Significance
Conventional dendritic cells (cDCs), cDC1s and cDC2s, are hematopoietic cells that play central roles in mounting T cell responses. It is important for host protection that cDCs are continuously replenished by bone marrow precursors. cDC1s and cDC2s rely on the cytokine FLT3L, and, because both subsets mount different types of immune responses, their relative abundance must be regulated and balanced. In this work, we demonstrated that cDC2s produced their own FLT3L and utilized the transmembrane metalloproteinase ADAM10 to shed membrane-bound FLT3L into soluble forms. This process was crucial for cDC2s to develop and survive in the spleen. These findings likely represent cell-autonomous developmental and survival processes that enable cDC2s to preferentially gain access to FLT3L.
Keywords: immunology, dendritic cells, ADAM10
Abstract
Conventional dendritic cells (cDCs) derive from bone marrow (BM) precursors that undergo cascades of developmental programs to terminally differentiate in peripheral tissues. Pre-cDC1s and pre-cDC2s commit in the BM to each differentiate into CD8α+/CD103+ cDC1s and CD11b+ cDC2s, respectively. Although both cDCs rely on the cytokine FLT3L during development, mechanisms that ensure cDC accessibility to FLT3L have yet to be elucidated. Here, we generated mice that lacked a disintegrin and metalloproteinase (ADAM) 10 in DCs (Itgax-cre × Adam10-fl/fl; ADAM10∆DC) and found that ADAM10 deletion markedly impacted splenic cDC2 development. Pre-cDC2s accumulated in the spleen with transcriptomic alterations that reflected their inability to differentiate and exhibited abrupt failure to survive as terminally differentiated cDC2s. Induced ADAM10 ablation also led to the reduction of terminally differentiated cDC2s, and restoration of Notch signaling, a major pathway downstream of ADAM10, only modestly rescued them. ADAM10∆DC BM failed to generate cDC2s in BM chimeric mice with or without cotransferred ADAM10-sufficient BM, indicating that cDC2 development required cell-autonomous ADAM10. We determined cDC2s to be sources of soluble FLT3L, as supported by decreased serum FLT3L concentration and the retention of membrane-bound FLT3L on cDC2 surfaces in ADAM10∆DC mice, and by demonstrating the release of soluble FLT3L by cDC2 in ex vivo culture supernatants. Through in vitro studies utilizing murine embryonic fibroblasts, we determined FLT3L to be a substrate for ADAM10. These data collectively reveal cDC2s as FLT3L sources and highlight a cell-autonomous mechanism that may enhance FLT3L accessibility for cDC2 development and survival.
Dendritic cells (DCs) are immunological sentinels that are crucial for the initiation and regulation of immune responses against pathogens and autoantigens, respectively (1). DCs are classically classified into 2 distinct subsets of conventional DCs (cDCs), referred to as cDC1s and cDC2s (2, 3). cDC1s express CD8α or CD103, the former expressed by migratory cDC1s and the latter by resident cDC1s, and are specialized in cross-presenting antigens to CD8+ T cells. The IL-12 they produce effectively promotes type I immune responses (4). cDC2s coexpress CD11b and CD4 as well as ESAM in the spleen, are efficient drivers of CD4+ T cell responses, and have been reported to be crucial for initiating type 2 and 17 immune responses, thereby targeting extracellular pathogens (5).
cDC1s and cDC2s exhibit distinct transcription factor dependencies for development, during which cDC1s depend on IRF8, Id2, and Batf3 (6, 7) and cDC2s on IRF4, KLF4, and Zeb2 (8–10). In the bone marrow (BM), the monocyte and DC precursor MDP differentiates into monocytes and common DC precursors, the latter further generating the immediate DC precursor, pre-DCs (11). Commitment of pre-DCs to differentiate into cDC1s or cDC2s takes place in the BM (3), and these pre-cDC1s and pre-cDC2s ultimately differentiate into cDC1s and cDC2s after migrating to nonlymphoid and lymphoid tissues.
cDCs are short-lived, and their homeostatic maintenance relies on constant replenishment from the BM precursors (5). The cytokine Fms-related tyrosine kinase 3 ligand (FLT3L) (12), by signaling through its receptor FLT3 expressed on DC precursors, is essential during the development of DCs (7, 13). In vitro culture of mouse BM with recombinant FLT3L (rFLT3L) is sufficient to generate cDCs (14). Furthermore, the administration of rFLT3L in mice results in massive expansion of cDCs and pDCs in vivo (13) and the genetic ablation of FLT3L results in marked loss of these subsets (7, 15). FLT3L is produced by a variety of cell lineages, both hematopoietic and nonhematopoietic (16). Although organ-specific FLT3L can guide the terminal differentiation of DCs in nonlymphoid tissues, FLT3L expressed by hematopoietic cells is sufficient to support the generation of cDCs in the spleen (17). Among hematopoietic cells, CD4+ T cells are reported to be major sources of FLT3L (16). Other source(s) of FLT3L that support cDC development in secondary lymphoid tissues, however, remain unclear.
A disintegrin and metalloproteinase (ADAM) is a large family of transmembrane proteinases that regulate cell differentiation, migration, adhesion, and signaling through ectodomain-mediated shedding of many different molecules, and are therefore critically involved in wide varieties of biological processes (18). The classic example of how ADAMs function is demonstrated through studies on ADAM17 (also known as TNF-alpha converting enzyme, or TACE), wherein it has been shown to shed membrane-bound forms of TNF (19) or epidermal growth factor receptor ligands (20) to release active forms. Along with ADAM17, ADAM10 has been extensively studied and has been shown to cleave a long list of substrates that range from cell adhesion molecules, such as classic cadherins, to extracellular matrix proteins, such as collagen XVII, as well as chemokines (18). ADAM10 and ADAM17 are reported to play redundant and nonredundant roles, and genetic ablation of either molecule is embryonic lethal in mice (21, 22). The finding that ADAM10 mediates Notch signaling highlights its importance in regulating cell fate during differentiation (21). Notably, Notch signaling has been reported to be crucial during CD8α− cDC differentiation (23). Furthermore, a recent report that studied mice in which DC-specific depletion of ADAM10 revealed that ADAM10-mediated Notch signaling was crucial for TH2 allergic lung inflammation (24). However, the role of ADAM10 during cDC differentiation has yet to be elucidated.
Herein, we generated Itgax-Cre × Adam10fl/fl (ADAM10ΔDC) mice to explore the role(s) of ADAM10 in DC physiology and found that the ablation of ADAM10 in DCs led to a marked loss of CD11b+ ESAM+ cDC2s specifically in the spleen. This was accompanied by an increase of pre-cDC2s in the spleen, RNA-sequencing analysis of which revealed broad transcriptomic alterations with down-regulation of lineage-determining transcription factors (LDTFs) that suggested their incapability to terminally differentiate as cDC2s. Induced ablation of ADAM10 resulted in a rapid decrease in splenic cDC2s, indicating that ADAM10 was also required for cDC2 maintenance, and cDC2s were incompletely rescued upon restoration of Notch signaling. Through in vivo and ex vivo studies, we determined that cDC2s produced FLT3L and that they accumulated membrane-bound FLT3L on their cell surfaces in the absence of ADAM10. By utilizing wild-type (WT) and ADAM10-deficient murine embryonic fibroblasts (MEFs), we determined FLT3L as a substrate for ADAM10, revealing an ADAM10–FLT3 signaling axis that was crucial for cDC2 differentiation.
Results
Deletion of ADAM10 in DCs Result in a Loss of Splenic cDC2.
To determine the roles of the 2 major ADAMs in DC physiology, we crossed Itgax-Cre mice to Adam10fl/fl or Adam17fl/fl mice to generate mice that lacked either ADAM10 or ADAM17 (ADAM17ΔDC) in DCs. Both lines of mice were viable with no gross abnormalities. Flow cytometry analysis of lineage− CD45+ viable cells in the spleen, plotted via Uniform Manifold Approximation and Projection (UMAP), revealed a selective reduction in the numbers of CD11b+ CD4+ ESAM+ cDC2s in ADAM10ΔDC, indicating that ADAM10 played a nonredundant role during splenic cDC2 differentiation, and an emergence of a CD11bhi ESAM− DC population (Fig. 1 A and B and SI Appendix, Fig. S1A). Plasmacytoid DCs exhibited a slight increase (Fig. 1C), and the numbers of T and B cells remained unaltered (SI Appendix, Fig. S1B).
Fig. 1.
Deletion of Adam10 in DCs primarily impacts splenic CDC2s. (A) Uniform Manifold Approximation and Projection (UMAP) analysis of flow cytometry data gated on DAPI−Lin−CD45+ cells from WT littermate (WT), ADAM10ΔDC, and ADAM17ΔDC mice. Gating and DC identities (Left) and plots for indicated markers (Right). (B) Quantification of the percentages of total cDCs, CD11b+ ESAM+ cDC2s, and CD11bhi ESAM− DCs in WT, ADAM10ΔDC, and ADAM17ΔDC mice. (C) Flow cytometry analysis (Left) and quantification of Ly6C+ siglecH+ pDCs gated on DAPI−Lin+CD45+ from WT, ADAM10ΔDC, and ADAM17ΔDC mice (Right). (D) Lethally irradiated C57BL/6 CD45.1 mice were reconstituted with bone marrow from ADAM10ΔDC (CD45.2) or WT mice and were analyzed for CD45.2+ splenic CD11bhi ESAM− DCs and cDC2s at week 8 after transplantation. (E) Lethally irradiated WT C57BL/6 CD45.2 mice were reconstituted with bone marrow from ADAM10ΔDC (CD45.2) and WT C57BL/6 CD45.1 mice at a 1:1 ratio and were analyzed for splenic CD11bhi ESAM− DCs and cDC2s of each congenic origin at week 8 after transplantation. Quantifications in D and E are shown as percentages of indicated DC subsets among splenic DCs of their respective origins. Experiments in A–C are representative of more than 5 independent experiments with n = 3 in each group. Experiments in D and E are representative of 2 independent experiments with 3 to 4 mice in each group. ANOVA with Tukey’s multiple comparison test was used to measure significance (*P < 0.05, ***P < 0.001, and ****P < 0.0001; n/s: not significant).
To determine if DCs in other secondary lymphoid tissues were affected by the loss of ADAM10, we studied lung- and skin-draining lymph nodes of ADAM10ΔDC mice and WT littermate controls. Migratory cDC2 numbers were modestly decreased in ADAM10ΔDC mouse lung-draining lymph nodes, whereas resident cDC2s were unaffected (SI Appendix, Fig. S1C). In contrast, in the skin-draining lymph nodes, migratory cDC numbers were unchanged, but resident cDC1 and cDC2 numbers were modestly increased (SI Appendix, Fig. S1D). cDC2s in the lung and skin remained unaltered in ADAM10ΔDC mice (SI Appendix, Fig. S1E). These data demonstrated that the critical requirement for ADAM10 was restricted to splenic cDC2s, further suggesting that tissue-specific mechanisms were operative during the terminal differentiation or maintenance of cDC2s.
Cell-Autonomous Requirement of ADAM10 for cDC2 Development.
Whereas it is established that ADAM17 operates in a cell-autonomous manner (25), ADAM10 has been demonstrated to be capable of exerting its effects in trans, mediating the shedding of ephrin on opposing cells (26). To investigate if ADAM10 was required for cDC2 differentiation in a cell-autonomous manner, we first generated BM chimeric mice by transplanting WT or ADAM10ΔDC BM (CD45.2) into lethally irradiated CD45.1 mice. Splenic DC population in ADAM10ΔDC BM-recipient mice recapitulated that of ADAM10ΔDC mice, with marked decreases in cDC2 numbers and increased CD11bhi ESAM− DC numbers (Fig. 1D), indicating that ADAM10 expression by hematopoietic cells was required for the development of cDC2s. To further determine whether the coexistence of ADAM10-expressing hematopoietic cells could restore ADAM10ΔDC BM-derived cDC2s, we generated mixed bone marrow chimeras by cotransferring CD45.1 and ADAM10ΔDC BM at a 1:1 ratio into lethally irradiated CD45.2 C57BL/6 mice. Notably, ADAM10ΔDC BM failed to generate cDC2s but gave rise to CD11bhi ESAM− DCs (Fig. 1E). These data demonstrated that any soluble factors that may derive from WT hematopoietic cells or activities of ADAM10 in trans were incapable of inducing ADAM10ΔDC cDC2s and that cell-autonomous ADAM10 was required for splenic cDC2 development.
Characterization of Emerging CD11bhi ESAM− DC Population in ADAM10ΔDC Mice.
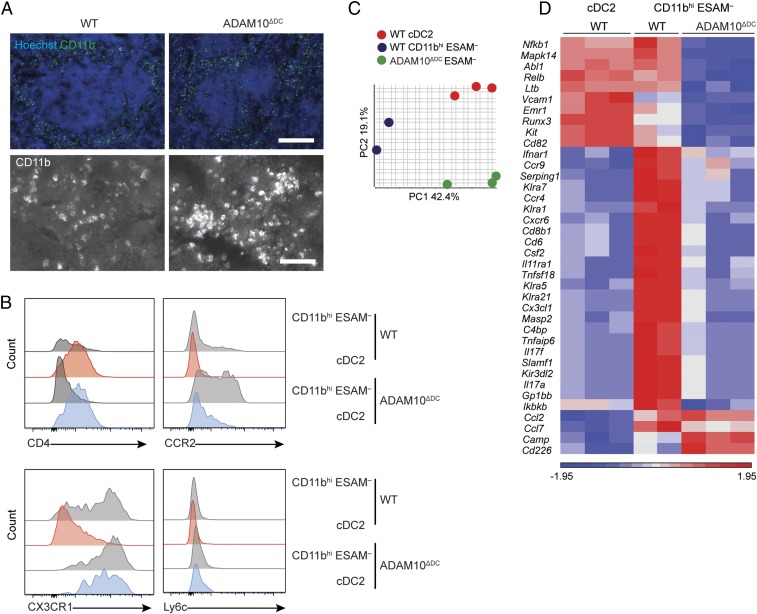
To characterize the emerging CD11bhi ESAM− DC population in ADAM10ΔDC mice, we first visualized CD11b via immunofluorescence microscopy. Staining of frozen spleen sections from ADAM10ΔDC mice and WT controls revealed similar distribution of CD11b+ cells surrounding the lymphoid follicles, with higher fluorescence intensity in ADAM10ΔDC mice (Fig. 2A). Flow cytometry analysis demonstrated that, in contrast to cDC2s in WT and remnant cDC2s in ADAM10ΔDC mice, CD11bhi ESAM− DCs lacked CD4 expression (Fig. 2B). CD11bhi ESAM− DCs in ADAM10ΔDC mice expressed the chemokine receptors CCR2 and CX3CR1, both of which are expressed by monocyte-derived cells (27), and showed slightly increased expression of Ly6c, also a monocyte marker (Fig. 2B). Gene expression analysis for 561 genes via Nanostring (mouse immunology panel) with unsupervised clustering analysis and principal component analysis plot of differentially expressed genes (P value with FDR < 0.05) revealed distinct transcriptomic profiles between cDC2 and CD11bhi ESAM– cells from WT mice, as well as CD11bhi ESAM– DCs from ADAM10ΔDC mice (Fig. 2 C and D). CD11bhi ESAM– DCs in ADAM10ΔDC mice exhibited a striking down-regulation of genes that were expressed by their WT counterparts and had up-regulated several genes that are commonly expressed by monocytes, such as Camp, which encodes cathelicidin; Cd226, an adhesion protein; and chemokines such as Ccl2 and Ccl7 (Fig. 2D and SI Appendix, Table S1). Differential expression of LDTFs was not detected. Taken together, these data suggested that emerging CD11bhi ESAM– cells in ADAM10ΔDC mice were distinct from cDC2s, and might represent CD11bhi ESAM− DCs that were altered in the absence of ADAM10 or monocyte-derived cells that emerged through ADAM10-independent mechanism(s).
Fig. 2.
Emerging CD11bhi ESAM− DCs in ADAM10ΔDC display unique transcriptomic signature. (A) Merged immunofluorescence images of anti-CD11b antibody and Hoechst staining in spleens from WT littermate and ADAM10ΔDC mice (Upper) and higher-magnification image for CD11b staining (Lower). (Scale bars: Upper, 200 µm; Lower, 50 µm.) (B) Flow cytometry analysis for cDC2 and monocyte markers expressed by indicated DC subsets in WT and ADAM10ΔDC mice. (C) PCA plot and (D) unsupervised heat map of Nanostring analysis of indicated cell subsets from WT and ADAM10ΔDC mouse spleens. A and B are representative of 3 independent experiments with n = 3 mice. C and D are representative of 2 experiments with 2 to 3 mice in each group.
ADAM10 Is Required for Both Terminal Differentiation and Maintenance of cDC2.
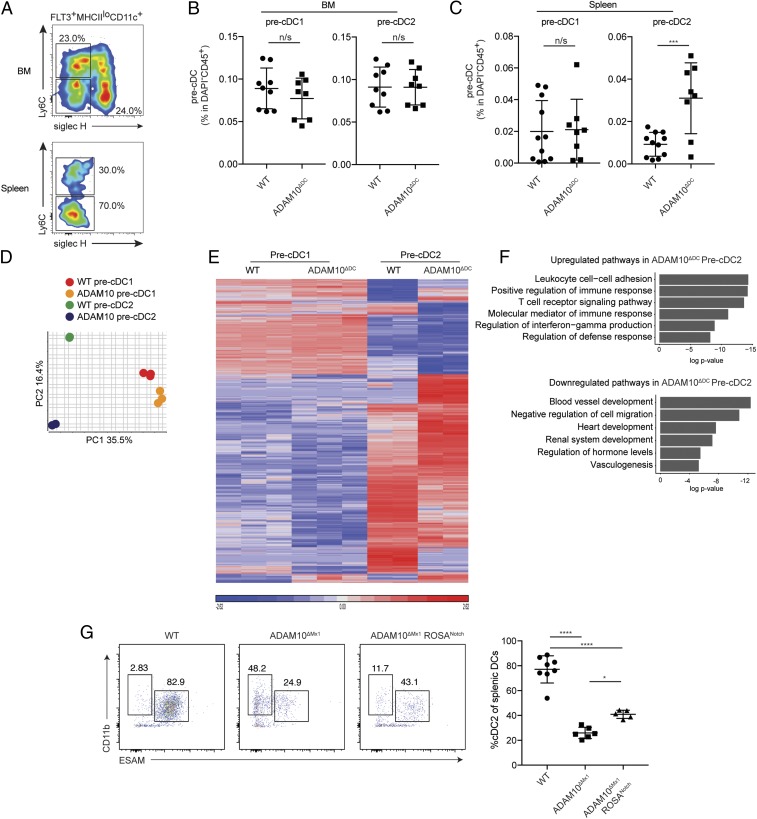
To determine the stage at which cDC2 differentiation was compromised by the loss of ADAM10, we studied pre-cDCs from the BM and spleen via flow cytometry. Pre-cDCs were defined as lineage– FLT3+ MHC IIlo CD11c+ Siglec H− cells and pre-cDC1s as Ly6c– and precDC2 as Ly6c+ (SI Appendix, Fig. S2 A and B and Fig. 3A). Pre-cDC1 numbers in the BM and spleens of ADAM10ΔDC mice were comparable to those of WT mice (Fig. 3 B and C). Whereas pre-cDC2 numbers were normal in ADAM10ΔDC mouse BM, they were increased in the spleen (Fig. 3C). Together with the lack of terminally differentiated cDC2s in the spleen (Fig. 1 A and B), these data suggested that cDC2 differentiation had halted at the splenic pre-cDC2 stage in ADAM10ΔDC mice.
Fig. 3.
ADAM10 is required during cDC2 development and maintenance in a Notch-independent manner. (A) Representative flow cytometry plot for pre-DC1s (Ly-6C− Siglec H−) and pre-DC2s (Ly-6C+ Siglec H−) among FLT3L+ MHCIIlo CD11c+ cells from BM or spleen in WT littermate mice. (B and C) The ratio of pre-DC1s and pre-DC2s among CD45+ cells in the BM and spleen from WT or ADAM10ΔDC mice. Student’s t test was used to measure significance. Values shown as mean ± SD (***P < 0.001; n/s: not significant). (D) PCA plot of RNA-seq data from sorted pre-cDC1s and pre-cDC2s from WT or ADAM10ΔDC mouse BM. (E) Unsupervised heat map of data from D. (F) Metascape pathway analysis of differentially expressed genes obtained by direct comparison of pre-cDC2s from WT and ADAM10ΔDC mice. (G) Flow cytometry analysis of splenic cDC2s (Left) and quantification (Right) in WT, ADAM10ΔMx1, and ADAM10ΔMx1 ROSANotch mice 24 h after poly(I:C) injection. Cells were gated for CD45+ MHC IIhi CD11c+ cells. Quantification shown as mean ± SD. ANOVA with Tukey’s multiple comparison test was used to measure significance (*P < 0.05 and ****P < 0.0001). A is representative of more than 3 experiments with n = 3 mice. B and C represent pooled data from 2 independent experiments. (D–F) Data are from 1 experiment with 2 to 3 mice from each group. G represents pooled data from 2 independent experiments with a total of 5 to 9 mice in each group.
To characterize the changes that had occurred in splenic pre-cDCs in the absence of ADAM10, we performed RNA sequencing on sorted pre-cDC1s and 2s from the spleens of WT and ADAM10ΔDC mice. Unsupervised hierarchical clustering and principal component analysis of all 4 groups demonstrated that distinct transcriptomic changes had taken place in ADAM10ΔDC pre-cDC2s, whereas minimal changes were observed for pre-cDC1s (Fig. 3 D and E). Direct comparison between pre-cDC2s from WT and ADAM10ΔDC mice revealed 4,805 differentially expressed genes (SI Appendix, Table S2). Metascape pathway analysis of 506 up-regulated and 296 down-regulated genes in ADAM10ΔDC pre-cDC2s revealed the up-regulation of pathways related to immune activation and the down-regulation of pathways involved during various developmental processes (Fig. 3F). Curiously, up-regulation of pathways was in part driven by the expression of T cell-associated genes such as Bcl2, Cd3d, Il4ra, and Tcf7 (SI Appendix, Table S2). Although the possibility of T cell precursor contamination cannot be completely excluded (mature T cells were excluded during sorting with an antibody against CD3e), this gene expression pattern is reminiscent of that reported for DC progenitors from mice that lack GATA2, a transcription factor crucial for DC development (28). Remarkably, comparison of LDTFs that were identified among differentially expressed genes revealed that ADAM10ΔDC pre-cDC2s up-regulated Id2, a cDC1 LDTF (6), and down-regulated Irf4, Klf4, Zeb2, and Gata2, all of which are established LDTFs for cDC2s (SI Appendix, Fig. S2 C and D and Tables S2 and S3) (9, 10, 28, 29). These data collectively demonstrated that pre-cDC2s from ADAM10ΔDC mice exhibited broadly altered transcriptomic landscape with dysregulation of LDTFs that are crucial for cDCs, which further suggested that ADAM10 was required as pre-DC2s terminally differentiated into cDC2s in the spleen.
It was also possible that cDC2s required ADAM10 immediately after terminal differentiation. To examine if induced ablation of ADAM10 also affected existing cDC2s, we generated Mx1-cre × Adam10fl/fl (ADAM10ΔMx1) mice, in which type I IFN-responsive cells would be ablated of ADAM10 (30). A single poly(I:C) injection into these mice and analysis 24 h later demonstrated a rapid and striking loss of cDC2s (Fig. 3G), demonstrating that ADAM10 was crucial not only for the development but also for the maintenance of terminally differentiated cDC2s.
It is well established that ADAM10 is critical for Notch signaling (21). Furthermore, Notch signaling has been demonstrated to be important for cDC2 differentiation (23). To examine if the failure of cDC2s to differentiate and survive was attributed to impaired Notch signaling, we restored Notch signaling in ADAM10ΔMx1 mice by crossing them with mice that express the intracellular portion of the mouse Notch1 gene upon Cre-mediated excision of a loxP-flanked STOP fragment (ROSANotch mice). Thus, Notch signaling would be operative in cDC2s in ADAM10ΔMx1 ROSANotch mice in the absence of ADAM10. Interestingly, restoration of Notch signaling only modestly rescued cDC2s (Fig. 3G), suggesting that another pathway could be involved during cDC2 development and maintenance.
CDC2s Are Sources for FLT3L.
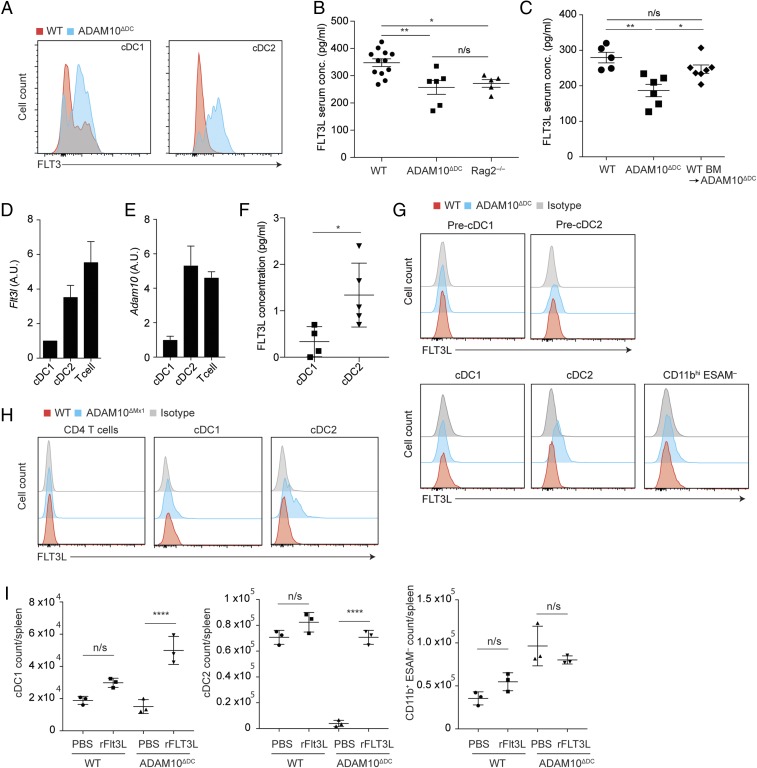
We explored other pathways downstream of ADAM10 that may support cDC2 development and maintenance. Interestingly, during the analysis of FLT3-expressing pre-DCs, we noticed that cDC1s and remnant cDC2s in the spleen of ADAM10ΔDC mice had up-regulated cell-surface FLT3, the receptor for FLT3L (Fig. 4A). We hypothesized that this up-regulation might reflect the deprivation of FLT3L in vivo. Indeed, quantification of soluble FLT3L in sera of ADAM10ΔDC mice via ELISA demonstrated that serum concentrations of FLT3L were decreased to levels that were comparable to Rag2−/− mice (Fig. 4B), which lack T cells, a major hematopoietic source for FLT3L (16), suggesting that cDCs might also be important sources for FLT3L. Reconstitution of lethally irradiated ADAM10ΔDC mice with WT BM restored serum FLT3L levels (Fig. 4C). These data confirmed that BM-derived cells were sources of FLT3L, and suggested that ADAM10 was required to release soluble FLT3L from its membrane-bound form.
Fig. 4.
ADAM10 deficiency leads to increased membrane-bound FLT3L on cDC2 cell surface. (A) Flow cytometry analysis for cell-surface Flt3 expression on splenic cDC1s and cDC2s from WT littermate and ADAM10ΔDC mice. (B) ELISA analysis for serum FLT3L concentrations in WT, ADAM10ΔDC, and Rag2−/− mice. (C) ELISA analysis for serum FLT3L concentrations in WT, ADAM10ΔDC, and ADAM10ΔDC mice that were reconstituted with WT BM. (D) Quantitative real-time PCR analysis for Flt3l expression in cDC1s, cDC2s, and T cells that were sorted from WT mouse spleens displayed as arbitrary units (A.U.). (E) Quantitative real-time PCR analysis of RT-PCR for Adam10 expression in splenic cDC1s, cDC2s, and T cells from WT mice. (F) WT cDC1s and cDC2s were sorted from the spleen and cultured for 2 d. Supernatants were collected and analyzed via ELISA for FLT3L concentrations. (G) Flow cytometry analysis for cell-surface FLT3L on splenic pre-DC1s, pre-DC2s, cDC1s, and cDC2s from WT and ADAM10ΔDC mice or (H) WT and ADAM10ΔMx1 mice. (I) Absolute numbers of cDC1s, cDC2s, and CD11bhi ESAM− DCs in spleens from WT or ADAM10ΔDC mice that received i.p. administration of rFlt3L or PBS solution. B and C are pooled data from 2 and 3 independent experiments, respectively. D–F are representative of 2 experiments. G and H are representative of 3 independent experiments with n = 3 in each group. I is representative of 2 independent experiments. B, F, and I are shown as mean ± SD. Statistical significance was measured by ANOVA with Tukey’s multiple comparison test in B, C, and I and by Student’s t test in F (*P < 0.05, **P < 0.01, and ****P < 0.0001; n/s: not significant).
Given that cDC2s were specifically decreased in ADAM10ΔDC mice, we explored the possibility that cDC2s might represent major FLT3L sources among cDCs. To address this, we performed real-time PCR using cDNA obtained from cDC1s, cDC2s, and T cells from WT mouse spleen. We found that cDC2s expressed FLT3L mRNA at levels that were comparable to T cells (Fig. 4D) and that cDC2s and T cells expressed ADAM10 mRNA at levels higher than in cDC1s (Fig. 4E), revealing spatial overlap of Flt3l and Adam10 expression in cDC2s and T cells. To definitively show that cDC2s produced FLT3L at the protein level, we sorted splenic cDC1s and cDC2s from WT mice and cultured them short-term for 2 d. Quantification of soluble FLT3L in the culture supernatants by ELISA revealed significantly higher concentrations of FLT3L in culture supernatants of cDC2s than those of cDC1s, demonstrating that cDC2s preferentially produced FLT3L (Fig. 4F).
The decreased serum levels of FLT3L in ADAM10ΔDC mice and the preferential production of FLT3L by cDC2s led us to hypothesize that ADAM10 cleaved membrane-bound FLT3L from cDC2 cell surface to release soluble forms. Indeed, flow cytometry analysis detected a retention of membrane-bound FLT3L on remnant cDC2s from ADAM10ΔDC mice but not on pre-DCs or cDC1s from WT or ADAM10ΔDC mice. (Fig. 4G). Taken together, these observations establish cDC2s as sources of FLT3L and suggest that ADAM10 was involved in mediating the shedding of membrane-bound FLT3L.
Despite the broad transcriptomic changes that took place in splenic pre-cDC2s from ADAM10ΔDC mice, they did not display a clear retention of membrane-bound FLT3L. We therefore examined WT pre-cDC RNAseq data and found that pre-cDC2s were more enriched in ADAM10 and FLT3L mRNA than pre-cDC1s (SI Appendix, Fig. S3A). Possibly, the relatively low levels of mRNA for both genes (<10 RPKM) has hampered FLT3L detection at the protein level. It also remains to be excluded that the lack of ADAM10 in pre-DC2s alters other pathways that may, in aggregate, be crucial for terminal differentiation.
To determine if the ADAM10–FLT3L axis was also operative in T cells, particularly CD4+ T cells because they preferentially produce FLT3L (16), we generated Mx1-Cre × ROSA26R YFP reporter mice. An i.p. injection of poly(I:C) led to YFP expression in the vast majority of cDCs and ∼25% of CD4+ T cells (SI Appendix, Fig. S3B). We then induced the ablation of ADAM10 by treating ADAM10 ΔMx1 mice with poly(I:C) and analyzed CD4+ T cells, cD1s, and cDC2s for membrane-bound FLT3L via flow cytometry. Interestingly, and consistent with our present observations in ADAM10ΔDC mice, FLT3L retention was specific to cDC2s in ADAM10 ΔMx1 mice. Although WT T cells expressed ADAM10 and FLT3L (Fig. 4 D and E), induced ablation of ADAM10 did not result in detectable retention of membrane-bound FLT3L (Fig. 4H). It is possible that ADAM10 substrate specificity is differentially regulated among different cell subsets (31, 32).
To examine if supplementation of FLT3L alone could rescue cDC2 differentiation in ADAM10ΔDC mice, we injected WT and ADAM10ΔDC mice with rFLT3L (10 µg per mouse) consecutively for 6 d and harvested their spleens for analysis 1 d after the last injection. Whereas rFLT3L only modestly increased cDC1s and 2s in WT mice, it rescued cDC2s in ADAM10ΔDC mice to numbers that were comparable to those in WT mice (Fig. 4I). This complete rescue of cDC2s identifies the ADAM10–FLT3 signaling axis as a crucial pathway for the development and maintenance of cDC2s, independent of Notch signaling. In contrast, CD11bhi ESAM− cell numbers neither increased nor decreased upon rFLT3 administration (Fig. 4I), consistent with our notion that they represent a cell lineage that is distinct from cDC2s.
ADAM10-Mediated Shedding of Membrane-Bound FLT3L.
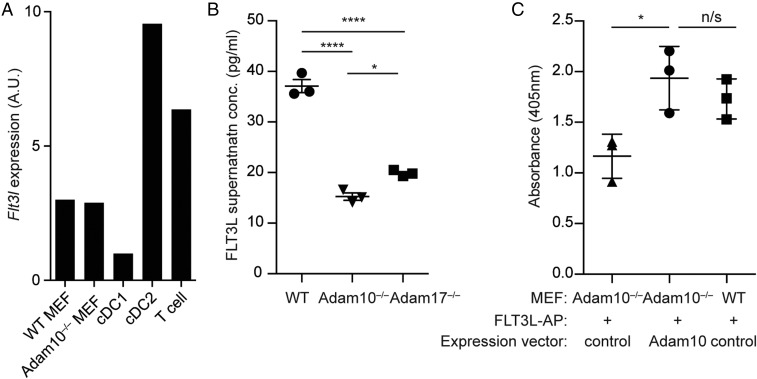
Previous studies have reported that ADAM17 and ADAM10 share common substrates (18). In addition, ADAM17 has been reported to cleave FLT3L (33). Together with the observation of FLT3L retention on cDC2 cell surface (Fig. 4G), we reasoned that ADAM10 might cleave membrane-bound FLT3L. To definitively determine this possibility, we utilized MEFs that were generated from WT and Adam10−/− mice (30, 33). WT and Adam10−/− MEFs expressed Flt3l transcripts at levels that were lower than cDC2s and T cells, but higher than that of cDC1s (Fig. 5A). MEFs were cultured for 2 d after achieving confluence, and supernatants were collected and analyzed for FLT3L concentration via ELISA. FLT3L concentrations were markedly decreased in Adam10−/− MEF supernatants compared with WT MEFs and were slightly, but significantly, lower than that of Adam17−/− MEF supernatants (Fig. 5B). To further solidify this finding, we transfected Adam10−/− or WT MEFs with a vector expressing FLT3L-alkaline phosphatase (AP), thereby excluding any contribution of endogenous FLT3L, and simultaneously transfected them with a vector encoding Adam10 or its control, thereby rescuing ADAM10 expression in Adam10−/− MEFs (Fig. 5C). Consistent with the results from unmanipulated MEFs, colorimetric analysis for alkaline phosphatase activity revealed that rescuing ADAM10 expression in Adam10−/− MEFs restored the levels of FLT3L-AP released into the supernatants to levels that were comparable to WT MEFs (Fig. 5C). These data establish FLT3L as a substrate of ADAM10 and suggest that cDC2s utilize ADAM10 to cleave FLT3L to support their development and maintenance in a cell-autonomous manner.
Fig. 5.
FLT3L is a substrate for ADAM10. (A) Quantitative real-time PCR analysis for Flt3l expression in MEFs from WT and ADAM10ΔDC mice and sorted cDC1s, cDC2s, and T cells from C57BL/6 WT mice. (B) FLT3L concentration (in picograms per milliliter) in supernatants of WT, Adam10−/−, and Adam17−/− MEFs that were cultured for 2 d after achieving confluence. (C) Control or expression vector for Adam10 was transfected into WT or Adam10−/− MEFs to rescue Adam10 expression. MEFs were also transfected with a vector expressing Flt3L-AP to exclude the contribution of endogenous FLT3L production. Cells were cultured as in B, and culture supernatants were analyzed for AP absorbance. B and C are shown as mean ± SD (*P < 0.05 and ****P < 0.0001, ANOVA with Tukey’s multiple comparison test; n/s: not significant). All data are representative of 2 independent experiments.
Discussion
Studies during the past decade have established DCs as a hematopoietic lineage distinct from macrophages, and, among the DC lineage, it has become clearer that distinct mechanisms, such as transcription factor dependencies, control the differentiation of cDC1s and cDC2s (2). All DC subsets develop from FLT3-expressing precursors and critically depend on FLT3L for differentiation (2). It is possible that DC subsets compete for FLT3L, but how they differentially gain access to FLT3L has not been addressed. In this study, we demonstrated that cDC2s critically relied on ADAM10 for differentiation and survival in a cell-autonomous manner. cDC2s expressed FLT3L at the mRNA and protein levels and accumulated membrane-bound form of FLT3L in the absence of ADAM10. By utilizing MEFs, we demonstrated that ADAM10 cleaves membrane-bound FLT3L to release soluble forms. These results suggested that cDC2s contribute as FLT3L sources by shedding it via ADAM10, which may enable cDC2s to gain preferential access to the cytokine.
FLT3L produced by nonlymphoid peripheral tissues supports terminal differentiation of tissue-specific DCs, and BM chimeric studies have demonstrated that tissue-derived or hematopoietic-derived FLT3L is capable of supporting DC differentiation (17), in agreement with the independency of peripheral tissue cDC2s on ADAM10 we have shown in this work. Among the hematopoietic lineage, T cells, particularly CD4+ T cells, have been reported to be a major source of FLT3L that enhances DCs differentiation (16). Consistent with this report, we detected soluble form of FLT3L in circulation in mouse sera and observed that FLT3L concentration was reduced by 2-fold in Rag2−/− mice, corroborating that T cells were a major source of FLT3L in circulation (Fig. 4B). Our data further indicated that conditional ablation of ADAM10 in DCs in ADAM10ΔDC mice led to decreases in serum concentrations of FLT3L, demonstrating DCs as an additional source of circulating FLT3L, a contribution that appears to be comparable to that of T cells (Fig. 4B). cDC2s, compared with cDC1s, preferentially produced FLT3L in vitro, and this was supported by the finding that cell-surface retention of FLT3L was specific to cDC2s in ADAM10ΔDC and ADAM10ΔMx1 mice. Despite the decreases in FLT3L in circulation, prominent cDC2 decreases in ADAM10ΔDC mice were limited to the spleen, highlighting the importance of FLT3L that is produced locally for cDC maintenance (17).
ADAM10 and ADAM17 are the most extensively studied members of the ADAMs family. Although some redundancy of the 2 ADAMs has been reported (34), germline ablation of ADAM10 or ADAM17 is embryonic-lethal, indicating that they do not completely compensate for the loss of each other during early development (21, 22). Among molecules that have been reported to be substrates of both ADAM10 and ADAM17, Notch is of interest because the disruption of Notch signaling via the conditional deletion of RBPJ in DCs is sufficient to impair splenic cDC2 development (23). Although ADAM10 is believed to be the primary ADAM that regulates Notch signaling (18), ADAM17 has also been reported to cleave Notch (35). Although the latter finding for ADAM17 is not undisputed, if true, the finding that cDC2 numbers were decreased in ADAM10ΔDC mice, but not in ADAM17ΔDC mice, suggests a mechanism involved downstream of ADAM10 that is independent of Notch signaling. Indeed, cDC2s were incompletely rescued by the restoration of the Notch signaling pathway, and the complete rescue of cDC2s by rFLT3L administration alone indicated that driving the FLT3 signaling pathway was sufficient for cDC2 terminal differentiation in the absence of ADAM10. Whether the Notch and FLT3L pathways intersect, particularly via ADAM10, has yet to be explored.
cDC2s expressed Flt3l and Adam10 at levels that were higher than those of cDC1s. Thus, this gene expression pattern can potentially explain the distinct dependency on the ADAM10–FLT3L axis between the 2 cDC subsets. However, if both Adam10 and Adam17 are expressed by cDC2s, and because both ADAMs are capable of cleaving FLT3L (33), spatial differences in gene expression may not sufficiently explain the selective dependence of cDC2s on the ADAM10–FLT3L axis and its independence on ADAM17. In addition, the induced ablation of ADAM10 in ADAM10ΔMx1 mice led to membrane-bound retention of FLT3L on cDC2s, but not on T cell surfaces, suggesting that FLT3L cleavage among different cell subsets might be mediated through distinct mechanisms.
The regulation of ADAMs is not yet fully understood. However, recent studies on the TspanC8 subgroup of tetraspanins and iRhom subgroup of protease-inactive rhomboids have begun to elucidate mechanisms by which localization and trafficking of ADAM10 and ADAM17 are regulated, respectively (36). TspanC8 subgroups Tspan5, 10, 14, 17, and 33 have been found to associate with ADAM10 to promote enzymatic maturation. Partnering of ADAM10 with Tspan5, 10, or 14 positively regulates Notch signaling (31), but partnering with Tspan15 or 33 negatively regulates it (32). Similarly, iRhom1 and iRhom2 mediate enzymatic maturation of ADAM17 (37). Thus, despite the widespread expression of ADAM10 and ADAM17, tetraspanin- and rhomboid-mediated regulation of ADAM activation and specificity of substrates may dictate proteolytic activity within or among different cell types. Expression of certain tetraspanins by DCs have been reported (38). It is therefore possible that cDCs utilize tetraspanin- and rhomboid-mediated mechanisms for the differential regulation of molecules and downstream pathways that are critically involved in cDC2 development and maintenance, such as FLT3 and Notch signaling pathways.
In conclusion, we have determined that constitutive or induced ablation of ADAM10 in DCs resulted in the striking loss of splenic DC2s through the disruption of a mechanism in which cell-autonomous ADAM10 cleaved cell-surface membrane-bound forms of FLT3L to release soluble FLT3L. The finding that cDC2 loss was specific to the spleen points to tissue-dependent mechanisms that support cDC2 maintenance and opens the door for studies on how cDC homeostasis is regulated in different organs. The ADAM10–FLT3 signaling axis we describe highlights a mechanism that might allow for cDC2s to gain prioritized access to FLT3L, which may be relevant for the development of novel strategies to harness immune responses in the contexts of vaccines, tumor immunity, and autoimmune diseases.
Materials and Methods
Mice.
C57BL/6 (CD45.2) mice were purchased from CLEA Japan, and C57BL/6J Rag2−/− mice were purchased from the Central Institute for Experimental Animals. Adam10fl/fl and Adam17fl/fl mice were generated previously (30, 39). B6.Cg-Tg (Itgax-cre)i-1Reiz/J (CD11c-cre mice), B6.Cg-Tg(Mx1-cre)1Cgn/J (Mx1-Cre mice), and Gt(ROSA)26Sortm1(Notch1)Dam/J (ROSANotch mice) were purchased from the Jackson Laboratory and were each bred to Adam10fl/fl and Adam17fl/fl mice to generate CD11c-cre × Adam10fl/fl (ADAM10ΔDC), CD11c-cre × Adam17fl/fl (ADAM17 ΔDC), Mx1-cre × Adam10fl/fl (ADAM10ΔMX1), and Mx1-cre × Adam10fl/fl × ROSANotch (ADAM10ΔMX1 ROSANotch; heterozygous for ROSANotch) mice. As WT controls, CD11c-cre × Adam10fl/wt mice or Adam10fl/fl mice were used unless stated otherwise. Male and female mice aged between 6 and 12 wk of age were used for experiments, with no apparent sex-related differences in reported results. All animal procedures and study protocols were approved by the Keio University School of Medicine Ethics Committee for Animal Experiments (Tokyo, Japan) or animal care and use committee of the National Cancer Institute, National Institutes of Health (Bethesda, MD).
Antibodies Utilized for Flow Cytometry Analysis.
The following fluorochrome-coupled monoclonal antibodies against indicated molecules were used for flow cytometry. CD11c (clone N418), CD11b (clone M1/70), ESAM (clone 1G8), I-A/I-E (clone M5/114.15.2), B220 (RA3-6B2), CD45.1 (A20), CD45.2 (104), CD45 (30-F11), SiglecH (clone 551), CD135 (clone A2F10), and CD16/32 (clone 93) were obtained from Biolegend. Anti-CD19 antibody (1D3) was from BD Biosciences. A biotinylated goat polyclonal antibody against FLT3L (cat. no. BAF427) was obtained from R&D Systems. For the detection of biotin, streptavidin-PE/Cy7 or Brilliant Violet 421 streptavidin (Biolegend) was used at 1:400 concentrations.
Preparation of Single Cell Suspensions.
BM was flushed from tibia and femur of experimental mice, pipetted, and filtered to generate single cell suspensions. Spleens were mashed against a 100-µm Cell Strainer (BD Falcon) and washed with 2% fetal calf serum (FCS) in PBS solution. Cells were lysed in ACK lysing buffer (Thermo Fisher) for 1 min at room temperature, washed, and suspended with 2% FCS in PBS solution.
For sorting pre-DCs, spleens were digested for 30 min in RPMI 10% FBS and collagenase type IV (0.2 mg/mL; working activity of 770 U/mg; Sigma). For sorting, spleen cell suspensions underwent preenrichment for CD135+ cells. Splenocytes were stained with anti-CD135 biotinilated antibody (eBioscence cat. no. 13–1351-85) and preenriched by using anti-biotin MicroBeads (Miltenyi cat. no. 130–090-485) and were separated on an autoMACS device (Miltenyi). After enrichment, cells were labeled with fluorochrome-conjugated monoclonal primary antibodies anti-CD45.2 (eBioscence cat. no. 45–0454-82), anti-CD19 (Biolegend cat. no. 115512), anti-CD3 (Biolegend cat. no. 100312), anti-CD49b (Biolegend cat. no. 108910), anti-Ly6G (Biolegend cat. no. 127614), anti-MHCII (eBioscence cat. no. 56–5321-82), anti-CD11c (Biolegend cat. no. 117308), anti-CD172a (Biolegend cat. no. 144006), anti-Ly6C (Biolegend cat. no. 128026), anti-SiglecH (eBioscence cat. no. 48–0333), or streptavidin (Biolegend cat. no. 405206) and sorted on a FACSAria II device (Becton Dickinson).
Flow Cytometry Analysis and Cell Sorting.
Data were acquired with FACS Canto II (BD Biosciences), FACS Diva (BD Biosciences), or LSRII (BD Biosciences) and were analyzed by using FlowJo version 10. Combination of antibodies to exclude lineage-positive cells in the bone marrow and spleen included those against I-A/I-E, B220, CD19, NK1.1, CD3ε, Ter119, CD11c, CD11b, Gr-1, Ly6G, and CD172a. Antibody against mouse CD16/32 was routinely used to block Fcγ receptors before staining with primary antibodies.
A nonviable cell was identified with propidium iodide (Sigma-Aldrich) after antibody staining or with LIVE/DEAD Fixable Aqua Dead cell stain kit (Thermo Fisher) before antibody staining. Fluorescence-activated cell sorting of DC subsets was performed with MoFlow (Beckman Coulter), during which cells were directly sorted into TRIzol LS (Invitrogen). A FACSAria II device (Becton Dickinson) was used for sorting splenic pre-DCs from WT C57BL/6 mice that were lethally irradiated and reconstituted with BM from ADAM10ΔDC or ADAM17 ΔDC mice, which were used for RNA-sequencing analysis.
Generation of Bone Marrow Chimeras.
C57BL/6J (CD45.2) WT female mice 6 to 10 wk of age were lethally irradiated (950 rad) and were reconstituted via i.v. injection of 2.0 × 106 total BM cells from WT (CD45.1) and ADAM10ΔDC mice (CD45.2). BM chimeric mice were analyzed 8 wk after reconstitution.
Immunofluorescence Microscopy.
Spleens from WT and ADAM10ΔDC mice were embedded in OCT compound (Sakura) to generate frozen sections. Sections (5 to 7 µm) were prepared and stored at −20 °C until further use. For staining, sections were air-dried and fixed in acetone (Wako Pure Chemical) for 5 min at −20 °C, air-dried, and rehydrated in PBS solution for 5 min. Before staining, they were blocked in 3% skim milk (Morinaga) in PBS solution and 5% goat serum (Dako) for 1 h at room temperature in a humidified chamber. Sections were then stained with an anti-CD11b antibody (M1/70) conjugated with FITC (diluted to 2 μg/mL in blocking buffer) and incubated overnight at 4 °C. After 3 washes with PBS solution, FITC was further detected by incubating sections with Alexa Fluor 488-conjugated anti-fluorescein/Oregon Green Antibody (A-11096; Thermo Fisher) at 1:400 dilution, and nuclei were visualized with Hoechst 33258 (Invitrogen). After washing 3 times in PBS solution, sections were embedded in mounting medium.
Stained sections were observed with a Zeiss Axio Observer.Z1 microscope (Carl Zeiss) and acquired with AxioVision software (version 4.8) with identical exposure time between experimental groups. Levels were adjusted equally among experimental and control samples in Photoshop CC 2015 (Adobe).
In Vivo rFlt3L Injection.
Recombinant mouse FLT3L (10 μg in 200 μL PBS solution; eBioscience) or PBS solution was injected intraperitoneally into WT and Adam10ΔDC mice daily for 6 d. Spleens of treated mice were analyzed on the day after the last injection.
Quantification of FLT3L Concentration.
Soluble FLT3L concentrations in mouse sera were measured by an ELISA system (R&D Systems) following the manufacturer’s instructions. Small incisions were made onto anesthetized WT, Rag2−/−, and ADAM10ΔDC mouse tails, and whole blood was collected by using capillaries, which were centrifuged at 5,000 rpm for 10 min to obtain sera. Fresh sera were utilized for analysis.
Sorted cDC1s and cDC2s from WT mice were cultured in 96-well plates in complete RPMI medium supplemented with 20 ng/mL of recombinant GM-CSF (Peprotech) for 48 h. Culture supernatants were then collected, and FLT3L concentrations were measured by ELISA as described earlier.
Quantification of FLT3L Concentration in MEF Culture Supernatants.
Fibroblasts collected from a 10.5-d-postconception WT, Adam10−/−, or Adam17−/− embryo were immortalized by introducing SV40 expression vector to generate MEFs as previously reported (33). MEFs were seeded onto 12-well plates and cultured at 37 °C in complete RPMI. After obtaining confluency, cells were further cultured for 2 d, and supernatants were then collected for analysis of FLT3L concentrations via ELISA.
The expression vectors for WT Adam10 and AP-tagged Flt3lg have been described previously (30, 33). To rescue ADAM10 expression in Adam10−/− MEFs, we utilized Adam10-encoding plasmid DNA or control pcDNA3.1 (33). Adam10−/− MEFs transfected with Adam10-encoding plasmid DNA or control pcDNA3.1, as well as WT MEFs transfected with control pcDNA3.1, were all simultaneously transfected with alkaline phosphatase-tagged Flt3L (Flt3L-AP) (33) by using the Fugene system (Roche) according to the manufacturer’s protocol. MEFs were cultured in fresh complete RPMI medium for 48 h, and supernatants were collected. p-Nitrophenyl phosphate (pNPP; Sigma) was added to each serum to detect alkaline phosphatase activity, which was measured by colorimetry read at 405 nm absorbance by using a SpectraMax Paradigm (Molecular Devices).
Real-Time PCR.
Splenocyte populations were sorted by FACS, and RNA was extracted by using an RNeasy Micro kit (Qiagen). Then, cDNA was generated by using SuperScript III First-Strand Synthesis SuperMix (Invitrogen). SYBR Green Master Mix (Applied Biosystems) and the StepOnePlus real-time PCR system with standard mode (Applied Biosystems) were used according to the manufacturer’s protocol. All primers were designed by Primer Express software (Applied Biosystems). Normalization of mRNA expression was performed on the basis on the expression of β-actin by using the threshold cycle (ΔCT) method, and the amount of PCR product was calculated based on 2−ΔCT. The following primers were used to amplify genes of interest: Flt3lg forward, 5′-GGGAACCAAAACAAGGAACAAG-3′; reverse, 5′-GTCCATCGCCATACCCAGA-3′; and Adam10 forward, 5′-GGTTTAAAGCGTACCCACGA-3′; reverse, 5′-GAACTGGGCGTCATCATTTT-3′. Cycle conditions were 10 min at 95 °C followed by 40 cycles of 15 s at 95 °C and 60 s at 60 °C.
Nanostring Gene Expression Analysis.
Gene expression analysis of extracted RNA from sorted cells from the spleen was performed by nCounter Immunology Panels (NanoString Technologies) according to the manufacturer’s protocol. Normalization of gene expression was performed by using nSolver Analysis Software (NanoString Technologies), and differentially expressed genes were generated by Partek Genomics Suite (Partek).
Bulk RNA Sequencing.
FACS-sorted pre-DCs from several animals were pooled. Total RNA was extracted by using an Arcturus PicoPure RNA Isolation kit (Applied Biosystems Thermo Fisher Scientific) according to the manufacturer’s protocol. All mouse RNAs were analyzed on an Agilent Bioanalyzer for quality assessment with RNA Integrity Number range of RIN 8.2 to 9.9 and a median RIN of 9.6. cDNA libraries were prepared by using 2 ng of total RNA and 1 µL of a 1:50,000 dilution of ERCC RNA Spike-In Controls (Ambion Thermo Fisher Scientific) using the SMARTSeq v2 protocol (40) with the following modifications: 1) addition of 20 µM TSO and 2) use of 200 pg cDNA with 1/5 reaction of Illumina Nextera XT kit (Illumina). The length distribution of the cDNA libraries was monitored by using a DNA High Sensitivity Reagent Kit on the Labchip system (Perkin–Elmer). All samples were subjected to an indexed paired-end sequencing run of 2 × 151 cycles on an Illumina HiSEq 4000 system (25 samples per lane). PCA plots and heat maps of the differential gene expression (P value with FDR < 0.05) were generated by using Partek Genomics Suite (Partek). Sequence data were deposited in the NCBI Gene Expression Omnibus database under accession number GSE131264.
Supplementary Material
Acknowledgments
K.F. and M.A. were supported by Ministry of Education, Culture, Sports, Science and Technology/Japan Society for the Promotion of Science KAKENHI. K.N. was supported by the Intramural Research Program of National Institute of Arthritis and Musculoskeletal and Skin Diseases and National Cancer Institute. F.G. was funded by the Singapore Immunology Network core funding, Agency for Science, Technology and Research (A*STAR) and Singapore National Research Foundation Senior Investigatorship (NRFI2017-02). The bioinformatics and immunogenomics platforms are part of the SIgN Immunomonitoring platform.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The data reported in this paper have been deposited in the Gene Expression Omnibus (GEO) database, https://www.ncbi.nlm.nih.gov/geo (accession no. GSE131264).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1818907116/-/DCSupplemental.
References
- 1.Steinman R. M., et al. , Dendritic cell function in vivo during the steady state: A role in peripheral tolerance. Ann. N. Y. Acad. Sci. 987, 15–25 (2003). [DOI] [PubMed] [Google Scholar]
- 2.Guilliams M., et al. , Dendritic cells, monocytes and macrophages: A unified nomenclature based on ontogeny. Nat. Rev. Immunol. 14, 571–578 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Schlitzer A., et al. , Identification of cDC1- and cDC2-committed DC progenitors reveals early lineage priming at the common DC progenitor stage in the bone marrow. Nat. Immunol. 16, 718–728 (2015). [DOI] [PubMed] [Google Scholar]
- 4.Thacker R. I., Janssen E. M., Cross-presentation of cell-associated antigens by mouse splenic dendritic cell populations. Front. Immunol. 3, 41 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Pakalniškytė D., Schraml B. U., Tissue-specific diversity and functions of conventional dendritic cells. Adv. Immunol. 134, 89–135 (2017). [DOI] [PubMed] [Google Scholar]
- 6.Jaiswal H., et al. , Batf3 and Id2 have a synergistic effect on Irf8-directed classical CD8α+ dendritic cell development. J. Immunol. 191, 5993–6001 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ginhoux F., et al. , The origin and development of nonlymphoid tissue CD103+ DCs. J. Exp. Med. 206, 3115–3130 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Suzuki S., et al. , Critical roles of interferon regulatory factor 4 in CD11bhighCD8α- dendritic cell development. Proc. Natl. Acad. Sci. U.S.A. 101, 8981–8986 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Schlitzer A., et al. , IRF4 transcription factor-dependent CD11b+ dendritic cells in human and mouse control mucosal IL-17 cytokine responses. Immunity 38, 970–983 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Scott C. L., et al. , The transcription factor Zeb2 regulates development of conventional and plasmacytoid DCs by repressing Id2. J. Exp. Med. 213, 897–911 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Schlitzer A., McGovern N., Ginhoux F., Dendritic cells and monocyte-derived cells: Two complementary and integrated functional systems. Semin. Cell Dev. Biol. 41, 9–22 (2015). [DOI] [PubMed] [Google Scholar]
- 12.Lyman S. D., et al. , Molecular cloning of a ligand for the flt3/flk-2 tyrosine kinase receptor: A proliferative factor for primitive hematopoietic cells. Cell 75, 1157–1167 (1993). [DOI] [PubMed] [Google Scholar]
- 13.Karsunky H., Merad M., Cozzio A., Weissman I. L., Manz M. G., Flt3 ligand regulates dendritic cell development from Flt3+ lymphoid and myeloid-committed progenitors to Flt3+ dendritic cells in vivo. J. Exp. Med. 198, 305–313 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Naik S. H., et al. , Cutting edge: Generation of splenic CD8+ and CD8- dendritic cell equivalents in Fms-like tyrosine kinase 3 ligand bone marrow cultures. J. Immunol. 174, 6592–6597 (2005). [DOI] [PubMed] [Google Scholar]
- 15.McKenna H. J., et al. , Mice lacking flt3 ligand have deficient hematopoiesis affecting hematopoietic progenitor cells, dendritic cells, and natural killer cells. Blood 95, 3489–3497 (2000). [PubMed] [Google Scholar]
- 16.Saito Y., Boddupalli C. S., Borsotti C., Manz M. G., Dendritic cell homeostasis is maintained by nonhematopoietic and T-cell-produced Flt3-ligand in steady state and during immune responses. Eur. J. Immunol. 43, 1651–1658 (2013). [DOI] [PubMed] [Google Scholar]
- 17.Miloud T., Fiegler N., Suffner J., Hämmerling G. J., Garbi N., Organ-specific cellular requirements for in vivo dendritic cell generation. J. Immunol. 188, 1125–1135 (2012). [DOI] [PubMed] [Google Scholar]
- 18.Edwards D. R., Handsley M. M., Pennington C. J., The ADAM metalloproteinases. Mol. Aspects Med. 29, 258–289 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Black R. A., et al. , A metalloproteinase disintegrin that releases tumour-necrosis factor-α from cells. Nature 385, 729–733 (1997). [DOI] [PubMed] [Google Scholar]
- 20.Sahin U., et al. , Distinct roles for ADAM10 and ADAM17 in ectodomain shedding of six EGFR ligands. J. Cell Biol. 164, 769–779 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hartmann D., et al. , The disintegrin/metalloprotease ADAM 10 is essential for Notch signalling but not for alpha-secretase activity in fibroblasts. Hum. Mol. Genet. 11, 2615–2624 (2002). [DOI] [PubMed] [Google Scholar]
- 22.Peschon J. J., et al. , An essential role for ectodomain shedding in mammalian development. Science 282, 1281–1284 (1998). [DOI] [PubMed] [Google Scholar]
- 23.Lewis K. L., et al. , Notch2 receptor signaling controls functional differentiation of dendritic cells in the spleen and intestine. Immunity 35, 780–791 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Damle S. R., et al. , ADAM10 and Notch1 on murine dendritic cells control the development of type 2 immunity and IgE production. Allergy 73, 125–136 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Uchikawa S., et al. , ADAM17 regulates IL-1 signaling by selectively releasing IL-1 receptor type 2 from the cell surface. Cytokine 71, 238–245 (2015). [DOI] [PubMed] [Google Scholar]
- 26.Janes P. W., et al. , Adam meets Eph: An ADAM substrate recognition module acts as a molecular switch for ephrin cleavage in trans. Cell 123, 291–304 (2005). [DOI] [PubMed] [Google Scholar]
- 27.Panek C. A., et al. , Differential expression of the fractalkine chemokine receptor (CX3CR1) in human monocytes during differentiation. Cell. Mol. Immunol. 12, 669–680 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Onodera K., et al. , GATA2 regulates dendritic cell differentiation. Blood 128, 508–518 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tussiwand R., et al. , Klf4 expression in conventional dendritic cells is required for T helper 2 cell responses. Immunity 42, 916–928 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yoda M., et al. , Dual functions of cell-autonomous and non-cell-autonomous ADAM10 activity in granulopoiesis. Blood 118, 6939–6942 (2011). [DOI] [PubMed] [Google Scholar]
- 31.Dornier E., et al. , TspanC8 tetraspanins regulate ADAM10/Kuzbanian trafficking and promote Notch activation in flies and mammals. J. Cell Biol. 199, 481–496 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Jouannet S., et al. , TspanC8 tetraspanins differentially regulate the cleavage of ADAM10 substrates, Notch activation and ADAM10 membrane compartmentalization. Cell. Mol. Life Sci. 73, 1895–1915 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Horiuchi K., et al. , Ectodomain shedding of FLT3 ligand is mediated by TNF-alpha converting enzyme. J. Immunol. 182, 7408–7414 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Pruessmeyer J., Ludwig A., The good, the bad and the ugly substrates for ADAM10 and ADAM17 in brain pathology, inflammation and cancer. Semin. Cell Dev. Biol. 20, 164–174 (2009). [DOI] [PubMed] [Google Scholar]
- 35.Brou C., et al. , A novel proteolytic cleavage involved in Notch signaling: The role of the disintegrin-metalloprotease TACE. Mol. Cell 5, 207–216 (2000). [DOI] [PubMed] [Google Scholar]
- 36.Matthews A. L., Noy P. J., Reyat J. S., Tomlinson M. G., Regulation of A disintegrin and metalloproteinase (ADAM) family sheddases ADAM10 and ADAM17: The emerging role of tetraspanins and rhomboids. Platelets 28, 333–341 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Adrain C., Zettl M., Christova Y., Taylor N., Freeman M., Tumor necrosis factor signaling requires iRhom2 to promote trafficking and activation of TACE. Science 335, 225–228 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zuidscherwoude M., Worah K., van der Schaaf A., Buschow S. I., van Spriel A. B., Differential expression of tetraspanin superfamily members in dendritic cell subsets. PLoS One 12, e0184317 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Horiuchi K., et al. , Cutting edge: TNF-alpha-converting enzyme (TACE/ADAM17) inactivation in mouse myeloid cells prevents lethality from endotoxin shock. J. Immunol. 179, 2686–2689 (2007). [DOI] [PubMed] [Google Scholar]
- 40.Picelli S., et al. , Full-length RNA-seq from single cells using Smart-seq2. Nat. Protoc. 9, 171–181 (2014). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.