Abstract
Background
High circulating low-density lipoprotein cholesterol (LDL-C) is a major risk factor for atherosclerosis and age-associated cardiovascular events. Long-term dyslipidaemia could contribute to the development of frailty in older individuals through its role in determining cardiovascular health and potentially other physiological pathways.
Methods
We conducted Mendelian randomization (MR) analyses using genetic variants to estimate the effects of long-term LDL-C modification on frailty in UK Biobank (n = 378,161). Frailty was derived from health questionnaire and interview responses at baseline when participants were aged 40 to 69 years, and calculated using an accumulation-of-deficits approach, i.e. the frailty index (FI). Several aggregated instrumental variables (IVs) using 50 and 274 genetic variants were constructed from independent single-nucleotide polymorphisms (SNPs) to instrument circulating LDL-C concentrations. Specific sets of variants in or near genes that encode six lipid-lowering drug targets (HMGCR, PCSK9, NPC1L1, APOB, APOC3, and LDLR) were used to index effects of exposure to related drug classes on frailty. SNP-LDL-C effects were available from previously published studies. SNP-FI effects were obtained using adjusted linear regression models. Two-sample MR analyses were performed with the IVs as instruments using inverse-variance weighted, MR-Egger, weighted median, and weighted mode methods. To address the stability of the findings, MR analyses were also performed using i) a modified FI excluding the cardiometabolic deficit items and ii) data from comparatively older individuals (aged ≥60 years) only. Several sensitivity analyses were also conducted.
Findings
On average 0.14% to 0.23% and 0.16% to 0.31% decrements in frailty were observed per standard deviation reduction in LDL-C exposure, instrumented by the general IVs consisting of 50 and 274 variants, respectively. Consistent, though less precise, associations were observed in the HMGCR-, APOC3-, NPC1L1-, and LDLR-specific IV analyses. In contrast, results for PCSK9 were in the same direction but more modest, and null for APOB. All sensitivity analyses produced similar findings.
Interpretation
A genetically-predicted life-long lowering of LDL-C is associated with decreased frailty in midlife and older age, representing supportive evidence for LDL-C's role in multiple health- and age-related pathways. The use of lipid-lowering therapeutics with varying mechanisms of action may differ by the extent to which they provide overall health benefits.
Keywords: Low-density lipoprotein cholesterol, Frailty, Mendelian randomization, UK biobank
Research in context.
Evidence before this study
High levels of low-density lipoprotein cholesterol (LDL-C) is a major risk factor for atherosclerosis and age-associated cardiovascular events. Long-term dyslipidaemia could contribute to the development of frailty in older individuals, either solely or beyond its role in determining cardiovascular health. We searched PubMed without language or publication date restrictions for (“low-density lipoprotein cholesterol” OR “LDL-C" OR “LDL”) AND (“frailty” or “frail”) through Mar 22, 2019. About 12 articles were retrieved. However, only one observational study evaluated the association between LDL-C and frailty directly, observing no association between them. Besides, no study using the Mendelian Randomization (MR) design, as in the current study, was reported.
Added value of this study
An MR design was used to analyze the non-confounded effect of genetically predicted low lipid levels on frailty. The European individuals enriched with lipid-lowering alleles from SNPs associated with LDL-C concentrations presented a lower risk of being frail as assessed by the frailty index (FI). The LDL-C and FI association was verified to be independent of cardiometabolic traits. Meanwhile, the effect on FI reduction in response to life-long lowering of LDL-C concentrations turned slightly larger when excluding the comparatively young participants aged <60 years, suggesting that genetic predisposition to low LDL-C concentrations decreases the risk of being frail later in life. We also profiled gene-specific effects from loci that index the modulation of existing and emerging lipid-lowering drug targets (e.g., HMGCR, APOC3, and LDLR), and found evidence that the on-target effects of classes used to lower LDL-C may contribute notable differences to the overall health of users.
Implications of all the available evidence
All available evidence highlights the importance of LDL-C monitoring during the ageing process, especially since the association with the FI was independent of any detected atherosclerotic pathogenesis. Genetically-predisposed low LDL-C concentration is associated with overall better health among the European ancestry population although more studies are still needed to evaluate the relationship between the life-long lowering of LDL-C concentrations and other geriatric diseases and/or traits. The implication that different LDL-C lowering therapeutics could affect frailty at differing degrees may also indicate need for pharmacovigilance regarding recently introduced drug classes, such as PCSK9 inhibitors and ApoB antisense therapeutics. All these results may provide some evidence for the efficacy of LDL-C lowering therapies in the treatment of age-related diseases other than CVDs.
Alt-text: Unlabelled Box
1. Introduction
High circulating low-density lipoprotein cholesterol (LDL-C) is a key driver of atherosclerosis—an ageing and cellular senescence-related process responsible for the high morbidity and mortality of cardiovascular diseases (CVDs) and other age-related diseases affecting older populations worldwide [[1], [2], [3]]. LDL-C has a pro-atherogenic role via several modified species, including oxidized LDL, acetylated LDL, ethylated, methylated, and glycated LDL, all of which promote vascular injury by increasing oxidative stress and accelerating senescence of endothelial progenitor cells through modifications and damage to DNA [4]. The critical contribution of LDL-C to atherosclerotic pathology is also underscored by accumulating epidemiologic evidence that the widely used statin therapy (with or without other non-statin lipid-modifying agents) with well-established efficacy in lowering LDL-C has achieved great success in reducing cardiovascular events [5]. Moreover, cumulative evidence suggests that there is not yet an ideal attainment of LDL-C lowering target for the cardiovascular risk reduction [6].
Frailty has gained scientific attention in gerontology and geriatrics in the past few decades due to its consistent association with all-cause mortality and various negative health outcomes that present in older individuals [7]. One commonly used frailty measure is the Fried frailty phenotype, classifying individuals as non-frail, pre-frail and frail with respect to the presence of five physical components (unintentional weight loss, self-reported exhaustion, weakness, slow walking speed and low physical activity) [8]. Another generally accepted operationalization of frailty is the frailty index (FI), which is a continuous measure and calculated as a ratio of the number of age-related health deficits to the number of total deficits considered [9]. As a proxy of overall health, the deficits constituting an FI can be symptoms, signs, diseases, and functional impairments. Hence, FI measurements consider different aspects of health simultaneously and are strongly predictive of many adverse health outcomes, including functional decline, disability, falls, fractures, morbidity and mortality [10].
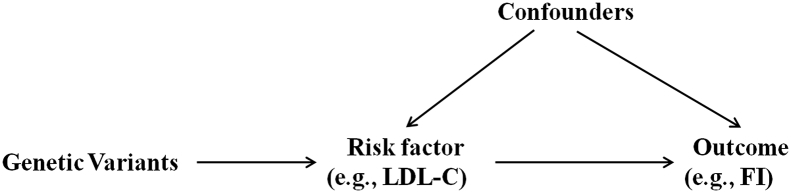
Mendelian Randomization (MR) is an approach which uses genetic variants to assess whether a risk factor has a causal effect on an outcome in a non-experimental (observational) setting [11,12]. MR relies on the natural, random assortment of genetic variants during meiosis yielding a random distribution of genetic variants in the population. In the MR design, genetic variants associated with a biomarker (e.g., LDL-C) as instrumental variables (IVs) are then proxies for the biomarker itself and can be used to determine whether the biomarker is causally associated with an outcome (e.g., FI) [13]. The MR framework is illustrated in Fig. 1. Three main assumptions are made for MR inference: [1] the genetic variant (e.g., SNP, single nucleotide polymorphism) has a robust causal association with the risk factor of interest; [2] the genetic variant (e.g., SNP) affects the outcome only through its effect on the risk factor; and [3] the genetic variant and the outcome do not have common causes. Because the genetic variants are typically unassociated with confounders due to the independent and random allocation of alleles at conception, differences in the outcome between those who carry the variant and those who do not can be attributed to the difference in the risk factor [14].
Fig. 1.
The framework of a Mendelian randomization analysis.
Considering the evidence that atherosclerosis is linked to the ageing process [15] and given the causal association between LDL-C and atherosclerosis, we hypothesize that long-term LDL-C lowering leads to improved overall health. However, LDL-C lowering can be achieved through several approved therapeutics classes, which have different mechanisms of action and may affect overall health to different degrees (encapsulating lipid-lowering effects along with any other on- and off-target physiological effects). In this study, we investigated the association between the genetically-predicted life-long lowering of LDL-C concentrations and frailty in the UK Biobank (UKB) study, a large-scale cohort with over 500,000 participants. Additional attention was paid to potential effects on the frailty index (FI) from existing LDL-C lowering therapeutics instrumented by genetic variants which index the modulation of several well-known drug targets.
2. Methods
2.1. Study population and design
The UKB is a multi-centre cohort study with over 500,000 British participants aged 40 to 69 years, enrolled at 22 assessment sites in England, Scotland and Wales between 2006 and 2010 [16]. All participants undertook a baseline assessment, relevant items of which were detailed in another report [16]. Genome-wide genotype data were available for all participants. SNP data were retained if they had a genotype call rate ≥ 0.8 and non-ambiguous alleles (not palindromic with allele frequencies of 0.4 to 0.6). We used data from European ancestry participants for analysis. We controlled for relatedness between individuals in the sample by randomly dropping one relative within pairs with kinship thresholds of 0.1768 or 0.0442, yielding corresponding sample sizes of 378,161 or 335,034 in our study, respectively.
A two-sample MR design was used to explore the associations between genetically-predicted lipid-lowering effects and the FI, details of which are presented below. Multiple SNPs were combined to evaluate the genetic LDL-C concentrations. Using multiple genetic variants increases the power of an MR investigation compared with an analysis based on a single variant [17]. However, when using multiple genetic variants from different gene regions in an MR analysis, it is highly implausible that all the genetic variants satisfy the instrumental variable assumptions [18]. To address the inflation effect of invalid instrumental variables, MR-Egger regression, weighted median, and weighted modal estimators can be used [[19], [20], [21], [22]]. The SNP-LDL-C effects were derived from the genome-wide associations based on 188,578 individuals of European ancestry and published by Global Lipids Genetics Consortium (GLGC) [23] and the variant-FI effects were estimated in UKB. We predicted the effects of LDL-C modification generally, using either independent SNPs not in linkage disequilibrium (LD) (pairwise r2 values <0.2), or that reported strong effect on LDL-C relative to other lipid effects [24], and via exposure to specific lipid-lowering drugs using variants in or near six genes (HMGCR, PCSK9, NPC1L1, APOB, APOC3, and LDLR) that encode representative drug targets.
2.2. Frailty index
A standard FI was developed as described previously [16], involving 49 items covering 11 types of deficits (sensory, cranial, mental wellbeing, infirmity, cardiometabolic, respiratory, musculoskeletal, immunological, cancer, pain, and gastrointestinal) indicating ill health across a variety of physiological and mental domains, symptoms, diagnosed diseases, and disabilities, assessed via questionnaire and interview at the cohort's baseline assessment. An FI value is composed as the sum of deficits accrued by an individual divided by the total number of deficits composing the FI, e.g. an individual with 10 deficits from a total of 50 items, would have an FI value of 0.2 (10/50). In the present study, we adopted a transformed FI equaling to the original FI value multiplied by 100. Hence, the size of LDL-C effect on FI was evaluated on a 100-percentage scale and can be regarded as a relative FI change in response to LDL-C exposure. Relevant details about the FI composition are provided in the appendix. Participants were excluded if they had missing data for 10 or more items (over 20%) or had death certificate dates that preceded dates of attendance at the UKB baseline assessment. The FI proportions used in this study were calculated to be expressed on a 100-point scale. The associations with age, sex and risk of all-cause mortality suggest FI is a valid measure of frailty in UKB [16].
2.3. Instrumental variables for LDL-C
A total of eight aggregated IV sets were adopted to indicate genetically-predicted LDL-C exposure, and the modulation of specific drug targets. For each SNP included in the aggregated IV sets, we defined the effect alleles as the allele associated with lower LDL-C concentrations. A ‘large’ IV set was constructed by combining all independent SNPs (n = 274) reported to be associated with LDL-C concentrations reaching the genome-wide significance (GWAS) level (P < 5.0 × 10−8) [23] and that were not in LD (pairwise r2 values <0.2) with all other variants included in it. A ‘small’ IV, which is more conservative, was constructed using 57 SNPs associated to LDL-C and without strong associations with other lipids (the list is available on http://www.mrbase.org/) [24]. Of those, seven SNPs were excluded due to low quality genotyping in the UKB sample leaving 50 SNPs. Additionally, six gene-specific IVs were constructed for genes that encode the targets of approved or emerging LDL-lowering drugs including HMGCR, PCSK9, NPC1L1, APOB, APOC3, and LDLR. For these IVs, we combined all independent SNPs (pairwise r2 values<0.2) within 100 kb of gene boundaries [25]. A total of 16, 34, 24, 30, 19, and 30 SNPs, respectively, were included for constructing the six IVs.
2.4. Statistical analyses
Selection of the independent SNPs not in LD was performed with the software Plink v.1.9 [26]. The list of SNPs involved in the IVs and relevant information are presented in the appendix (Tables S1 and S2). Using Plink, the composed FI was linearly regressed on the variants included in each of the IVs to obtain their effect estimators [26]. We included age, sex, and the first 15 principal components (PCs) as covariates. Stata version 15.1 (StataCorp, College Station, Texas, USA) was used for statistical analyses. The ivreg2 and ivpois commands and the mrrobust package [27] were used to carry out the MR analyses. To address the possibility that the LDL-C may be associated with FI due to underlying associations between LDL-C IVs and cardiometabolic traits (e.g., cardiovascular diseases, type-2 diabetes, metabolic syndrome, etc.), a modified FI (MFI) excluding the seven items related to cardiometabolic health was also used for MR analyses. Moreover, considering that frailty at younger ages is more attributable to non-ageing-related severe diseases or traits, we also investigated the effect of genetically predicted LDL-C on frailty exclusively for participants aged ≥60 years (FI60). The inverse-variance weighted (IVW), MR-Egger regression, weighted median and weighted mode estimators were mainly reported in the MR analyses [19,21]. All statistical analyses used a two-sided p < .05 threshold to indicate statistical significance.
2.5. Sensitivity analyses
To evaluate the stability of our findings we performed several sensitivity analyses. First, given that variants located in the APOE gene (and other loci on chromosome 19 that might be in LD with APOE variants) have particularly pleiotropic effects on multiple lipoproteins, we modified the large and small IVs excluding variants on chromosome 19 [28]. Then we performed MR analyses for the FI effect of genetic LDL-C concentrations instrumented by the modified ones. Second, we performed additional sensitivity analyses on individual-level data, in which we calculated individual genetic risk scores (GRS) for each participant by summing up the number of LDL-C effect alleles at each variant included in each score, weighted by the effect of each variant on LDL-C concentrations measured in per standard deviation (SD) change units. Linear regression models regressing FI on individual GRSs were performed adjusting for covariates age, sex, and 15 PCs in the individual-level analyses. Third, we also adopted more stringent pairwise r2 thresholds (0.1 and 0.01) in the selection of independent SNPs for constructing the aggregated IVs (the large and six gene-specific ones) to remove nominally correlated estimates among the genetic variants in the aggregated LDL-C IV sets.
3. Results
3.1. LDL-C effects on the FI instrumented by the aggregated IVs
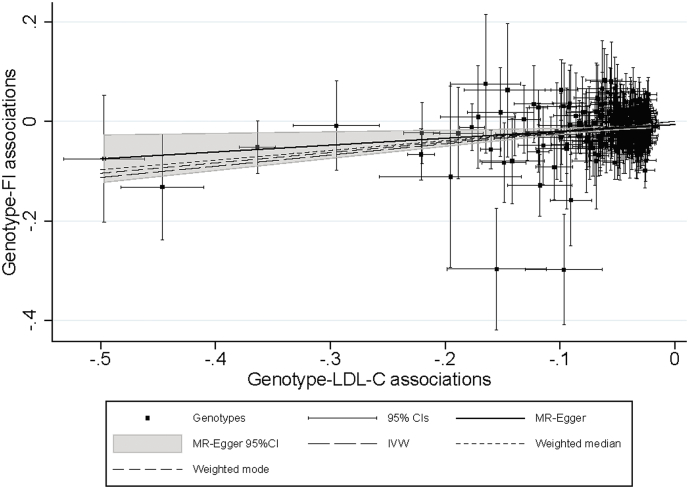
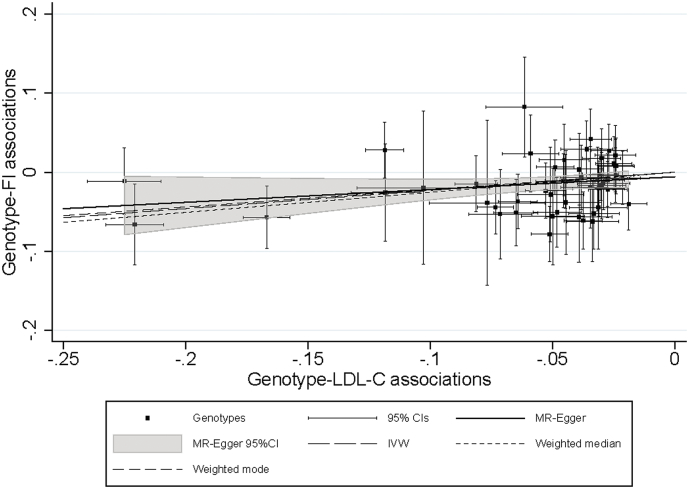
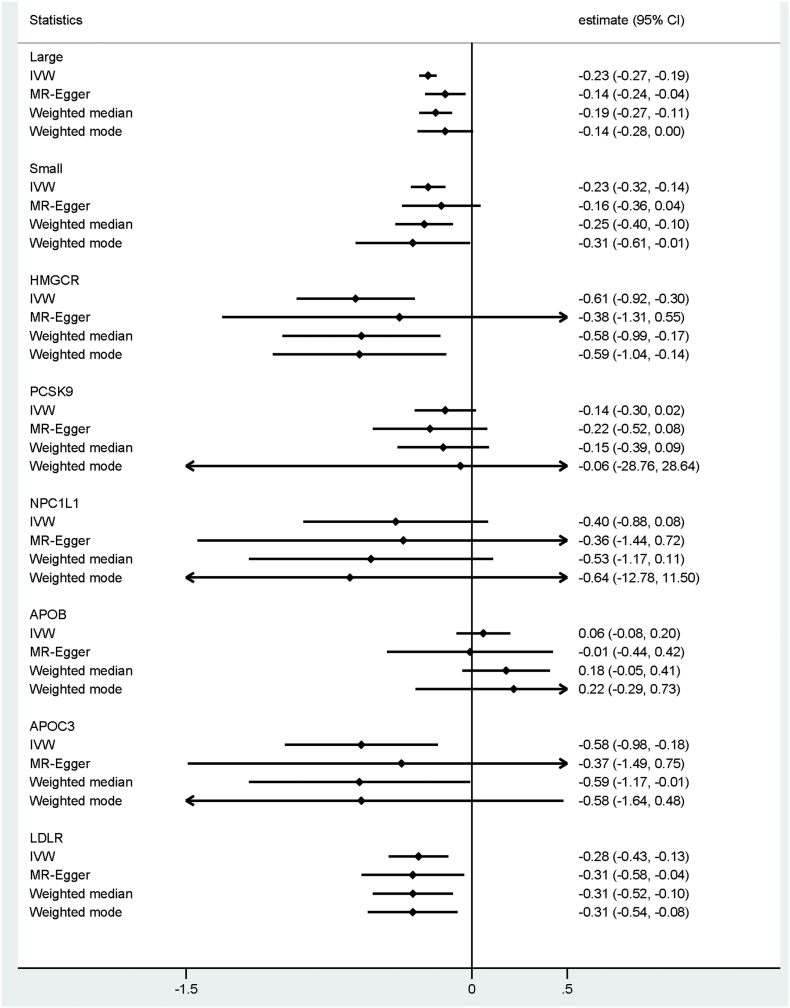
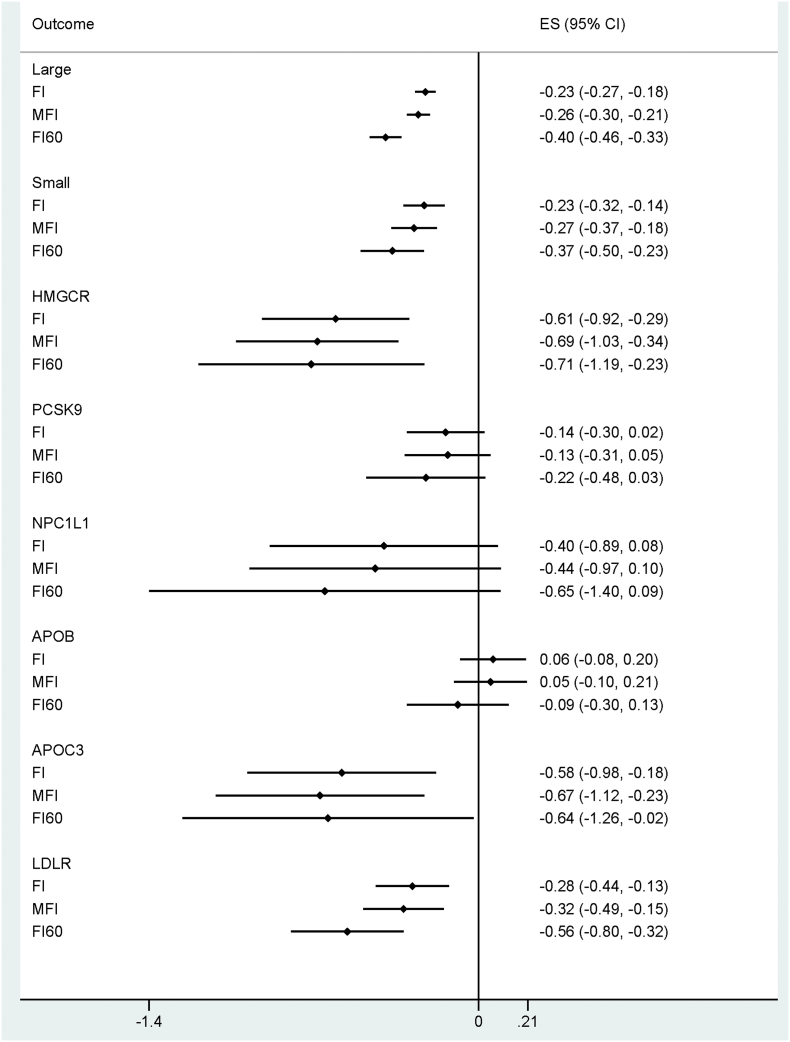
The number of unrelated individuals of European ancestry who contributed to the main analyses was 378,161. In general, protective alleles in LDL-C linked variants were associated with lower FI values (Table 1). Using the large IV set (including 274 SNPs), the FI decreased by 0.23% [95% CI: −0.18 to −0.27], 0.17% [−0.24 to −0.03], 0.19% [−0.28 to −0.11], and 0.14% [−0.29 to −0.00] in response to one SD decrease in LDL-C in the IVW, MR-Egger, weighted median and weighted mode MR analyses, respectively. Similar estimates were also observed when using the small IV set (including 50 SNPs) for predicting the genetic effect of LDL-C on the FI (−0.23% [95%CI: −0.32 to −0.14], −0.16% [−0.37 to 0.04], −0.25% [−0.40 to −0.10], and −0.31% [−0.61 to −0.01] sequentially per SD decrease in LDL-C). For drug-target specific loci, HMGCR, APOC3 and LDLR showed protective effects for the FI (Table 1). Interestingly, APOB variants demonstrated a consistent null association with the FI throughout the various analyses. Effects from alleles within the PCSK9 and NPC1L1 loci were inconclusive. Pleiotropy was observed for several variants constructing the large and small IVs (Fig. 2, Fig. 3). However, the results from the MR-Egger, weighted median, and weighted mode estimates—which can address the pleiotropy in MR analyses—showed great consistency with the IVW estimates (Fig. 4).
Table 1.
IVW estimates for the FI effect of life-long lowering of LDL-C concentrations indicated by the constructed IVs; independent individuals were involved by randomly dropping the related ones reaching a kinship threshold of 0.1768.
| IV set (number of SNPs) | Main analyses (n = 378,161) |
Modified FI excluding cardiometabolic deficit items (n = 378,161) |
FI in participants ≥60 years (n = 167,013) |
|||
|---|---|---|---|---|---|---|
| Effect (95% CI) | p | Effect (%95 CI) | p | Effect (%95 CI) | p | |
| Large (274) | −0.23 (−0.27, −0.18) | <0.0005 | −0.26 (−0.30, −0.21) | <0.0005 | −0.40 (−0.46, −0.33) | <0.0005 |
| Small (50) | −0.23 (−0.32, −0.14) | <0.0005 | −0.27 (−0.37, −0.18) | <0.0005 | −0.37 (−0.50, −0.23) | <0.0005 |
| HMGCR (16) | −0.61 (−0.92, −0.30) | <0.0005 | −0.68 (−1.03, −0.34) | <0.0005 | −0.71 (−1.19, −0.23) | 0.004 |
| PCSK9 (34) | −0.14 (−0.30, 0.02) | 0.097 | −0.13 (−0.31, 0.05) | 0.159 | −0.22 (−0.48, 0.03) | 0.083 |
| NPC1L1 (24) | −0.40 (−0.89, 0.08) | 0.104 | −0.44 (−0.97, 0.10) | 0.107 | −0.65 (−1.40, 0.09) | 0.086 |
| APOB (30) | 0.06 (−0.08, 0.20) | 0.388 | 0.05 (−0.10, 0.20) | 0.521 | −0.09 (−0.30, 0.13) | 0.426 |
| APOC3 (19) | −0.58 (−0.98, −0.18) | 0.004 | −0.67 (−1.12, −0.23) | 0.003 | −0.64 (−1.26, −0.02) | 0.043 |
| LDLR (30) | −0.28 (−0.44, −0.13) | <0.0005 | −0.32 (−0.49, −0.15) | <0.0005 | −0.56 (−0.80, −0.32) | <0.0005 |
Bold indicates reaching statistical significance of P < .05. IVW, inverse-variance weighted; CI, confidence interval; LDL-C, low-density lipoprotein cholesterol level in serum; FI, frailty index; IV, instrumental variable; SNPs, single-nucleotide polymorphisms.
Fig. 2.
MR-Egger plot with IVW, weighted median, and weighted mode lines for pleiotropy investigation and estimate comparison; variants used for instrumental analysis are independent SNPs (n = 274) constructing the large LDL-C IV. The results of per LDL-C-lowering allele have been coded so that trends correspond to the effects on frailty expected from the lowering of the biomarker. MR, Mendelian Randomization; IVW, inverse-variance weighted; LDL-C, low-density lipoprotein cholesterol; SNP, single-nucleotide polymorphism; IV, instrumental variable; FI, frailty index; CI, confidence interval.
Fig. 3.
MR-Egger plot with IVW, weighted median, and weighted mode lines for pleiotropy investigation and estimate comparison; variants used for instrumental analysis are independent SNPs (n = 50) constructing the small LDL-C IV. The results of per LDL-C-lowering allele have been coded so that trends correspond to the effects on frailty expected from the lowering of the biomarker. MR, Mendelian Randomization; IVW, inverse-variance weighted; LDL-C, low-density lipoprotein cholesterol; SNP, single-nucleotide polymorphism; IV, instrumental variable; FI, frailty index; CI, confidence interval.
Fig. 4.
The IVW, MR-Egger, weighted median, and weighted modal estimates for the FI effects of genetically predicted LDL-C concentrations, stratified by the aggregated IVs. MR, Mendelian randomization; IVW, inverse-variance weighted; LDL-C, low-density lipoprotein cholesterol; IV, instrumental variable; FI, frailty index.
3.2. LDL-C effects on the modified FI excluding cardiometabolic deficits
Similar effects of genetically predicted LDL-C concentrations were found for the modified FI (MFI) excluding seven items related to cardiometabolic deficits. The IVW estimates for the MFI effects of LDL-C concentrations instrumented by all eight aggregated IVs showed great consistency with those for the FI effects (Fig. 5). Corresponding MR-Egger, weighted median, and weighted modal estimates were also presented (Table S3), to address potential pleiotropy of the IVs. In general, effect sizes changed slightly but not dramatically.
Fig. 5.
IVW estimates for the association between genetically predicted LDL-C concentration and FI/MFI/FI60, stratified by the IV versions. MFI indicated the modified FI excluding the items related to cardiometabolic deficit; FI60 indicated that estimates were obtained with data from participants ≥60 years of age. IVW, inverse variance weighted; LDL-C, low-density lipoprotein cholesterol; IV, instrumental variable; FI, frailty index.
3.3. Genetic effects of LDL-C on FI among participants aged ≥60 years
Larger effects were observed when excluding younger participants <60 years old (Fig. 5 and Table S4). In general, the effect sizes changed by a factor of 0.1 in all estimates indicating greater protective effects from LDL-C on the FI in the older group. Corresponding MR-Egger, weighted median, and weighted mode estimates were also presented, and estimates were to the most parts consistent with the IVW ones (Table S4).
3.4. Results of sensitivity analyses
When excluding the genetic variants located in chromosome 19 from the large and small IVs, the effects from LDL-C on the FI, instrumented by the modified IVs, were consistent with those observed in the main analyses (Table S5).
To test how relatedness affected the results, a more stringent cut-off (0.0442) for kinship relationship was applied to the data, reducing the number of individuals in the main analysis to 335,034. Most effect sizes remained similar, with the overall conclusion from the main results unchanged (Table S6). However, gene-specific variants from the NPC1L1 loci presented a somewhat higher effect size, indicative of a protective effect of NPC1L1 inhibition on frailty, which was also confirmed for the modified FI score and in individuals ≥60 years.
To further confirm our findings, age-, sex-, and PC-adjusted linear regression models using individual level GRSs in UKB were applied. Results were concordant with the main analyses, revealing protective effects from genetically determined LDL-C on the FI with statistically significant effects both instrumented by the large and small GRS (Table S7). Moreover, for the drug-target specific loci, statistically significant protective effects were also observed in the adjusted linear regression modeling for individual HMGCR-, APOC3-, and LDLR-specific GRSs, but not for PCSK9-, NPC1L1-, and APOB-specific GRSs (Table S7). The results from the two sensitivity analyses, using a modified FI excluding seven items related to cardiometabolic deficit as the dependent variable in one and excluding the data from relatively younger participants (<60 years) in the other, differed from those in the main analysis to some extent, but remained statistically significant.
Finally when more stringent pairwise r2 thresholds (0.1 and 0.01) were used for selecting independent genetic variants to construct the aggregated IVs, the results were still concordant with the main observations (Tables S8–S11).
4. Discussion
In this study, we used an MR design to estimate the effect of long-term lipid-lowering on frailty measured by the FI—akin to overall health profile—accrued by mid-life and late middle-age. We found that individuals predicted to have life-long lowering LDL-C concentrations had substantially lower frailty scores, consistent with, and perhaps exceeding, the degree of cardioprotection expected from lipid lowering. We also profiled gene-specific effects from loci that index the modulation of existing and emerging lipid-lowering drug targets (e.g., HMGCR, APOC3, and LDLR), and found evidence that the on-target effects of classes used to lower LDL-C may contribute notable differences to the overall health of users.
Our work highlights the importance of LDL-C monitoring during the ageing process, especially since the association with the FI was independent of any detected atherosclerotic pathogenesis. To our knowledge, this is the first study to use MR to quantify the effect of lipid lowering on frailty. Some observational studies—results of which are prone to confounding—observed a null or an opposite association between the two [29,30]. Hypercholesterolaemia may influence frailty via several underlying mechanisms, including the promotion of endothelial dysfunction and inflammation [31,32]. As an oxidative stress biomarker, modified LDL-C is a pro-inflammatory stimulant present in circulation, affecting local sites in various tissues. Hence it has the capacity to induce inflammation in a variety of tissues [33]. Our inference is also supported to some extent by a randomized clinical trial study demonstrating that regular flavonoids consumption positively affects blood oxidative stress and inflammation end points, etc., resulting in improved LDL-C concentrations and frailty status in parallel [34].
Epidemiologic evidence also demonstrates that decreasing LDL-C concentrations may promote healthy ageing. Besides lowering CVD risk, other age-related diseases might be mitigated through the lowering of LDL-C concentrations. Geriatric traits and/or diseases with elevated age-related incident risk are often concurrent and correlate, possibly due to shared underlying mechanisms, e.g., oxidative stress, low-grade level inflammation, cell senescence, etc. [10,35]. Atkins et al. reported that persons aged 60–69 years with more benign cardiovascular risk factor profiles (smoking status, LDL-C, blood pressure, etc.) have substantially lower incidence of geriatric conditions (e.g., chronic pain, incontinence, falls, fragility fractures, and dementia) and frailty [36]. The authors inferred that optimizing CVD risk factors may substantially reduce the burden of morbidity in later life [36], which is supported by our finding that genetically-predicted LDL-C exposure also associated with the modified FI after excluding cardiometabolic deficit items.
The more pronounced association of LDL-C with frailty in those aged 60–69 years, compared to the association in younger participants, may reflect cumulative and more impactful benefits of LDL-C reductions achieved earlier in the life course [37]. Similarly, another study has observed that maintenance of optimal risk factor profiles in middle-age rather than in later life stages, was associated with substantially longer CVD morbidity-free survival [38]. However, we note that the UKB cohort is not representative of older individuals and were evaluated to have a good overall health status on average [16]. Considering the inconsistency in the effects of LDL-C lowering therapeutics (e.g. statin use) on CVD risk events, all-cause mortality, etc. among the older adults (e.g., those aged ≥75) [[39], [40], [41], [42]], it is not clear whether our findings would extend to show benefits of LDL-C reduction for overall health this far into the life course.
In gene-specific models, which to some extent mimic the effects of long-term exposure of individuals to the modulation of lipid-lowering drug targets, most of the corresponding therapeutics (including statins) would be predicted to reduce frailty. Owing to the analogy of an MR analysis and a randomized clinical trial [14], our gene-specific IV findings also suggested the several instrumental LDL-C lowering therapeutics affect frailty of different magnitudes. However, this cannot be verified due to unavailable RCT evidence. Prior observational evidence along these lines has been inconsistent. A prospective cohort study of 383 residents aged ≥65 years found that the risk of mortality, all-cause hospitalizations, and incidence of falls during the 12-month follow-up were lower among statin users than non-users [43]. In contrast, an observational study of statin use and incident frailty in women aged 65 years old or older failed to observe associations between current statin use, duration and potency of statin use and incident frailty [30]. However, among users of low potency statins, longer duration of use was associated with reduced risk of frailty [30]. Some other observational studies have suggested effects of LDL-C lowering therapeutic exposure on other diseases or traits besides CVDs although inconsistency exists [1,2,44,45]. On the other hand, the relative magnitude of FI decrement showed differences when using different gene-specific IVs. For instance, the PCSK9 and APOB estimates were more modest or null compared to the HMGCR estimates, despite each being estimated to confer approximately the same degree of protection from CAD per unit lowering of LDL-C. This implies that modulating these targets could have detrimental effects on overall health via other (i.e. non-LDL related) pathways compared to statin use [46].
There are several strengths in our study. First, genetically-predicted LDL-C concentration is observed to associate with the overall health of European population for the first time in UKB. The association is independent of cardiometabolic traits, and stronger among older individuals. These findings emphasize LDL-C's role in biological ageing besides atherosclerosis processing. Second, studying these hypotheses with MR means that findings are far less likely to be prone to confounding than conventional observational epidemiology, and not subject to reverse causation (that disease processes have influenced exposure status). Third, the substantial sample with detailed health and genetic data in UK Biobank provided our study with good precision for estimates, and enabled us to present results using both summary and individual-level data, as well as several sensitivity analyses to scrutinize the genetic associations in several ways.
Several limitations are also worth noting. First, directly detected serum LDL-C concentrations were not available for the individuals in the UKB cohort at the time of the study conduct. Hence, effects of the selected variants for LDL-C IV construction could not be verified directly in this study. Second, the UKB is subject to healthy cohort effect. That may limit the extensionality of our findings to other population with a different health status. Third, just as drug target models only encompass on-target (while not off-target) effects of using the related therapeutics, our genetic results for drug targets cannot reflect the pharmacokinetics of drug use. Our estimates from the gene-specific models cannot incorporate secondary effects on health outcomes of using these classes (i.e. where a drug affects other proteins besides the primary target, which are ‘off-target’).
In conclusion, our results may provide evidence for the efficacy of LDL-C lowering (and the specific benefits of several related therapeutic classes) in the maintenance of overall health in ageing, and perhaps for the prevention of treatment of age-related diseases other than CVDs. Future studies should help to evaluate the relationship between the life-long lowering of LDL-C concentrations and other specific geriatric diseases and/or traits. The implication that different LDL-C lowering therapeutics could affect frailty at differing degrees may also indicate need for pharmacovigilance regarding recently introduced drug classes, such as PCSK9 inhibitors and ApoB antisense therapeutics. All these results may provide some evidence for the efficacy of LDL-C lowering therapies in the treatment of age-related diseases other than CVDs. Continued follow-up of clinical trials of lipid-lowering medications, including an array of health indicators for multiple health domains, may help to confirm both beneficial and any detrimental effects of lipid-lowering by specific means on frailty.
Contributors
Study design: SH, DW, QW. Study conduct: QW. Data analysis, and data interpretation: YW, KL, QW. Draft manuscript: QW. Revising manuscript content: YW, KL, NP, DW, SH. Approving final version of manuscript: QW, YW, KL, NP, DW, SH. SH takes responsibility for the integrity of the data analysis.
Declaration of interests
We declare no competing interests.
Acknowledgments
This work was supported by the Erik Rönnberg award for scientific studies on ageing and age-related diseases, China Scholarship Council (State Scholarship Fund 2017), National Natural Science Foundation of China (grant number: 81573235), Karolinska Institutet Delfinansiering (KID), Loo and Hans Osterman Foundation (grant number: 2017-00103 to KL), Foundation for Geriatric Diseases, Magnus Bergwall foundation, Foundation for Gamla Tjänarinnor, Karolinska Institutet (Strategic Research Area in Epidemiology), the Swedish Research Council (521-2013-8689 and 2015-03255) and the Swedish Council for Working Life and Social Research (FORTE) (2013-2292).
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.ebiom.2019.07.007.
Appendix A. Supplementary data
Supplementary material
References
- 1.Benn M., Nordestgaard B.G., Frikke-Schmidt R., Tybjaerg-Hansen A. Low LDL cholesterol, PCSK9 and HMGCR genetic variation, and risk of Alzheimer's disease and Parkinson's disease: Mendelian randomisation study. BMJ. 2017;357 doi: 10.1136/bmj.j1648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chen P.H., Wang J.S., Lin S.Y., Li C.H., Wang C.Y., Hu C.Y. Effects of statins on all-cause mortality at different low-density-lipoprotein cholesterol levels in Asian patients with type 2 diabetes. Curr Med Res Opin. 2018;34(11):1–8. doi: 10.1080/03007995.2018.1439829. [DOI] [PubMed] [Google Scholar]
- 3.White J., Swerdlow D.I., Preiss D., Fairhurst-Hunter Z., Keating B.J., Asselbergs F.W. Association of lipid fractions with risks for coronary artery disease and diabetes. JAMA Cardiol. 2016;1(6):692–699. doi: 10.1001/jamacardio.2016.1884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Alique M., Luna C., Carracedo J., Ramirez R. LDL biochemical modifications: a link between atherosclerosis and aging. Food Nutr Res. 2015;59 doi: 10.3402/fnr.v59.29240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Fulcher J., O'Connell R., Voysey M., Emberson J., Blackwell L., Mihaylova B. Efficacy and safety of LDL-lowering therapy among men and women: meta-analysis of individual data from 174,000 participants in 27 randomised trials. Lancet. 2015;385(9976):1397–1405. doi: 10.1016/S0140-6736(14)61368-4. [DOI] [PubMed] [Google Scholar]
- 6.Masana L., Girona J., Ibarretxe D., Rodriguez-Calvo R., Rosales R., Vallve J.C. Clinical and pathophysiological evidence supporting the safety of extremely low LDL levels-the zero-LDL hypothesis. J Clin Lipidol. 2018;12(2):292–9 e3. doi: 10.1016/j.jacl.2017.12.018. [DOI] [PubMed] [Google Scholar]
- 7.Kojima G., Iliffe S., Walters K. Frailty index as a predictor of mortality: a systematic review and meta-analysis. Age Ageing. 2018;47(2):193–200. doi: 10.1093/ageing/afx162. [DOI] [PubMed] [Google Scholar]
- 8.Fried L.P., Tangen C.M., Walston J., Newman A.B., Hirsch C., Gottdiener J. Frailty in older adults: evidence for a phenotype. J Gerontol A Biol Sci Med Sci. 2001;56(3):M146–M156. doi: 10.1093/gerona/56.3.m146. [DOI] [PubMed] [Google Scholar]
- 9.Mitnitski A.B., Mogilner A.J., Rockwood K. Accumulation of deficits as a proxy measure of aging. Sci World J. 2001;1:323–336. doi: 10.1100/tsw.2001.58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Theou O., Tan E.C., Bell J.S., Emery T., Robson L., Morley J.E. Frailty levels in residential aged care facilities measured using the frailty index and FRAIL-NH scale. J Am Geriatr Soc. 2016;64(11):e207–e212. doi: 10.1111/jgs.14490. [DOI] [PubMed] [Google Scholar]
- 11.Lawlor D.A., Harbord R.M., Sterne J.A., Timpson N., Davey Smith G. Mendelian randomization: using genes as instruments for making causal inferences in epidemiology. Stat Med. 2008;27(8):1133–1163. doi: 10.1002/sim.3034. [DOI] [PubMed] [Google Scholar]
- 12.Smith G.D., Ebrahim S. 'Mendelian randomization': can genetic epidemiology contribute to understanding environmental determinants of disease? Int J Epidemiol. 2003;32(1):1–22. doi: 10.1093/ije/dyg070. [DOI] [PubMed] [Google Scholar]
- 13.Burgess S., Scott R.A., Timpson N.J., Davey Smith G., Thompson S.G. Using published data in Mendelian randomization: a blueprint for efficient identification of causal risk factors. Eur J Epidemiol. 2015;30(7):543–552. doi: 10.1007/s10654-015-0011-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Emdin C.A., Khera A.V., Kathiresan S. Mendelian randomization. JAMA. 2017;318(19):1925–1926. doi: 10.1001/jama.2017.17219. [DOI] [PubMed] [Google Scholar]
- 15.Wang J.C., Bennett M. Aging and atherosclerosis: mechanisms, functional consequences, and potential therapeutics for cellular senescence. Circ Res. 2012;111(2):245–259. doi: 10.1161/CIRCRESAHA.111.261388. [DOI] [PubMed] [Google Scholar]
- 16.Williams D.M., Jylhava J., Pedersen N.L., Hagg S. A frailty index for UK biobank participants. J Gerontol A Biol Sci Med Sci. 2019;74(4):582–587. doi: 10.1093/gerona/gly094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Pierce B.L., Ahsan H., Vanderweele T.J. Power and instrument strength requirements for Mendelian randomization studies using multiple genetic variants. Int J Epidemiol. 2011;40(3):740–752. doi: 10.1093/ije/dyq151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Clarke P.S., Windmeijer F. Instrumental variable estimators for binary outcomes. J Am Stat Assoc. 2012;107:1638–1652. [Google Scholar]
- 19.Bowden J., Davey Smith G., Burgess S. Mendelian randomization with invalid instruments: effect estimation and bias detection through Egger regression. Int J Epidemiol. 2015;44(2):512–525. doi: 10.1093/ije/dyv080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bowden J., Davey Smith G., Haycock P.C., Burgess S. Consistent estimation in Mendelian randomization with some invalid instruments using a weighted median estimator. Genet Epidemiol. 2016;40(4):304–314. doi: 10.1002/gepi.21965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Burgess S., Bowden J., Fall T., Ingelsson E., Thompson S.G. Sensitivity analyses for robust causal inference from Mendelian randomization analyses with multiple genetic variants. Epidemiology. 2017;28(1):30–42. doi: 10.1097/EDE.0000000000000559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hartwig F.P., Davey Smith G., Bowden J. Robust inference in summary data Mendelian randomization via the zero modal pleiotropy assumption. Int J Epidemiol. 2017;46(6):1985–1998. doi: 10.1093/ije/dyx102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Willer C.J., Schmidt E.M., Sengupta S., Peloso G.M., Gustafsson S., Kanoni S. Discovery and refinement of loci associated with lipid levels. Nat Genet. 2013;45(11):1274–1283. doi: 10.1038/ng.2797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hemani G., Zheng J., Elsworth B., Wade K.H., Haberland V., Baird D. The MR-base platform supports systematic causal inference across the human phenome. Elife. 2018;7 doi: 10.7554/eLife.34408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ference B.A., Yoo W., Alesh I., Mahajan N., Mirowska K.K., Mewada A. Effect of long-term exposure to lower low-density lipoprotein cholesterol beginning early in life on the risk of coronary heart disease:a Mendelian randomization analysis. J Am Coll Cardiol. 2012;60(25):2631–2639. doi: 10.1016/j.jacc.2012.09.017. [DOI] [PubMed] [Google Scholar]
- 26.Purcell S., Neale B., Todd-Brown K., Thomas L., Ferreira M.A., Bender D. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet. 2007;81(3):559–575. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Spiller W., Davies N.M., Palmer T.M. Software application profile: mrrobust-a tool for performing two-sample summary Mendelian randomization analyses. Int J Epidemiol. 2018 [Google Scholar]
- 28.Marais A.D. Apolipoprotein E in lipoprotein metabolism, health and cardiovascular disease. Pathology. 2019;51(2):165–176. doi: 10.1016/j.pathol.2018.11.002. [DOI] [PubMed] [Google Scholar]
- 29.Jayanama K., Theou O., Blodgett J.M., Cahill L., Rockwood K. Frailty, nutrition-related parameters, and mortality across the adult age spectrum. BMC Med. 2018;16(1):188. doi: 10.1186/s12916-018-1176-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.LaCroix A.Z., Gray S.L., Aragaki A., Cochrane B.B., Newman A.B., Kooperberg C.L. Statin use and incident frailty in women aged 65 years or older: prospective findings from the Women's Health Initiative Observational Study. J Gerontol A Biol Sci Med Sci. 2008;63(4):369–375. doi: 10.1093/gerona/63.4.369. [DOI] [PubMed] [Google Scholar]
- 31.McEwen J.E., Zimniak P., Mehta J.L., Shmookler Reis R.J. Molecular pathology of aging and its implications for senescent coronary atherosclerosis. Curr Opin Cardiol. 2005;20(5):399–406. doi: 10.1097/01.hco.0000175517.50181.89. [DOI] [PubMed] [Google Scholar]
- 32.Ross R. Atherosclerosis--an inflammatory disease. N Engl J Med. 1999;340(2):115–126. doi: 10.1056/NEJM199901143400207. [DOI] [PubMed] [Google Scholar]
- 33.Itabe H. Oxidized low-density lipoprotein as a biomarker of in vivo oxidative stress: from atherosclerosis to periodontitis. J Clin Biochem Nutr. 2012;51(1):1–8. doi: 10.3164/jcbn.11-00020R1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Munguia L., Rubio-Gayosso I., Ramirez-Sanchez I., Ortiz A., Hidalgoa I., Gonzalez C. High flavonoid cocoa supplement ameliorates plasma oxidative stress and inflammation levels while improving mobility and quality of life in older subjects: a double blind randomized clinical trial. J Gerontol En Biol Sci Med Sci. May 6 2019 doi: 10.1093/gerona/glz107. (Pii: glz107) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Edo M.D., Andres V. Aging, telomeres, and atherosclerosis. Cardiovasc Res. 2005;66(2):213–221. doi: 10.1016/j.cardiores.2004.09.007. [DOI] [PubMed] [Google Scholar]
- 36.Atkins J.L., Delgado J., Pilling L.C., Bowman K., Masoli J.A.H., Kuchel G.A. Impact of low cardiovascular risk profiles on geriatric outcomes: evidence from 421,000 participants in two cohorts. J Gerontol A Biol Sci Med Sci. 2018;74(3):350–357. doi: 10.1093/gerona/gly083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Strandberg T.E., Kolehmainen L., Vuorio A. Evaluation and treatment of older patients with hypercholesterolemia: a clinical review. JAMA. 2014;312(11):1136–1144. doi: 10.1001/jama.2014.10924. [DOI] [PubMed] [Google Scholar]
- 38.Wilkins J.T., Ning H., Berry J., Zhao L., Dyer A.R., Lloyd-Jones D.M. Lifetime risk and years lived free of total cardiovascular disease. JAMA. 2012;308(17):1795–1801. doi: 10.1001/jama.2012.14312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Charlton J., Ravindrarajah R., Hamada S., Jackson S.H., Gulliford M.C. Trajectory of Total cholesterol in the last years of life over age 80 years: cohort study of 99,758 participants. J Gerontol A Biol Sci Med Sci. 2018;73(8):1083–1089. doi: 10.1093/gerona/glx184. [DOI] [PubMed] [Google Scholar]
- 40.Gnjidic D., Le Couteur D.G., Blyth F.M., Travison T., Rogers K., Naganathan V. Statin use and clinical outcomes in older men: a prospective population-based study. BMJ Open. 2013;3(3) doi: 10.1136/bmjopen-2012-002333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Singh S., Zieman S., Go A.S., Fortmann S.P., Wenger N.K., Fleg J.L. Statins for primary prevention in older adults-moving toward evidence-based decision-making. J Am Geriatr Soc. 2018;66(11):2188–2196. doi: 10.1111/jgs.15449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Strandberg T.E. Role of statin therapy in primary prevention of cardiovascular disease in elderly patients. Curr Atheroscler Rep. 2019;21(8):28. doi: 10.1007/s11883-019-0793-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Korhonen M.J., Ilomaki J., Sluggett J.K., Brookhart M.A., Visvanathan R., Cooper T. Selective prescribing of statins and the risk of mortality, hospitalizations, and falls in aged care services. J Clin Lipidol. 2018;12(3):652–661. doi: 10.1016/j.jacl.2018.02.012. [DOI] [PubMed] [Google Scholar]
- 44.Robinson J.G., Farnier M., Krempf M., Bergeron J., Luc G., Averna M. Efficacy and safety of alirocumab in reducing lipids and cardiovascular events. N Engl J Med. 2015;372(16):1489–1499. doi: 10.1056/NEJMoa1501031. [DOI] [PubMed] [Google Scholar]
- 45.Sabatine M.S., Giugliano R.P., Keech A.C., Honarpour N., Wiviott S.D., Murphy S.A. Evolocumab and clinical outcomes in patients with cardiovascular disease. N Engl J Med. 2017;376(18):1713–1722. doi: 10.1056/NEJMoa1615664. [DOI] [PubMed] [Google Scholar]
- 46.Bandyopadhyay D., Ashish K., Hajra A., Qureshi A., Ghosh R.K. Cardiovascular outcomes of PCSK9 inhibitors: with special emphasis on its effect beyond LDL-cholesterol lowering. J Lipids. 2018;2018 doi: 10.1155/2018/3179201. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material