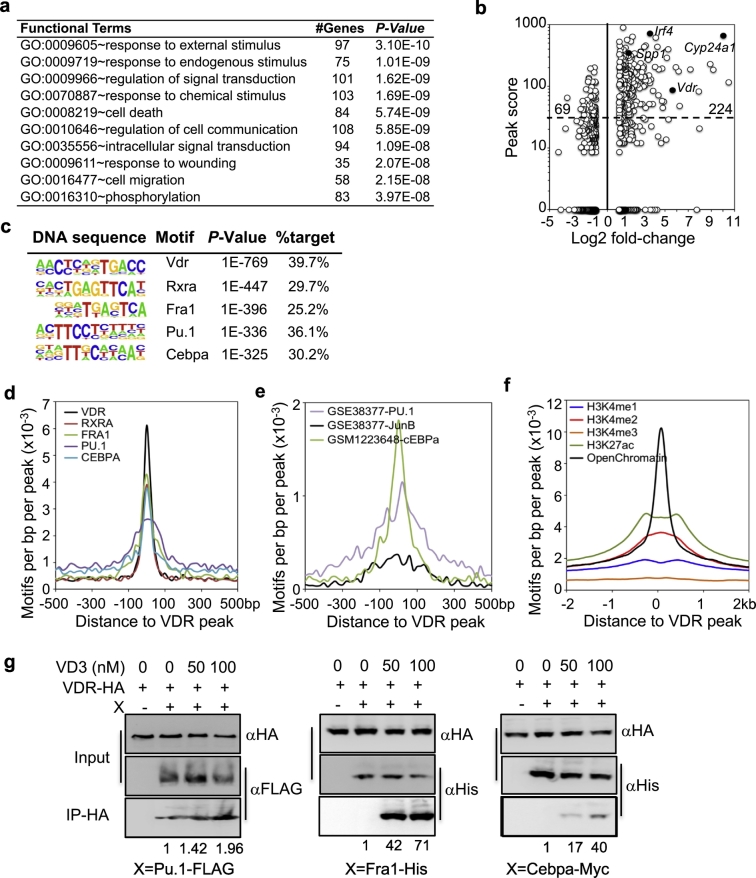
Fig. 3.
Transcriptional regulation by VDR in AM. a, GO enrichment analysis of 500 genes with top-ranked peak scores. GO sets of biological process, number of genes and P-value are shown. b, Comparison of ChIP-seq peak scores of VDR-binding genes and the fold-changes of the same genes induced by VD3 in AM. Each dot represents one gene. Selected genes (solid dots) are indicated. The number indicate the number of genes in the gated regions. c, Enriched sequence motifs in VDR ChIP-seq peaks by de novo motif analysis. d, Comparison of relative positions of VDR, Rxra, Fra1, PU.1 and Cebpa motifs from ChIP-seq. e, Comparison of DNA occupancies of VDR to those published for PU.1, JunB and Cebpa in AM. f, DNA occupancies of epigenetic marks in macrophages around VDR-binding peaks. g. Co-IP of VDR with Pu.1, Fra1 and Cebpa. 293FT cells were transiently transfected with HA-tagged VDR and FLAG-tagged Pu.1 (Spi1), His-tagged Fra1 (Fosl2) or Myc-tagged Cebpa as indicated. Cell lysates were precipitated with anti-HA in the presence or absence of VD3, followed by Western blotting with anti-HA, anti-FLAG, anti-His or anti-Myc antibodies. Input is the total cell lysate. The pull-down levels were quantified densitometry and normalized to the input and then the pull-down blots without VD3. Shown are representative gel image and average of pull-down level from three independent experiments.