Significance
From an interspecies study, we identified and validated downstream targets of the MYC oncogene and demonstrate the conserved role of aminoacyl-tRNA synthetases (aaRSs) in MYC-driven growth. We also tested aaRS inhibitors and describe their potential as therapeutic agents to selectively target MYC-driven cancers. By interfering with protein synthesis, we create a catastrophic situation in which the translation machinery is incapable of handling the increased demand triggered by the pro-growth oncogenic program. This provides a general principle for the design of therapeutic approaches to tumorigenesis.
Keywords: MYC, tRNA synthetase, cancer, nucleolus, Drosophila
Abstract
Aberrant MYC oncogene activation is one of the most prevalent characteristics of cancer. By overlapping datasets of Drosophila genes that are insulin-responsive and also regulate nucleolus size, we enriched for Myc target genes required for cellular biosynthesis. Among these, we identified the aminoacyl tRNA synthetases (aaRSs) as essential mediators of Myc growth control in Drosophila and found that their pharmacologic inhibition is sufficient to kill MYC-overexpressing human cells, indicating that aaRS inhibitors might be used to selectively target MYC-driven cancers. We suggest a general principle in which oncogenic increases in cellular biosynthesis sensitize cells to disruption of protein homeostasis.
An increase in MYC oncogene level drives tumor formation and is associated with poor prognosis (1, 2). Unfortunately, MYC has often been classified as undruggable due to the absence of a ligand-binding domain or a hydrophobic pocket suitable for a small-molecule inhibitor (3, 4). Thus, research to date has focused on synthetic lethal approaches, identifying MYC coactivators and downstream targets that mediate its role in tumorigenesis (5–10).
MYC is a transcription factor that controls a core set of target genes involved in ribosome biogenesis and protein synthesis (11, 12). Deregulation of these processes leads to excessive cell growth/proliferation, suggesting that targeting anabolic pathways downstream of MYC might effectively kill cancer cells. In fact, many chemotherapy drugs function by inhibiting ribosome biogenesis (13). Interestingly, in cancer cells with oncogenic activation of the similarly growth-promoting PI3K pathway, disruption of either anabolic or catabolic pathways, but not both simultaneously, was seen to selectively kill the tumor cells (14). This suggests that oncogenic up-regulation of cellular biosynthesis may render cells generally susceptible to disruption of homeostasis.
Drosophila has a single MYC gene (Myc), which functions downstream of insulin signaling, controlling nucleolus size and tissue growth (15–18). Here we describe our effort to identify genes downstream of insulin that are required for Myc control of growth. By overlapping lists of genes that are insulin-responsive and regulate nucleolus size, we enriched for Myc target genes that are required for Myc function in vivo. We identify the aminoacyl-tRNA synthetases (aaRSs) as essential mediators of Myc growth control in Drosophila and show that their inhibition is sufficient to kill MYC-overexpressing human cells. We propose a general principle in which disruption of homeostasis in an otherwise balanced progrowth oncogenic program can be selectively toxic to cells with excessive growth.
Results and Discussion
Identification of Insulin-Responsive Nucleolar Regulators.
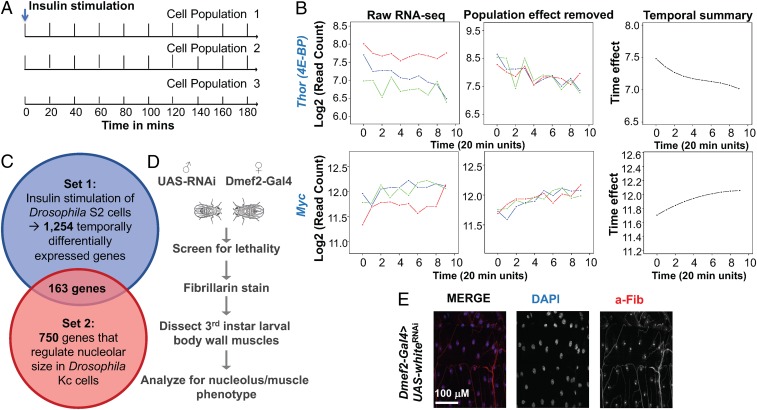
Because Myc activity regulates ribosome biogenesis in response to insulin signaling, we hypothesized that overlapping genes regulating ribosome biogenesis (RiBi) and genes responsive to insulin pathway would enrich for Myc targets involved in RiBi-mediated growth. We first evaluated the gene expression response to insulin stimulation in Drosophila S2R+ cells by RNA-seq. We selected 10 different time points at 20-min intervals (20, 40, 60, 80, 100, 120, 140, and 180 min) after insulin stimulation and performed RNA-seq on 3 biological replicates for each time point (Fig. 1A) (19). The short time frame of sample collection enabled us to tease apart secondary responses arising from changes in protein translation and to focus on the direct transcriptional response. To capture the overall temporal gene expression pattern instead of differential gene expression between time points, we applied time series statistical modeling to the RNA-seq dataset and identified approximately 1,254 insulin-responsive temporally differentially expressed genes (Datasets S1 and S2). Known targets of insulin signaling, such as Thor (4E-BP) and Myc, were identified as down-regulated and up-regulated over time, respectively, consistent with their previously reported transcriptional response (Fig. 1B).
Fig. 1.
Insulin-responsive temporally differentially expressed genes. (A) RNA-seq of 10 different time points at 20-min intervals (3 biological replicates per time point) after insulin stimulation of Drosophila S2R+ cells. (B) RNA-seq data for Thor and Myc, both known to be transcriptionally regulated by insulin signaling in Drosophila. (C) 163 genes overlapped between 1,254 insulin-regulated genes and 750 previously identified nucleolus regulators. (D) Scheme for identification of genes affecting nucleolar morphology/growth. (E) Control ventrolateral muscles from third instar larvae stained for DAPI (nucleus) in blue and anti-Fibrillarin (nucleolus) in red.
We then overlapped the set of 1,254 insulin-responsive genes with a set of 750 genes previously identified as important for nucleolus size in Drosophila Kc cells (Fig. 1C and Dataset S2) (20). We found 163 genes were shared in the two sets, a highly statistically significant overlap (P = 5.4e-28). From this overlap set, we performed an in vivo screen for lethality and nucleolus phenotypes in larval muscle, a tissue that we previously used as an effective readout of insulin/Myc activity (Fig. 1D and Dataset S3) (15). We dissected muscles from larvae in which gene knockdown generated a lethal phenotype, reasoning that these would have the most significant effect on the nucleolus. Indeed, we did not observe nucleolar morphology phenotypes in any nonlethal knockdown crosses. Lethality of the knockdowns ranged from third instar to pharate lethal.
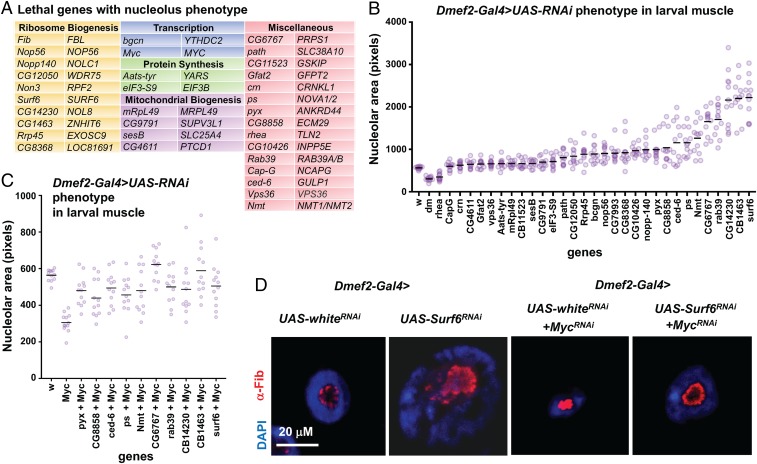
Dissection and staining of the larval muscles from lethal RNAi knockdowns revealed 33 genes required for normal nucleolus morphology (Fig. 2A). The largest subset comprised genes involved in ribonucleoprotein complex biogenesis (12.65-fold enrichment; P = 2.55e-8; false discovery rate [FDR] = 3.96e-4) and RiBi (15.75-fold enrichment; P = 3.32e-8; FDR = 2.57e-4) based on Gene Ontology biological process assignments. Other multiple gene categories were transcription, protein synthesis, and mitochondrial biogenesis. Fifteen genes could not be categorized together. All of the 33 genes have conserved human orthologs, with several previously identified in analyses of Myc/MYC target genes (21–30). We take the high number of previously reported Myc/MYC targets in our dataset as confirming the effectiveness of our screening approach.
Fig. 2.
Identification of Myc-dependent nucleolus regulators in Drosophila. (A) List of 33 genes that were lethal when knocked down and were required for normal nucleolus morphology. (B and C) Area of α-Fibrillarin (nucleolus) stain following knockdown of each gene by Dmef2-Gal4; UAS-RNAi (B) or simultaneous knockdown of Myc and the 10 genes with the highest-scoring nucleolar area phenotype (C). Circles represent the total areas from single VL4 muscles from individual larvae. Horizontal lines denote the grand mean. (D) Dmef2-Gal4 knockdown of Surf6 causes enlarged nucleolus and nucleus compared with control white knockdown. Concurrent knockdown of Myc blocks the Surf6 nucleolus phenotype. DAPI (nucleus) is in blue, and α-Fibrillarin (nucleolus) is in red.
We next scored the mean nucleolar area (α-Fibrillarin stain) for 12 individual larval muscles from each knockdown experiment (Fig. 2B and Dataset S4). For all genes except Myc and rhea, the nucleolar area resulting from knockdown was greater than that in controls. This is likely due to increased ribosomal nucleolar stress, as evidenced by the uneven DAPI and Fibrillarin staining and consistent with a previous report (31). Concurrent knockdown of Myc and the 10 genes with the highest-scoring nucleolar area phenotypes dramatically reduced the area compared with each gene alone (Fig. 2 C and D), indicating that the function of these genes with respect to nucleolar size depends on Myc expression level.
Insulin/Nucleolus Gene Set Is Enriched for Myc Targets.
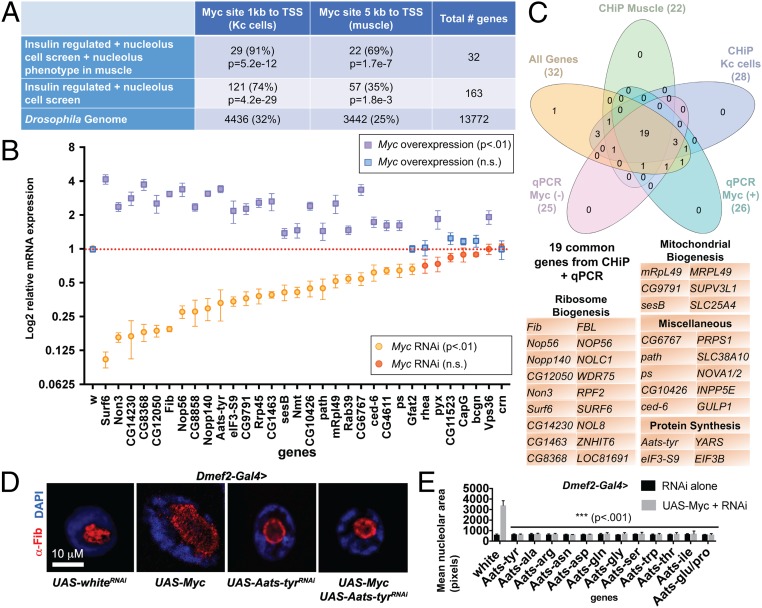
To determine which of the 33 genes might be direct downstream targets of Myc, we performed chromatin immunoprecipitation sequencing (ChIP-seq) with a Myc antibody on dissected larval cuticle/muscle preparations (SI Appendix, Fig. S1A). Data from 3 biological replicates were combined to yield better peaks (SI Appendix, Fig. S1B). A total of 3,442 genes had a Myc-binding site within 5 kb of their transcription start site (MACS; P < 0.0001), representing approximately 25% of the genome (Fig. 3A and Dataset S5). Considering only the 32 genes, other than Myc itself, that were included in the overlap set of insulin and nucleolus datasets and had nucleolus phenotypes, 22 (69%) had a Myc site within 5 kb of the transcription start site, a 2.8-fold enrichment (P = 1.7e-7). We observed a similar 2.86-fold enrichment (P = 5.2e-12) of insulin regulated/nucleolus overlap genes using ChIP data previously generated from Drosophila Kc cells (Fig. 3A) (32). These data suggest that our approach is indeed able to enrich for Myc-regulated genes.
Fig. 3.
Enrichment of Myc targets reveals aaRSs as mediators of nucleolar hypertrophy. (A) Enrichment of Myc-binding sites in muscle ChIP-seq data and previously published data from Kc cells. The percentage of genes with a Myc site is lowest in the total genome, increases in the nucleolus/insulin overlap gene set, and is highest in the overlap gene set with a knockdown phenotype. P values are shown for overenrichment based on the hypergeometric distribution. (B) qPCR of putative Myc targets from Dmef2-Gal4 > UAS-MycRNAi larval muscle or Dmef2-Gal4 > UAS-Myc overexpression muscle normalized to Dmef2-Gal4 > UAS-whiteRNAi control. Significant changes in gene expression are indicated by purple squares for Myc overexpression and by orange circles for Myc knockdown. (C) Venn diagram showing overlap of the nucleolus/insulin overlap gene set with positive scoring genes from qPCR of Myc knockdown, qPCR of Myc overexpression, Myc ChIP-seq of larval muscles, and Myc ChIP-seq of Kc cells. The 19 common genes are listed. (D) Aats-tyr knockdown suppresses nucleolar hypertrophy induced by Myc overexpression. Dmef2-Gal4 was used to drive expression of UAS transgenes in larval muscle. Larvae were stained for DAPI (nucleus) in blue and α-Fibrillarin (nucleolus) in red. (E) Knockdown of 8 additional aaRS genes was able to suppress Myc-induced hypertrophy to wild-type levels. Mean area of α-Fibrillarin stain (nucleolus) from single VL4 muscles (>6 larvae) following knockdown of each gene by Dmef2-Gal4+/− UAS-Myc.
We next knocked down or overexpressed Myc using the Dmef2-Gal4 driver line and examined expression levels in the third instar larval muscle of the 32 genes (other than Myc) with a nucleolus phenotype (Fig. 3B). Of these genes, 25 were significantly down-regulated in Myc RNAi muscles relative to control, while 26 were significantly up-regulated in Myc-overexpression muscles (P < 0.01) relative to control. Thus, Myc modulates the expression of approximately 75% of the putative Myc target genes present in our insulin/nucleolus dataset, with 19 of the 32 genes found in all 4 datasets (Myc RNAi qPCR, Myc overexpression qPCR, larval muscle Myc ChIP-seq, and Kc cell ChIP-seq) (Fig. 3C and Dataset S6).
aaRS Knockdown/Inhibition Blocks Myc-Induced Nucleolar Hypertrophy and Cell Proliferation.
To identify Myc targets that could be pharmacologically targeted, we focused our attention on Aats-tyr, the Drosophila tyrosyl aaRS. aaRSs are essential enzymes for all cellular life and have been pursued as drug targets in bacteria, fungi, and eukaryotic parasites (33, 34). Dmef2-Gal4–driven knockdown of Aats-tyr in the larval muscle resulted in a slight increase in nucleolar size (Fig. 2B), accompanied by disrupted nuclear DAPI staining and nucleolar α-Fibrillarin staining (Fig. 3D). However, Aats-tyr knockdown strongly suppressed the nucleolar hypertrophy resulting from Myc overexpression in the larval muscle (Fig. 3D). Importantly, we tested all the aaRS genes identified by Myc ChIP-seq (Fig. 3E) and found that knockdown of each was able to suppress Myc-induced nucleolar hypertrophy. This supports the idea that their canonical role in charging tRNAs with amino acids is what determines the suppression of Myc-induced hypertrophy.
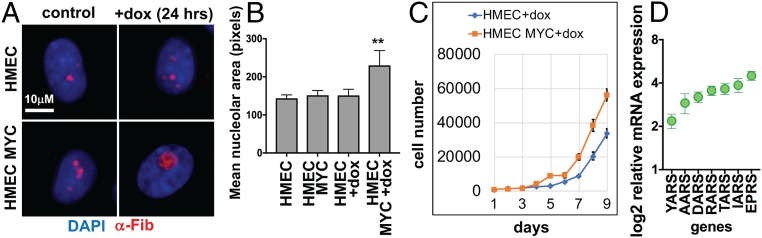
We next explored the effect of aaRS inhibition in a human mammary epithelial cell (HMEC) line in which the presence of doxycycline (dox) induces MYC expression from a tet-inducible promoter (SI Appendix, Fig. S2) (35, 36). The addition of dox to HMEC-MYC cells for 24 h induced strong MYC up-regulation (SI Appendix, Fig. S2), increasing the nucleolar area and cell proliferation relative to control (Fig. 4 A–C). Consistent with our observations in Drosophila, we also observed increased expression of several aaRS genes in dox-treated HMEC-MYC cells relative to control (Fig. 4D).
Fig. 4.
MYC induces nucleolar hypertrophy, proliferation, and aaRS expression in HMECs. (A) HMEC MYC + dox (10 ng/mL) cells show larger nucleolar size compared with control HMEC cells after 24 h. Cells are stained with DAPI (blue) and α-Fibrillarin (red). (B) Quantitation of the increased mean nucleolar area in HMEC MYC + dox cells (P < 0.01). (C) Growth curves of HMEC MYC (orange line) vs. control cells (blue line) treated with 10 ng/mL dox. MYC expression increases the rate of growth. (D) qPCR of aaRS genes in dox-treated HMEC MYC normalized to dox-treated control HMECs. MYC induction significantly increased aaRS expression (P < 0.01).
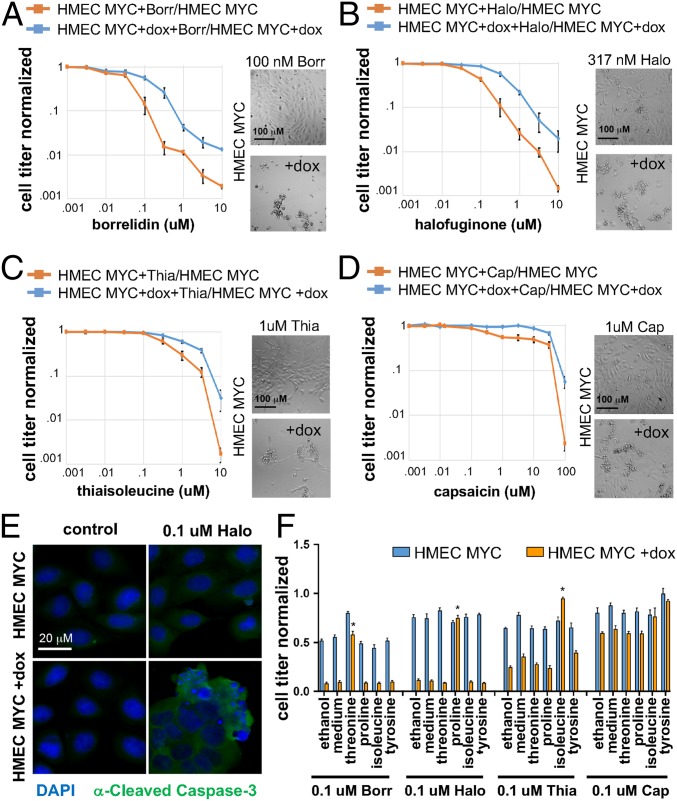
To test the effect of aaRS inhibitors for their ability to selectively target MYC-overexpressing cells, we compared noninduced control HMEC-MYC cells and dox-treated HMEC-MYC cells after 24 h of aaRS inhibition. Cell number/viability was assayed with CellTiter-Glo luminescent cell viability assay reagent (Promega). Borrelidin blocks most bacterial and eukaryotic threonyl-tRNA synthetases with sub-nM affinity (37). At concentrations of 100 nM and above, borrelidin treatment selectively killed HMEC-MYC + dox cells compared with control HMECs (Fig. 5A). Halofuginone inhibits dual glutamyl-prolyl-tRNA synthetase and has been designated an orphan drug for the treatment of scleroderma (38). Similar to borrelidin, at concentrations 100 nM and above, halofuginone selectively killed HMEC-MYC + dox cells compared with control HMECs (Fig. 5B). The amino acid isoleucine analog thiaisoleucine competes with isoleucine for binding to isoleucyl-tRNA synthetase (39, 40), and this compound selectively killed HMEC-MYC + dox cells compared with control HMECs at concentrations of 1 μM and above (Fig. 5C). Finally, we tested capsaicin, which had previously been identified as an analog of tyrosine that competes for binding to tyrosyl-tRNA synthetase (41). Of the 4 compounds, this was the least effective at selective killing of HMEC-MYC + dox cells compared with control HMECs (Fig. 5D). Halofuginone, borrelidin, and thiaisoleucine treatment triggered strong caspase staining and nuclear fragmentation in HMEC-MYC + dox cells, but not in control HMEC-MYC cells (Fig. 5E and SI Appendix, Fig. S3), indicating that the combination of MYC overexpression and aaRS inhibition causes apoptotic cell death.
Fig. 5.
aaRS inhibitors selectively kill c-Myc–overexpressing cells. (A–D) Viability curves of control HMEC-MYC cells vs. dox-induced HMEC-MYC cells after 24 h of aaRS inhibition. Cells treated with aaRS inhibitors were normalized to control untreated cells. Representative cell culture images are shown adjacent to curves. (A) At 100 nM and above, borrelidin (Borr) selectively kills dox-induced compared with uninduced HMEC-MYC cells. (B) At 100 nM and above, halofuginone (Halo) selectively kills dox-induced compared with uninduced HMEC-MYC cells. (C) At 1 uM and above, thiaisoleucine (Thia) selectively kills dox-induced compared with uninduced HMEC-MYC cells. (D) Capsaicin does not selectively kill dox-induced compared with uninduced HMEC-MYC cells. (E) Halo treatment triggers cell death of dox-induced HMEC-MYC cells, but not control HMEC-MYC cells. Cells are stained with DAPI (blue) and α-cleaved caspase-3 (green). (F) Viability of HMEC-MYC ± dox cells treated with aaRS inhibitors and supplemented with either controls (ethanol and medium) or amino acids (2 mM). Only the cognate amino acid for the aaRSs targeted by the drugs were able to rescue cell viability (Thr/Borr, Pro/Halo, Ile/Thia) P < 0.01.
Finally, to determine whether the effects of the compounds were due to competitive inhibition of the aaRSs, we tested whether addition of the cognate amino acid for each targeted aaRS could inhibit HMEC-MYC killing. Cell viability was rescued by the addition of 2 mM threonine, proline, and isoleucine to borrelidin-, halofuginone-, and thiaisoleucine-treated cells, respectively (Fig. 5F). Importantly, this rescue was specific to the amino acid and compound, demonstrating that the cellular effects of the compounds are the result of direct inhibition of a specific aaRS.
To identify druggable MYC downstream targets that mediate its role in tumorigenesis, we used nucleolus size as a proxy for Myc-driven growth in Drosophila larval muscles and identified 33 potential Myc target genes involved in growth regulation. The largest subset of these comprised genes involved in RiBi, and when knocked down, most produced abnormally enlarged nucleoli with uneven DAPI and Fibrillarin staining. One explanation for this phenotype is that loss of a single component of the translation machinery causes defective RiBi, loss of cell homeostasis, and nucleolar stress, as is seen in ribosomapathies (42). In contrast, Myc knockdown leads to regulated global reduction in RiBi and the protein synthesis machinery, which we hypothesize can attenuate the stress caused by KD of a single component. This phenotype could be exacerbated in the rapidly growing Drosophila larval muscle cells.
One of the less studied aspect of MYC control of protein synthesis is the regulation of aaRSs. Expression profiles of aaRSs indicate up-regulation in several cancers (43). Importantly, these proteins have been successfully targeted by drugs. In the present study, the most effective compounds were halofuginone and borrelidin, and the antiproliferative effects of both of these drugs in HMECs were heightened by increased MYC levels.
The MYC oncogene promotes growth by initiating a strong and balanced program of protein and RNA synthesis, with increased RNA synthesis driving a demand for increased protein synthesis capacity. The importance of tight control of RiBi and translation is evident in the coregulation of these processes by the transcriptional repressor MNT. Previous analysis of Myc-regulated genes in Drosophila showed that expression of several genes predicted to be involved in RiBi, including several genes identified in the present study, were partially rescued in Myc;Mnt double mutants (44). Thus, a balanced expression of RiBi components is maintained by input from both the transcriptional activator Myc and the repressor Mnt.
By generating an imbalance through interfering with protein synthesis, we can create a catastrophic situation in which the translation machinery is incapable of handling the increased demand, disrupting protein synthesis homeostasis in a toxic manner. A similar situation is thought to occur with PI3 kinase-driven cell growth, in which the increased protein synthesis is balanced with an increase in protein degradation. We recently performed a synthetic lethal screen for PI3K activation and found that interference with either protein synthesis or protein catabolism was synthetically lethal, but simultaneous impairment of both pathways caused no lethality, as the balance of competing needs was restored (14). In another study, we found splicing interference in MYC-overexpressing cells (6, 45), which would impair the mRNA component of the balanced program driven by MYC, much like our tRNA synthesis inhibition. Based on these findings, we believe that we have uncovered a general principle whereby the generation of an imbalance in an otherwise balanced progrowth oncogenic program can be selectively toxic to cancer cells. This may provide a general principle by which therapeutic approaches to tumorigenesis can be designed.
Materials and Methods
Immunostaining and Antibodies.
Third instar larval body wall muscles were dissected and fixed for 20 min in phosphate-buffered saline (PBS) with 4% formaldehyde. Cultured cells were fixed in 2% formaldehyde for 1 h. After washing in PBS containing 0.1% Tween-20 (PBT), samples were incubated overnight at 4 °C with rabbit α-Fibrillarin (1:300, ab5821; Abcam). The samples were then washed in PBT and incubated with Alexa Fluor-conjugated secondary antibodies (1:1,000; Molecular Probes) and DAPI (1 μg/mL; Sigma-Aldrich). Larval tissues were washed in PBT and mounted in 1:1 glycerol/PBS, and images were acquired with a Leica SP2 laser scanning confocal microscope. Cultured cells were washed in PBT and then PBS, and imaged with a GE IN Cell Analyzer 6000.
Nucleolus Size Analysis.
Z-stacks at 40× of single ventral longitudinal (VL4) muscles were obtained by confocal microscopy. Maximum intensity projections of the stacks were done with ImageJ. The threshold of the Fibrillarin channel was produced with the RATS Plugin (noise threshold = 20, lambda factor = 3, minimum leaf size = 100), and the total nucleolar area per muscle (n = 12 VL4 muscles) was measured. For cultured cells, ImageJ was used to threshold the Fibrillarin channel as above, and the total nucleolar area, number of nuclei, and mean nucleolus size were calculated for 4 replicates. Mean values were calculated for all measurements, with error bars indicating SEM. P values were calculated using Student’s t test.
qPCR.
Muscles from 10 third instar animals were dissected off of the cuticle, and RNA was prepared with TRIzol (Invitrogen), followed by purification with the RNeasy Kit (Qiagen). For cell cultures, 5 × 106 HMECs were seeded in T75 flasks and incubated for 48 h. MYC expression was induced by the addition of doxycycline (10 ng/mL) for 24 h, and RNA was produced as above. cDNA was synthesized with the iScript cDNA Synthesis kit (Bio-Rad), and qPCR was performed with iQ SYBR Green Supermix (Bio-Rad) with tubulin and GAPDH as an internal reference gene control. All experiments were conducted in triplicate qPCR reactions. Relative mRNA expression was calculated using the comparative CT method. Primers are listed in SI Appendix.
RNAi.
Details of fly strains are provided in SI Appendix. Dmef2-Gal4 females were crossed to UAS-RNAi males at 27 °C, and progeny were screened for lethality. For dissections, early third instar (∼72 h after egg laying) larvae were hand-picked and transferred to new food vials. The larvae were allowed to forage for an additional 24 h, after which the feeding larvae, not wandering, were dissected and immunostained (see above). For epistasis, Dmef2-Gal4 females were crossed to UAS-RNAi + UAS-Myc males.
Identification of Insulin-Responsive Nucleolar Regulators.
Drosophila S2R+ cells were incubated 12 h in serum-free Schneider’s Drosophila medium (21720–024; Thermo Fisher Scientific). Cells were then treated with 25 mg/mL insulin from bovine pancreas (I6634; Sigma-Aldrich) and lysed at 10 different time points at 20-min intervals (3 biological replicates per time point) after stimulation. RNA-seq was performed with the Illumina Hi-Seq platform with 100-bp single-end sequencing. Sequencing reads were mapped back to the Drosophila genome using TopHat, with ∼70% of the sequences uniquely mapped. Next, expression levels were summarized for each gene from the sequencing data with read count values (i.e., number of sequenced reads per exon model) and a >0.97 Spearman’s correlation coefficient between any 2 biological replicates. We modeled the expression levels of each gene as the added sum of 3 parts: (i) the time effect after insulin stimulation, modeled as a cubic polynomial of time t; (ii) potential confounding components, such as batch effects, extracted using surrogate variables; and (iii) random white noise, modeled as a mean 0 normal distribution. We applied an F-test statistical framework to test each gene for the null hypothesis that the gene is not temporally differentially expressed vs. the alternative hypothesis that the gene is temporally differentially expressed. After correcting for multiple hypothesis testing, we obtained ∼1,250 temporally differentially expressed genes with different summary values at P < 0.01. This list was then overlapped with a list of 750 high-confidence nucleolar size regulators (20).
ChIP-Seq.
Somatic muscles plus cuticle from 30 third instar larvae were dissected in cold PBS, fixed and cross-linked in PBS plus 2% formaldehyde for 10 min, after which the reaction was stopped with glycine (final concentration of 50 mM). The tissue was dounce homogenized in nuclear lysis buffer plus protease inhibitors. Washes and sonication followed the standard mod-ENCODE ChIP-seq protocol. ChIP was performed with a commercially available Drosophila Myc antibody (sc-28208; Santa Cruz Biotechnology) following standard protocol using magnetic protein A and G mixed beads with input samples as control. Sequencing libraries were prepared with the Wafergen Apollo 324 system. To obtain the required amount of library material, we performed an additional round of 10 PCR amplification cycles. Then 50-bp single-end reads were generated with the Illumina HiSeq 2500 platform.
Identification of Myc-Binding Peaks.
Sequence reads were trimmed to remove 3 base pairs at the 5′ end, where the relative sequencing quality score is significantly lower. Trimmed reads were then mapped back to Drosophila genome assembly dmel 6 with BWA, and uniquely mapped reads were kept for analyses. Owing to the low volume of starting materials, a high percentage of sequence duplicates were detected in the data. We performed “de-duplication” for each sample and pooled the reads together as a “pool” ChIP sample, randomly sampling the same number of reads from their corresponding input control sample to form a pool control sample. MACS2 was used to identify binding peaks in the “pool” ChIP sample compared with pool control sample. We also called peaks in each of the 3 biological replicates, and high signal consistency between each biological replicate and the pool ChIP sample was observed. As a sanity check, we performed a quick Ebox motif sequence search and found that 75% of the identified Myc-binding peaks contained an Ebox motif within 200 bp of the peak summit coordinates.
Human Cell Culture.
Inducible MYC cells were derived from HMECs (CC-2551B; Lonza) immortalized with telomerase and stably transduced with lentivirus carrying rtTA and MYC under the control of the TRE promoter, as described previously (35, 36). Cells were maintained in MEGM medium (CC3150; Lonza) and selected with 2 μg/mL puromycin (Clontech) for 3–4 d following infection. HMECs lacking inducible MYC served as a control. For proliferation and drug screening experiments, cells were seeded in 384-well plates in the presence/absence of 1 ug/mL dox for the indicated times. Cell counts for proliferation rates were obtained with a hemocytometer. For viability curves, cells were seeded in the presence/absence of dox for 24 h, followed by the addition of the indicated amount of drug or ethanol control. Following 24 h of treatment, CellTiter-Glo reagent (Promega) was added to each well, and luminescence was measured with a SpectraMax Paradigm Microplate Detection Platform (Molecular Probes). All experiments were performed in triplicate.
Statistical Analyses.
Statistical analyses were performed using Microsoft Excel or Bio-Rad CFX Manager software (for qPCR). Unless noted otherwise, figures show SEM, and asterisks denote P values < 0.01 (Student’s 2-tailed t test). A hypergeometric test was used to calculate the significance of enrichment between 2 datasets.
Supplementary Material
Acknowledgments
We thank the Bloomington Drosophila Stock Center, Japanese National Institute of Genetics, Vienna Drosophila Resource Center, and Transgenic RNAi Project at Harvard Medical School (supported by National Institute of General Medical Sciences Grant R01 GM084947) for fly stocks. This work was funded by grants from the National Institutes of Health (R01 AR057352 and P01 CA120964, to N.P.) and the Ludwig Foundation to (S.J.E.). N.P. and S.J.E. are investigators with the Howard Hughes Medical Institute.
Footnotes
The authors declare no conflict of interest.
Data deposition: The data reported in this paper have been deposited in the Gene Expression Omnibus (GEO) database, https://www.ncbi.nlm.nih.gov/geo (accession no. GSE129292).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1821863116/-/DCSupplemental.
References
- 1.Beroukhim R., et al. , The landscape of somatic copy-number alteration across human cancers. Nature 463, 899–905 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Nesbit C. E., Tersak J. M., Prochownik E. V., MYC oncogenes and human neoplastic disease. Oncogene 18, 3004–3016 (1999). [DOI] [PubMed] [Google Scholar]
- 3.Darnell J. E., Jr, Transcription factors as targets for cancer therapy. Nat. Rev. Cancer 2, 740–749 (2002). [DOI] [PubMed] [Google Scholar]
- 4.Nair S. K., Burley S. K., X-ray structures of Myc-Max and Mad-Max recognizing DNA. Molecular bases of regulation by proto-oncogenic transcription factors. Cell 112, 193–205 (2003). [DOI] [PubMed] [Google Scholar]
- 5.Delmore J. E., et al. , BET bromodomain inhibition as a therapeutic strategy to target c-Myc. Cell 146, 904–917 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kessler J. D., et al. , A SUMOylation-dependent transcriptional subprogram is required for Myc-driven tumorigenesis. Science 335, 348–353 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Toyoshima M., et al. , Functional genomics identifies therapeutic targets for MYC-driven cancer. Proc. Natl. Acad. Sci. U.S.A. 109, 9545–9550 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Liu L., et al. , Deregulated MYC expression induces dependence upon AMPK-related kinase 5. Nature 483, 608–612 (2012). [DOI] [PubMed] [Google Scholar]
- 9.Li Y., Zhu Y., Prochownik E. V., MicroRNA-based screens for synthetic lethal interactions with c-Myc. RNA Dis. 3, e1330 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.D’Andrea A., et al. , The mitochondrial translation machinery as a therapeutic target in Myc-driven lymphomas. Oncotarget 7, 72415–72430 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dang C. V., MYC, metabolism, cell growth, and tumorigenesis. Cold Spring Harb. Perspect. Med. 3, a014217 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ji H., et al. , Cell-type independent MYC target genes reveal a primordial signature involved in biomass accumulation. PLoS One 6, e26057 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Burger K., et al. , Chemotherapeutic drugs inhibit ribosome biogenesis at various levels. J. Biol. Chem. 285, 12416–12425 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Davoli T., et al. , Functional genomics reveals that tumors with activating phosphoinositide 3-kinase mutations are dependent on accelerated protein turnover. Genes Dev. 30, 2684–2695 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Demontis F., Perrimon N., Integration of Insulin receptor/Foxo signaling and dMyc activity during muscle growth regulates body size in Drosophila. Development 136, 983–993 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Parisi F., et al. , Drosophila insulin and target of rapamycin (TOR) pathways regulate GSK3 beta activity to control Myc stability and determine Myc expression in vivo. BMC Biol. 9, 65 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Teleman A. A., Hietakangas V., Sayadian A. C., Cohen S. M., Nutritional control of protein biosynthetic capacity by insulin via Myc in Drosophila. Cell Metab. 7, 21–32 (2008). [DOI] [PubMed] [Google Scholar]
- 18.Johnston L. A., Prober D. A., Edgar B. A., Eisenman R. N., Gallant P., Drosophila myc regulates cellular growth during development. Cell 98, 779–790 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ni X., Bulyk M. L., Zirin J., Hu Y., Perrimon N., Time series RNA-seq analyses of Drosophila S2R+ cells after insulin stimulation. Gene Expression Omnibus. https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE129292. Deposited 3 April 2019.
- 20.Neumüller R. A., et al. , Conserved regulators of nucleolar size revealed by global phenotypic analyses. Sci. Signal. 6, ra70 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Coller H. A., et al. , Expression analysis with oligonucleotide microarrays reveals that MYC regulates genes involved in growth, cell cycle, signaling, and adhesion. Proc. Natl. Acad. Sci. U.S.A. 97, 3260–3265 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cowling V. H., Turner S. A., Cole M. D., Burkitt’s lymphoma-associated c-Myc mutations converge on a dramatically altered target gene response and implicate Nol5a/Nop56 in oncogenesis. Oncogene 33, 3519–3527 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Fernandez P. C., et al. , Genomic targets of the human c-Myc protein. Genes Dev. 17, 1115–1129 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Herter E. K., et al. , snoRNAs are a novel class of biologically relevant Myc targets. BMC Biol. 13, 25 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Schlosser I., et al. , A role for c-Myc in the regulation of ribosomal RNA processing. Nucleic Acids Res. 31, 6148–6156 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kim J., Lee J. H., Iyer V. R., Global identification of Myc target genes reveals its direct role in mitochondrial biogenesis and its E-box usage in vivo. PLoS One 3, e1798 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Li F., et al. , Myc stimulates nuclearly encoded mitochondrial genes and mitochondrial biogenesis. Mol. Cell. Biol. 25, 6225–6234 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Morrish F., Hockenbery D., MYC and mitochondrial biogenesis. Cold Spring Harb. Perspect. Med. 4, a014225 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Seitz V., et al. , Deep sequencing of MYC DNA-binding sites in Burkitt lymphoma. PLoS One 6, e26837 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Cunningham J. T., Moreno M. V., Lodi A., Ronen S. M., Ruggero D., Protein and nucleotide biosynthesis are coupled by a single rate-limiting enzyme, PRPS2, to drive cancer. Cell 157, 1088–1103 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gerstberger S., et al. , The conserved RNA exonuclease Rexo5 is required for 3′ end maturation of 28S rRNA, 5S rRNA, and snoRNAs. Cell Rep. 21, 758–772 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yang J., Sung E., Donlin-Asp P. G., Corces V. G., A subset of Drosophila Myc sites remain associated with mitotic chromosomes colocalized with insulator proteins. Nat. Commun. 4, 1464 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Pham J. S., et al. , Aminoacyl-tRNA synthetases as drug targets in eukaryotic parasites. Int. J. Parasitol. Drugs Drug Resist. 4, 1–13 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yao P., Fox P. L., Aminoacyl-tRNA synthetases in medicine and disease. EMBO Mol. Med. 5, 332–343 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sack L. M., et al. , Profound tissue specificity in proliferation control underlies cancer drivers and aneuploidy patterns. Cell 173, 499–514.e23 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sack L. M., Davoli T., Xu Q., Li M. Z., Elledge S. J., Sources of error in mammalian genetic screens. G3 (Bethesda) 6, 2781–2790 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Fang P., et al. , Structural basis for full-spectrum inhibition of translational functions on a tRNA synthetase. Nat. Commun. 6, 6402 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Keller T. L., et al. , Halofuginone and other febrifugine derivatives inhibit prolyl-tRNA synthetase. Nat. Chem. Biol. 8, 311–317 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Busiello V., Di Girolamo M., De Marco C., Thiaisoleucine and protein synthesis. Biochim. Biophys. Acta 561, 206–214 (1979). [DOI] [PubMed] [Google Scholar]
- 40.Istvan E. S., et al. , Validation of isoleucine utilization targets in Plasmodium falciparum. Proc. Natl. Acad. Sci. U.S.A. 108, 1627–1632 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Cochereau C., Sanchez D., Bourhaoui A., Creppy E. E., Capsaicin, a structural analog of tyrosine, inhibits the aminoacylation of tRNA(Tyr). Toxicol. Appl. Pharmacol. 141, 133–137 (1996). [DOI] [PubMed] [Google Scholar]
- 42.Sulima S. O., Hofman I. J. F., De Keersmaecker K., Dinman J. D., How ribosomes translate cancer. Cancer Discov. 7, 1069–1087 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kim S., You S., Hwang D., Aminoacyl-tRNA synthetases and tumorigenesis: More than housekeeping. Nat. Rev. Cancer 11, 708–718 (2011). [DOI] [PubMed] [Google Scholar]
- 44.Pierce S. B., et al. , Drosophila growth and development in the absence of dMyc and dMnt. Dev. Biol. 315, 303–316 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hsu T. Y., et al. , The spliceosome is a therapeutic vulnerability in MYC-driven cancer. Nature 525, 384–388 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.