Significance
With >10,000 living species, squamate reptiles (lizards, snakes) exhibit enormous phenotypic variation reflecting their incredible range in ecology and developmental strategies. What drove this exceptional diversity? We analyze high-density surface morphometric data for skulls representing ∼200 modern and extinct species to provide a comprehensive, clade-wide investigation of how ecological and developmental factors contributed to cranial evolution across geologic time, skull regions, and taxa. Although diet and habitat have had an overarching impact on skull evolution (e.g., herbivory, along with aquatic and fossorial habitats, are associated with rapid evolution), lizards and snakes surprisingly share a common pattern of trait correlations (integration). The remarkable ecological and morphological diversity in squamates thus arose from selection acting on a conserved architecture of phenotypic integration.
Keywords: geometric morphometrics, integration and modularity, macroevolution, skull, Squamata
Abstract
Factors intrinsic and extrinsic to organisms dictate the course of morphological evolution but are seldom considered together in comparative analyses. Among vertebrates, squamates (lizards and snakes) exhibit remarkable morphological and developmental variations that parallel their incredible ecological spectrum. However, this exceptional diversity also makes systematic quantification and analysis of their morphological evolution challenging. We present a squamate-wide, high-density morphometric analysis of the skull across 181 modern and extinct species to identify the primary drivers of their cranial evolution within a unified, quantitative framework. Diet and habitat preferences, but not reproductive mode, are major influences on skull-shape evolution across squamates, with fossorial and aquatic taxa exhibiting convergent and rapid changes in skull shape. In lizards, diet is associated with the shape of the rostrum, reflecting its use in grasping prey, whereas snakes show a correlation between diet and the shape of posterior skull bones important for gape widening. Similarly, we observe the highest rates of evolution and greatest disparity in regions associated with jaw musculature in lizards, whereas those forming the jaw articulation evolve faster in snakes. In addition, high-resolution ancestral cranial reconstructions from these data support a terrestrial, nonfossorial origin for snakes. Despite their disparate evolutionary trends, lizards and snakes unexpectedly share a common pattern of trait integration, with the highest correlations in the occiput, jaw articulation, and palate. We thus demonstrate that highly diverse phenotypes, exemplified by lizards and snakes, can and do arise from differential selection acting on conserved patterns of phenotypic integration.
What are the factors that dictate phenotypic evolution? Van Valen (1) famously stated that “evolution is the control of development by ecology,” underscoring the importance of intrinsic (“development”) and extrinsic (“ecology”) factors in shaping the course of evolutionary trajectories. Although numerous studies have examined the impact of intrinsic or extrinsic factors on morphological evolution separately, a comprehensive evaluation of how these factors interact remains elusive. With >10,000 known extant species, Squamata (snakes and paraphyletic “lizards”) is an excellent clade in which to investigate this topic because of their remarkable variation in morphology and reproductive strategies that mirrors their vast ecological spectrum. For instance, the group spans 6 orders of magnitude in body length from the <30-mm-long chameleon Brookesia (2) and the gecko Sphaerodactylus (3) to extinct mosasaurs exceeding 17 m in length (4), and ranges in cranial architecture from the highly kinetic skulls of macrostomatan snakes to the heavily ossified skulls of burrowing taxa (5). Additionally, squamates have undergone numerous independent origins of viviparity and oviparity (6, 7). This morphological, ecological, and developmental diversity permits a greater opportunity to disentangle the effects of intrinsic and extrinsic factors compared with other well-studied vertebrate clades such as birds and mammals.
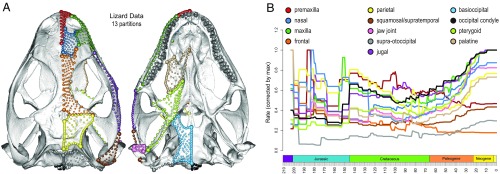
Many large-scale studies on squamates have investigated evolutionary dynamics of size (8, 9). Although size is an important metric, it is only one of many aspects of form, and elucidating the macroevolutionary patterns of such an exceptionally diverse group requires a robust and comprehensive characterization of morphology. Some recent studies have applied landmark-based geometric morphometric (GM) methods to characterize the overall shape of the skull—an information-rich structure to address questions pertaining to the ecological and developmental influences on morphology (10–15). Although they provide important data on shape evolution, these approaches cannot adequately characterize many aspects of skull morphology, as landmarks are generally restricted to sutures and the edges of structures. To surmount these limitations of previous studies, we implement a single framework to extract high-density, 3D cranial shape data across squamates (181 extant and extinct spp.), using more than 1,000 landmarks and sliding semilandmarks to robustly capture the shape of 13 cranial partitions, primarily at the level of individual bones (SI Appendix, Table S1 and Fig. S1). These partitions include the premaxilla, maxilla, jugal (in lizards), nasal, frontal, parietal, squamosal (supratemporal in snakes), jaw joint of quadrate, supraoccipital + otoccipital (supraotoccipital), occipital condyle, basioccipital + basisphenoid, pterygoid, and palatine. Because of the varying number of partitions across specimens, we performed separate alignments and analyses on 4 different datasets: an extant lizard dataset (Fig. 1A) with 109 taxa and all 13 partitions with 1,222 landmarks and sliding semilandmarks (47 fixed, 595 curve, and 580 patch points); an extant snake dataset (SI Appendix, Fig. S1G) with 65 taxa and 12 partitions (as a result of the absence of the jugal) with 1,056 landmarks and sliding semilandmarks (44 fixed, 535 curve, and 477 patch points); an extant squamate dataset with 174 taxa and 12 partitions with the same 1,056 landmarks and sliding semilandmarks as the snake dataset; and a combined extant and extinct dataset (SI Appendix, Fig. S1H) with all 181 taxa (SI Appendix, Table S2), 605 landmarks and sliding semilandmarks (27 fixed, 260 curve, and 318 patch points), and 5 partitions because many regions were not exposed in all of the fossil specimens included in the study. These rich morphological data provide an opportunity to assess ecological and developmental influences on cranial shape and its individual components.
Fig. 1.
Cranial partitions in the lizard, snake, and combined datasets. (A) Skull of Sceloporus variabilis (FMNH 122866) reconstructed from µCT imaging in dorsal and ventral views. (B) Rate-through-time plot of each partition corrected by maximum rate value within each module for comparability.
By using a suite of computational methods, we elucidate macroevolutionary trends in cranial disparity and evolutionary rates across individual cranial regions and across the whole skull, and test which ecological and developmental traits have influenced cranial evolution across the entire clade. Although the relevance of ecological factors to phenotypic variation is intuitive, reproductive strategy can also significantly bias phenotypic variation and thus could also constrain or promote phenotypic evolution (16) and drive patterns of trait covariation (17). A high-density ancestral skull shape was created to infer the long-debated ecological origin of snakes. In addition, we investigate whether the marked differences in cranial architecture between lizards and snakes arose from divergences in (i) evolutionary rates, (ii) the relative influences of ecological and developmental factors underlying overall skull-shape evolution or the evolution of specific cranial regions, and (iii) patterns of cranial integration that are generated from developmental and functional linkages among traits (13, 18–20). Identification of the patterns of integration among cranial regions has enormous potential to illuminate the factors that dictate the path of morphological evolution because the interplay among traits has the capacity to constrain the evolution of individual traits and promote the coordinated evolution of correlated traits (21, 22). In sum, this study provides a squamate-wide, high-density morphometric analysis to quantify the effects of developmental and ecological factors, demonstrating the power and validity of this emerging approach to understanding the drivers of phenotypic evolution.
Results
Morphospace.
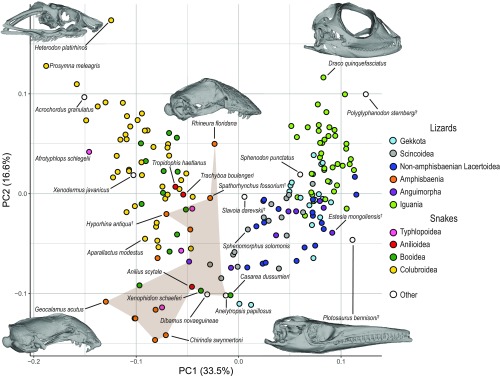
To visualize the distribution of skull-shape variation, a morphospace was constructed from the first 3 principal components (PCs) of shape data that account for 61.8% of the total cranial shape variation in the combined extant and fossil dataset (Fig. 2 and SI Appendix, Fig. S2). PC1, which accounts for 33.5% of shape variation, broadly separates lizards and snakes, as suggested by previous studies on skull shape of squamates (12, 23), underscoring their distinct cranial morphotypes. Notably, the intermediate space is occupied by a polyphyletic group of fossorial lizards that includes amphisbaenians, annielids, and dibamids. Within lizards, there is a broad separation between Iguania and noniguanian taxa (“Scleroglossa”), although partial overlap exists. The pygopodids (Delma, Lialis) occupy a unique area of the morphospace likely associated with ecomorphological adaptations related to their unique limbless body among gekkotans. The overall distribution of shape variation, including this taxonomic separation, is similar to that of the extant-only dataset (SI Appendix, Fig. S2). PC2 and PC3, corresponding to 16.6% and 10.6% of the shape variation, respectively, are both associated with the narrowness of the skull: PC2 relates to the anteroposterior elongation of the parietal, whereas PC3 relates to that of the rostrum (SI Appendix, Figs. S2 and S3). Taxonomic groups at and above the family level tend to cluster morphologically (Blomberg’s Kcombined = 0.675, Kextant = 0.889), congruent with results from 2D skull-shape data in lizards (23). With PC2 and PC3, the phylogenetic structure is less clear (SI Appendix, Fig. S3B), suggesting multiple convergences in the evolution of narrow and broad skulls in Squamata.
Fig. 2.
Morphospace of skull shape constructed from first two PCs of combined extant and fossil skull-shape data. Data points color-coded by taxonomic group, as indicated by key. Brown polygon spans fossorial lizards sampled in this study. Morphospace with all taxonomic labels in SI Appendix, Figs. S2 and S3.
Shape Disparity and Evolutionary Rates.
We fit variable-rates evolutionary models to the data to assess evolutionary mode by using PC scores encompassing 95% of the shape variation. A λ model of trait evolution was supported, albeit weakly, for the extant squamate dataset (SI Appendix, Table S3). However, Ornstein–Uhlenbeck (OU) models were favored for the lizard and snake datasets when analyzed separately, suggesting that each group exhibits different optimum trait values for skull shape. Model fitting supports the λ model for every partition, with the exception of the δ model for the squamosal (supratemporal) and jaw joint (SI Appendix, Table S3).
Estimated disparity and evolutionary rates also display a mosaic pattern across partitions. The early history of squamates is characterized by elevated rates of shape evolution in the premaxilla, nasal, jugal, frontal, parietal, and palatine (Fig. 1B), mirroring their relatively high disparity in the Jurassic except for the premaxilla (SI Appendix, Fig. S4). At the Cretaceous–Paleogene (K-Pg) boundary, the evolutionary rate of the parietal bone noticeably dips (Fig. 1B), although a coincident decrease in disparity is absent (SI Appendix, Fig. S4). Throughout the Cenozoic, most partitions underwent faster evolution toward the Recent, whereas the premaxilla, frontal, squamosal (supratemporal), and supraotoccipital regions showed a constant or decreasing rate of evolution (Fig. 1B). When comparing the mean rates estimated from the full shape data (i.e., not PC scores) under Brownian motion (BM) models for comparability, the jaw joint and pterygoid exhibit the fastest rates of evolution, largely driven by snakes (SI Appendix, Table S4). The morphological rates of different cranial regions remained within an order of magnitude of each other in lizards, whereas proportionately high rates are observed in the maxilla, jaw joint, and pterygoid in snakes (SI Appendix, Table S4).
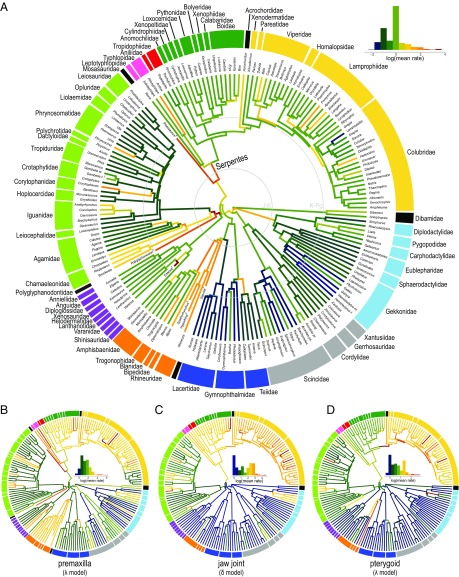
Mapping estimated rates according to the best-fit model onto time-calibrated phylogenies illustrates how evolutionary rates within each partition differ across taxonomic groups (Fig. 3 and SI Appendix, Fig. S5). Although the distribution of evolutionary rates is highly heterogeneous, several large-scale patterns are evident. First, higher rates of cranial evolution are concentrated among snakes, fossorial lizards (e.g., amphisbaenians, dibamids), and iguanian taxa with elaborate cranial ornamentations (e.g., Corytophanes, Phrynosoma, Chamaeleonidae; Fig. 3A and SI Appendix, Fig. S5). Among fossorial groups, amphisbaenians collectively sustain high rates of evolution in several regions, including the premaxilla, nasal, frontal, parietal, and occiput (supraotoccipital, basioccipital, occipital condyle). Snakes are characterized by elevated evolutionary rates in the nasal, maxilla, pterygoid, and especially the jaw joint (SI Appendix, Figs. S5 and S6B). Additionally, we observe rapid evolution of many regions along extinct lineages because fossil specimens contribute considerable morphological variation toward the base of major subclades (Fig. 3A and SI Appendix, Fig. S5).
Fig. 3.
Estimated rates of cranial shape evolution mapped on time-calibrated phylogeny of squamates under best-supported model of trait evolution. (A) Evolutionary rates of overall skull shape of combined dataset under the OU model. Gray rings signify Jurassic–Cretaceous (J-K) and K-Pg boundaries. (B) Evolutionary rates of premaxilla bone under λ model. (C) Evolutionary rates of jaw joint under δ model. (D) Evolutionary rates of pterygoid bone under λ model. Color gradient on branches indicates the rate of shape evolution as indicated by histogram (Insets). Color of circular band denotes taxonomic group as specified in Fig. 2. Rates estimated by using BayesTraitsV3 (78).
Estimated evolutionary rates also exhibit localized patterns. Within snakes, the premaxilla and nasal evolved quickly (Fig. 3B and SI Appendix, Fig. S5E), enabled possibly by rhinokinesis (24). Likewise, the jaw joint and pterygoid show accelerated evolution in shape and relative position (Fig. 3 C and D), reflecting greater feeding-related kinesis. Rapid evolution of the basioccipital and occipital condyle (SI Appendix, Fig. S5) also characterizes skull evolution in snakes, potentially related to shifts in braincase ossification (25). Likewise, the occiput (supraotoccipital, basioccipital, occipital condyle) evolved at an elevated pace in the fossorial amphisbaenians and dibamids, likely related to the constraint associated with head-first burrowing. The polyphyletic group of fossorial taxa (e.g., amphisbaenians, annelids, dibamids) is also characterized by rapid evolution of the frontal and parietal bones, leading to the convergent anteroposteriorly elongated skull roof observed in these taxa (SI Appendix, Fig. S5).
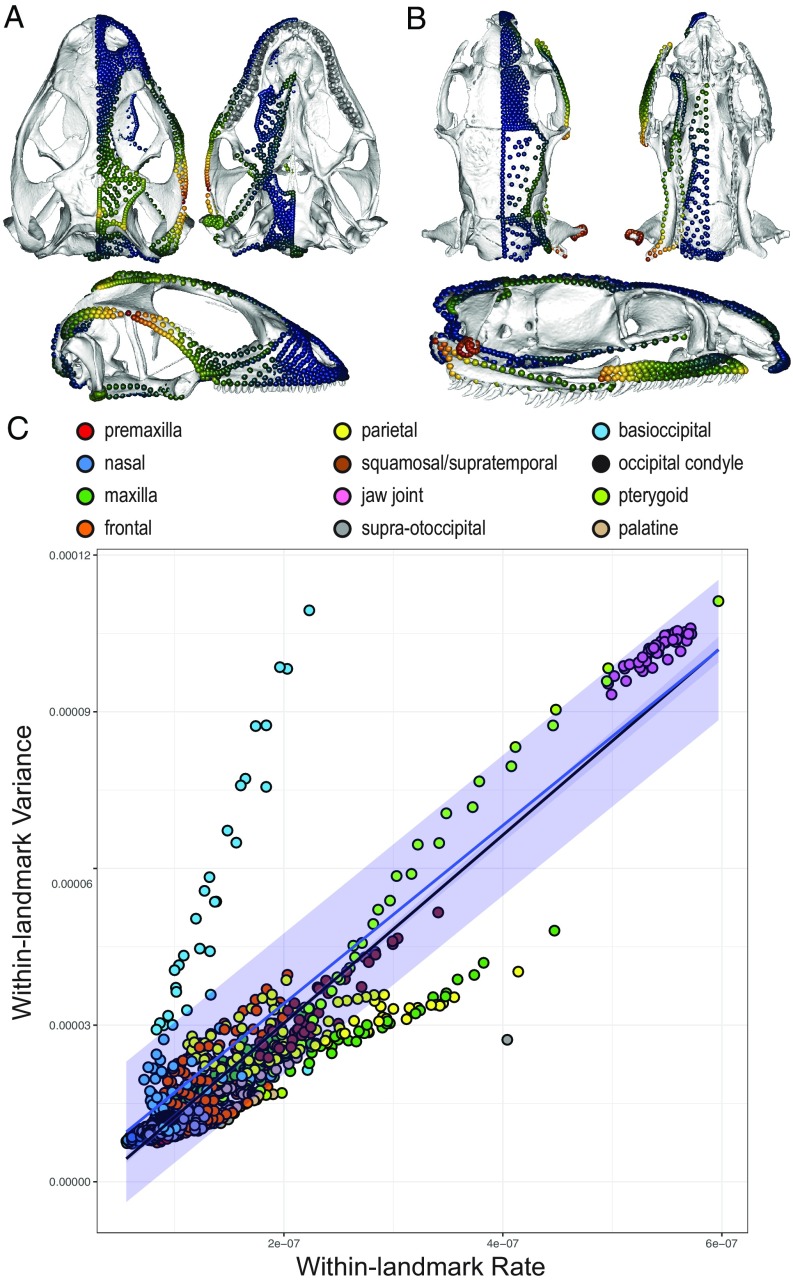
To more closely examine the relationship between disparity and rate, we also calculated the Procrustes variance and mean rate for each landmark and semilandmark according to BM model of trait evolution. When these values are mapped onto individual landmarks and sliding semilandmarks (Fig. 4 and SI Appendix, Fig. S6), we find that high disparity and rates are concentrated on restricted parts of the skull, including the posterior regions of the maxilla, parietal, jaw joint, basisphenoid, and pterygoid. When examined in lizards and snakes separately, there are notable differences in the distribution of variation and rates. In lizards, the parietal and squamosal bones exhibit high variance and rate, whereas snakes show higher disparity and rates in the maxilla, pterygoid, and jaw joint. A bivariate plot of within-landmark variance and rates (Fig. 4C) illustrates that strong, yet different, relationships between disparity and rates have directed the phenotypic evolution of each partition. Furthermore, the maxilla and parietal bones show particularly low disparity relative to their rates of evolution, suggesting more convergent or constrained evolution in these cranial regions. Conversely, the basioccipital + basisphenoid region demonstrates greater variance than expected for its rate of evolution, potentially reflecting greater divergence in the evolution of this region across squamates.
Fig. 4.
Per-landmark Procrustes variance and mean evolutionary rates. Landmarks and sliding semilandmarks on skull reconstruction in dorsal (Top Left), ventral (Top Right), and lateral (Bottom) views indicating Procrustes variance in lizards (A) and snakes (B), where a warmer color indicates greater value. (C) Bivariate plot of within-landmark variance against rate for each landmark, where the black line and gray band denote the regression line and 95% CI. Blue line and band indicate expected correlation between within-landmark rate and variance given Brownian motion model of trait evolution and its 95% CI.
Ancestral Shape Reconstruction.
Taking advantage of our high-dimensional representation of cranial morphology, we reconstructed the ancestral 3D skull morphology of crown squamates and snakes. The ancestral squamate skull (SI Appendix, Fig. S7 A and C), based on extant-only and combined datasets, features subrectangular frontals and parietals, most closely resembling the scincid Sphenomorphus solomonis and the lacertid Lacerta with regard to Procrustes distance. The estimated ancestral snake skull (SI Appendix, Fig. S7B) exhibits a broad nasal articulated posteriorly with the frontal, a relatively deep maxilla, subrectangular frontal and parietal, triangular supratemporal, and a relatively flat mandibular articular surface of the quadrate that is positioned immediately lateral to the otoccipital–basioccipital boundary (i.e., not far posterior from the occiput). Among sampled taxa, it is closest to the terrestrial lamprophiid Aparallactus modestus in shape.
Ecology and Developmental Covariates.
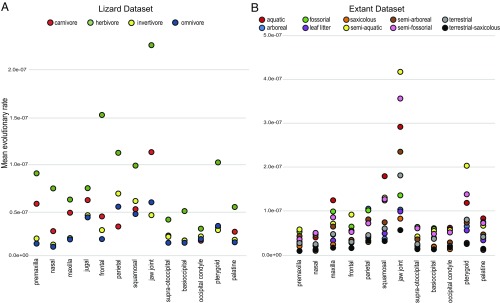
To explicitly test how ecological and developmental factors have driven skull-shape evolution in squamates, we examined the link between disparity and evolutionary rates to key ecological and developmental variables, including habitat, diet, locomotion, and reproductive mode (Materials and Methods and SI Appendix, Table S5). For these analyses, we analyzed the extant-only dataset because of the need for data on ecology and reproductive modes. Reproductive mode is not a clear predictor of cranial shape or mean evolutionary rates (SI Appendix, Fig. S8A), although weak effect is seen on the occiput in lizards (R2 < 0.04; P ≈ 0.05). In both lizards and snakes, an aquatic lifestyle is associated with higher rates of evolution (Fig. 5B and SI Appendix, Fig. S8). Fossorial and herbivorous lizards also generally exhibit elevated rates of skull evolution (Fig. 5A and SI Appendix, Fig. S8). Performing phylogenetically informed MANOVA on skull-shape data indicates that habitat and locomotion are correlated with the shape of all cranial partitions in lizards (SI Appendix, Table S6). Among lizards, diet is significantly correlated with the shape of premaxilla, nasal, and jaw joint, which are important structures for feeding. In contrast, snakes show strong correlation between diet and posterior cranial regions, such as the supratemporal, jaw joint, supraotoccipital, occipital condyle, and pterygoid.
Fig. 5.
Plots of mean evolutionary rates for each partition colored by trait values. (A) Mean evolutionary rates of partitions in extant lizards colored by diet preference. (B) Mean evolutionary rates of partitions in extant squamates colored by habitat preference. Mean rates calculated with the compare.multi.evol.rates function in geomorph R package (79).
Cranial Integration.
To determine the pattern of cranial integration across squamates and within lizards and snakes separately, we compared multiple a priori hypotheses of modular organization based on previous studies (10, 14) by using phylogenetically informed analyses of trait correlation matrices: EMMLi (26) and covariance ratio (CR) analysis (27). These analyses supported the most complex model tested, which considers each characterized partition as a module. As previously described (28), we further inspected the estimated correlation (ρ) values within each partition and between each pair of partitions (SI Appendix, Table S7). For the extant dataset with 12 partitions, these criteria pointed to an “occiput” module consisting of supraotoccipital, occipital condyle, and basioccipital + basisphenoid partitions, a “palate” module comprising the pterygoid and palatine, and all other partitions each forming separate modules, for a total of 10 modules in lizards and 9 modules in snakes.
Remarkably, results from extant lizard and snake datasets supported the same pattern of cranial integration despite fundamental differences in their cranial architecture as demonstrated by the morphospace analyses. In snakes, the jaw joint on the quadrate yielded stronger correlations with the supraotoccipital and occipital condyle (ρbetween = 0.59 and 0.67, respectively) than in lizards, but these partitions were not coalesced into a single module because of their extremely high within-partition correlations (ρjaw joint = 0.99, ρoccipital condyle = 0.97). Phylogenetically naïve EMMLi and CR analysis (27) and analyses of landmark-only and subsampled landmark + sliding semilandmark datasets produced largely congruent results that support these cranial modules (Materials and Methods and SI Appendix, Tables S7 and S8 and Fig. S9). For the combined dataset with 5 regions, EMMLi and CR analysis supported the partitioning of the premaxilla, maxilla, frontal, parietal, and supraotoccipital as separate anatomical modules (SI Appendix, Tables S7 and S8).
To assess whether integrated structures constrain or promote phenotypic evolution in squamates, we performed least-squares regression analyses on disparity (Procrustes variance), mean evolutionary rates, and the degree of integration within each module. For comparability across partitions, we corrected the Procrustes variance and mean rates by the number of landmarks and sliding semilandmarks within each partition. Results indicate that the total disparity and mean evolutionary rates of skull regions are highly correlated across squamates and within lizards and snakes (SI Appendix, Table S9 and Fig. S10), meaning that faster-evolving regions are generally also the most variable in morphology (R2 > 0.781; P ≤ 0.005). In contrast, the magnitude of integration within partitions does not appear to be closely linked to shape disparity or evolutionary rates (R2 < 0.665; P > 0.05; SI Appendix, Table S9).
Discussion
Skull-Shape Evolution in Squamates.
How have disparate cranial morphologies evolved in squamates? Comparative analyses of our high-dimensional cranial data demonstrate mosaic patterns across time, taxa, and cranial regions. Early diversification of the squamate cranium in the Jurassic is driven by elevated rates in the frontal bone of dibamids, iguanians, and snakes, as well as the jaw joint at the origin of snakes (Figs. 1B and 3 and SI Appendix, Figs. S4 and S5). These initial pulses of skull evolution correspond to the radiation of squamate lineages following a niche vacancy from the Permian–Triassic mass extinction event (29). Proximal to the K-Pg boundary, slower rates of evolution are observed in the premaxilla and frontal, but disparity remains largely unaffected with the exception of the premaxilla, maxilla, and nasal. These results suggest that the end-Cretaceous mass extinction event did not reduce diversity in skull form even as it dramatically reduced taxonomic diversity (8, 30). This discordance suggests a nonselective extinction with respect to squamate cranial morphology (31).
Beyond temporal dynamics, drivers of skull-shape evolution are heterogeneous across squamates. Habitat preference and locomotion (i.e., fossorial, aquatic), but not reproductive mode, had substantial influence on cranial shape evolution across squamates. Faster evolutionary rates also reflect major changes in diet and ecology. Iguanids show increased rates in the rostrum (premaxilla, nasal) and the pterygoid relative to other iguanian lizards (Fig. 3B and SI Appendix, Fig. S5), a pattern compatible with significant differences in rate of skull evolution associated with herbivory in lizards (SI Appendix, Table S6). Notably, the premaxilla and nasal show elevated rates across snakes compared with lizards overall, which is possibly a result of rhinokinesis where the rostral elements are only loosely connected (24). Although they were not characterized in the present study, the septomaxilla and vomer also form moveable elements of rhinokinesis and are expected to have evolved quickly in snakes. Rapid evolution of most cranial regions is also observed along the lineage to the unusual myrmecophagous genus Phrynosoma (Fig. 3 and SI Appendix, Fig. S5). Among snakes, Dipsas and Heterodon exhibit particularly fast cranial evolution that is mostly attributed to the palate in both taxa and the nasal and maxilla in the latter (SI Appendix, Fig. S5). Dipsas possesses relatively short pterygoids that are free from articulation with the quadrate, which are adapted for consuming snails (32). In Heterodon (Hognose snake), higher rates in the nasal and maxilla are unsurprising given its eponymous snout and large posterior maxillary teeth. The palate of Heterodon comprises pterygoids that become transversely constricted posteriorly and palatines that are anteroposteriorly short compared with closely related taxa. These adaptations permit extreme mobility in the palate critical for consuming large prey and flattening their skull for defensive behavior (33). We demonstrate, with morphometric and taxonomic scale, that diet and habitat are primary drivers of skull evolution across squamates as proposed previously (23, 34, 35), in contrast to recent studies of other major amniote clades such as birds (36).
Elevated rates are also observed in terminal branches of fossil taxa sampled in this study (Fig. 3 and SI Appendix, Fig. S5), reflecting the unusual morphologies of several extinct clades relative to extant squamates (Fig. 2). The phylogenetic relationship of some extinct taxa to other major squamate clades remains unclear, including Slavoia and mosasaurs (37–41). Although the placement of these fossils could affect the results of comparative analyses, the extent of this impact on macroevolutionary reconstructions would likely be restricted to their immediate lineages. For instance, rapid rates of evolution tend to be limited to the branches leading to extinct taxa (Fig. 3A), whereas estimated rates remain similar for other branches compared with the extant-only dataset (SI Appendix, Fig. S5A). Furthermore, we find congruence between results based on extant-only and combined extant and fossil datasets. As such, alternative placements of the fossil taxa sampled in this study are not expected to substantially alter the overall results and conclusions of this study, as supported by our analyses of cranial integration and evolutionary rates using alternative phylogenetic placements of Estesia, Polyglyphanodon, and the mosasaur Plotosaurus based on Conrad (39), which yielded nearly identical results to the original topology (SI Appendix, Table S7 and Fig. S5 N–S).
Given the dearth of complete, fully articulated, and minimally deformed skull material of early squamates, 3D reconstructions of estimated ancestral skull shape can provide additional insights into the morphological and ecological origins of lizards and snakes. The ancestral skull reconstruction for crown-group squamates resembles the scincid Sphenomorphus and the gymnophthalmid Colobosaura based on the extant-only dataset, and the lacertid Lacerta based on the combined dataset. Notably, these taxa are insectivorous forest dwellers (42, 43). Compared with the oldest known squamate Megachirella from the Middle Triassic (29), the estimated ancestral skull exhibits a similar cranial aspect ratio, with an anteroposteriorly narrow frontal and a triangular squamosal. The estimated skull for the ancestor of snakes is largely congruent with the cranial morphology of the basally diverging Dinilysia patagonica from the Upper Cretaceous of Argentina (44) in possessing relatively deep maxillae and a jaw joint in close proximity to the occiput. The ecological origin of snakes has been a subject of much discussion for centuries (45–50). Here, the resemblance of the ancestral reconstruction with the genus Aparallactus suggests a nonfossorial, terrestrial origin for snakes, corroborating previous studies employing multiple ancestral reconstruction methods on habitat character (47) and analysis of snake skull shape (50).
Commonalities and Divergence in Lizard and Snake Skull Evolution.
Our analyses reveal the large-scale evolutionary trends leading to distinct morphologies between lizards and snakes. Shape variation and fast rates of evolution are concentrated in regions that are critical to feeding biomechanics. In lizards, the posterior area of the parietal and jugal, as well as the anterior end of the squamosal, are regions of elevated disparity and rates (Fig. 4A and SI Appendix, Fig. S6A). These cranial regions are important sites for the attachment of jaw opening and closing muscles, such as adductor mandibulae, depressor mandibulae, and pterygoid muscles (51). Intriguingly, these are also locations of concentrated mechanical stress during biting based on finite element analysis on Uromastyx (52). Snakes, in contrast, exhibit high disparity and rates in the posterior end of the maxilla and the pterygoid, as well as the jaw joint (Fig. 4B and SI Appendix, Fig. S6B). Again, these regions are highly mobile elements in snakes that are crucial in predation as they directly interact with prey (53). Therefore, the differences in areas of elevated rates and variance between lizards and snakes strongly suggests that disparate cranial morphotypes exemplified by these two groups arose from differences in the areas that are under strong functional selection. MANOVA suggests that diet is a key factor shaping cranial morphology in divergent ways in these groups. In lizards, many of which use rostral prey capture, diet is correlated with shape variation in the premaxilla, nasal, and jaw joint. In contrast, snakes, known for their bulk feeding approach via increasing gape, exhibit significant correlations in posterior cranial regions, including the supratemporal, jaw joint, supraotoccipital, occipital condyle, and pterygoid.
Despite fundamental differences in regions under ecological selection, lizards and snakes share a pattern of cranial integration that differs from that of other vertebrate clades based on analysis of high-density morphometric data (16, 28). Conserved patterns of cranial integration and modularity have been previously identified in mammals (54, 55) and caecilians (16), but the conservation of these patterns of integration has never been tested in a clade as diverse, and as divergent, as Squamata. This result thus strengthens the case that the covariation pattern in the skull are highly conserved within major vertebrate clades and have directed their phenotypic evolution through deep time. A squamate-wide pattern seemingly contrasts with a previous study proposing that species of Anolis exhibit different covariation patterns in the skull, particularly within lineages that exhibit more extreme morphologies (14). In addition, the pattern of cranial integration identified here differs from previous analyses with far fewer landmarks and limited to single genus or family (10, 14, 15). Although the pattern within species or genera has not been assessed with comparable high-dimensional morphometric analyses, small-scale variation under localized selection pressures does not contradict large-scale conservation of overall pattern. In addition, cranial modularity and integration clearly evolve across vertebrate clades, as differences across mammals (54, 55), birds (28), and now squamates indicate. Even within squamates, snakes possess, on average, stronger correlation between the jaw joint of the quadrate and the occipital elements than is observed in lizards. Across squamates, however, we find that a shared set of integrated cranial regions has shaped their large-scale phenotypic diversification. Coupled with results from analysis of ecological and reproductive correlates of skull shape and evolutionary rates, we find that the evolution of exceptionally disparate skull shapes in squamates, exemplified by lizards and snakes, resulted from ecological specializations structured by a shared pattern of cranial integration.
The Causes and Consequences of Cranial Integration in Squamates.
Does integration within anatomical structures constrain or promote phenotypic evolution? Regression analyses of within-partition correlation (ρwithin), Procrustes variance, and evolutionary rates indicate that, whereas disparity and rates are strongly correlated across taxonomic and anatomical scales, trait integration is not consistently associated with shape variance or rate across deep time in squamates (SI Appendix, Table S9 and Fig. S10). The absence of a clear relationship counters previous studies showing that integration could constrain (28, 56) or promote (57–59) morphological evolution, although nonconforming modules were also observed in some of these studies, including nares (28), molar, and palate (56). Overall, this outcome echoes a previous study on the cranial modularity of Anolis lizards in which there was no relationship observed between cranial integration and disparity (14). These results suggest that, rather than consistently influencing the total amount or rate of variation, trait integration structures the direction of variation, with the total amount of change also reflecting the direction and magnitude of selection, a factor that cannot be directly captured in studies at this scale.
The linkage system crucial for feeding in many squamates may induce certain patterns of trait correlations. For instance, partitions that form a kinetic joint may be modular because each moveable unit evolves largely independently. Alternatively, these partitions could be integrated as a result of the need for coordinated maintenance of the linkage system (5, 60). The cranial modules identified in this study suggest that individual mobile components of the linkage system form separate modules. Compared with lizards, however, the highly kinetic skulls of snakes indicate a slightly more integrated structure characterized by greater correlations between the jaw joint and occiput and stronger between-partition correlations on average (SI Appendix, Table S8). Accordingly, the greater magnitude of integration may indicate increased kinetic properties of the skull, although this hypothesis remains to be tested further. In addition to functional demands, developmental processes may enforce a modular structure in the skull. Employing an equivalent high-dimensional morphometric approach, a recent study on avian skulls proposed that complexity in developmental origin corresponds to lower within-partition trait correlation (28). In squamates, however, no clear association emerges between embryonic tissue origin and within-partition integration, disparity, or evolutionary rates.
Beyond macroevolutionary insights, the results of our study inform character construction for phylogenetic systematics. Simões et al. (41) expressed the need for broad taxonomic investigation of trait dependencies to mitigate redundant information and character dependencies in phylogenetic analyses, particularly in squamates. Here, the results show that the occiput and the palate are each an integrated structure, suggesting that characters within these regions are nonindependent. In contrast, the rostrum, often used as unit of character coding (e.g., “skull, rostrum anterior to the bony external nares” [39]), is highly modular, indicating that its treatment as a single character may poorly reflect biological reality. Similarly, the shapes of certain sets of cranial regions are strongly dependent on ecological factors (SI Appendix, Table S6). In snakes, for example, the shape of pterygoid and occipital condyle is strongly associated with diet (R2 > 0.09), whereas the jaw joint, basioccipital, and pterygoid exhibit greater degrees of phylogenetic signal (Kmult > 1.0; SI Appendix, Table S11). These bones with higher phylogenetic signal may thus be hypothesized to be a more reliable source for phylogenetic characters across squamates.
A major issue in squamate systematics has been the disagreement between molecular and morphological phylogenies with respect to fossorial taxa. As expected, morphological data typically unite fossorial taxa that molecular data consider phylogenetically distant (e.g., ref. 61). Our study demonstrates this extreme convergence in shape among fossorial taxa, with these taxa occupying a distinct region of PC2 (Fig. 1 and SI Appendix, Figs. S2 and S3). Shape changes along PC2 indicate that fossorial taxa are generally associated with (i) anteroposteriorly elongated skull, particularly the frontal and basioccipital; (ii) subrectangular premaxilla and nasal; (iii) small maxilla; (iv) anteriorly positioned and oriented jaw joint of the quadrate; (v) globular supraotoccipital; (vi) expansive foramen magnum; (vii) transversely narrow pterygoid and palate; and (vii) hypotrophied jugal. Because of their strong link with fossorial habitat, these characters may obscure the phylogenetic relationships of fossorial taxa. As illustrated here, large-scale phenome-level analyses reveal trait dependencies that could facilitate the construction or selection of reliable phylogenetic characters.
High-Density Morphometric Data Improve Macroevolutionary Analyses.
In this study, we present a squamate-wide analysis of cranial evolution with high-density morphometric data, far surpassing previous studies in shape characterization and taxonomic breadth. Although larger data do not necessarily equate to “better” data (62), we find that our high-dimensional GM approach combining landmarks and sliding semilandmarks is a marked improvement over analyses of landmarks alone beyond visual fidelity. First, performing LaSEC (63) on shape data of each partition indicates that, for many regions, 10–25 landmarks and sliding semilandmarks are needed to adequately capture the shape variation of each partition (SI Appendix, Table S10 and Fig. S11). Second, the fit between our full landmark + sliding semilandmark dataset to a landmark-only dataset is fairly poor across regions (SI Appendix, Table S10), suggesting that the use of few to several landmarks in each partition is insufficient for characterizing shape variation at this taxonomic scale. Moreover, integration analysis on landmark-only data results in greater correlations between adjacent regions, likely caused by landmarks being largely limited to boundaries between partitions (SI Appendix, Fig. S9). Meanwhile, subsampling our full dataset down to 10% of landmarks and sliding semilandmarks within each partition still recovers results that are congruent with our full shape data (SI Appendix, Fig. S9). Therefore, we advocate the collection and analysis of high-dimensional GM approach for phenotypic studies, especially when investigating shape variation and covariation across anatomical regions.
These data reveal a wealth of insights into skull evolution in squamates, including (i) evolutionary bursts associated with fossoriality, major changes in diet, and sexually selected traits such as elaborate cranial ornamentation; (ii) intense evolutionary changes in snakes and lizards occurring in completely different regions critical to feeding biomechanics; (iii) a shared pattern of cranial integration underlying fundamentally disparate crania of lizards and snakes; (iv) a tight correspondence between evolutionary rates and disparity across partitions; (v) an important exception to recent studies suggesting high trait integration constrains (or facilitates) phenotypic evolution; and (vi) corroborating evidence of a terrestrial, nonburrowing origin for snakes. Collectively, this study marks a new frontier in phenomic studies that couples high-density morphometry with modern comparative methods, opening new avenues to elucidate the drivers of phenotypic evolution in unprecedented detail and scale across disparate organisms.
Methods
Morphometric Data.
The taxonomic sampling represents our aim to capture the morphological, ecological, and phylogenetic breadth of Squamata, while considering available computed tomographic (CT) data to facilitate data collection (SI Appendix, Table S2). Our goal was to sample 1 to 3 genera for each extant subfamily, with consideration for including species that exhibit disparate morphological and ecological characteristics (SI Appendix, SI Text). We collected 3D landmark and semilandmark data from high-resolution digital reconstructions of skulls (SI Appendix, SI Text). All mesh files were constructed from CT imaging data or surface scan data (SI Appendix, Table S2). The meshes underwent a systematic protocol in the program GeoMagic Wrap (3D Systems), including virtual filling of small holes, centering, and smoothing to facilitate the placement of surface sliding semilandmarks. The meshes are freely available for download on Phenome10k (phenome10k.org) and MorphoSource (morphosource.org).
We then used Landmark Editor (64) to virtually place and export landmarks and curve semilandmarks on the right side of the skull to delineate the outlines of a priori partitions (SI Appendix, Table S1), including the premaxilla, nasal, maxilla, jugal, frontal, parietal, squamosal (supratemporal), jaw joint of the quadrate, supraotoccipital, occipital condyle, basioccipital + basisphenoid, pterygoid, and palatine (SI Appendix, Fig. S1A). Coordinate data were not collected from taxa in which cranial osteoderms are inseparable from the skull bones. By using the Morpho R package v2.5.1 (65), template mesh and coordinate data were used to conduct a semiautomated procedure to place surface semilandmarks within these partitions (SI Appendix, Fig. S1 C and D). The template was a hemispheric mesh with simplified geometric representations of partitions defined by corresponding landmarks and curve semilandmarks, as well as regularly distributed surface semilandmarks within these partitions. When they had been mapped onto the meshes of actual specimens, the coordinate data comprising landmarks and curve and surface sliding semilandmarks were mirrored along the median plane to temporarily create a bilateral dataset (SI Appendix, SI Text). Curve and surface sliding semilandmarks on the right side were then slid to minimize bending energy (66, 67), and the resulting bilateral landmark + sliding semilandmark data were then subjected to generalized Procrustes alignment (68, 69). The mirrored left landmarks and sliding semilandmarks were then removed from further analyses. Snake and extant datasets excluded the jugal partition and surface semilandmarks on the pterygoid and palatine because of the absence of the jugal bone in snakes and the prominent teeth on the latter 2 bones that impeded the placement of surface semilandmarks on these bones. For this study, the supratemporal bone in snakes was considered to be homologous to the squamosal bone in nonsnake squamates, at least functionally (but see ref. 70). In the combined extant and fossil datasets, 5 of the 13 partitions (premaxilla, maxilla, frontal, parietal, and supraotoccipital) that are exposed and preserved across all fossil specimens were included. Although effort was made to use predominantly type I landmarks to define partitions that are strictly homologous, this was not possible for such a broad taxonomic sampling. For this reason, some partitions were delineated by using type II and III landmarks sensu Bookstein (71) as functionally homologous regions.
Phylogenetic Tree.
To conduct comparative phylogenetic methods, we constructed a time-calibrated phylogenetic tree by using a published time-calibrated molecular phylogeny of extant squamates (72) and incorporating extinct taxa based on previous systematic work (39–41, 73) and fossil occurrence data from the Paleobiology Database (paleobiodb.org). We grafted extinct taxa onto the extant tree by applying the equal-branching method (74) based on the mean of first occurrence age range. Although the phylogenetic placement of Mosasauria within squamates remains ambiguous (37–39, 61), Plotosaurus was placed within the molecular phylogeny as a sister taxon to Serpentes (“Pythonomorpha hypothesis”) and Polyglyphanodon as sister to iguanians in accordance with the phylogeny based on combined molecular and morphological data (40). Although uncertainty in topology and branch lengths may alter the results of comparative analyses, we find that large-scale patterns are robust to alternative phylogenetic positions of extinct taxa, which generally have greater uncertainty in terms of position and thus may be expected to have the greatest potential to impact the outcome of analyses (SI Appendix, SI Text). Furthermore, congruent results between extant-only and combined datasets, as well as alternative placements of fossil taxa (SI Appendix, Table S7 and Fig. S5), suggest that the results and conclusions are robust to the inclusion, removal, and alternative phylogenetic positions of fossil specimens.
Ecological and Life History Data.
Existing literature was used to collect ecological and developmental data for extant taxa (SI Appendix, Table S5). We collected data on locomotion (ground dweller, swimmer, climber, climber/glider, digger, litter dweller, and combinations of these categories), habitat (terrestrial, fossorial, semifossorial, aquatic, semiaquatic, arboreal, semiarboreal, saxicolous, leaf litter, and combinations of these categories), diet (carnivorous, herbivorous, invertivorous, omnivorous), and reproductive mode (oviparous, viviparous). Because of the small number of ovoviviparous taxa, they were considered to be viviparous for regression analysis of reproductive mode. The definitions for these categories are provided in the SI Appendix. Although assigning categorical variables to a continuous trait is difficult, we used established categories known for species sampled in this study. Foraging and stomach content data for several taxa are unknown (SI Appendix, Table S5), and these taxa were excluded from MANOVA and rates analysis on diet preference.
Data Analysis.
Analyses were largely conducted in R v3.4.1 (75). First, a disparity-through-time plot was created for the individual partitions by using the dtt function in the GEIGER R package v2.0.6 (76, 77). Second, we used BayesTraitsV3 (78) to estimate evolutionary rates for the overall skull and its partitions based on PCs accounting for >95% of total shape variation (SI Appendix, Table S3). All available evolutionary models were tested, including BM, OU, δ, κ, and λ models. Bayes Factor was used to determine the best-supported evolutionary model. Estimated rates based on the preferred model were used to generate a rate-through-time plot for each partition. Total disparity (Procrustes variance) and mean evolutionary rates under the BM model were calculated within each model and for each landmark and sliding semilandmark by using the morphol.disparity and compare.multi.evol.rates functions in the geomorph R package v.3.0.5 (79, 80), respectively. We simulated shape evolution under BM model by using the sim.char function in GEIGER R package. Ancestral shapes were calculated by using the anc.recon function in the Rphylopars R package v0.2.9 (81) on extant-only and combined datasets.
The geomorph R package was used to assess allometry, phylogenetic signal, evolutionary allometry, and phylogenetically corrected correlations with ecological and reproductive variables in partition-specific and whole skull-shape data by using the functions procD.allometry, physignal, and procD.pgls (SI Appendix, Tables S6, S9, and S11). Because of the proportionately low contribution of (evolutionary) allometric signal, we did not correct for allometry in the datasets for other analyses. To identify cranial modules from partitions, we employed EMMLi (26), a maximum-likelihood approach, and CR analysis (27), which is implemented in geomorph. By using the results output by EMMLi, we further assessed the pattern of integration by comparing estimated within-module correlations (ρwithin) with the estimated between-module correlation (ρbetween) for each pair of partitions (SI Appendix, SI Text). We applied a subsampling approach to ascertain the robustness of these results by running 100 repetitions of EMMLi with 10% of the total landmark dataset while keeping a minimum of three landmarks + sliding semilandmarks per partition, which consistently returned the same pattern of cranial modules, as did analyses limited to landmarks only (SI Appendix, Fig. S9). Results from EMMLi and CR analyses with and without phylogenetic correction yielded largely congruent results (SI Appendix, Tables S7 and S8). Least-squares linear regression was conducted on these values to determine any relationships between disparity, mean evolutionary rates, and within-module correlation (SI Appendix, Table S9). Integration and macroevolutionary patterns were assessed separately for extant lizards, extant snakes, all extant squamates, and combined extant and extinct squamates.
Supplementary Material
Acknowledgments
We thank Kristin Mahlow and Erin Westeen for data collection assistance; Eve Noirault for reconstruction assistance; Henry Ferguson-Gow and Léo Botton-Divet for methodological assistance; and 2 anonymous reviewers for their constructive comments. European Research Council (Grant STG-2014-637171 to A.G.), National Science Foundation (EF-0334961 to J.A.M.), Deutsche Forschungsgemeinschaft (Mu 1760/7-1 to J.M.), European Council (Synthesis of Systematic Resources [SYNTHESYS] Grant DE-TAF-6532 to A.W.), and Paleontological Society (Newell Grant to A.W.) funded this study and the data collection.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1820967116/-/DCSupplemental.
References
- 1.Van Valen L., Multivariate structural statistics in natural history. J. Theor. Biol. 45, 235–247 (1974). [DOI] [PubMed] [Google Scholar]
- 2.Glaw F., Köhler J., Townsend T. M., Vences M., Rivaling the world’s smallest reptiles: Discovery of miniaturized and microendemic new species of leaf chameleons (Brookesia) from northern Madagascar. PLoS One 7, e31314 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hedges S. B., Thomas R., At the lower size limit in amniote vertebrates: A new diminutive lizard from the West Indies. Caribb. J. Sci. 37, 168–173 (2001). [Google Scholar]
- 4.Lingham-Soliar T., Anatomy and functional morphology of the largest marine reptile known, Mosasaurus hoffmanni (Mosasauridae, Reptilia) from the Upper Cretaceous, Upper Maastrichtian of The Netherlands. Phil. Trans. R. Soc. B 347, 155–172 (1995). [Google Scholar]
- 5.Evans S., “The skull of lizards and tuatara” in Biology of the Reptilia, The Skull of Lepidosauria, Gans C., Gaunt A. S., Adler K., Eds. (Society for the Study of Amphibians and Reptiles, Ithaca, NY, 2008), vol. 20, pp. 1–347. [Google Scholar]
- 6.Blackburn D. G., Squamate reptiles as model organisms for the evolution of viviparity. Herpetol. Monogr. 20, 131–146 (2006). [Google Scholar]
- 7.Pyron R. A., Burbrink F. T., Early origin of viviparity and multiple reversions to oviparity in squamate reptiles. Ecol. Lett. 17, 13–21 (2014). [DOI] [PubMed] [Google Scholar]
- 8.Longrich N. R., Bhullar B.-A. S., Gauthier J. A., Mass extinction of lizards and snakes at the Cretaceous-Paleogene boundary. Proc. Natl. Acad. Sci. U.S.A. 109, 21396–21401 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Feldman A., Sabath N., Pyron R. A., Mayrose I., Meiri S., Body sizes and diversification rates of lizards, snakes, amphisbaenians and the tuatara. Glob. Ecol. Biogeogr. 25, 187–197 (2016). [Google Scholar]
- 10.Andjelković M., Tomović L., Ivanović A., Morphological integration of the kinetic skull in Natrix snakes. J. Zool. 303, 188–198 (2017). [Google Scholar]
- 11.Hipsley C. A., Müller J., Developmental dynamics of ecomorphological convergence in a transcontinental lizard radiation. Evolution 71, 936–948 (2017). [DOI] [PubMed] [Google Scholar]
- 12.Ollonen J., Da Silva F. O., Mahlow K., Di-Poï N., Skull development, ossification pattern, and adult shape in the emerging lizard model organism Pogona vitticeps: A comparative analysis with other squamates. Front. Physiol. 9, 278 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Klingenberg C. P., Cranial integration and modularity: Insights into evolution and development from morphometric data. Hystrix Ital. J. Zool. (Modena) 24, 43–58 (2013). [Google Scholar]
- 14.Sanger T. J., Mahler D. L., Abzhanov A., Losos J. B., Roles for modularity and constraint in the evolution of cranial diversity among Anolis lizards. Evolution 66, 1525–1542 (2012). [DOI] [PubMed] [Google Scholar]
- 15.Urošević A., Ljubisavljević K., Ivanović A., Multilevel assessment of the Lacertid lizard cranial modularity. J. Zool. Syst. Evol. Res. 57, 145–158 (2019). [Google Scholar]
- 16.Bardua C., Wilkinson M., Gower D. J., Sherratt E., Goswami A., Morphological evolution and modularity of the caecilian skull. BMC Evol. Biol. 19, 30 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Goswami A., Polly P. D., Mock O. B., Sánchez-Villagra M. R., Shape, variance and integration during craniogenesis: Contrasting marsupial and placental mammals. J. Evol. Biol. 25, 862–872 (2012). [DOI] [PubMed] [Google Scholar]
- 18.Wagner G. P., Homologues, natural kinds and the evolution of modularity. Am. Zool. 36, 36–43 (1996). [Google Scholar]
- 19.Wagner G. P., Altenberg L., Perspective: Complex adaptations and the evolution of evolvability. Evolution 50, 967–976 (1996). [DOI] [PubMed] [Google Scholar]
- 20.Klingenberg C. P., Morphological integration and developmental modularity. Annu. Rev. Ecol. Evol. Syst. 39, 115–132 (2008). [Google Scholar]
- 21.Klingenberg C. P., “Developmental constraints, modules, and evolvability” in Variation, Hallgrímsson B., Hall B. K., Eds. (Academic Press, Cambridge, MA, 2005), pp. 219–247. [Google Scholar]
- 22.Goswami A., Smaers J. B., Soligo C., Polly P. D., The macroevolutionary consequences of phenotypic integration: From development to deep time. Philos. Trans. R. Soc. Lond. B Biol. Sci. 369, 20130254 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Stayton C. T. Morphological evolution of the lizard skull: A geometric morphometrics survey. J. Morphol. 263, 47–59 (2005). [DOI] [PubMed] [Google Scholar]
- 24.Cundall D., Shardo J., Rhinokinetic snout of thamnophiine snakes. J. Morphol. 225, 31–50 (1995). [DOI] [PubMed] [Google Scholar]
- 25.Cundall D., Irish F., “The snake skull” Biology of the Reptilia, The Skull of Lepidosauria, Gans C., Gaunt A. S., Adler K., Eds. (2008), vol. 20, pp 349–692. [Google Scholar]
- 26.Goswami A., Finarelli J. A., EMMLi: A maximum likelihood approach to the analysis of modularity. Evolution 70, 1622–1637 (2016). [DOI] [PubMed] [Google Scholar]
- 27.Adams D. C., Evaluating modularity in morphometric data: Challenges with the RV coefficient and a new test measure. Methods Ecol. Evol. 7, 565–572 (2016). [Google Scholar]
- 28.Felice R. N., Goswami A., Developmental origins of mosaic evolution in the avian cranium. Proc. Natl. Acad. Sci. U.S.A. 115, 555–560 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Simões T. R., et al. , The origin of squamates revealed by a Middle Triassic lizard from the Italian Alps. Nature 557, 706–709 (2018). [DOI] [PubMed] [Google Scholar]
- 30.Cleary T. J., Benson R. B. J., Evans S. E., Barrett P. M., Lepidosaurian diversity in the Mesozoic-Palaeogene: The potential roles of sampling biases and environmental drivers. R. Soc. Open Sci. 5, 171830 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Foote M., Discordance and concordance between morphological and taxonomic diversity. Paleobiology 19, 185–204 (1993). [Google Scholar]
- 32.Peters J. A., The snakes of the subfamily Dipsadinae. Misc Publ Museum Zool Univ Michigan 114, 1–224 (1960). [Google Scholar]
- 33.Platt D. R., Natural history of the Hognose snakes Heterodon platyrhinos and Heterodon nasicus. Univ. Kans. Publ. Mus. Nat. Hist. 18, 253–420 (1969). [Google Scholar]
- 34.Fabre A.-C., Bickford D., Segall M., Herrel A., The impact of diet, habitat use, and behaviour on head shape evolution in homalopsid snakes. Biol. J. Linn. Soc. Lond. 118, 634–647 (2016). [Google Scholar]
- 35.Klaczko J., Sherratt E., Setz E. Z. F., Are diet preferences associated to skulls shape diversification in xenodontine snakes? PLoS One 11, e0148375 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Felice R. N., Tobias J. A., Pigot A. L., Goswami A., Dietary niche and the evolution of cranial morphology in birds. Proc. Biol. Sci. 286, 20182677 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Camp L., Classification of the lizards. Bull. Am. Mus. Nat. Hist. 48, 289–481 (1923). [Google Scholar]
- 38.Lee M. S. Y., Caldwell M. W., Adriosaurus and the affinities of mosasaurs, dolichosaurs, and snakes. J. Paleontol. 74, 915–937 (2000). [Google Scholar]
- 39.Conrad J., Phylogeny and systematics of Squamata (Reptilia) based on morphology. Bull. Am. Mus. Nat. Hist. 310, 1–182 (2008). [Google Scholar]
- 40.Reeder T. W., et al. , Integrated analyses resolve conflicts over squamate reptile phylogeny and reveal unexpected placements for fossil taxa. PLoS One 10, e0118199 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Simões T. R., Caldwell M. W., Palci A., Nydam R. L., Giant taxon-character matrices: Quality of character constructions remains critical regardless of size. Cladistics 33, 198–219 (2017). [DOI] [PubMed] [Google Scholar]
- 42.Zug G. R., Reptiles and Amphibians of the Pacific Islands: A Comprehensive Guide (University of California Press, 2013). [Google Scholar]
- 43.Ávila-Pires T. C., Lizards of Brazilian Amazonia (Reptilia: Squamata). Zool. Verhandel. 299, 1–706 (1995). [Google Scholar]
- 44.Zaher H., Scanferla C. A., The skull of the Upper Cretaceous snake Dinilysia patagonica Smith-Woodward, 1901, and its phylogenetic position revisited. Zool. J. Linn. Soc. 164, 194–238 (2012). [Google Scholar]
- 45.Lee M. S. Y., Bell G. L. Jr, Caldwell M. W., The origin of snake feeding. Nature 400, 655–659 (1999). [Google Scholar]
- 46.Lee M. S. Y., Molecular evidence and marine snake origins. Biol. Lett. 1, 227–230 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Hsiang A. Y., et al. , The origin of snakes: Revealing the ecology, behavior, and evolutionary history of early snakes using genomics, phenomics, and the fossil record. BMC Evol. Biol. 15, 87 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Yi H., Norell M. A., The burrowing origin of modern snakes. Sci. Adv. 1, e1500743 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Palci A., Hutchinson M. N., Caldwell M. W., Lee M. S. Y., The morphology of the inner ear of squamate reptiles and its bearing on the origin of snakes. R. Soc. Open Sci. 4, 170685 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Da Silva F. O., et al. , The ecological origins of snakes as revealed by skull evolution. Nat. Commun. 9, 376 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Haas G., “Muscle of the jaws and associated structures in the Rhynchocephalia and Squamata” in Biology of the Reptilia, Gans C., Parsons T., Eds. (Academic Press, London, 1973), vol. 4, pp. 285–490. [Google Scholar]
- 52.Moazen M., Curtis N., Evans S. E., O’Higgins P., Fagan M. J., Combined finite element and multibody dynamics analysis of biting in a Uromastyx hardwickii lizard skull. J. Anat. 213, 499–508 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.McDowell S. B., “The skull of Serpentes” in Biology of Reptilia, Vol. 21, Morphology I, The Skull and Appendicular Locomotor Apparatus of Lepidosauria, Gans C., Gaunt A. S., Adler K., Eds. (2008), pp. 467–620. [Google Scholar]
- 54.Goswami A., Cranial modularity shifts during mammalian evolution. Am. Nat. 168, 270–280 (2006). [DOI] [PubMed] [Google Scholar]
- 55.Porto A., de Oliveira F. B., Shirai L. T., de Conto V., Marroig G., The evolution of modularity in the mammalian skull I: Morphological integration patterns and magnitudes. Evol. Biol. 36, 118–135 (2009). [Google Scholar]
- 56.Goswami A., Polly P. D., Methods for studying morphological integration and modularity. Quant Methods Paleobiol 16, 213–243 (2010). [Google Scholar]
- 57.Bright J. A., Marugán-Lobón J., Cobb S. N., Rayfield E. J., The shapes of bird beaks are highly controlled by nondietary factors. Proc. Natl. Acad. Sci. U.S.A. 113, 5352–5357 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Randau M., Goswami A., Shape covariation (or the lack thereof) between vertebrae and other skeletal traits in felids: The whole is not always greater than the sum of parts. Evol. Biol. 45, 196–210 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Larouche O., Zelditch M. L., Cloutier R., Modularity promotes morphological divergence in ray-finned fishes. Sci. Rep. 8, 7278 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Frazzetta T. H., A functional consideration of cranial kinesis in lizards. J. Morphol. 111, 287–319 (1962). [DOI] [PubMed] [Google Scholar]
- 61.Gauthier J. A., Kearney M., Maisano J. A., Rieppel O., Behlke A. D. B., Assembling the squamate tree of life: Perspectives from the phenotype and the fossil record. Bull. Peabody Mus. Nat. Hist. 53, 3–308 (2012). [Google Scholar]
- 62.Cardini A., Integration and modularity in Procrustes shape data: Is there a risk of spurious results? Evol. Biol. 46, 90–105 (2018). [Google Scholar]
- 63.Watanabe A. How many landmarks are enough to characterize shape and size variation? PLoS One 13, e0198341 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Wiley D. F., et al. , Evolutionary Morphing (IEEE, Washington, DC, 2005), pp. 432–438. [Google Scholar]
- 65.Schlager S., “Morpho and Rvcg - shape analysis in R: R-Packages for geometric morphometrics, shape analysis and surface manipulations” in Statistical Shape and Deformation Analysis: Methods, Implementation and Applications, Zhen G., Li S., Szekely G., Eds. (Academic Press, 2017), pp. 217–256. [Google Scholar]
- 66.Gower J. C., Generalized Procrustes analysis. Psychometrika 40, 33–51 (1975). [Google Scholar]
- 67.Rohlf F. J., Slice D., Extensions of the Procrustes method for the optimal superimposition of landmarks. Syst. Zool. 39, 40–59 (1990). [Google Scholar]
- 68.Gunz P., Mitteroecker P., Bookstein F. L., “Semilandmarks in three dimensions” in Modern Morphometrics in Physical Anthropology, Slice D. E., Ed. (Kluwer Academic/Plenum Publishers, New York, 2005), pp. 73–98. [Google Scholar]
- 69.Gunz P., Mitteroecker P., Semilandmarks: A method for quantifying curves and surfaces. Hystrix Ital. J. Zool. (Modena) 24, 103–109 (2013). [Google Scholar]
- 70.Werneburg I., Sánchez-Villagra M. R., Skeletal heterochrony is associated with the anatomical specializations of snakes among squamate reptiles. Evolution 69, 254–263 (2015). [DOI] [PubMed] [Google Scholar]
- 71.Bookstein F. L., Morphometric Tools for Landmark Data: Geometry and Biology (Cambridge University Press, Cambridge, United Kingdom, 1991). [Google Scholar]
- 72.Zheng Y., Wiens J. J., Combining phylogenomic and supermatrix approaches, and a time-calibrated phylogeny for squamate reptiles (lizards and snakes) based on 52 genes and 4162 species. Mol. Phylogenet. Evol. 94, 537–547 (2016). [DOI] [PubMed] [Google Scholar]
- 73.Hipsley C. A., Müller J., Relict endemism of extant Rhineuridae (Amphisbaenia): Testing for phylogenetic niche conservatism in the fossil record. Anat. Rec. 297, 473–481 (2014). [DOI] [PubMed] [Google Scholar]
- 74.Brusatte S. L., Lloyd G. T., Wang S. C., Norell M. A., Gradual assembly of avian body plan culminated in rapid rates of evolution across the dinosaur-bird transition. Curr. Biol. 24, 2386–2392 (2014). [DOI] [PubMed] [Google Scholar]
- 75.R Core Development Team , R: A Language and Environment for Statistical Computing, Version 3.4.1. https://www.r-project.org. Accessed 4 December 2015.
- 76.Harmon L. J., Weir J. T., Brock C. D., Glor R. E., Challenger W., GEIGER: Investigating evolutionary radiations. Bioinformatics 24, 129–131 (2008). [DOI] [PubMed] [Google Scholar]
- 77.Pennell M. W., et al. , geiger v2.0: An expanded suite of methods for fitting macroevolutionary models to phylogenetic trees. Bioinformatics 30, 2216–2218 (2014). [DOI] [PubMed] [Google Scholar]
- 78.Meade A., Pagel M., BayesTraitsV3. http://www.evolution.rdg.ac.uk/BayesTraitsV3.0.1/BayesTraitsV3.0.1.html. Accessed 23 November 2017.
- 79.Adams D. C., Otárola-Castillo E., geomorph: An R Package for the collection and analysis of geometric morphometric shape data. Methods Ecol. Evol. 4, 393–399 (2013). [Google Scholar]
- 80.Adams A. D., Collyer M., Otárola-Castillo E., Sherratt E., Adams M. D., Package ‘geomorph’ Version 3.0.5. https://cran.r-project.org/web/packages/geomorph/index.html. Accessed 1 March 2018.
- 81.Goolsby E. W., Likelihood-based parameter estimation for high-dimensional phylogenetic comparative models: Overcoming the limitations of “distance-based” methods. Syst. Biol. 65, 852–870 (2016). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.