Significance
The nuclear pore complex (NPC) is a huge protein assembly that selectively transports macromolecules across the nuclear envelope (NE) of eukaryotes. Determining the precise organization of that machinery has been a long-standing goal of structural biology. Here, we introduce a methodology called NuRIM that can map the average position of NE proteins in vivo based on the analysis of intensity patterns in fluorescence micrographs. This generally applicable technique delivers a precise positional map of the native yeast NPC and associated factors. Further, it allows us to investigate the structural consequences of NPC compositional perturbations, and to orient specific protein segments that play an essential role in NPC transport and assembly.
Keywords: nuclear pore complex, superresolution, nucleoporins, quantitative fluorescence microscopy, FG repeats
Abstract
Selective transport across the nuclear envelope (NE) is mediated by the nuclear pore complex (NPC), a massive ∼100-MDa assembly composed of multiple copies of ∼30 nuclear pore proteins (Nups). Recent advances have shed light on the composition and structure of NPCs, but approaches that could map their organization in live cells are still lacking. Here, we introduce an in vivo method to perform nuclear radial intensity measurements (NuRIM) using fluorescence microscopy to determine the average position of NE-localized proteins along the nucleocytoplasmic transport axis. We apply NuRIM to study the organization of the NPC and the mobile transport machinery in budding yeast. This reveals a unique snapshot of the intact yeast NPC and identifies distinct steady-state localizations for various NE-associated proteins and nuclear transport factors. We find that the NPC architecture is robust against compositional changes and could also confirm that in contrast to Chlamydomonas reinhardtii, the scaffold Y complex is arranged symmetrically in the yeast NPC. Furthermore, NuRIM was applied to probe the orientation of intrinsically disordered FG-repeat segments, providing insight into their roles in selective NPC permeability and structure.
The nuclear pore complex (NPC) provides the essential passageway for proteins and RNAs across the nuclear envelope (NE), which separates the nucleus from the cytoplasm in eukaryotes (1, 2). NPC malfunctions are associated with a spectrum of detrimental conditions including aging-related disorders, infectious diseases, neurodegeneration, and cancer (3, 4). In recent years, electron microscopy (EM) coupled to integrative modeling has greatly contributed to delineating the architecture of the NPC despite its sheer size and compositional heterogeneity (5–8). Current structural NPC models reveal the double-layered lipid pore membrane, overall 8-fold rotational symmetry of the NPC, and finer anatomical details including the nuclear basket, cytoplasmic export platform, and core scaffold where distinct ring-like features are recognized (Fig. 1A) (9–12). By relying on a powerful set of inferential methods (aka “integrative modeling”), high-resolution models of the NPC were presented that account for most known NPC features. However, given the limited resolution of current NPC tomograms (∼3 nm) and the high structural similarity among nucleoporins (Nups), the fitting of individual protein structures into the EM density of entire NPCs has been a formidable challenge, critically dependent on orthogonal input such as restraints from chemical cross-linking data, symmetry considerations, and accurate stoichiometry (5, 13–16).
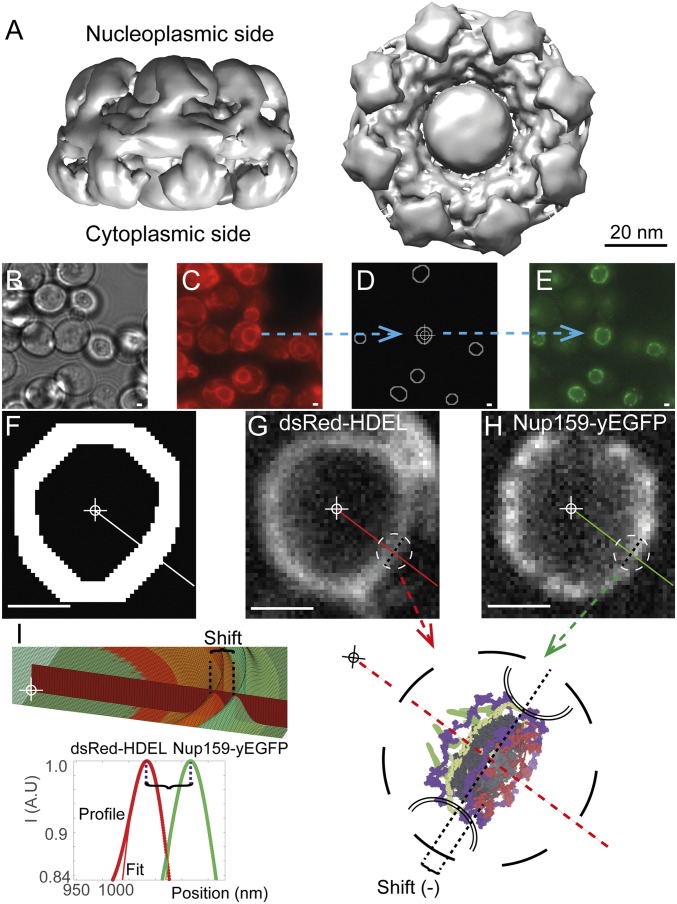
Fig. 1.
Description of the NuRIM method. (A) State-of-the-art electron density map of the yeast NPC adapted from the work of Kim et al. (5) introduces salient features of the NPC including a central channel permeated by a central “transporter,” framed by the NPC core. UCSF Chimera software was used to display a density map at the recommended threshold of 0.015 (64). (B) Bright-field image of S. cerevisiae cells. (C) The luminal dsRed-HDEL reference is used for NE tracing. (D) Using automated image analysis, NEs are precisely outlined and their center is determined (cross-hair). (E) Nup-yEGFP channel allows quantifying Nup abundance and radial shift relative to the dsRed-HDEL reference channel. (F) Dilated NE traces restrict analysis to informative area only. (G–I) Lines from the nuclear center intersect the NE. Intensity profiles along these lines are then fitted with Gaussian functions, allowing the measurement of the radial shift of a given Nup, for example, Nup159-yEGFP (green) against the dsRed-HDEL reference profile along the same ray (red). Averaging thousands of such differential measurements delivers structural information on NPC architecture with nanoscale accuracy (SI Appendix). (Scale bars, 1 μm.)
We previously introduced technology to precisely determine the stoichiometry of molecular complexes in the yeast NE by fluorescence intensity measurements (Fig. 1 B–E) (16). That method was applied to the yeast NPC, revealing 16 copies per NPC for most Nups as well as a capacity for NPCs to remain functional despite alterations in their composition, a type of robustness termed “compositional plasticity” (1, 9, 10, 16–18). However, whether compositional plasticity is also reflected by structural plasticity is unexplored. Also, it has proven difficult to obtain positional information for the intrinsically disordered Phe-Gly (FG) repeats. Yet these are essential functional elements that endow the NPC with the capacity to interact selectively and transiently with cognate transporters as well as the NPC core (19). It has been suggested that FG repeats play an additional role in NPC assembly and stability via multivalent interactions with scaffold Nups (20), and that nuclear transport receptors form a pool at the NPC, influencing the permeability properties of the NPC (Fig. 1A) (21–23). However, because of the intrinsically disordered nature of FG repeats, these essential functional elements remain a challenge for current structural approaches (5, 14, 20, 24–27).
In this study, we develop an in vivo quantification tool named nuclear radial intensity measurements (NuRIM) to extract the average position of Nups and associated transport factors along the nucleocytoplasmic transport axis. This provides us with a tool to comprehensively reexamine the organization of the native yeast NPC and the nucleocytoplasmic transport machinery.
Results
NuRIM: A Superresolution Technique to Map GFP-Tagged Nucleoporins in Native NPCs.
NPCs are ∼100 nm in size, suggesting that it is impossible to resolve subfeatures under optical microscopy (Abbe’s diffraction limit is ∼200 nm for fluorescence microscopy) (9, 11, 17, 28). Superresolution may nonetheless be reached by single-molecule excitation or by averaging many noisy measurements of the same quantity (28, 29). One fruitful application of the latter principle has been single-particle EM, whereby thousands of noisy images are averaged into structures reaching subnanometer resolution (12, 30). We set out to utilize similar principles under fluorescence microscopy to map the relative positions of Nups (31). To implement such an approach, we took advantage of yeast strains coexpressing yeast-enhanced GFP (yEGFP)-tagged Nups together with dsRed-HDEL serving as a fluorescent NE fiduciary marker (16, 32). The NE signal in the immediate proximity of the NPCs allows differential measurement of small-scale radial shifts (called “heights” for brevity) of Nups with respect to the NE marker, and thus to systematically arrange most Nups according to their radial position within the NPC (Fig. 1). By convention, the origin of coordinates lies at the NPC center of mass; negative coordinates are directed toward the cytoplasm and positive coordinates toward the nucleus. For example, we would obtain a positive value for the position of the nuclear basket component Nup60-yEGFP (Dataset S1, sheet 1).
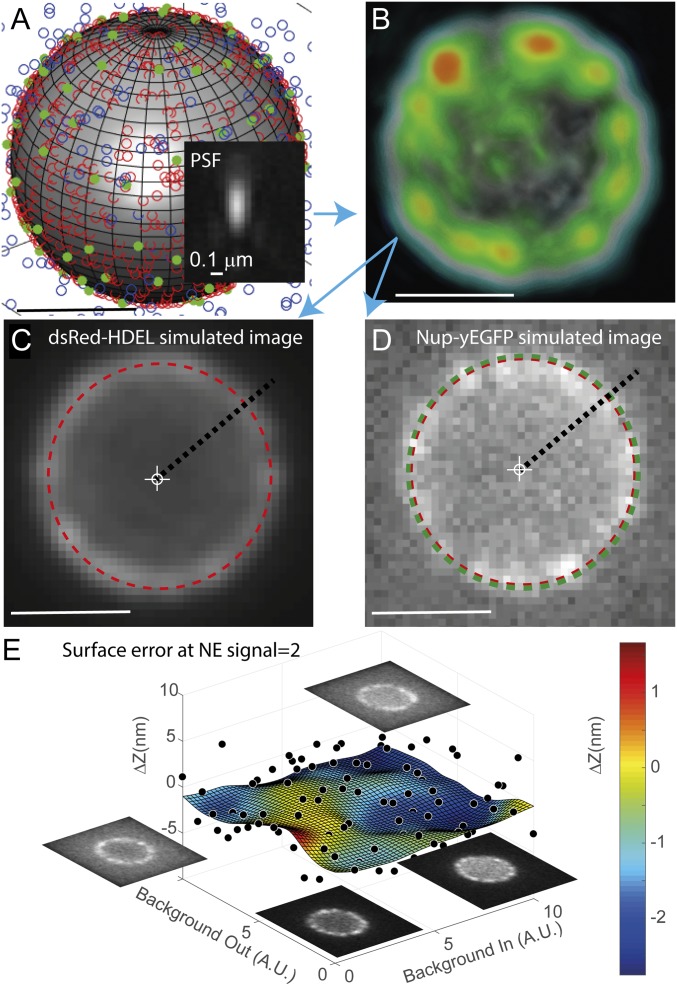
To gauge the theoretical accuracy of NuRIM (see SI Appendix for accuracy and precision notions), we generated 160,000 simulated 3D image stacks of NEs under variable conditions of NPC signal intensity and confounding background fluorescence in the cytoplasm and nucleus (Fig. 2 and SI Appendix, Fig. S1). Errors were characterized extensively by comparing the positions recovered by NuRIM against a known ground truth (e.g., 0 nm when no shift was imposed). For typical signal conditions encountered during Nup imaging, errors remained below 5 nm (Fig. 2E). A major benefit of that analysis is that one is able to subsequently subtract these small biases from the values measured experimentally to obtain adjusted coordinates, referred to as “position” (SI Appendix and SI Appendix, Fig. S1). Altogether, these simulations suggest that gathering structural information for the Nups along the nucleocytoplasmic axis using wide-field microscopy is possible.
Fig. 2.
Testing NuRIM’s theoretical accuracy using simulated data. (A) Point-like NPCs (green solid dots) and reference fiduciary markers (red open dots) are distributed on the NE, itself modeled as a sphere. Variable levels of confounding fluorescence are introduced inside and outside the NE (blue open dots). (B) Convolution with a realistic point spread function (PSF) yields 3D diffraction-limited stacks (a 3D rendering of the NE is shown). (C and D) Single Z slices sampled from the simulated image volumes. Small transverse shifts in the distributions of NPCs away from the NE lead to measurable subpixel shifts in the simulated images (red and green dashed circles). (E) Over 10 million simulated images were generated under variable background conditions. The plot shows the error made by NuRIM when recovering the ground-truth shift for an NE signal level corresponding to nucleoporins. Black dots represent average shift error for batches of 64 simulated images. The fitting surface was obtained using neural network training, thus yielding estimates of bias error in any background conditions. These small bias errors are subsequently subtracted from experimental values to obtain adjusted positions (SI Appendix and SI Appendix, Fig. S1). (Scale bars, 1 μm.)
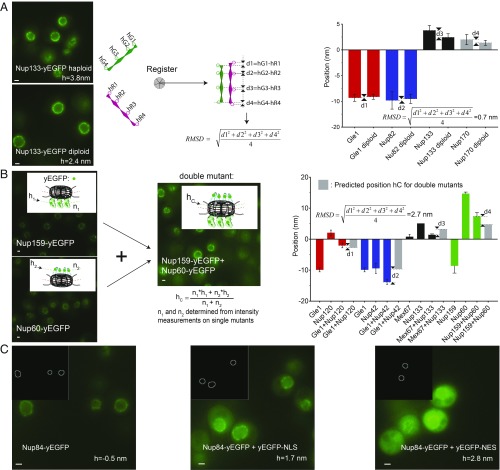
To assess experimental accuracy in ultramicroscopy, one typically performs 2 independent structure determinations of the same object (e.g., by dividing one’s dataset into 2 halves, or by relying on 2 independent techniques) and then measures by how much they differ after rigid registration, using the pairwise rmsd criterion (33, 34) (Fig. 3A and SI Appendix). To apply that protocol to NuRIM, we independently collected structural information in haploid and diploid strains, registered the 2 corresponding 1D structures (by iterative minimization of the rmsd), and obtained a pairwise rmsd accuracy of 0.7 nm (Fig. 3A and Dataset S1, sheet 2). Diploid cells are larger than haploid cells, so the experiment also served to test accuracy against variations in the nuclear diameter (35). Assuming that the NPC structure is identical in haploid and diploid cells (SI Appendix, Fig. S3), we concluded that our method can map the average position of individual Nups with nanometer accuracy.
Fig. 3.
Testing the accuracy of NuRIM. (A) Haploid nuclei are smaller than diploid nuclei, but the recovered Nup positions in both cases were highly consistent (pairwise rmsd = 0.7 nm; error bars always represent the SEM except when stated). Schematics illustrates the general notion of the pairwise rmsd serving to assess the accuracy of structural methods. (B) Another strategy to test NuRIM accuracy was to compare the predicted average positions for double mutants with the positions actually measured (pairwise rmsd = 2.7 nm). (C) To test NuRIM’s robustness against variations in background fluorescence, soluble yEGFP-NES and yEGFP-NLS were overexpressed on top of Nup84-yEGFP (pairwise rmsd = 2.4 nm) (36). (Scale bars, 1 μm.)
To provide an independent estimate of accuracy in a more complex setting, we used strains simultaneously expressing 2 different yEGFP-tagged Nups (Fig. 3B). These cannot be resolved as they occupy the same diffraction-limited volume, but their simultaneous presence is expected to lead to a radial shift of their combined signal compared with the singly labeled strains. The predicted position hC for the double mutant is the center of mass of the combined yEGFP distributions: hC = (n1 × h1 + n2 × h2)/(n1 + n2), where the copy numbers n1 and n2 and positions h1 and h2 are obtained experimentally from the singly labeled strains. By comparing those predicted values with the experimental ones, a pairwise rmsd accuracy of 2.7 nm was obtained (Dataset S1, sheet 3).
The presence of nuclear or cytoplasmic background fluorescence is expected to affect the distribution of the measured Nup signal at the NE and could thus introduce errors into our positional assignments. To test robustness against background fluorescence under challenging conditions, we selected 2 fluorescently tagged Nup strains (Nup84-yEGFP and Nup159-yEGFP) and overexpressed free yEGFP fused either to a nuclear export sequence (NES) or a nuclear localization sequence (NLS) (36) (Fig. 3C). Despite prominent background fluorescence, positions remained very consistent (pairwise rmsd = 2.4 nm; Dataset S1, sheet 4).
It is widely appreciated that fluorescence-based measurements can be affected by the environment, for example by changes in pH or polarity (37). To assess potential interferences within the NPC environment, we increased the length of the flexible linkers between the yEGFP and its Nup to a total of 102 amino acids, thereby attempting to release the yEGFPs from any local effects (16). This only led to minimal positional changes (pairwise rmsd = 1.6 nm; Dataset S1, sheet 5). These accuracy results compare favorably with state-of-the-art positional uncertainties of ∼6 nm in immunogold electron microscopy (IEM) (9, 38, 39).
We also examined whether the maturation kinetics of yEGFP coupled to variable turnover at the NPC could possibly affect our measurements. Thus, we generated strains tagged with “superfolder GFP,” a variant specially developed for rapid folding (40). These measurements led to an rmsd of 1.1 nm by pairwise comparison with the original yEGFP-based results (Dataset S1, sheet 6).
Finally, we also wanted to assess how signal strength and bleaching influence our measurements. To this end, we continuously photobleached a single field of view until the fluorescence intensity decreased to less than 10% of the original signal. The measured Nup positions remained surprisingly stable even at the lowest signal intensities (SI Appendix, Fig. S2).
Additional tests to appreciate the role of chromatic aberrations, NE size (SI Appendix, Fig. S3), and projection effects (SI Appendix, Fig. S5) are deferred to SI Appendix. It turns out that projection effects can be compensated by the introduction of a “projection factor” slightly larger than 1, and that chromatic effects are minimal and can be eliminated by performing color registration. Altogether, our tests indicate that NuRIM offers an average accuracy of ∼2 nm, which is on par with that of IEM (Table 1).
Table 1.
Accuracy of NuRIM evaluated using various approaches
| Approach | Rmsd accuracy, nm | Comments |
| Nup positions in haploid strains compared with diploid strains (Fig. 3A) | 0.7 | 4 haploid Nup-yEGFP strains versus matching diploids (Dataset S1, sheet 2) |
| Positions in double mutants compared with predicted values based on data from single mutants (Fig. 3B) | 2.7 | 5 double-mutant combinations (Dataset S1, sheet 3) |
| Positions in base strain compared with strains where soluble yEGFP is enriched in the cytoplasm or nucleoplasm (Fig. 3C) | 2.4 | 2 Nup-yEGFP base strains, 6 Nup-yEGFP strains in total (Dataset S1, sheet 4) |
| Positions in base strain compared with strains with 102-aa linkers to yEGFP | 1.6 | 3 Nup-yEGFP base strains (Dataset S1, sheet 5) |
| Positions in base strain compared with superfolder GFP strains | 1.1 | 4 Nup-yEGFP base strains (Dataset S1, sheet 6) |
| Positions in Y complex obtained using 2 different NE reference probes (Fig. 4C) | 1.9 | 5 Nup-yEGFP strains from the Y complex (Dataset S1, sheet 7) |
| Average accuracy | 1.55 |
NuRIM Reveals the Position of Stably Anchored Nup Domains Along the Nucleocytoplasmic Transport Axis and Demonstrates Robustness of the NPC Structure Against Compositional Changes.
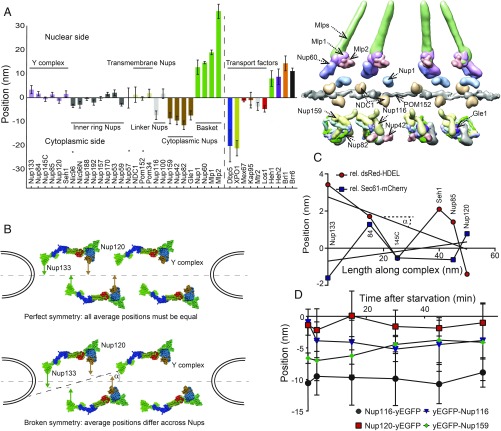
Owing to extensive averaging, systematic position measurements produced by NuRIM for yeast Nups had high precision—providing a 1D map with an average SEM equal to 1.5 nm across 28 Nups (Fig. 4A and Dataset S1, sheet 1). The NPC is akin to a modular assembly of stable subcomplexes (2). The majority of yeast Nups are believed to be symmetrically organized along the NPC transport axis, with half of the copies facing the nucleus and the other half facing the cytoplasm (2). Since the symmetric units occupy the same diffraction-limited volume they cannot be resolved under fluorescence microscopy, and we can only determine their average position in between the 2 symmetric units along the transport axis. By contrast, Nup82 and Nup159, that both belong to the mRNA export platform, exclusively face the cytoplasm (12, 41). Thus, considerations of symmetry do not play a role for these Nups. NuRIM measured average positions of −8.6 ± 2.3 nm for Nup159 compared with −11.5 ± 1.8 nm for Nup82 (Dataset S1, sheet 1). Further, Gle1, which is known to interact with the same subcomplex, was mapped at −7.4 ± 1.9 nm. Interestingly, the only other Nups seen within a range of 3 nm were Nup116 (h = −7.3 ± 1.8 nm), consistent with its known interaction with Nup82 (42), as well as Nup42 (h = −9.5 ± 1.9 nm), another documented interactor (43). Altogether, this is consistent with previous reports that depict the export platform hovering over NPCs (41). Additional Nups are likely to localize nearby but, owing to symmetry, their average location is close to the midplane (Fig. 4A).
Fig. 4.
NuRIM delivers structural information on native pores. (A) The average positions along the nucleocytoplasmic axis were determined for most Nups and transport factors. Strains that showed any evidence of tag interference are indicated by an asterisk. For comparison, selected Nup probability densities from the integrative NPC model of Kim et al. (5) are displayed using UCSF Chimera software (see also Table 2). (B) A fully symmetric arrangement of the Y complex across the NE plane must lead to equal average values for all Y-complex Nups (hypothetical arrangements are shown). (C) Successive NuRIM positions of the Nups along the long axis of the Y complex resulted in a linear regression angle α of only 3 ± 2°, thus providing little evidence for asymmetry. (D) Upon acute glucose starvation, no large-scale changes were observed for either Nup116-yEGFP or Nup120-yEGFP but yEGFP tags at the extremity of FG repeats appeared to converge. Error bars represent the SEM.
Table 2.
Comparison of NuRIM results with reference values from the literature
| Nucleoporin | NuRIM, nm | Kim et al. (5),* nm | Alber et al. (9),† nm | Rout et al. (2),‡ nm |
| Nup133 | 3.4 | −0.3 | 0.0 | 0.0 |
| Nup84 | 1.7 | 1.7 | 0.0 | 0.0 |
| Nup145C | −0.6 | −1.7 | 0.0 | 0.0 |
| Nup85 | 1.4 | 0.4 | 0.0 | 0.0 |
| Nup120 | −1.4 | −0.2 | 0.0 | 0.0 |
| Seh1 | 2.1 | 0.2 | 0.0 | 0.0 |
| Nic96 | −2.9 | 0.7 | 0.0 | 0.0 |
| Nup188 | −0.5 | −0.3 | 0.0 | 0.0 |
| Nup192 | −2 | 0.9 | 0.0 | 0.0 |
| Nup157 | −0.3 | 0.5 | 0.0 | 0.0 |
| Nup170 | 1.5 | 0.7 | 0.0 | 0.0 |
| Nup53 | 2.0 | −0.5 | 0.0 | 0.0 |
| Nup59 | −2.8 | 1.7 | 0.0 | 0.0 |
| Nup57 | −0.1 | 0.9 | 0.0 | 0.0 |
| NDC1 | 1.3 | 1.3 | 0.0 | |
| Pom152 | −0.31 | 0.1 | 0.0 | 0.0 |
| Pom34 | 1.9 | 1.1 | 0.0 | |
| Nup116 | −7.3 | −19.6 | −13 | −28 |
| Nup100 | 1.7 | −9.5 | −8 | −32 |
| Nup159 | −8.6 | −16.5 | −13 | −27 |
| Nup42 | −9.5 | −21.9 | −13 | −35 |
| Nup82 | −11.5 | −20.7 | −13 | −52 |
| Gle1 | −7.4 | −17.2 | −11 | −22 |
| Nup1 | 12.8 | 7.2 | 12 | 53 |
| Nup60 | 14.7 | 14.8 | 14 | 41 |
| Mlp1 | 19.7 | 36.2 | ||
| Mlp2 | 17 | 12.9 |
Values were sourced from the reference probability density maps using UCSF Chimera software with the “measure center” command (5). Thus, by virtue of averaging across symmetric units, many Nups are close to 0 nm. This facilitates comparison with NuRIM results where such averaging cannot be avoided.
Values were sourced from reference supplementary figure 25 in ref. 9. Nups considered “symmetric” were rewritten here as “0.0” nm to facilitate comparison with NuRIM results.
Values were sourced from reference table II in ref. 2. Nups considered symmetric were rewritten here as 0.0 nm to facilitate comparison with NuRIM results.
On the other side of the NPC, the asymmetric nuclear basket nucleoporins comprising Nup1 (12.8 ± 3.0 nm), Nup60 (14.7 ± 0.5 nm), Mlp1 (36.2 ± 1.1 nm), and Mlp2 (13.0 ± 1.5 nm) were in agreement with integrative models as well (Fig. 4A) (5, 9).
The symmetrical organization of scaffold Nups was recently shown not to hold for the NPCs of Chlamydomonas reinhardtii, where an asymmetry of the Y complex (also called the Nup84 subcomplex) was demonstrated (44). For budding yeast, however, it is generally accepted that such bilateral symmetry does hold (2, 5, 9). Therefore, one would expect to obtain identical NuRIM average positions for all symmetric Nups, as illustrated in Fig. 4B. Conversely, the observation of any difference in the average height of Nups from the Y complex could indicate that such a symmetry holds only approximately.
The coordinates of the Y complex members determined by NuRIM did not differ strongly and overall coincided with those of other “symmetric Nups” (Fig. 4 A and C). By linear fitting, we determined that the long axis of the Y complex is on average slanted by as little as 3°, a result that could be reproduced independently using Sec61-mCherry instead of dsRed-HDEL as the NE fiduciary marker (Fig. 4C and Dataset S1, sheet 7). Thus, NuRIM does not provide evidence of asymmetry in the organization of the yeast Y complex.
It was previously shown that some NPCs with altered compositions are still functional, namely they do exhibit an intriguing plastic character (16, 18, 20, 45, 46). We therefore wanted to test whether such “plastic” NPC variants would also exhibit structural plasticity, that is, whether structural anomalies could be detected. In one such instance, deletion of Nup170 in a Nup157-yEGFP strain led to functional NPCs, where the missing Nup170 was not substituted by its paralog, Nup157-yEGFP (16). NuRIM measured an average position of −0.3 ± 0.8 nm for the Nup157-yEGFP strain and 0.5 ± 0.5 nm in nup170Δ, suggesting that no major structural changes had occurred despite the compositional changes (Dataset S1, sheet 9).
The reverse scenario whereby NUP157 was deleted also produced a viable strain but, in this case, overexpression of an additional copy of Nup170-yEGFP from the URA3 locus was shown to generate functional pores displaying twice the stoichiometry of Nup170 compared with the wild-type pores (16). Again, positional changes were quite small: from h = 1.5 ± 0.7 to −0.9 ± 0.7 nm upon deletion of Nup157, and from 1.5 ± 1.5 to 2.9 ± 0.5 nm upon overexpression of Nup170-yEGFP. In this latter case, the lack of significant changes may be explained by the similarity of the average position for both Nups involved.
Likewise, for Nup192-yEGFP, deletion of its ancient paralog Nup188 brought the initial position of Nup192-yEGFP from −2.0 ± 1.1 to −1.2 ± 0.3 nm (47), and overexpression of Nup192-yEGFP from the URA3 locus which was previously shown to lead to partial substitution led to a position of −3.9 ± 0.2 nm (16).
Finally, we examined the case of Nic96-yEGFP. When tagged C-terminally, that Nup was only present on average in 24 copies per NPC, but when tagged N-terminally, 32 Nic96-yEGFP copies were counted (16). We applied NuRIM to both strains and measured a change from −3.4 ± 0.6 nm for the C-terminal strain to −2.9 ± 1.8 nm for the N-terminal strain.
None of the changes reported in these plasticity experiments were significant at a 5% level. Thus, the NPC structure appears to be robust against compositional changes that do not affect biological function (48).
We then asked whether environmental changes could affect the NPC organization. Thus, we monitored the positions of various yEGFP-tagged strains upon acute glucose starvation. The overall organization of the NPC did not change significantly, as shown by relatively constant positions for Nup120-yEGFP or Nup116-yEGFP (Fig. 4D), for example. However, Nups that were tagged N-terminally at the extremity of their intrinsically disordered FG repeats showed more variability: It appeared that the GFP tags of yEGFP-Nup116 and yEGFP-Nup159 both moved toward each other by approximately 3 nm (Fig. 4D). Although this change was not significant at a 5% level, the experiment illustrates how NuRIM could be employed to track changes in the distribution of Nups over time.
NuRIM Reveals Steady-State Localization for the Mobile Phase of the Nucleocytoplasmic Transport Machinery.
NuRIM is not limited to the analysis of nucleoporins. We thus examined a panel of NE-enriched proteins consisting mainly of karyopherins and mRNA transport factors integral to the function of the transport machinery (43) (Fig. 4A). An average position of −20.1 ± 6.7 nm was obtained for Dbp5, in agreement with its documented interactions with Nup159 and Gle1 and its role in remodeling mRNPs at the terminal stage of the export process (49). The exportin XPO1 (CRM1) was found in close proximity (−21.1 ± 3.2 nm) and had almost the same abundance (16.1 ± 5.0, versus 16.8 ± 3.0 copies for Dbp5). In contrast to these cytoplasmic-facing factors, the majority of transport factors tested were located on average medially in the NPC. For example, Kap95 was found at −1.0 ± 3.3 nm, while Mex67 was at −1.2 ± 0.5 nm on average (Fig. 4A and Dataset S1, sheet 10). Interestingly, when applied to purified nuclei that were gently released by osmotic shock in an excess of buffer (no detergents or fixatives were used), NuRIM analysis showed that a significant fraction of Mex67-yEGFP and Kap95-yEGFP remained at the NPC and that these factors retained their position relative to Nup84 (50) (Dataset S1, sheet 12).
To test NuRIM beyond the NPC architecture, we next attempted to determine the localization of transmembrane proteins found at the NE. Consistent with previous reports (26, 51), we found Heh1 (8.1 ± 3.3 nm) and Heh2 (8.7 ± 3.3 nm) biased toward the nuclear side (Dataset S1, sheet 10). Likewise, and despite weak expression levels, the paralogs Brl1 (14.4 ± 3.1 nm) and Brr6 (11.1 ± 1.3 nm) were clearly nuclear-facing (52) (SI Appendix, Fig. S4). We conclude that NuRIM may be applied to address other NE-related questions that hitherto demanded time-consuming IEM measurements.
NuRIM Allows the Study of Intrinsically Disordered FG Repeats.
The ability of NPCs to transport larger substrates only when these are bound to transport factors remains one of the most enigmatic features of NPCs (53). Most models that have been put forth to account for this selective behavior have focused on the role of FG repeats. For example, these were proposed to form a labile and dynamic polymer brush or phase providing a set of transient binding sites for transport factors (23, 54).
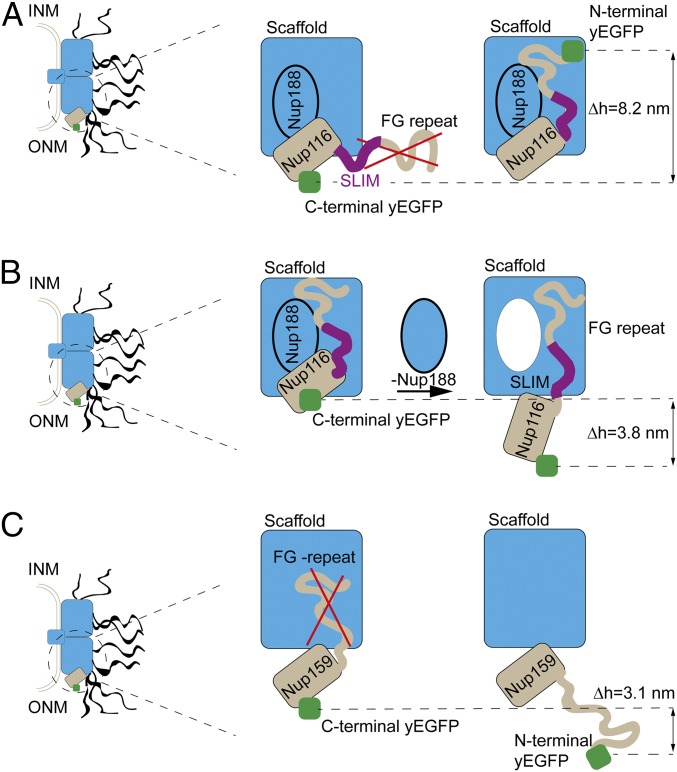
The NPC model of Kim et al. (5) represents the FG repeats as dynamic polymer brushes protruding from their anchor sites and projecting on average toward the NPC 8-fold axis. When we tagged Nup116 N-terminally with yEGFP at the extremity of its FG repeats, we measured a position 8.2 ± 3.1 nm deeper than the Nup116 variant tagged at the C terminus, and docked to the outer ring (Dataset S1, sheet 8). Thus, our data suggest that these FG repeats point on average toward the inner-scaffold Nups (Fig. 5A). This orientation is consistent with the presence of cohesive interactions between the FG repeats of Nup116 and inner-scaffold Nups (20, 55). It is also consistent with other types of interactions that have been detected between those scaffold Nups and a short linear interaction motif (SLIM) in Nup116 adjacent to its FG-repeat stretch (5, 7, 56). Indeed, in a mutant where the SLIM was deleted, the abundance of Nup116-yEGFP at the NE decreased by 55% (Dataset S1, sheet 8). To address whether the FG repeats of Nup116 contribute to interactions with scaffold Nups beyond those already provided by the SLIM, we exploited 2 otherwise identical versions of Nup116-yEGFP comprising the same native SLIM and the same truncated C-terminal domain. The first version kept its native FG repeat but, in the second one, all phenylalanines were replaced by alanines to abolish putative interactions mediated specifically by the FG repeats (20). Consistent with a distinct role for those FG repeats, the GFP on the second Phe-to-Ala Nup116-yEGFP version variant shifted on average by 6.5 ± 3.5 nm farther into the cytoplasm (Dataset S1, sheet 8).
Fig. 5.
Characterizing FG repeats using NuRIM. (A) Comparing results from C-terminal versus N-terminal tagging of Nup116 revealed that the flexible region of Nup116 (including both its FG-repeat region and a short linear interaction motif [SLIM]) are directed on average toward the core scaffold Nup188 (Δh = 8.2 ± 3.1 nm). INM and ONM indicate the inner and outer nuclear membranes, respectively. (B) Repressing the scaffold Nup188 led to a partial release of Nup116 toward the cytoplasm (Δh = 3.8 ± 0.5 nm) (see also figure 4G in ref. 20). (C) Comparing the positions of C-terminally versus N-terminally tagged Nup159 confirms the view that cytoplasmic-facing FG Nups more typically protrude into the cytoplasm (Δh = 3.1 ± 2.2 nm).
To further probe the interactions between the cognate domains of Nup116 and the scaffold Nup188, we depleted Nup188 via a methionine-repressible PMET3 promoter controlling its expression (20). Following repression of Nup188, Nup116-yEGFP moved by 3.8 ± 0.5 nm farther into the cytoplasm, compatible with the notion that Nup188 indeed contributes to anchoring Nup116 to the NPC scaffold (Fig. 5B and Dataset S1, sheet 8). This effect was specific: For example, as shown earlier, the deletion of Nup188 did not lead to significant positional shifts for Nup192-yEGFP.
Cytoplasm-facing FG NUPs are typically depicted as hairbrush structures protruding randomly into the cytoplasm, a picture at odds with Nup116 as was just shown. We thus labeled Nup159 N-terminally with yEGFP at the extremity of its FG repeats. Compared with the C-terminal counterpart, yEGFP was 3.1 ± 2.2 nm farther into the cytoplasm, this time consistent with the dynamic hairbrush picture (57) (Fig. 5C).
Altogether, these results are revealing of the multifaceted roles played by FG repeats and establish NuRIM as a suitable tool for their study.
Discussion
We have developed an in vivo wide-field microscopy technique, NuRIM, which provides insight into the organization of native yeast NPCs at a precision and accuracy of ∼2 nm. We have also demonstrated that NuRIM is a generally applicable method that simplifies localization analysis of NE-associated proteins and complexes. As a 1D methodology that necessitates fluorescent tagging, NuRIM will not replace EM or X-ray diffraction but provides a complementary viewpoint: In contrast to those techniques, it operates on live cells and conclusive analyses may be conducted within a few hours, compared with weeks or months (58). One should bear in mind, however, that NuRIM is not a single-molecule technique: It measures the full complement of proteins intimately associated with the NE rather than only those exclusively present in the NPC. This might possibly explain some of our observations—for example, the fact that the paralogs Mlp1 and Mlp2 did not colocalize exactly (Fig. 4A). Indeed, it has been shown previously that these Nups can also be present at the NE independent of their structural role within NPCs (16, 59). This might also explain our finding that Nup100 is on average found symmetrically whereas its paralog Nup116 is not. However, an alternative explanation is that Nup116 is asymmetrically localized as part of the cytoplasmic RNA export platform, whereas Nup100 is not a member of this complex (60).
Similar to a recently introduced technique named “surface-induced FRET,” NuRIM can deliver estimates of distances between proteins positioned distally at the NE, yet it does not depend on engineered surfaces nor does it require complex calibration (24). In addition, several other techniques have been developed recently to study NE protein localization, including single-point FRAP, bimolecular fluorescence complementation, FRET, and enzymatic-based methods (14, 27, 61, 62). Due to NuRIM’s accuracy and simplicity, we anticipate that it will be widely used for such localization studies. Another exciting prospect for NuRIM will be to characterize NPCs in various conditions of cell stress, cell cycle, age, and assembly stages, thus promising to capture the wide gamut of NPC plasticity.
In this contribution, we have mainly examined the native NPC architecture in the presence of all accessory proteins. Our results are largely consistent with data obtained previously using IEM, although they portray a somewhat flatter NPC (2, 9, 39). NuRIM also corroborates recent integrative models of entire NPCs (5). In particular, the Y complex appeared symmetric on average (44). By contrast, an interesting example of asymmetry was revealed by the analysis of the FG repeats of Nup116, which—contrary to those of Nup159—were found to project on average into the NPC channel, possibly serving a cohesive function in addition to their role in transport (20). NuRIM appears unique as a technique that can provide positional information on intrinsically disordered regions present in as much as ∼30% of the human genome (63).
Applying NuRIM to functional NPCs off-balance in their composition revealed that their morphology was quite robust against such manipulations. In keeping with the structure–function paradigm, it appears that compositional changes in NPCs are tolerated if they preserve the structural integrity of the complex. In conclusion, NuRIM represents a timely addition to the toolkit of biologists interested in the structure and assembly of membrane-associated complexes.
Materials and Methods
Yeast cells expressing yEGFP-tagged Nups were grown in synthetic complete media to log phase (optical density 0.7) and imaged in concavalin A (Sigma-Aldrich)-coated MatriPlates (Matrical Technologies) using an epifluorescence microscope (Nikon Ti-E) equipped with an sCMOS camera (Hamamatsu Flash 4.0). A Nikon 100× Plan-Apo objective N.A. 1.4 was used. Pixel size was 65 nm in object space. NIS Element software controlled image acquisition and the GFP/RFP filter settings were used. All yeast strains used for quantitative analysis of Nup-yEGFP intensity also expressed dsRed-HDEL (or Sec61-mCherry when stated). Exposure time was 500 ms and LED power was set to 50% except when indicated otherwise. Further information for researchers who would like to apply NuRIM analysis to their own data is presented in SI Appendix.
The Saccharomyces cerevisiae strains and plasmids used in this study were produced using standard yeast and molecular cloning protocols and are listed in SI Appendix, Tables S1 and S2.
Supplementary Material
Acknowledgments
We are grateful to members of the K.W. lab for helpful discussions, suggestions, and sharing various experimental materials. E. Dultz is acknowledged for providing feedback on the manuscript. This work is supported by a grant of the Swiss National Science Foundation to K.W. (SNF 179275).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1903764116/-/DCSupplemental.
References
- 1.Beck M., Hurt E., The nuclear pore complex: Understanding its function through structural insight. Nat. Rev. Mol. Cell Biol. 18, 73–89 (2017). [DOI] [PubMed] [Google Scholar]
- 2.Rout M. P., et al. , The yeast nuclear pore complex: Composition, architecture, and transport mechanism. J. Cell Biol. 148, 635–651 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Simon D. N., Rout M. P., Cancer and the nuclear pore complex. Adv. Exp. Med. Biol. 773, 285–307 (2014). [DOI] [PubMed] [Google Scholar]
- 4.Sakuma S., D’Angelo M. A., The roles of the nuclear pore complex in cellular dysfunction, aging and disease. Semin. Cell Dev. Biol. 68, 72–84 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kim S. J., et al. , Integrative structure and functional anatomy of a nuclear pore complex. Nature 555, 475–482 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kosinski J., et al. , Molecular architecture of the inner ring scaffold of the human nuclear pore complex. Science 352, 363–365 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lin D. H., et al. , Architecture of the symmetric core of the nuclear pore. Science 352, aaf1015 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Maimon T., Elad N., Dahan I., Medalia O., The human nuclear pore complex as revealed by cryo-electron tomography. Structure 20, 998–1006 (2012). [DOI] [PubMed] [Google Scholar]
- 9.Alber F., et al. , The molecular architecture of the nuclear pore complex. Nature 450, 695–701 (2007). [DOI] [PubMed] [Google Scholar]
- 10.Bui K. H., et al. , Integrated structural analysis of the human nuclear pore complex scaffold. Cell 155, 1233–1243 (2013). [DOI] [PubMed] [Google Scholar]
- 11.Eibauer M., et al. , Structure and gating of the nuclear pore complex. Nat. Commun. 6, 7532 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gaik M., et al. , Structural basis for assembly and function of the Nup82 complex in the nuclear pore scaffold. J. Cell Biol. 208, 283–297 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lasker K., Sali A., Wolfson H. J., Determining macromolecular assembly structures by molecular docking and fitting into an electron density map. Proteins 78, 3205–3211 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kim D. I., et al. , Probing nuclear pore complex architecture with proximity-dependent biotinylation. Proc. Natl. Acad. Sci. U.S.A. 111, E2453–E2461 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mi L., Goryaynov A., Lindquist A., Rexach M., Yang W., Quantifying nucleoporin stoichiometry inside single nuclear pore complexes in vivo. Sci. Rep. 5, 9372 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rajoo S., Vallotton P., Onischenko E., Weis K., Stoichiometry and compositional plasticity of the yeast nuclear pore complex revealed by quantitative fluorescence microscopy. Proc. Natl. Acad. Sci. U.S.A. 115, E3969–E3977 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Aitchison J. D., Rout M. P., The yeast nuclear pore complex and transport through it. Genetics 190, 855–883 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Akey C. W., Structural plasticity of the nuclear pore complex. J. Mol. Biol. 248, 273–293 (1995). [DOI] [PubMed] [Google Scholar]
- 19.Smith C., et al. , In vivo single-particle imaging of nuclear mRNA export in budding yeast demonstrates an essential role for Mex67p. J. Cell Biol. 211, 1121–1130 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Onischenko E., et al. , Natively unfolded FG repeats stabilize the structure of the nuclear pore complex. Cell 171, 904–917.e19 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kapinos L. E., Huang B., Rencurel C., Lim R. Y. H., Karyopherins regulate nuclear pore complex barrier and transport function. J. Cell Biol. 216, 3609–3624 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lowe A. R., et al. , Importin-β modulates the permeability of the nuclear pore complex in a Ran-dependent manner. eLife 4, e04052 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Timney B. L., et al. , Simple rules for passive diffusion through the nuclear pore complex. J. Cell Biol. 215, 57–76 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chizhik A. M., et al. , Three-dimensional reconstruction of nuclear envelope architecture using dual-color metal-induced energy transfer imaging. ACS Nano 11, 11839–11846 (2017). [DOI] [PubMed] [Google Scholar]
- 25.Fiserova J., Spink M., Richards S. A., Saunter C., Goldberg M. W., Entry into the nuclear pore complex is controlled by a cytoplasmic exclusion zone containing dynamic GLFG-repeat nucleoporin domains. J. Cell Sci. 127, 124–136 (2014). [DOI] [PubMed] [Google Scholar]
- 26.King M. C., Lusk C. P., Blobel G., Karyopherin-mediated import of integral inner nuclear membrane proteins. Nature 442, 1003–1007 (2006). [DOI] [PubMed] [Google Scholar]
- 27.Smoyer C. J., et al. , Analysis of membrane proteins localizing to the inner nuclear envelope in living cells. J. Cell Biol. 215, 575–590 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Heintzmann R., Ficz G., Breaking the resolution limit in light microscopy. Methods Cell Biol. 114, 525–544 (2013). [DOI] [PubMed] [Google Scholar]
- 29.Hoffman M. T., Sheung J., Selvin P. R., Fluorescence imaging with one nanometer accuracy: In vitro and in vivo studies of molecular motors. Methods Mol. Biol. 778, 33–56 (2011). [DOI] [PubMed] [Google Scholar]
- 30.Park W., Madden D. R., Rockmore D. N., Chirikjian G. S., Statistical analysis and deblurring of class averages in single-particle electron microscopy. Biophys. J. 98, 384a (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Szymborska A., et al. , Nuclear pore scaffold structure analyzed by super-resolution microscopy and particle averaging. Science 341, 655–658 (2013). [DOI] [PubMed] [Google Scholar]
- 32.Madrid A. S., Mancuso J., Cande W. Z., Weis K., The role of the integral membrane nucleoporins Ndc1p and Pom152p in nuclear pore complex assembly and function. J. Cell Biol. 173, 361–371 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hryc C. F., et al. , Accurate model annotation of a near-atomic resolution cryo-EM map. Proc. Natl. Acad. Sci. U.S.A. 114, 3103–3108 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kuzmanic A., Zagrovic B., Determination of ensemble-average pairwise root mean-square deviation from experimental B-factors. Biophys. J. 98, 861–871 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wang R., et al. , High resolution microscopy reveals the nuclear shape of budding yeast during cell cycle and in various biological states. J. Cell Sci. 129, 4480–4495 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kalderon D., Roberts B. L., Richardson W. D., Smith A. E., A short amino acid sequence able to specify nuclear location. Cell 39, 499–509 (1984). [DOI] [PubMed] [Google Scholar]
- 37.Vámosi G., et al. , EGFP oligomers as natural fluorescence and hydrodynamic standards. Sci. Rep. 6, 33022 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Shida H., A study of protein A-gold resolution for immunoelectron microscopy. J. Electron. Microsc. Tech. 18, 291–295 (1991). [DOI] [PubMed] [Google Scholar]
- 39.Kosova B., Panté N., Rollenhagen C., Hurt E., Nup192p is a conserved nucleoporin with a preferential location at the inner site of the nuclear membrane. J. Biol. Chem. 274, 22646–22651 (1999). [DOI] [PubMed] [Google Scholar]
- 40.Pédelacq J. D., Cabantous S., Tran T., Terwilliger T. C., Waldo G. S., Engineering and characterization of a superfolder green fluorescent protein. Nat. Biotechnol. 24, 79–88 (2006). Erratum in: Nat. Biotechnol.24, 1170 (2006). [DOI] [PubMed] [Google Scholar]
- 41.Fernandez-Martinez J., et al. , Structure and function of the nuclear pore complex cytoplasmic mRNA export platform. Cell 167, 1215–1228.e25 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yoshida K., Seo H. S., Debler E. W., Blobel G., Hoelz A., Structural and functional analysis of an essential nucleoporin heterotrimer on the cytoplasmic face of the nuclear pore complex. Proc. Natl. Acad. Sci. U.S.A. 108, 16571–16576 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Adams R. L., Mason A. C., Glass L., Aditi, Wente S. R., Nup42 and IP6 coordinate Gle1 stimulation of Dbp5/DDX19B for mRNA export in yeast and human cells. Traffic 18, 776–790 (2017). Erratum in: Traffic19, 650 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Mosalaganti S., et al. , In situ architecture of the algal nuclear pore complex. Nat. Commun. 9, 2361 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Jovanovic-Talisman T., et al. , Artificial nanopores that mimic the transport selectivity of the nuclear pore complex. Nature 457, 1023–1027 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Niño C. A., et al. , Posttranslational marks control architectural and functional plasticity of the nuclear pore complex basket. J. Cell Biol. 212, 167–180 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Andersen K. R., et al. , Scaffold nucleoporins Nup188 and Nup192 share structural and functional properties with nuclear transport receptors. eLife 2, e00745 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kenna M. A., Petranka J. G., Reilly J. L., Davis L. I., Yeast N1e3p/Nup170p is required for normal stoichiometry of FG nucleoporins within the nuclear pore complex. Mol. Cell. Biol. 16, 2025–2036 (1996). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Fiserova J., Richards S. A., Wente S. R., Goldberg M. W., Facilitated transport and diffusion take distinct spatial routes through the nuclear pore complex. J. Cell Sci. 123, 2773–2780 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kiseleva E., et al. , Yeast nuclear pore complexes have a cytoplasmic ring and internal filaments. J. Struct. Biol. 145, 272–288 (2004). [DOI] [PubMed] [Google Scholar]
- 51.Lokareddy R. K., et al. , Distinctive properties of the nuclear localization signals of inner nuclear membrane proteins Heh1 and Heh2. Structure 23, 1305–1316 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Zhang W., et al. , Brr6 and Brl1 locate to nuclear pore complex assembly sites to promote their biogenesis. J. Cell Biol. 217, 877–894 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Weis K., The nuclear pore complex: Oily spaghetti or gummy bear? Cell 130, 405–407 (2007). [DOI] [PubMed] [Google Scholar]
- 54.Rout M. P., Aitchison J. D., Magnasco M. O., Chait B. T., Virtual gating and nuclear transport: The hole picture. Trends Cell Biol. 13, 622–628 (2003). [DOI] [PubMed] [Google Scholar]
- 55.Patel S. S., Belmont B. J., Sante J. M., Rexach M. F., Natively unfolded nucleoporins gate protein diffusion across the nuclear pore complex. Cell 129, 83–96 (2007). [DOI] [PubMed] [Google Scholar]
- 56.Fischer J., Teimer R., Amlacher S., Kunze R., Hurt E., Linker Nups connect the nuclear pore complex inner ring with the outer ring and transport channel. Nat. Struct. Mol. Biol. 22, 774–781 (2015). [DOI] [PubMed] [Google Scholar]
- 57.Atkinson C. E., Mattheyses A. L., Kampmann M., Simon S. M., Conserved spatial organization of FG domains in the nuclear pore complex. Biophys. J. 104, 37–50 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Perutz M. F., et al. , Structure of haemoglobin: A three-dimensional Fourier synthesis at 5.5-Å resolution, obtained by X-ray analysis. Nature 185, 416–422 (1960). [DOI] [PubMed] [Google Scholar]
- 59.Niepel M., et al. , The nuclear basket proteins Mlp1p and Mlp2p are part of a dynamic interactome including Esc1p and the proteasome. Mol. Biol. Cell 24, 3920–3938 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Bailer S. M., et al. , Nup116p associates with the Nup82p-Nsp1p-Nup159p nucleoporin complex. J. Biol. Chem. 275, 23540–23548 (2000). [DOI] [PubMed] [Google Scholar]
- 61.Damelin M., Silver P. A., In situ analysis of spatial relationships between proteins of the nuclear pore complex. Biophys. J. 83, 3626–3636 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Mudumbi K. C., Schirmer E. C., Yang W., Single-point single-molecule FRAP distinguishes inner and outer nuclear membrane protein distribution. Nat. Commun. 7, 12562 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Dunker A. K., et al. , The unfoldomics decade: An update on intrinsically disordered proteins. BMC Genomics 9 (suppl. 2), S1 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Pettersen E. F., et al. , UCSF Chimera—A visualization system for exploratory research and analysis. J. Comput. Chem. 25, 1605–1612 (2004). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.