Significance
Secondary amplification of small RNA triggers is necessary to silence repetitive transposable elements (TEs) and thus maintain genome integrity. Despite their importance, these secondary small RNAs are produced by various mechanisms in different eukaryotes. How such a variety of mechanisms has evolved has not been thoroughly elucidated. In this report, we show that in the ciliate Tetrahymena, secondary small RNA production occurs from the eliminated sequences prior to their excision during the small RNA-directed programmed DNA elimination of TE-related sequences. This situation contrasts with the excision-dependent secondary small RNA production observed in another ciliate, Paramecium. This study therefore demonstrates the possibility of tracing the evolution of TE-targeting small RNA amplification by investigating DNA elimination mechanisms of ciliates.
Keywords: small RNA, transposon silencing, DNA elimination, ciliate, Tetrahymena
Abstract
The silencing of repetitive transposable elements (TEs) is ensured by signal amplification of the initial small RNA trigger, which occurs at distinct steps of TE silencing in different eukaryotes. How such a variety of secondary small RNA biogenesis mechanisms has evolved has not been thoroughly elucidated. Ciliated protozoa perform small RNA-directed programmed DNA elimination of thousands of TE-related internal eliminated sequences (IESs) in the newly developed somatic nucleus. In the ciliate Paramecium, secondary small RNAs are produced after the excision of IESs. In this study, we show that in another ciliate, Tetrahymena, secondary small RNAs accumulate at least a few hours before their derived IESs are excised. We also demonstrate that DNA excision is dispensable for their biogenesis in this ciliate. Therefore, unlike in Paramecium, small RNA amplification occurs before IES excision in Tetrahymena. This study reveals the remarkable diversity of secondary small RNA biogenesis mechanisms, even among ciliates with similar DNA elimination processes, and thus raises the possibility that the evolution of TE-targeting small RNA amplification can be traced by investigating the DNA elimination mechanisms of ciliates.
Signal amplification of the initial small RNA trigger is important to ensure the silencing of repetitive transposable elements (TEs). Curiously, secondary small RNA biogenesis occurs via various mechanisms that are coupled with distinct steps of TE silencing in different eukaryotes, such as nucleolytic cleavage of TE transcripts, recruitment of RNA-dependent RNA polymerase, and heterochromatin-directed transcription (1–5). To understand how such a variety of small RNA amplification mechanisms have evolved, it is important to compare evolutionarily closely related TE-silencing processes that have distinct secondary small RNA biogenesis mechanisms. This study raises the possibility that the programmed DNA elimination mechanisms of ciliated protozoa can be used for such a comparative evolutionary biology approach.
Like most other ciliates, each cell of Tetrahymena thermophila (hereinafter Tetrahymena) contains 2 types of nuclei: the transcriptionally inactive germ line micronucleus (MIC) and the transcriptionally active somatic macronucleus (MAC). During the sexual process of conjugation, the MIC produces new MICs and MACs, while the parental MAC is discarded. The MIC genome includes ∼12,000 internal eliminated sequences (IESs), which are removed from the newly formed MAC. IESs contain many potentially active TEs and their remnants (6). Each IES can be excised in linear or circular form. Because excised IESs in linear form are very short-lived (7, 8) while circularized IESs are stable for more than several hours (9), circularized IESs should be produced either directly during the excision process or from linear IESs immediately after their excision.
Small RNAs play a key role in the DNA elimination in Tetrahymena (10–12). At early conjugation stages (∼2–3 h postmixing [hpm]), ∼60% of IESs, known as type A IESs, and their genomic surroundings are bidirectionally transcribed in the MIC (13, 14), and the transcripts are processed to ∼26- to 32-nt primary small RNAs known as early-scnRNAs (15, 16). Early-scnRNAs are loaded into the Piwi-clade Argonaute protein Twi1p (17, 18) and move into the parental MAC, where those complementary to the MAC genome (i.e., non-IES sequences) are degraded by a process known as scnRNA selection (14, 19). The remaining IES-specific early-scnRNAs are then shuttled into the developing new MACs (∼7–8 hpm) and are believed to base pair with nascent transcripts from type A and type B IESs, the latter of which constitute ∼40% of all IESs and share repetitive sequences with type A IESs (20). These interactions recruit the Enhancer of zeste [E(z)]-like methyltransferase Ezl1p, which mediates the accumulation of trimethylations of histone H3 at lysine 9 (H3K9me3) and lysine 27 (H3K27me3) and the HP1-like protein Pdd1p to IESs (21, 22). This heterochromatin nucleation induces the production of ∼26- to 32-nt secondary small RNAs, known as late-scnRNAs (20), and the excision of IESs by the domesticated piggyBac transposase Tpb2p (23, 24). The flanking sequences are religated by nonhomologous end-joining (NHEJ) (25). Besides type A and type B IESs, there are ∼20 IESs in the genome that produce few scnRNAs and are removed by the other piggyBac transposases, Tpb1p and Tpb6p (26, 27).
The other ciliate, Paramecium tetraurelia (hereinafter Paramecium), exhibits a very similar DNA elimination process, including Dicer-dependent scnRNA production (28), loading of small RNAs into Piwi-clade Argonaute proteins (29), scnRNA selection (28, 30), scnRNA- and an E(z)-like protein-dependent accumulation of H3K9me3 and H3K27me3 on IESs (31), IES excision by a piggyBac transposase (32, 33), and repair of the excision sites by NHEJ (34). It was recently reported that the secondary small RNAs, known as iesRNAs, in Paramecium are produced after the excision of IESs (35). The excised IESs are concatenated and circularized and are proposed to serve as templates for the production of precursors for iesRNAs in this ciliate (35, 36). On the other hand, although heterochromatin formation has been shown to be essential for the biogenesis of late-scnRNAs in Tetrahymena (20), it remains to be determined if the late-scnRNA production is dependent on IES excision. To examine whether the postexcision production of secondary small RNAs is conserved, we investigated the causality between the excision of IESs and the biogenesis of secondary small RNAs in Tetrahymena.
Results
Highly Sensitive Assay for Determining the Timing of De Novo IES Excision.
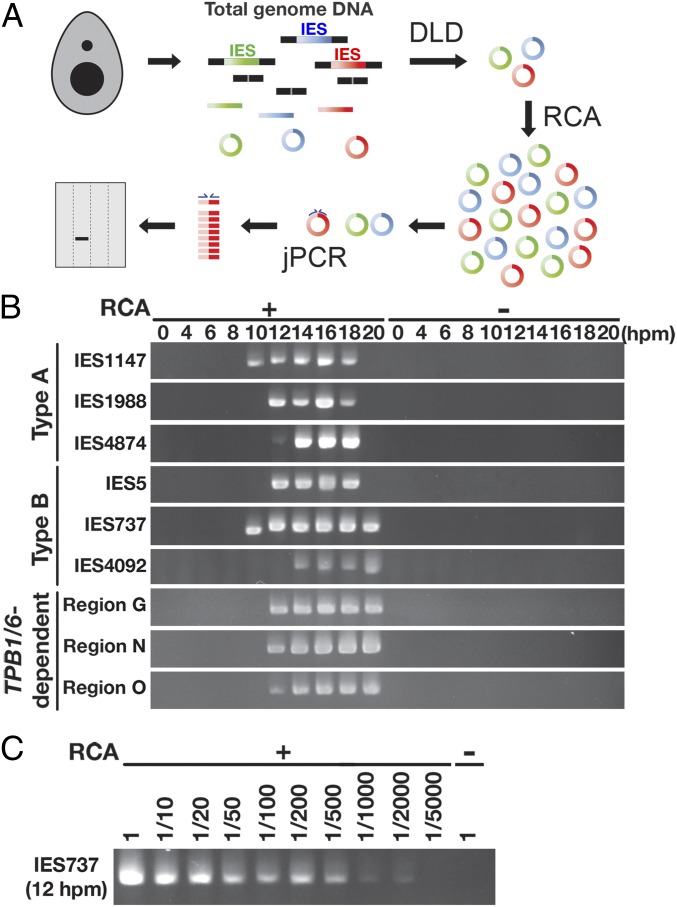
To understand the temporal relationship between the excision of IESs and the production of late-scnRNAs in Tetrahymena, we first aimed to determine the timing of IES excision. It has been reported that de novo IES excision can be detected by polymerase chain reaction (PCR) amplification of the ligation junctions of excised IES circles (jPCR) (9, 37). However, the sensitivity of this detection has not been examined. To detect de novo DNA elimination events with high sensitivity, we have established a method combining jPCR with the digestion of linear DNA (DLD) and rolling circular amplification (RCA), which we term DLR-jPCR (Fig. 1A).
Fig. 1.
Detection of de novo IES excision events. (A) Linear DNAs in total genomic DNA were digested with exonucleases (digestion of linear DNA [DLD]). Then the remaining circular DNAs, including IES circles produced by de novo IES excision in the new MAC, were amplified by RCA, and the junction of the DNA circle from an IES locus was detected by jPCR. The entire procedure is referred to as DLR-jPCR. (B) WT strains were mated, and genomic DNA was extracted at the indicated hpm. The circles excised from the indicated IES loci were analyzed by DLR-jPCR with (+) or without (−) RCA. (C) Genomic DNA from 12 hpm was treated as in B, but the sample was diluted by the indicated factors before jPCR to detect the IES737 locus.
Two wild-type (WT) Tetrahymena cultures with different mating types were mixed to initiate conjugation, and total genomic DNA was extracted at the onset of mixing and subsequently at 2-h intervals from 4 to 20 hpm. The genomic DNA was then used in DLR-jPCR assays. We chose 9 representative IESs, including 3 each of the major IES types, type A, type B, and TPB1/6-dependent IESs, to investigate the appearances of IES circles. As shown in Fig. 1B, IES circles were specifically detected in the late stages of conjugation (Fig. 1B, RCA+), consistent with the fact that DNA elimination occurs during development of the new MAC, which is formed at ∼7 hpm. IES circles were detected for all 9 tested IES loci, indicating that DLR-jPCR can detect de novo IES excision events regardless of the type of IES.
The timing of the appearance of IES circles varied among different IESs. For example, circles of IES737 and IES1147 were detected at 10 hpm, whereas those of the other IESs appeared only after 12 hpm (Fig. 1B). Although to date we have failed to find any obvious rule defining the timing of DNA excision for different IESs, these results are consistent with the previously proposed idea that some IESs are eliminated earlier than others (7). IES circles were detected continuously for at least several hours. A previous report (9) suggested that this situation is caused, at least in part, by the persistence of each IES circle for several hours before it is degraded, although cell-to-cell variability of the timing of IES excision may also contribute to the continuous detection of IES circles.
We failed to detect any IES circles when we performed similar reactions without RCA (Fig. 1B, RCA−), indicating that the RCA step significantly sensitizes the detection of IES circles. To further assess the sensitivity of DLR-jPCR, we prepared serial dilutions of genomic DNAs after the RCA reaction and performed jPCR. As shown in Fig. 1C, we found that DNA circles from IES737 could be detected even when genomic DNA from 12 hpm was diluted 2,000-fold. This result indicates that DLR-jPCR is likely >2,000-fold more sensitive than the previous IES circle detection methods, which do not include the RCA step, and the dynamic range of the detection sensitivity of the DLR-jPCR assay spans more than 3 orders of magnitude.
Late-scnRNA Production Is Initiated before IES Excision Becomes Detectable.
We next sought to compare the timing of the appearance of IES circles with that of late-scnRNAs. When late-scnRNAs are produced in the new MAC, early-scnRNAs expressed from the MIC at earlier stages of conjugation persist. Early- and late-scnRNAs cannot be distinguished by simple biochemical properties, such as length and base composition (20). Therefore, to specifically investigate late-scnRNAs, we focused on type B IES-derived small RNAs, which are predominantly, if not exclusively, late-scnRNAs (20).
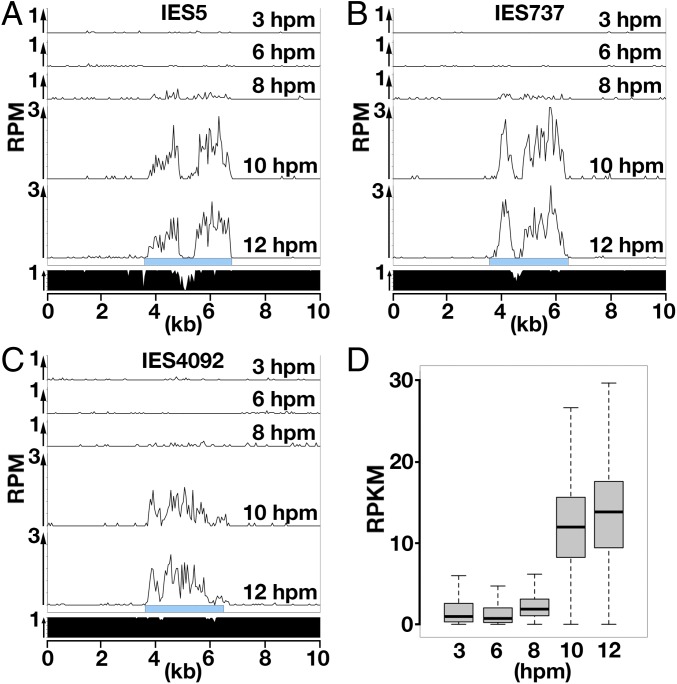
Small RNAs extracted from mating WT cells at 3, 6, 8, 10, or 12 hpm were sequenced (small RNA-seq) (38), and those that uniquely mapped to the 3 studied type B IESs indicated above were counted. As shown in Fig. 2 A–C, small RNAs derived from these type B IESs remained at a low level till 6 hpm and clearly increased after 8 hpm, indicating that late-scnRNAs production from these IESs began at 8 hpm at the latest. A similar result was obtained when small RNAs were mapped to each of the 3,293 type B IESs (Fig. 2D). The earliest time point at which IES circles were detected for these IESs was 10 hpm (IES737) or later (IES5, IES4092) (Fig. 1B). Therefore, late-scnRNAs expressed from an IES can be detected by small RNA-seq before the excision of that IES becomes detectable by DLR-jPCR.
Fig. 2.
Accumulation of late-scnRNAs in WT cells. (A–C) Normalized numbers (reads per million [RPM]) of sequenced 26- to 32-nt small RNAs from WT cells at indicated time points uniquely mapping to the representative type B IES loci are shown as histograms with 50-nt bins. Sky-blue boxes indicate IESs. The fraction of unique sequences that can be mapped by small RNAs in each 50-nt bin is shown at the bottom. (D) Normalized numbers (reads per kilobase of unique sequences per million [RKPM]) of 26- to 32-nt small RNAs from WT cells at indicated time points that uniquely matched the 3,293 type B IESs are shown as box plots. The horizontal bar in the box indicates the median value. The bars on the top and bottom of the box indicate the minimum and maximum values, respectively, within 1.5 times the interquartile range.
Although these results most likely indicate that the production of late-scnRNAs occurs before the excision of IESs, the foregoing data do not exclude the possibility that some IES excision occurs before the late-scnRNA production, but the detection of late-scnRNAs by small RNA-Seq is more sensitive than that of IES circles by DLR-jPCR. Therefore, we next aimed to use a genetic mutant defective in IES excision to further investigate the relationship between the production of late-scnRNAs and IES excision.
TPB2-Null Mutants Block Accumulation of the Tpb2p Excisase.
Tpb2p is important for the DNA elimination of type A and type B IESs in vivo (23) and catalyzes DNA double-strand breaks to form a 4-base 5′ overhang in vitro (24) that is identical to the intermediate structure observed in DNA elimination in vivo (9). Therefore, Tpb2p is believed to be the enzyme catalyzing the final excision step of type A and type B IESs. Previous loss-of-function studies of TPB2 were conducted using RNAi (23) or conditional (24) knockdown. In those studies, IES elimination was not completely blocked, probably due to incomplete loss of TPB2 expression. Therefore, the conditions applied in these previous genetic studies cannot be used to explicitly determine the requirement for TPB2 in late-scnRNA biogenesis.
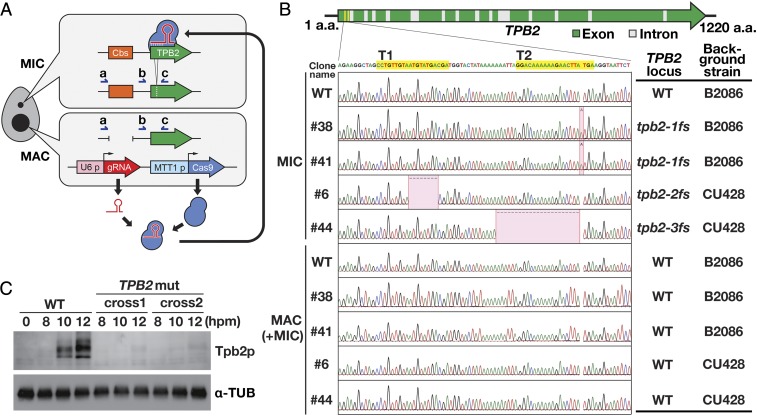
To produce TPB2-null mutants, we used CRISPR-based mutagenesis technology. Although CRISPR technology was used for mutagenesis of Tetrahymena in 2 previous reports (39, 40), we made several modifications to the previously reported protocols to efficiently obtain Tetrahymena mutants (SI Appendix, SI Materials and Methods). We introduced a construct expressing Cas9 and gRNAs into the MAC of vegetative cells of 2 WT strains (B2086 and CU428) with different mating types. We expected the Cas9-gRNA complex to introduce a mutation in the MIC (Fig. 3A). We used 2 gRNAs that target slightly different positions of the first exon of TPB2 (Fig. 3B). After the induction of Cas9 expression, the cells were cloned, and the TPB2 locus in the MIC was amplified by PCR using primers “a” and “c” shown in Fig. 3A. The products were then sequenced. The PCR amplicon contains a chromosome breakage sequence (Cbs) that remains intact in the MIC but is severed to form the ends of 2 separate chromosomes in the MAC (Fig. 3A). Therefore, this PCR assay specifically amplifies the TPB2 MIC locus. Approximately one-half of the clones examined contained insertions or deletions around the targeted region. We chose 2 clones each from the B2086 and CU428 strains containing 1 of the 3 frameshift mutations in TPB2, referred to as tpb2-1fs, tpb2-2fs, and tpb2-3fs (Fig. 3B). We did not detect any copies of WT TPB2 in the MIC in these cell lines, indicating that both copies of TPB2 in the diploid MIC were disrupted in these TPB2 mutants.
Fig. 3.
Production of TPB2 mutants. (A) A construct expressing Cas9 from the cadmium-inducible MTT1 promoter and TPB2-targeting gRNAs from the constitutive U6 promoter was introduced into the MAC. Some of the Cas9-gRNA complexes are expected to enter the MIC and introduce a mutation at the TPB2 locus. Immediately next to the TPB2 locus is a chromosome breakage sequence (Cbs), which remains intact in the MIC but is severed to form the ends of 2 separate chromosomes in the MAC. PCR with primers “a” and “c” specifically amplifies the TPB2 MIC locus, while PCR with primers “b” and “c” amplifies both the TPB2 MAC and MIC loci. (B) Two gRNAs (T1 and T2) were used to target the sequences in the first exon of TPB2 (in yellow). The TPB2 MIC and MAC loci were amplified, respectively, with the primers “a” and “c” and the primers “b” and “c” shown in A. Although primers “b” and “c” could amplify both the MAC and MIC loci, polyploid MAC copies were predominantly represented in this PCR assay. The detected insertions and deletions are indicated in red. (C) Total cell proteins were extracted at the indicated time points from conjugation cultures of either WT cells or TPB2 mutants (TPB2 mut; cross 1: #6 × #38; cross 2: #41 × #44) and analyzed by Western blotting using an anti-Tpb2p antibody (Top) and an anti–α-tubulin antibody (Bottom).
The TPB2 MAC locus of the TPB2 mutants was also analyzed by PCR using primers “b” and “c” shown in Fig. 3A. Although this PCR assay was designed to amplify both the MAC and MIC loci, the MIC locus was underrepresented because of the difference in ploidy between the MAC (∼50n) and the MIC (2n). We detected only the WT TPB2 locus in these PCR products, indicating that the CRISPR system used here did not efficiently induce mutations in the MAC (Fig. 3B). This result might have occurred because Cas9 was fused to part of the MIC-localized protein MLH1 in our experiment to enhance the MIC localization of Cas9 and/or because DNA double-strand breaks might have been repaired predominantly by homologous recombination with the sister copies in the polyploid MAC. Nevertheless, Tpb2p detected by Western blot analysis was reduced to background levels in the TPB2 mutants (Fig. 3C), indicating that TPB2 was exclusively expressed zygotically from the new MAC in the WT cells. We concluded that the mutants were TPB2-null mutants.
TPB2 Mutants Block DNA Elimination of Type A and Type B IESs.
H3K9me3 and H3K27me3, scnRNA-dependent heterochromatic modifications, specifically occur on IESs in WT cells (21). Consistent with a previous report describing RNAi knockdown of TPB2 (23), these modifications accumulated in the new MAC in the TPB2 mutants at 8 hpm (SI Appendix, Fig. S1). In the WT cells at 12 hpm, these heterochromatic marks accumulated in nuclear foci known as heterochromatin bodies (HBs). The formation of HBs is believed to be triggered by Tpb2p-induced DNA double-strand breaks (41). Consistently, HBs were not detected in the TPB2 mutants at 12 hpm (SI Appendix, Fig. S1). Therefore, the presence of H3K9/K27 me3 and the absence of HBs in the TPB2 mutants indicate that heterochromatin is formed but IES excision is generally blocked in these mutants.
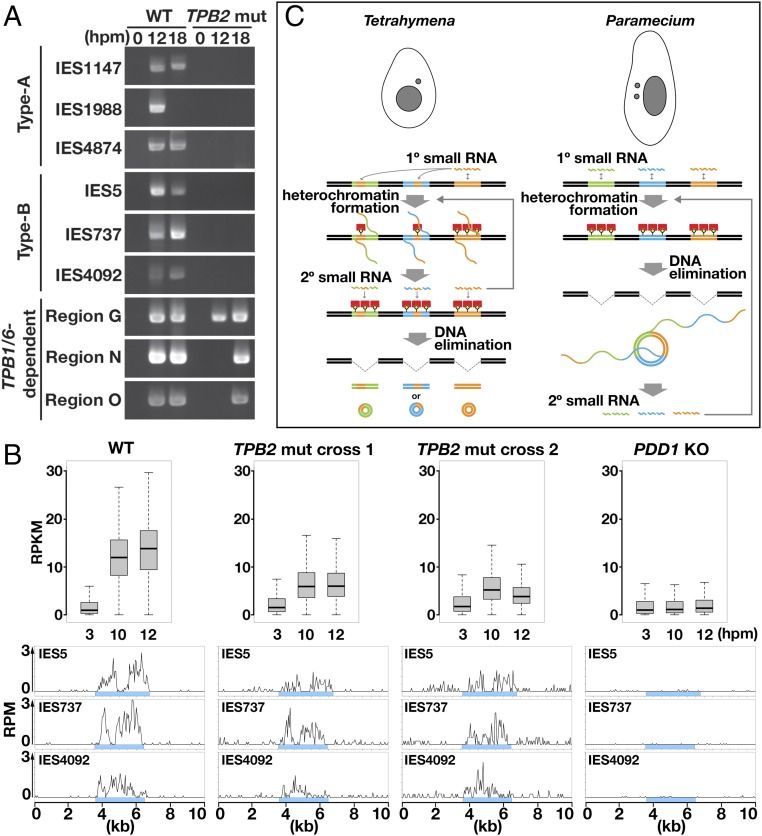
We next directly investigated DNA elimination in the TPB2 mutants by DLR-jPCR. As shown in Fig. 4, we could not detect IES circles from any of the tested type A and type B IESs at 12 and 18 hpm in the absence of Tpb2p (Fig. 4A, TPB2 mut), whereas IES circles from all of these IESs were detected at 1 or both of these time points in WT cells (Fig. 4A, WT). In contrast, IES circles from the 3 tested TPB1/6-dependent IES loci were detected in the TPB2 mutants, although their appearance was delayed compared with that in WT cells (Fig. 4A), Therefore, DNA elimination of all studied type A and type B IESs was reduced to undetectable levels in the TPB2 mutants. According to the results of sensitivity testing of DLR-jPCR shown in Fig. 1C, we estimate that the frequency of the excision of these IESs in the TPB2 mutants was reduced to less than ∼1/2,000 of that in the WT cells.
Fig. 4.
Analyses of DNA elimination and late-scnRNA accumulation in TPB2 mutants. (A) WT and TPB2 mutant (TPB2 mut; mating of #41 and #44) strains were mated, and their genomic DNA was extracted at 0, 12, and 18 hpm. The excised circles from the indicated IES loci were detected by DLR-jPCR. (B) WT, TPB2 mut (cross 1: #6 × #38; cross 2: #41 × #44) and PDD1 KO (∆PDD1) strains were mated, and their small RNAs were sequenced. (Top) Normalized numbers (in RPKM) of 26- to 32-nt small RNAs at 3, 10, and 12 hpm that uniquely matched the 3,293 type B IESs are shown as box plots as in Fig. 2D. (Bottom) Normalized numbers (in RPM) of sequenced 26- to 32-nt small RNAs at 10 hpm uniquely mapping to the indicated type B IES loci are shown as histograms with 50-nt bins. Sky-blue boxes indicate IESs. (C) Comparison of the timing of secondary small RNA biogenesis. (Left) In Tetrahymena, primary (1°) small RNAs fully or partially cover IESs and induce heterochromatin formation, which triggers the production of secondary (2°) small RNAs. Secondary small RNAs further induce heterochromatin formation and small RNA production. Eventually, heterochromatinized IESs are excised. (Right) In Paramecium, primary small RNAs cover IESs homogeneously. The excised IESs form concatenated circles, which are transcribed to produce secondary small RNAs. Secondary small RNAs further induce IES elimination and small RNA production.
Blocking DNA Elimination Does Not Result in Loss of Late-scnRNAs.
Finally, to investigate the accumulation of late-scnRNAs in the absence of IES excision, the TPB2 mutants were mated and their 26- to 32-nt small RNAs were analyzed (38). The total amount of small RNAs detected by denaturing gel electrophoresis was similar in the WT cells and TPB2 mutants at both early (3 hpm) and late (10 and 12 hpm) conjugation stages (SI Appendix, Fig. S2A). Small RNAs that uniquely mapped to the 3,293 type B IESs were increased between 3 and 10 hpm in the TPB2 mutants (Fig. 4B, Top), and small RNAs derived from the 3 representative type B IESs were detected in the TPB2 mutants at 10 hpm (Fig. 4B, Bottom) and 12 hpm. In contrast, the levels of type B IES-mapping small RNAs remained low at 10 and 12 hpm in PDD1 knockout (KO) cells, which lacked the core heterochromatin component (42) (Fig. 4B). Because early-scnRNAs are produced only at an early conjugation stage (∼3 hpm) (20), any increase in small RNAs at later stages should be attributed to the production of late-scnRNAs. Therefore, the foregoing results indicate that IES excision, but not heterochromatin formation, is dispensable for the production of late-scnRNAs.
The amounts of type B IES-mapping small RNAs at 10 and 12 hpm in the TPB2 mutants were lower than those in WT cells (Fig. 4 B, Top). In contrast, type A IES-mapping small RNAs were not obviously affected in the TPB2 mutants (SI Appendix, Fig. S2B). It is highly likely that the reduction of type B IES-mapping small RNAs, as well as the delayed elimination of the TPB1/6-dependent IESs (Fig. 4A), in the TPB2 mutants is indirectly caused by the genome-wide retention of IESs (23), which should disturb the expression of numerous genes. Alternatively, given our observation that small RNAs, which are normally produced predominantly from IESs, were also produced from non-IES sequences in the TPB2 mutants (Fig. 4 B, Bottom), we suggest that Tpb2p may have a noncatalytic function to confine late-scnRNA production to IESs, and that the reduction in late-scnRNAs that map type B IESs might be due to the gain of late-scnRNA production out of IESs. Nevertheless, the reduction of late-scnRNAs from type B IESs in the TPB2 mutants was approximately 2-fold, in contrast to the >2,000-fold reduction of IES excision in these mutant cells.
The results obtained with the TPB2 mutants do not exclude the possibility that some late-scnRNA biogenesis occurs after IES excision in WT cells and/or that late-scnRNA biogenesis normally occurs exclusively after IES excision in WT cells, but some compensatory mechanism allows the TPB2 mutants to produce late-scnRNAs without IES excision. However, together with the fact that late-scnRNAs were detected even at the time when elimination of their derived IESs was not detectable in WT cells (Fig. 1), the results shown in this study led us to conclude that the biogenesis of late-scnRNAs occurs before the excision of IESs.
Discussion
Two technical advances made in this study—the sensitive detection of de novo DNA elimination (Fig. 1) and the production of TPB2-null mutants (Fig. 3)—enabled us to show that late-scnRNAs accumulate at least a few hours before the elimination of their derived IESs becomes detectable (Fig. 2) and that IES excision is dispensable for their biogenesis (Fig. 4). Therefore, secondary small RNAs are produced from IESs before their excision in Tetrahymena, at least in part. This situation contrasts considerably with the excision-dependent secondary small RNA production observed in another ciliate, Paramecium, in which concatenated circles formed from excised IESs are suggested to be the templates for the transcription of precursors of iesRNAs (secondary small RNAs in this ciliate) (28, 35) (Fig. 4C). The postexcision production of secondary small RNAs in Paramecium is further supported by the fact that injection of DNA fragments into cells during sexual reproduction can induce ectopic DNA elimination at complementary genomic loci (35). In contrast, our attempts to induce ectopic DNA elimination in Tetrahymena by introducing linear and circular DNAs have failed. Considering all these findings together, we conclude that the modes of secondary small RNA biogenesis are fundamentally different in Paramecium and Tetrahymena. This difference was unexpected, given that these 2 ciliates use otherwise very similar molecular mechanisms for DNA elimination.
The postexcision production of iesRNAs from concatenated IES circles in Paramecium might allow “rolling circle transcription” to produce a large amount of small RNAs sufficient to target all copies of IESs in the highly polyploid (∼800 copies) somatic MAC genome. Such massive amplification of the small RNA signal probably is not necessary in Tetrahymena, which exhibits only ∼50 copies in the MAC in the vegetative stage and 4–8 copies when IES elimination occurs. Instead, because the IESs of Tetrahymena are much longer (median, 2.8 kb) than those of Paramecium (median, 51 bp) (6, 43), secondary small RNA production in Tetrahymena is necessary to cover IES regions that are not complementary to the primary small RNAs. The fact that heterochromatin formation, but not downstream IES excision, is necessary for the secondary small RNA biogenesis in Tetrahymena (Fig. 4B) strongly indicates that the secondary small RNA production mechanism is directly linked to a heterochromatin machinery. Indeed, the cis-spreading of late-scnRNA biogenesis via an RNAi-heterochromatin feedback loop plays an important role in IES elimination in Tetrahymena (20, 39), reminiscent of the heterochromatin-driven production of small RNAs observed in fungi (44), plants (45), and animals (46, 47) and thus likely originated from an ancient TE silencing mechanism.
If the foregoing speculations are correct, then the remarkable difference in secondary small RNA biogenesis reflects the evolutionary history of genome organization after the 2 ciliate linages diverged several million years ago. In this context, it would be interesting to explore the genomes and DNA elimination mechanisms of other ciliates to trace the evolution of TE-targeting small RNA amplification. Such observations would also provide unique views of how TEs and their silencing mechanisms drive the evolution of genome structures and vice versa.
Materials and Methods
Strains.
The WT Tetrahymena thermophila strains B2086 and CU428 were obtained from the Tetrahymena Stock Center. The PDD1 KO strains (W3-3 and W39-1) have been described previously (42). Construction of the TPB2 mutant strains is described in SI Appendix, Materials and Methods.
DLR-jPCR Assay.
Total genomic DNA was extracted using a NucleoSpin Tissue Kit (Macherey-Nagel). Here 5 µg of the genomic DNA was treated with exonuclease V (New England BioLabs) for 1 h at 37 °C, purified using the QIAquick Gel Extraction kit (QIAGEN), and then treated with Plasmid-Safe DNase (Epicentre) for 16 h at 37 °C. The remaining circular DNAs were ethanol-precipitated and amplified by RCA using the illustra TempliPhi Amplification kit (GE Healthcare) according to the manufacturer’s instructions, except that the 30 °C incubation was performed for >18 h. The junctions of circularized IESs were amplified by PCR using the primers listed in SI Appendix, Table S1. The positions of the type A and type B IESs investigated in this study have been described previously (20), as have the positions of TPB1/6-dependent IESs (26).
Small RNA Analyses.
High-throughput sequencing and analyses of small RNAs were performed as described previously (14), except that radiolabeled 16-nt and 32-nt RNA oligos (5′- GAAUAGUUUAAACUGU-3′ and 5′-AUCUUGGUCGUACGCGGAAUAGUUUAAACUGU-3′) were used to excise small RNAs and their intermediate products from gels. The raw sequencing data have been deposited in the Gene Expression Omnibus database under accession no. GSE125297.
Western Blotting and Immunofluorescent Staining.
Detailed procedures for SDS/PAGE sample preparation, Western blotting, and immunofluorescent staining are described in SI Appendix, Materials and Methods.
Supplementary Material
Acknowledgments
We acknowledge the Montpellier Ressources Imagerie facility, a member of the France-BioImaging national infrastructure supported by the French National Research Agency (ANR-10-INBS-04), and the Next-Generation Sequencing unit of Vienna BioCenter Core Facilities. This work was supported by an Advanced Grant from the Investissements d’avenir program Labex EpiGenMed (ANR-10-LABX-12-01) and an Accueil de Chercheurs de Haut Niveau grant (ANR-16-ACHN-0017) from the French National Research Agency (to K.M.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The data reported in this paper have been deposited in the Gene Expression Omnibus (GEO) database, https://www.ncbi.nlm.nih.gov/geo (accession no. GSE125297).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1903491116/-/DCSupplemental.
References
- 1.Czech B., et al. , piRNA-guided genome defense: From biogenesis to silencing. Annu. Rev. Genet. 52, 131–157 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Gebert D., Rosenkranz D., RNA-based regulation of transposon expression. Wiley Interdiscip. Rev. RNA 6, 687–708 (2015). [DOI] [PubMed] [Google Scholar]
- 3.Castel S. E., Martienssen R. A., RNA interference in the nucleus: Roles for small RNAs in transcription, epigenetics and beyond. Nat. Rev. Genet. 14, 100–112 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zhou M., Law J. A., RNA Pol IV and V in gene silencing: Rebel polymerases evolving away from Pol II’s rules. Curr. Opin. Plant Biol. 27, 154–164 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Luteijn M. J., Ketting R. F., PIWI-interacting RNAs: From generation to transgenerational epigenetics. Nat. Rev. Genet. 14, 523–534 (2013). [DOI] [PubMed] [Google Scholar]
- 6.Hamilton E. P., et al. , Structure of the germline genome of Tetrahymena thermophila and relationship to the massively rearranged somatic genome. eLife 5, e19090 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Austerberry C. F., Allis C. D., Yao M. C., Specific DNA rearrangements in synchronously developing nuclei of Tetrahymena. Proc. Natl. Acad. Sci. U.S.A. 81, 7383–7387 (1984). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Saveliev S. V., Cox M. M., Product analysis illuminates the final steps of IES deletion in Tetrahymena thermophila. EMBO J. 20, 3251–3261 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Saveliev S. V., Cox M. M., The fate of deleted DNA produced during programmed genomic deletion events in Tetrahymena thermophila. Nucleic Acids Res. 22, 5695–5701 (1994). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chalker D. L., Meyer E., Mochizuki K., Epigenetics of ciliates. Cold Spring Harb. Perspect. Biol. 5, a017764 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Noto T., Mochizuki K., Whats, hows and whys of programmed DNA elimination in Tetrahymena. Open Biol. 7, 170172 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yao M.-C., Fuller P., Xi X., Programmed DNA deletion as an RNA-guided system of genome defense. Science 300, 1581–1584 (2003). [DOI] [PubMed] [Google Scholar]
- 13.Chalker D. L., Yao M. C., Nongenic, bidirectional transcription precedes and may promote developmental DNA deletion in Tetrahymena thermophila. Genes Dev. 15, 1287–1298 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Schoeberl U. E., Kurth H. M., Noto T., Mochizuki K., Biased transcription and selective degradation of small RNAs shape the pattern of DNA elimination in Tetrahymena. Genes Dev. 26, 1729–1742 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mochizuki K., Gorovsky M. A., A Dicer-like protein in Tetrahymena has distinct functions in genome rearrangement, chromosome segregation, and meiotic prophase. Genes Dev. 19, 77–89 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Malone C. D., Anderson A. M., Motl J. A., Rexer C. H., Chalker D. L., Germ line transcripts are processed by a Dicer-like protein that is essential for developmentally programmed genome rearrangements of Tetrahymena thermophila. Mol. Cell. Biol. 25, 9151–9164 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mochizuki K., Fine N. A., Fujisawa T., Gorovsky M. A., Analysis of a piwi-related gene implicates small RNAs in genome rearrangement in tetrahymena. Cell 110, 689–699 (2002). [DOI] [PubMed] [Google Scholar]
- 18.Noto T., et al. , The Tetrahymena argonaute-binding protein Giw1p directs a mature argonaute-siRNA complex to the nucleus. Cell 140, 692–703 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Noto T., Mochizuki K., Small RNA-mediated trans-nuclear and trans-element communications in Tetrahymena DNA elimination. Curr. Biol. 28, 1938–1949.e5 (2018). [DOI] [PubMed] [Google Scholar]
- 20.Noto T., et al. , Small-RNA-mediated genome-wide trans-recognition network in Tetrahymena DNA elimination. Mol. Cell 59, 229–242 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Liu Y., et al. , RNAi-dependent H3K27 methylation is required for heterochromatin formation and DNA elimination in Tetrahymena. Genes Dev. 21, 1530–1545 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Taverna S. D., Coyne R. S., Allis C. D., Methylation of histone h3 at lysine 9 targets programmed DNA elimination in tetrahymena. Cell 110, 701–711 (2002). [DOI] [PubMed] [Google Scholar]
- 23.Cheng C.-Y., Vogt A., Mochizuki K., Yao M.-C., A domesticated piggyBac transposase plays key roles in heterochromatin dynamics and DNA cleavage during programmed DNA deletion in Tetrahymena thermophila. Mol. Biol. Cell 21, 1753–1762 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Vogt A., Mochizuki K., A domesticated PiggyBac transposase interacts with heterochromatin and catalyzes reproducible DNA elimination in Tetrahymena. PLoS Genet. 9, e1004032 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lin I.-T., Chao J.-L., Yao M.-C., An essential role for the DNA breakage-repair protein Ku80 in programmed DNA rearrangements in Tetrahymena thermophila. Mol. Biol. Cell 23, 2213–2225 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cheng C.-Y., et al. , The piggyBac transposon-derived genes TPB1 and TPB6 mediate essential transposon-like excision during the developmental rearrangement of key genes in Tetrahymena thermophila. Genes Dev. 30, 2724–2736 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Feng L., et al. , A germline-limited piggyBac transposase gene is required for precise excision in Tetrahymena genome rearrangement. Nucleic Acids Res. 45, 9481–9502 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sandoval P. Y., Swart E. C., Arambasic M., Nowacki M., Functional diversification of Dicer-like proteins and small RNAs required for genome sculpting. Dev. Cell 28, 174–188 (2014). [DOI] [PubMed] [Google Scholar]
- 29.Bouhouche K., Gout J.-F., Kapusta A., Bétermier M., Meyer E., Functional specialization of Piwi proteins in Paramecium tetraurelia from post-transcriptional gene silencing to genome remodelling. Nucleic Acids Res. 39, 4249–4264 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lepère G., Bétermier M., Meyer E., Duharcourt S., Maternal noncoding transcripts antagonize the targeting of DNA elimination by scanRNAs in Paramecium tetraurelia. Genes Dev. 22, 1501–1512 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lhuillier-Akakpo M., et al. , Local effect of enhancer of zeste-like reveals cooperation of epigenetic and cis-acting determinants for zygotic genome rearrangements. PLoS Genet. 10, e1004665 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Baudry C., et al. , PiggyMac, a domesticated piggyBac transposase involved in programmed genome rearrangements in the ciliate Paramecium tetraurelia. Genes Dev. 23, 2478–2483 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bischerour J., et al. , Six domesticated PiggyBac transposases together carry out programmed DNA elimination in Paramecium. eLife 7, e37927 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kapusta A., et al. , Highly precise and developmentally programmed genome assembly in Paramecium requires ligase IV-dependent end joining. PLoS Genet. 7, e1002049 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Allen S. E., et al. , Circular concatemers of ultra-short DNA segments produce regulatory RNAs. Cell 168, 990–999.e7 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hoehener C., Hug I., Nowacki M., Dicer-like enzymes with sequence cleavage preferences. Cell 173, 234–247.e7 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yao M. C., Yao C. H., Detection of circular excised DNA deletion elements in Tetrahymena thermophila during development. Nucleic Acids Res. 22, 5702–5708 (1994). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mutazono M., Noto T., Mochizuki K., Investigation of the timing of secondary small RNA biogenesis during programmed DNA elimination in Tetrahymena. Gene Expression Omnibus database. https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE125297. Deposited 17 January 2019.
- 39.Suhren J. H., et al. , Negative regulators of an RNAi-heterochromatin positive feedback loop safeguard somatic genome integrity in Tetrahymena. Cell Rep. 18, 2494–2507 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Howard-Till R., Tian M., Loidl J., A specialized condensin complex participates in somatic nuclear maturation in Tetrahymena thermophila. Mol. Biol. Cell 30, 1326–1338 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Shieh A. W. Y., Chalker D. L., LIA5 is required for nuclear reorganization and programmed DNA rearrangements occurring during tetrahymena macronuclear differentiation. PLoS One 8, e75337 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Schwope R. M., Chalker D. L., Mutations in Pdd1 reveal distinct requirements for its chromodomain and chromoshadow domain in directing histone methylation and heterochromatin elimination. Eukaryot. Cell 13, 190–201 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Guérin F., et al. , Flow cytometry sorting of nuclei enables the first global characterization of Paramecium germline DNA and transposable elements. BMC Genomics 18, 327 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Yamanaka S., et al. , RNAi triggered by specialized machinery silences developmental genes and retrotransposons. Nature 493, 557–560 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Law J. A., et al. , Polymerase IV occupancy at RNA-directed DNA methylation sites requires SHH1. Nature 498, 385–389 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Klattenhoff C., et al. , The Drosophila HP1 homolog Rhino is required for transposon silencing and piRNA production by dual-strand clusters. Cell 138, 1137–1149 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Andersen P. R., Tirian L., Vunjak M., Brennecke J., A heterochromatin-dependent transcription machinery drives piRNA expression. Nature 549, 54–59 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.