Abstract
Background
Chronic obstructive pulmonary disease (COPD) is characterized by emphysema and/or obstructive bronchiolitis. Deficiency in vitamin D3 (VD3), which regulates gene expression through binding to vitamin D receptor (VDR), is associated with high risks of COPD susceptibility. Alveolar macrophages (AM), which are generated during early ontogeny and maintained in alveoli by self-renewal in response to cytokine GM-CSF, are positively correlated with severity of emphysema. However, whether and how VD3, VDR and AM interact to contribute to COPD pathogenesis at the molecular and cellular levels are largely unknown.
Methods
We used systems biology approaches to analyze gene expression in mouse macrophages from different tissues to identify key transcription factors (TF) for AM and infer COPD disease genes. We used RNA-seq and ChIP-seq to identify genes that are regulated by VD3 in AM. We used VDR-deficient (Vdr−/−) mice to investigate the role of VD3-VDR axis in the pathogenesis of COPD and characterized the transcriptional and functional alterations of Vdr−/− AM.
Findings
We find that VDR is a key TF for AM and a COPD disease gene. VDR is highly expressed in AM and in response to VD3 inhibits GM-CSF-induced AM proliferation. In Vdr−/− AM, genes involved in proliferation and immune response are upregulated. Consistently, Vdr−/− mice progressively accumulate AM and concomitantly develop emphysema without apparent infiltration of immune cells into the lung tissue. Intratracheal transfer of Vdr−/− AM into wildtype mice readily induces emphysema. The production of reactive oxygen species at basal level and in response to heme or lipopolysaccharide is elevated in Vdr−/− AM and suppressed by VD3 in wildtype AM.
Interpretation
These results show that the VD3-VDR axis is critical to counteract GM-CSF-induced AM proliferation and defect in this regulation leads to altered AM homeostasis and function. Our findings identify that VD3 deficiency contributes to emphysema by altering AM function without contributing to bronchiolitis. Our findings also suggest possibilities of modulating the VD3-VDR axis for inhibiting emphysema in COPD patients.
Keywords: Vitamin D3, Vitamin D receptor, Transcriptional regulation, Alveolar macrophage, Emphysema, Chronic obstructive pulmonary disease
Research in context.
Evidence before this study
Both VD3 and AM have been implicated in COPD pathogenesis. However, it is not known whether and how they interact to contribute to COPD pathogenesis.
Added value of this study
We find that VDR is a key transcription factor for AM and a COPD disease gene. We identify that VD3-VDR axis regulates AM homeostasis and function and defect in this regulation results in gradual accumulation of AM, leading to emphysema without overt bronchiolitis.
Implications of all the available evidence
Our findings identify the molecular mechanisms by which VD3-VDR regulates AM homeostasis and function, and how the defect in this regulation contributes to specific COPD pathogenesis. Our findings suggest possibilities of inhibiting emphysema in COPD patients by modulating the VD3-VDR axis.
Alt-text: Unlabelled Box
1. Introduction
Chronic obstructive pulmonary disease (COPD) is a complex and progressive lung disease, characterized by poor airflow and breathing problems [1]. The main pathological features of COPD are emphysema due to destruction of alveolar walls and enlargement of alveoli, and obstructive bronchiolitis (or small airway disease) due to chronic inflammation of peripheral airways and lung parenchyma. Although some COPD patients predominantly have emphysema or obstructive bronchiolitis, patients with severe COPD often exhibit overlapping pathologies. Evidence suggests that the different pathologies may have different underlying mechanisms and possibly different causes [1]. For example, obstructive bronchiolitis is predominant in COPD patients associated with inhaling household wood smoke whereas obstructive bronchiolitis and emphysema coexist in COPD patients associated with deeply inhaled cigarette smoke. Globally, the most common cause of COPD is tobacco smoke, but only about 20–50% of smokers ever develop COPD [[2], [3], [4]], suggesting contribution of genetic, epigenetic and environmental factors in the development and progression of COPD. Supporting a critical role of environmental factors, most COPD exacerbations are triggered by respiratory infections [5]. However, how different factors interact and contribute to the different COPD pathologies at the molecular and cellular levels is largely unknown.
The lung tissue contains two types of macrophages. Alveolar macrophages (AM), which reside in alveoli and function to clear apoptotic cells and inhaled substance and particles, are derived from erythro-myeloid progenitors in the yolk sac and maintained by self-renewal in response to cytokine GM-CSF [6]. During lung inflammation, monocytes are also recruited into the lung tissue to generate inflammatory macrophages, which contribute to immune responses and tissue repair and remodeling [7]. Macrophages, especially AM, are known to play important roles in COPD pathogenesis [8,9]. Macrophage numbers are increased by 5-10-fold in bronchoalveolar lavage (BAL) fluid as well as in the airways, parenchyma, and sputum of patients with COPD [10,11]. Macrophages are often found at the sites of alveolar wall destruction in patients with emphysema, and macrophage numbers in the parenchyma are positively correlated with the severity of emphysema [12], raising the possibility that dysfunction of AM is responsible for emphysema. Consistent with this possibility, AM are activated by cigarette smoke, pollutants, and microbes to secrete matrix metalloproteinases (MMPs) 2, 9 and 12, and cathepsins K, L and S, which are directly involved in the destruction of alveolar walls [13]. Transcriptional analysis showed that AM in COPD patients are abnormally polarized to express genes involved in tissue remodeling [14,15]. Following stimulation, AM also secrete cytokines and chemokines, which in turn induce immune cell recruitment, inflammation, mucus hypersecretion, lung tissue damages and remodeling. Furthermore, both AM and monocyte-derived macrophages from COPD patients show reduced phagocytosis of bacteria [16]. The impaired elimination of bacteria by AM likely contributes to chronic colonization of low airways by bacteria and therefore increased frequencies of acute exacerbation [17]. Despite these observations, the precise role of AM and how their dysfunction contributes to COPD pathogenesis are not fully understood.
1,25-dihydroxyvitamin D3 (VD3) is the active ligand of vitamin D receptor (VDR). Upon binding of VD3 to VDR in the cytosol, the heterodimer further interacts with the retinoid X receptor (RXR). The trimeric complex is then translocated into the nucleus to regulate gene transcription through vitamin D response elements (VDRE) present in the enhancers and promoters of responsive genes [18]. The VD3-VDR axis has been shown to regulate both innate and adaptive immune responses [19] and exert a significant impact on COPD pathogenesis [20,21]. VD3 deficiency causes impairment of lung structure and function and increased severity of COPD-like diseases in the cigarette smoke-exposed mice whereas VD3 supplementation readily attenuates the disease symptoms and pulmonary inflammation [[22], [23], [24]]. In human, VD3-deficiency is also associated with higher risks of COPD susceptibility and more severe impairment of lung function in COPD patients [[25], [26], [27], [28]]. Although conflicting results have been reported on the efficacy of VD3 supplements for treating COPD, large cross-sectional and prospective studies have shown that VD3 deficiency is prevalent in patients with COPD and VD3 supplementation improves lung function and prevents exacerbation of COPD [29,30]. A possible confounding factor is that COPD is a complex disease with different pathologies and possible underlying mechanisms, but most large population-based studies and clinical cohort studies of VD3 supplements on COPD did not make these distinctions. A mechanistic understanding of how VD3 deficiency contributes to COPD pathogenesis is urgently needed for rationale use of VD3 supplements for managing COPD.
Vdr is highly expressed in AM comparing to other tissue macrophages [31]. Macrophages are particularly responsive to VD3 to downregulate expression of inflammatory cytokines and chemokines. VDR-deficient (Vdr−/−) mice are viable but exhibit severe bone defects and die prematurely around 16 weeks of age, which could be extended by giving mice calcium and phosphorus supplements [[32], [33], [34]]. Although Vdr−/− mice have no obvious alterations in the immune system in lymphoid organs and peripheral blood [34], VDR-deficient macrophages display defects in cellular function and immunity both in vitro and in vivo [[34], [35], [36]]. More recently, Vdr−/− mice at 16-weeks of age were reported to exhibit premature emphysema with increased secretion of MMPs [37], suggesting a role of VDR deficiency in the pathogenesis of emphysema. However, because the mice were deficient in VDR in germline (every cell type) and were given calcium and phosphorus supplements, the cellular and molecular mechanisms underlying the development of emphysema in Vdr−/− mice are not fully understood.
In this study, we have investigated the mechanisms by which VD3, VDR and AM interact to contribute to COPD pathogenesis. We show: i) VDR is a key transcriptional factor (TF) for AM and a COPD disease gene; ii) VD3-VDR axis inhibits AM proliferation and inflammatory responses; iii) Vdr−/− mice exhibit a progressive accumulation of AM and concomitant lung emphysema, without gross immune cell infiltration into the lung; iv) intratracheal transfer of Vdr−/− AM into wildtype mice is sufficient to induce lung emphysema; and v) VD3 inhibits inflammatory responses and oxidative stress of AM induced by heme and LPS. These results show that VD3-VDR axis regulates AM homeostasis and function and defect in this pathway results in emphysema. Thus, our findings identify the molecular and cellular mechanisms by which VD3, VDR, and AM interact to contribute to specific aspect of COPD pathogenesis and provide new insights into the use of vitamin D supplements for treating emphysema.
2. Material and methods
2.1. Systems biology approaches to identify key transcription factors and key disease genes
The methodological framework to identify key TFs for a particular gene set have been established in our previous studies [38]. A genome-wide regulatory network of mouse macrophage was assembled based on 1378 gene expression profiles of tissue macrophages, bone marrow-derived macrophages and macrophage cell lines collected from 124 independent GEO data sets (Table 1S) using a reverse-engineering algorithm CLR [39]. Microarray datasets from the same platform were uniformly normalized using Robust Multichip Averaging (RMA) and samples from different platforms were integrated and normalized with total intensity. The signature gene sets of different tissue macrophages (Tables S2) were extracted based on a previous study [31]. The adipose macrophage signature was extracted from the expression profiles of adipose macrophages from WT and obesity mice [40]. Master regulator analysis (MRA) is applied to identify TFs to regulate expression of signature genes of particular tissue macrophages by computing the statistical significance of overlaps of all interactions of each TF (inferred by CLR) with each set of signature or a background gene set by a binomial test. The set of 314 background genes (Tables S2) was identified from the 1378 gene expression profiles based on high levels of gene expression (RMA value >8.5) with minimal variation among all samples (variation from mean < 1).
To explore the key disease genes in macrophages for COPD, a high-confident gene functional network of macrophages was assembled using the Bayesian likelihood integration method based on four different whole-genome datasets: 1889 gene expression profiles, protein-protein interaction data (PPI) downloaded from eight databases (Biogrid, BIND, DIP, Complex, HPRD, InAct, MINT, MPPI), protein phylogenetic profiling based on all mouse proteins blastn scores to all other 59 ENSEMBL genomes (http://ensemblgenomes.org/) and gene phenotypic data from MGI (http://www.informatics.jax.org/phenotypes.shtml). For each gene pair, Pearson correlation coefficient (PCC) based on gene expression profiles, number of interaction observations in 8 PPI datasets, mutual information scores based on phylogenetic profiles, phenotypic similarity based on gene phenotypic data. The likelihood ratios for any gene pairs of each dataset were computed based on the overlap with positive and negative gold standard of the 8th grade GO biology process as previously described [41]. 10-fold cross validation was used to determine the cutoff of an integrated likelihood ratio with 90% network confidence (Fig. S1 and Table S3). The training COPD disease genes were extracted from OMINI database and curated from previous publications (Table S4). Network-based multiple algorithms including Random Walk [42], flow-based propagation [43], Nearest Neighbor [44], Short Path [45] and Markov chain clustering (MCL) [46] were used to prioritize the disease genes based on the functional network and known disease genes. The performance of the predictions was evaluated by plotting precision against recall over various thresholds as described [41]. A community-based method by combining multiple algorithms had higher sensitivity and specificity to identify disease genes than any single algorithms.
2.2. Mice, antibodies and flow cytometry
Vdr−/− mice, CD45.2 and CD45.1 B6 mice were purchased from the Jackson Laboratory and maintained in the animal facility at the Massachusetts Institute of Technology. All animal studies and procedures were approved by the Massachusetts Institute of Technology's Committee for Animal Care. Antibodies specific for CD11c (N418), CD11b (M1/70), F4/80 (BM8), MHC-II (M5/114.15.2), CD45.2 (104), CD45.1 (A20), CD4 (HK1.4), CD8 (53–6.7), CD19 (1D3) for flow cytometry were from Biolegend. Antibody for Irf4 (3E4) was from eBioscience. Anti-VDR (D2K6W) for immunochemistry staining was from Cell Signaling Technology. Single cell preparation from different organs, staining of cells with fluorophore-conjugated antibodies and analysis of the stained cells using flow cytometry are as described [38]. Briefly, cells in single cell suspension were incubated with specific antibodies at 4 °C for 20 min, washed twice, and re-suspended in FACS buffer containing DAPI. Cells were run on BD-LSRII, collecting 20,000 to 100,000 live cells per sample. The data were analyzed by FlowJo.
2.3. Histopathology and immunochemical staining
Mice were euthanized and lungs tissues were inflated and fixed with 10% neutral-buffered formalin solution (Sigma-Aldrich) for 24 h. The tissues were processed with Tissue Processor (Leica Microsystems) and embedded in paraffin. Sections were cut at 5 μm thickness, mounted on polylysine-coated slides (Thermo Fisher Scientific), de-waxed, rehydrated, and processed for hematoxylin and eosin (H&E) staining according to standard protocol. For immunochemical staining, antigen retrieval was carried out by either microwaving the slides in 0.01 M sodium citric acid buffer (pH 6.0) for 30 min. Sections were then immersed for 1 h in blocking buffer (3% BSA, 0.2% Triton X-100 in PBS), then incubated in primary antibody (made up in blocking buffer) at 4 °C overnight, followed by incubation with secondary antibody conjugated HRP at 4 °C for 1 h. All lung stained sections were scanned with a high-resolution Leica Aperio Slide Scanner. Images were analyzed by WebScope software.
2.4. Alveolar macrophage isolation, culture and adoptive transfer
GM-CSF is known to promote proliferation and maintain the global gene transcription and function of AM in vitro and in vivo [[47], [48], [49]]. In our study, AM were isolated from mice as described [50]. To expand AM numbers, freshly isolated AM were cultured in complete RPMI (RPMI 1640 supplemented with 10% FBS, 5 mM HEPES, 2 mM glutamine, 100 U/mL penicillin and 100 μg/mL streptomycin (Invitrogen)) supplemented with 25 ng/mL murine GM-CSF (Peprotech). Detailed culture and treatment conditions of AM with or without 50 nM VD3 for other assays were described in the text. For adoptive transfer experiment, 5 × 105 Vdr−/− AM (CD45.2+) after expansion in vitro for one week were adoptively transferred into 8-week-old CD45.1+ C57BL/6 (B6) mice intratracheally every week for a consecutive four weeks. Mice were analyzed at the sixth weeks following transfer. Unless specified, AM refers to mouse alveolar macrophages throughout the text.
2.5. Human alveolar macrophage isolation
Human alveolar macrophages were isolated by bronchoalveolar lavage (BAL) from normal volunteers with a lifetime non-smoking history, no acute or chronic illness, and no current medications. The lavage procedure used two or three 20-ml aliquots of sterile, warmed saline in right middle or left lingular lobe of the lung. The lavage fluid was filtered through two layers of gauze and centrifuged at 500 g for 5 min. The cell pellet was washed twice in PBS and suspended in complete RPMI. Cells were seeded on cell culture plates for 20 min and non-adherent cells were collected with purity of alveolar macrophages ranging from 90 to close to 100% based on Giemsa staining and flow cytometry. All protocols were approved by the Ethics Committee of The First Affiliated Hospital, Jilin University. All experiments were performed in accordance with the relevant guidelines and regulations. In addition, written informed consent was obtained from each subject.
2.6. Heme measurement and cellular ROS
Heme concentration in BAL was quantified using a fluorescence assay as previously described [51]. Briefly, BAL was collected by one-time washing with 1 mL PBS supplement with 2 mM EDTA for one mouse lung. BAL was cleaned by centrifuge at 1500 g for 5 min. 50 μl of BAL fluid was mixed with 200 μL 2 M oxalic acid. Samples as well as heme standards (0 to 100 nM) were heated at 95 °C for 30 min to remove iron from heme. The resultant protoporphyrin was detected using a fluorescence microplate reader (Tecan Infinite M200 Pro). Heme concentration in each sample was calculated based on the heme standards. Intracellular total ROS were quantified directly by flow cytometry using the broad free radical probe CM-H2DCFDA (Molecular Probes). Cells were incubated with 5 mM H2DCFDA in complete RPMI at 37 °C for 30 min.
2.7. RNA isolation, RNA sequencing and data analysis
AM were isolated from Vdr−/− mice and wild type littermates. Freshly isolated wildtype AM were treated with or without 50 nM VD3 for 24 h in the complete RPMI medium supplemented with 25 ng/mL murine GM-CSF (Peprotech). RNAs were extracted with RNeasy MiniElute kit (Qiagen), converted into cDNA and sequenced using Next-Generation Sequencing (Illumina). Raw sequences are deposited in the database of Gene Expression Omnibus (GEO) with accession ID: GSE124725. RNAseq data was aligned to the mouse genome (version mm10) and raw counts of each gene of each sample were calculated with bowtie2 2.2.3 [52] and RSEM 1.2.15 [53]. Differential expression analysis was performed using the program edgeR at P < .05 with a 2 fold-change [54]. The gene expression level across different samples was normalized and quantified using the function of cpm. Differentially expressed genes were annotated using online functional enrichment analysis tool DAVID (http://david.ncifcrf.gov/) [55]. Gene set enrichment analysis were performed with GSEA [56] with FDR q-value<0.05. The heatmap was visualized with MeV [57]. To validate and quantify the levels of RNA transcripts, total RNA was extracted from various cells and reverse transcribed by TaqMan® Reverse Transcription Reagents Kit (ABI Catalog No. N8080234), followed by amplification with Sybr Green Master Mix (Roche Catalog No. 04707516001) with specific primers (Table S5) and detected by Roche LightCycler 480. The Ct values were normalized with housekeeping gene GAPDH for comparison.
2.8. ChIP, sequencing and analysis
Freshly isolated AM (1 × 106) were cultured and expanded in complete RPMI supplemented with 25 ng/mL murine GM-CSF (Peprotech) for 2 weeks. Chromatin immunoprecipitation (ChIP) was performed using the SimpleChIP (Cell Signaling Technology) following the manufacture's manual. Briefly, 20 × 106 AM were treated with formaldehyde at a final concentration of 1% to cross-link DNA-protein complexes. Cells were lysed and DNA-protein complexes were sheared by micrococcal nuclease, followed by precipitation with nonspecific rabbit anti-IgG or Anti-VDR (D2K6W) (Cell Signaling Technology. Eluted and purified ChIP DNA was used to prepare the DNA-sequencing libraries with the NEBnext ChIPSeq library Prep kit (NEB) for Next-Generation Sequencing (Illumina). Raw sequencing data was deposited in GEO with accession ID: GSE124725. ChIP-seq were analyzed with the software package HOMER [58].
2.9. Co-immunoprecipitation
Plasmids of pCDNA3.1 with murine Vdr-HA, Pu.1-FLAG, Fra1-6xHis, Cebpa-Myc and Pparg-FLAG were purchased from GeneScript (USA). For the co-immunoprecipitation, Pu.1-FLAG, Fra1-6xHis, Cebpa-Myc and Pparg-FLAG were co-transfected with HA-tagged Vdr into 293 T cells using TransIT®-LT1 Transfection Reagent (Mirus). 48 h after transfection, the cells were lysed with cold Lysis Buffer containing 20 mM Tris-HCl (PH 7.4), 150 mM NaCl, 0.1% NP-40, 10% glycerol, proteinase inhibitor (Roche Catalog No. 11836153001), and phosphatase inhibitors (Roche Catalog No. 04906845001). The clear supernatants from the lysate were incubated with anti-HA magnetic beads (Thermo Fisher, #88836) at room temperature for 30 min in the presence or absence of VD3. Then the beads were washed four times and eluted with loading buffer for Western blotting. Eluted proteins were detected by anti-HA (CST, #2999S), anti-FLAG (Sigma, #M8823), anti-His (CST, #2365) and anti-Myc (Genetex, GT0002).
2.10. Statistic methods
Statistical significance was determined with the two-tailed unpaired or paired Student's t-test. The FDRs were computed with q = p*n/i, (p = P value, n = total number of tests, i = sorted rank of P value).
3. Results
3.1. Systems biology analyses predict vitamin D receptor as a key transcriptional factor for alveolar macrophages and a COPD disease gene
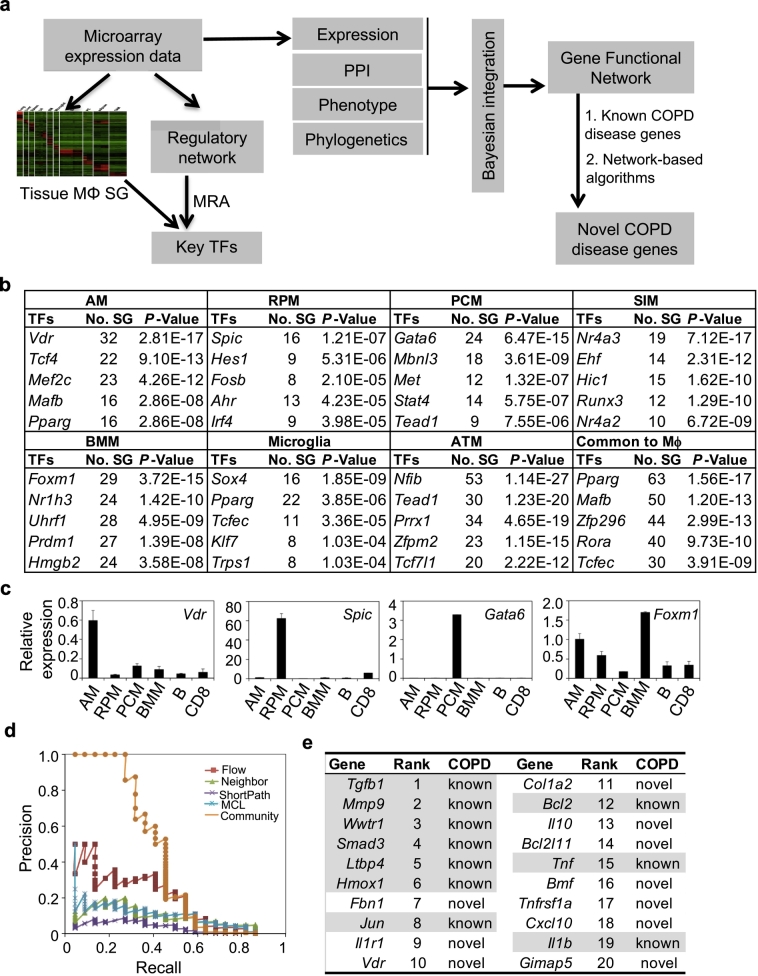
We have previously developed a network approach to identify key TFs for a particular cell state [38] and used the same approach to identify key TFs for macrophages in different tissues. A genome-wide regulatory network of mouse macrophages was assembled based on 1378 transcription profiles of 124 independent macrophage GEO data sets (Table S1) using a reverse-engineering algorithm CLR [39]. We identified lineage-specific key TFs for macrophages from seven different tissues using the regulatory network and the signature gene sets from public data (Fig. 1a, Table S2). Vdr, Spic, Nr4a3, Gata6, Foxm1, Sox4 and Nfib were identified as the top key TFs for alveolar macrophages (AM), spleen red-pulp macrophages (RPM), small intestine macrophages (SIM), peritoneal cavity macrophages (PCM), bone marrow macrophages (BMM), microglia, and adipose tissue macrophages (ATM), respectively (Fig. 1b, see Table S6 for a complete list). Spic and Gata6 are known lineage-specific key TFs for the development and function of RPM and PCM, respectively [[59], [60], [61], [62]]. We confirmed the expression of Vdr, Spic, Gata6, and Foxm1 in freshly purified AM, RPM, PCM and BMM, respectively, by quantitative RT-PCR (qPCR) (Fig. 1c and Fig. S2). VDR protein was also highly expressed in AM comparing to other cell types in the lung (Fig. S3). Mafb and Pparg were identified as top key TFs for the macrophage consensus signature (Fig. 1b), which is either required for the terminal differentiation of monocytic progenitors to macrophages [63,64] or a master regulator to maintain the anti-inflammatory phenotype of tissue macrophages [65,66]. Together, these findings support our network approach to identify key TFs for tissue-resident macrophages.
Fig. 1.
VDR is predicted as a key TF in AM and COPD disease gene. a, schematic diagram depicting systems biology approaches to identify the key TFs in tissue-resident macrophages and COPD-disease genes in AM. MRA: master regulatory analysis; PPI: protein-protein interaction; SG: signature genes. b, top-ranked key TFs for each of the seven tissue macrophages and for all seven tissue macrophages. #SG, number of signature genes; AM, alveolar macrophages; RPM, red pulp macrophages; PCM, peritoneal cavity macrophages; SIM, small intestine macrophages; BMM, bone marrow macrophages; ATM, adipose tissue macrophages; Common to Mϕ, common TF for all seven types of tissue macrophages. c, Comparison of the transcript levels of Vdr, Spic, Gata6 and Foxm1 in freshly purified AM, RPM, PCM, BMM, B cells (B) and CD8+ T cells (CD8) by qPCR. Data were average of 3 biological replicates per group. d, Comparison of the precision rates of community algorithm versus other algorithms for identifying COPD-disease genes. e, Top ranked key COPD-disease genes in AM. Shaded ones are known COPD disease genes and the others are newly identified in this study. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
Because AM play critical roles in the pathogenesis of COPD [1,8,9], we investigated whether the key TFs for AM are potential COPD disease genes using a systems biology approach. We constructed a high-confident gene functional network of macrophages by a Bayesian integration method [41] using mouse macrophage expression data, protein-protein interaction (PPI) data, gene phenotype data and phylogenetic data (Fig. 1S and Table S3). The functional network was then applied to prioritize potential novel COPD disease genes using a community method by integrating five different algorithms with 19 known COPD disease genes curated from OMINI database and previous publications (Fig. 1a and Table S4). This method had a significantly higher precision than other widely used algorithms (Fig. 1d). Seven of the top 10 genes identified are known COPD disease genes (Fig. 1e, see Table S7 for a complete list), supporting the validity of our approach. Vdr, the key TF for AM, is inferred as one of novel COPD disease genes.
3.2. VD3-VDR regulates diverse sets of gene expression in AM
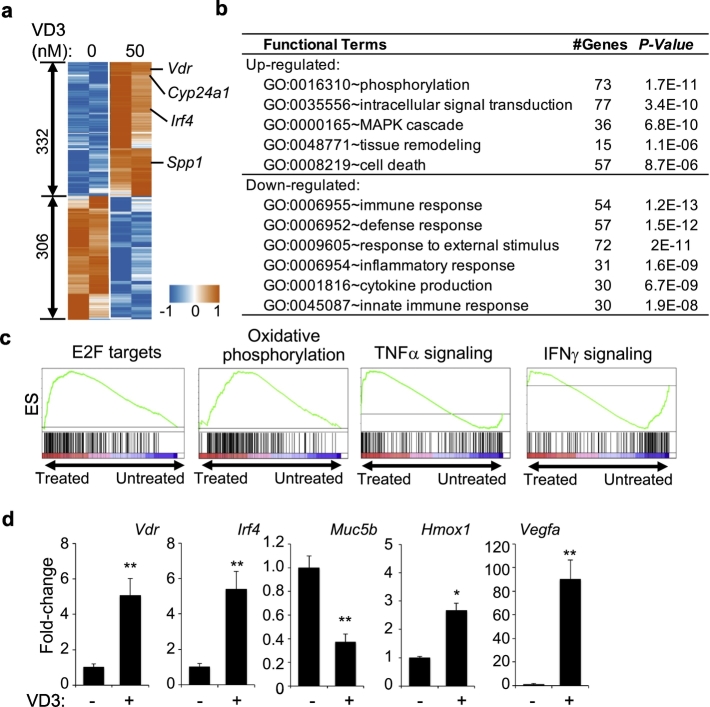
We determined the effect of VD3 on AM at transcription level by RNA-seq. AM were purified from C57BL/6 (B6) mice and cultured for 24 h in the presence or absence of 50 nM VD3, followed by RNA isolation and sequencing. Differential expression analysis revealed that 332 genes were up-regulated and 306 genes were down-regulated (Fig. 2a and Table S8), including known VD3-induced genes Vdr, Cyp24a1 and Spp1. The up-regulated genes were enriched with phosphorylation (73 genes, P = 1.7e-11), signal transduction (77 genes, P = 3.4e-10), tissue remodeling (15 genes, P = 1.1e-6) and cell death (57 genes, P = 8.7e-6), while the down-regulated genes were enriched with immune pathways, including immune response (54 genes, P = 1.2e-13), defense response (57 genes, P = 1.5e-12), and cytokine production (30 genes, P = 6.7e-9) (Fig. 2b). Gene set enrichment analysis (GSEA) revealed that up-regulated genes were enriched in E2F targets and oxidative phosphorylation and down-regulated genes were enriched in TNFα and IFNγ signaling pathways (Fig. 2c). VD3-induced transcription changes of selected genes, including Vdr, Irf4, Hmox1, Muc5b and Vegfa, were verified by qPCR (Fig. 2d).
Fig. 2.
Transcriptional regulation by VD3 in AM. a, Heatmap of gene expression of 332 up-regulated and 306 down-regulated genes induced by VD3 in AM from two independent experiments. Shown is centralized CPM (counts per million reads). b, GO enrichment analysis showing enrichment of certain pathways in the up-regulated and down-regulated genes. GO sets of biological process, number of genes and P-value are shown. c, GSEA showing samples of enriched gene sets in VD3-treated and untreated AM (q-value <0.05). d, Transcript levels of Vdr, Irf4, Muc5b, Hmox1 and Vegfa in AM with or without VD3 treatment for 24 h. Data were collected from two independent experiments with 4 biological replicates per group. Error bars indicate standard deviation (SD). P value was calculated by t-test. * P < .05, ** P < .01.
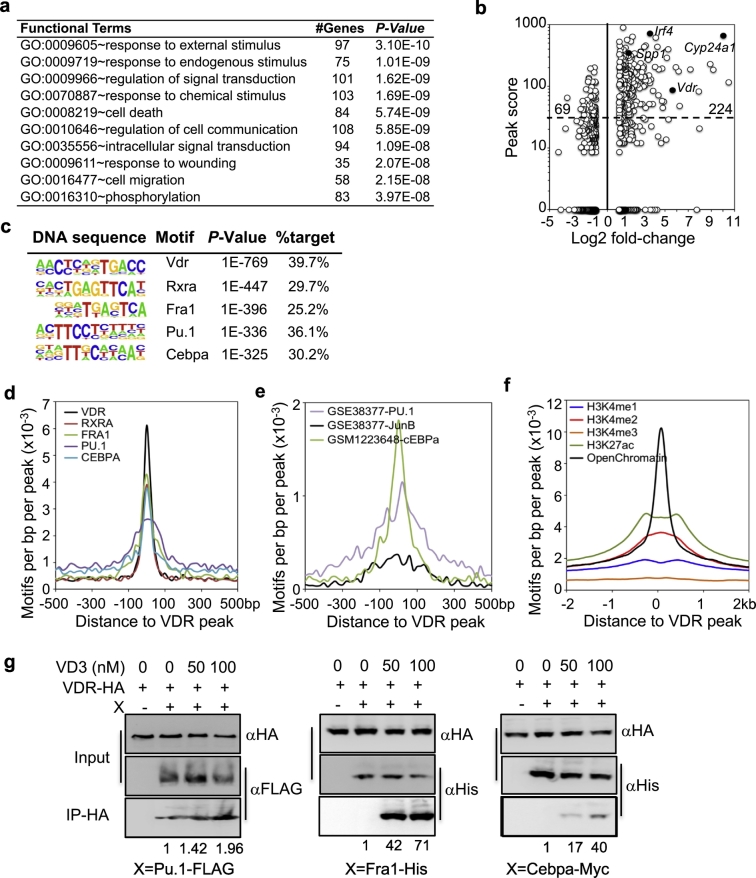
To further confirm the transcriptional regulation by VD3-VDR in AM, we assayed the genome-wide VDR binding sites in AM by ChIP-seq. AM were purified from B6 mice, expanded and then cultured for 24 h in the presence or absence of 50 nM VD3. ChIP-seq identified a total of 6080 peaks associated with 3428 genes (peakscore >30 & P ≤1e-10, Table S9). Functional enrichment analysis of 500 genes with top peak scores showed that similar gene pathways, such as phosphorylation, cell death, signal transduction, immune response were enriched (Fig. 3a), consistent with GO enrichment analysis of differentially expressed genes (Fig. 2b). 70% of the VD3 up-regulated genes, including Vdr, Cyp24a1 and Spp1, had a peak score larger than 30 (P ≤1e-10), whereas only 22% of the VD3 down-regulated genes had a peak score larger than 30 (Fig. 3b). De novo motif analysis showed that the VDR-binding motif was the most abundant and significantly enriched binding motif and present in ~40% peaks (Fig. 3c). The second most significantly enriched motif (based on P value) was for Rxra, a known TF that forms complex with VDR [67]. Binding motifs for PU.1, Fra1 (component of AP1 complex) and Cebpa were also found in high frequencies (Fig. 3c). All the identified VDR, Rxra, Fra1, PU.1, and Cebpa binding motifs were mapped to within 200 bp to each other (Fig. 3d). Similarly, the VDR peaks overlapped with published binding peaks [68] for PU.1, JunB and Cebpa in macrophages (Fig. 3e).
Fig. 3.
Transcriptional regulation by VDR in AM. a, GO enrichment analysis of 500 genes with top-ranked peak scores. GO sets of biological process, number of genes and P-value are shown. b, Comparison of ChIP-seq peak scores of VDR-binding genes and the fold-changes of the same genes induced by VD3 in AM. Each dot represents one gene. Selected genes (solid dots) are indicated. The number indicate the number of genes in the gated regions. c, Enriched sequence motifs in VDR ChIP-seq peaks by de novo motif analysis. d, Comparison of relative positions of VDR, Rxra, Fra1, PU.1 and Cebpa motifs from ChIP-seq. e, Comparison of DNA occupancies of VDR to those published for PU.1, JunB and Cebpa in AM. f, DNA occupancies of epigenetic marks in macrophages around VDR-binding peaks. g. Co-IP of VDR with Pu.1, Fra1 and Cebpa. 293FT cells were transiently transfected with HA-tagged VDR and FLAG-tagged Pu.1 (Spi1), His-tagged Fra1 (Fosl2) or Myc-tagged Cebpa as indicated. Cell lysates were precipitated with anti-HA in the presence or absence of VD3, followed by Western blotting with anti-HA, anti-FLAG, anti-His or anti-Myc antibodies. Input is the total cell lysate. The pull-down levels were quantified densitometry and normalized to the input and then the pull-down blots without VD3. Shown are representative gel image and average of pull-down level from three independent experiments.
PU.1 and AP-1 are known key TFs for maintaining macrophage identity through controlling the global macrophage-specific enhancer repertoire [[69], [70], [71]]. Cebpa and its cofactor Cebpb bind to enhancers and are key TFs required for regulating macrophage polarization [72,73]. Since ~40% of VDR binding peaks in AM resided in the intergenic regions, but only ~6% in the promoter region (Fig. S4), we tested whether VDR binding motifs are localized in the enhancer regions. We mapped VDR peaks to the published epigenomic tracks of macrophages [49]. As shown in Fig. 3f, VDR peaks were localized in open chromatins of macrophages with high intensity of active transcription (H3K4me2 and H3K27ac) but not in enriched promoter region (H3K4me1). Moreover, VDR constitutively interacted with Pu.1, Fra1 and Cebpa; and the interactions were enhanced by VD3 (Fig. 3g). Interestingly, VD3 also promoted the interaction between VDR and Pparg (Fig. S5), which is Rxra competitor but a key TF for the maturation and function of AM [[74], [75], [76], [77]]. Together, these results suggest that in response to VD3, VDR regulates gene expression in AM, partly by forming complexes with Rxra, Pu.1 and Cebpa in the transcription active enhancer regions.
3.3. Mice deficient of VDR develop lung emphysema
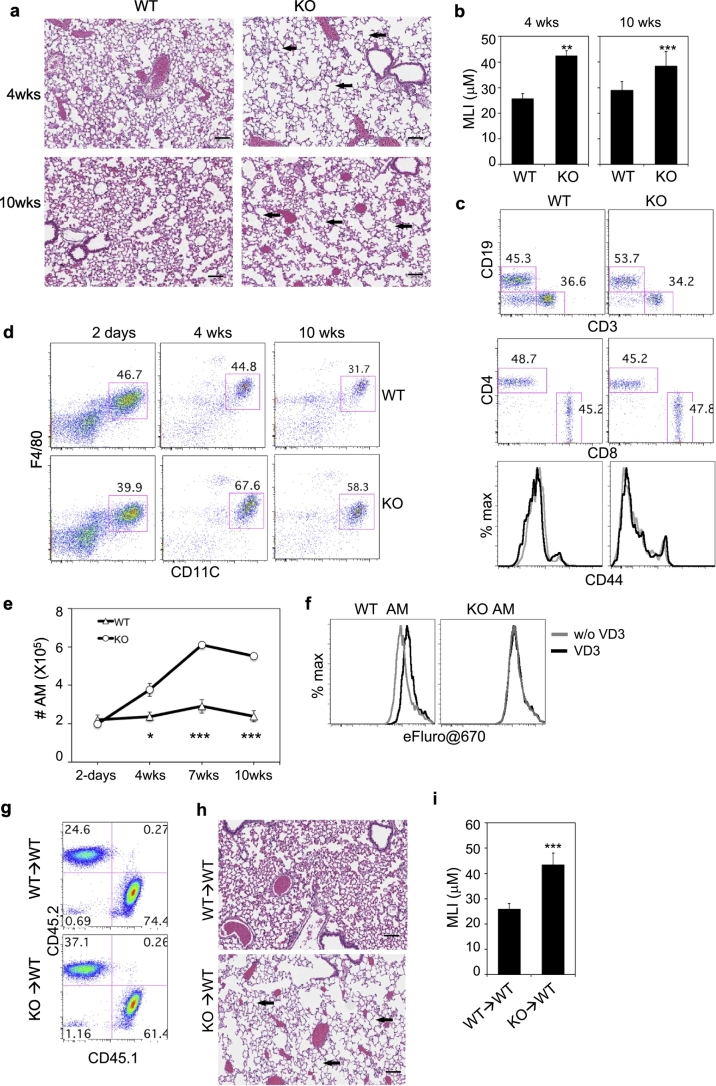
To investigate the physiological roles of VDR in AM, we analyzed AM and lung pathology in Vdr−/− mice. Hematoxylin and eosin (H&E) staining of lung sections revealed that Vdr−/− mice had significant enlargement of airspace (emphysema) as indicated by increase in the mean linear intercept (MLI) as early as 4 weeks of age as compared to age-matched wildtype and heterozygous littermates (Fig. 4a-b). However, there was no obvious infiltration of immune cells in the lung (Fig. 4a) and no significant difference in the frequency and CD44 expression of CD4+ T cells, CD8+ T cells and B cells in the lung tissue between wildtype and Vdr−/− mice (Fig. 4c and Fig. S6a). Similarly, expression of F4/80, CD11b, CD11c and Siglec-F were similar on AM from both wildtype and Vdr−/− mice (Fig. 4d and Fig. S6b), although MHCII expression was up-regulated on AM from Vdr−/− mice at 4 weeks of age (Fig. S6b). The number of AM was similar 2 days after birth between wildtype and Vdr−/− mice, but was significantly increased in Vdr−/− mice by 4 weeks of age and kept on increasing until 7–10 weeks of age (Fig. 4d-e). AM proliferated in response to GM-CSF and this proliferation was inhibited by addition of VD3 and abolished by Vdr deletion (Fig. 4f). Remarkably, intratracheal transfer of CD45.2+ Vdr−/− AM into wildtype mice (CD45.1+, once per week for four weeks) also led to significant enlargement of airspace (Fig. 4g-i and Fig. S7), although a single adoptive transfer of Vdr−/− AM into wildtype mice failed to induce emphysema probably due to low persistence of the transferred AM (Fig. S7). Together, these results show that by regulating gene expression in AM, VD3-VDR axis plays a critical role in maintaining the homeostasis of AM and VDR-deficient AM are sufficient to induce pulmonary emphysema in wildtype mice.
Fig. 4.
Accumulation of AM and development of emphysema in Vdr−/− mice. a, Comparison of H&E staining of lung sections from 4- and 10-weeks old wildtype (WT) and Vdr−/− littermates. b, Quantification of mean linear intercept (MLI) of alveolar airspace in WT and Vdr−/− littermates. n = 3 per genotype and age group. c, Flow cytometry analysis of T and B cells from the lung tissues of WT and Vdr−/− littermates at 4 weeks of age. Single cell suspension was prepared by digesting the lung tissue and stained for CD45, CD19, CD3, CD4, CD8, NK1.1 and CD44. Shown are representative CD19 versus CD3 staining profiles gating on CD45+ cells, CD4 versus CD8 staining profiles gating on CD3+ cells. Histograms show CD44 expression by CD4 T cells (left) and CD8 T cells (right) from WT (black) and Vdr−/− (grey) mice. The results from the same staining of 10-week-old WT and Vdr−/− mice are shown in Fig. S6a. d, Cells from BAL were stained for CD45, F4/80, CD11c, CD11b, MHCII and SiglecF. Shown are representative CD11c versus F4/80 staining profiles of CD45+ cells from WT and Vdr−/− littermates at different ages. Expression of F4/80, MHCII, CD11b, CD11c and SiglecF by AM are shown in Fig. S6b. e, Comparison of AM numbers in the BAL of WT and Vdr−/− mice at different ages (n = 6 mice each per genotype and age group). f, Inhibition of GM-CSF-induced AM proliferation by VD3. WT and Vdr−/− AM were labeled with eFluor®670 and cultured in the presence of GM-CSF for two days with or without VD3. Shown are representative eFluor®670 histograms from one of the three experiments. A decrease in eFluor®670 intensity indicates cell proliferation. g-i, Induction of pulmonary emphysema by intratracheal transfer of Vdr−/− AM into WT B6 mice. 5 × 105 WT or Vdr−/− AM (CD45.2+) were adoptively transferred into CD45.1+ B6 recipient mice every week for four weeks (n = 4 per group). At the 6th week, recipient mice were euthanized for flow cytometry to confirm the presence of the transferred cells in BAL (g) or H&E staining of lung sections (h) to quantify the mean linear intercept (MLI) of alveolar airspace (i). Scale bars in a and h: 100 μm. The numbers in c, d and g indicate percentages of cells in the gated regions. The arrows in a and h indicate enlarged alveolar space. Error bars indicate standard deviation (SD). P value was calculated by t-test. * P < .05, ** P < .01, *** P < .001.
3.4. VDR deficiency alters AM function
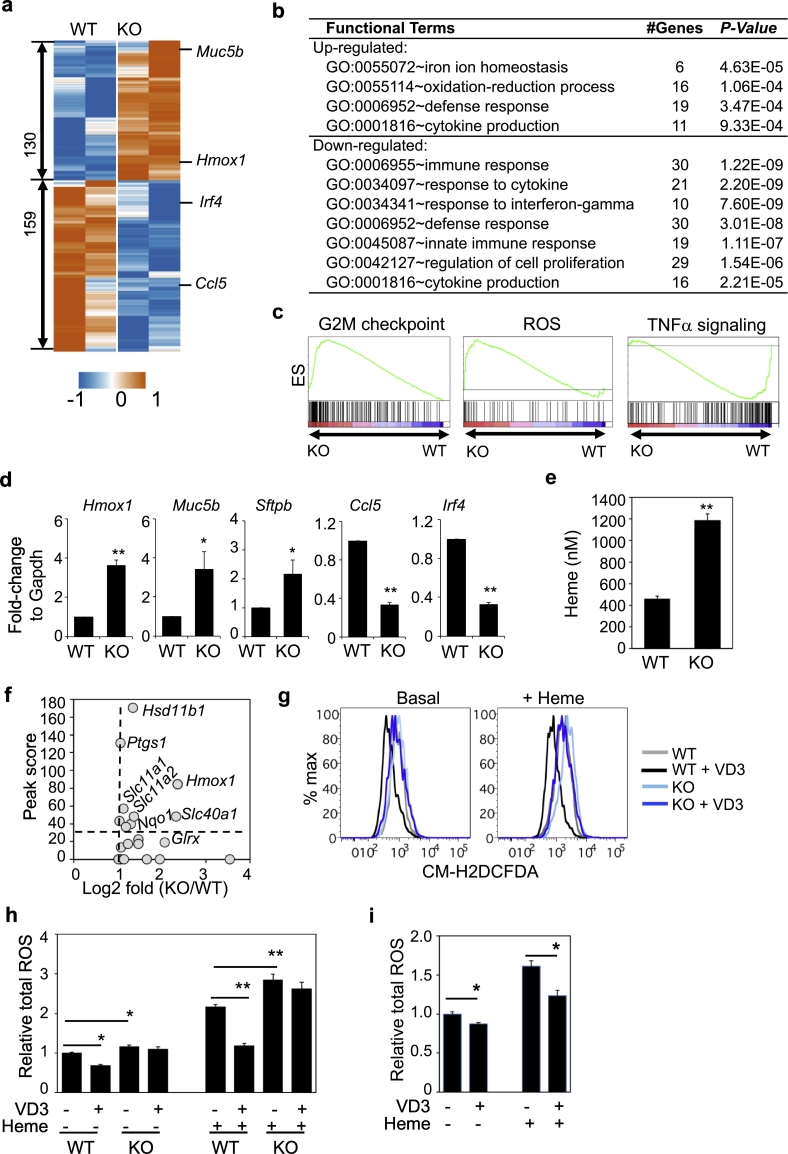
To investigate the molecular basis underlying the deregulated AM homeostasis in Vdr−/− mice, we assayed the gene expression of AM from 4-week-old mice by RNA-seq. Differential expression analysis identified 130 up-regulated and 159 down-regulated genes in Vdr−/− AM as compared to wildtype AM at a cutoff of 2-fold change with P-value <.05 (Fig. 5a and Table S10). Functional enrichment analysis showed that altered genes in Vdr−/− AM were involved in cell proliferation, ROS production, heme response, and cell immunity (Fig. 5b), consistent with the GESA results (Fig. 5c). The observation of up-regulation of genes involved in cell proliferation was consistent with the increased numbers of AM in Vdr−/− mice. Both upregulation and downregulation of immune response genes were observed in Vdr−/− AM while the expression of genes involved in response to IFNγ pathway were downregulated (Fig. S8), consistent with observations in AM from COPD patients [78]. Several COPD disease genes were also altered, including Hmox1, Muc5b, Sftpb and Ccl5 (Fig. 5a), which were further validated by qPCR (Fig. 5d). Expression changes of several ECM-remodeling genes were also observed, including Ctsk and Serpine1 (Fig. S9). Additional genes involved in COPD, fibrosis and tissue-remodeling were up-regulated in AM from 10 weeks old Vdr−/− mice (Fig. S10). These results are consistent with a key role of AM in pulmonary emphysema in Vdr−/− mice.
Fig. 5.
VD3-VDR suppresses the basal and heme-dependent ROS in AM. a, Heatmap showing gene expression of 130 up-regulated and 159 down-regulated genes in Vdr−/− AM as compared to WT AM from 4 weeks old mice. Shown is centralized CPM. b, GO enrichment analysis showing enrichment of certain pathways in the up-regulated and down-regulated genes. GO sets of biological process, number of genes and P-value are shown. c, GSEA showing enriched gene sets in Vdr−/− AM (q-value <0.05). d, Comparison of the transcript levels of Hmox1, Muc5b, Sftpb, Ccl5 and Irf4 in WT and Vdr−/− AM by qPCR. e, Heme level in BAL of WT and Vdr−/− mice at 5–7 weeks of age. n = 6–8 mice per group. f, Plot of VDR-binding scores of ROS-related genes and their expression change in Vdr−/− AM. g-h, Comparison of ROS levels in WT and Vdr−/− AM. WT and Vdr−/− AM were cultured for 1 h in the absence or presence of heme and/or VD3. Intracellular ROS level was detected by CM-H2DCFDA and flow cytometry. Shown are representative histograms (g) of basal (left) and heme-induced (right) ROS level and summarized data (h) from three independent experiments. i, Comparison of ROS levels in human AM. AM from healthy individuals (n = 10) were cultured for 1 h in the absence or presence of heme and/or VD3. ROS level was assayed as in g. Statistical significance is calculated by t-test. * P < .05, **P < .01.
3.5. VD3-VDR axis suppresses heme-induced ROS in AM
Oxidative stress is known to play a significant role in COPD pathogenesis [79,80]. Heme and iron are strong ROS inducers and are significantly elevated in BAL or serum of COPD patients and smokers [81,82]. Similarly, heme level was significantly higher in BAL of Vdr−/− mice than WT mice (Fig. 5e). We found that half of the genes involved in ROS and heme responsive pathways were directly bound by VDR in AM based on ChIP-seq (Fig. 5f). Consistently, VD3 suppressed the basal level as well as heme-induced ROS in AM and this effect was abolished in Vdr−/− AM (Fig. 5g-h). Abundant ROS-induced DNA damage was detected in the lung sections of Vdr−/− mice by anti-8OHdG staining (Fig. S11). Moreover, VD3 similarly suppressed heme-induced ROS in human AM (Fig. 5i). Taken together, these results suggest that the VD3-VDR axis suppresses both the basal and heme-induced ROS production in AM, consistent with the observed effect of VD3 in inhibition of COPD exacerbation.
3.6. VDR modulates LPS-induced gene expression in AM
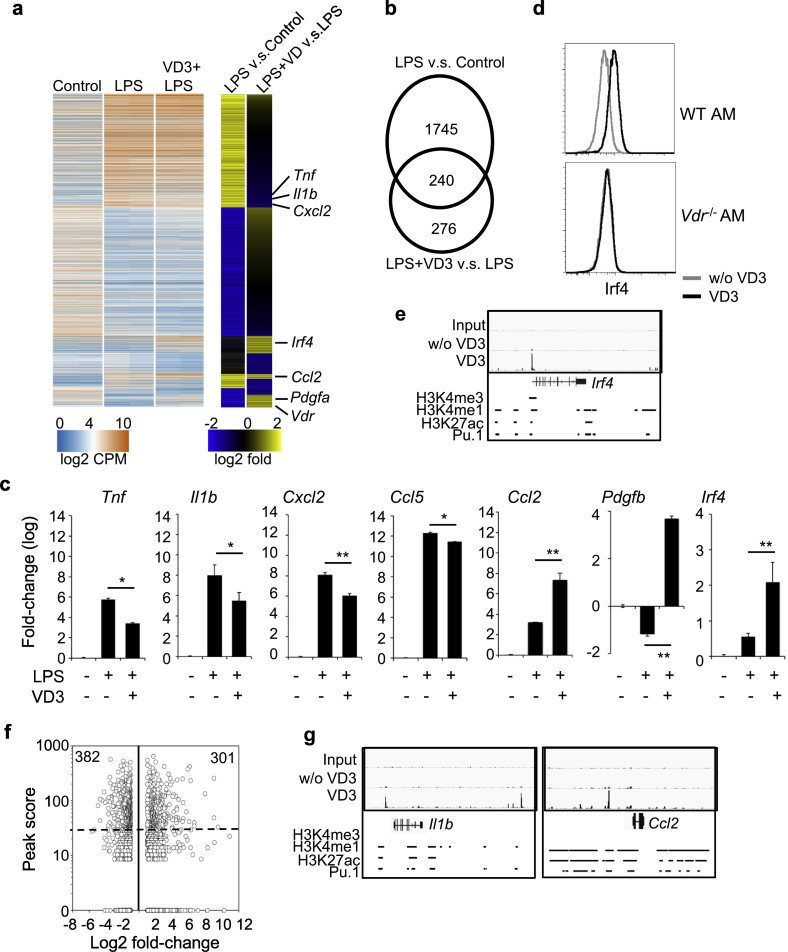
Because of their anatomical location, AM are constantly exposed to external stimuli. We therefore investigated the role of the VD3-VDR axis in modulating gene expression in AM following LPS stimulation. Purified AM from B6 mice were stimulated with LPS for 4 h in the presence or absence of VD3, followed by RNA-seq. Differential expression analysis revealed 1985 genes with 2-fold or more change following LPS stimulation (LPS vs. control) and 516 genes with 2-fold or more change following LPS + VD3 stimulation (LPS + VD3 vs. LPS) (Fig. 6a and Table S11). Among the 516 genes, 276 genes were regulated by VD3 alone whereas the remaining 240 were regulated by both LPS and VD3 (Fig. 6b). To validate the regulation of LPS-induced genes by VDR, freshly isolated AM were stimulated with LPS for 4 h in the presence or absence of VD3. Quantitative RT-PCR showed that the transcript levels of pro-inflammatory genes, such as Tnfa, Il1b, Cxcl2 and Ccl5 were induced by LPS (Fig. 6c); but the induction was significantly inhibited by VD3. In contrast, anti-inflammatory genes, such as Ccl2, Pdgfb and Irf4, were up-regulated by VD3. Upregulation of Irf4 by VD3 was further confirmed by flow cytometry and ChiP-seq in wildtype AM, whereas the upregulation was abolished in Vdr−/− AM (Fig. 6d-e). VD3 also suppressed LPS-induced ROS in AM and this effect was abolished in Vdr−/− AM (Fig. S12). Furthermore, we analyzed whether LPS-induced genes had VDR binding peaks in ChIP-seq data. Notably, ~35% (683 of 1985) of LPS-induced genes (P < .05 plus >2-fold change) in AM had VDR-binding peaks (P < 1e-10 and peak score > 30, Fig. 6f), including typical LPS-response genes, such as Il1b and Ccl2 (Fig. 6a, g). Together, these data show that the VD3-VDR axis modulates AM response to LPS, by suppressing the expression of pro-inflammatory genes and up-regulating the expression of anti-inflammatory genes.
Fig. 6.
VD3-VDR axis modulates gene expression in AM following LPS stimulation. a-b, Genome-wide transcriptional analysis of AM stimulated with LPS or LPS plus VD3 for 4 h. Heatmap (a) and overlap (b) of LPS-induced genes and VD3-induced genes in the presence LPS. LPS-induced genes were identified by comparison of LPS stimulation to control. VD3-induced genes were identified by comparison of LPS plus VD3 to LPS. c, Comparison of Tnfa, Il1b, Cxcl2, Ccl5, Ccl2, Pdgfb and Irf4 transcript levels by qPCR in AM without any stimulation, with LPS or LPS plus VD3 stimulation for 4 h. d. Flow cytometry analysis of Irf4 intracellular staining in wildtype (WT) and Vdr−/− AM with or without VD3 treatment for 24 h. e, Genome-browse of VDR-binding peaks in Irf4 by IGV. f, Comparison of ChIP-seq peak scores of VDR-binding genes and LPS-induced genes (>2-fold changes) in AM. g, Genome-browse of VDR-binding peaks in Il1b and Ccl2 by IGV.
4. Discussion
COPD is a complex disease with overlapping clinical phenotypes and pathologies, and poorly understood molecular and cellular mechanisms. In this study, we have examined the role of AM in COPD pathogenesis and the underlying molecular mechanisms. We show that VD3-VDR axis is critical for maintaining AM homeostasis and function and defect in this axis results in emphysema. First, VDR is highly expressed in AM (Fig. S3) and AM accumulate in Vdr−/− mice (Fig. 4e). The accumulation is likely driven by GM-CSF induced proliferation because VD3 inhibited GM-CSF induced proliferation of wildtype but not Vdr−/− AM (Fig. 4f). At molecular level, VD3 stimulated AM to express E2F-related cell cycle genes (Fig. 2b-c), which are known to inhibit cell proliferation. Thus, our finding identifies VD3 as a natural antagonist of GM-CSF for controlling AM homeostasis. Second, concomitant with the accumulation of AM, Vdr−/− mice develop emphysema as early as 4 weeks of age. The critical role of Vdr−/− AM in the observed pathogenesis was directly tested by intratracheal transfer of AM from Vdr−/− mice into wildtype mice. The induction of emphysema in the recipient mice after 4 weeks suggests that dysfunctional macrophages alone are sufficient to induce the observed pathology. Notably, the development of emphysema in Vdr−/− mice or recipient wildtype mice was not accompanied by apparent infiltration of immune cells into the lung tissue (Fig. 4), suggesting that the two key COPD pathologies - emphysema and bronchiolitis – can be separated from each other. Our findings are consistent with observations that patients with emphysema and COPD exhibit dramatic increases in AM as compared to healthy individuals [[9], [10], [11]]. Development of emphysema has also been reported in old Vdr−/− mice [37], which survived for >16 weeks because of supplementation with calcium and phosphorus. What distinguish our study from the previous studies is that we delineated the causal effect between dysfunctional AM and development of emphysema.
Our comprehensive analyses of gene expression in wildtype and Vdr−/− AM with or without VD3 treatment further reveal possible molecular mechanisms by which Vdr−/− AM cause emphysema. VD3-VDR axis regulates the expression of over six hundred genes, including several COPD biomarkers such as Hmox1 [83], Muc5b [84] and Clec5a [85]. Deficiency in VD3-VDR axis results in dramatic changes in gene expression. For example, extracellular matrix (ECM) remodeling enzymes play important roles in alveolar wall destruction during the pathogenesis of COPD [13]. Several of these enzymes are regulated by the VD3-VDR axis (Fig. S9). PAI-1 (Serpine1) and cathepsins K (Ctsk) were up-regulated in Vdr−/− AM but down-regulated by VD3 in wildtype AM. PAI-1 is elevated in COPD patients and its polymorphism is associated with susceptibility to COPD [86,87]. The cysteine protease cathepsins K, which is potent elastase and collagenase and capable to degrade ECM, is also up-regulated in emphysema patients and animal models [88]. Interestingly, another elastase MMP-12 was downregulated in Vdr−/− AM but up-regulated by VD3 treatment of wildtype AM. MMP-12 is up-regulated in COPD patients [15,89] and essential for the development of emphysema in mice following exposure to cigarette smoke [90]. However, MMP-12 is also involved in inflammation either directly or through the activation of TNFα and IFNα or inactivation of IFNγ [[91], [92], [93], [94]]. Oxidative stress is another major factor in the pathogenesis of COPD [80]. Vdr−/− AM produced higher levels of ROS at the basal level or in response to heme or LPS and Vdr−/− mice had higher levels of heme in BAL and ROS-induced lung damage, while VD3 treatment suppressed the ROS production of AM (Fig. 6 and Fig. S12). The increased level of heme in BAL of Vdr−/− mice is likely due to leakage and lysis of red blood cells in the lung due to tissue damage. Heme released from red blood cells could stimulate inflammatory response, including ROS production by AMs. The feedback between heme and ROS release could positively enforce each other in lung tissue damage. In addition, Vdr−/− AM showed down-regulation of genes involved in pathways of immune response and response to IFNγ (Fig. 5b and Fig. S8), similar to alternatively activated AM from COPD patients [78]. Previous studies have also shown that the immune response pathways in human AM are modulated by VD3 in vivo and in vitro in the context of bacterial infection [95,96], including some of validated genes (CCL2 and TNF) in mouse AM. Together, the increased numbers and dysfunction of AM could mediate the development of emphysema in Vdr−/− mice.
Our study sheds light on the conflicting results from longitudinal and cohort studies of VD3 supplements in COPD patients. Although the large population-based studies of VD3 supplements on COPD measured the levels of vitamin D3 in the serum, none of the studies made the fine distinction between COPD patients with emphysema, obstructive bronchiolitis or both. Based on our finding that deficiency in the VD3-VDR axis leads to development of emphysema but not bronchiolitis, VD3 supplements would be expected to exert most benefit in COPD patients with VD3 deficiency and emphysema, whereas VD3 supplements may not have much effect on obstructive bronchiolitis. Lumping different patients with different COPD pathologies together may have masked the efficacy of VD3 supplements on emphysema. While our prediction needs to be verified, our study suggests new direction for longitudinal and cohort studies.
Our systems biology analyses identify key transcription factors (TF) for tissue-resident macrophages and disease genes. We developed regulatory network-based approaches to identify key TFs to understand tissue-specific programming of gene expression for seven different tissue macrophages, and functional network-based approaches to prioritize key disease genes associated with particular tissue macrophages. Vdr was predicted as top-1 key TF for AM and a key COPD disease gene (ranked 10th, Fig. 1e). Our follow-up studies confirmed that Vdr is required for AM homeostasis and function and dysregulation of this pathway specifically results in emphysema. Besides heterodimerization with RXR, VDR also interacts with Pu.1, Fra1 and Cebpa, and VD3 strongly promotes these interactions (Fig. 3g). Pu.1 and Fra1 are essential coactivators to maintain macrophage identity and phenotype [6,69,73], our results would suggest VDR recruits macrophage-specific coactivators to regulate AM homeostasis and function. Pparg, top 5th predicted key TF for AM, is induced by GM-CSF in fetal monocytes for their differentiation to AM, but not in mature AM [75,97]. Previous studies have shown Pparg plays important roles in maintaining the phenotype and function of AM [76,77] and activation of PPARG in AM inhibits inflammation in the cigarette-smoke exposed mice [98]. Similar to VDR, PPARG also forms a complex with RXR and binds to different DNA motifs to regulate gene expression in response to different ligands [99]. Interestingly, VD3 promotes the interaction between VDR and PPARG as shown by co-immunoprecipitation (Fig. S5). These findings suggest that VDR and PPARG may regulate the phenotype and function of AM in different stages through competing for their heterodimer RXR in the presence of different ligands. In addition, studies have shown that Irf4 is an essential regulator required for maintaining M2 phenotype of tissue macrophages [100]. The expression of Irf4 was significantly induced by VD3 through VDR binding to the Irf4 promoter. Consistently, Irf4 was down-regulated in Vdr−/− AM as compared to wildtype AM (Fig. 2, Fig. 3, Fig. 4, Fig. 5). Thus, VD3-VDR axis also maintains the M2 phenotype and function of AM partly through up-regulating expression of Irf4 directly.
Our studies also identify the top-1 key TFs Spic for red pulp macrophages and Gata6 for peritoneal macrophages. Both are known as the lineage-specific key TFs for the respective macrophage development and function [[59], [60], [61], [62]]. Sox4 was predicted as top-1 key TF for microglia in our study. Sox4 is a target of TGF-β signaling [101] and TGF-β plays a critical role in microglia development as its deletion leads to the absence of microglia in mice [102]. Interestingly, microglia are present in Csf1op/op mice but absent from Csf1r−/− mice [103]. Deficiency of IL-34, another ligand of CSF1R, reduces microglia numbers [104]. These observations suggest Sox4 is likely a key regulator for microglia and it would be interesting to investigate the roles of Sox4 in development and function of microglia and how it interacts with signals from TGF-β and IL-34. Tissue macrophages play critical roles in homeostasis and exert diverse functions. Abnormal functional changes of tissue macrophages contribute to the development and progression of autoimmune disease, fibrosis and cancer [105,106]. Identification of key TFs for tissue macrophages would help to understand the mechanisms by which they contribute to health and diseases.
Funding sources
This work was supported in part by Grants AI69208, CA197605 and NS104315 from the National Institutes of Health of United States of America, Ivan R. Cottrell Professorship and Research Fund, the Koch Institute Support (core) Grant P30-CA14051 from the National Cancer Institute of Unites States of America, and the National Research Foundation of Singapore through the Singapore–MIT Alliance for Research and Technology's (SMART) Interdisciplinary Research Group in Antimicrobial Resistance Research Program. The funders of this work had no role in study design, data collection, data analysis, data interpretation, writing of the report, or decision to submit the article for publication.
Author contributions
G.H. and J.C. designed the research, interpreted the data and wrote the manuscript. G.H. performed the experiments and analyzed the data. T.D. performed the co-IP experiment. S.W. and H.J. performed the experiments of human AM.
Declaration of Competing Interest
The authors declare no competing interests.
Acknowledgements
The authors thank Starsha Kolodziej and Megan Kaiser for technique assistance and Koch Institute Swanson Biotechnology Center and core facilities in MIT for assistance with flow cytometry, histology, and RNAseq data acquisition and analysis. We also thank members of Chen lab for their suggestions.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.ebiom.2019.06.039.
Contributor Information
Guangan Hu, Email: gahu@mit.edu.
Jianzhu Chen, Email: jchen@mit.edu.
Appendix A. Supplementary data
Supplementary figures
Supplementary Table S1
Supplementary Table S2-S11
References
- 1.Barnes P.J., Burney P.G., Silverman E.K., Celli B.R., Vestbo J., Wedzicha J.A. Chronic obstructive pulmonary disease. Nat Rev Dis Primers. 2015;1:15076. doi: 10.1038/nrdp.2015.76. [DOI] [PubMed] [Google Scholar]
- 2.Al Omari M., Khassawneh B.Y., Khader Y., Dauod A.S., Bergus G. Prevalence of chronic obstructive pulmonary disease among adult male cigarettes smokers: a community-based study in Jordan. Int J Chron Obstruct Pulmon Dis. 2014;9:753–758. doi: 10.2147/COPD.S62898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Teramoto S. 1. COPD pathogenesis from the viewpoint of risk factors. Intern Med. 2007;46:77–79. doi: 10.2169/internalmedicine.46.1775. [DOI] [PubMed] [Google Scholar]
- 4.Yawn B.P., Keenan J.M. COPD--the primary care perspective: addressing epidemiology, pathology, diagnosis, treatment of smoking's multiple morbidities and the patient's perspective. COPD. 2007;4:67–83. doi: 10.1080/15412550601169562. [DOI] [PubMed] [Google Scholar]
- 5.Wedzicha J.A., Seemungal T.A. COPD exacerbations: defining their cause and prevention. Lancet. 2007;370:786–796. doi: 10.1016/S0140-6736(07)61382-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Okabe Y., Medzhitov R. Tissue biology perspective on macrophages. Nat Immunol. 2016;17:9–17. doi: 10.1038/ni.3320. [DOI] [PubMed] [Google Scholar]
- 7.Hussell T., Bell T.J. Alveolar macrophages: plasticity in a tissue-specific context. Nat Rev Immunol. 2014;14:81–93. doi: 10.1038/nri3600. [DOI] [PubMed] [Google Scholar]
- 8.Shapiro S.D. The macrophage in chronic obstructive pulmonary disease. Am J Respir Crit Care Med. 1999;160:S29–S32. doi: 10.1164/ajrccm.160.supplement_1.9. [DOI] [PubMed] [Google Scholar]
- 9.Vlahos R., Bozinovski S. Role of alveolar macrophages in chronic obstructive pulmonary disease. Front Immunol. 2014;5:435. doi: 10.3389/fimmu.2014.00435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Keatings V.M., Collins P.D., Scott D.M., Barnes P.J. Differences in interleukin-8 and tumor necrosis factor-alpha in induced sputum from patients with chronic obstructive pulmonary disease or asthma. Am J Respir Crit Care Med. 1996;153:530–534. doi: 10.1164/ajrccm.153.2.8564092. [DOI] [PubMed] [Google Scholar]
- 11.Pesci A., Balbi B., Majori M., Cacciani G., Bertacco S., Alciato P. Inflammatory cells and mediators in bronchial lavage of patients with chronic obstructive pulmonary disease. Eur Respir J. 1998;12:380–386. doi: 10.1183/09031936.98.12020380. [DOI] [PubMed] [Google Scholar]
- 12.Di Stefano A., Capelli A., Lusuardi M., Balbo P., Vecchio C., Maestrelli P. Severity of airflow limitation is associated with severity of airway inflammation in smokers. Am J Respir Crit Care Med. 1998;158:1277–1285. doi: 10.1164/ajrccm.158.4.9802078. [DOI] [PubMed] [Google Scholar]
- 13.Dey T., Kalita J., Weldon S., Taggart C.C. Proteases and their inhibitors in chronic obstructive pulmonary disease. J Clin Med. 2018;7 doi: 10.3390/jcm7090244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Shaykhiev R., Krause A., Salit J., Strulovici-Barel Y., Harvey B.-G., O'Connor T.P. Smoking-dependent reprogramming of alveolar macrophage polarization: implication for pathogenesis of chronic obstructive pulmonary disease. J Immunol. 2009;183:2867–2883. doi: 10.4049/jimmunol.0900473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Woodruff P.G., Koth L.L., Yang Y.H., Rodriguez M.W., Favoreto S., Dolganov G.M. A distinctive alveolar macrophage activation state induced by cigarette smoking. Am J Respir Crit Care Med. 2005;172:1383–1392. doi: 10.1164/rccm.200505-686OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kapellos T.S., Bassler K., Aschenbrenner A.C., Fujii W., Schultze J.L. Dysregulated functions of lung macrophage populations in COPD. J Immunol Res. 2018;2018:2349045. doi: 10.1155/2018/2349045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Berenson C.S., Kruzel R.L., Eberhardt E., Dolnick R., Minderman H., Wallace P.K. Impaired innate immune alveolar macrophage response and the predilection for COPD exacerbations. Thorax. 2014;69:811–818. doi: 10.1136/thoraxjnl-2013-203669. [DOI] [PubMed] [Google Scholar]
- 18.Zhang J., Chalmers M.J., Stayrook K.R., Burris L.L., Wang Y., Busby S.A. DNA binding alters coactivator interaction surfaces of the intact VDR-RXR complex. Nat Struct Mol Biol. 2011;18:556–563. doi: 10.1038/nsmb.2046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Adams J.S., Hewison M. Unexpected actions of vitamin D: new perspectives on the regulation of innate and adaptive immunity. Nat Clin Pract Endocrinol Metab. 2008;4:80. doi: 10.1038/ncpendmet0716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Janssens W., Bouillon R., Claes B., Carremans C., Lehouck A., Buysschaert I. Vitamin D deficiency is highly prevalent in COPD and correlates with variants in the vitamin D-binding gene. Thorax. 2010;65:215–220. doi: 10.1136/thx.2009.120659. [DOI] [PubMed] [Google Scholar]
- 21.Janssens W., Decramer M., Mathieu C., Korf H. Vitamin D and chronic obstructive pulmonary disease: hype or reality? Lancet Respir Med. 2013;1:804–812. doi: 10.1016/S2213-2600(13)70102-4. [DOI] [PubMed] [Google Scholar]
- 22.Foong R.E., Shaw N.C., Berry L.J., Hart P.H., Gorman S., Zosky G.R. Vitamin D deficiency causes airway hyperresponsiveness, increases airway smooth muscle mass, and reduces TGF-β expression in the lungs of female BALB/c mice. Phys Rep. 2014;2 doi: 10.1002/phy2.276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Heulens N., Korf H., Cielen N., De Smidt E., Maes K., Gysemans C. Vitamin D deficiency exacerbates COPD-like characteristics in the lungs of cigarette smoke-exposed mice. Respir Res. 2015;16:110. doi: 10.1186/s12931-015-0271-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zosky G.R., Berry L.J., Elliot J.G., James A.L., Gorman S., Hart P.H. Vitamin D deficiency causes deficits in lung function and alters lung structure. Am J Respir Crit Care Med. 2011;183:1336–1343. doi: 10.1164/rccm.201010-1596OC. [DOI] [PubMed] [Google Scholar]
- 25.Bouillon R., Carmeliet G., Verlinden L., van Etten E., Verstuyf A., Luderer H.F. Vitamin D and human health: lessons from vitamin D receptor null mice. Endocr Rev. 2008;29:726–776. doi: 10.1210/er.2008-0004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Holick M.F. Vitamin D deficiency. N Engl J Med. 2007;357:266–281. doi: 10.1056/NEJMra070553. [DOI] [PubMed] [Google Scholar]
- 27.Kunisaki K.M., Niewoehner D.E., Connett J.E., Clinical Research Network C.O.P.D. Vitamin D levels and risk of acute exacerbations of chronic obstructive pulmonary disease: a prospective cohort study. Am J Respir Crit Care Med. 2012;185:286–290. doi: 10.1164/rccm.201109-1644OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Persson L.J.P., Aanerud M., Hiemstra P.S., Hardie J.A., Bakke P.S., Eagan T.M.L. Chronic obstructive pulmonary disease is associated with low levels of vitamin D. PLoS One. 2012;7 doi: 10.1371/journal.pone.0038934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Martineau A.R., James W.Y., Hooper R.L., Barnes N.C., Jolliffe D.A., Greiller C.L. Vitamin D3 supplementation in patients with chronic obstructive pulmonary disease (ViDiCO): a multicentre, double-blind, randomised controlled trial. Lancet Respir Med. 2015;3:120–130. doi: 10.1016/S2213-2600(14)70255-3. [DOI] [PubMed] [Google Scholar]
- 30.Sanjari M., Soltani A., Habibi Khorasani A., Zareinejad M. The effect of vitamin D on COPD exacerbation: a double blind randomized placebo-controlled parallel clinical trial. J Diabetes Metab Disord. 2015;15:33. doi: 10.1186/s40200-016-0257-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gautier E.L., Shay T., Miller J., Greter M., Jakubzick C., Ivanov S. Gene-expression profiles and transcriptional regulatory pathways that underlie the identity and diversity of mouse tissue macrophages. Nat Immunol. 2012;13:1118–1128. doi: 10.1038/ni.2419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Li Y.C., Pirro A.E., Amling M., Delling G., Baron R., Bronson R. Targeted ablation of the vitamin D receptor: an animal model of vitamin D-dependent rickets type II with alopecia. Proc Natl Acad Sci U S A. 1997;94:9831–9835. doi: 10.1073/pnas.94.18.9831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Li Y.C., Amling M., Pirro A.E., Priemel M., Meuse J., Baron R. Normalization of mineral ion homeostasis by dietary means prevents hyperparathyroidism, rickets, and osteomalacia, but not alopecia in vitamin D receptor-ablated mice. Endocrinology. 1998;139:4391–4396. doi: 10.1210/endo.139.10.6262. [DOI] [PubMed] [Google Scholar]
- 34.Mathieu C., Van Etten E., Gysemans C., Decallonne B., Kato S., Laureys J. In vitro and in vivo analysis of the immune system of vitamin D receptor knockout mice. J Bone Miner Res. 2001;16:2057–2065. doi: 10.1359/jbmr.2001.16.11.2057. [DOI] [PubMed] [Google Scholar]
- 35.Kong J., Zhu X., Shi Y., Liu T., Chen Y., Bhan I. VDR attenuates acute lung injury by blocking Ang-2-Tie-2 pathway and renin-angiotensin system. Mol Endocrinol. 2013;27:2116–2125. doi: 10.1210/me.2013-1146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Shi Y.Y., Liu T.J., Fu J.H., Xu W., Wu L.L., Hou A.N. Vitamin D/VDR signaling attenuates lipopolysaccharideinduced acute lung injury by maintaining the integrity of the pulmonary epithelial barrier. Mol Med Rep. 2016;13:1186–1194. doi: 10.3892/mmr.2015.4685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sundar I.K., Hwang J.W., Wu S., Sun J., Rahman I. Deletion of vitamin D receptor leads to premature emphysema/COPD by increased matrix metalloproteinases and lymphoid aggregates formation. Biochem Biophys Res Commun. 2011;406:127–133. doi: 10.1016/j.bbrc.2011.02.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hu G., Chen J. A genome-wide regulatory network identifies key transcription factors for memory CD8+ T cell development. Nat Commun. 2013;4:2830. doi: 10.1038/ncomms3830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Faith J.J., Hayete B., Thaden J.T., Mogno I., Wierzbowski J., Cottarel G. Large-scale mapping and validation of Escherichia coli transcriptional regulation from a compendium of expression profiles. PLoS Biol. 2007;5 doi: 10.1371/journal.pbio.0050008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Prieur X., Mok C.Y.L., Velagapudi V.R., Núñez V., Fuentes L., Montaner D. Differential lipid partitioning between adipocytes and tissue macrophages modulates macrophage lipotoxicity and M2/M1 polarization in obese mice. Diabetes. 2011;60:797–809. doi: 10.2337/db10-0705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hu G., Cabrera A., Kono M., Mok S., Chaal B.K., Haase S. Transcriptional profiling of growth perturbations of the human malaria parasite plasmodium falciparum. Nat Biotechnol. 2010;28:91–98. doi: 10.1038/nbt.1597. [DOI] [PubMed] [Google Scholar]
- 42.Köhler S., Doelken S.C., Mungall C.J., Bauer S., Firth H.V., Bailleul-Forestier I. The human phenotype ontology project: linking molecular biology and disease through phenotype data. Nucleic Acids Res. 2014;42:D966–D974. doi: 10.1093/nar/gkt1026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Vanunu O., Magger O., Ruppin E., Shlomi T., Sharan R. Associating genes and protein complexes with disease via network propagation. PLoS Comput Biol. 2010;6 doi: 10.1371/journal.pcbi.1000641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Xu J., Li Y. Discovering disease-genes by topological features in human protein-protein interaction network. Bioinformatics. 2006;22:2800–2805. doi: 10.1093/bioinformatics/btl467. [DOI] [PubMed] [Google Scholar]
- 45.Franke L., van Bakel H., Fokkens L., de Jong E.D., Egmont-Petersen M., Wijmenga C. Reconstruction of a functional human gene network, with an application for prioritizing positional candidate genes. Am J Hum Genet. 2006;78:1011–1025. doi: 10.1086/504300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Enright A.J., Van Dongen S., Ouzounis C.A. An efficient algorithm for large-scale detection of protein families. Nucleic Acids Res. 2002;30:1575–1584. doi: 10.1093/nar/30.7.1575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Akagawa K.S., Kamoshita K., Tokunaga T. Effects of granulocyte-macrophage colony-stimulating factor and colony-stimulating factor-1 on the proliferation and differentiation of murine alveolar macrophages. J Immunol. 1988;141:3383–3390. [PubMed] [Google Scholar]
- 48.Fejer G., Wegner M.D., Györy I., Cohen I., Engelhard P., Voronov E. Nontransformed, GM-CSF-dependent macrophage lines are a unique model to study tissue macrophage functions. Proc Natl Acad Sci U S A. 2013;110:E2191–E2198. doi: 10.1073/pnas.1302877110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lavin Y., Winter D., Blecher-Gonen R., David E., Keren-Shaul H., Merad M. Tissue-resident macrophage enhancer landscapes are shaped by the local microenvironment. Cell. 2014;159:1312–1326. doi: 10.1016/j.cell.2014.11.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Zhang X., Goncalves R., Mosser D.M. The isolation and characterization of murine macrophages. Curr Protoc Immunol. 2008 doi: 10.1002/0471142735.im1401s83. CHAPTER:Unit-14.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Sinclair P.R., Gorman N., Jacobs J.M. Measurement of heme concentration. Curr Protoc Toxicol. 2001 doi: 10.1002/0471140856.tx0803s00. Chapter 8:Unit 8.3. [DOI] [PubMed] [Google Scholar]
- 52.Langmead B., Trapnell C., Pop M., Salzberg S.L. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 2009;10:R25. doi: 10.1186/gb-2009-10-3-r25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Li B., Dewey C.N. RSEM: accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinformatics. 2011;12:323. doi: 10.1186/1471-2105-12-323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Robinson M.D., McCarthy D.J., Smyth G.K. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics. 2010;26:139–140. doi: 10.1093/bioinformatics/btp616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Huang D.W., Sherman B.T., Tan Q., Collins J.R., Alvord W.G., Roayaei J. The DAVID gene functional classification tool: a novel biological module-centric algorithm to functionally analyze large gene lists. Genome Biol. 2007;8:R183. doi: 10.1186/gb-2007-8-9-r183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Subramanian A., Tamayo P., Mootha V.K., Mukherjee S., Ebert B.L., Gillette M.A. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci U S A. 2005;102:15545. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Saeed A.I., Sharov V., White J., Li J., Liang W., Bhagabati N. TM4: a free, open-source system for microarray data management and analysis. BioTechniques. 2003;34:374–378. doi: 10.2144/03342mt01. [DOI] [PubMed] [Google Scholar]
- 58.Heinz S., Benner C., Spann N., Bertolino E., Lin Y.C., Laslo P. Simple combinations of lineage-determining transcription factors prime cis-regulatory elements required for macrophage and B cell identities. Mol Cell. 2010;38:576–589. doi: 10.1016/j.molcel.2010.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Haldar M., Kohyama M., So A.Y.-L., Kc W., Wu X., Briseño C.G. Heme-mediated SPI-C induction promotes monocyte differentiation into iron-recycling macrophages. Cell. 2014;156:1223–1234. doi: 10.1016/j.cell.2014.01.069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Kohyama M., Ise W., Edelson B.T., Wilker P.R., Hildner K., Mejia C. Role for Spi-C in the development of red pulp macrophages and splenic iron homeostasis. Nature. 2009;457:318–321. doi: 10.1038/nature07472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Okabe Y., Medzhitov R. Tissue-specific signals control reversible program of localization and functional polarization of macrophages. Cell. 2014;157:832–844. doi: 10.1016/j.cell.2014.04.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Rosas M., Davies L.C., Giles P.J., Liao C.-T., Kharfan B., Stone T.C. The transcription factor Gata6 links tissue macrophage phenotype and proliferative renewal. Science. 2014;344:645–648. doi: 10.1126/science.1251414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Aziz A., Soucie E., Sarrazin S., Sieweke M.H. MafB/c-Maf deficiency enables self-renewal of differentiated functional macrophages. Science. 2009;326:867–871. doi: 10.1126/science.1176056. [DOI] [PubMed] [Google Scholar]
- 64.Bakri Y., Sarrazin S., Mayer U.P., Tillmanns S., Nerlov C., Boned A. Balance of MafB and PU.1 specifies alternative macrophage or dendritic cell fate. Blood. 2005;105:2707–2716. doi: 10.1182/blood-2004-04-1448. [DOI] [PubMed] [Google Scholar]
- 65.Bouhlel M.A., Derudas B., Rigamonti E., Dièvart R., Brozek J., Haulon S. PPARgamma activation primes human monocytes into alternative M2 macrophages with anti-inflammatory properties. Cell Metab. 2007;6:137–143. doi: 10.1016/j.cmet.2007.06.010. [DOI] [PubMed] [Google Scholar]
- 66.Ricote M., Li A.C., Willson T.M., Kelly C.J., Glass C.K. The peroxisome proliferator-activated receptor-γ is a negative regulator of macrophage activation. Nature. 1998;391:79. doi: 10.1038/34178. [DOI] [PubMed] [Google Scholar]
- 67.Kimmel-Jehan C., Jehan F., DeLuca H.F. Salt concentration determines 1,25-dihydroxyvitamin D3 dependency of vitamin D receptor-retinoid X receptor--vitamin D-responsive element complex formation. Arch Biochem Biophys. 1997;341:75–80. doi: 10.1006/abbi.1997.9952. [DOI] [PubMed] [Google Scholar]
- 68.Ostuni R., Piccolo V., Barozzi I., Polletti S., Termanini A., Bonifacio S. Latent enhancers activated by stimulation in differentiated cells. Cell. 2013;152:157–171. doi: 10.1016/j.cell.2012.12.018. [DOI] [PubMed] [Google Scholar]
- 69.Lawrence T., Natoli G. Transcriptional regulation of macrophage polarization: enabling diversity with identity. Nat Rev Immunol. 2011;11:750–761. doi: 10.1038/nri3088. [DOI] [PubMed] [Google Scholar]
- 70.Phanstiel D.H., Van Bortle K., Spacek D., Hess G.T., Shamim M.S., Machol I. Static and dynamic DNA loops form AP-1-bound activation hubs during macrophage development. Mol Cell. 2017;67:1037–1048. doi: 10.1016/j.molcel.2017.08.006. e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Srivastava P.K., Hull R.P., Behmoaras J., Petretto E., Aitman T.J. JunD/AP1 regulatory network analysis during macrophage activation in a rat model of crescentic glomerulonephritis. BMC Syst Biol. 2013;7:93. doi: 10.1186/1752-0509-7-93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Cain D.W., O'Koren E.G., Kan M.J., Womble M., Sempowski G.D., Hopper K. Identification of a tissue-specific, C/EBPβ-dependent pathway of differentiation for murine peritoneal macrophages. J Immunol. 2013;191:4665–4675. doi: 10.4049/jimmunol.1300581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Lee B., Qiao L., Lu M., Yoo H.S., Cheung W., Mak R. C/EBPα regulates macrophage activation and systemic metabolism. Am J Physiol Endocrinol Metab. 2014;306:E1144–E1154. doi: 10.1152/ajpendo.00002.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Guilliams M., De Kleer I., Henri S., Post S., Vanhoutte L., De Prijck S. Alveolar macrophages develop from fetal monocytes that differentiate into long-lived cells in the first week of life via GM-CSF. J Exp Med. 2013;210:1977–1992. doi: 10.1084/jem.20131199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Schneider C., Nobs S.P., Kurrer M., Rehrauer H., Thiele C., Kopf M. Induction of the nuclear receptor PPAR-γ by the cytokine GM-CSF is critical for the differentiation of fetal monocytes into alveolar macrophages. Nat Immunol. 2014;15:1026–1037. doi: 10.1038/ni.3005. [DOI] [PubMed] [Google Scholar]
- 76.Asada K., Sasaki S., Suda T., Chida K., Nakamura H. Antiinflammatory roles of peroxisome proliferator-activated receptor gamma in human alveolar macrophages. Am J Respir Crit Care Med. 2004;169:195–200. doi: 10.1164/rccm.200207-740OC. [DOI] [PubMed] [Google Scholar]
- 77.Malur A., McCoy A.J., Arce S., Barna B.P., Kavuru M.S., Malur A.G. Deletion of PPAR gamma in alveolar macrophages is associated with a Th-1 pulmonary inflammatory response. J Immunol. 2009;182:5816–5822. doi: 10.4049/jimmunol.0803504. [DOI] [PubMed] [Google Scholar]
- 78.Xue J., Schmidt S.V., Sander J., Draffehn A., Krebs W., Quester I. Transcriptome-based network analysis reveals a spectrum model of human macrophage activation. Immunity. 2014;40:274–288. doi: 10.1016/j.immuni.2014.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Domej W., Oettl K., Renner W. Oxidative stress and free radicals in COPD – implications and relevance for treatment. Int J Chron Obstruct Pulmon Dis. 2014;9:1207–1224. doi: 10.2147/COPD.S51226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Kirkham P.A., Barnes P.J. Oxidative stress in COPD. Chest. 2013;144:266–273. doi: 10.1378/chest.12-2664. [DOI] [PubMed] [Google Scholar]
- 81.Aggarwal S., Ahmad I., Lam A., Carlisle M.A., Li C., Wells J.M. Heme scavenging reduces pulmonary endoplasmic reticulum stress, fibrosis, and emphysema. JCI Insight. 2018;3 doi: 10.1172/jci.insight.120694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Cloonan S.M., Mumby S., Adcock I.M., Choi A.M.K., Chung K.F., Quinlan G.J. The “Iron”-y of iron overload and Iron deficiency in chronic obstructive pulmonary disease. Am J Respir Crit Care Med. 2017;196:1103–1112. doi: 10.1164/rccm.201702-0311PP. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Slebos D.J., Kerstjens H.A.M., Rutgers S.R., Kauffman H.F., Choi A.M.K., Postma D.S. Haem oxygenase-1 expression is diminished in alveolar macrophages of patients with COPD. Eur Respir J. 2004;23:652–653. doi: 10.1183/09031936.04.00127904. [DOI] [PubMed] [Google Scholar]
- 84.Kirkham S., Kolsum U., Rousseau K., Singh D., Vestbo J., Thornton D. MUC5B is the major mucin in the gel phase of sputum in chronic obstructive pulmonary disease. Am J Respir Crit Care Med. 2008;178:1033–1039. doi: 10.1164/rccm.200803-391OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Wortham B.W., Eppert B.L., Flury J.L., Garcia S.M., Donica W.R., Osterburg A. Cutting edge: CLEC5A mediates macrophage function and chronic obstructive pulmonary disease pathologies. J Immunol. 2016;196:3227–3231. doi: 10.4049/jimmunol.1500978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Waschki B., Watz H., Holz O., Magnussen H., Olejnicka B., Welte T. Plasminogen activator inhibitor-1 is elevated in patients with COPD independent of metabolic and cardiovascular function. Int J Chron Obstruct Pulmon Dis. 2017;12:981–987. doi: 10.2147/COPD.S128689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Xu X., Wang H., Li H., Cui X., Zhang H. SERPINE1 -844 and −675 polymorphisms and chronic obstructive pulmonary disease in a Chinese Han population. J Int Med Res. 2016;44:1292–1301. doi: 10.1177/0300060516664270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Golovatch P., Mercer B.A., Lemaitre V., Wallace A., Foronjy R.F., D'Armiento J. Role for cathepsin K in emphysema in smoke-exposed guinea pigs. Exp Lung Res. 2009;35:631–645. doi: 10.3109/01902140902822304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Molet S., Belleguic C., Lena H., Germain N., Bertrand C.P., Shapiro S.D. Increase in macrophage elastase (MMP-12) in lungs from patients with chronic obstructive pulmonary disease. Inflamm Res. 2005;54:31–36. doi: 10.1007/s00011-004-1319-4. [DOI] [PubMed] [Google Scholar]
- 90.Hautamaki R.D., Kobayashi D.K., Senior R.M., Shapiro S.D. Requirement for macrophage elastase for cigarette smoke-induced emphysema in mice. Science. 1997;277:2002–2004. doi: 10.1126/science.277.5334.2002. [DOI] [PubMed] [Google Scholar]
- 91.Churg A., Wang R.D., Tai H., Wang X., Xie C., Wright J.L. Tumor necrosis factor-alpha drives 70% of cigarette smoke-induced emphysema in the mouse. Am J Respir Crit Care Med. 2004;170:492–498. doi: 10.1164/rccm.200404-511OC. [DOI] [PubMed] [Google Scholar]
- 92.Dufour A., Bellac C.L., Eckhard U., Solis N., Klein T., Kappelhoff R. C-terminal truncation of IFN-gamma inhibits proinflammatory macrophage responses and is deficient in autoimmune disease. Nat Commun. 2018;9:2416. doi: 10.1038/s41467-018-04717-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Marchant D.J., Bellac C.L., Moraes T.J., Wadsworth S.J., Dufour A., Butler G.S. A new transcriptional role for matrix metalloproteinase-12 in antiviral immunity. Nat Med. 2014;20:493–502. doi: 10.1038/nm.3508. [DOI] [PubMed] [Google Scholar]
- 94.Nenan S., Planquois J.M., Berna P., De Mendez I., Hitier S., Shapiro S.D. Analysis of the inflammatory response induced by rhMMP-12 catalytic domain instilled in mouse airways. Int Immunopharmacol. 2005;5:511–524. doi: 10.1016/j.intimp.2004.10.011. [DOI] [PubMed] [Google Scholar]
- 95.Gerke A.K., Pezzulo A.A., Tang F., Cavanaugh J.E., Bair T.B., Phillips E. Effects of vitamin D supplementation on alveolar macrophage gene expression: preliminary results of a randomized, controlled trial. Multidiscip Respir Med. 2014;9:18. doi: 10.1186/2049-6958-9-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Heulens N., Korf H., Mathyssen C., Everaerts S., De Smidt E., Dooms C. 1,25-Dihydroxyvitamin D modulates antibacterial and inflammatory response in human cigarette smoke-exposed macrophages. PLoS One. 2016;11 doi: 10.1371/journal.pone.0160482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Guilliams M., De Kleer I., Henri S., Post S., Vanhoutte L., De Prijck S. Alveolar macrophages develop from fetal monocytes that differentiate into long-lived cells in the first week of life via GM-CSF. J Exp Med. 2013;210:1977. doi: 10.1084/jem.20131199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Yin Y., Hou G., Li E., Wang Q., Kang J. PPARgamma agonists regulate tobacco smoke-induced toll like receptor 4 expression in alveolar macrophages. Respir Res. 2014;15:28. doi: 10.1186/1465-9921-15-28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Matsuda S., Kitagishi Y. Peroxisome proliferator-activated receptor and vitamin d receptor signaling pathways in cancer cells. Cancers (Basel) 2013;5:1261–1270. doi: 10.3390/cancers5041261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Satoh T., Takeuchi O., Vandenbon A., Yasuda K., Tanaka Y., Kumagai Y. The Jmjd3-Irf4 axis regulates M2 macrophage polarization and host responses against helminth infection. Nat Immunol. 2010;11:936–944. doi: 10.1038/ni.1920. [DOI] [PubMed] [Google Scholar]
- 101.Kuwahara M., Yamashita M., Shinoda K., Tofukuji S., Onodera A., Shinnakasu R. The transcription factor Sox4 is a downstream target of signaling by the cytokine TGF-β and suppresses TH2 differentiation. Nat Immunol. 2012;13:778–786. doi: 10.1038/ni.2362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Butovsky O., Jedrychowski M.P., Moore C.S., Cialic R., Lanser A.J., Gabriely G. Identification of a unique TGF-β dependent molecular and functional signature in microglia. Nat Neurosci. 2014;17:131–143. doi: 10.1038/nn.3599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Ginhoux F., Greter M., Leboeuf M., Nandi S., See P., Gokhan S. Fate mapping analysis reveals that adult microglia derive from primitive macrophages. Science. 2010;330:841–845. doi: 10.1126/science.1194637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Wang Y., Szretter K.J., Vermi W., Gilfillan S., Rossini C., Cella M. IL-34 is a tissue-restricted ligand of CSF1R required for the development of Langerhans cells and microglia. Nat Immunol. 2012;13:753–760. doi: 10.1038/ni.2360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Gordon S., Plüddemann A. Tissue macrophages: heterogeneity and functions. BMC Biol. 2017;15:53. doi: 10.1186/s12915-017-0392-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Murray P.J., Wynn T.A. Protective and pathogenic functions of macrophage subsets. Nat Rev Immunol. 2011;11:723–737. doi: 10.1038/nri3073. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary figures
Supplementary Table S1
Supplementary Table S2-S11