Significance
Photosensitive G protein-coupled receptors (GPCRs) have been used as model systems in the study of class A GPCRs. These are classified into mono- and bistable rhodopsins based on the thermal stability of their active states. Although much is known about monostable rhodopsins, their bistable counterparts remain structurally elusive. The crystal structure of the bistable jumping spider rhodopsin-1 lays the basis in the understanding of molecular mechanistic differences between these 2 subclasses. Furthermore, bistable rhodopsins conform to an “activation-ready” architecture bearing similarities to nonphotosensitive class A GPCRs when compared to monostable rhodopsins. These insights provide a valuable basis for understanding bistable rhodopsin functionality and help establish future light-driven applications for the broader family of class A GPCRs.
Keywords: rhodopsin, bistability, G protein-coupled receptors, retinal, X-ray crystallography
Abstract
Light-sensitive G protein-coupled receptors (GPCRs)—rhodopsins—absorb photons to isomerize their covalently bound retinal, triggering conformational changes that result in downstream signaling cascades. Monostable rhodopsins release retinal upon isomerization as opposed to the retinal in bistable rhodopsins that “reisomerize” upon absorption of a second photon. Understanding the mechanistic differences between these light-sensitive GPCRs has been hindered by the scarcity of recombinant models of the latter. Here, we reveal the high-resolution crystal structure of a recombinant bistable rhodopsin, jumping spider rhodopsin-1, bound to the inverse agonist 9-cis retinal. We observe a water-mediated network around the ligand hinting toward the basis of their bistable nature. In contrast to bovine rhodopsin (monostable), the transmembrane bundle of jumping spider rhodopsin-1 as well that of the bistable squid rhodopsin adopts a more “activation-ready” conformation often observed in other nonphotosensitive class A GPCRs. These similarities suggest the role of jumping spider rhodopsin-1 as a potential model system in the study of the structure–function relationship of both photosensitive and nonphotosensitive class A GPCRs.
G protein-coupled receptors (GPCRs) act as signal transducers converting external stimuli into cellular responses. Among these, rhodopsins are light-sensitive class A GPCRs operating in visual and nonvisual processes (1). Photosensivity is rendered by the chromophore retinal, covalently bound via a protonated Schiff base to a lysine in the orthosteric ligand-binding site. A nearby negatively charged residue, the counterion, stabilizes the protonated Schiff base (2–4). Upon illumination, the retinal isomerizes from cis to trans, triggering conformational changes that culminate in the formation of an activated form of the receptor (Meta), ready for G protein binding (5).
Based on the thermal stability of these active states, rhodopsins can be classified as monostable and bistable. Illumination eventually bleaches monostable rhodopsins by deprotonation and hydrolysis of the Schiff base and subsequent release of the chromophore (6). Bistable rhodopsins, on the other hand, undergo a 2-photon bidirectional photoreaction (cis-trans/trans-cis) in which the Schiff base remains protonated and the retinal bound throughout the photocycle (4, 7, 8). Over the years, biophysical and structural characterization of bovine rhodopsin (monostable) and squid rhodopsin (bistable) have provided insights into these mechanistic differences. For instance, in monostable rhodopsins a glutamate on the third transmembrane domain (Glu-1133.28 in TM3 of bovine rhodopsin; SI Appendix, Table S1) is the proximal counterion in the inactive conformation and mediates the charge stabilization of the protonated Schiff base via a salt bridge (2, 3). Upon activation, these rhodopsins transiently adopt a distal counterion on the extracellular loop 2 (ECL2; Glu-18145.44) before deprotonation and hydrolysis of the Schiff base (5, 9). In contrast, bistable rhodopsins employ the equivalent glutamate on ECL2 (45.44) (10) as their sole distal counterion for the entire photocycle (4). Nevertheless, the counterion position alone does not explain the mechanistic differences between the 2 receptor types. Structural and Fourier transform infrared (FTIR) spectroscopic analysis of squid rhodopsin (11) suggested the presence of a hydrogen bond network between its protonated Schiff base and the distal counterion (12, 13). However, the exact architecture of this network remains elusive, posing a challenge to our understanding of the functional differences between mono- and bistable rhodopsins. In addition, the lack of a recombinant model system for the study of bistable rhodopsins impedes further advances that would shed new light on this topic.
In this work, we present the high-resolution crystal structure of the bistable jumping spider rhodopsin-1 (JSR1) in its ground (inactive) state. This structure provides concrete insights into the molecular architecture of the retinal-binding pocket that lay the basis for understanding the activation mechanism around the protonated Schiff base in bistable opsins. In comparison with squid rhodopsin, the structure of JSR1 shows a large number of tightly coordinated water molecules that span the transmembrane region. Interestingly, the orientation of key residue side chains in this region of both bistable receptors resemble that of the inactive conformation of other class A GPCRs, in contrast to those of the monostable bovine rhodopsin (14). Furthermore, our functional data also indicate a promiscuity of JSR1 in G protein recognition, as often observed in other class A GPCRs (15). Altogether, our findings highlight remarkable similarities between bistable rhodopsins and nonphotosensitive class A GPCRs, in contrast to the functionally distinct and highly specialized bovine rhodopsin system (16, 17). Hence, we propose JSR1 as a suitable photosensitive target to study activation dynamics of bistable rhodopsins and, potentially, the broader class A G protein-coupled receptors applying light-driven methods by the development of new optogenetic tools (18–20).
Results
JSR1 Isorhodopsin and Overall Structure.
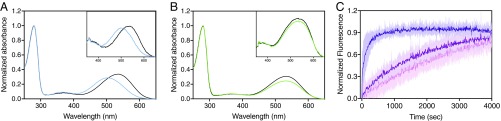
Wild-type JSR1 was recombinantly produced in mammalian HEK293 GnTI− cells (21), reconstituted with the inverse agonist 9-cis retinal analog, and purified in 3 chromatographic steps (see Methods). Jumping spider isorhodopsin-1 (JSiR1, 9-cis retinal bound) shows an absorbance maximum at 505 nm, which is blue shifted from the 535-nm absorbance maximum of the native 11-cis retinal-bound form (JSR1) (22) (ground states; Fig. 1 A and B). Nevertheless, upon activation, isomerization of either of these isoforms yields an all-trans retinal-bound photoproduct with an absorbance maxima at 535 nm (active conformation, a-Meta) (22). Therefore, the spectral separation provided by the 9-cis retinal facilitates in vitro handling of the sample by allowing the detection of light contamination.
Fig. 1.
In vitro activation of recombinantly produced JSiR1 and JSR1. (A and B) Normalized UV-vis spectra of JSiR1 (9-cis retinal, λmax 505 nm, A) and JSR1 (11-cis retinal, λmax 535 nm, B) recorded in solution. Ground states (inactive) are represented by blue (A) and green (B) curves, respectively. Photoproducts (λmax 535 nm) are indicated in black. The Insets of A and B represent normalized UV-vis spectra of JSiR1 (A) and JSR1 (B) recorded in LCP. (C) G protein activation assay, where intrinsic tryptophan fluorescence from the Gα subunit is recorded upon addition of GTPγS (24). Photoactivated recombinant JSiR1 triggers Gi protein dissociation (dark purple curve; K = 5.65 ± 0.24 × 10−3) The dark pink curve indicates inactive JSiR1 whose activity is slightly higher (K = 0.51 ± 0.002 × 10−3) than that of the control (no JSiR1 in light pink; K = 0.32 ± 0.001 × 10−3). The value after the ± sign indicates the SE mean (SEM). All data are measured in triplicates at 20 °C at a pH of 7.3. A concentration of 600 nM of heterotrimeric Gi was used for the assay.
To ensure the full functionality of the receptor, we measured the ability of JSiR1 to activate G proteins using a Gi-based activity assay (23–25). We observed that recombinant JSiR1 triggers Gi activation in vitro (Fig. 1C). Interestingly, JSR1 was previously suggested to activate Gq in vivo (22), which suggests that JSR1 exhibits an inherent promiscuity in G protein activation as observed in many class A GPCRs (15). Finally, recombinant wild-type JSiR1 shows high thermostability (26) in a range of detergents and pH conditions (SI Appendix, Fig. S1), thus being an ideal target for structural and biophysical characterization.
Purified JSiR1 was incorporated into a lipidic cubic phase (LCP) to facilitate its crystallization. Analysis by UV-visible (UV-vis) spectroscopy reveals a similar spectrum to the detergent-solubilized protein, indicating that the receptor reconstituted in LCP is qualitatively identical (Fig. 1 A and B). Diffraction datasets from the crystals were collected at the Swiss Light Source (SLS, Paul Scherrer Institut [PSI]), and the structure was solved by molecular replacement using squid rhodopsin as a template (PDB ID code 2Z73) (12).
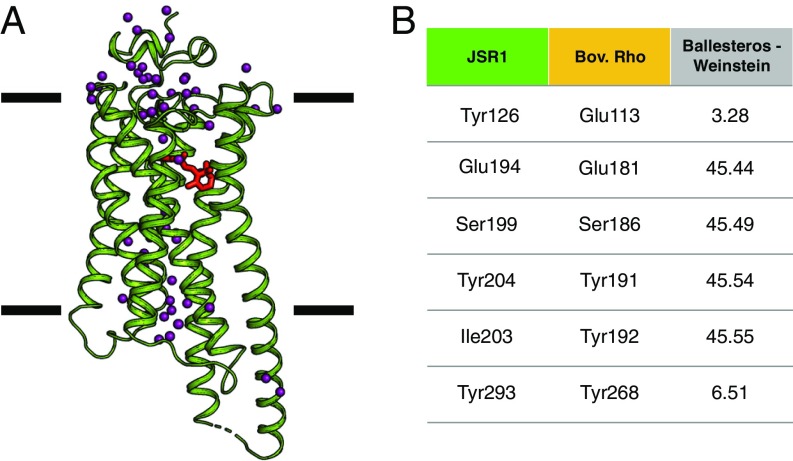
Diffraction data were processed to a high-resolution limit of 2.5 Å, 2.1 Å, and 2.3 Å along the a*, b*, and c* axes. The structure lacks part of the N terminus (residues 1–19), intracellular loop 3 (ICL3; residues 257–262), and C terminus (including TM8; residues 336–380) that have poor electron density. The rest of the structure is very-well resolved; in particular, TM5 and TM6 are well ordered and extend well beyond the transmembrane bundle (Fig. 2 and SI Appendix, Fig. S2). While such features are also present in squid rhodopsin (12), these regions are seldom observed in other class A GPCR structures, usually due to protein engineering schemes to remove flexible regions (27). However, these extensions in TM5/6 are very relevant to GPCR function as they have been suggested to partake in G protein and arrestin binding (16).
Fig. 2.
Overall JSR1 structure in complex with 9-cis retinal. (A) Overall structure of JSiR1 (green, 6I9K; ref. 63) bound to 9-cis retinal (red). The crystal structure reveals 7 transmembrane helices with TM5 and 6 extending well beyond the lipid bilayer. (Membrane boundaries drawn based on OPM65 server calculations.) Ordered water molecules (purple spheres) span the length of the receptor. (B) Nomenclature of the main residues in the retinal-binding pocket (see SI Appendix, Table S1 for an extended version) (9).
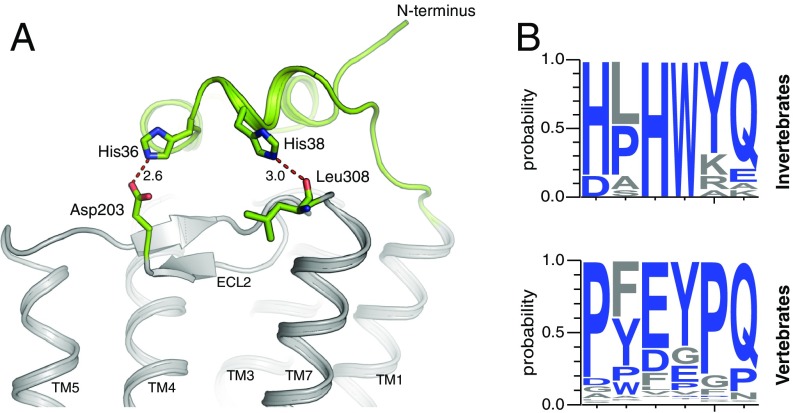
The Extracellular N-Terminal Domain.
The N-terminal region of JSiR1 caps the extracellular domains of the receptor stacking over the second extracellular loop (ECL2) (Fig. 3). This fold, observed with some variations in all opsin crystal structures (12, 28) (SI Appendix, Fig. S3), occludes the orthosteric ligand-binding site from the extracellular milieu, contributing to the stability of the ligand-binding pocket and possibly of the overall receptor (28). In JSiR1, the N-terminal domain folds into 3 short hydrophobic helices anchored to the transmembrane domains through histidine-mediated contacts with residues in ECL2 and ECL3 (Fig. 3A). This fold resembles that of squid rhodopsin, but clearly differs from that of bovine rhodopsin (6, 28, 29). Analysis of structure-based sequence alignments of rhodopsins from different organisms suggests the presence of 2 distinct patterns in their N terminus (10, 30, 31); while invertebrate rhodopsins present an HxHW helical motif, as seen in the crystal structures of JSiR1 and squid rhodopsin, vertebrates rhodopsins show instead a Px(E/D)xPQ motif in the same region (Fig. 3B).
Fig. 3.
The N-terminal domain is anchored to the tranmsmebrane bundle by conserved histidines. (A) The N-terminal domain (green, 6I9K; ref. 63) of JSiR1 folds into 3 ordered short helices forming a cap over the transmembrane bundle (gray) anchored by residues His-36 and His-38 through polar contacts with the side chain of Asp-203 (ECL2) and the backbone carbonyl of Leu-308 (ECL3). (B) Sequence analysis of the N-terminal region of 143 opsins (see Methods) reveals that invertebrate rhodopsins feature an HxHWYQ motif—as observed in the crystal structures of JSiR1 and squid rhodopsin—while rhodopsins of the chordata family (including vertebrates) have instead a Px(E/D)xPQ motif at a similar position.
The Ligand-Binding Pocket.
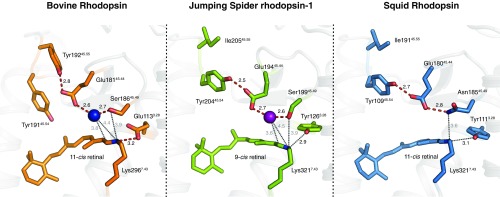
In the orthosteric ligand-binding site, electron density before refinement confirmed the presence of 9-cis retinal covalently bound to Lys-3217.43 via a protonated Schiff base (Fig. 4 and SI Appendix, Fig. S4). We observed that the 9-cis retinal bends out of absolute planarity (−46.7° torsional twist at the C9–C10 cis bond, see SI Appendix, Fig. S4), similarly to bovine rhodopsin bound to 11-cis retinal (−36.1° torsional twist at the C11–C12 cis bond; 1U19) (29). It has been shown that such a “pretwist” of retinal reduces the energy barrier for isomerization of the cis bond and enables a more efficient activation of the receptor (32). However, 9-cis retinal bound to bovine and squid rhodopsins (2PED, 3AYN) (33, 34) did not show this higher torsional twist.
Fig. 4.
Comparisons of the retinal binding pockets between mono- and bistable rhodopsins. Comparison of the retinal binding pockets of bovine rhodopsin (1GZM, orange) (28), JSiR1 (6I9K, green; ref. 63), and squid rhodopsin (PDB ID code 2Z73, blue) (11) sheds light on the common elements of their architecture. The counterion system of JSiR1 involves water-mediated (purple sphere) interactions (red dashes) bridging Glu-19445.44 to Ser-19945.49 and to the Schiff base. This counterion triad is observed in squid rhodopsin as well with Asn-18545.49 playing the role of the serine in JSiR1. Structural similarities can also be observed between JSiR1 and bovine rhodopsin including the presence of an ordered water molecule (blue sphere) in the region. In contrast, the Schiff base of bovine rhodopsin is stabilized by the proximal Glu-1133.28 by forming a direct salt bridge. Other key distances within the binding pocket are depicted by black dotted lines.
The physicochemical environment around the Schiff base is crucial in determining its protonation state and thus, the ultimate fate of retinal (i.e., hydrolysis and release in monostable rhodopsins or the ability to isomerize again in bistable rhodopsins). The counterion, a conserved negative charge in the ligand pocket, is fundamental for the stabilization of the charge distribution in this region. In monostable rhodopsins, such as bovine rhodopsin, the role of counterion in the inactive state is performed by the proximal Glu-1133.28. Additionally, in bovine rhodopsin a highly conserved “glutamate cage” of tyrosines (Tyr-19145.54, Tyr19245.55, and Tyr-2686.51) restrains the conformation of the Glu-18145.44 side chain. As it is the case in other bistable rhodopsins, the proximal counterion position of JSiR1 is occupied by a conserved tyrosine (Tyr-1263.28) and the role of the counterion is instead played by the distal Glu-19445.44 (2–4, 35). In our crystal structure, Glu-19445.44 is too far from the Schiff base (Glu-19445.44[Cδ]–Lys-3217.43[NΖ] = 7.6 Å) for a direct salt bridge (Fig. 4). Instead, the Schiff base and Glu-19445.44 are linked by a water-mediated hydrogen bond network. Interestingly, the adjacent Ser-19945.49 is also involved in this network. A similar architecture of this extended Schiff base/counterion system is seen in bovine rhodopsin, while squid rhodopsin has an asparagine in the place of serine (Fig. 4). In JSiR1, Glu-19445.44 also interacts with a nearby “glutamate cage” formed in this case by only 2 residues, Tyr-2936.51 and Tyr-20445.54. A structure-based sequence analysis (31) of monostable and bistable rhodopsins reveals a connection between the nature of the residue at position 3.28 and the glutamate cage. Receptors with a proximal counterion (i.e., a glutamate at position 45.44) have an additional tyrosine at the 45.55 position in the second extracellular loop (SI Appendix, Fig. S5), oriented toward Glu-18145.44 in bovine rhodopsin, while Tyr-19145.54 is oriented toward the C19 of retinal and is involved in its deprotonation (5). Interestingly, position 45.55 is not a tyrosine in bistable rhodopsins. In JSiR1, Tyr-20445.54 appears to take on the role of Tyr-19245.55 and orients toward Glu-18145.44 and away from retinal (Fig. 4 and SI Appendix, Fig. S8A).
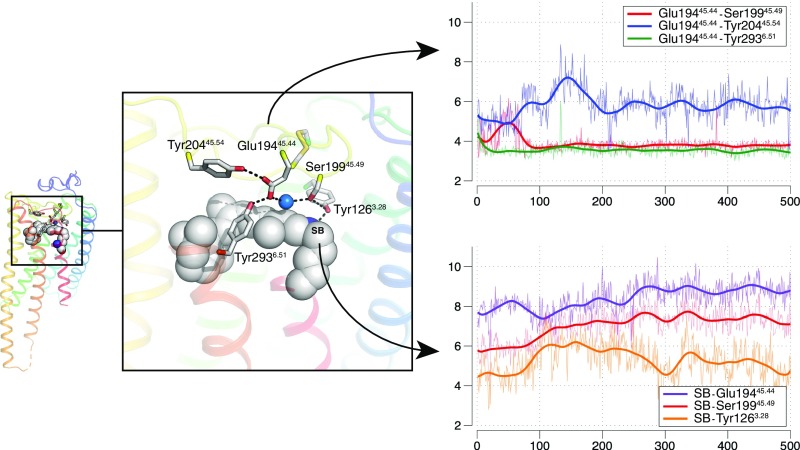
To assess the dynamic nature of this extended water-mediated hydrogen bond network around the Schiff base, we performed molecular dynamics simulations of JSiR1 embedded in a lipid bilayer (Fig. 5; see Methods). Our results indicate that the counterion Glu-19445.44 can maintain stable direct hydrogen bonds with Ser-19945.49 and Tyr-2936.51 (Fig. 5B, red and green traces) and indirect water-mediated interactions with Tyr-20445.54 (Fig. 5B, blue trace). Thus, Ser-19945.49, Tyr-2936.51, and Tyr-20445.54 restrain the conformation of the Glu-19445.44 side chain. The Schiff base itself is strongly solvated (4–5 water molecules during the simulation, see SI Appendix, Fig. S6A) and forms indirect water-mediated interactions with Glu-19445.44, Ser-19945.49, and Tyr-1263.28 in the extended counterion system (Fig. 5C). In summary, our simulations show that transiently ordered water molecules around the Schiff base, in particular in the triad Glu-19445.44–water–Ser-19945.49, are involved in key polar interactions that are likely to play a functional role in regulating the protonation state of the Schiff base (SI Appendix, Fig. S8A).
Fig. 5.
Dynamics of the extended water-mediated hydrogen bond network around the Schiff base and Glu-19445.44. The retinal-binding pocket of JSiR1 indicates an extended Schiff base/counterion system. The Top Left graph depicts temporal evolution of distances between pairs of residues involving Glu-19445.44. In our simulations, the interaction to Tyr-20445.54 is mediated by a bridging water molecule. The Bottom Left graph depicts temporal evolution of distances between the Schiff base (SB) and nearby residues. In our simulations, all these interactions are mediated by water molecules.
The Transmembrane Hydrogen Bonding Network.
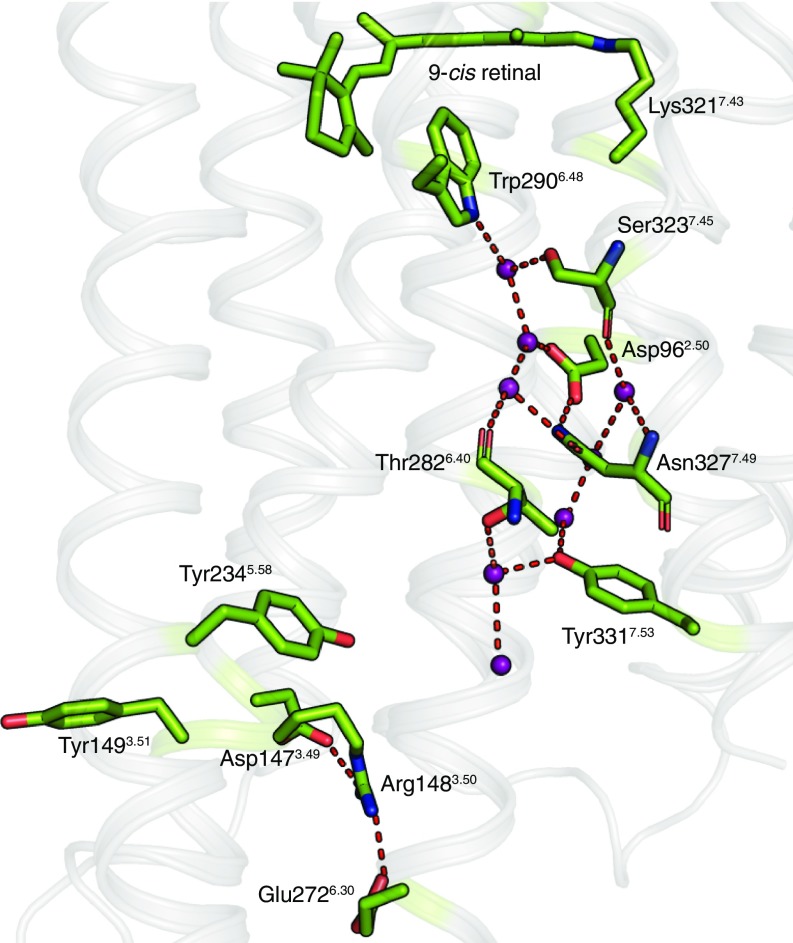
Our structure also reveals the presence of a set of highly ordered and coordinated waters forming a polar network from the retinal-binding site to the G protein-binding site (Fig. 6). In class A GPCRs—including JSR1—the highly conserved Trp2906.48 acts as a hydrophobic gate that separates the ligand-binding pocket and the receptor core between TM2, TM3, TM6, and TM7 (17, 36). Similar to other rhodopsins, JSR1 does not have a Na+-binding site in this region, which has been observed in some other class A GPCRs (17). Instead, a trail of waters connect the polar residues around this pocket: Asp-962.50 in TM2 to Ser-3237.45, Asn-3277.49, and Tyr-3317.53 in the (S/N)xxxNPxxY motif of TM7. These residues are highly conserved in class A GPCRs (17) and are involved in structural rearrangements that lead to receptor activation (37). In addition to hydration of this interhelical interface, a few specific waters have a structural role, such as stabilizing a 310 helical segment in TM7.
Fig. 6.
Water trail in the transmembrane region. A network of water-mediated interactions (purple spheres and red dashes) connect the ligand-binding pocket to the G protein-binding site in JSiR1 (green, 6I9K; ref. 63). The “locked” conformation of the DRY motif indicates the inactive conformation of the receptor. Interestingly, Arg-1483.50 is held in place by both Asp-1473.49 (through an intrahelical salt bridge) and Glu-2726.30 (interhelical salt bridge).
Interestingly, there are specific characteristics around the G protein-binding site of JSR1 that resemble those of ligand-activated class A GPCRs (Fig. 7 and SI Appendix, Fig. S7). For instance, Tyr-2345.58 in TM5 and Tyr-3317.53 in the (S/N)xxxNPxxY motif of TM7 face toward the transmembrane core (Fig. 7A)—in this last case, with the possible aid of 2 linking waters. This conformation contrasts with the ground state/inactive bovine rhodopsin, where the side chains of the equivalent residues orient away from the receptor core and toward the surrounding membrane. Thus, we conclude that the transmembrane region of JSR1 differs in a few key aspects from that of the monostable bovine rhodopsin and resembles instead the inactive state structures of the majority of ligand-binding class A GPCRs.
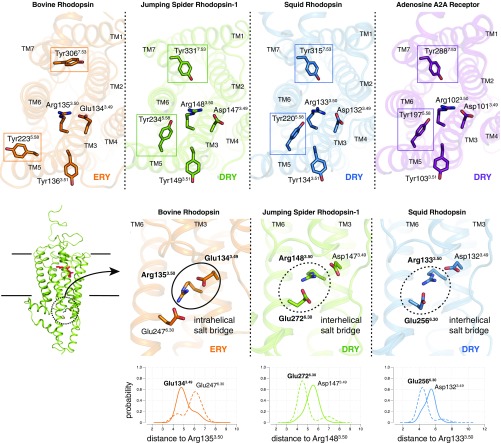
Fig. 7.
Bistable receptors share common structural features with ligand-binding class A GPCRs. The Top represents comparisons between bovine rhodopsin (1GZM, orange) 724 (28), JSiR1 (6I9K, green; ref. 63), squid rhodopsin (2Z73, blue) (11), and the adenosine A2A receptor (5OM4, purple) (26) indicating structural similarities of bistable rhodopsins to class A GPCRs. First, the conserved tyrosines on TM5 (5.58) and TM7 [7.53, in the (S/N)xxxNPxxY motif] orient toward the DRY motif on JSiR1, squid rhodopsin, and the adenosine A2A receptor; however, in bovine rhodopsin these tyrosines orient away from the transmembrane bundle. Second, bistable rhodopsins have an aspartate at position 3.49, forming part of the DRY observed in a majority of class A GPCRs, whereas bovine rhodopsin holds a glutamate at the same position. The Bottom depicts comparisons between the extended ERY/DRY motifs (i.e., including the nearby glutamate at position 6.30) of bovine, spider, and squid rhodopsin. Molecular dynamics simulations (see Methods) were used to estimate the relative 729 populations of the intrahelical (Glu/Asp-3.49–Arg-3.50) and interhelical (Glu-6.30–Arg-3.50) salt bridges.
Furthermore, JSR1 contains a DRY motif at the cytoplasmic side of TM3, which is highly conserved in ligand-binding class A GPCRs (17, 36), in contrast to the ERY motif present in vertebrate visual opsins (Fig. 7B). In a few class A GPCR subfamilies (e.g., amine, prostanoid, adenosine receptors, and opsins), the (D/E)RY motif is part of a more extensive polar network involving Glu-2726.30 in TM6. Similarly, in JSR1 Arg-1483.50 is held in its warped inactive conformation by Asp-1473.49 and Glu-2726.30 (Fig. 7 A and B). This double ionic lock (36, 38), with both intra- and interhelical components, is involved in the stabilization of the inactive state of the receptor. Our simulations allowed us to estimate the dynamics of these interactions in the context of a solvated bilayer. Interestingly, we observe that in JSR1 the interhelical salt bridge (Arg-1483.50–Glu-2726.30) is more stable than the intrahelical component (the “canonical” ionic lock between Arg-1483.50 and Asp-1473.49) (Fig. 7B), which is weakened by the large solvation of the negatively charged aspartate (SI Appendix, Fig. S6B).
Discussion
In rhodopsins, the retinal chromophore is covalently bound to a lysine in the ligand pocket via a protonated Schiff base that is stabilized by a negative counterion, usually a nearby glutamate (4). Photon absorption triggers retinal isomerization and drives subsequent structural changes that lead to receptor activation (i.e., formation of the Meta states). Monostable rhodopsins, e.g., bovine rhodopsin, differ from bistable rhodopsins, e.g., squid and jumping spider rhodopsins, as well as the vertebrate melanopsin involved in the circadian clock regulation, by having a unidirectional isomerization, followed by deprotonation and hydrolysis of the Schiff base. Conversely, in bistable rhodopsins a second photon reisomerizes the retinal while the Schiff base remains protonated throughout the photocycle (8).
Our crystal structure of jumping spider rhodopsin-1 (JSR1) bound to 9-cis retinal sheds light on the molecular architecture of the inactive state of bistable rhodopsins. Analysis of the retinal-binding pocket indicates that the stabilization of the protonated Schiff base involves a water-mediated hydrogen bond network including Ser-19945.49 (Figs. 4 and 5). Our structural data explain recent mutational studies on JSR1 and highlight the role of Glu-19445.44 and Ser-19945.49 as part of a complex hydrogen bond-based counterion system (39).
The retinal-binding pocket architectures of JSR1 and bovine rhodopsin are similar, where the “Glu–water–Ser” triad sits in proximity to the protonated Schiff base. In JSR1, a distal counterion (Glu-19445.44) stabilizes the protonated Schiff base through the above-mentioned water-mediated network. Interestingly, this position (45.44) also serves as a counterion during bovine rhodopsin activation (in the Meta I state) (5, 9). However, 2 main structural elements differentiate the 2 rhodopsin systems. Firstly, in bovine rhodopsin the protonated Schiff base is maintained by the proximal counterion Glu-1133.28 through a direct ionic interaction (28). Conversely, in JSR1 this position is held by a neutral and highly conserved tyrosine (Fig. 4). Secondly, compared with bovine rhodopsin, the “glutamate cage” of JSR1 lacks 1 of the 3 tyrosines (at position 45.55), allowing for a higher flexibility of its counterion Glu-19445.44. Finally, while JSR1 and squid rhodopsin share a similar hydrogen-bond network, key differences set it apart. In squid rhodopsin, position 45.49 is occupied by Asn-18545.49 instead of the Ser45.49 observed in JSR1 and bovine rhodopsin. Furthermore, the squid rhodopsin structures did not reveal the presence of a water molecule in this region (12, 13), possibly implying that the NH2 group of the asparagine functions as a surrogate for the “water–Ser” duo seen in JSR1. However, it is also possible that a mobile water molecule mediates the hydrogen bond network in this region, as suggested by FTIR data (11).
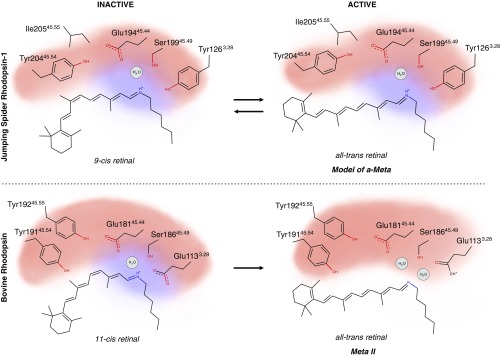
We propose that one of the major differences between mono- and bistable rhodopsins lies in the distribution of charge around the Schiff base (Fig. 8). In the monostable bovine rhodopsin, the positive charge of the Schiff base is more localized due to the presence of the proximal Glu-1133.28, while the distal Glu-18145.44 is trapped by the “glutamate polar cage.” Upon photoactivation, the positive charge is ultimately transferred onto Glu-1133.28, allowing for the subsequent hydrolysis of the deprotonated Schiff base. A structure-based sequence analysis suggests that vertebrate monostable rhodopsins may require a stronger polar cage where 3 tyrosines engage Glu-18145.44 (SI Appendix, Fig. S5). On the other hand, in JSR1, Tyr-1263.28 cannot assist in the deprotonation of the Schiff base upon illumination, and the members of the extended counterion system maintain their protonation state. This is supported by our spectroscopic analyses on JSR1, which confirm the protonated state of the Schiff base throughout the whole photocycle (8). The existence of a similar water-mediated network around the extended Schiff base/counterion system may facilitate the bidirectional reaction in bistable rhodopsins (i.e., a second isomerization).
Fig. 8.
Comparison of activation models between different photosensitive GPCRs. The retinal binding pocket is represented schematically through its main components in JSR1 (Top) and bovine rhodopsin (Bottom). The red and blue clouds represent the distribution of the negative and positive charge, respectively. Upon illumination, the charge distribution changes due to the different physicochemical environment created by differing polar networks in the 2 receptors. The photoproduct of JSR1 is a schematic representation of a potential model.
The crystal structure of JSR1 indicates that its cytoplasmic transmembrane domain conforms to the general architecture of ligand-binding class A GPCRs rather than to bovine rhodopsin (16). Structural similarities can be observed in key residues such as Tyr-2235.58 on TM5 and Tyr-3317.53 on TM7 (S/N)xxxNPxxY motif), which resemble the A2A adenosine receptor (5OM4) (27) or the β-adrenergic receptor (2VT4) (40) rather than bovine rhodopsin (1GZM) (28) (Fig. 7). These residues point toward the transmembrane core of the receptor and are involved in the water-mediated hydrogen bond network in the G protein-binding site. This architecture contrasts to that observed in the inactive conformation of bovine rhodopsin where Tyr-3067.53 and Tyr-2235.58 are oriented away from the receptor core and toward the membrane (28, 41). In both scenarios, these residues reposition themselves upon activation to adopt the general architecture of the G protein-binding site in class A GPCRs (41–43). Hence, we suggest that specific transmembrane structural elements of JSR1 are positioned into an activation-ready conformation, as observed in the bistable squid rhodopsin and in ligand-binding class A GPCRs (Fig. 7 and SI Appendix, Fig. S7). Finally, we hypothesize that the malleable water-mediated networks observed in the binding pocket as well as in the transmembrane domain contribute together to the unique functionality of bistable rhodopsins, allowing them to stably switch between the ground state and the active photoproduct. Thus, internal water-mediated hydrogen bond networks are an important factor in shaping the conformational landscape of GPCR activation, providing an additional mechanism to regulate functional aspects such as basal activity and signaling efficacy.
Methods
Expression and Purification.
Wild-type jumping spider rhodopsin isoform-1 (JSR1) from Hasarius adansoni was recombinantly expressed in suspension cultures of HEK293 GnTI− cells as described elsewhere (8). Harvested cells were pelleted and stored at −80 °C before purification. Frozen cell pellets were thawed and mechanically lysed in buffer A (50 mM Hepes pH 6.5, 150 mM NaCl, 3 mM MgCl2, cOmplete EDTA-free Protease Inhibitor Mixture tablets; Roche) using a handheld dounce homogenizer. All steps from this point onwards were carried out under 640 nm (dim red) light. The cell lysate was incubated with 30 µM 9-cis retinal (98% grade, Sigma-Aldrich) overnight at 4 °C (identical procedures were followed for the 11-cis retinal sample). This suspension was initially centrifuged at 500 × g for 10 min at 4 °C separating the membrane faction (supernatant) from the cell debris pellet. To ensure maximum recovery of membrane factions the pellet was resuspended in buffer A, homogenized, and spun down at 500 × g for 10 min at 4 °C. The supernatants were combined for the following separation of membrane fractions (repeated thrice): first, the new supernatant was spun down at 100,000 × g for 40 min; second, the resultant pellet was resuspended in buffer B (50 mM Hepes pH 6.5, 150 mM NaCl, 3 mM MgCl2) using an Ultra-Turrax (IKA). The final membrane fraction was solubilized using 19.5 mM with n-dodecyl-β-d-maltopyranoside (DDM-Anagrade; Anatrace) and incubated for 2 h at 4 °C. The solubilized membrane suspension was centrifuged at 100,000 × g for 50 min after which the supernatant was mixed with CNBr-activated Sepharose 4B resin (GE Healthcare Life Science) conjugated with anti-1D4 antibody and incubated overnight at 4 °C. The resin was collected in a BioRad Glass Econo-Column and washed with buffer C (50 mM Hepes pH 6.5, 150 mM NaCl, 3 mM MgCl2, 0.195 mM DDM). The sample was eluted overnight at 4 °C in buffer D (50 mM Hepes pH 6.5, 150 mM NaCl, 3 mM MgCl2, 0.195 mM DDM, 0.8 mM 1D4 peptide). Concavalin A resin (GE Healthcare Life Science) was preactivated with buffer E (50 mM Hepes pH 6.5, 150 mM NaCl, 3 mM MgCl2, 0.195 mM DDM, 1 mM CaCl2, 1 mM MnCl2). The collected protein sample was then treated with 1 mM CaCl2 and 1 mM MnCl2 and incubated with the preactivated Concavalin A resin for 30 min. The incubated resin was collected and packed into a 5-mL cartridge column (Qiagen) and washed with buffer E using an Äkta FPLC system (GE Healthcare Life Science). A linear gradient of buffer F (50 mM Hepes pH 6.5, 150 mM NaCl, 3 mM MgCl2, 0.195 mM DDM, 1 mM CaCl2, 1 mM MnCl2, 500 mM methyl-α-d-mannopyranoside; Sigma-Aldrich) was used for elution. This concavalin A-based affinity chromatography step was found to be necessary to ensure that misfolded/nonglycosylated receptor was removed. The eluted protein was assessed and quantified using UV-vis spectroscopy and SDS/PAGE. The sample was finally treated with 1/500× Endoglycosidase H (2.5 U stock; Roche Diagnostics) and incubated at 4 °C overnight.
UV-Visible Spectroscopy.
UV-vis protein spectra were recorded in a spectrophotometer (UV-2401PC, Shimadzu), against the appropriate buffer. Purity was assessed using the OD λmax 280/505 nm ratio, which yielded a value between 2.8 and 3.5. Protein concentration was calculated by Lambert–Beer law, where εJSiR1 is ∼32.660 M−1⋅cm−1 and the molecular weight was calculated from the protein sequence as 44 kDa (8). Illumination was carried out in the dark with 495 nm (short-pass filter) mounted on a projector lamp at maximum intensity for 10 min (lamp parameters 150 W, 2A, 50/60 Hz, 220–240 V, irradiance 5 W/cm2). Spectral characterization of JSR1 and JSiR1 reconstituted LCP was performed in a cuvette with a 0.05-cm path. In this case, buffer H (see below Crystallization) reconstituted in LCP was used as the baseline.
Thermostability Assay by CPM Fluorescence.
JSiR1 in buffer C was diluted to a final concentration of 0.01 mg/mL in detergents with varied chemical properties: n-decyl-β-d-maltopyranoside (DM), n-dodecyl-β-d-maltopyranoside (DDM), 2,2- dihexylpropane-1,3-bis-β-d-glucopyranoside (OGNG), 2,2-didecylpropane-1,3-bis-β-d-maltopyranoside (LMNG), n-nonyl-β-d-glucoside (b-NG), n-octyl-β-d-glucoside (b-OG), n-octyl-β-d-thioglucoside (OTG), 6-cyclohexyl-1-hexyl-β-d-maltoside (Cymal-6), and nonanoyl-N-methylglucamide (MEGA-9). The detergents were used at 5 times critical micelle concentration (CMC), and samples were incubated overnight at 4 °C to facilitate maximum detergent micelle exchange. The thermostable engineered β1-adrenergic receptor in complex with 100 mM cyanopindolol (44) in 2× CMC DM was used as control in addition to purified JSiR1 in 1× CMC DDM. Similarly, JSiR1 was diluted in diverse pH conditions of buffer C: pH from 5 to 6.5 with MES, 7–8 Hepes, 8–10 in CHES buffers. All reaction mixtures were supplemented with 1 μL of 0.15 mg/mL of CPM fluorophore dye (n-[4-(7-diethylamino- 4-methyl-3-coumarinyl)phenyl]maleimide) (26) in a final volume of 20 μL. A Rotor-Gene Q qPCR machine (Qiagen) was used to ramp the temperature from 25 °C to 95 °C during which the receptor starts to unfold, allowing the CPM dye to bind to buried cysteines. This binding results in an increase in fluorescence which was recorded as a function of temperature. The resultant sigmoidal curves were fitted by Boltzmann equation and a melting temperature (Tm) was obtained in GraphPad Prism.
G Protein-Based Activity Assay.
Heterotrimeric G protein was assembled using recombinant Gαi and βγ dimer from bovine retina in the presence of buffer G (50 mM Bis-Tris, pH 7.3, 130 mM NaCl, 1 mM MgCl2, 1 mM DTT) as shown by Maeda et al. (25). A total of 50 nM JSiR1 was incubated in a 1-mL quartz cuvette (path length 1 cm), with 600 nM heterotrimeric Gi in the dark for 2 min in buffer H (50 mM Bis-Tris, pH 7.3, 130 mM NaCl, 1 mM MgCl2, 1 mM DTT, 0.196 mM DDM) on ice. This mixture was irradiated with 495 nm for 3 min so as to form a JSiR1/Gi complex. The cuvette was returned to the dark (on ice) for 2 additional minutes after which a baseline fluorescence was recorded using a Cary Eclispse Fluorescence Spectophotometer (Agilent) with a stirring cell chamber (excitation [ex.] wavelength: 295 nm; emission [em.] wavelength: 340 nm; ex. slit width: 5; em. slit width: 20; average time 1 s; 20 °C). The reading was paused and 10 μM of GTPγS was added to the mixture after which data acquisition was continued for 1.5 h. Data were analyzed using a 1 phase association fit (Y = Y0 + (Ymax-Y0)*[1-exp(-K*x)]) to obtain a rate constant. All statistical analyses were performed on GraphPad Prism (25).
Crystallization.
Deglycosylated JSiR1 was concentrated using a 50-kDa Amicon Ultra concentrator (Merck Millipore) at 1,363 × g to facilitate buffer exchange into buffer H (50 mM Hepes pH 6.5, 150 mM NaCl, 3 mM MgCl2, 0.195 mM DDM, 1 mM CaCl2, 1 mM MnCl2, 280 mM methyl α-d-mannopyranoside) on a PD-10 Desalting Column (GE Healthcare Life Science). The eluent was further concentrated using a 100-kDa Amicon Ultra concentrator at 6,797 × g for 10 min per run. Final concentrations between 20 and 30 mg/mL were used for crystallization. Concentrated JSiR1 was mixed with monoolein (9.9 MAG, Nu-check) in a 40:60 ratio to form LCP at room temperature (45) under a 640-nn LED lamp. This mixture was used to screen for crystal conditions using a mosquito LCP robot (TTP Labtech) at 4 °C using 100 µm glass on glass Laminex plates (Molecular Dimensions). The plates were then incubated at 20 °C in the dark. Successful initial hits were further optimized. Diffracting crystals were obtained from 31% to 36% PEG400 and 100 mM Bis-Tris pH 6.5.
Data Collection and Processing.
Diffraction data were collected at 100 K using a wavelength of 1,000 Å on a 16 M Eiger detector at the ×06SA (PX1) beamline, Swiss Light Source (SLS, PSI). Collected datasets were individually indexed and integrated using XDS (46) (Table 1). Two datasets were merged and scaled with Aimless (47) (CCP4 suite) (48, 49). Merging statistics indicated that the overall resolution distribution was anisotropic with resolution leading up to 2.1 Å along the b* reciprocal axis at a CC1/2 of 0.3. To include these regions, and to exclude background noise, the data were reprocessed to obtain unmerged reflections [Pointless (50, 51) and CCP4 suite] and further run on the STARANISO server (52, 53). This resulted in an elliptically truncated, scaled, and merged dataset [performed by Aimless (47) on the STARANISO server]. It has to be noted that the elliptical truncation reduces the completeness in the low-resolution shell although the CC1/2 is maintained (0.537). Before truncation, the data appeared to be ∼90% complete in the low-resolution shell. The final resolution limits along the 3 reciprocal axes were identified to be 2.5 Å (0.525 a* − 0.851 c*), 2.1 Å (b*), and 2.4 Å (2.366 a* + 0.810 c*).
Table 1.
Crystallographic table
| Protein | JSiR1 |
| Data collection | |
| Space group | I 121 |
| Cell dimensions | |
| a, b, c (Å) | 50.95, 130.63, 77.30 |
| α, β, γ (°) | 90, 100.55, 90 |
| Resolution (Å) | 46.78–2.145 (2.39–2.145) |
| Ellipsoidal high-resolution limit | 2.1, 2.3, 2.6 |
| Rmerge | 0.142 (4.12) |
| Rpim | 0.092 (0.693) |
| CC1/2 | 0.995 (0.537) |
| I/Sigma | 7.2 (1.4) |
| Completeness (%) | 70.5 (12.9) (spherical) 90.6 (51.7) (ellipsoidal) |
| Multiplicity | 6.1 (3.5) |
| Refinement | |
| Resolution (Å) | 46.78–2.145 |
| No. reflections | 18,977 |
| Rwork/Rfree | 0.215/0.255 |
| No. atoms | |
| Protein | 2,417 |
| Ligand/ion | 20 (9-cis retinal)/133 (OLC) |
| Water | 54 |
| B-factors | |
| Protein | 38.5 |
| Ligand/ion | 36.46 (9-cis retinal)/51.7 (OLC) |
| Water | 39.75 |
| R.M.S. deviations | |
| Bond lengths (Å) | 0.008 |
| Bond angles (°) | 1.010 |
| Ramachandran favored | 97.7% |
| Ramachandran accepted | 2.3% |
| Ramachandran outliers | 0% |
Phasing and Refinement.
Phase information was obtained via molecular replacement with Phaser-MR (54) (Phenix, version 1.13–2998) (55). An apomonomer of squid rhodopsin (PDB ID code 2Z73) (12) was used as the search model. A preliminary model was generated from the output of the molecular replacement model by manual amino acid substitution from squid rhodopsin to that of JSR1 using Coot (56, 57). This model served as a basis for the Autobuild (58) (Phenix) which generated the initial apomodel of JSiR1, for the resulting model was corrected manually in Coot (56, 57). Iterative refinement was carried out in phenix.refine (59) (Phenix). Toward the final stages of model refinement, the option to optimize weightage between experimental electron data and stereochemistry was enabled. A large unknown density (in both 2Fo-Fc and Fo-Fc) was observed to be anchored to His-244 near ICL3. The N-terminal region, parts of ICL3 and the C terminus appeared to be highly disordered and hence were not modeled due to lack of electron density. Polder maps (60) were generated to ascertain the quality of the electron density in the ligand-binding pocket (SI Appendix, Fig. S4). Structure validation was carried out by the MolProbity server (61, 62), showing 97.70% of residues in the Ramachandran favored region with 2.3% in the allowed regions and 0% outliers. Rotamer outliers were observed to be at 1.98%. The JSR1 structure was deposited with the PDB ID code 6I9K (63).
Structural Modeling.
To add the missing residues in ICL3 and helix 8 and C-terminal of JSiR1, homology modeling was performed using the crystal structure of squid rhodopsin (PDB ID code 2Z73) (12) as a template. First, the sequences of JSR1 (UniProt ID code B1B1U5) and squid rhodopsin (UniProt ID code P31356; ∼38.14% sequence identity) were aligned using MUSCLE (64). This initial alignment was manually refined using Chimera (65) to adjust the gaps in the loop regions. Using this alignment and the squid rhodopsin template, a 3D model of JSiR1 was built using the multitemplate modeling method of Modeler v9.14 (66). All models were subjected to 300 iterations of variable target function method optimization and thorough molecular dynamics and simulated annealing optimization, and scored using the discrete optimized protein energy potential. The 20 best-scoring models were analyzed visually, and a suitable model (in terms of low score and structure of the loops) was selected for the next step. We added to this preliminary model the crystallographic waters resolved within the transmembrane bundle of JSiR1 crystal structure. We then used PROPKA at pH 7.0 as implemented in PDB ID code 2PQR (67) to determine protonation states of titratable groups, to add hydrogens to the structure, and to optimize the hydrogen bond network. The counterion Glu-19445.44 was kept charged and disulphide bond between Cys-1233.25 and Cys-20045.50 was retained. The model was superimposed according to Orientation of Proteins in Membranes (OPM) (68) coordinates for proper assignment of membrane boundaries. The model was embedded in a preequilibrated lipid bilayer consisting of 137 molecules of 1-palmitoyl-2-oleoyl-sn-glycerol-3-phosphatidylcholine (POPC), which was hydrated with a layer of approximately 45 Å of water molecules on each side. Sodium and chloride ions were added to a concentration of 150 mM NaCl, and then additional ions were added to achieve charge neutrality. The dimensions of the final tetragonal box were ∼90 × 90 × 144 Å, containing a total of ∼68,000 atoms. This system was equilibrated as follows: first a short (0.5 ns) simulation was performed in which only the lipid tails were allowed to move, to induce the appropriate disorder of a fluid-like bilayer. Then, the geometry of the entire system was optimized by 1,000 steps of energy minimization, followed by 2 equilibration steps with the protein constrained (0.5 ns) and without constraints (0.5 ns). This equilibrated system was used as a starting point to perform unrestrained molecular dynamics simulations. All simulations were carried out with NAMD 2.10 (69) with the c36 CHARMM force field (70) in the NPT ensemble, using Langevin dynamics to control temperature at 300 K, and with a time step of 2 fs, while constraining all bonds between hydrogen and heavy atoms. Simulations were carried out on a Euler supercomputer (ETH Zürich). The above molecular dynamics setup was used also for bovine rhodopsin. Highly resolved crystallographic waters of mutant bovine rhodopsin (5DYS) (71) was added to the structure of bovine rhodopsin (1GZM) (28). Analysis of the simulation trajectories were done using VMD (72) and MD analysis (73) toolkit.
Sequence Conservation Analysis.
We measured the sequence variability at N-terminal and “ERY” motif positions in an alignment of ∼900 sequences from a recent large-scale analysis of opsin evolution (74), in which the 26 outgroups had been removed. The sequence variability was created using Berkeley WebLogo3 (75).
Supplementary Material
Acknowledgments
We thank Dr. Pikyee Ma, Thomas Gruhl, and Petr Skopintsev for crystallization inputs. We also thank Dr. Ching-Ju Tsai for providing G protein components. We thank the MX group (Swiss Light Source, PSI facility) for support during beamtimes and the Laboratory of Biomolecular Research (PSI) for overall project support. This project was funded by the following agencies: National Centres of Competence in Research Molecular Systems Engineering 2015–2017 and Swiss National Science Foundation for Grant 173335 (to G.F.X.S.); Swiss National Science Foundation for Grant PZ00P3_174169 (to P.N.); X-Probe ITN 637295 (to N.V.); Peter und Traudl Engelhorn Stiftung (E.L.); European Community’s Seventh Framework Program (FP7/2007-2013) under Grant Agreement 290605 (COFUND: PSI-FELLOW) (to E.M.); the Swiss Nanoscience Institute-Nanoargovia Grant A13.12 (to J.M.); Japanese Ministry of Education, Culture, Sports, Science and Technology Grants-in-Aid for Scientific Research 15H05777; and Technology Agency (JST) Core Research for Evolutional Science and Technology Grant JPMJCR1753 (to A.T.).
Footnotes
Conflict of interest statement: G.F.X.S. declares that he is a cofounder and scientific advisor of the companies leadXpro AG and InterAx Biotech AG.
This article is a PNAS Direct Submission.
Data deposition: The atomic coordinates and structure factors have been deposited in the Protein Data Bank, www.wwpdb.org (PBD ID 6I9K).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1902192116/-/DCSupplemental.
References
- 1.Terakita A., Kawano-Yamashita E., Koyanagi M., Evolution and diversity of opsins. Wiley Interdiscip. Rev. Membr. Transp. Signal. 1, 104–111 (2012). [Google Scholar]
- 2.Sakmar T. P., Franke R. R., Khorana H. G., Glutamic acid-113 serves as the retinylidene Schiff base counterion in bovine rhodopsin. Proc. Natl. Acad. Sci. U.S.A. 86, 8309–8313 (1989). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sakmar T. P., Franke R. R., Khorana H. G., The role of the retinylidene Schiff base counterion in rhodopsin in determining wavelength absorbance and Schiff base pKa. Proc. Natl. Acad. Sci. U.S.A. 88, 3079–3083 (1991). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Terakita A., et al. , Counterion displacement in the molecular evolution of the rhodopsin family. Nat. Struct. Mol. Biol. 11, 284–289 (2004). [DOI] [PubMed] [Google Scholar]
- 5.Kimata N., et al. , Retinal orientation and interactions in rhodopsin reveal a two-stage trigger mechanism for activation. Nat. Commun. 7, 12683 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Palczewski K., G protein-coupled receptor rhodopsin. Annu. Rev. Biochem. 75, 743–767 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Tsukamoto H., Terakita A., Diversity and functional properties of bistable pigments. Photochem. Photobiol. Sci. 9, 1435–1443 (2010). [DOI] [PubMed] [Google Scholar]
- 8.Ehrenberg D., et al. , The two-photon reversible reaction of the bistable jumping spider rhodopsin-1. Biophys. J. 116, 1248–1258 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yan E. C. Y., et al. , Retinal counterion switch in the photoactivation of the G protein-coupled receptor rhodopsin. Proc. Natl. Acad. Sci. U.S.A. 100, 9262–9267 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Isberg V., et al. , Generic GPCR residue numbers–Aligning topology maps while minding the gaps. Trends Pharmacol. Sci. 36, 22–31 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ota T., Furutani Y., Terakita A., Shichida Y., Kandori H., Structural changes in the Schiff base region of squid rhodopsin upon photoisomerization studied by low-temperature FTIR spectroscopy. Biochemistry 45, 2845–2851 (2006). [DOI] [PubMed] [Google Scholar]
- 12.Murakami M., Kouyama T., Crystal structure of squid rhodopsin. Nature 453, 363–367 (2008). [DOI] [PubMed] [Google Scholar]
- 13.Shimamura T., et al. , Crystal structure of squid rhodopsin with intracellularly extended cytoplasmic region. J. Biol. Chem. 283, 17753–17756 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Angel T. E., Chance M. R., Palczewski K., Conserved waters mediate structural and functional activation of family A (rhodopsin-like) G protein-coupled receptors. Proc. Natl. Acad. Sci. U.S.A. 106, 8555–8560 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Flock T., et al. , Selectivity determinants of GPCR-G-protein binding. Nature 545, 317–322 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Deupi X., Relevance of rhodopsin studies for GPCR activation. Biochim. Biophys. Acta 1837, 674–682 (2014). [DOI] [PubMed] [Google Scholar]
- 17.Venkatakrishnan A. J., et al. , Molecular signatures of G-protein-coupled receptors. Nature 494, 185–194 (2013). [DOI] [PubMed] [Google Scholar]
- 18.Deisseroth K., Optogenetics: 10 years of microbial opsins in neuroscience. Nat. Neurosci. 18, 1213–1225 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ye H., Daoud-El Baba M., Peng R. W., Fussenegger M., A synthetic optogenetic transcription device enhances blood-glucose homeostasis in Mice. Science 332, 1565–1568 (2011). [DOI] [PubMed] [Google Scholar]
- 20.Koyanagi M., Terakita A., Diversity of animal opsin-based pigments and their optogenetic potential. Biochim. Biophys. Acta 1837, 710–716 (2014). [DOI] [PubMed] [Google Scholar]
- 21.Reeves P. J., Callewaert N., Contreras R., Khorana H. G., Structure and function in rhodopsin: High-level expression of rhodopsin with restricted and homogeneous N-glycosylation by a tetracycline-inducible N-acetylglucosaminyltransferase I-negative HEK293S stable mammalian cell line. Proc. Natl. Acad. Sci. U.S.A. 99, 13419–13424 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Nagata T., et al. , Depth perception from image defocus in a jumping spider. Science 335, 469–471 (2012). [DOI] [PubMed] [Google Scholar]
- 23.Faurobert E., Otto-Bruc A., Chardin P., Chabre M., Tryptophan W207 in transducin T alpha is the fluorescence sensor of the G protein activation switch and is involved in the effector binding. EMBO J. 12, 4191–4198 (1993). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Muller R. E., Siderovski D. P., Kimple A. J., Carolina N., Hill C., Using intrinsic tryptophan fluorescence to measure heterotrimeric G-protein activation. BMG Labtech 7, AN196 (2009). [Google Scholar]
- 25.Maeda S., et al. , Crystallization scale preparation of a stable GPCR signaling complex between constitutively active rhodopsin and G-protein. PLoS One 9, e98714 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Alexandrov A. I., Mileni M., Chien E. Y. T., Hanson M. A., Stevens R. C., Microscale fluorescent thermal stability assay for membrane proteins. Structure 16, 351–359 (2008). [DOI] [PubMed] [Google Scholar]
- 27.Rucktooa P., et al. , Towards high throughput GPCR crystallography: In Meso soaking of Adenosine A2A receptor crystals. Sci. Rep. 8, 41 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Li J., Edwards P. C., Burghammer M., Villa C., Schertler G. F. X., Structure of bovine rhodopsin in a trigonal crystal form. J. Mol. Biol. 343, 1409–1438 (2004). [DOI] [PubMed] [Google Scholar]
- 29.Okada T., et al. , The retinal conformation and its environment in rhodopsin in light of a new 2.2 A crystal structure. J. Mol. Biol. 342, 571–583 (2004). [DOI] [PubMed] [Google Scholar]
- 30.van der Kant R., Vriend G., Alpha-bulges in G protein-coupled receptors. Int. J. Mol. Sci. 15, 7841–7864 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pándy-Szekeres G., et al. , GPCRdb in 2018: Adding GPCR structure models and ligands. Nucleic Acids Res. 46, D440–D446 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Saam J., Tajkhorshid E., Hayashi S., Schulten K., Molecular dynamics investigation of primary photoinduced events in the activation of rhodopsin. Biophys. J. 83, 3097–3112 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Nakamichi H., Okada T., X-ray crystallographic analysis of 9-cis-rhodopsin, a model analogue visual pigment. Photochem. Photobiol. 83, 232–235 (2007). [DOI] [PubMed] [Google Scholar]
- 34.Murakami M., Kouyama T., Crystallographic analysis of the primary photochemical reaction of squid rhodopsin. J. Mol. Biol. 413, 615–627 (2011). [DOI] [PubMed] [Google Scholar]
- 35.Birge R. R., Hubbard L. M., Molecular dynamics of cis-trans isomerization in rhodopsin. J. Am. Chem. Soc. 102, 2195–2205 (1980). [Google Scholar]
- 36.Rosenbaum D. M., Rasmussen S. G. F., Kobilka B. K., The structure and function of G-protein-coupled receptors. Nature 459, 356–363 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yuan S., Filipek S., Palczewski K., Vogel H., Activation of G-protein-coupled receptors correlates with the formation of a continuous internal water pathway. Nat. Commun. 5, 4733 (2014). [DOI] [PubMed] [Google Scholar]
- 38.Kobilka B. K., G protein coupled receptor structure and activation. Biochim. Biophys. Acta 1768, 794–807 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Nagata T., et al. , The counterion–Retinylidene Schiff base interaction of an invertebrate rhodopsin rearranges upon light activation. Commun. Biol. 2, 180 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Warne T., et al. , Structure of a β1-adrenergic G-protein-coupled receptor. Nature 454, 486–491 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Goncalves J. A., et al. , Highly conserved tyrosine stabilizes the active state of rhodopsin. Proc. Natl. Acad. Sci. U.S.A. 107, 19861–19866 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Deupi X., et al. , Stabilized G protein binding site in the structure of constitutively active metarhodopsin-II. Proc. Natl. Acad. Sci. U.S.A. 109, 119–124 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Choe H.-W., et al. , Crystal structure of metarhodopsin II. Nature 471, 651–655 (2011). [DOI] [PubMed] [Google Scholar]
- 44.Isogai S., et al. , Backbone NMR reveals allosteric signal transduction networks in the β1-adrenergic receptor. Nature 530, 237–241 (2016). [DOI] [PubMed] [Google Scholar]
- 45.Caffrey M., Cherezov V., Crystallizing membrane proteins using lipidic mesophases. Nat. Protoc. 4, 706–731 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kabsch W., XDS. Acta Crystallogr. D Biol. Crystallogr. 66, 125–132 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Evans P. R., Murshudov G. N., How good are my data and what is the resolution? Acta Crystallogr. D Biol. Crystallogr. 69, 1204–1214 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Winn M. D., et al. , Overview of the CCP4 suite and current developments. Acta Crystallogr. D Biol. Crystallogr. 67, 235–242 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Potterton E., Briggs P., Turkenburg M., Dodson E., A graphical user interface to the CCP4 program suite. Acta Crystallogr. D Biol. Crystallogr. 59, 1131–1137 (2003). [DOI] [PubMed] [Google Scholar]
- 50.Evans P. R., An introduction to data reduction: Space-group determination, scaling and intensity statistics. Acta Crystallogr. D Biol. Crystallogr. 67, 282–292 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Evans P., Scaling and assessment of data quality. Acta Crystallogr. D Biol. Crystallogr. 62, 72–82 (2006). [DOI] [PubMed] [Google Scholar]
- 52.Vonrhein C., et al. , Data processing and analysis with the autoPROC toolbox. Acta Crystallogr. D Biol. Crystallogr. 67, 293–302 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Tickle I. J., et al. , STARANISO (United Kingdom Glob Phasing Ltd, Cambrige, 2018). [Google Scholar]
- 54.McCoy A. J., et al. , Phaser crystallographic software. J. Appl. Cryst. 40, 658–674 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Adams P. D., et al. , PHENIX: A comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D Biol. Crystallogr. 66, 213–221 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Emsley P., Cowtan K., Coot: Model-building tools for molecular graphics. Acta Crystallogr. D Biol. Crystallogr. 60, 2126–2132 (2004). [DOI] [PubMed] [Google Scholar]
- 57.Emsley P., Lohkamp B., Scott W. G., Cowtan K., Features and development of Coot. Acta Crystallogr. D Biol. Crystallogr. 66, 486–501 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Terwilliger T. C., et al. , Iterative model building, structure refinement and density modification with the PHENIX AutoBuild wizard. Acta Crystallogr. D Biol. Crystallogr. 64, 61–69 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Afonine P. V., et al. , Towards automated crystallographic structure refinement with phenix.refine. Acta Crystallogr. D Biol. Crystallogr. 68, 352–367 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Liebschner D., et al. , Polder maps: Improving OMIT maps by excluding bulk solvent. Acta Crystallogr. D Struct Biol. 73, 148–157 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Chen V. B., et al. , MolProbity: All-atom structure validation for macromolecular crystallography. Acta Crystallogr. D Biol. Crystallogr. 66, 12–21 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Davis I. W., et al. , MolProbity: All-atom contacts and structure validation for proteins and nucleic acids. Nucleic Acids Res. 35, W375–W383(2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Schertler G., et al. , Crystal structure of jumping spider rhodopsin-1 bound to 9-cis retinal. Protein Data Bank. https://www.rcsb.org/structure/6I9K. Deposited 7 December 2018.
- 64.Edgar R. C., MUSCLE: Multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 32, 1792–1797 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Pettersen E. F., et al. , UCSF Chimera–A visualization system for exploratory research and analysis. J. Comput. Chem. 25, 1605–1612 (2004). [DOI] [PubMed] [Google Scholar]
- 66.Webb B., Sali A., Comparative protein structure modeling using MODELLER. Curr. Protoc. Bioinforma. 54, 5.6.1–5.6.37 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Dolinsky T. J., Nielsen J. E., McCammon J. A., Baker N. A., PDB2PQR: An automated pipeline for the setup of Poisson-Boltzmann electrostatics calculations. Nucleic Acids Res. 32, W665–W667 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Lomize M. A., Pogozheva I. D., Joo H., Mosberg H. I., Lomize A. L., OPM database and PPM web server: Resources for positioning of proteins in membranes. Nucleic Acids Res. 40, D370–D376 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Phillips J. C., et al. , Scalable molecular dynamics with NAMD. J. Comput. Chem. 26, 1781–1802 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Best R. B., et al. , Optimization of the additive CHARMM all-atom protein force field targeting improved sampling of the backbone φ, ψ and side-chain χ(1) and χ(2) dihedral angles. J. Chem. Theory Comput. 8, 3257–3273 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Singhal A., et al. , Structural role of the T94I rhodopsin mutation in congenital stationary night blindness. EMBO Rep. 17, 1431–1440 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Humphrey W., Dalke A., Schulten K., VMD: Visual molecular dynamics. J. Mol. Graph. 14, 33–38, 27-8 (1996). [DOI] [PubMed] [Google Scholar]
- 73.Michaud-Agrawal N., Denning E. J., Woolf T. B., Beckstein O., MDAnalysis: A toolkit for the analysis of molecular dynamics simulations. J. Comput. Chem. 32, 2319–2327 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Porter M. L., et al. , Shedding new light on opsin evolution. Proc. Biol. Sci. 279, 3–14 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Crooks G. E., Hon G., Chandonia J.-M., Brenner S. E., WebLogo: A sequence logo generator. Genome Res. 14, 1188–1190 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.