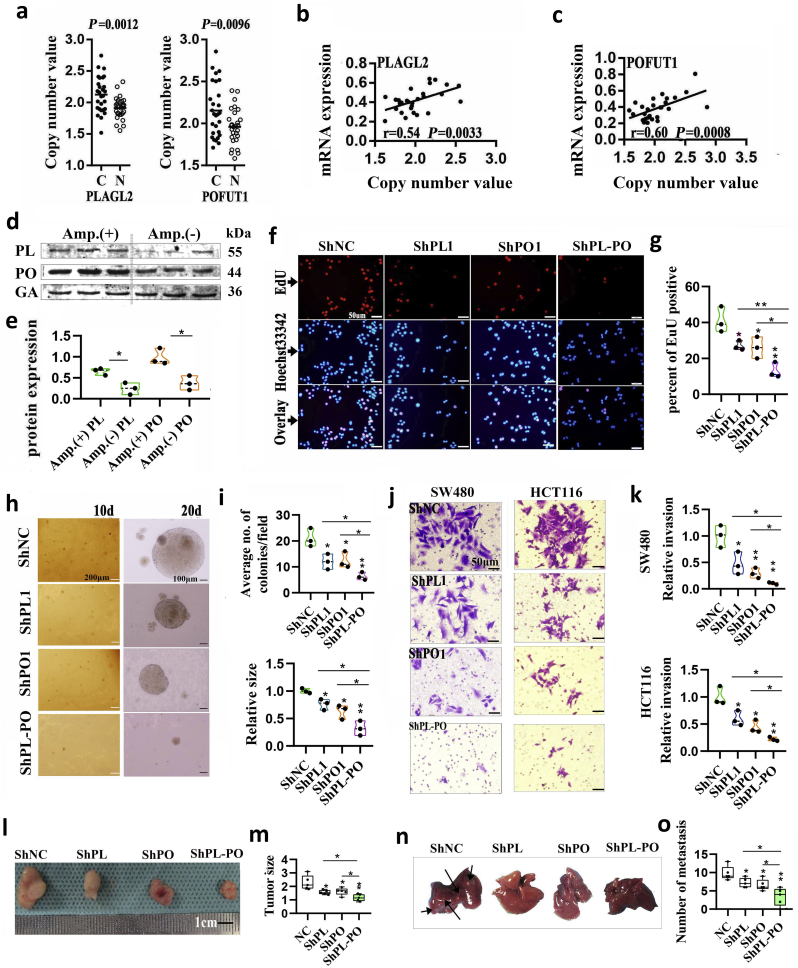
Fig. 4.
PLAGL2 and POFUT1 owned overexpression driven by CNA and combined PLAGL2 /POFUT1 inhibition have greater effects on opposing tumors.
(a) AccuCopy copy number analysis of copy number of PLAGL2 (including PLAGL2–1 and PLAGL2–2) and POFUT1 (including POFUT1–1 and POFUT1–2) in 14 pair colorectal tissues.
(b and c) The correlation between copy number value of PLAGL2–2 and PLAGL2 mRNA expression and POFUT1–2 and POFUT1 mRNA expression.
(d) Comparison of PLAGL2 and POFUT1 protein expression between CRC tissues without CNA (Amp. (−)) and CRC tissues with amplified (Amp (+)). PL: PLAGL2; PO: POFUT1; GA: GAPDH.
(e) Quantification of (d).
(f) The EdU proliferation assay was performed after shRNA-mediated suppression of POFUT1 or/and PLAGL2 in SW480 cells; shown here are the results for the ShPLAGL2–1 and ShPOFUT1–1 sequences (scale bar:50 μM).
(g) Quantification of (B).
(h) Suppressing PLAGL2 or/and POFUT1 expression reduce anchorage-independent growth in HCT116 cells, shown here are the results for the ShPLAGL2–1 and ShPOFUT1–1 sequences. (scale bar:200 μm (left), 100 μm (right)); d: day.
(i) Quantification of (D), upper showing average number of colonies after cultivation for 10 days and down showing relative size of colonies after cultivation for 20 days; the shNC group (NC) was set to 1.
(j) Representative images of invasion assay showing control group (NC) and shRNA-mediated suppression of POFUT1 or/and PLAGL2 in SW480 and HCT116 cells; shown here are the results for the ShPLAGL2–1 and ShPOFUT1–1 sequences (scale bar:50 μm).
(k) Quantification of (j)
(l and m) shRNA-mediated suppression of POFUT1 or/and PLAGL2 can decelerated growth of HCT116 xenografts in nude mice (n = 5).
(n and o) Representative macroscopic appearances of liver metastasis are shown (some tumor nodules were indicated by black arrows). (HCT116 cells was used in this assay). (scale bar:100 μm) (n = 5).
Data in (d)–(o) presented as violin plots from at least two independent experiments with triplicates. Each plot represents the mean from one independent experiment with triplicates. *p < .05, **p < .01. The *at the vertex of the bar graph is compared to the control group.