Abstract
Safer and more efficient methods for directing therapeutic genes to specific sequences could increase the repertoire of treatable conditions. Many current approaches act passively, first initiating a double-stranded break, then relying on host repair to uptake donor DNA. Alternatively, we delivered an actively integrating transposase to the target sequence to initiate gene insertion. We fused the hyperactive piggyBac transposase to the highly specific, catalytically dead SpCas9-HF1 (dCas9) and designed guide RNAs (gRNAs) to the CCR5 safe harbor sequence. We introduced mutations to the native DNA-binding domain of piggyBac to reduce non-specific binding of the transposase and cause the fusion protein to favor binding by dCas9. This strategy enabled us, for the first time, to direct transposition to the genome using RNA. We showed that increasing the number of gRNAs improved targeting efficiency. Interestingly, over half of the recovered insertions were found at a single TTAA hotspot. We also found that the fusion increased the error rate at the genome-transposon junction. We isolated clonal cell lines containing a single insertion at CCR5 and demonstrated long-term expression from this locus. These vectors expand the utility of the piggyBac system for applications in targeted gene addition for biomedical research and gene therapy.
Keywords: transposon, piggyBac, gene therapy, CRISPR Cas9
Introduction
Novel gene replacement technologies have the potential to safely and efficiently reverse genetic defects underlying many diseases (1, 2). Sequence-specific approaches offer key advantages over traditional virus-based integrating vectors by avoiding insertion into unwanted regions (3–6). clustered regularly interspaced short palindromic repeats (CRISPR) nucleases are excellent at intentionally mutating the genome at specific sites (7). However, homology-directed repair (HDR) for inserting DNA is far less efficient (8). This is because current gene-targeting technologies are passive; following the DNA break, rate-limiting host factors are needed for the addition of new sequence, often resulting in a mutation without the desired insert (9). Furthermore, because HDR factors are present during cell division, HDR is inefficient in non-dividing cells that make up the vast majority of the tissues (9, 10).
Double-stranded breaks induced by CRISPR nucleases can have undesirable outcomes. Studies have found that CRISPR nucleases induce a p53-mediated DNA damage response (11, 12). p53-dependent toxicity resulting from CRISPR nuclease gene editing can lead to unwanted selection for cells with an impaired p53 pathway. Additionally, a long-range sequence analysis of CRISPR nuclease cut sites reported a high frequency of large deletions extending over many kilobases as well as complex genomic rearrangements (13). The authors warned that such rearrangements could join transcriptionally active regions with cancer-related genes.
The challenges of nuclease-based gene delivery may be overcome by alternative technologies. Transposase enzymes are capable of actively inserting DNA into the genome. During integration, DNA is protected during cleavage, strand exchange and religation (14, 15). This process does not rely on rate-limiting host factors to repair DNA breaks and, importantly, does not provoke error-prone DNA repair processes resulting in indel formation that can be caused by unprotected cleavage by nucleases (16). The piggyBac transposase specifically, has been shown to be highly efficient at integration across species and cell types (17–20). The piggyBac system has a large cargo capacity of >100 kb and all components can be delivered as DNA plasmids, obviating the need for viral proteins that may cause unwanted immune response in vivo (21–26). This system is ideally suited for non-viral approaches, such as nanoparticle technologies, that have been shown to deliver plasmids both ex vivo and in vivo (27–33). Many of these approaches currently use transient expression plasmids and could be improved by a stably inserting vector (32–35).
We previously fused a transcription activator like effector (TALE) DNA-binding domain (DBD) to piggyBac to direct the transposase to a desired sequence in the genome (36). This relocalization of piggyBac promoted insertional activity at the intended target sequence in a subset of cells. This approach enabled the isolation of clones harboring single-copy insertions at the CCR5 safe harbor locus. Subsequently, Luo et al. successfully directed piggyBac insertion to the endogenous hypoxanthine-guanine phosphoribosyltransferase (HPRT) locus using both zinc finger and TALE DBDs (37). Mapping of transcription factor binding locations has been performed by using piggyBac tethered to DNA-binding proteins. Transposon insertions are first directed by the transcription factor of interest to the genome. This is followed by sequencing of the DNA flanking the transposon to identify the transcription factor binding sites (38, 39).
The CRISPR Cas9 DNA-binding protein with a catalytically dead nuclease domain (dCas9) has been fused to a variety of effector domains including transcription activators and repressors, epigenetic modifiers, and base editing enzymes, among others (40, 41). Unlike zinc finger and TALE DBDs, generation of a dCas9 DBD does not require time-consuming protein design, instead requiring only a 20 bp target sequence expressed as a gRNA (42). This ease-of-construction has increased the availability of CRISPR Cas9 technologies to numerous research labs (43). An RNA-guided transposase would represent a simple-to-use tool potentially enabling efficient gene delivery to flexible cell types. Although several programmable DBD platforms have previously been tethered to piggyBac, including dCas9, to date, no active dCas9-piggyBac has been reported (37).
A challenge to previous attempts at generating a targetable piggyBac vector has been high levels of off-target integration (36, 37, 44–46). This could be due to the presence of the native piggyBac DBD which may enable the transposase to bind and integrate at off-target sites before the additional custom DBD is able to bind the target sequence. Li et al. previously identified piggyBac mutations that attenuated DNA binding (47). We reasoned that mutations in the native DBD may inhibit off-target binding and cause the transposase to require the binding of dCas9 to the target sequence as a prerequisite for insertion.
We tested a panel of RNA-guided transposase vectors containing mutations in the native piggyBac DBD for their ability to target a single sequence in the CCR5 gene. Here, we report the first evidence of RNA-guided transposition in human cells. This proof-of-concept establishes a framework for improved targeting vectors with potential applications in genome editing research and gene therapy.
Materials and methods
Plasmid development
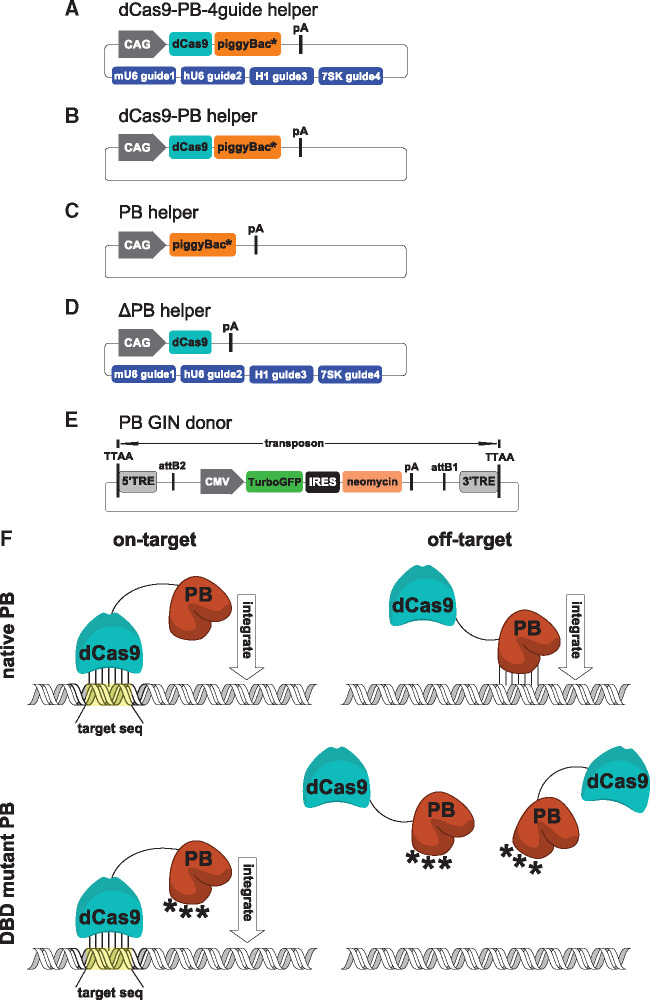
Illustrations of targeting plasmids are depicted in Figure 1A–E. The SpCas9-HF1 gene (48) was mutated at the D10A and H840A residues to inactivate the catalytic domain and generate dCas9 (42, 49). The dCas9-PB helper plasmid was generated using Gibson assembly by fusing the hyperactive piggyBac transposase gene (17) to the dCas9 DNA-binding protein (48) using a flexible linker described previously (36). The fusion protein was placed under the CAG [cytomegalovirus (CMV) immediate early enhancer, chicken β-actin promoter and β-globin intron] promoter. Two mutant piggyBac transposase helper plasmids containing codon changes in the presumed piggyBac DBD originally identified by Li et al. were generated using Gibson assembly (47). First, the hyperactive piggyBac gene was human codon optimized and synthesized by Genscript. Next, mutations R372A and D450N were introduced to generate the dCas9-H2 helper plasmid and a third K375A mutation was introduced to generate the dCas9-H3 helper plasmid. Four gRNAs were appended to the helper plasmid backbone using Golden Gate assembly as previously described (50). Briefly, single stranded oligos containing the guide sequence were annealed and ligated into BbsI linearized expression plasmids containing either the hU6, mU6, H1 or 7SK promoter. One of each of the four resulting guide expression plasmids were first digested with BsmBI and then assembled into a single BsmBI-linearized helper plasmid in a single step. For experiments requiring eight guides, two plasmids each containing four guides were co-transfected in equal amounts. Guide sequences are listed in the Supplementary Data. Negative control helper plasmids lacked gRNAs. Control helper plasmids that contained either the PB, H2 or H3 transposase but lacking a DBD were also generated using Gibson assembly. To generate the non-integrating ΔPB control, the entire piggyBac coding sequence was removed from the dCas9-PB helper plasmid using Gibson assembly. To generate the donor plasmid, Gateway cloning (Thermo Fisher) was used to recombine a pENTR plasmid featuring the CMV promoter driving TurboGFP, internal ribosome entry site (IRES) and neomycin (GIN) gene with a pDONR plasmid containing piggyBac terminal repeat elements (TREs) flanking the transgene (36). Plasmid maps can be found in the Supplementary Data.
Figure 1.
Helper and donor plasmids. (A) Catalytically inactive dCas9 was fused to the hyperactive piggyBac transposase via a flexible linker and placed under the CAG promoter. *Three different piggyBac genes were tested: (i) PB, the original hyperactive piggyBac, (ii) H2, a human codon-optimized hyperactive piggyBac with two mutations in the presumed DBD and (iii) H3, a human codon-optimized hyperactive piggyBac with three mutations in the presumed DBD. Four guide RNAs under the mU6, hU6, H1 and 7SK promoters were cloned into the plasmid backbone. (B) Control helper devoid of guide RNA. (C) Control helper devoid of the dCas9 DNA-binding protein. (D) Non-insertional control helper devoid of the piggyBac transposase (ΔPB). (E) The donor plasmid contained the TurboGFP IRES neomycin transgene under the CMV promoter and flanked by the piggyBac transposon TREs. (F) Proposed model for improvement of specificity by disruption of the piggyBac DBD. The native PB transposase retains full DNA-binding capability and can either integrate following dCas9 targeting (on-target) or integrate following binding to off-target sequences without dCas9 targeting (off-target). Similar to PB, the H2 and H3 mutant transposase variants can integrate following dCas9 targeting (on-target). However, off-target binding of the transposase is inhibited due to mutations in the DBD.
Cell transfections
Human embryonic kidney (HEK293) cells were maintained in complete Dulbecco's modified Eagle's medium (DMEM) supplemented with 10% heat inactivated fetal bovine serum. Prior to transfection, 4 × 105 cells per well were seeded in 6-well plates. Cells at ∼80% confluency were transfected with 2 μg of plasmid DNA using X-tremeGENE 9 (Sigma Aldrich). Twenty-four hours after transfection, cells were resuspended and 10% of cells were removed for flow cytometry analysis to measure transfection efficiency. Forty-eight hours after transfection, 90% of the cells were transferred to a T75 flask and cultured for 3 weeks under 200 µg/ml G418 at which point the cells were pelleted for lysis and genomic polymerase chain reaction (PCR) analysis. The remaining 10% of cells in the 6-well dish were cultured without antibiotic for 3 weeks and analyzed by flow cytometry to measure stable insertion efficiency. For single-cell isolation, two dCas9-H2-8guide transfections were repeated. The G418-selected polyclonal populations were each plated into a 96-well poly-d-lysine coated plate (BD Biosciences) resulting in an average of 50 colonies per well. After wells became greater than 40% confluent, media was aspirated, and the cells were manually resuspended in 30 µl of phosphate-buffered saline. A volume of 20 µl of the resuspension was removed and mixed with 30 µl of the DirectPCR Lysis Reagent (Viagen Biotech) for analysis. The remaining cells were cultured further. Two wells identified to contain targeted clones by genomic PCR were expanded and single-cell sorted using serial dilution. Wells were visually monitored until 157 single-cell expansions were obtained. Clonally expanded cells were subsequently resuspended by manual pipetting and lysed for analysis. Two positive clonal lines, 293-c1 and 293-c2, containing targeted insertions to CCR5 were expanded for flow cytometry analysis to detect potential silencing of the transgene. Genomic DNA was isolated from the 293-c1 and 293-c2 lines using the NucleoSpin Tissue kit (Machery-Nagel) and used as template for copy number assay.
Copy number assay
In order to determine the number of transposons present in CCR5-targeted single clones, a copy number assay was performed as previously described (44). Briefly, TaqMan quantitative PCR was used to estimate the number of neomycin genes present in the genome. The human RNase P gene was used to normalize the total genomes per sample. Templates included: genomic DNA from clonal lines 293-c1 and 293-c2, negative control untransfected human genomic DNA and reference control genomic DNA from a clonal cell line with a single neomycin gene insertion. Quantitative PCR using the QuantStudio 12K Flex thermocycler (Applied Biosystems) was performed using the TaqPath ProAmp Master Mix reagent (Thermo Fisher) according to the manufacturer’s instructions. Primers and probes were included in the TaqMan Copy Number Reference Assay for human RNase P and the TaqMan NeoR Assay ID: Mr00299300_cn (Thermo Fisher). CopyCaller Software v2.1 was used to predict the number of insertions for each sample.
Flow cytometry
Green fluorescent protein (GFP) expression of 20 000 live cells from CCR5-targeted single-cell expansions was analyzed using a FACSAria III cytometer (BD Biosciences) after 13 weeks of culture, following transfection with dCas9-H2-8guide.
T7 endonuclease I assay
In 12-well plates, HEK293 cells at 80% confluency in DMEM supplemented with 10% heat inactivated fetal bovine serum, were co-transfected with 500 ng of SpCas9-HF1 expression plasmid and 500 ng of one of eight CCR5 directed gRNA or negative control gRNA expression plasmids, using X-tremeGENE 9 (Sigma Aldrich). Seventy-two hours later, cells were pelleted and lysed using DirectPCR Cell lysis buffer (Viagen Biotech). Genomic PCR using the KOD Xtreme Hot Start DNA Polymerase (Novagen) was performed using primers designed to flank all eight guide binding sites. Products were purified with the PureLink PCR Micro Kit (Invitrogen) and melted and reannealed to form heteroduplexes. For each sample, identical incubations with or without T7 endonuclease I (T7E1) (New England Biolabs) were performed to cut DNA containing mismatched sequences. Products were separated on a 2% gel for gel imaging. A 2100 Bioanalyzer (Agilent) was used to measure the concentration of products obtained by the T7E1 assay. The fraction of cleaved products was calculated by dividing the total pg/μl of the two expected cleavage products by the total pg/μl of the two expected cleavage products and uncleaved product. Percent of indel occurrence was calculated using the formula = 100 × (1 − (1 − fraction cleaved)1/2) (51).
Targeted genomic integration site recovery
Pellets from stable transfections of HEK293 cells were lysed using the DirectPCR Cell lysis buffer (Viagen Biotech) for use as template for nested PCR to identify targeted transposon insertions. In order to optimize the PCR, the lysate template was used at three dilutions, 1:1, 1:4 and 1:8. Forward primers were designed to extend outward from the transposon whereas reverse primers were designed to extend from the CCR5 target sequence. A 10 μl primary PCR was performed using the KOD Xtreme Hot Start DNA Polymerase (Novagen) that was diluted 1:50 in H2O and used as template for a 20 μl nested PCR using PrimeSTAR GXL DNA Polymerase (Clontech). Amplification products were gel purified with the Zymoclean Gel DNA Recovery Kit (Zymo Research) and sequenced directly or cloned into pJet1.2 (Thermo Fisher) for sequencing. Sequences were aligned against the piggyBac transposon sequence using BLAST and against the human reference genome (hg38) using BLAT to identify insertion site locations. All primer sequences are listed in the Supplementary Data.
Statistical analysis
We performed ANalysis Of VAriance (ANOVA) analysis of variance followed by a Bonferroni post-test using the two-tailed, two-sample t-test assuming equal variances to determine if the difference between integration efficiencies was significant with a confidence interval of 95% (P < 0.05).
Results
Fusion of dCas9 to the piggyBac transposase
To generate an RNA-guided transposase, we fused the RNA-guided dCas9 DNA-binding protein to the piggyBac transposase. We hypothesized that binding of dCas9 to the target genomic sequence would physically sequester the piggyBac transposase to the same location and promote transposition to nearby sequences. During Cas9 recognition of its target sequence, the DNA strands are separated by forming an R-loop structure, which allows the target strand to base-pair to the guide RNA. DNA recognition is strictly dependent on the presence of a cognate PAM downstream of the target site. These events enable the HNH and RuvC nuclease domains to catalyze the cleavage of the target and non-target DNA strands, respectively (49). Inactivating mutations H840A and D10A prevent the HNH and RuvC domains from cleaving their respective DNA strands (42). These mutations disrupt the cutting but not the binding of dCas9. We used the high-fidelity Streptococcus pyogenes Cas9 (SpCas9-HF1) that has been designed to reduce non-specific DNA contacts (48). To make helper plasmids, SpCas9-HF1 was catalytically inactivated by introducing mutations D10A and H840A, then fused to three unique piggyBac transposase variants, referred to as PB, H2 and H3. Plasmid maps are depicted in Figure 1A–E. The PB variant contains seven mutations that increase the excision efficiency by 17-fold over wildtype (17). The H2 and H3 variants contain additional mutations in residues that are believed to play a role in DNA binding (47). We reasoned that disrupting the piggyBac DBD may cause the fusion molecule to preferentially use dCas9 for binding (Figure 1F). H2 had two mutations, R372A/D450N, aimed at moderately disrupting DNA binding. H3 had three mutations, R372A/K375A/D450N, aimed at a high level of disruption of DNA binding. A flexible 23 amino acid (aa) linker containing 11 glycines was used to fuse dCas9 to the transposase (36). We engineered a simplified Golden Gate cloning strategy enabling simultaneous addition of multiple gRNAs in a single, efficient step (50). We designed eight total gRNAs tiled across the first intron of the human CCR5 gene. Combinations of guides 1–4 or guides 5–8 were cloned into the backbone of each of the PB, H2 and H3 helper plasmids. For experiments requiring four guides, a single plasmid containing guides 1–4 was used. For experiments using eight guides, two helper plasmids containing guides 1–4 or guides 5–8 were combined. We reasoned that multiple guides directed to the CCR5 target site would provide added opportunities for favorable insertion sequences. Additionally, multiple guides could sequester an increased number of transposase molecules at the target site.
Controls included helper plasmids devoid of gRNAs (Figure 1B), devoid of dCas9 (Figure 1C), or devoid of transposase (Figure 1D). The donor plasmid featured a bicistronic TurboGFP-IRES-neomycin gene for visualization and selection within the transposon (Figure 1E). Successful targeting was expected to result in the excision of the transposon from the donor plasmid by the transposase encoded on the helper plasmid. This would be followed by permanent introduction of the transposon into the CCR5 sequence and enable TurboGFP visualization or neomycin selection.
Transposase integration and gRNA efficiency
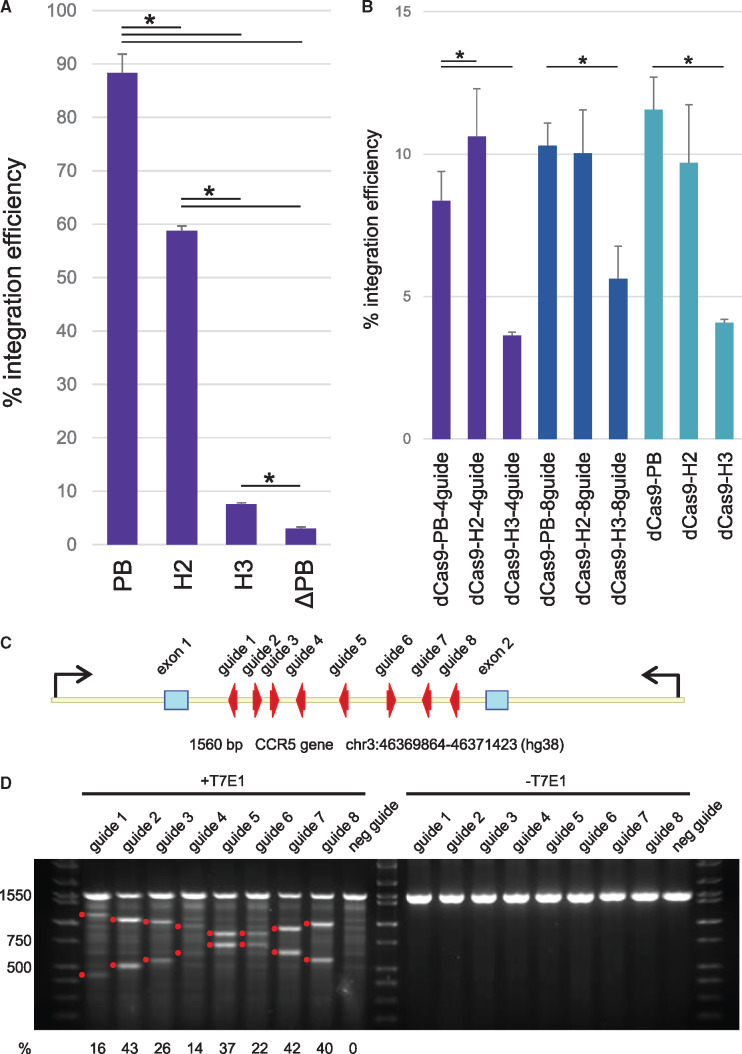
We first determined the integration efficiency of PB, H2 and H3 independent of dCas9. HEK293 cells were cotransfected with 500 ng of helper plasmid and 1500 ng of donor plasmid in 6-well dishes. After 24 h, TurboGFP expression was measured using flow cytometry to determine transfection efficiency. Expression from transient plasmid transfection is typically undetectable after 2 weeks of culture. Therefore, we measured the percentage of TurboGFP after 3 weeks of culture and corrected for the transfection efficiency to calculate the integration efficiency. We measured high levels of integration for the hyperactive piggyBac helper, confirming previous reports (17–19) (Figure 2A). H2 efficiency was reduced by 33% compared to PB. H3 efficiency was dramatically reduced by 87% and 91% compared to H2 and PB, respectively. The reduction in integration observed for H2 and H3 follows the expected pattern for a transposase with impaired ability to bind DNA.
Figure 2.
Transposase integration and guide RNA efficiency. (A) Integration efficiency of helper plasmids devoid of dCas9. TurboGFP expression was measured by flow cytometry 24 h and 3 weeks after transfection (n ≥ 3, ±SEM). Integration efficiency was calculated by dividing the percentage of cells glowing after 3 weeks by the percentage of cells glowing after 24 h. ANOVA followed by Bonferroni post-test analysis revealed the difference between each sample was significant (*P < 0.05). (B) Comparison of integration efficiency between dCas9 fusion constructs containing either 4, 8 or no guides (n ≥ 3, ±SEM). ANOVA followed by Bonferroni post-test analysis revealed that the dCas9-H3 variant was significantly different than the dCas9-PB variant for each guide combination (*P < 0.05). (C) The location and orientation of guide RNA binding sites in the CCR5 gene. Arrows indicate the primers used to amplify the target region for the T7E1 assay. (D) Guide efficiency was determined by T7E1 assay. Individual guide RNA expression plasmids were each cotransfected with a catalytically active Cas9 expression plasmid. Products from genomic PCR were used to form heteroduplexes and were digested with T7E1. Digested products were gel imaged and quantified using a bioanalyzer. Red dots indicate the expected size for each digest. Percent of indel occurrence for each guide is listed below the gel image.
We next compared integration efficiencies between PB, H2 and H3 fused to dCas9. The presence or absence of guides did not impact efficiency (Figure 2B). Overall, fusion of dCas9 resulted in average efficiencies of ∼10%, 8% and 4% for PB, H2 and H3, respectively.
We designed eight gRNAs with binding sites tiled across the first intron of the CCR5 gene (Figure 2C and Supplementary Data). Efficiencies of the gRNAs were determined by the T7E1 assay. Guide expression plasmids were individually cotransfected with a catalytically active Cas9 expression plasmid in HEK293 cells. Products obtained from genomic PCR were reannealed and digested with T7EI. The ratio of resulting products was used to calculate the percent of indel occurrence for each gRNA. All guides were found to be active on their target sequence with efficiencies varying from 14% to 43% (Figure 2D). Taken together, our transposase vectors actively integrate into the genome and our gRNAs target their intended sequences.
RNA-guided transposition to the genome
We tested the ability of our dCas9-piggyBac fusion constructs to deliver a transgene to the CCR5 safe harbor locus. The donor plasmid was cotransfected with dCas9-PB, dCas9-H2 or dCas9-H3 each with 0, 4 or 8 guides, in duplicate. Following 3 weeks of antibiotic selection, the cultures were lysed for use as template for genomic PCR. To improve the chances of recovering insertions, three dilutions of the lysate template were used. Primary PCR primers were designed to extend out from each side of the transposon. Four additional primary PCR primers were designed to extend towards the target site in CCR5 (two on each side). Individual PCR reactions were performed using all pair-wise primer combinations (eight total). Products arising from the primary PCR reactions were used as template for nested PCR. Sequenced products included the flanking TRE of the transposon, the canonical TTAA sequence at the junction and the genomic sequence flanking the insertion site.
A total of 23 insert junctions were recovered (Table 1 and Supplementary Data). Each inserted transposon is expected to be flanked on either side by a junction with the genome. We recovered six transposon insertions that contained junctions on either side (12 junctions total) as well as another 11 transposon insertions that contained one of the two junctions (11 junctions total). Therefore, a minimum of 17 independent insertion sites were confirmed. No insertions were recovered from control transfections devoid of guides. Transfections of dCas9-PB-4guide and dCas9-H2-4guide also did not result in insertions. Across the different transfections, the number of insertions in the forward or reverse orientation was similar. Over half the insertions originated from the dCas9-H2-8guide transfection. All but one insertion occurred within 400 bp of the guide target sequence (Figure 3A and Supplementary Data). Surprisingly, 59% of recovered insertions occurred at a single TTAA hotspot. Because two identical insertions at the same TTAA would be counted as a single insertion, the frequency of insertions at this hotspot could in fact be higher. Interestingly, over one-third of the insertion junctions were imprecise. Errors included small deletions within the TTAA or edge of the TRE as well as duplications of the TTAA (Table 1). This represents the first evidence that RNA can be used to direct transposition to a desired sequence.
Table 1.
Transposon junctions recovered in CCR5
| Genome | TTAA | Transposon | Junction | Helper plasmid | Insertion | T RE | Opposite side sequenced? | Orientation | bp from hotspot | Location on ch3 |
|---|---|---|---|---|---|---|---|---|---|---|
| aactcttaagataatcagaattttc | ttaa | ccctagaaagataatcatattgtga | 1 | dCas9-H3-4guide (a) | Precise | 3' | No | For | 0 | 46370544 |
| aactcttaagataatcagaattttc | ttaa | ccctagaaagataatcatattgtga | 2 | dCas9-H3-4guide (b) | Precise | 3' | Yes | For | 0 | 46370544 |
| gcttttcaacagtaaggctaaaagg | ttaa | ccctagaaagatagtctgcgtaaaa | 3 | dCas9-H3-4guide (b) | Precise | 5' | Yes | For | 0 | 46370544 |
| gcttttcaacagtaaggctaaaagg | ttaa | ccctagaaagatagtctgcgtaaaa | 4 | dCas9-PB-8guide (b) | Precise | 5' | No | For | 0 | 46370544 |
| gcttttcaacagtaaggctaaaagg | tta- | ccctagaaagatagtctgcgtaaaa | 5 | dCas9-H2-8guide (a) | Imprecise | 5' | No | For | 0 | 46370544 |
| agttggctttaattaatagcaactc | t‐‐‐ | ccctagaaagatagtctgcgtaaaa | 6 | dCas9-H2-8guide (a) | Imprecise | 5' | Yes | Rev | Upstream 20 | 46370522 |
| aaaggttaagaaaattctgattatc | ttaa | ccctagaaagataatcatattgtga | 7 | dCas9-H2-8guide (a) | Precise | 3' | Yes | Rev | Upstream 20 | 46370522 |
| ctgagctgcaccatgcttgacccag | ‐taa | ccctagaaagatagtctgcgtaaaa | 8 | dCas9-H2-8guide (a) | Imprecise | 5' | No | Rev | Downstream 2037 | 46372581 |
| aacaccaagtgctcatacaattatc | ttaa | ccctagaaagataatcatattgtga | 9 | dCas9-H2-8guide (a) | Precise | 3' | No | For | Downstream 560 | 46371103 |
| gcttttcaacagtaaggctaaaagg | ttaa | ccctagaaagatagtctgcgtaaaa | 10 | dCas9-H2-8guide (b) | Precise | 5' | Yes | For | 0 | 46370544 |
| aactcttaagataatcagaattttc | ttaa | ‐‐‐‐‐‐aaagatagtctgcgtaaaa | 11 | dCas9-H2-8guide (b) | Imprecise | 5' | No | Rev | 0 | 46370544 |
| aactcttaagataatcagaattttc | ttaa | ccctagaaagatagtctgcgtaaaa | 12 | dCas9-H2-8guide (b) | Precise | 5' | Yes | Rev | 0 | 46370544 |
| gcttttcaacagtaaggctaaaagg | ttaa | ccctagaaagataatcatattgtga | 13 | dCas9-H2-8guide (b) | Precise | 3' | Yes | Rev | 0 | 46370544 |
| aactcttaagataatcagaattttc | ttaa | ccctagaaagataatcatattgtga | 14 | dCas9-H2-8guide (b) | Precise | 3' | Yes | For | 0 | 46370544 |
| ccagagatctattctctagcttatt | ttaa | ccctagaaagatagtctgcgtaaaa | 15 | dCas9-H2-8guide (b) | Precise | 5' | Yes | Rev | Downstream 258 | 46370802 |
| aacagttcttccttttaagttgagc | ttaa | ccctagaaagataatcatattgtga | 16 | dCas9-H2-8guide (b) | Precise | 3' | Yes | Rev | Downstream 258 | 46370802 |
| atacaattatcttaaaatataatct | ttaaa | ccctagaaagatagtctgcgtaaaa | 17 | dCas9-H2-8guide (b) | Imprecise | 5' | No | Rev | Downstream 574 | 46371118 |
| aactcttaagataatcagaattttc | ‐aaa | ccctagaaagatagtctgcgtaaaa | 18 | dCas9-H3-8guide (a) | Imprecise | 5' | No | Rev | 0 | 46370544 |
| cagctaagactcatctctctgcaaa | ‐aga | ‐‐ctagaaagatagtctgcgtaaaa | 19 | dCas9-H3-8guide (a) | Imprecise | 5' | No | For | Downstream 716 | 46371259 |
| aactcttaagataatcagaattttc | ttaa | ccctagaaagatagtctgcgtaaaa | 20 | dCas9-H3-8guide (b) | Precise | 5' | Yes | Rev | 0 | 46370544 |
| gcttttcaacagtaaggctaaaagg | ttaa | ccctagaaagataatcatattgtga | 21 | dCas9-H3-8guide (b) | Precise | 3' | Yes | Rev | 0 | 46370544 |
| gcttttcaacagtaaggctaaaagg | ttaa | ccctagaaagatagtctgcgtaaaa | 22 | dCas9-H3-8guide (b) | Precise | 5' | No | For | 0 | 46370544 |
| gtagagggggatcctggacttcaca | ttaattaa | ccctagaaagataatcatattgtga | 23 | dCas9-H3-8guide (b) | Imprecise | 3' | No | Rev | Upstream 90 | 46370454 |
| aactcttaagataatcagaattttc | ttaattaa | ccctagaaagatagtctgcgtaaaa | 24 | 293-c1 | Imprecise | 5' | No | Rev | 0 | 46370544 |
| dCas9-H2-8guide | ||||||||||
| agttggctttaattaatagcaactc | ttaa | ccctagaaagatagtctgcgtaaaa | 25 | 293-c2 | Precise | 5' | No | Rev | Upstream 20 | 46370522 |
| dCas9-H2-8guide |
Figure 3.
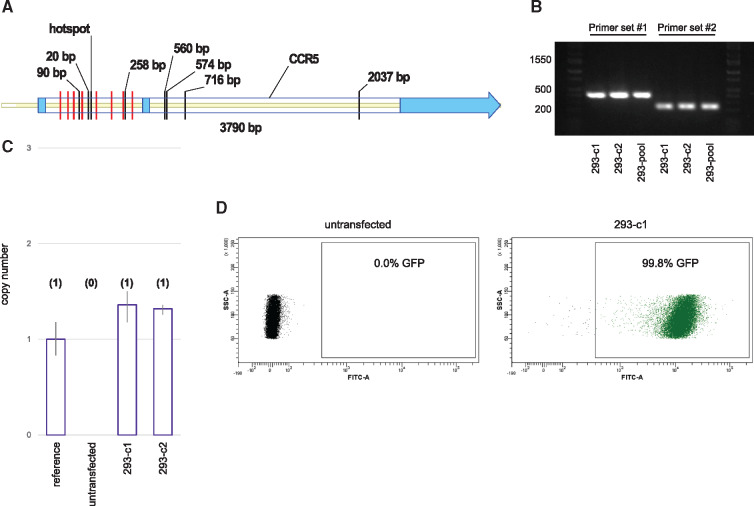
RNA-guided transposition to the genome. (A) Recovered insertion sites in the CCR5 gene. Helper and donor plasmids were transfected into HEK293 cells and selected for 3 weeks. Genomic PCR was used to recover insertion sites. Ten independent insertions were found at a single TTAA hotspot. Labeled black lines indicate the location and distance from the hotspot for alternate insertion sites. Multiple independent insertions at the same TTAA for a given transfection would appear identical and be counted as a single insertion. Red lines indicate guide target sequences. CCR5 exons are shown in light blue. (B) Genomic PCR demonstrating clonal cell lines 293-c1 and 293-c2 are positive for targeted insertion to CCR5. The cell lines were derived from a positively identified well containing about 50 colonies called 293-pool. Two CCR5-directed primer sets were used. The expected sizes for a hotspot insertion using primer sets #1 and #2 were 392 and 229 bp, respectively. (C) Transposon copy number for clones 293-c1 and 293-c2. Quantitative PCR predictions were calibrated using a reference HEK293 cell line known to contain a single-copy transposon. Predicted copy number is shown in parenthesis. (D) CCR5 targeted cell lines maintained stable transgene expression following 13 weeks of culture. Flow cytometry analysis displaying GFP positive events for both untransfected HEK293 cells and an expansion of clone 293-c1.
Isolation of CCR5 targeted clonal cell lines
We chose dCas9-H2-8guide as the favored helper plasmid based on the high number of insertions recovered compared to the other five vectors tested. In order to demonstrate that we were capable of deriving clonal cell lines containing a single targeted insertion to CCR5 using dCas9-H2-8guide, we repeated the helper/donor transfections in HEK293 cells in duplicate. Following 3 weeks of antibiotic selection, cells derived from the two independent transfections were each plated into 96 individual wells (192 wells total). One week later, colonies were counted, and each well was found to contain an average of 50 colonies. We estimated 9600 total colonies between the two transfections. The 192 wells from the two plates were each resuspended, and a fraction of the cells were removed and analyzed by genomic PCR for the presence of insertions to CCR5. The first plate was found to have five positive wells and the second plate was found to have a single positive well. In total, six positive wells were identified, and we assumed that each positive well contained a single positive colony. Therefore, 6/9600 or 0.06% of total selected cells contained targeted insertions to CCR5 (Table 2). Because the integration efficiency of dCas9-H2-8guide is 10% (Figure 2B), the efficiency for total transfected cells is 10-fold less. Sequencing of the genomic PCR products (junctions 24 and 25) revealed that the insertions occurred at or near the hotspot (Table 1).
Table 2.
Targeted cells recovered from repeat dCas9-H2-8guide transfections
| dCas9-H2-8guide transfection #1 | dCas9-H2-8guide transfection #2 | dCas9-H2-8guide combined | |
|---|---|---|---|
| Total cells screened | 4800 | 4800 | 9600 |
| Positive wells | 5 | 1 | 6 |
| % Targeted cells | 0.1 | 0.02 | 0.06 |
Next, two of the positive wells from the first plate were single-cell sorted and 157 single-cell expansions were screened by genomic PCR. We estimated that 1/50 of the cells within the positive wells contained the insert. By screening 157 clones originating from pools of 50 cells, we estimated that three positive wells would be identified. In practice, we successfully isolated two positive clonal lines, referred to as 293-c1 and 293-c2 (Figure 3B).
Additional, unidentified transposon insertions at off-target sequences would not be detected using CCR5-specific PCR primers. We reasoned that cells containing a single insertion, for which the location is known, would be absent for insertions at undesired sequences. In order to determine the presence or absence of off-target insertions, we performed a quantitative PCR-based copy number assay and detected a single insertion for each of our clonal lines (Figure 3C).
It is possible that chromosomal position effects may silence the transgene at the CCR5 locus. To assess if TurboGFP expression was stable in our targeted clonal lines, the cells were cultured for 13 weeks and analyzed by flow cytometry (Figure 3D). Populations expanded from clones 293-c1 and 293-c2 were found to be 99.8% and 99.6% positive, respectively. Together, these data indicate that RNA-guided transposition can be used to obtain clonal cell lines containing a targeted single insertion of a stably expressed transgene.
Discussion
The ability to manipulate genomes in a customizable fashion requires a toolbox of technologies capable of different functions. Presently, targeted integration of large DNA sequences is inefficient, even with CRISPR genome editing. One potential solution to this problem is to engineer targetable transposases capable of directing insertions to defined genomic sequences. The characterization of novel RNA-guided piggyBac vectors described here represents an important step towards the generation of improved targeting vectors. For our experiments, the dCas9 DBD was fused to the carboxyl terminus of three piggyBac variants: PB, H2 and H3. PB contained seven mutations designed to increase the efficiency of insertion and has been shown to be the most efficient transposase available for use in human cells (17, 18). We human codon-optimized PB and introduced either two or three mutations in the native DBD to generate H2 (R372A/D450N) or H3 (R372A/K375A/D450N), respectively. A total of eight unique gRNAs designed to the human CCR5 safe harbor sequence were used to direct insertions of the dCas9-piggyBac fusion constructs.
We stably transfected helper transposase expression plasmids containing either 4-guide or 8-guide combinations with a transposon donor plasmid and used genomic PCR to recover targeted insertions to CCR5. Despite millions of potential TTAA sequences available for insertion throughout the genome, we were able to recover inserted transposons adjacent to the gRNA target sequence. Control transfections without gRNA did not result in any targeted insertions. This was expected, given that the likelihood of an insertion randomly occurring at the target sequence is extremely low (1.5 × 10−6 for a 5 kb potential target sequence within a 3234 mb genome). We were only able to recover one junction of the transposon for several insertions, and we found it necessary to perform nested PCR on various dilutions of template, indicating that the PCR was underrepresenting the true number of insertions. Interestingly, we recovered the greatest number of insertions for the dCas9-H2-8guide and dCas9-H3-8guide variants. This may be due to reduced binding at off-target sequences by the native DBD which would be expected to reduce off-target insertions. Because the number of transposons for a given cell is limited, reduced off-target insertion could make available more transposons for on-target insertion. Moreover, transposases structurally related to piggyBac act as dimers and the orientation of piggyBac DBDs suggests that the transposase also acts as a dimer (15). The clustering of individual piggyBac monomers may support dimerization and facilitate integration at the target sequence. While this may be achieved with a minimum of two guide binding sites, an increased number of binding sites may contribute to clustering of monomers near the target sequence and increase the likelihood for a given transposase to encounter other monomers. An alternate explanation for why the 8-guide vectors outcompeted the 4-guide vectors is that the wider spacing across the target sequence for the 8-guide vectors may have provided more opportunities for a favorable orientation of the tethered transposase to perform insertion. The close proximity of the 4-guide binding sites may have been too restrictive. Future studies are warranted to determine the ideal spacing of gRNA binding sites.
We did not recover insertions from either the dCas9-PB-4guide or dCas9-H2-4guide helpers. The reason for this is unclear. One explanation is that the dCas9-PB-4guide helper retains full ability of the transposase to bind independently, without the need for dCas9, which may lead to higher number of insertions off-target. Both helpers used less guides (4 instead of 8), which appeared to reduce the number of targeted insertions for all helpers tested.
Over half of the insertions recovered were located at a single TTAA hotspot. Future studies are needed to determine what characteristics of both the helper protein and target sequence support this interaction. Because multiple insertions to the same site appear identical, hotspot insertions may have been more frequent than reported. This hotspot was not targeted in a previous study using a TALE to direct piggyBac to CCR5 (36). Because the TALE DBD bound only 2 bp from the hotspot, it is possible that steric constraints would not allow for an insertion within such close proximity. The present study focused on a single target sequence. The relative availability of this site compared to other sequences remains unknown. Future studies are warranted to determine if alternate target sites support RNA-guided transposition.
Repeat transfections using dCas9-H2-8guide were performed to demonstrate the ability of this approach at isolating single clones containing single, targeted insertions. For each of the two transfections, a two-step pooling approach was used in which a single 96-well plate containing 50 colonies per well was first screened. Each of the plates contained at least one positive well. Overall, six pooled wells were positively identified from the two 96-well plates. This corresponds to approximately 0.06% of total cells. Clonal lines were derived during a second step by plating single cells from the positively identified pooled wells. Two clonal cell lines were identified using this pooling/screening approach. We determined that each of these lines contained a single copy of the transposon. These results are based on a quantitative PCR assay used to detect the number of genomic copies of the neomycin gene. It is possible that detection by this method is inaccurate or that a partial insert devoid of neomycin could occur, which would underestimate the number of insertions. Finally, we demonstrated that each of these cell lines expressed the transgene long-term.
Previously, Li et al. attempted to use both R372A/D450N and R372A/K375A/D450N mutations to improve the specificity of a zinc-finger/piggyBac fusion (47). The group showed that R372A/K375A/D450N mutations completely inhibited integration of both unfused piggyBac and piggyBac fused to the zinc finger. Interestingly, they showed that R372A/D450N mutations dramatically reduced integration of unfused piggyBac but that integration activity was rescued by fusion of the zinc finger. A potential mechanism for this rescue was proposed in which piggyBac alone is inhibited from integrating due to the R372A/D450N mutations and that the fused zinc finger promotes DNA binding and subsequent insertion to nearby sequences. Unfortunately, a genome-wide search did not recover insertions occurring near the zinc finger binding sites. The reason for this is unknown. Because genome targeting was not yet demonstrated with a native piggyBac fusion to these zinc fingers, there may be several explanations unrelated to the DBD mutations that could account for the inability of this vector to target the genome. For example, the linker between the DBD and transposase may not support the needed steric alignment required for piggyBac catalytic activity. Additionally, the zinc finger DBD might not have had the required level of specificity to the target sequence or sufficient affinity to the DNA.
Previously, Luo et al. (37) compared genome targeting efficiencies between zinc finger, TALE and dCas9 DBD fusions to piggyBac. The group used a system for optimizing targeting technologies in which the vectors intentionally disrupt the endogenous HPRT gene located on chrX. Selection with 6-thioguanine (6-TG) kills cells expressing HPRT, thus successful targeting and disruption of HPRT leads to cell survival. They were successful at targeting HPRT with both zinc finger and TALE piggyBac fusion vectors but not with the dCas9 piggyBac fusion. The 1377 aa dCas9 protein is larger than both zinc finger (176 aa) and TALE DBDs (776 aa), necessitating careful choice of the linker between the dCas9 and piggyBac. In our hands, shorter linkers than the 23 aa linker we used in this work failed to mediate genome targeting (unpublished observations). Steric hindrance may impede the transposase’s ability to integrate and may limit potential target sequences.
Yusa et al. (17) reported that the hyperactive variant used in our PB helper has an error frequency of 0.74%. We observed an approximate 47-fold increase in errors for our dCas9-fused transposase. These errors are likely caused by physical constraints imposed by the linker and dCas9 DBD. By tethering piggyBac to the target sequence, free movement of the transposase might be disrupted, leading to impairment of integration. Tethering might also result in the repeated excision and reintegration of the transposon which may increase the frequency of errors. In general, excision of the transposon, especially a transposon located at the target sequence, is undesirable. A potential improvement to the system is a re-excision deficient piggyBac vector. By moving the majority of the TRE sequence outside of the transposon to the non-inserted backbone of the donor plasmid, the size of the inserted TRE can be reduced with minimal loss of transposition efficiency (52, 53). Once inserted into the genome, the transposon lacks full-length TREs and cannot be efficiently excised.
Because our approach requires the insertion of the flanking TREs, it cannot be used for precise sequence exchange and would not be suitable for correcting coding sequences. Instead, this method would be best suited for delivering large inserts away from undesirable sequences. Insertions into intronic sequences could also be feasible. The errors that we detected at the junction sites were confined to the terminal edge of the TREs. Therefore, although unpredictable, these modifications are not expected to negatively impact the inserted DNA within the transposon.
Gogol-Doring et al. (54) demonstrated that piggyBac interacts with BRD4 to bias insertions near BRD4 binding sites in the genome. It is possible that BRD4 competes with dCas9 in relocating piggyBac in our experiments. Future studies are needed to identify the BRD4 binding residues and to determine if disruption of BRD4 binding reduces off-target insertions.
The low specificity of targeting represents the greatest challenge to this technology. Future studies focusing on improving the specificity of RNA-guided transposition are warranted. Despite adding R372A/D450N and R372A/K375A/D450N mutations, the transposase in these experiments remained capable of DNA binding and did not solely utilize dCas9 to locate to the genome. It is likely that native DNA-binding activity of piggyBac permits off-target insertion of the majority of available transposons. Future steps for this technology include extensive characterization of potential mutations aimed at generating a novel transposase devoid of autonomous binding. This might be achieved by a directed evolution strategy aimed at screening for random or rationally designed mutations that encourage the co-dependence of dCas9 and piggyBac for insertion. Therefore, although we measured low specificity, these vectors exhibit the baseline activity that is necessary for future improvements.
Yusa et al. (17) used directed evolution to identify seven mutations that improved piggyBac excision by 17-fold and integration by 9-fold over wildtype. It is likely that hyperactive variants exceeding this efficiency are possible. This may be especially beneficial to a site-specific transposase because fewer insertion sites are available. Additionally, a hyperactive variant may overcome steric hindrance imposed by the linker and dCas9.
The dCas9 DBD can easily be designed to bind nearly any target sequence, facilitating applications by an expanded number of research groups. Ultimately, we hope to develop an improved vector that exclusively integrates at the target sequence. This would enable targeted delivery of large cargos such as multiple gene cassettes or long endogenous sequences. A strictly site-specific, RNA-guided transposase would have important applications including transgenic animal generation, modification of cell lines for research and diagnostics or delivery of therapeutic cargo to a designated location in the genome. The proof-of-concept that dCas9 can direct piggyBac genome insertions is a crucial step for this early stage technology.
Supplementary Material
Acknowledgments
We thank Dr Stefan Moisyadi for expert advice in the design and trouble-shooting of the experiments. We thank Dr David J. Segal, UC Davis, for expert advice on experimental design and for critical reading of the manuscript. We would also like to thank Dr Allan Bradley and the Sanger Institute for the hyperactive piggyBac plasmid.
Funding
The National Institutes of Health [8P20GM103457 and 1R21GM132779-01 to J.B.O.]. Funding for open access charge: National Institutes of Health.
Conflict of interest statement. J.B.O. has filed a patent based on the findings of this study.
References
- 1. Naldini L. (2015) Gene therapy returns to centre stage. Nature, 526, 351–360. [DOI] [PubMed] [Google Scholar]
- 2. Dunbar C.E., High K.A., Joung J.K., Kohn D.B., Ozawa K., Sadelain M. (2018) Gene therapy comes of age. Science, 359, eaan4672. [DOI] [PubMed] [Google Scholar]
- 3. Daniel R., Smith J.A. (2008) Integration site selection by retroviral vectors: molecular mechanism and clinical consequences. Hum. Gene Ther. 19, 557–568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Mitchell R.S., Beitzel B.F., Schroder A.R., Shinn P., Chen H., Berry C.C., Ecker J.R., Bushman F.D. (2004) Retroviral DNA integration: ASLV, HIV, and MLV show distinct target site preferences. PLoS Biol., 2, E234.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Hacein-Bey-Abina S., Garrigue A., Wang G.P., Soulier J., Lim A., Morillon E., Clappier E., Caccavelli L., Delabesse E., Beldjord K.. et al. (2008) Insertional oncogenesis in 4 patients after retrovirus-mediated gene therapy of SCID-X1. J. Clin. Invest., 118, 3132–3142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Howe S.J., Mansour M.R., Schwarzwaelder K., Bartholomae C., Hubank M., Kempski H., Brugman M.H., Pike-Overzet K., Chatters S.J., de Ridder D.. et al. (2008) Insertional mutagenesis combined with acquired somatic mutations causes leukemogenesis following gene therapy of SCID-X1 patients. J. Clin. Invest., 118, 3143–3150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Cox D.B., Platt R.J., Zhang F. (2015) Therapeutic genome editing: prospects and challenges. Nat. Med., 21, 121–131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Lieber M.R. (2010) The mechanism of double-strand DNA break repair by the nonhomologous DNA end-joining pathway. Annu. Rev. Biochem., 79, 181–211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Fung H., Weinstock D.M. (2011) Repair at single targeted DNA double-strand breaks in pluripotent and differentiated human cells. PloS One, 6, e20514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Orthwein A., Noordermeer S.M., Wilson M.D., Landry S., Enchev R.I., Sherker A., Munro M., Pinder J., Salsman J., Dellaire G.. et al. (2015) A mechanism for the suppression of homologous recombination in G1 cells. Nature, 528, 422–426. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 11. Haapaniemi E., Botla S., Persson J., Schmierer B., Taipale J. (2018) CRISPR-Cas9 genome editing induces a p53-mediated DNA damage response. Nat. Med., 24, 927–930. [DOI] [PubMed] [Google Scholar]
- 12. Ihry R.J., Worringer K.A., Salick M.R., Frias E., Ho D., Theriault K., Kommineni S., Chen J., Sondey M., Ye C.. et al. (2018) p53 inhibits CRISPR-Cas9 engineering in human pluripotent stem cells. Nat. Med., 24, 939–946. [DOI] [PubMed] [Google Scholar]
- 13. Kosicki M., Tomberg K., Bradley A. (2018) Repair of double-strand breaks induced by CRISPR-Cas9 leads to large deletions and complex rearrangements. Nat. Biotechnol., 36, 765–771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Mitra R., Fain-Thornton J., Craig N.L. (2008) piggyBac can bypass DNA synthesis during cut and paste transposition. EMBO J., 27, 1097–1109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Morellet N., Li X., Wieninger S.A., Taylor J.L., Bischerour J., Moriau S., Lescop E., Bardiaux B., Mathy N., Assrir N.. et al. (2018) Sequence-specific DNA binding activity of the cross-brace zinc finger motif of the piggyBac transposase. Nucleic Acids Res., 46, 2660–2677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Woodard L.E., Wilson M.H. (2015) piggyBac-ing models and new therapeutic strategies. Trends Biotechnol., 33, 525–533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Yusa K., Zhou L., Li M.A., Bradley A., Craig N.L. (2011) A hyperactive piggyBac transposase for mammalian applications. Proc. Natl. Acad Sci. USA, 108, 1531–1536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Doherty J.E., Huye L.E., Yusa K., Zhou L., Craig N.L., Wilson M.H. (2012) Hyperactive piggyBac gene transfer in human cells and in vivo. Hum. Gene Ther., 23, 311–320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Marh J., Stoytcheva Z., Urschitz J., Sugawara A., Yamashiro H., Owens J.B., Stoytchev I., Pelczar P., Yanagimachi R., Moisyadi S. (2012) Hyperactive self-inactivating piggyBac for transposase-enhanced pronuclear microinjection transgenesis. Proc. Natl. Acad Sci. USA, 109, 19184–19189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Eckermann K.N., Ahmed H.M.M., KaramiNejadRanjbar M., Dippel S., Ogaugwu C.E., Kitzmann P., Isah M.D., Wimmer E.A. (2018) Hyperactive piggyBac transposase improves transformation efficiency in diverse insect species. Insect Biochem. Mol. Biol., 98, 16–24. [DOI] [PubMed] [Google Scholar]
- 21. Hareendran S., Balakrishnan B., Sen D., Kumar S., Srivastava A., Jayandharan G.R. (2013) Adeno-associated virus (AAV) vectors in gene therapy: immune challenges and strategies to circumvent them. Rev. Med. Virol., 23, 399–413. [DOI] [PubMed] [Google Scholar]
- 22. Basner-Tschakarjan E., Mingozzi F. (2014) Cell-mediated immunity to AAV vectors, evolving concepts and potential solutions. Front. Immunol., 5, 350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Rogers G.L., Martino A.T., Aslanidi G.V., Jayandharan G.R., Srivastava A., Herzog R.W. (2011) Innate immune responses to AAV vectors. Front. Microbiol., 2, 194.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Boutin S., Monteilhet V., Veron P., Leborgne C., Benveniste O., Montus M.F., Masurier C. (2010) Prevalence of serum IgG and neutralizing factors against adeno-associated virus (AAV) types 1, 2, 5, 6, 8, and 9 in the healthy population: implications for gene therapy using AAV vectors. Hum. Gene Ther., 21, 704–712. [DOI] [PubMed] [Google Scholar]
- 25. Mingozzi F., High K.A. (2011) Therapeutic in vivo gene transfer for genetic disease using AAV: progress and challenges. Nat. Rev. Genet., 12, 341–355. [DOI] [PubMed] [Google Scholar]
- 26. Li M.A., Turner D.J., Ning Z., Yusa K., Liang Q., Eckert S., Rad L., Fitzgerald T.W., Craig N.L., Bradley A. (2011) Mobilization of giant piggyBac transposons in the mouse genome. Nucleic Acids Res., 39, e148.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Cullis P.R., Hope M.J. (2017) Lipid nanoparticle systems for enabling gene therapies. Mol. Ther., 25, 1467–1475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Foldvari M., Chen D.W., Nafissi N., Calderon D., Narsineni L., Rafiee A. (2016) Non-viral gene therapy: gains and challenges of non-invasive administration methods. J. Control. Release, 240, 165–190. [DOI] [PubMed] [Google Scholar]
- 29. Slivac I., Guay D., Mangion M., Champeil J., Gaillet B. (2017) Non-viral nucleic acid delivery methods. Expert Opin. Biol. Ther., 17, 105–118. [DOI] [PubMed] [Google Scholar]
- 30. Yin H., Kanasty R.L., Eltoukhy A.A., Vegas A.J., Dorkin J.R., Anderson D.G. (2014) Non-viral vectors for gene-based therapy. Nat. Rev. Genet., 15, 541–555. [DOI] [PubMed] [Google Scholar]
- 31. Zylberberg C., Gaskill K., Pasley S., Matosevic S. (2017) Engineering liposomal nanoparticles for targeted gene therapy. Gene Ther., 24, 441–452. [DOI] [PubMed] [Google Scholar]
- 32. Anderson C.D., Urschitz J., Khemmani M., Owens J.B., Moisyadi S., Shohet R.V., Walton C.B. (2013) Ultrasound directs a transposase system for durable hepatic gene delivery in mice. Ultrasound Med. Biol., 39, 2351–2361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Tipanee J., Chai Y.C., VandenDriessche T., Chuah M.K. (2017) Preclinical and clinical advances in transposon-based gene therapy. Biosci. Rep., 37, BSR20160614.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Nakamura S., Ishihara M., Watanabe S., Ando N., Ohtsuka M., Sato M. (2018) Intravenous delivery of piggyBac transposons as a useful tool for liver-specific gene-switching. Int. J. Mol. Sci., 19, 3452.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Skipper K.A., Nielsen M.G., Andersen S., Ryo L.B., Bak R.O., Mikkelsen J.G. (2018) Time-restricted piggyBac DNA transposition by transposase protein delivery using lentivirus-derived nanoparticles. Mol. Ther. Nucleic Acids, 11, 253–262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Owens J.B., Mauro D., Stoytchev I., Bhakta M.S., Kim M.S., Segal D.J., Moisyadi S. (2013) Transcription activator like effector (TALE)-directed piggyBac transposition in human cells. Nucleic Acids Res., 41, 9197–9207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Luo W., Galvan D.L., Woodard L.E., Dorset D., Levy S., Wilson M.H. (2017) Comparative analysis of chimeric ZFP-, TALE- and Cas9-piggyBac transposases for integration into a single locus in human cells. Nucleic Acids Res., 45, 8411–8422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Wang H., Mayhew D., Chen X., Johnston M., Mitra R.D. (2011) Calling Cards enable multiplexed identification of the genomic targets of DNA-binding proteins. Genome Res., 21, 748–755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Wang H., Mayhew D., Chen X., Johnston M., Mitra R.D. (2012) “Calling cards” for DNA-binding proteins in mammalian cells. Genetics, 190, 941–949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Komor A.C., Badran A.H., Liu D.R. (2017) CRISPR-based technologies for the manipulation of eukaryotic genomes. Cell, 169, 559.. [DOI] [PubMed] [Google Scholar]
- 41. Qi L.S., Larson M.H., Gilbert L.A., Doudna J.A., Weissman J.S., Arkin A.P., Lim W.A. (2013) Repurposing CRISPR as an RNA-guided platform for sequence-specific control of gene expression. Cell, 152, 1173–1183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Jinek M., Chylinski K., Fonfara I., Hauer M., Doudna J.A., Charpentier E. (2012) A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity. Science, 337, 816–821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Knott G.J., Doudna J.A. (2018) CRISPR-Cas guides the future of genetic engineering. Science, 361, 866–869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Owens J.B., Urschitz J., Stoytchev I., Dang N.C., Stoytcheva Z., Belcaid M., Maragathavally K.J., Coates C.J., Segal D.J., Moisyadi S. (2012) Chimeric piggyBac transposases for genomic targeting in human cells. Nucleic Acids Res., 40, 6978–6991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Kettlun C., Galvan D.L., George A.L. Jr., Kaja A., Wilson M.H. (2011) Manipulating piggyBac transposon chromosomal integration site selection in human cells. Mol. Ther., 19, 1636–1644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Ammar I., Gogol-Doring A., Miskey C., Chen W., Cathomen T., Izsvak Z., Ivics Z. (2012) Retargeting transposon insertions by the adeno-associated virus Rep protein. Nucleic Acids Res., 40, 6693–6712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Li X., Burnight E.R., Cooney A.L., Malani N., Brady T., Sander J.D., Staber J., Wheelan S.J., Joung J.K., McCray P.B. Jr.. et al. (2013) piggyBac transposase tools for genome engineering. Proc. Natl. Acad. Sci. USA, 110, E2279–2287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Kleinstiver B.P., Pattanayak V., Prew M.S., Tsai S.Q., Nguyen N.T., Zheng Z., Joung J.K. (2016) High-fidelity CRISPR-Cas9 nucleases with no detectable genome-wide off-target effects. Nature, 529, 490–495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Garcia-Doval C., Jinek M. (2017) Molecular architectures and mechanisms of Class 2 CRISPR-associated nucleases. Curr. Opin. Struct. Biol., 47, 157–166. [DOI] [PubMed] [Google Scholar]
- 50. Kabadi A.M., Ousterout D.G., Hilton I.B., Gersbach C.A. (2014) Multiplex CRISPR/Cas9-based genome engineering from a single lentiviral vector. Nucleic Acids Res., 42, e147.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Guschin D.Y., Waite A.J., Katibah G.E., Miller J.C., Holmes M.C., Rebar E.J. (2010) A rapid and general assay for monitoring endogenous gene modification. Methods Mol. Biol., 649, 247–256. [DOI] [PubMed] [Google Scholar]
- 52. Troyanovsky B., Bitko V., Pastukh V., Fouty B., Solodushko V. (2016) The functionality of minimal piggyBac transposons in mammalian cells. Mol. Ther. Nucleic Acids, 5, e369.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Solodushko V., Bitko V., Fouty B. (2014) Minimal piggyBac vectors for chromatin integration. Gene Ther., 21, 1–9. [DOI] [PubMed] [Google Scholar]
- 54. Gogol-Doring A., Ammar I., Gupta S., Bunse M., Miskey C., Chen W., Uckert W., Schulz T.F., Izsvak Z., Ivics Z. (2016) Genome-wide profiling reveals remarkable parallels between insertion site selection properties of the MLV retrovirus and the piggyBac transposon in primary human CD4(+) T cells. Mol. Ther., 24, 592–606. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.