.pone.0002196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Griffin J.T., Hollingsworth T.D., Reyburn H., Drakeley C.J., Riley E.M., Ghani A.C. Gradual acquisition of immunity to severe malaria with increasing exposure. Proc R Soc B. 2015;282(1801) doi: 10.1098/rspb.2014.2657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kenya National Bureau of Statistics (KNBS) 2011. United nations children's fund (UNICEF). Multiple indicator cluster survey. [Google Scholar]
- 45.Saito F., Hirayasu K., Satoh T. Immune evasion of Plasmodium falciparum by RIFIN via inhibitory receptors. Nature. 2017;552(7683):101. doi: 10.1038/nature24994. [DOI] [PMC free article] [PubMed] [Google Scholar]