Abstract
Background
Estrogen‐related receptor gamma (ESRRG) has been identified as a tumor suppressor gene in several cancers. We aimed to evaluate ESRRG promoter methylation in laryngeal squamous cell carcinoma (LSCC) and its relative clinical value in LSCC.
Methods
Bisulfite pyrosequencing assays were performed on 91 pairs of tumor and paracancer tissues from LSCC patients in China. The diagnostic value and overall survival (OS) were analyzed descriptively by receiver operating characteristic (ROC) curves and the Kaplan‐Meier methods, respectively.
Results
The ESRRG promoter was more frequently hypermethylated in tumor tissues than in adjacent tissues (P < 0.01). ESRRG promoter methylation was significantly increased in advanced T stage tumors (P < 0.01) and advanced clinical stage patients (P < 0.01). Moreover, the area under the ROC curve (AUC) value (0.81) indicated high discrimination accuracy. Furthermore, ESRRG hypermethylation was associated with poor OS, as confirmed by Kaplan‐Meier survival curves (P < 0.01).
Conclusion
Our study indicated that ESRRG promoter hypermethylation contributed to LSCC‐related risks, primarily tumor progression and survival prognosis, in patients. ESRRG promoter methylation could, therefore, be a diagnostic and prognostic biomarker in LSCC.
Keywords: diagnostics, DNA methylation, estrogen‐related receptor gamma, laryngeal squamous cell carcinoma, prognostics
1. INTRODUCTION
The estrogen‐related receptor gamma (ESRRG) gene, also known as ERRγ, locating at the chromosome 1q41 region, is a member of the estrogen‐related receptor (ERR) family, an orphan nuclear receptor subgroup.1 ESRRG has been identified as a tumor suppressor gene in several cancers, especially sex‐related tumors. ESRRG protein expression is downregulated in several types of female cancers, such as breast cancer, cervical cancer, and endometrial cancer.2, 3, 4, 5 In addition, in prostate cancer, a male cancer, patients with low ESRRG immunoreactivity had poor cancer‐specific survival.6
Laryngeal carcinomas are the most common malignant otorhinolaryngological carcinomas.7 However, laryngeal squamous cell carcinoma (LSCC) accounts for the majority of laryngeal carcinomas.8 Surgery, radiotherapy, and chemotherapy remain the major common treatments for LSCC,9 but the optimal strategy is still unclear.10 Therefore, the identification of the molecular mechanisms underlying the development and progression of LSCC is urgently needed for exploring novel therapeutic strategies for LSCC. According to the Global Cancer Statistics, the incidence of laryngeal cancers was much higher in males than in females.11 Although collected evidence showed a role for epigenetic mechanisms in LSCC, a detailed exploration of the sex difference is lacking.
DNA hypermethylation is a basic epigenetic modification known to be involved in the regulation of gene expression, mRNA splicing, and genomic stability,12 and it has been intensively and widely studied in cancer epigenetics.13 Hypermethylation of the tumor suppressor gene (TSG) promoter region is an important mechanism in several types of cancers, such as colorectal cancer14 and hepatocellular carcinoma.15 In recent studies, the potential significance of gene methylation for the diagnosis of LSCC has been evaluated.16, 17
In this study, we assessed the ESRRG promoter methylation status in LSCC by measuring ESRRG promoter methylation in LSCC tissue samples. Moreover, we evaluated the clinical value of the ESRRG promoter methylation status by correlating it to the clinical characteristics and survival of LSCC patients.
2. MATERIALS AND METHODS
2.1. Patients and tissue samples
A total of 91 pairs of tumor and adjacent nontumor tissue specimens were obtained from LSCC patients at the Department of Otolaryngology‐Head and Neck Surgery at Lihuili Hospital of Ningbo University in China between 2011 and 2015. All patients were subjected to surgery and diagnosed according to the criteria of the World Health Organization before chemoradiotherapy. All specimens were stored at −80°C immediately after being harvested from fresh tissues during surgery. The adjacent nontumor tissues were dissected at >0.5 cm from the margin of the neoplastic lesion and confirmed histopathologically. The pathological stage was determined by the TNM classification (7th edition) of the Union for International Cancer Control (29 stage I cases, 16 stage II cases, 12 stage III cases, and 34 stage IV cases). The ages of the patients ranged from 40 to 86 (59.98 ± 9.10) years, and the majority were males (98%). The follow‐up time for the LSCC patients after surgery ranged from 2 to 60 (41.70 ± 14.77) months. Five patients were lost to follow‐up, 32 patients died during the follow‐up period, and 54 patients were still alive after 5 years of follow‐up. This study was approved by the Human Research Ethical Committee of Lihuili Hospital of Ningbo University in China.
2.2. DNA extraction and bisulfite modification
Genomic DNA was extracted from tissue samples in accordance with the QIAamp DNA Mini Kit (Qiagen, Hilden, Germany) instructions. Subsequently, the DNA concentrations and quality were evaluated by a NanoDrop 1000 spectrophotometer (Thermo Fisher Scientific Co. Ltd., Wilmington, DE, USA). All DNA absorbance ratios at 260/280 nm were between 1.8 and 2.0. The extracted DNA was chemically modified with a ZYMO EZ DNA Methylation‐Gold Kit (Zymo Research, Orange, CA, USA).
2.3. Pyrosequencing assay
Bisulfate‐treated DNA was amplified using polymerase chain reaction (PCR) with a PyroMark PCR Kit (Qiagen) and the forward primer 5′‐TAGAGTTAGAGGGAGATGAATTG‐3′ and the reverse primer 5′‐biotin‐TCTTTTCAAATCCATCACTAA‐3′. The PCR conditions were as follows: denaturation at 95°C for 10 minutes, 45 cycles at 95°C for 30 seconds, 50°C for 30 seconds, and 72°C for 1 minutes, and final extension at 72°C for 10 minutes. Then, the methylation levels of six CpG sites in the ESRRG promoter were measured via bisulfite pyrosequencing; the sequencing primer used was 5′‐GGGAGATGAATTGGG‐3′. The pyrosequencing results were confirmed by repeating the tests at least twice.
2.4. Statistical analysis
All statistical analyses were performed by SPSS v22.0 (IBM SPSS Statistics, IBM®, IL, USA). The data in this study are expressed as the means ± standard deviations (SDs). The Pearson correlation coefficient was calculated to estimate the correlation among each of the six CpG methylation sites. The comparison of the methylation status between the tumor and paired normal tissue samples and the association between the methylation status of the tumor samples and the clinical characteristics were analyzed by t tests. Receiver operating characteristic (ROC) curve analyses were conducted to assess the diagnostic value of ESRRG promoter methylation for LSCC.18 The area under the ROC curve (AUC) was determined with a 95% CI. The overall survival rate (OSR) of LSCC patients according to the ESRRG methylation status was compared using the Kaplan‐Meier survival method and the log‐rank test. Statistical significance was considered at a P value of <0.05. All figures were generated using GraphPad Prism 6 software (GraphPad Inc, San Diego, CA, USA).
3. RESULTS
3.1. Hypermethylation of the ESRRG promoter in tumor samples
In our study, bisulfite pyrosequencing was performed to evaluate the ESRRG promoter methylation status in 91 primary LSCC tissues and paired adjacent nontumor tissues. There are three CpG islands in the ESRRG promoter, Chr1: 217307745‐217309179, Chr1: 217310750‐217311178, and Chr1: 217311468‐217311773. We used Pyromark Q96 Pyrosequencing System software to score the sites of these CpG islands (CGIs) on the ESRRG promoter. Six sites selected from a fragment in a CGI of the ESRRG promoter were detected, as shown in Figure 1. As shown in Table 1, the methylation levels of each of the six CpG sites were significantly higher in the tumor samples than in the paired samples (CpG1‐6: all P < 0.01). Representative pyrograms are shown in Figure 2A. In addition, all correlation comparisons among the methylation levels of the six CpGs were significant (Figure 1, r > 0.8, P < 0.01). Thus, the methylation index (MI) was calculated as the mean percent methylation across the six CpG sites in the ESRRG promoter.19 The MI was significantly higher for the tumor samples (36.18 ± 14.54%) than for the adjacent nontumor samples (21.11 ± 10.45%; P < 0.01; Figure 2B).
Figure 1.
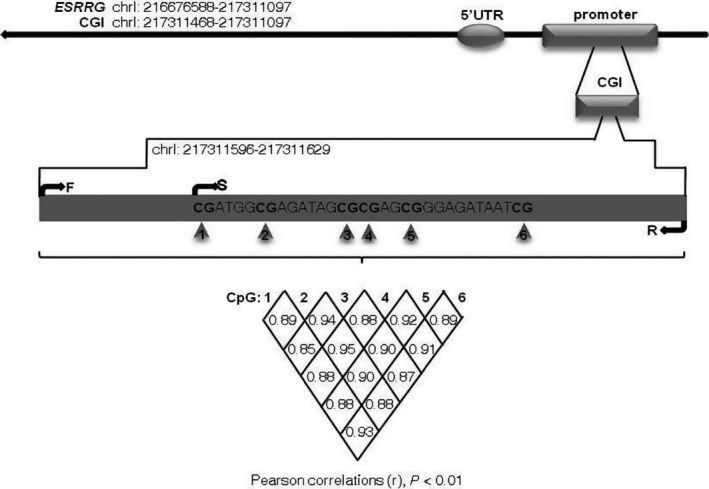
Correlation among the methylation status of six CpG sites in the ESRRG promoter. Six CpG sites were selected from a fragment (chrl: 217311596‐217311629) in a CGI of the ESRRG promoter. The correlation among the methylation status of the sites was calculated (Pearson correlation, all r > 0.8, P < 0.01). CGI, CpG island; F, forward primer; S, sequencing primer; R, reverse primer; C, G, T and A, nucleotides
Table 1.
Analysis of the methylation status of six CpG sites in the ESRRG promoter for laryngeal squamous cell carcinoma (LSCC) tumor and normal tissues
| CpG sites | Tumor tissue (Mean ± SD) | Normal tissue (Mean ± SD) | P value |
|---|---|---|---|
| CpG1 (%) | 40.47 ± 14.23 | 24.25 ± 10.81 | <0.01 |
| CpG2 (%) | 34.16 ± 14.25 | 20.99 ± 12.72 | <0.01 |
| CpG3 (%) | 34.81 ± 15.07 | 21.58 ± 11.97 | <0.01 |
| CpG4 (%) | 34.88 ± 15.54 | 19.34 ± 11.30 | <0.01 |
| CpG5 (%) | 39.88 ± 14.81 | 25.97 ± 11.76 | <0.01 |
| CpG6 (%) | 32.88 ± 17.07 | 14.52 ± 6.87 | <0.01 |
| MIa (%) | 36.18 ± 14.54 | 21.11 ± 10.45 | <0.01 |
MI, methylation index, calculated as the mean methylation status of the six CpG sites analyzed.
Figure 2.
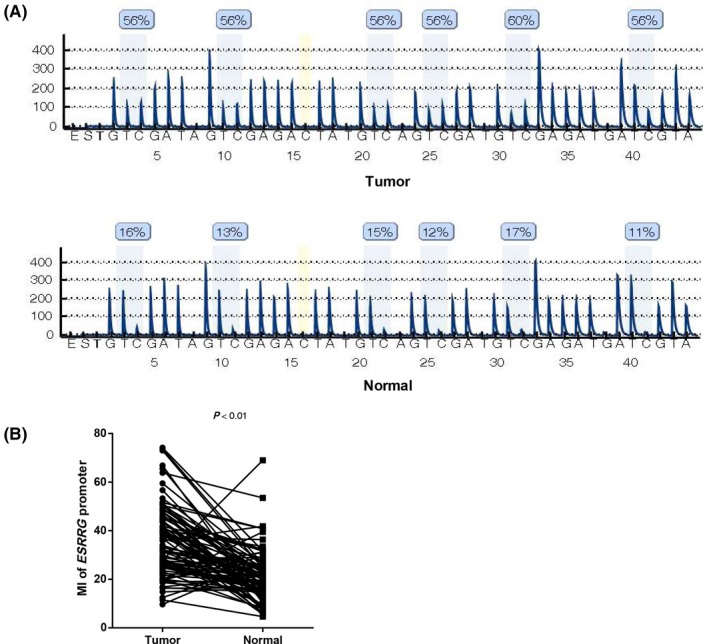
ESRRG promoter methylation status in laryngeal squamous cell carcinoma (LSCC) tumor and nontumor adjacent normal tissues. A, Representative pyrogram of the ESRRG promoter in tumor and nontumor adjacent tissue samples. The blue shaded bars enclosing T/C pairs represent the six CpG sites, and the yellow bar enclosing a C nucleotide indicates the internal reference. The methylation value of each CpG site is expressed as a percentage of C incorporation above the shaded bars. B, Comparison of the ESRRG promoter MI between LSCC tumor and nontumor adjacent normal tissues (n = 91, paired t test, P < 0.01). E, enzyme mix; S, substrate; A, G, C, and T, nucleotides; MI, methylation index
3.2. Association of ESRRG promoter methylation with clinical characteristics
In addition, we explored the association of ESRRG promoter methylation levels with clinical characteristics, including age, smoking behavior (patients who never smoked were defined as nonsmokers), differentiation, T stage, N stage, and clinical stage. LSCC patients were divided into two groups by each clinical characteristic for methylation status comparison. As shown in Table 2, the ESRRG promoter methylation levels were markedly increased in patients with advanced T stage tumors (P < 0.01) and advanced clinical stage disease (P < 0.01), while no significant correlation was observed between methylation and any other clinical characteristic (P > 0.05).
Table 2.
Association between the methylation status of ESRRG promoter and the clinical characteristics of laryngeal squamous cell carcinoma (LSCC) patients
| Clinical characteristics | n | MIa of ESRRG (Mean ± SD)% | P value |
|---|---|---|---|
| Age (y) | |||
| <60 | 49 | 36.79 ± 14.33 | |
| ≥60 | 42 | 35.47 ± 14.93 | 0.67 |
| Smoking | |||
| No | 17 | 33.04 ± 14.00 | |
| Yes | 74 | 36.90 ± 14.66 | 0.33 |
| Differentiation | |||
| Well | 48 | 38.85 ± 15.43 | |
| Moderate/poor | 43 | 33.21 ± 13.03 | 0.06 |
| T stage | |||
| T1‐2 | 56 | 33.06 ± 13.58 | |
| T3‐4 | 35 | 41.17 ± 14.82 | <0.01 |
| N stage | |||
| N0 | 64 | 34.93 ± 13.37 | |
| N1‐2 | 27 | 39.15 ± 16.91 | 0.21 |
| Clinical stage | |||
| I, II | 45 | 31.96 ± 11.61 | |
| III, IV | 46 | 40.31 ± 16.00 | <0.01 |
MI, methylation index, calculated as the mean methylation status of the six CpG sites analyzed.
3.3. Diagnostic value of ESRRG promoter methylation
A ROC curve analysis of ESRRG promoter methylation was performed to evaluate the diagnostic potential of ESRRG methylation levels for LSCC by calculating the AUC for nontumor samples as the control, as shown in Figure 3A. At a diagnostic threshold value (cut‐off value) of 26.58% for the ESRRG promoter MI, the AUC, sensitivity, and specificity were 0.81 (95% CI = 0.75‐0.88, P < 0.01), 70.33%, and 80.22%, respectively. Artificially, an MI above the cut‐off value was defined as a positive diagnostic indicator, while an MI below the cut‐off value was defined as a negative indicator. Then, the false‐negative and false‐positive rates were 19.78% and 29.67%, respectively. The positive and negative predictive values were 78.05% and 73.00%, respectively. The diagnostic concordance rate was 75.27%.
Figure 3.
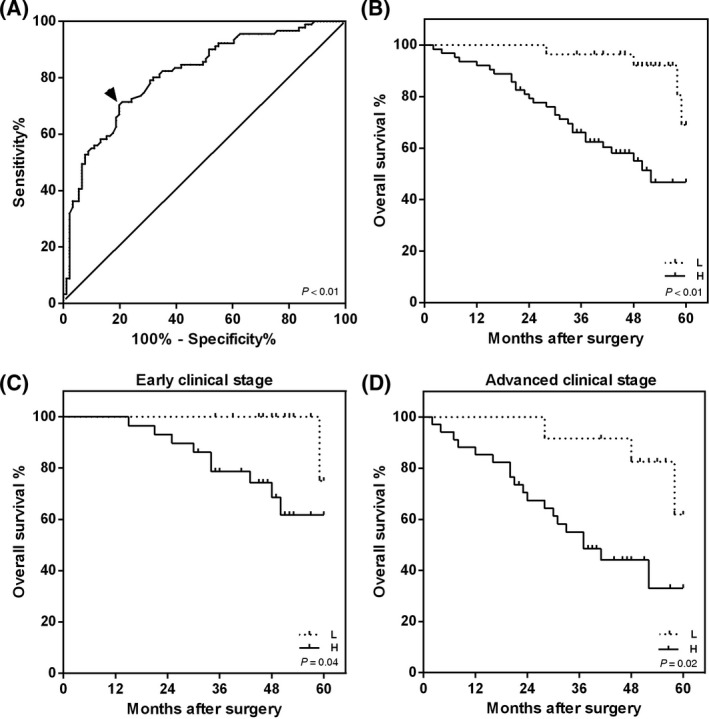
Receiver operating characteristic (ROC) curve and overall survival (OS) analysis stratified by the ESRRG promoter methylation status in patients with laryngeal squamous cell carcinoma (LSCC). A, ROC curve analysis for the ESRRG promoter methylation status in LSCC patients (n = 91). The black arrow indicates the best cut‐off point for the ESRRG methylation status, which is 26.58% (AUC = 0.81, 95% CI = 0.75‐0.88, P < 0.01, sensitivity = 70.33% and specificity = 80.22%). B‐D, The OS time of LSCC patients with ESRRG promoter hypomethylation was longer than that of patients with ESRRG promoter hypermethylation. The Kaplan‐Meier survival curves of (B) all LSCC patients (n = 91, P < 0.01), (C) patients with early clinical stage disease (I and II, n = 45, P = 0.04) and (D) patients with advanced clinical stage disease (III and IV, n = 46, P = 0.02) indicated an effect of the ESRRG promoter methylation status on 5‐year survival by the log‐rank test. A hypomethylation group (MI < 26.58%) and hypermethylation group (MI > 26.58%) were defined. H, hypermethylation group; L, hypomethylation group
3.4. Association of ESRRG promoter methylation with LSCC patient prognosis
Kaplan‐Meier survival analysis was performed to evaluate the prognostic potential of ESRRG methylation for LSCC patients who survived for 5 years. Based on the previously described ROC curve results, the patients were divided into two groups by using a mean MI of 26.58% for the 91 LSCC patient samples as a cut‐off value. The patients were divided into two groups: a hypermethylation group (n = 63) and a hypomethylation group (n = 28). In addition, we analyzed the overall survival (OS) of LSCC patients in early clinical stages (n = 45, 16 in the hypomethylation group and 29 in the hypermethylation group) and advanced clinical stages (n = 46, 12 in the hypomethylation group and 34 in the hypermethylation group). During follow‐up, five patients were lost to follow‐up (three patients were in an early clinical stage and two patients were in an advanced clinical stage), all of whom were in the hypomethylation group. Additionally, four patients in the hypomethylation group and 28 patients in the hypermethylation group died. A log‐rank test demonstrated that patients in the hypermethylation group, whether in an early or advanced clinical stage, tended to have a shorter OS than those in the hypomethylation group (all patients, P < 0.01; early clinical stage, P = 0.04; and advanced clinical stage, P = 0.02; Figure 3B‐D).
4. DISCUSSION
Recently, vast findings have been made regarding the mechanisms of aberrent gene methylation in LSCC.20 For instance, aberrant hypermethylation of CpG sites close to the transcriptional start site (TSS) of ZNF667‐AS1, a long noncoding RNA was critical for gene silencing, and associated with moderate/poor pathological differentiation in LSCC patients.21 ZNF667‐AS1 may be associated with the epithelial‐mesenchymal transition (EMT) process. Similarly, hypermethylation of the miR‐145‐5p promoter inhibits the expression of miR‐145‐5p which is a negative regulator of fascin actin‐bundling protein 1 (FSCN1), this effect is associated with the migration, invasion, and growth of LSCC because EMT is suppressed, and cell‐cycle arrest and apoptosis are induced.22 Promoter methylation of both ZNF667‐AS1 and miR‐145‐5p is an important potential prognostic marker and therapeutic target for LSCC. Targeted therapies for nonsmall cell lung cancer (NSCLC), an incurable disease, have improved the survival of NSCLC patients.23 In addition to targeted therapies for LSCC, early‐stage diagnosis of LSCC is needed. LSCC is an aggressive malignancy, and abundant evidence has shown that most LSCC patients are male.24 An understanding of the mechanisms underlying the sex difference in LSCC could lead to therapeutic targets, which are urgently needed. ESRRG has been studied extensively in female tumors and has been shown to play a tumor‐suppressive role. However, recent studies reported that ESRRG exerted oncogenic potential in breast cancer by increasing the expression of E‐cadherin, which promoted mesenchymal‐to‐epithelial transition (MET) in vivo.25 Indirectly, ESRRG could inhibit tumor growth. Moreover, in endometrial carcinoma, ESRRG is inhibited by miR‐205 resulting to promote tumor proliferation, migration, and invasion.2 Similar to its role in female tumors, ESRRG has been reported to be a tumor suppressor in prostate cancer, a male cancer, inducing p21 and p27 expression to arrest the cell cycle.26 Recently, Nam et al27 reported that ESRRG plays a canonical role in renal cell carcinoma (RCC). The study found that ESRRG loss occurred via DNA methylation and histone repressive silencing mediated by the polycomb repressor complex 2 (PRC2), and ESRRG restoration in RCC lines suppressed the migratory and invasive phenotypes. Additionally, clinicopathological analyses revealed that prostate cancer patients with high ESRRG protein expression level tended to show great cancer‐specific survival.6 In conclusion, ESRRG plays a role in inhibiting tumor growth, metastasis and invasion and could be a potential diagnostic and prognostic biomarker for these cancers. However, the roles of the ESRRG in LSCC progression are unclear. Therefore, the purpose of the study was to explore whether ESRRG plays a role in inhibiting tumor growth, metastasis, and invasion in LSCC, and whether ESRRG could be a potential diagnostic and prognostic biomarker for LSCC. As used in the current study, identifying epigenetic modifications in genes is an important approach to understand.
We therefore validated the methylation status of the ESRRG promoter in LSCC tumor samples relative to that in tumor‐adjacent normal tissues by pyrosequencing and further analyzed the clinical utility of the ESRRG promoter methylation status. In the present study, six sites were identified in a CGI of the ESRRG promoter in Figure 1. The mean methylation rate of the six sites was determined to be representative of ESRRG protomer methylation by a Pearson correlation analysis. Surprisingly, our results showed that ESRRG promoter methylation was significantly higher in LSCC tissues than in noncancerous tissues, suggesting that ESRRG promoter methylation is a tumor‐associated event during LSCC tumorigenesis. This result is the first to show hypermethylation of the ESRRG promoter in LSCC. However, we could not compare the gene and protein expression of ESRRG in LSCC because of the limited amount of tissue samples.
Furthermore, by determining the clinical significance of ESRRG promoter methylation, we showed that it was significantly increased in patients with advanced T stage tumors and advanced clinical stage disease. These results were similar to those found in previous tumor studies. However, ESRRG promoter methylation was not significantly related to tumor metastasis. Although smoking is a negative risk factor for LSCC and we previously found a link between smoking and gene methylation in LSCC,28 no significant correlation was found between methylation and the environmental risk factors that we studied here. Thus, these present observations encourage us to seek other environmental risk or protective factors for ESRRG methylation.
Our strategy used ROC curves to evaluate the value of ESRRG promoter methylation as a potential biomarker for LSCC. Obtaining normal laryngeal tissue from noncancer patients is not practical. Therefore, we used adjacent nontumor tissue as normal samples to evaluate the diagnostic value as described in a previous study.18 Several studies have established the optimal methylation threshold to diagnose cancers and assess prognosis by ROC curve analysis,29 highlighting the importance of methylation as a biomarker in tumor management. We calculated the AUC of ESRRG methylation as 0.81. Thus, these data clarified the high diagnostic value of ESRRG methylation for LSCC. In the present study, promoter methylation in a series of genes was reported to be a potential biomarker in LSCC (PCDH17,16 CMTM3,28 and SSTR2 30). Taken together, these results indicate that the detection of ESRRG promoter methylation could be included in a diagnostic panel of multiple gene methylation statuses.
In addition, DNA methylation levels have been reported to be biomarkers for assessing cancer prognosis.31 Based on the SEER Cancer Statistics Review, the 5‐year OSR is approximately 60% in the United States.32 Concerningly, CMTM3 was reported to increase cancer risk among patients with early clinical stage disease. We performed a Kaplan‐Meier analysis to explore whether ESRRG promoter methylation is associated with LSCC prognosis. In a previous study, a series of methylation statuses was measured by pyrosequencing to assess the OSR by the log‐rank test.33 Using clinical data, we demonstrated that the methylation status of the ESRRG promoter differed in early and advanced clinical stages. Clinical stage is considered a perfect predictor of long‐term survival.34 To eliminate the influence of clinical stage on ESRRG promoter methylation when analyzing the survival status, we adjusted the clinical stage by dividing the samples into two groups, the early and advanced clinical stage groups. When stratified according to clinical stage, ESRRG promoter hypermethylation was significantly associated with poor survival in patients with both early and advanced clinical stage disease. Thus, we showed that hypermethylation of the ESRRG promoter is an adverse factor affecting the prognosis of patients with LSCC.
5. CONCLUSIONS
In conclusion, the current study showed that the ESRRG promoter was highly methylated in LSCC tumor tissues and that its hypermethylation was significantly associated with advanced T stage tumors and advanced clinical stage patients. In addition, the ESRRG promoter methylation status could be a biomarker to diagnose LSCC and assess prognosis.
ACKNOWLEDGMENTS
The research was supported by the Zhejiang Provincial Natural Science Foundation of China (No. LY14H160003, LH19H160014); the Ningbo Health Branding Subject Fund (No. PPXK2018‐02); the Medical and Health Training Project of Zhejiang Province (No. 2015RCB025); the Scientific Innovation Team Project of Ningbo (No. 2012B82019); the Ningbo Social Developmental Key Research Project (No. 2012C5015); the Medical and Health Research Project of Zhejiang Province(No. 2012ZDA042, No. 2014PYA017, No. 2018RC063, No. 2019ZD018); the Natural Science Foundation of Ningbo (No. 2012A610208, No.2013A610217, No.2017A610236, No. 2018A610361) and the Leading and Top Talents Training Projects of Ningbo (No. NBLJ201801032).
Shen Z, Hu Y, Zhou C, et al. ESRRG promoter hypermethylation as a diagnostic and prognostic biomarker in laryngeal squamous cell carcinoma. J Clin Lab Anal. 2019;33:e22899 10.1002/jcla.22899
REFERENCES
- 1. Kim JH, Choi YK, Byun JK, et al. Estrogen‐related receptor gamma is upregulated in liver cancer and its inhibition suppresses liver cancer cell proliferation via induction of p21 and p27. Exp Mol Med. 2016;48:e213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Su N, Qiu H, Chen Y, Yang T, Yan Q, Wan X. miR‐205 promotes tumor proliferation and invasion through targeting ESRRG in endometrial carcinoma. Oncol Rep. 2013;29:2297‐2302. [DOI] [PubMed] [Google Scholar]
- 3. Heckler MM, Thakor H, Schafer CC, et al. ERK/MAPK regulates ERRgamma expression, transcriptional activity and receptor‐mediated tamoxifen resistance in ER+ breast cancer. FEBS J. 2014;281:2431‐2442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Eichner LJ, Perry MC, Dufour CR, et al. miR‐378( *) mediates metabolic shift in breast cancer cells via the PGC‐1beta/ERRgamma transcriptional pathway. Cell Metab. 2010;12:352‐361. [DOI] [PubMed] [Google Scholar]
- 5. Zhang Y, Wang L. Nuclear receptor SHP inhibition of Dnmt1 expression via ERRgamma. FEBS Lett. 2011;585:1269‐1275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Fujimura T, Takahashi S, Urano T, et al. Differential expression of estrogen‐related receptors beta and gamma (ERRbeta and ERRgamma) and their clinical significance in human prostate cancer. Cancer Sci. 2010;101:646‐651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Siegel RL, Miller KD, Jemal A. Cancer statistics, 2018. CA Cancer J Clin. 2018;68:7‐30. [DOI] [PubMed] [Google Scholar]
- 8. Stembalska A, Leszczyński P, Gil J, et al. Global DNA methylation status in laryngeal cancer. Head Neck. 2014;36:419‐424. [DOI] [PubMed] [Google Scholar]
- 9. Huang P‐W, Lin C‐Y, Hsieh C‐H, et al. A phase II randomized trial comparing neoadjuvant chemotherapy followed by concurrent chemoradiotherapy versus concurrent chemoradiotherapy alone in advanced squamous cell carcinoma of the pharynx or larynx. Biomed J. 2018;41:129‐136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Ma C‐H, Zhang Y‐X, Tang L‐H, et al. MicroRNA‐1469, a p53‐responsive microRNA promotes Genistein induced apoptosis by targeting Mcl1 in human laryngeal cancer cells. Biomed Pharmacother. 2018;106:665‐671. [DOI] [PubMed] [Google Scholar]
- 11. Siegel RL, Miller KD, Jemal A. Cancer statistics, 2017. CA Cancer J Clin. 2017;67:7‐30. [DOI] [PubMed] [Google Scholar]
- 12. Jones PA. Functions of DNA methylation: islands, start sites, gene bodies and beyond. Nat Rev Genet. 2012;13:484‐492. [DOI] [PubMed] [Google Scholar]
- 13. Gyobu K, Yamashita S, Matsuda Y, et al. Identification and validation of DNA methylation markers to predict lymph node metastasis of esophageal squamous cell carcinomas. Ann Surg Oncol. 2011;18:1185‐1194. [DOI] [PubMed] [Google Scholar]
- 14. Mojtabanezhad Shariatpanahi A, Yassi M, Nouraie M, et al. The importance of stool DNA methylation in colorectal cancer diagnosis: a meta‐analysis. PLoS ONE. 2018;13:e0200735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Yu J, Yuan X, Sjöholm L, et al. Telomerase reverse transcriptase regulates DNMT3B expression/aberrant DNA methylation phenotype and AKT activation in hepatocellular carcinoma. Cancer Lett. 2018;434:33‐41. [DOI] [PubMed] [Google Scholar]
- 16. Byzia E, Soloch N, Bodnar M, et al. Recurrent transcriptional loss of the PCDH17 tumor suppressor in laryngeal squamous cell carcinoma is partially mediated by aberrant promoter DNA methylation. Mol Carcinog. 2018;57:878‐885. [DOI] [PubMed] [Google Scholar]
- 17. Bednarek K, Kostrzewska‐Poczekaj M, Szaumkessel M, et al. Downregulation of CEACAM6 gene expression in laryngeal squamous cell carcinoma is an effect of DNA hypermethylation and correlates with disease progression. Am J Cancer Res. 2018;8:1249‐1261. [PMC free article] [PubMed] [Google Scholar]
- 18. Foteinou E, Kontos CK, Giotakis AI, Scorilas A. Low mRNA expression levels of kallikrein‐related peptidase 4 (KLK4) predict short‐term relapse in patients with laryngeal squamous cell carcinoma. Biol Chem. 2014;395:1051‐1062. [DOI] [PubMed] [Google Scholar]
- 19. Dankova Z, Brany D, Dvorska D, et al. Methylation status of KLF4 and HS3ST2 genes as predictors of endometrial cancer and hyperplastic endometrial lesions. Int J Mol Med. 2018;42:3318‐3328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Bednarek K, Kostrzewska‐Poczekaj M, Szaumkessel M, et al. Downregulation of gene expression in laryngeal squamous cell carcinoma is an effect of DNA hypermethylation and correlates with disease progression. Am J Cancer Res. 2018;8:1249‐1261. [PMC free article] [PubMed] [Google Scholar]
- 21. Meng W, Cui W, Zhao L, Chi W, Cao H, Wang B. Aberrant methylation and downregulation of ZNF667‐AS1 and ZNF667 promote the malignant progression of laryngeal squamous cell carcinoma. J Biomed Sci. 2019;26:13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Gao W, Zhang C, Li W, et al. Promoter methylation‐regulated miR‐145‐5p inhibits laryngeal squamous cell carcinoma progression by targeting FSCN1. Mol Ther. 2019;27:365‐379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Black A, Morris D. Personalized medicine in metastatic non‐small‐cell lung cancer: promising targets and current clinical trials. Curr Oncol. 2012;19:S73‐S85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Starska K, Forma E, Lewy‐Trenda I, Stasikowska‐Kanicka O, Skóra M, Bryś M. Fibroblast growth factor receptor 1 and 3 expression is associated with regulatory PI3K/AKT kinase activity, as well as invasion and prognosis, in human laryngeal cancer. Cell Oncol (Dordr). 2018;41:253‐268. [DOI] [PubMed] [Google Scholar]
- 25. Tiraby C, Hazen BC, Gantner ML, Kralli A. Estrogen‐related receptor gamma promotes mesenchymal‐to‐epithelial transition and suppresses breast tumor growth. Cancer Res. 2011;71:2518‐2528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Yu S, Wang X, Ng CF, et al. ERRgamma suppresses cell proliferation and tumor growth of androgen‐sensitive and androgen‐insensitive prostate cancer cells and its implication as a therapeutic target for prostate cancer. Cancer Res. 2007;67:4904‐4914. [DOI] [PubMed] [Google Scholar]
- 27. Nam H‐Y, Chandrashekar DS, Kundu A, et al. Integrative epigenetic and gene expression analysis of renal tumor progression to metastasis. Mol Cancer Res. 2019;17:84‐96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Shen Z, Chen X, Li Q, et al. Elevated methylation of CMTM3 promoter in the male laryngeal squamous cell carcinoma patients. Clin Biochem. 2016;49:1278‐1282. [DOI] [PubMed] [Google Scholar]
- 29. Dong X, Hou Q, Chen Y, Wang X. Diagnostic value of the methylation of multiple gene promoters in serum in hepatitis B virus‐related hepatocellular carcinoma. Dis Markers. 2017;2017:2929381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Shen Z, Chen X, Li Q, et al. SSTR2 promoter hypermethylation is associated with the risk and progression of laryngeal squamous cell carcinoma in males. Diagn Pathol. 2016;11:10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Paluszczak J, Misiak P, Wierzbicka M, Woźniak A, Baer‐Dubowska W. Frequent hypermethylation of DAPK, RARbeta, MGMT, RASSF1A and FHIT in laryngeal squamous cell carcinomas and adjacent normal mucosa. Oral Oncol. 2011;47:104‐107. [DOI] [PubMed] [Google Scholar]
- 32. Chen L, Zeng H, Yang J, et al. Survival and prognostic analysis of preoperative inflammatory markers in patients undergoing surgical resection for laryngeal squamous cell carcinoma. BMC Cancer. 2018;18:816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Lee SM, Lee WK, Kim DS, et al. Quantitative promoter hypermethylation analysis of RASSF1A in lung cancer: comparison with methylation‐specific PCR technique and clinical significance. Mol Med Rep. 2012;5:239‐244. [DOI] [PubMed] [Google Scholar]
- 34. Balch CM, Soong S‐J, Gershenwald JE, et al. Prognostic factors analysis of 17,600 melanoma patients: validation of the American Joint Committee on cancer melanoma staging system. J Clin Oncol. 2001;19:3622‐3634. [DOI] [PubMed] [Google Scholar]


