Abstract
Backgrounds
One of the limitations of somatic cell nuclear transfer (SCNT) strategy to generate genetically modified offspring is the low birth rate. Placental dysfunction is one of the causes of abortion. Circular RNA (circRNA) is noncoding RNA which functions as microRNA (miRNA) sponges in biological processes.
Methods
Two aberrant pregnant placenta (aberrant group, AG) and three normal pregnant placenta (normal group, NG) during late gestation (180‐210 days) with bovine SCNT fetus were collected for high‐throughput sequencing and analyzed. The host genes of differentially expressed (DE) circRNAs were predicted. And the microRNAs (miRNAs) which could interact with DE circRNAs were analyzed. Then, the expressional level of partial DE circRNAs and corresponding host genes was verified through qRT‐PCR. At last, the function of host genes was analyzed through Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG).
Results
Altogether 123 differentially expressed circRNAs between two groups were identified, which were found related to 60 host genes and 32 miRNAs. The top 10 upregulated circRNAs were bta_circ_0012985, bta_circ_0013071, bta_circ_0013074, bta_circ_0016024, bta_circ_0013068, bta_circ_0008816, bta_circ_0012982, bta_circ_0013072, bta_circ_0019285, and bta_circ_0013067. The top 10 downregulated circRNAs were bta_circ_0024234, bta_circ_0017528, bta_circ_0008077, bta_circ_0003222, bta_circ_0007500, bta_circ_0020328, bta_circ_0011001, bta_circ_0016364, bta_circ_0008839, and bta_circ_0016049. The qRT‐PCR results showed consistent trend with sequencing analysis result, while host genes had no statistic difference. The GO and KEGG analyses of the host genes suggested that abnormal circRNA expression may play multiple roles in placental structure and dysfunction.
Conclusion
The abnormal circRNA expression may be one of reasons of placental dysfunction, leads to abortion of bovine SCNT fetus.
Keywords: aberrant development, bovine, circular RNAs, placenta, somatic cell nuclear transfer
1. INTRODUCTION
During the past two decades, tremendous progress has been achieved in animal cloning since the birth of Dolly. One major breakthrough in the field, somatic cell nuclear transfer (SCNT), has given birth to a barnyard of livestock animals, including cattle, pig, sheep, and goat.1 Combined with gene editing technology, this technique had proven valid in developing genetically modified livestock. However, one of the bottlenecks of SCNT is low birth rate. Only 6% of transferred cloned embryos result healthy offspring in cattle.2 According to our previous research, the survival rate of genetically modified cloned cattle was below 5%. Incomplete reprogramming is amenable to the developmental failure of cloned embryo.3, 4 Except for the fetal aberrant development, placental dysfunction, such as reduced vascularization, placentomegaly, hypoplasia of trophoblastic epithelium, and altered basement membrane, was another cause to lead pregnancy losses.2, 4 We found that a fairly large number of cloned cattle aborted during late gestation (180‐210 days). The abnormal pregnant recipient showed engorged uterus and enlarged umbilical vessels. Coincidentally, a equine clone research depicted similar symptoms.5 It indicates that this is a relative common abnormality during SCNT fetal pregnancy, while the causes are ambiguous.
Placenta is a circular organ which temporarily exists in placental mammals during gestation. It not only supplies the space for fetus with protection and nutrition metabolism, but also secretes multiple growth factors and hormones to maintain gestation. In addition, it is the only pathway to connect the mother and the fetus. Placental research by RNA‐seq for abortion and aberrant pregnancy in livestock mainly focused on early gestation or postnatal.6, 7, 8, 9 little about which However, few studies have looked at the placenta during the third trimester, when large quantity of SCNT fetal abortion occur.
Circular RNAs (circRNAs) were first discovered in RNA viruses as early as the 1970s.10 It formed as covalently closed loop structures with neither 5′‐3′ polarities nor polyadenylated tails and more stable than linear RNA.11 Serious reports showed that circRNAs could function as miRNA sponges, regulate alternative splicing, and modulate the expression of mRNAs.12, 13, 14, 15 The different types of RNAs serve different roles and form a network called the competing endogenous RNAs (ceRNAs).16 Like other noncoding RNAs, circRNAs have been associated with a particular role in biological development and disease initiation and progression.17 They have been found implicated with various cancers, including colorectal, lung, and cervical cancer.18, 19, 20 Hitherto noticed features of circRNA are mainly based on evidence gathered from human, and studies on other species are insufficient.21, 22, 23, 24
This study aims to explore the multiple factors that potentially lead to high abortion frequency exist in SCNT fetus generation during late gestation. To this end, we collected two aberrant pregnant placenta (abnormal group, AG) and three normal pregnant placenta (normal group, NG) at late gestation (180‐210 days) of bovine SCNT fetus. We acquired five bovine late gestational placental circRNA expression profiles and analyzed its differentiation. We sought to uncover the mechanism associated with this phenomenon. The discovery may provide a new insight for SCNT fetal aberrant development and improve the SCNT efficiency.
2. MATERIALS AND METHODS
2.1. Ethics statement
All experimental procedures and sample collections were conducted in accordance with the Regulations for the Administration of Affairs Concerning Experimental Animals (Ministry of Science and Technology, China; revised in August 2011) and were approved by the Institutional Animal Care and Use Committee of Inner Mongolia University, Hohhot, China.
2.2. Sample information and collection
Cloned embryo, embryo transfer, and recipient cow experimental work were supplied by Inner Mongolia University. Briefly, the donor cell was fetal skin fibroblast. The recipients were 2‐5 years. The procedure was followed as Wu et al.25
A total of five late pregnant cows were used in the present study from two groups, that is, the aberrant pregnant cows (aberrant group, AG: n = 2) and normal pregnant cows (Normal group, NG: n = 3). All of the selected cows were at late pregnancy stage (180‐210 days). After the pregnant cows were slaughtered, the placenta was rapidly harvested and immediately frozen in liquid nitrogen and stored for use toward the subsequent generation of circle RNA libraries.
2.3. RNA preparation
The total RNA was extracted using TRIzol™ reagent (Invitrogen) following the manufacturer's procedure.26 Briefly, 50‐100 mg of tissues was lysed by 1 mL of TRIzol™ reagent. 0.2 mL of chloroform per 1 mL of TRIzol™ Reagent was added after 5 minutes of incubation. Then, the samples were centrifuged for 15 minutes at 12 000 g at 4°C after 2‐3 minutes incubation. The mixture separated into a lower red phenol‐chloroform, and interphase, and a colorless upper aqueous phase. The RNA was contained in the aqueous phase. The aqueous phase was transferred to a new tube and added 0.5 mL of isopropanol. After incubation of 10 minutes, centrifuge for 10 minutes at 12 000 g at 4°C. The supernatant was discarded and 1 mL of 75% ethanol was added to wash RNA. Centrifuge for 5 minutes at 7500 g at 4°C. The supernatant was discarded and air‐dried the RNA pellet for 5‐10 minutes. At last, the RNA was resuspended in 20‐50 µL of RNase‐free water. The quantity and purity of total RNA were analyzed using the Bioanalyzer 2100 (Agilent) with RIN number >7.0.
2.4. Library synthesis and high‐throughput sequencing
Approximately 3 µg of total RNA was used to prepare the circRNA library. Ribo‐Zero™ Gold Kits were used to degrade rRNA, and linear RNA was degraded by RNase R. Then, RNA libraries were generated according to the protocol outlined for NEBNext Ultra Directional RNA Library Prep Kit for Illumina (NEB). We then performed the single‐end sequencing on an Illumina Hiseq2500 at the ANOROAD GENOME Co., Ltd. (Beijing, CN) following the vendor's recommended protocol.
2.5. Differentially expressed circRNA analyses
The differentially expressed circRNAs between AG and NG were calculated by edge R using the likelihood ratio test (LRT) based on generalized linear model which estimates probability distributions according to mean‐variance relationship of each gene.27 Only transcripts with expression greater than 0.1 count per million (CPM) in at least one samples were selected for differential testing. Transcripts with P < 0.05 and |log2 ratio| ≥ 1 were considered differentially expressed.
2.6. Validation of differentially expressed circRNAs through qRT‐PCR
Eight differentially expressed circRNAs and relative host genes were selected for validation. Total RNA was extracted as previous. PrimeScript™ RT reagent kit (TAKARA) was used to cDNA synthesis, and only random 6‐mers were added. TB Green™ Premix Ex Taq™ II was used to qRT‐PCR. The procedure was followed as the manufacturer's instruction book. The primer sequences were listed at supplemental Table S1. The GAPDH was used as reference gene. The relative expression level of each circRNA and host gene was calculated using the 2−ΔΔCt method. The data are indicated as the means ± SE (n = 3). The significance of the expression in two samples was calculated using a two sample t test in SPSS statistical software (Version17.0), whose difference was considered as significant when P < 0.05.
2.7. Functional enrichment analysis of host genes of differentially expressed circRNAs
The enrichment analyses of KEGG (Kyoto Encyclopedia of Genes and Genomes) and GO (Gene Ontology) were performed using DAVID (The Database for Annotation, Visualization, and Integrated Discovery) with the default parameters.
2.8. Target miRNAs of differentially expressed circRNA prediction and co‐expression network analysis
The target miRNAs of differentially expressed circRNAs were evaluated using miRanda (3.3a), investigating only perfect seed matching without gap of wobble pairing (“strict” parameter).28 A hit between any expressed miRNA (including the new predicted miRNA) and a target circRNA was considered for a miRanda score of 140 or higher, corresponding to at least a perfect seed match.
3. RESULTS
3.1. Characteristics of bovine placental circRNA expression pattern
In this study, we analyzed two aberrant pregnant placenta (AG) and three normal pregnant placenta (NG) at late gestation (180‐210 days) with bovine SCNT fetus. To study the general characteristics of all circRNAs in bovine placenta, we performed a preliminary analysis of all these sequencing results. A total of 12 454 circRNAs were evaluated, 6161 and 10 544 circRNAs of AG and NG, respectively.
These circRNAs were widely scattered on almost all bovine chromosomes, and chromosome 1 was the most abundant, followed by chromosome X and 2 (Figure 1A). The properties of circRNAs contain classic, alter exon, intron, overlap exon, antisense, and intergenic. The compositional type of each sample is shown in Figure 1B. In total, the ratio of classic was the largest, exceeding 60% in each sample. CircRNAs transcribed from three exons (3‐exon circRNAs) were the most abundant circRNAs in all samples, followed by 2‐exon and 4‐exon circRNAs (Figure 1C).
Figure 1.
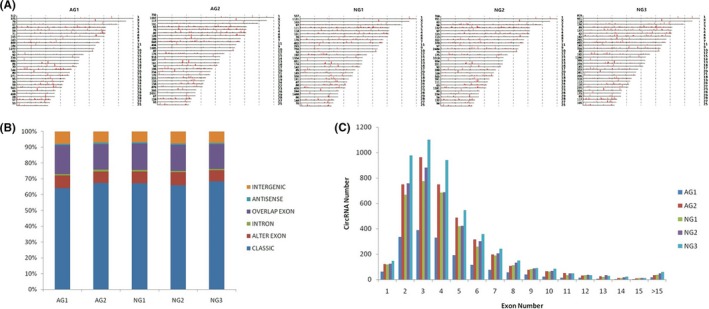
Characteristics of genomic location and classification of circRNAs expressed in bovine late gestational placenta. A, The chromosome distributions of circRNAs. B, Classification of circRNAs. C, Distribution of exon composition. AG: abnormal group; NG: normal group
3.2. Identification validation of differentially expressed circRNAs between AG and NG
Hierarchical cluster analysis was used to reveal the circRNA expression levels in AG and NG (Figure 2A), which showed that these levels were distinguishable between two groups. The significantly differentially expressed (DE) circRNAs between two groups were shown in the volcano plot (Figure 2B). In total, 123 circRNAs were identified as differentially expressed circRNAs by the filter criteria of fold change (FC) ≥2.0, P value <0.05. Among these, 49 circRNAs were upregulated, and 74 circRNAs were downregulated (Figure 2C). The differentially expressed circRNAs were listed in supplemental Table S2. The top 10 upregulated circRNAs were bta_circ_0012985, bta_circ_0013071, bta_circ_0013074, bta_circ_0016024, bta_circ_0013068, bta_circ_0008816, bta_circ_0012982, bta_circ_0013072, bta_circ_0019285, and bta_circ_0013067. The top 10 downregulated circRNAs were bta_circ_0024234, bta_circ_0017528, bta_circ_0008077, bta_circ_0003222, bta_circ_0007500, bta_circ_0020328, bta_circ_0011001, bta_circ_0016364, bta_circ_0008839, and bta_circ_0016049. Eight DE circRNAs and relative host genes were validated by qRT‐PCR. The results of DE circRNAs showed similar trend with sequencing result, while their host genes with no significant difference (Figure 2D).
Figure 2.
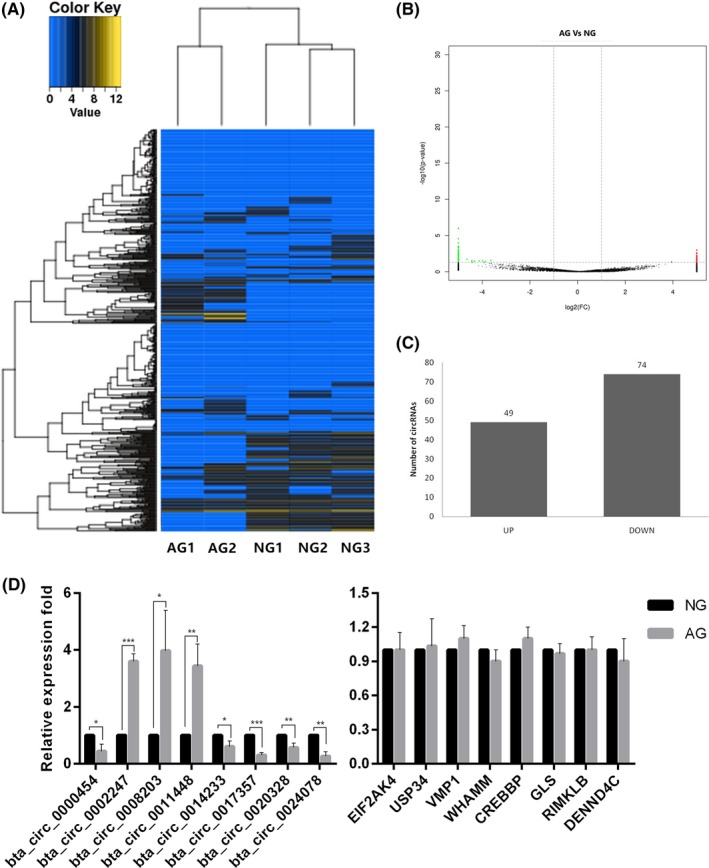
Differential expression of circRNAs between AG and NG A, Hierarchical cluster analysis of circRNAs. The color scale of the strips runs from blue (low relative expression) through black (medium relative expression) to yellow (high relative expression). B, Volcano plots visualize the differentially expressed (DE) circRNAs. The green and red plots represent the significantly DE circRNAs (FC ≥ 2.0, P value <0.05). C, Number of upregulated and downregulated circRNAs of DE circRNAs. D, Validation of DE circRNA expression level by qRT‐PCR. Eight DE circRNAs (left) and their host genes (right) were selected to validate the expression level (N = 3. *P < 0.05; **P < 0.01; ***P < 0.001)
3.3. Host gene enrichment of DE circRNAs and functional analysis
Through edge R analysis, 60 host genes of DE circRNAs were enriched (Table 1). Gene ontology (GO) analysis of these genes showed that there were two genes related to protein K48‐linked deubiquitination, two genes related to retromer complex, and two genes related to endoplasmic reticulum‐Golgi intermediate compartment membrane (Table 2). However, none of the suggested correlations was significant (P > 0.05). These result indicated that the DE circRNAs may play a role in placental protein metabolism and transport. Kyoto Encyclopedia of Genes and Genomes (KEGG) analysis showed that the predicted host genes were associated with virus invasion and amine acid metabolism (Table 3). Unfortunately, the result was also not significant (P > 0.05). Combined with these results, we surmised that DE circRNAs may affect placental protein metabolism.
Table 1.
Host gene enrichment of differentially expressed circRNAs
| CircRNA_ID | Host gene | Aberrant group | Normal group | |||||||
|---|---|---|---|---|---|---|---|---|---|---|
| AG1_CPM | AG2_CPM | NG1_CPM | NG2_CPM | NG3_CPM | AG_mean | NG_mean | log2(fc) | P value | ||
| bta_circ_0000010 | SPATA7 | 0 | 0 | 77.95 | 234.05 | 148.25 | 0.001 | 153.417 | 17.2271 | 0.043291 |
| bta_circ_0000454 | EIF2AK4 | 0 | 0 | 116.92 | 306.06 | 164.72 | 0.001 | 195.9 | 17.5798 | 0.025803 |
| bta_circ_0001735 | RMDN2 | 779.12 | 355.49 | 0 | 108.02 | 0 | 567.305 | 36.0067 | −3.9778 | 0.030563 |
| bta_circ_0001852 | PLEKHH2 | 467.47 | 0 | 0 | 0 | 0 | 233.735 | 0.001 | −17.835 | 0.03198 |
| bta_circ_0002247 | USP34 | 272.69 | 71.1 | 0 | 0 | 0 | 171.895 | 0.001 | −17.391 | 0.023302 |
| bta_circ_0002631 | ASAP2 | 0 | 0 | 97.43 | 126.03 | 164.72 | 0.001 | 129.393 | 16.9814 | 0.049523 |
| bta_circ_0003287 | ATP12A | 233.74 | 284.39 | 0 | 36.01 | 0 | 259.065 | 12.0033 | −4.4318 | 0.028896 |
| bta_circ_0005735 | TRIM6 | 155.82 | 106.65 | 0 | 0 | 0 | 131.235 | 0.001 | −17.002 | 0.048234 |
| bta_circ_0006537 | PRDM2 | 662.25 | 0 | 0 | 0 | 0 | 331.125 | 0.001 | −18.337 | 0.018117 |
| bta_circ_0007503 | OAS1Z | 506.43 | 355.49 | 0 | 0 | 0 | 430.96 | 0.001 | −18.717 | 0.000867 |
| bta_circ_0007867 | TANGO6 | 311.65 | 53.32 | 0 | 0 | 0 | 182.485 | 0.001 | −17.477 | 0.021199 |
| bta_circ_0008203 | VMP1 | 311.65 | 35.55 | 0 | 0 | 0 | 173.6 | 0.001 | −17.405 | 0.03001 |
| bta_circ_0008450 | ANKFY1 | 233.74 | 71.1 | 0 | 0 | 0 | 152.42 | 0.001 | −17.218 | 0.034744 |
| bta_circ_0008540 | AKAP10 | 623.3 | 0 | 0 | 0 | 0 | 311.65 | 0.001 | −18.25 | 0.019989 |
| bta_circ_0008616 | B4GALNT2 | 311.65 | 533.23 | 0 | 36.01 | 65.89 | 422.44 | 33.9667 | −3.6366 | 0.02648 |
| bta_circ_0009422 | RYK | 0 | 0 | 331.27 | 54.01 | 131.77 | 0.001 | 172.35 | 17.395 | 0.04259 |
| bta_circ_0009546 | BRWD1 | 0 | 0 | 233.84 | 306.06 | 148.25 | 0.001 | 229.383 | 17.8074 | 0.016827 |
| bta_circ_0009559 | BRWD1 | 389.56 | 53.32 | 0 | 0 | 0 | 221.44 | 0.001 | −17.757 | 0.012467 |
| bta_circ_0010335 | PAK2 | 389.56 | 142.19 | 0 | 0 | 0 | 265.875 | 0.001 | −18.02 | 0.004782 |
| bta_circ_0010808 | SLC38A9 | 194.78 | 106.65 | 0 | 0 | 0 | 150.715 | 0.001 | −17.201 | 0.030874 |
| bta_circ_0010882 | PARP8 | 0 | 53.32 | 272.81 | 396.08 | 560.04 | 26.66 | 409.643 | 3.94162 | 0.046802 |
| bta_circ_0011448 | WHAMM | 389.56 | 675.42 | 0 | 0 | 0 | 532.49 | 0.001 | −19.022 | 0.000423 |
| bta_circ_0011569 | SCAPER | 467.47 | 0 | 0 | 0 | 0 | 233.735 | 0.001 | −17.835 | 0.032032 |
| bta_circ_0012875 | UBR2 | 0 | 0 | 311.78 | 54.01 | 164.72 | 0.001 | 176.837 | 17.4321 | 0.0407 |
| bta_circ_0013512 | OSBPL1A | 0 | 0 | 350.75 | 198.04 | 98.83 | 0.001 | 215.873 | 17.7198 | 0.023113 |
| bta_circ_0013840 | WDR7 | 0 | 0 | 97.43 | 360.07 | 115.3 | 0.001 | 190.933 | 17.5427 | 0.032938 |
| bta_circ_0013949 | LITAF | 233.74 | 248.84 | 0 | 36.01 | 0 | 241.29 | 12.0033 | −4.3293 | 0.035077 |
| bta_circ_0014233 | CREBBP | 0 | 0 | 253.32 | 342.07 | 263.55 | 0.001 | 286.313 | 18.1272 | 0.009391 |
| bta_circ_0015895 | RSF1 | 0 | 0 | 194.86 | 234.05 | 131.77 | 0.001 | 186.893 | 17.5119 | 0.025366 |
| bta_circ_0016364 | ACSL3 | 0 | 0 | 915.86 | 0 | 1729.53 | 0.001 | 881.797 | 19.7501 | 0.015514 |
| bta_circ_0016481 | DIS3L2 | 233.74 | 106.65 | 0 | 0 | 0 | 170.195 | 0.001 | −17.377 | 0.020055 |
| bta_circ_0016740 | PLEKHB2 | 194.78 | 319.94 | 0 | 0 | 0 | 257.36 | 0.001 | −17.973 | 0.004105 |
| bta_circ_0017357 | GLS | 0 | 0 | 253.32 | 108.02 | 181.19 | 0.001 | 180.843 | 17.4644 | 0.02894 |
| bta_circ_0017447 | AOX1 | 0 | 0 | 175.38 | 90.02 | 164.72 | 0.001 | 143.373 | 17.1294 | 0.042112 |
| bta_circ_0017528 | RAPH1 | 0 | 0 | 545.62 | 126.03 | 691.81 | 0.001 | 454.487 | 18.7939 | 0.005775 |
| bta_circ_0017631 | FAAH | 272.69 | 71.1 | 0 | 0 | 0 | 171.895 | 0.001 | −17.391 | 0.023247 |
| bta_circ_0017877 | TRAF3IP1 | 0 | 0 | 175.38 | 288.06 | 115.3 | 0.001 | 192.913 | 17.5576 | 0.025511 |
| bta_circ_0017956 | UBAP2L | 155.82 | 124.42 | 0 | 0 | 0 | 140.12 | 0.001 | −17.096 | 0.03783 |
| bta_circ_0017979 | POGZ | 155.82 | 142.19 | 0 | 0 | 0 | 149.005 | 0.001 | −17.185 | 0.02946 |
| bta_circ_0017986 | MINDY1 | 506.43 | 0 | 0 | 0 | 0 | 253.215 | 0.001 | −17.95 | 0.029053 |
| bta_circ_0018723 | DNAJC6 | 272.69 | 124.42 | 0 | 0 | 0 | 198.555 | 0.001 | −17.599 | 0.011159 |
| bta_circ_0018735 | RAVER2 | 194.78 | 106.65 | 0 | 0 | 0 | 150.715 | 0.001 | −17.201 | 0.030878 |
| bta_circ_0019344 | UBE3C | 77.91 | 213.29 | 0 | 0 | 0 | 145.6 | 0.001 | −17.152 | 0.032971 |
| bta_circ_0019346 | DNAJB6 | 0 | 0 | 155.89 | 126.03 | 181.19 | 0.001 | 154.37 | 17.236 | 0.037511 |
| bta_circ_0020328 | RIMKLB | 0 | 0 | 0 | 828.17 | 1317.74 | 0.001 | 715.303 | 19.4482 | 0.018351 |
| bta_circ_0020647 | PLXNC1 | 545.38 | 0 | 0 | 0 | 0 | 272.69 | 0.001 | −18.057 | 0.025205 |
| bta_circ_0020821 | PPHLN1 | 0 | 0 | 116.92 | 306.06 | 345.91 | 0.001 | 256.297 | 17.9675 | 0.015023 |
| bta_circ_0021640 | CDS1 | 0 | 0 | 233.84 | 90.02 | 197.66 | 0.001 | 173.84 | 17.4074 | 0.032148 |
| bta_circ_0022053 | CCSER1 | 0 | 0 | 272.81 | 108.02 | 280.02 | 0.001 | 220.283 | 17.749 | 0.019935 |
| bta_circ_0022077 | HERC6 | 0 | 0 | 77.95 | 216.04 | 164.72 | 0.001 | 152.903 | 17.2223 | 0.042512 |
| bta_circ_0022579 | SULT1E1 | 584.34 | 373.26 | 0 | 0 | 98.83 | 478.8 | 32.9433 | −3.8614 | 0.040189 |
| bta_circ_0023166 | PRRC1 | 0 | 0 | 409.21 | 180.04 | 214.13 | 0.001 | 267.793 | 18.0308 | 0.012955 |
| bta_circ_0023610 | SSBP2 | 545.38 | 551 | 0 | 0 | 0 | 548.19 | 0.001 | −19.064 | 0.000351 |
| bta_circ_0024078 | DENND4C | 0 | 0 | 194.86 | 126.03 | 98.83 | 0.001 | 139.907 | 17.0941 | 0.046141 |
| bta_circ_0024594 | STC1 | 0 | 622.1 | 0 | 0 | 0 | 311.05 | 0.001 | −18.247 | 0.015471 |
| bta_circ_0024720 | NTRK2 | 0 | 0 | 97.43 | 360.07 | 280.02 | 0.001 | 245.84 | 17.9074 | 0.018113 |
| bta_circ_0025499 | COQ3 | 311.65 | 213.29 | 0 | 0 | 0 | 262.47 | 0.001 | −18.002 | 0.004004 |
| bta_circ_0026690 | SYTL4 | 0 | 0 | 97.43 | 126.03 | 181.19 | 0.001 | 134.883 | 17.0414 | 0.049087 |
| bta_circ_0026843 | CHM | 0 | 0 | 155.89 | 180.04 | 230.6 | 0.001 | 188.843 | 17.5268 | 0.024553 |
Table 2.
GO annotations of differentially expressed circRNA host genes
| Category | Term | Count | P value | Genes |
|---|---|---|---|---|
| GOTERM_BP | Protein K48‐linked deubiquitination | 2 | 0.060108 | MINDY1; USP34 |
| GOTERM_CC | Retromer complex | 2 | 0.0515133 | ANKFY1; DENND4C |
| GOTERM_CC | Endoplasmic reticulum‐Golgi intermediate compartment membrane | 2 | 0.0544612 | WHAMM; VMP1 |
Table 3.
KEGG analysis of differentially expressed circRNA host genes
| Category | Term | Count | P value | Genes |
|---|---|---|---|---|
| KEGG_PATHWAY | Influenza A | 3 | 0.062348 | CREBBP; EIF2AK4; OAS1Z |
| KEGG_PATHWAY | Herpes simplex infection | 3 | 0.07349 | CREBBP; EIF2AK4; OAS1Z |
| KEGG_PATHWAY | Alanine, aspartate, and glutamate metabolism | 2 | 0.08218 | GLS; RIMKLB |
3.4. Prediction of differentially expressed circRNA‐miRNA interaction
CircRNAs act as miRNA sponges and exert their effects via the circRNA‐miRNA‐mRNA axis.29 Through miRanda based on the MREs, interaction between DE circRNAs and miRNAs was theoretically predicted. We found that 32 miRNAs could be paired with eight DE circRNAs (Table 4), with the criteria of a max score ≥140 and a max energy ≤−25 (a lower max energy is indicative of a stronger correlation). The result suggested that circRNAs may play a part in causing placental dysfunction via interaction with miRNAs.
Table 4.
miRNA prediction which interact with differentially expressed circRNAs
| CircRNA_ID | miRNA_Name |
|---|---|
| bta_circ_0006612 | bta‐miR‐153; bta‐miR‐2325c; bta‐miR‐2340; bta‐miR‐2346; bta‐miR‐2897; bta‐miR‐383; bta‐miR‐544a; bta‐miR‐544b; bta‐miR‐545‐3p; bta‐miR‐574 |
| bta_circ_0008203 | bta‐miR‐200c |
| bta_circ_0008839 | bta‐miR‐2285g; bta‐miR‐2285z; bta‐miR‐2399‐3p |
| bta_circ_0010876 | bta‐miR‐1248 |
| bta_circ_0013512 | bta‐miR‐148b; bta‐miR‐152 |
| bta_circ_0019285 | bta‐miR‐145; bta‐miR‐181b; bta‐miR‐2285ad; bta‐miR‐2285n; bta‐miR‐2305; bta‐miR‐2411‐3p; bta‐miR‐342 |
| bta_circ_0022053 | bta‐miR‐29b; bta‐miR‐29c; bta‐miR‐29d‐3p |
| bta_circ_0026700 | bta‐miR‐146b; bta‐miR‐2340; bta‐miR‐2355‐5p; bta‐miR‐544a; bta‐miR‐544b; bta‐miR‐574; bta‐miR‐6531; bta‐miR‐671 |
4. DISCUSSION
Compare with in vitro fertilized (IVF) embryo, SCNT embryo showed lower developmental efficiency. Due to the oocyte's microenvironment is suitable for gamete epigenetic reprogramming, somatic cell nucleus reprogramming in SCNT embryo is incomplete.30 Low birth rate and birth deficiency could be mainly ascribed to incomplete epigenetic reprogramming. Except fetus, extraembryonic tissue is also harmed by incorrect reprogramming, which leads to pregnancy failure. One symptom is cloned fetus aborted during late gestation, accompany with engorged uterus. We surmise that it is related to material transportation dysfunction. We observed that placental cotyledon present as different size in aberrant gestation recipient, which is uniform in normal gestation recipient. It may be compensatory hypertrophy. A equine clone research reported similar symptom,5 but no deep research. All of these symptoms were caused by abnormal gene expression. However, less of research focus on these abnormal SCNT fetal development which caused by placental dysfunction.
In this study, a total of 12 454 circRNAs were obtained. Yan et al23 obtained 48 270 circRNAs at human placental research. In other three reports of placental circRNA research, the number of sequenced circRNAs was similar as our study. In bovine, the study related to circRNA was not many. In circRNA expression study of bovine mammary glands, more than 6000 circRNAs were identified.31 In the research of genome‐wide analysis of circRNAs in bovine cumulus cells, 1706 circRNAs were identified.32 In another research, circular RNA profiling during myoblasts differentiation, 12 981 circRNAs were sequenced.33 In total, our data size was comparable with other circRNA research.
We predicted 60 target genes of differentially expressed circRNAs. The GO analysis indicated that MINDY1 and USP34 matched with protein K48‐linked deubiquitination, ANKFY1 and DENND4C matched with retromer complex, and WHAMM and VMP1 matched with endoplasmic reticulum‐Golgi intermediate compartment membrane. The results indicated aberrant bovine placenta may have dysfunctional endoplasmic reticulum‐Golgi intermediate material translation. KEGG analysis reflected that virus infection and alanine, aspartate and glutamate metabolism pathway‐related genes were involved. OAS1 has been found to be related to gestation.34, 35 EIF2AK4 belongs to a family of kinases that regulate angiogenesis in response to cellular stress, the mutation of which is likely to cause pulmonary capillary hemangiomatosis (PCH).36 CREBBP mutation is found accountable for a high incidence of preeclampsia.37 Glutamine plays a vital role in carbon and nitrogen metabolism of the fetus and exhibits the highest fetal‐maternal plasma ratio among all amino acids in pigs.38 These results indicated that differentially expressed circRNAs may have multiple effects in placental both structure and function.
CircRNAs could function with miRNAs and co‐regulate target genes' expression. We predicted 32 miRNAs which can pair with eight differentially expressed circRNAs. Among these miRNAs, miR‐145 was reported to be related to abnormal placental development in transgenic cloned cattle.39 Our results indicated that circRNAs may play a role in abnormal bovine fetus development in late gestation through interactions with miRNAs.
For it was one type of pregnancy familiar of bovine SCNT research and occurs randomly, the sample was not sufficient. CircRNAs as noncoding RNA need to contact with other RNA to act biological function. Further study is needed to explore the mechanism. Low birth rate of SCNT is a complicated question, and relative research should pay attention to placental dysfunction. Combined with multiple strategies, such as RNA expression, protein expression, histological and hormone analysis, the mechanism of bovine SCNT fetal abortion‐related placental dysfunction will be discovered. It is also helpful to improve the SCNT efficiency.
5. CONCLUSION
In this study, we acquired five circRNA expression profiles of SCNT bovine placentas (two abnormal and three normal) during late gestation. We identified 123 circRNAs were DE circRNAs between AG and NG. 60 target genes and 32 miRNAs were related to DE circRNAs. Through GO and KEGG analyses, we surmise that abnormal circRNA expression may play multiple roles in placental both structure and dysfunction. In the future, we would detect related mRNA and miRNA expression profiles to further explore its mechanism.
Supporting information
ACKNOWLEDGMENTS
This work was funded by the National Natural Science Foundation of China (31460599). The authors would like to thank Professor Guangpeng Li of Inner Mongolia University for providing the recipient heifers and technical assistance.
Su X, Gao G, Wang S, et al. CircRNA expression profile of bovine placentas in late gestation with aberrant SCNT fetus. J Clin Lab Anal. 2019;33:e22918 10.1002/jcla.22918
Su and Gao contributed equally to this project.
REFERENCES
- 1. Edwards JL, Schrick FN, McCracken MD, et al. Cloning adult farm animals: a review of the possibilities and problems associated with somatic cell nuclear transfer. Am J Reprod Immunol. 2003;50:113‐123. [DOI] [PubMed] [Google Scholar]
- 2. Wells DN. Animal cloning: problems and prospects. Rev Sci Tech. 2005;24:251‐264. [PubMed] [Google Scholar]
- 3. Wang F, Kou Z, Zhang Y, Gao S. Dynamic reprogramming of histone acetylation and methylation in the first cell cycle of cloned mouse embryos. Biol Reprod. 2007;77:1007‐1016. [DOI] [PubMed] [Google Scholar]
- 4. Palmieri C, Loi P, Ptak G, Della Salda L. Review paper: a review of the pathology of abnormal placentae of somatic cell nuclear transfer clone pregnancies in cattle, sheep, and mice. Vet Pathol. 2008;45:865‐880. [DOI] [PubMed] [Google Scholar]
- 5. Pozor MA, Sheppard B, Hinrichs K, et al. Placental abnormalities in equine pregnancies generated by SCNT from one donor horse. Theriogenology. 2016;86:1573‐1582. [DOI] [PubMed] [Google Scholar]
- 6. Everts RE, Chavatte‐Palmer P, Razzak A, et al. Aberrant gene expression patterns in placentomes are associated with phenotypically normal and abnormal cattle cloned by somatic cell nuclear transfer. Physiol Genomics. 2008;33:65‐77. [DOI] [PubMed] [Google Scholar]
- 7. Salilew‐Wondim D, Tesfaye D, Hossain M, et al. Aberrant placenta gene expression pattern in bovine pregnancies established after transfer of cloned or in vitro produced embryos. Physiol Genomics. 2013;45:28‐46. [DOI] [PubMed] [Google Scholar]
- 8. Ko YG, Hwang S, Kim S, et al. Proteomic analysis of the extraembryonic tissues from cloned porcine fetus at day 35 of pregnancy. BMC Res Notes. 2014;7:861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Czernik M, Toschi P, Zacchini F, Iuso D, Ptak GE. Deregulated expression of mitochondrial proteins Mfn2 and Bcnl3L in placentae from sheep somatic cell nuclear transfer (SCNT) conceptuses. PLoS ONE. 2017;12:e0169579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Sanger HL, Klotz G, Riesner D, Gross HJ, Kleinschmidt AK. Viroids are single‐stranded covalently closed circular RNA molecules existing as highly base‐paired rod‐like structures. Proc Natl Acad Sci U S A. 1976;73:3852‐3856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Chen LL, Yang L. Regulation of circRNA biogenesis. RNA Biol. 2015;12:381‐388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Hansen TB, Jensen TI, Clausen BH, et al. Natural RNA circles function as efficient microRNA sponges. Nature. 2013;495:384‐388. [DOI] [PubMed] [Google Scholar]
- 13. Memczak S, Jens M, Elefsinioti A, et al. Circular RNAs are a large class of animal RNAs with regulatory potency. Nature. 2013;495:333‐338. [DOI] [PubMed] [Google Scholar]
- 14. Zhang Y, Zhang X‐O, Chen T, et al. Circular intronic long noncoding RNAs. Mol Cell. 2013;51:792‐806. [DOI] [PubMed] [Google Scholar]
- 15. Li Z, Huang C, Bao C, et al. Exon‐intron circular RNAs regulate transcription in the nucleus. Nat Struct Mol Biol. 2015;22:256‐264. [DOI] [PubMed] [Google Scholar]
- 16. Kartha RV, Subramanian S. Competing endogenous RNAs (ceRNAs): new entrants to the intricacies of gene regulation. Front Genet. 2014;5:8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Qu S, Yang X, Li X, et al. Circular RNA: a new star of noncoding RNAs. Cancer Lett. 2015;365:141‐148. [DOI] [PubMed] [Google Scholar]
- 18. Bachmayr‐Heyda A, Reiner AT, Auer K, et al. Correlation of circular RNA abundance with proliferation–exemplified with colorectal and ovarian cancer, idiopathic lung fibrosis, and normal human tissues. Sci Rep. 2015;5:8057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Ma X, Yang X, Bao W, et al. Circular RNA circMAN2B2 facilitates lung cancer cell proliferation and invasion via miR‐1275/FOXK1 axis. Biochem Biophys Res Commun. 2018;498:1009‐1015. [DOI] [PubMed] [Google Scholar]
- 20. Ma HB, Yao YN, Yu JJ, Chen XX, Li HF. Extensive profiling of circular RNAs and the potential regulatory role of circRNA‐000284 in cell proliferation and invasion of cervical cancer via sponging miR‐506. Am J Transl Res. 2018;10:592‐604. [PMC free article] [PubMed] [Google Scholar]
- 21. Qian Y, Lu Y, Rui C, Qian Y, Cai M, Jia R. Potential significance of circular RNA in human placental tissue for patients with preeclampsia. Cell Physiol Biochem. 2016;39:1380‐1390. [DOI] [PubMed] [Google Scholar]
- 22. Maass PG, Glažar P, Memczak S, et al. A map of human circular RNAs in clinically relevant tissues. J Mol Med (Berl). 2017;95:1179‐1189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Yan L, Feng J, Cheng F, et al. Circular RNA expression profiles in placental villi from women with gestational diabetes mellitus. Biochem Biophys Res Commun. 2018;498:743‐750. [DOI] [PubMed] [Google Scholar]
- 24. Hu X, Ao J, Li X, Zhang H, Wu J, Cheng W. Competing endogenous RNA expression profiling in pre‐eclampsia identifies hsa_circ_0036877 as a potential novel blood biomarker for early pre‐eclampsia. Clin Epigenetics. 2018;10:48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Wu X, Ouyang H, Duan B, et al. Production of cloned transgenic cow expressing omega‐3 fatty acids. Transgenic Res. 2012;21(3):537‐543. [DOI] [PubMed] [Google Scholar]
- 26. Chomczynski P. A reagent for the single‐step simultaneous isolation of RNA, DNA and proteins from cell and tissue samples. Biotechniques. 1993;15:532‐537. [PubMed] [Google Scholar]
- 27. Robinson MD, McCarthy DJ, Smyth GK. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics. 2010;26:139‐140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. John B, Enright AJ, Aravin A, Tuschl T, Sander C, Marks DS. Human microRNA targets. PLoS Biol. 2004;2:e363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Jeck WR, Sharpless NE. Detecting and characterizing circular RNAs. Nat Biotechnol. 2014;32:453‐461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Niemann H. Epigenetic reprogramming in mammalian species after SCNT‐based cloning. Theriogenology. 2016;86:80‐90. [DOI] [PubMed] [Google Scholar]
- 31. Zhang C, Wu H, Wang Y, et al. Circular RNA of cattle casein genes are highly expressed in bovine mammary gland. J Dairy Sci. 2016;99:4750‐4760. [DOI] [PubMed] [Google Scholar]
- 32. Fu Y, Jiang H, Liu JB, et al. Genome‐wide analysis of circular RNAs in bovine cumulus cells treated with BMP15 and GDF9. Sci Rep. 2018;8:7944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Wei X, Li H, Yang J, et al. Circular RNA profiling reveals an abundant circLMO7 that regulates myoblasts differentiation and survival by sponging miR‐378a‐3p. Cell Death Dis. 2017;8:e3153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Han H, Austin KJ, Rempel LA, Hansen TR. Low blood ISG15 mRNA and progesterone levels are predictive of non‐pregnant dairy cows. J Endocrinol. 2006;191:505‐512. [DOI] [PubMed] [Google Scholar]
- 35. Green JC, Okamura CS, Poock SE, Lucy MC. Measurement of interferon‐tau (IFN‐tau) stimulated gene expression in blood leukocytes for pregnancy diagnosis within 18‐20d after insemination in dairy cattle. Anim Reprod Sci. 2010;121:24‐33. [DOI] [PubMed] [Google Scholar]
- 36. Best DH, Sumner KL, Austin ED, et al. EIF2AK4 mutations in pulmonary capillary hemangiomatosis. Chest. 2014;145:231‐236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. van Uitert M, Moerland PD, Enquobahrie DA, et al. Meta‐analysis of placental transcriptome data identifies a novel molecular pathway related to preeclampsia. PLoS ONE. 2015;10:e0132468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Self JT, Spencer TE, Johnson GA, Hu J, Bazer FW, Wu G. Glutamine synthesis in the developing porcine placenta. Biol Reprod. 2004;70:1444‐1451. [DOI] [PubMed] [Google Scholar]
- 39. Liu F‐J, Jin L‐J, Ma X‐G, et al. Differentially expressed microRNAs and affected signaling pathways in placentae of transgenic cloned cattle. Theriogenology. 2014;82(2):338‐346.e3. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials


