Significance
As one of the most important oncogenes, c-Myc is activated in over half of human cancers. The oncogenic function of c-Myc has been largely attributed to its intrinsic nature as a master transcription factor. Dysregulation of long noncoding RNAs (lncRNAs) has been implicated in a variety of human diseases, including cancer. However, the function of lncRNA in the regulation of c-Myc oncogenic activity is still not well understood. Here, we report that as a transcriptional target of c-Myc, lncRNA E2F1 messenger RNA (mRNA) stabilizing factor regulates c-Myc function via modulating E2F1 mRNA stability. This study provides insights into the mechanisms of how c-Myc promotes tumorigenesis.
Keywords: c-Myc, lncRNA, E2F1, tumorigenesis
Abstract
Deregulated expression of c-Myc is an important molecular hallmark of cancer. The oncogenic function of c-Myc has been largely attributed to its intrinsic nature as a master transcription factor. Here, we report the long noncoding RNA (lncRNA) E2F1 messenger RNA (mRNA) stabilizing factor (EMS) as a direct c-Myc transcriptional target. EMS functions as an oncogenic molecule by promoting G1/S cell cycle progression. Mechanistically, EMS cooperates with the RNA binding protein RALY to stabilize E2F1 mRNA, and thereby increases E2F1 expression. Furthermore, EMS is able to connect c-Myc to cell cycle control and tumorigenesis via modulating E2F1 mRNA stability. Together, these findings reveal a previously unappreciated mechanism through which c-Myc induces E2F1 expression and also implicate EMS as an important player in the regulation of c-Myc function.
The c-Myc oncoprotein plays a prominent role in tumorigenesis (1, 2). In normal cells, the expression of c-Myc is tightly controlled by both transcriptional and posttranscriptional mechanisms (3). Through gene amplification, chromosomal translocations, or insertional mutagenesis, c-Myc becomes activated in more than half of human cancers, including lung, breast, and colon carcinomas (4). The potent oncogenic activity of c-Myc has been well characterized by extensive studies using both cell culture and mouse models (4, 5). As a transcription factor, c-Myc, with its partner protein Max, is able to transcriptionally regulate target gene expression (6, 7). It has been previously shown that c-Myc controls the expression of 10 to 15% of genes in the genome. Recent studies also suggest c-Myc as a global amplifier of already active promoters (8, 9). By modulating target gene expression, c-Myc regulates a variety of cellular processes, including cell proliferation, differentiation, metabolism, and genome instability (10–12).
Regulation of cell proliferation by c-Myc has been attributed to its ability to transcribe a number of genes involved in G1/S cell cycle progression, such as CDK4 and cdc25A (13–15). The activation of CDK4 by c-Myc results in Rb hyperphosphorylation and subsequent release of E2F from Rb. E2F, in turn, is capable of activating cyclin E and other target genes required for S phase entry (16, 17). In addition to this indirect activation of E2F, c-Myc is able to induce E2F expression, which provides an additional means for bypassing the inhibitory function of Rb (18–21). The E2F family of transcription factors consists of 8 proteins, E2F1 through E2F8, among which E2F1, E2F2, and E2F3a are associated with the positive regulation of G1/S cell cycle transition (22, 23). E2F1, in particular, is essential for cells to enter the S phase from a quiescent state and is required for the activation of other E2F genes (24, 25). It has been recognized that c-Myc induces the expression of E2F1 (12, 19). However, it is not known whether c-Myc could also regulate E2F1 expression via modulating its messenger RNA (mRNA) stability.
Although previous studies on c-Myc’s transcriptional network have mainly focused on protein-coding genes, it has been increasingly recognized that c-Myc is also capable of regulating noncoding RNAs (26). These include both microRNAs (miRNAs) and long noncoding RNAs (lncRNAs) (27–35). The lncRNAs are defined as non–protein-coding RNA transcripts longer than 200 nucleotides (nt) (36). To date, more than 16,000 human lncRNA genes have been identified. Compared with protein-coding genes, the understanding of lncRNAs is relatively limited; nevertheless, lncRNAs are emerging as important regulatory molecules of gene expression at different levels (37). The lncRNAs have been functionally implicated in the regulation of multiple cellular processes, such as X chromosome inactivation, cell metabolism, and cell differentiation (38). Dysregulation of lncRNAs has also been linked to a variety of human cancers (39, 40). However, the function of lncRNA in the regulation of c-Myc oncogenic activity remains obscure. More specifically, it has not yet been characterized whether lncRNA is involved in the regulation of c-Myc function via modulating E2F1 mRNA stability.
In the present study, we report a c-Myc–inducible lncRNA that we named E2F1 mRNA stabilizing factor (EMS). As a direct transcriptional target of c-Myc, EMS functions as an oncogenic lncRNA by inducing E2F1 expression. Mechanistically, EMS associates with the RNA binding protein RALY to stabilize E2F1 mRNA, thereby promoting G1/S cell cycle progression. Furthermore, EMS is shown to regulate c-Myc function via modulating E2F1 mRNA stability. Our study provides insights into the mechanisms of how c-Myc promotes tumorigenesis and leads us to propose that EMS may be an important player in mediating c-Myc oncogenic function.
Results
EMS Functions as an Oncogenic lncRNA by Promoting G1/S Cell Cycle Progression.
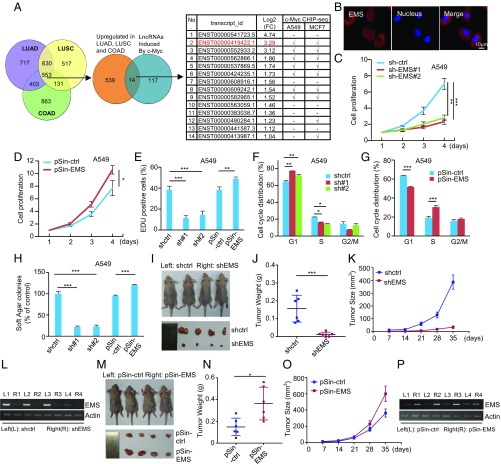
To generate new insights into the pivotal role of c-Myc in tumorigenesis, we sought to interrogate c-Myc–responsive lncRNAs with oncogenic functions. We first analyzed the aberrantly expressed lncRNAs in lung adenocarcinoma (LUAD), lung squamous cell carcinoma (LUSC), and colon adenocarcinoma (COAD) using The Cancer Genome Atlas (TCGA) database (41). A total of 553 lncRNAs were found to be overexpressed in these 3 cancer types (Fig. 1A and Dataset S1). By performing RNA sequencing analysis, 131 lncRNAs were shown to be induced by ectopic expression of c-Myc in LUAD A549 cells (Fig. 1A and Dataset S2). From these data, we selected 14 overlapping lncRNAs to examine whether c-Myc was associated with the promoter regions of these lncRNAs. Analysis of ENCODE c-Myc chromatin immunoprecipitation sequencing (ChIP-seq) datasets revealed that the promoters of 7 indicated lncRNAs were indeed occupied by c-Myc in both A549 and MCF7 cells, implying they are potential transcriptional targets of c-Myc (Fig. 1A). Of these 7 lncRNAs, lncRNA-2 (RP11-132A1.4, ENST00000419422.1) was found to be most significantly up-regulated by c-Myc based on our RNA sequencing data. Therefore, lncRNA-2 was chosen for further investigation in this study. As will be shown in the following sections, lncRNA-2 is able to promote E2F1 mRNA stability; we therefore named it EMS.
Fig. 1.
EMS functions as an oncogenic lncRNA. (A) Aberrantly expressed lncRNAs were analyzed in LUAD, LUSC, and COAD using TCGA database. A total of 553 lncRNAs overexpressed in these 3 cancer types (fold change [FC] ≥ 2, P < 0.05) were intersected with 131 c-Myc–induced lncRNAs in A549 cells (FC ≥ 2, P < 0.05) identified by RNA sequencing. Fourteen overlapping lncRNAs were then analyzed for potential association of c-Myc with their promoter regions using ENCODE c-Myc ChIP-seq datasets. (B) Localization of EMS in A549 cells was determined by RNA-fluorescence in situ hybridization analysis. (C) A549 cells were infected with lentiviruses expressing control shRNA (sh-ctrl), EMS shRNA#1, or EMS shRNA#2. Forty-eight hours later, the cell growth curves were measured by counting cell numbers at the indicated time points. Data shown are mean ± SD (n = 3). **P < 0.01; ***P < 0.001. The knockdown efficiency of EMS is shown in SI Appendix, Fig. S1D. (D) A549 cells were infected with lentiviruses expressing control (pSin-ctrl) or EMS (pSin-EMS). Forty-eight hours later, the cell growth curves were measured. Data shown are mean ± SD (n = 3). *P < 0.05. The successful overexpression of EMS is shown in SI Appendix, Fig. S1E. (E) A549 cells were infected with lentiviruses expressing control shRNA (shctrl), EMS shRNA#1, EMS shRNA#2, control (pSin-ctrl), or EMS (pSin-EMS). Forty-eight hours later, cells were subjected to EdU incorporation assay. Data shown are mean ± SD (n = 3). **P < 0.01; ***P < 0.001. (F) A549 cells were infected with lentiviruses expressing control shRNA, EMS shRNA#1, or EMS shRNA#2. Seventy-two hours later, cells were subjected to flow cytometry analysis. Data shown are mean ± SD (n = 3). *P < 0.05; **P < 0.01. (G) A549 cells were infected with lentiviruses expressing either control (pSin-ctrl) or EMS (pSin-EMS). Seventy-two hours later, cells were subjected to flow cytometry analysis. Data shown are mean ± SD (n = 3). ***P < 0.001. (H) A549 cells were infected with lentiviruses expressing control shRNA, EMS shRNA#1, EMS shRNA#2, control (pSin-ctrl), or EMS (pSin-EMS). Forty-eight hours later, cells were assayed for their ability to form colonies in soft agar. Data shown are mean ± SD (n = 3). ***P < 0.001. (I–L) Total of 2 × 106 A549 cells transduced with lentiviruses expressing either control shRNA or EMS shRNA (shEMS) were individually injected into the left flank and right flank of nude mice as indicated (n = 6 for each group). (I) Representative photographs of mice and xenograft tumors were taken 5 wk after injection. (J) Excised tumors were weighed. ***P < 0.001. (K) Tumor sizes were measured at the indicated time points. (L) RNA extracts from the excised xenografts were also analyzed by RT-PCR. (M–P) Total of 2 × 106 A549 cells transduced with lentiviruses expressing either control (pSin-ctrl) or EMS (pSin-EMS) were individually injected into the left flank and right flank of nude mice as indicated (n = 6 for each group). (M) Representative photographs of mice and xenograft tumors were taken 5 wk after injection. (N) Excised tumors were weighed. *P < 0.05. (O) Tumor sizes were measured at the indicated time points. (P) RNA extracts from the excised xenografts were also analyzed by RT-PCR.
EMS is located between genes encoding IFT22 and LncPRESS1 (42). By performing rapid amplification of complementary DNA (cDNA) ends (RACE) experiments, EMS was revealed as an RNA transcript with a molecular size of 1,098 nt (SI Appendix, Fig. S1 A and B), which is identical to the transcript described in the National Center for Biotechnology Information (reference sequence: NR_110115.2). EMS was predominantly localized in the cytoplasm (Fig. 1B and SI Appendix, Fig. S1C). To explore the functional role of EMS, we knocked down EMS in both A549 and PC9 cells. Short hairpin RNA (shRNA)-mediated knockdown of EMS resulted in a dramatic decrease in the proliferation of A549 and PC9 cells (Fig. 1C and SI Appendix, Fig. S1 D, F, and G). Similarly, antisense oligonucleotide (ASO)-mediated knockdown of EMS also greatly decreased cell proliferation (SI Appendix, Fig. S1 H and I). In contrast, ectopic expression of EMS led to a marked increase in the proliferation of A549 and PC9 cells (Fig. 1D and SI Appendix, Fig. S1 E, J, and K). Moreover, the EMS knockdown-decreased cell proliferation could be rescued by ectopic expression of shRNA-resistant EMS (SI Appendix, Fig. S1 L and M). These data suggest that EMS promotes cell proliferation. In support of this, EMS knockdown reduced, whereas EMS overexpression increased, the number of Edu-positive cells from both A549 and PC9 cells (Fig. 1E and SI Appendix, Fig. S1N). To determine how EMS promotes cell proliferation, we evaluated the effect of EMS on cell cycle distribution. Both shRNA- and ASO-mediated knockdown of EMS increased the percentage of cells in G1 phase and decreased the percentage of cells in S phase (Fig. 1F and SI Appendix, Fig. S1 O and P). In contrast, EMS-overexpressing cells displayed the decrease in G1 phase and the increase in S phase (Fig. 1G and SI Appendix, Fig. S1Q). Moreover, the EMS knockdown-induced G1 cell cycle arrest could be reversed by ectopic expression of shRNA-resistant EMS (SI Appendix, Fig. S1R). These data indicate that EMS promotes cell proliferation by accelerating G1/S cell cycle progression.
To further determine the oncogenic role of EMS in cancer, we first performed a soft agar colony formation assay. Knockdown of EMS dramatically reduced, whereas ectopic expression of EMS promoted, anchorage-independent cell growth (Fig. 1H and SI Appendix, Fig. S1S). By using a xenograft mouse model, EMS knockdown was shown to strongly inhibit in vivo xenograft tumor growth of A549 cells (Fig. 1 I–L). In contrast, EMS overexpression in A549 cells obviously promoted in vivo xenograft tumor growth (Fig. 1 M–P). Together, these findings suggest that EMS functions as an oncogenic lncRNA. In support, analysis of TCGA database showed that EMS expression levels were elevated in LUAD, LUSC, COAD, and breast invasive carcinoma (BRCA) compared with their paired normal tissues (SI Appendix, Fig. S1 T–W). Moreover, compared with nontransformed human adult foreskin fibroblast and IMR90 cells, tumor cell lines A549, PC9, and H1299 exhibited much higher copy numbers of EMS (SI Appendix, Fig. S1X).
EMS Is a Direct Transcriptional Target of c-Myc.
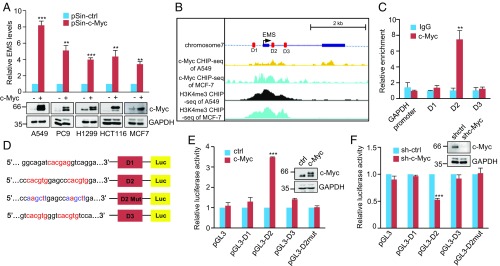
As mentioned above, EMS was identified as a c-Myc–responsive lncRNA by RNA sequencing analysis. We therefore sought to further confirm the effect of c-Myc on EMS expression. Induction of c-Myc increased, whereas knockdown of c-Myc decreased, EMS expression in A549, PC9, H1299, HCT116, and MCF7 cells (Fig. 2A and SI Appendix, Fig. S2A). However, neither overexpression nor knockdown of c-Myc affected expression of IFT22 and lncPRESS1 (SI Appendix, Fig. S2 B–E), the neighboring genes of EMS, indicating the specific effect of c-Myc on EMS expression. In accordance with the relationship between c-Myc and EMS cellular expression, analysis of TCGA database showed that the expression levels of c-Myc and EMS are positively correlated in LUAD, LUSC, COAD, and BRCA (SI Appendix, Fig. S2 F–I).
Fig. 2.
EMS is a direct c-Myc transcriptional target. (A) Indicated cells were infected with lentiviruses expressing control (pSin-ctrl) or c-Myc (pSin-c-Myc) protein. Forty-eight hours later, total RNA was analyzed by real-time RT-PCR. Data shown are mean ± SD (n = 3). **P < 0.01; ***P < 0.001. (B) Encode ChIP-seq data for c-Myc and H3K4me3 are displayed in the UCSC Genome Browser illustrations. Black boxes indicate 2 exons of EMS. D1, D2, and D3 represent 3 putative c-Myc binding sites predicted by the JASPAR database. (C) Lysates from A549 cells were subjected to ChIP assay using anti–c-Myc antibody or an isotype-matched control immunoglobulin G (IgG). ChIP products were amplified by real-time PCR. Data shown are mean ± SD (n = 3). **P < 0.01. (D) Shown are the pGL3-based wild-type and mutant reporter constructs used for luciferase (Luc) assay. (E) A549 cells were cotransfected with control vector (ctrl), Flag-c-Myc, or together with the reporter constructs in the indicated combination. Twenty-four hours after transfection, reporter activity was measured and plotted after normalizing with respect to Renilla luciferase activity. Data shown are mean ± SD (n = 3). ***P < 0.001. (F) A549 cells transduced with lentiviruses expressing control shRNA (sh-Ctrl) or c-Myc shRNA (sh-c-Myc) were cotransfected with the indicated reporter constructs. Reporter activity was then measured and plotted after normalizing with respect to Renilla luciferase activity. Data shown are mean ± SD (n = 3). ***P < 0.001.
We next explored whether c-Myc regulates EMS expression at the transcriptional level. We used the JASPAR database to inspect the upstream and intronic regions of the EMS gene (43). Three putative c-Myc binding sites (D1, D2, and D3) were identified (Fig. 2B). In addition, analysis of ENCODE ChIP-seq datasets revealed the presence of c-Myc and the active histone marker H3K4me3 around the transcription start site of EMS (Fig. 2B). The ChIP assay verified the interaction of c-Myc with the chromatin fragment comprising the D2 site (Fig. 2C). To further determine whether the D2 site confers c-Myc–dependent transcriptional activity, pGL3-based luciferase reporter constructs were used (Fig. 2D). Luciferase expression from the reporter construct containing the wild-type, but not the mutant D2, site was markedly induced by c-Myc overexpression and reduced by c-Myc knockdown (Fig. 2 E and F). Taken together, these data indicate that EMS is transcriptionally regulated by c-Myc.
EMS Exerts Its Oncogenic Role by Increasing E2F1 Expression.
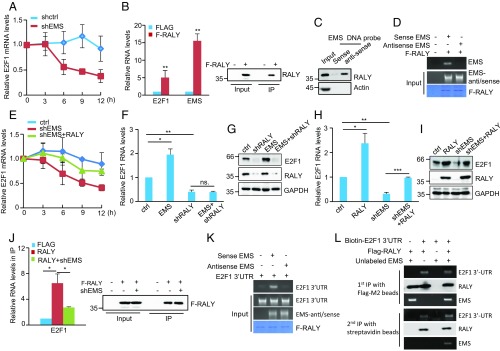
To explore the molecular mechanisms whereby EMS exerts its oncogenic function, we first examined whether EMS could regulate expression of its neighboring genes IFT22 and lncPRESS1. Neither knockdown nor overexpression of EMS was shown to affect expression levels of IFT22 and lncPRESS1 (SI Appendix, Fig. S3 A and B). We next performed gene expression profiling of control and EMS knockdown A549 cells. Knockdown of EMS resulted in the differential expression of 987 genes (down-regulation of 681 genes and up-regulation of 306 genes) (SI Appendix, Fig. S3C and Dataset S3). These differentially expressed genes were then subjected to gene ontology pathway enrichment analysis. Genes down-regulated in EMS knockdown cells were indeed enriched for regulators of cell cycle (SI Appendix, Fig. S3 D and E). Intriguingly, gene set enrichment analysis showed that E2F targets were significantly enriched among the genes down-regulated upon EMS silencing (SI Appendix, Fig. S3F). Among 66 cell cycle-related genes down-regulated in EMS knockdown cells, 30 genes are E2F1 targets (SI Appendix, Fig. S3G), indicating that EMS positively affects the E2F1 pathway. By performing real-time RT-PCR analysis, we validated the positive effect of EMS on mRNA expression of E2F1 and its target genes in both EMS-overexpressing and knockdown cells (Fig. 3A and SI Appendix, Fig. S3H). In addition, ectopic expression of EMS increased, whereas knockdown of EMS decreased protein levels of E2F1 (Fig. 3B). Moreover, analysis of TCGA database showed that the expression levels of EMS and E2F1 are positively correlated in LUAD, LUSC, COAD, and BRCA (SI Appendix, Fig. S3 I–L). These data suggest that EMS is able to positively regulate E2F1 expression.
Fig. 3.
EMS exerts its oncogenic role by increasing E2F1 expression. (A) A549 cells were infected with lentiviruses expressing control (pSin-ctrl) or EMS (pSin-EMS). Forty-eight hours later, total RNA was subjected to real-time RT-PCR analysis to evaluate mRNA levels of E2F1 and its target genes as indicated. (B) A549 cells were infected with lentiviruses expressing control (ctrl) shRNA (shctrl), EMS shRNA#1, EMS shRNA#2, control (pSin), or EMS (pSin-EMS). Forty-eight hours later, cell lysates were analyzed by Western blotting. (C) A549 cells were infected with lentiviruses expressing control, EMS, E2F1 shRNA, or both EMS and E2F1 shRNA. Forty-eight hours later, the cell growth curves were measured. Data shown are mean ± SD (n = 3). *P < 0.05. ns., no significance. The successful EMS overexpression and E2F1 knockdown are shown in SI Appendix, Fig. S4A. (D) A549 cells were infected with lentiviruses expressing control, EMS, E2F1 shRNA, or both EMS and E2F1 shRNA. Forty-eight hours later, cells were subjected to EdU incorporation assay. Data shown are mean ± SD (n = 3). ***P < 0.001. (E) A549 cells were infected with lentiviruses expressing control, EMS, E2F1 shRNA, or both EMS and E2F1 shRNA. Seventy-two hours later, cells were subjected to flow cytometry analysis. Data shown are mean ± SD (n = 3). *P < 0.05; **P < 0.01; ***P < 0.001. (F) A549 cells were infected with lentiviruses expressing control, EMS, E2F1 shRNA, or both EMS and E2F1 shRNA. Forty-eight hours later, cells were assayed for their ability to form colonies in soft agar. Data shown are mean ± SD (n = 3). **P < 0.01. (G–J) Total of 2 × 106 A549 cells transduced with lentiviruses expressing control, EMS, E2F1 shRNA, or both EMS and E2F1shRNA were individually injected into the left flank and right flank of nude mice as indicated (n = 6 for each group). (G) Representative photographs of mice and xenograft tumors were taken 5 wk after injection. (H) Excised tumors were weighed. *P < 0.05. (I) Tumor sizes were measured at the indicated time points. (J) RNA and protein extracts from the excised xenografts were also analyzed by RT-PCR (RT) and Western blotting (WB), respectively.
We next examined whether EMS exerts its oncogenic function via regulation of E2F1. Ectopic expression of EMS in control A549 cells was consistently shown to promote cell proliferation (Fig. 3C and SI Appendix, Fig. S4A), raise the numbers of Edu-positive cells (Fig. 3D), accelerate G1/S cell cycle progression (Fig. 3E), and increase anchorage-independent cell growth (Fig. 3F). However, EMS failed to show any of these effects when E2F1 was knocked down in A549 cells (Fig. 3 C–F and SI Appendix, Fig. S4A). In addition, the inhibitory effects of EMS knockdown on cell proliferation, the numbers of Edu-positive cells, G1/S cell cycle progression, and anchorage-independent cell growth could be markedly reversed by ectopically expressed E2F1 (SI Appendix, Fig. S4 B–F). By using a xenograft mouse model, we showed that ectopic expression of EMS in control cells, but not in E2F1 knockdown cells, promoted xenograft tumor growth in nude mice (Fig. 3 G–J). Moreover, EMS knockdown-inhibited xenograft tumor growth was able to be greatly restored by E2F1 induction (SI Appendix, Fig. S4 G–J). Taken together, these findings suggest that EMS exerts its oncogenic function by increasing E2F1 expression.
EMS Cooperates with RALY to Promote E2F1 mRNA Stability.
We next explored how EMS increases E2F1 expression. Considering the predominant localization of EMS in the cytoplasm, we asked whether EMS could regulate E2F1 mRNA stability. A549 cells with knockdown or overexpression of EMS were treated with actinomycin D for different periods of time to measure the decay of E2F1 mRNA. Knockdown of EMS resulted in a decrease in the half-life of E2F1 mRNA, whereas overexpression of EMS increased its half-life (Fig. 4A and SI Appendix, Fig. S5A), indicating that EMS stabilizes E2F1 mRNA. To further determine how EMS promotes E2F1 mRNA stability, we first examined whether EMS could function as a miRNA sponge. A miRNA sponge is expected to form a complex with Ago2, an integral component of the RNA-induced silencing complex (RISC) (44, 45). As a positive control, lncRNA H19 was indeed present in the Ago2 immunoprecipitates (46) (SI Appendix, Fig. S5B). Unlike H19, EMS was not enriched in the Ago2 immunoprecipitates (SI Appendix, Fig. S5B), making it less likely that EMS acts as a miRNA sponge.
Fig. 4.
EMS cooperates with RALY to promote E2F1 mRNA stability. (A) A549 cells were infected with lentiviruses expressing either control shRNA (shctrl) or EMS shRNA (shEMS). Forty-eight hours later, cells were incubated with actinomycin D (2 μg/mL) for the indicated periods of time. Total RNA was then analyzed by real-time RT-PCR to examine E2F1 mRNA stability. Data shown are mean ± SD (n = 3). (B) A549 cells were infected with lentiviruses expressing pSin-control or pSin-Flag-RALY. Forty-eight hours later, cell lysates were immunoprecipitated with anti-Flag antibody. RNAs in immunoprecipitates (IP) were then analyzed by real-time RT-PCR to examine EMS and E2F1 mRNA levels. Data shown are mean ± SD (n = 3). **P < 0.01. The input and immunoprecipitates were also analyzed by Western blotting. (C) Lysates from A549 cells were incubated with either sense or antisense biotin-labeled DNA oligomers corresponding to EMS, followed by the pull-down experiments using streptavidin-coated beads. The pull-down complexes were analyzed by Western blotting. (D) In vitro-synthesized EMS or its antisense RNA was incubated with purified recombinant Flag-RALY bound with M2 beads. The bead-bound RNAs were then eluted as templates for RT-PCR analysis. (E) A549 cells were infected with lentiviruses expressing control, EMS shRNA, or both EMS shRNA and RALY. Forty-eight hours later, cells were incubated with actinomycin D (2 μg/mL) for the indicated periods of time. Total RNA was then analyzed by real-time RT-PCR. Data shown are mean ± SD (n = 3). (F) A549 cells were infected with lentiviruses expressing control (ctrl), EMS, RALY shRNA (shRALY), or both EMS and RALY shRNA. Forty-eight hours later, total RNA was analyzed by real-time RT-PCR. Data shown are mean ± SD (n = 3). *P < 0.05; **P < 0.01, ns., no significance. (G) A549 cells were infected with lentiviruses expressing ctrl, EMS, RALY shRNA, or both EMS and RALY shRNA. Forty-eight hours later, cell lysates were analyzed by Western blotting. (H) A549 cells were infected with lentiviruses expressing control, EMS shRNA, RALY, or both EMS shRNA and RALY. Forty-eight hours later, total RNA was analyzed by real-time RT-PCR. Data shown are mean ± SD (n = 3). *P < 0.05; **P < 0.01; ***P < 0.001. (I) A549 cells were infected with lentiviruses expressing control, EMS shRNA, RALY, or both EMS shRNA and RALY. Forty-eight hours later, cell lysates were analyzed by Western blotting. (J) A549 cells transduced with lentiviruses expressing either control shRNA or EMS shRNA were transfected with or without Flag-RALY as indicated. Twenty-four hours after transfection, cell lysates were immunoprecipitated with anti-Flag antibody. RNAs present in immunoprecipitates (IP) were then analyzed by real-time RT-PCR. Data shown are mean ± SD (n = 3). *P < 0.05. The input and immunoprecipitates were also analyzed by Western blotting. (K) Purified recombinant Flag-RALY bound with M2 beads was incubated with in vitro-synthesized E2F1 3′-UTR, EMS, and its antisense RNA in the indicated combination. The bead-bound RNAs were then eluted as templates for RT-PCR analysis. (L) In vitro-synthesized biotin-labeled E2F1 3′-UTR and unlabeled EMS plus Flag-RALY were incubated at room temperature for 30 min. The mixtures were first immunoprecipitated with Flag-M2 beads, followed by the elution step with 3 × FLAG peptides. Half of the eluent was subjected to Western blot and RT-PCR analysis. The rest of the eluent was further immunoprecipitated with streptavidin beads. After extensive washing, the immunoprecipitates were analyzed by Western blotting and RT-PCR.
Because many lncRNAs exert their functions through interaction with their target proteins (47), we hypothesized that EMS could regulate E2F1 mRNA stability in a similar manner. It has been recently shown that the heterogeneous nuclear ribonucleoprotein RALY, a poly-U binding protein, is able to stabilize E2F1 mRNA by binding to its 3′-untranscribed region (UTR) (48). Analysis of the EMS sequence revealed that EMS contains a poly-U stretch with 22 uridines (SI Appendix, Fig. S1B), indicating that RALY may also interact with EMS. To test this, an RNA immunoprecipitation experiment was performed. EMS was indeed enriched in the Flag-RALY immunoprecipitates compared with the control immunoprecipitates (Fig. 4B). The RALY–EMS interaction was also validated by biotin pull-down assay using a biotin-labeled antisense DNA oligomer against EMS (Fig. 4C). In addition, an in vitro binding assay showed that RALY directly interacted with EMS, but not its antisense RNA (Fig. 4D). Moreover, compared with wild-type EMS, mutant EMS with deletion of 22 uridines (EMS-ΔpolyU) showed decreased binding to RALY (SI Appendix, Fig. S5C). These data support that RALY is an interacting partner for EMS.
To further determine whether EMS promotes E2F1 mRNA stability via RALY, E2F1 mRNA half-life was measured using actinomycin D. The results showed that EMS knockdown-decreased E2F1 mRNA stability could be restored by ectopic expression of RALY (Fig. 4E). In addition, unlike the stabilizing effect of EMS on E2F1 mRNA in control cells (SI Appendix, Fig. S5A), EMS failed to promote E2F1 mRNA stability in RALY knockdown cells (SI Appendix, Fig. S5D). In accordance, EMS overexpression was able to increase both mRNA and protein levels of E2F1 in control cells, but not in RALY knockdown cells (Fig. 4 F and G). Moreover, EMS knockdown-caused decrease in mRNA and protein expression of E2F1 could be markedly restored by ectopic expression of RALY (Fig. 4 H and I). Correlated with the decreased binding to RALY (SI Appendix, Fig. S5C), mutant EMS with deletion of 22 uridines exhibited no obvious effects on E2F1 expression and cell proliferation compared with wild-type EMS (SI Appendix, Fig. S5 E–G). Collectively, these data indicate that the stabilizing effect of EMS on E2F1 mRNA is dependent on RALY. They also prompted us to ask whether EMS could increase the binding of RALY to E2F1 mRNA, thereby leading to E2F1 mRNA stabilization. An RNA immunoprecipitation experiment revealed that knockdown of EMS substantially decreased the binding of RALY to E2F1 mRNA (Fig. 4J), indicating the promoting effect of EMS on the RALY-E2F1 mRNA binding. In support, an in vitro binding assay showed that EMS, but not its antisense RNA, was capable of increasing the interaction between RALY and E2F1 3′-UTR (Fig. 4K). The subsequent sequential immunoprecipitation experiments revealed that EMS, RALY, and E2F1 3′-UTR could form a ternary complex (Fig. 4L). Taken together, these data suggest that EMS cooperates with RALY to promote E2F1 mRNA stability.
EMS Regulates Tumor Cell Growth via RALY.
Given that the effect of EMS on E2F1 mRNA stability is RALY-dependent as shown above, we sought to evaluate whether EMS regulates tumor cell growth via RALY. As was expected, RALY knockdown strongly decreased, whereas RALY overexpression substantially increased, cell proliferation, the numbers of Edu-positive cells, and anchorage-independent cell growth (SI Appendix, Fig. S6 A–F). Ectopic expression of EMS consistently promoted cell proliferation (SI Appendix, Fig. S6A), increased the numbers of Edu-positive cells (SI Appendix, Fig. S6B), and accelerated anchorage-independent cell growth (SI Appendix, Fig. S6C). However, EMS failed to show any of these effects when RALY was knocked down (SI Appendix, Fig. S6 A–C), indicating EMS promotes tumor cell growth via RALY. In support, the inhibitory effects of EMS knockdown on cell proliferation, the numbers of Edu-positive cells, and anchorage-independent cell growth could be greatly reversed by ectopically expressed RALY (SI Appendix, Fig. S6 D–F). We next determined whether RALY mediates the promoting effect of EMS on G1/S cell cycle progression. Ectopic expression of EMS was shown to accelerate G1/S cell cycle progression in control cells, but not in RALY knockdown cells (SI Appendix, Fig. S6G). In addition, EMS knockdown-inhibited G1/S cell cycle progression was significantly reversed by RALY overexpression (SI Appendix, Fig. S6H). Together, these data suggest that EMS regulates tumor cell growth via RALY.
EMS Connects c-Myc to Cell Cycle Control and Tumorigenesis.
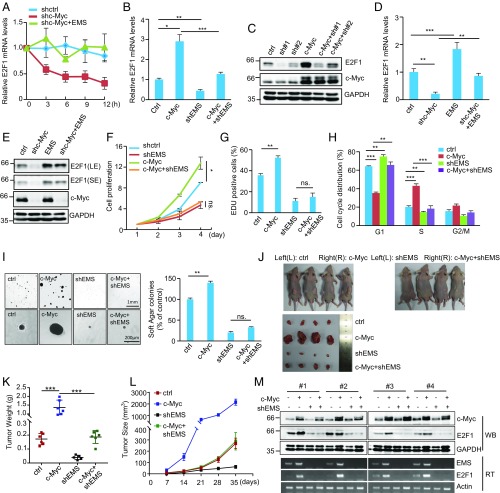
It has been previously shown that E2F1 expression is positively regulated by c-Myc (12, 19). Given the above findings that EMS is a transcriptional target of c-Myc and EMS is able to stabilize E2F1 mRNA, we asked whether c-Myc could regulate E2F1 mRNA stability via EMS. The results showed that knockdown of c-Myc led to a strong decrease in E2F1 mRNA half-life, however, which was reversed by ectopic expression of EMS (Fig. 5A), implying that c-Myc indeed promotes EMS-mediated E2F1 mRNA stabilization. Induction of c-Myc also greatly increased both mRNA and protein levels of E2F1 (Fig. 5 B and C). However, the promoting effect of c-Myc on E2F1 expression was minimized by EMS knockdown (Fig. 5 B and C). In addition, the decreased mRNA and protein levels of E2F1 caused by c-Myc knockdown were able to be markedly restored by EMS overexpression (Fig. 5 D and E). Together, these data indicate that c-Myc positively regulates E2F1 expression by at least partially promoting EMS-mediated stabilization of E2F1 mRNA.
Fig. 5.
EMS connects c-Myc to cell cycle control and tumorigenesis. (A) A549 cells were infected with lentiviruses expressing control shRNA (shctrl), c-Myc shRNA (shc-Myc), or both c-Myc shRNA and EMS. Forty-eight hours later, cells were incubated with actinomycin D (2 μg/mL) for the indicated periods of time. Total RNA was then analyzed by real-time RT-PCR. Data shown are mean ± SD (n = 3). (B) A549 cells were infected with lentiviruses expressing control (Ctrl), c-Myc, EMS shRNA (shEMS), or both c-Myc and EMS shRNA. Forty-eight hours later, total RNA was analyzed by real-time RT-PCR. Data shown are mean ± SD (n = 3). *P < 0.05; **P < 0.01; ***P < 0.001. (C) A549 cells were infected with lentiviruses expressing control, EMS shRNA#1, EMS shRNA#2, c-Myc, both c-Myc and EMS shRNA#1, or both c-Myc and EMS shRNA#2. Forty-eight hours later, cell lysates were analyzed by Western blotting. (D) A549 cells were infected with lentiviruses expressing control, c-Myc shRNA, EMS, or both c-Myc shRNA and EMS. Forty-eight hours later, total RNA was analyzed by real-time RT-PCR. Data shown are mean ± SD (n = 3). **P < 0.01; ***P < 0.001. (E) A549 cells were infected with lentiviruses expressing control, c-Myc shRNA, EMS, or both c-Myc shRNA and EMS. Forty-eight hours later, cell lysates were analyzed by Western blotting. (F) A549 cells were infected with lentiviruses expressing control, c-Myc, EMS shRNA, or both c-Myc and EMS shRNA. Forty-eight hours later, the cell growth curves were measured. Data shown are mean ± SD (n = 3). *P < 0.05. ns., no significance. (G) A549 cells were infected with lentiviruses expressing control, c-Myc, EMS shRNA, or both c-Myc and EMS shRNA. Forty-eight hours later, cells were subjected to EdU incorporation assay. Data shown are mean ± SD (n = 3). **P < 0.01. (H) A549 cells were infected with lentiviruses expressing control, c-Myc, EMS shRNA, or both c-Myc and EMS shRNA. Seventy-two hours later, cells were subjected to flow cytometry analysis. Data shown are mean ± SD (n = 3). **P < 0.01; ***P < 0.001. (I) A549 cells were infected with lentiviruses expressing control, c-Myc, EMS shRNA, or both c-Myc and EMS shRNA. Forty-eight hours later, cells were assayed for their ability to form colonies in soft agar. The shown images are representative of 3 independent experiments. Data shown are mean ± SD (n = 3). **P < 0.01. (J–M) Total of 2 × 106 A549 cells transduced with lentiviruses expressing control, c-Myc, EMS shRNA, or both c-Myc and EMS shRNA were individually injected into the left flank and right flank of nude mice as indicated (n = 6 for each group). (J) Representative photographs of mice and xenograft tumors were taken 5 wk after injection. (K) Excised tumors were weighed. ***P < 0.001. (L) Tumor sizes were measured at the indicated time points. (M) RNA and protein extracts from the excised xenografts were also analyzed by RT-PCR (RT) and Western blotting (WB), respectively.
We next investigated whether EMS could mediate the oncogenic function of c-Myc. Consistent with the promoting effect of c-Myc on tumor cell growth, ectopic expression of c-Myc enhanced cell proliferation and increased the numbers of Edu-positive cells in control cells (Fig. 5 F and G). However, c-Myc failed to show any of these effects when EMS was knocked down (Fig. 5 F and G). Analysis of cell cycle distribution revealed that c-Myc–accelerated G1/S cell cycle progression was able to be reversed by the simultaneous EMS knockdown (Fig. 5H). These data indicate that c-Myc could increase tumor cell growth by promoting EMS-mediated G1/S cell cycle progression. By performing a soft agar colony formation assay, we showed that ectopic expression of c-Myc only promoted anchorage-independent cell growth in control cells, but not in EMS knockdown cells (Fig. 5I). Moreover, by using a xenograft mouse model, ectopically expressed c-Myc in A549 cells was expectedly shown to dramatically increase xenograft tumor growth (Fig. 5 J–L). Accompanying the increased tumor growth by c-Myc overexpression, expression levels of both EMS and E2F1 were elevated in xenografts from c-Myc–overexpressing A549 cells (Fig. 5M), indicating that c-Myc may promote tumor growth via EMS. In support of this, the promoting effect of c-Myc on xenograft tumor growth was significantly minimized by EMS knockdown (Fig. 5 J–M). Taken together, these data suggest that EMS connects c-Myc to cell cycle control and tumorigenesis via modulating E2F1 mRNA stability.
Discussion
c-Myc is one of the most commonly activated oncogenes, and deregulated expression of this oncogene has been linked to a variety of human cancers (2, 4). It has been well recognized that c-Myc regulates a variety of cellular processes by modulating expression of a number of protein-coding genes (10, 26). Increasing evidence suggests that lncRNAs are involved in the regulation of the c-Myc signaling pathway (7, 26, 27, 49–53). Dysregulation of lncRNAs has also been implicated in tumorigenesis (54). Therefore, identification of new c-Myc–responsive lncRNA with oncogenic function is of great importance to the comprehensive understanding of c-Myc biology. In this study, we report that EMS, as a c-Myc–inducible lncRNA, is able to connect c-Myc to cell cycle control and tumorigenesis by promoting E2F1 mRNA stability. Therefore, EMS is an important player in the regulation of c-Myc function.
EMS expression is elevated broadly in different types of human cancers, including LUAD, LUSC, COAD, and BRCA, indicating that EMS is an oncogenic molecule. In support of this, knockdown of EMS decreases, whereas ectopic expression of EMS increases, cell proliferation. In addition, EMS knockdown strongly inhibits in vivo xenograft tumor growth, while EMS overexpression shows the opposite effect. Here, we rigorously document EMS as a bona fide transcriptional target of c-Myc. The importance of c-Myc–regulated EMS expression is also supported by the observation that the expression levels of c-Myc and EMS are positively correlated in LUAD, LUSC, COAD, and BRCA.
Through RNA sequencing and functional analyses, we demonstrate that EMS is able to positively regulate E2F1 expression, thereby promoting G1/S cell cycle progression. The findings of the positive correlation between EMS and E2F1 expression in LUAD, LUSC, COAD, and BRCA further indicate the physiological importance of EMS-regulated E2F1 expression. Although E2F1 is particularly known as a positive regulator of G1/S cell cycle progression, E2F1 can also trigger apoptosis (55). In line with the effects of E2F1 on both cell proliferation and apoptosis, E2F1 functions in vivo as an oncogene or a tumor suppressor in a context-dependent manner (56). In this study, we clearly show that EMS exerts its oncogenic function via regulation of E2F1. Collectively, we establish EMS as a direct c-Myc target that induces E2F1 expression to promote tumorigenesis. It is notable that c-Myc and E2F1 can activate each other’s transcription (57), thereby establishing a possible positive feedback loop. However, this feedback loop appears to be disrupted by miRNAs, because c-Myc–induced miR-17-5p and miR-20a are shown to inhibit E2F1 translation (58). We here show that, correlating with the predominant cytoplasmic localization, c-Myc–induced EMS is able to promote E2F1 mRNA stability, implicating EMS as an important factor to finely control E2F1 expression. These findings also indicate the complexity of the c-Myc–E2F1 network.
Various mechanisms have been proposed for lncRNA-regulated gene expression. One of the well-characterized mechanisms is the lncRNA-mediated gene regulation through interaction with miRNA or protein (47, 59). For example, long intergenic noncoding RNA (lincRNA) linc-MD1 controls muscle differentiation by sponging miR-133 and miR-135 (60). In addition, lincRNA-p21 exerts its diverse functions through binding to different target proteins (61–63). Because a miRNA sponge is expected to form a complex with Ago2, a key component of the RISC (44, 45), the observation of no interaction between EMS and Ago2 implies that EMS may not act as a miRNA sponge. Intriguingly, EMS is shown to cooperate with the RNA binding protein RALY to stabilize E2F1 mRNA. RALY belongs to the heterogeneous nuclear ribonucleoprotein family that binds to poly-U elements within several RNAs (64, 65). It has been recently shown that RALY is able to stabilize E2F1 mRNA by binding to its 3′-UTR (48). Here, we identify EMS as a RALY binding partner. The poly-U stretch with 22 uridines within EMS is required for the interaction with RALY, as demonstrated by the finding that mutant EMS with deletion of 22 uridines exhibits decreased binding to RALY. We also show that by binding to RALY, EMS increases the interaction between RALY and E2F1 3′-UTR. In addition, EMS, RALY, and E2F1 3′-UTR could form a ternary complex. These data indicate that EMS may act as an interacting scaffold to promote the RALY/E2F1/3′-UTR interaction, although the detailed underlying mechanisms await further investigation. Functionally, EMS indeed promotes E2F1 mRNA stability and increases tumor cell growth via RALY. Together with the recent finding that the lncRNA LeXis interacts with RALY and regulates expression of genes involved in the cholesterol biosynthesis pathway (66), our data therefore indicate RALY as an important player in the regulation of lncRNA’s cellular function.
It has been well accepted that c-Myc activation is not only able to drive tumor initiation and progression but is also essential for the maintenance of an established tumor (2), making c-Myc an attractive target for cancer therapy. Unfortunately, c-Myc itself is widely considered “undruggable” because c-Myc lacks a specific active site for small molecules. Essential targets involved in c-Myc deregulation and downstream effectors of c-Myc signaling have been exploited as new approaches to treat c-Myc–driven cancers (67–69). In this study, we show that as a direct c-Myc transcriptional target, EMS functions as an oncogenic molecule. In addition, EMS is able to mediate the tumor-promoting effect of c-Myc. These data therefore suggest that EMS may represent a potential therapeutic target for cancer.
Materials and Methods
Reagents and Antibodies.
The following reagents and antibodies were used in this study: DAPI (Sigma), Lipofectamine 2000 (Invitrogen), complete ethylenediaminetetraacetic acid (EDTA)-free protease inhibitor mixture (Roche Applied Science), actinomycin D (ab141058; Abcam), propidium iodide (ST511; Beyotime), streptavidin-coated agarose beads (Thermo Fisher Scientific), antibodies against actin (sc-47778,1:1,000; Santa Cruz Biotechnology), glyceraldehyde-3-phosphate dehydrogenase (GAPDH; sc-166545, 1:5,000; Santa Cruz Biotechnology), Flag (F3165, 1:4,000; Sigma), E2F1 (3742S, 1:1,000; Cell Signaling), c-Myc for ChIP assay (9402S; Cell Signaling), c-Myc for Western blotting (9402S, 1:1,000; Cell Signaling), RALY (A302-070A, 1:1,000; Bethyl), RARP (sc-8007, 1:1,000; Santa Cruz Biotechnology), and horseradish peroxidase-conjugated secondary antibodies against rabbit (111-035-144, 1:10,000; Jackson Immunoresearach) and mouse (115-035-062, 1:10,000; Jackson ImmunoResearch).
Generation of the Lentiviral Expression System.
To generate lentiviruses expressing the indicated shRNAs, HEK293T cells were transfected with shRNAs (cloned in PLKO.1), pREV, pGag/Pol/PRE, and pVSVG with a ratio of 2:2:2:1. To generate lentiviruses expressing control, EMS, or the indicated proteins, HEK293T cells were transfected with pSin-EF1α–based construct, pmd2.g, and pspax2 with a ratio of 2:1:2. For generation of the control virus, PLKO.1 or pSin empty vector was used. Twelve hours after transfection, cells were cultured with fresh medium for another 24 h. The culture medium containing lentivirus particles was then collected and filtered through a 0.45-μm poly(vinylidene difluoride) filter (Millipore). The cleared culture medium was used for infection. The successful lentivirus-mediated knockdown and overexpression were verified by real-time RT-PCR or Western blot analysis in each experiment. To avoid the off-target effect, 2 different EMS-targeting shRNAs (sh-EMS#1 and sh-EMS#2) were used in this study. Because sh-EMS#1 consistently achieved better knockdown efficiency, unless specified otherwise, sh-EMS#1 was used. The shRNA target sequences used in the study are listed in SI Appendix, Table S1.
Transfection of ASOs.
The 2-O-methyl RNA/DNA ASOs, which were modified by changing the 5 nt at the 5′ and 3′ ends into 2′-O-methyl ribonucleotides, were synthesized by RiboBio. All bases of ASOs were converted into phosphorothioate oligonucleotides. Transfection of ASOs was conducted with Lipofectamine 2000 (Invitrogen) according to the manufacturer’s instructions. The ASO sequences are as follows: NC-ASO, mU*mU*mC*mU*mC*CGAACGTGTCmA*mC*mG*mU*mU*; EMS-ASO, mU*mU*mC*UmG*mC*TTGATCTAGTmC*mU*mA*mU*mC*.
Real-Time RT-PCR.
Real-time RT-PCR was performed as previously described (70). Total RNA was isolated using TRIzol (Invitrogen). One microgram of total RNA was used for cDNA synthesis using the PrimeScriptTM RT Reagent Kit (DRR037A; TaKaRa) according to the instructions provided by the manufacturer. Real-time PCR was performed using SYBR premix EX Taq (TaKaRa) and analyzed with the StepOnePlus real-time PCR system (Thermo Fisher Scientific). The PCR results, recorded as threshold cycle (Ct) numbers, were normalized against an internal control (GAPDH). The expression data were analyzed using the 2-ΔΔCT method described by Livak and Schmittgen (71). The primer sequences are shown in SI Appendix, Table S1.
5′ RACE and 3′ RACE.
5′ RACE and 3′ RACE were performed using the SMARTer 5′/3′ RACE Kit (Clontech) according to the manufacturer’s instructions. The primers P1 and P2 were used for 5′-RACE. The primers P3 and P4 were used for 3′-RACE. P1 and P4 were supplied in the kit. The primer sequences of P2 and P3 are shown in SI Appendix, Table S1.
RNA In Situ Hybridization.
To determine cellular distribution of EMS, RNA in situ hybridization was carried out as previously described (30) using in vitro-transcribed Alexa Fluor 488-labeled antisense probes (Nucleic Acid Labeling Kit; Invitrogen). The nuclei were also visualized by DAPI staining. The sequence of the RNA probe is shown in SI Appendix, Table S1.
RNA Immunoprecipitation.
RNA immunoprecipitation was performed as we previously described (70). Briefly, 1 × 107 cells were lysed in immunoprecipitation lysis buffer (50 mM Tris⋅HCl [pH 7.4], 0.5% Nonidet P-40, 150 mM NaCl, 1.5 mM MgCl2, 1 mM EDTA) supplemented with RNase A inhibitor, DNase I, and 1 × protease inhibitor mixture. After preclearing with protein A/G beads (Pierce), cell lysates were incubated with antibody-coated protein A/G beads at 4 °C for 8 h. The immunocomplexes were then eluted from the beads using the elution buffer (50 mM Tris⋅HCl [pH 8.0], 10 mM EDTA, 1% sodium dodecyl sulfate) at 65 °C for 10 min. To isolate RNAs from the eluted immunocomplexes, proteinase K was first used for protein digestion, followed by RNA extraction by the phenol/chloroform method. Purified RNAs were then analyzed by real-time RT-PCR.
In Vitro Transcription of EMS and Its Antisense RNA.
To synthesize EMS and its antisense RNA, an in vitro transcription assay was performed using the MaxiScript T7 Kit (Ambion) according to the manufacturer’s instructions. The DNA templates used in the transcription assay were obtained by RT-PCR using the specific primers (SI Appendix, Table S1), followed by purification using the DNA Gel Extraction Kit (AxyPrep). To verify the interaction of EMS with RALY, in vitro-synthesized EMS or its antisense RNA was incubated with M2 bead-bound purified Flag-RALY. After incubation and washing, the bead-bound RNAs were analyzed by RT-PCR.
Biotin Pull-Down Assay.
The biotin pull-down assay was performed as we previously described (70). The whole process was carried out under RNase-free conditions. To examine the interaction between EMS and RALY, lysates from 5 × 106 A549 cells were incubated with 1 μg of sense or antisense biotin-labeled DNA oligomers corresponding to EMS. After 1 h of incubation, streptavidin-coated beads (Thermo Fisher Scientific) were added to the reaction mix to isolate the RNA–protein complex, followed by Western blot and real-time RT-PCR analyses.
ChIP Assay.
The ChIP assay was performed as we previously described (70). Briefly, A549 cells were cross-linked with 1% formaldehyde for 10 min. The ChIP assay was then performed using the ChIP Assay Kit (Beyotime Biotechnology) with anti–c-Myc antibody according to the instructions provided by the manufacturer. Normal rabbit immunoglobulin G was included as a negative control. The bound DNA fragments were analyzed by real-time PCR using the specific primers (SI Appendix, Table S1).
Xenograft Mouse Model.
Studies on mice were conducted with approval from the Animal Research Ethics Committee of the University of Science and Technology of China. For xenograft experiments, 2 × 106 A549 cells were injected into the left flank or right flank of 4-wk-old male athymic nude mice (Shanghai SLAC Laboratory Animal Co. Ltd.) (n = 6 for each group). Mice were used in the experiment at random. Severn days after injection, tumor volumes were measured every 7 d with a caliper and calculated using the following equation: volume = length × width2 × 0.52. Five weeks after injection, mice were killed and subjected to tumor excision. The experimentalists were blinded to the information of the excised tumors while testing the tumors’ weight. The extracted proteins and RNAs from the excised tumors were also subjected to Western blot and real-time RT-PCR analyses, respectively.
Statistical Analysis.
Statistical analysis was performed using Microsoft Excel software and GraphPad Prism. Statistical significance was analyzed by a 2-tailed Student’s t test. P values less than 0.05 were considered to be statistically significant (*P < 0.05; **P < 0.01; ***P < 0.001).
Data Availability.
The RNA sequencing data have been deposited in the National Center for Biotechnology Information Sequence Read Archive with accession codes SRP171977 and SRP171802.
Supplementary Material
Acknowledgments
This work was supported by the Ministry of Science and Technology of China (Grant 2015CB553800), National Natural Science Foundation of China (Grants 31671487 and 31871440), and Fundamental Research Funds for Central Universities (Grants WK2070000106 and WK9110000007).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The RNA sequencing data reported in this paper have been deposited in the National Center for Biotechnology Information Sequence Read Archive (SRA), https://www.ncbi.nlm.nih.gov/sra (SRA accession nos. SRP171977 and SRP171802).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1903432116/-/DCSupplemental.
References
- 1.Stine Z. E., Walton Z. E., Altman B. J., Hsieh A. L., Dang C. V., MYC, metabolism, and cancer. Cancer Discov. 5, 1024–1039 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Gabay M., Li Y., Felsher D. W., MYC activation is a hallmark of cancer initiation and maintenance. Cold Spring Harb. Perspect. Med. 4, a014241 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Meyer N., Penn L. Z., Reflecting on 25 years with MYC. Nat. Rev. Cancer 8, 976–990 (2008). [DOI] [PubMed] [Google Scholar]
- 4.Dang C. V., MYC on the path to cancer. Cell 149, 22–35 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Morton J. P., Sansom O. J., MYC-y mice: From tumour initiation to therapeutic targeting of endogenous MYC. Mol. Oncol. 7, 248–258 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Blackwood E. M., Eisenman R. N., Max: A helix-loop-helix zipper protein that forms a sequence-specific DNA-binding complex with myc. Science 251, 1211–1217 (1991). [DOI] [PubMed] [Google Scholar]
- 7.Zhang P., Cao L., Fan P., Mei Y., Wu M., LncRNA-MIF, a c-Myc-activated long non-coding RNA, suppresses glycolysis by promoting Fbxw7-mediated c-Myc degradation. EMBO Rep. 17, 1204–1220 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lin C. Y., et al. , Transcriptional amplification in tumor cells with elevated c-Myc. Cell 151, 56–67 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Nie Z., et al. , c-Myc is a universal amplifier of expressed genes in lymphocytes and embryonic stem cells. Cell 151, 68–79 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Carroll P. A., Freie B. W., Mathsyaraja H., Eisenman R. N., The MYC transcription factor network: Balancing metabolism, proliferation and oncogenesis. Front. Med. 12, 412–425 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kuzyk A., Mai S., c-MYC-induced genomic instability. Cold Spring Harb. Perspect. Med. 4, a014373 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fernandez P. C., et al. , Genomic targets of the human c-Myc protein. Genes Dev. 17, 1115–1129 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hermeking H., et al. , Identification of CDK4 as a target of c-MYC. Proc. Natl. Acad. Sci. U.S.A. 97, 2229–2234 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Galaktionov K., Chen X., Beach D., Cdc25 cell-cycle phosphatase as a target of c-myc. Nature 382, 511–517 (1996). [DOI] [PubMed] [Google Scholar]
- 15.Bretones G., Delgado M. D., León J., Myc and cell cycle control. Biochim. Biophys. Acta 1849, 506–516 (2015). [DOI] [PubMed] [Google Scholar]
- 16.Dyson N., The regulation of E2F by pRB-family proteins. Genes Dev. 12, 2245–2262 (1998). [DOI] [PubMed] [Google Scholar]
- 17.Bracken A. P., Ciro M., Cocito A., Helin K., E2F target genes: Unraveling the biology. Trends Biochem. Sci. 29, 409–417 (2004). [DOI] [PubMed] [Google Scholar]
- 18.Dong P., et al. , Division of labour between Myc and G1 cyclins in cell cycle commitment and pace control. Nat. Commun. 5, 4750 (2014). Erratum in: Nat. Commun. 9, 4766 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Leone G., DeGregori J., Sears R., Jakoi L., Nevins J. R., Myc and Ras collaborate in inducing accumulation of active cyclin E/Cdk2 and E2F. Nature 387, 422–426 (1997). [DOI] [PubMed] [Google Scholar]
- 20.Sears R., Ohtani K., Nevins J. R., Identification of positively and negatively acting elements regulating expression of the E2F2 gene in response to cell growth signals. Mol. Cell. Biol. 17, 5227–5235 (1997). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Adams M. R., Sears R., Nuckolls F., Leone G., Nevins J. R., Complex transcriptional regulatory mechanisms control expression of the E2F3 locus. Mol. Cell. Biol. 20, 3633–3639 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Attwooll C., Lazzerini Denchi E., Helin K., The E2F family: Specific functions and overlapping interests. EMBO J. 23, 4709–4716 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Trimarchi J. M., Lees J. A., Sibling rivalry in the E2F family. Nat. Rev. Mol. Cell Biol. 3, 11–20 (2002). [DOI] [PubMed] [Google Scholar]
- 24.Kong L. J., Chang J. T., Bild A. H., Nevins J. R., Compensation and specificity of function within the E2F family. Oncogene 26, 321–327 (2007). [DOI] [PubMed] [Google Scholar]
- 25.Leung J. Y., Ehmann G. L., Giangrande P. H., Nevins J. R., A role for Myc in facilitating transcription activation by E2F1. Oncogene 27, 4172–4179 (2008). [DOI] [PubMed] [Google Scholar]
- 26.Mei Y., Wu M., Noncoding RNAs regulating p53 and c-Myc signaling. Adv. Exp. Med. Biol. 927, 337–365 (2016). [DOI] [PubMed] [Google Scholar]
- 27.Winkle M., et al. , Long noncoding RNAs as a novel component of the Myc transcriptional network. FASEB J. 29, 2338–2346 (2015). [DOI] [PubMed] [Google Scholar]
- 28.Kim T., et al. , Role of MYC-regulated long noncoding RNAs in cell cycle regulation and tumorigenesis. J. Natl. Cancer Inst. 107, dju505 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kawasaki Y., et al. , MYU, a target lncRNA for Wnt/c-Myc signaling, mediates induction of CDK6 to promote cell cycle progression. Cell Rep. 16, 2554–2564 (2016). [DOI] [PubMed] [Google Scholar]
- 30.Cao L., Zhang P., Li J., Wu M., LAST, a c-Myc-inducible long noncoding RNA, cooperates with CNBP to promote CCND1 mRNA stability in human cells. eLife 6, e30433 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lu Y., et al. , MYC targeted long noncoding RNA DANCR promotes cancer in part by reducing p21 levels. Cancer Res. 78, 64–74 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Jackstadt R., Hermeking H., MicroRNAs as regulators and mediators of c-MYC function. Biochim. Biophys. Acta 1849, 544–553 (2015). [DOI] [PubMed] [Google Scholar]
- 33.Hart J. R., Roberts T. C., Weinberg M. S., Morris K. V., Vogt P. K., MYC regulates the non-coding transcriptome. Oncotarget 5, 12543–12554 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hamilton M. J., Young M. D., Sauer S., Martinez E., The interplay of long non-coding RNAs and MYC in cancer. AIMS Biophys. 2, 794–809 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sang B., et al. , Dual functions for OVAAL in initiation of RAF/MEK/ERK prosurvival signals and evasion of p27-mediated cellular senescence. Proc. Natl. Acad. Sci. U.S.A. 115, E11661–E11670 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Quinn J. J., Chang H. Y., Unique features of long non-coding RNA biogenesis and function. Nat. Rev. Genet. 17, 47–62 (2016). [DOI] [PubMed] [Google Scholar]
- 37.Bonasio R., Shiekhattar R., Regulation of transcription by long noncoding RNAs. Annu. Rev. Genet. 48, 433–455 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Salviano-Silva A., Lobo-Alves S. C., Almeida R. C., Malheiros D., Petzl-Erler M. L., Besides pathology: Long non-coding RNA in cell and tissue homeostasis. Noncoding RNA 4, E3 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sanchez Calle A., Kawamura Y., Yamamoto Y., Takeshita F., Ochiya T., Emerging roles of long non-coding RNA in cancer. Cancer Sci. 109, 2093–2100 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Fang Y., Fullwood M. J., Roles, functions, and mechanisms of long non-coding RNAs in cancer. Genomics Proteomics Bioinformatics 14, 42–54 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Weinstein J. N., et al. ; Cancer Genome Atlas Research Network , The cancer genome atlas pan-cancer analysis project. Nat. Genet. 45, 1113–1120 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Jain A. K., et al. , LncPRESS1 is a p53-regulated LncRNA that safeguards pluripotency by disrupting SIRT6-mediated de-acetylation of histone H3K56. Mol. Cell 64, 967–981 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Mathelier A., et al. , JASPAR 2016: A major expansion and update of the open-access database of transcription factor binding profiles. Nucleic Acids Res. 44, D110–D115 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Gregory R. I., Chendrimada T. P., Cooch N., Shiekhattar R., Human RISC couples microRNA biogenesis and posttranscriptional gene silencing. Cell 123, 631–640 (2005). [DOI] [PubMed] [Google Scholar]
- 45.Meister G., et al. , Human Argonaute2 mediates RNA cleavage targeted by miRNAs and siRNAs. Mol. Cell 15, 185–197 (2004). [DOI] [PubMed] [Google Scholar]
- 46.Kallen A. N., et al. , The imprinted H19 lncRNA antagonizes let-7 microRNAs. Mol. Cell 52, 101–112 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wang K. C., Chang H. Y., Molecular mechanisms of long noncoding RNAs. Mol. Cell 43, 904–914 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Cornella N., et al. , The hnRNP RALY regulates transcription and cell proliferation by modulating the expression of specific factors including the proliferation marker E2F1. J. Biol. Chem. 292, 19674–19692 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Tseng Y. Y., et al. , PVT1 dependence in cancer with MYC copy-number increase. Nature 512, 82–86 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Hung C. L., et al. , A long noncoding RNA connects c-Myc to tumor metabolism. Proc. Natl. Acad. Sci. U.S.A. 111, 18697–18702 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Xiao Z. D., et al. , Energy stress-induced lncRNA FILNC1 represses c-Myc-mediated energy metabolism and inhibits renal tumor development. Nat. Commun. 8, 783 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kim T., et al. , Long-range interaction and correlation between MYC enhancer and oncogenic long noncoding RNA CARLo-5. Proc. Natl. Acad. Sci. U.S.A. 111, 4173–4178 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Iaccarino I., lncRNAs and MYC: An intricate relationship. Int. J. Mol. Sci. 18, E1497 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Gutschner T., Diederichs S., The hallmarks of cancer: A long non-coding RNA point of view. RNA Biol. 9, 703–719 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Ginsberg D., E2F1 pathways to apoptosis. FEBS Lett. 529, 122–125 (2002). [DOI] [PubMed] [Google Scholar]
- 56.Johnson D. G., The paradox of E2F1: Oncogene and tumor suppressor gene. Mol. Carcinog. 27, 151–157 (2000). [DOI] [PubMed] [Google Scholar]
- 57.Matsumura I., Tanaka H., Kanakura Y., E2F1 and c-Myc in cell growth and death. Cell Cycle 2, 333–338 (2003). [PubMed] [Google Scholar]
- 58.O’Donnell K. A., Wentzel E. A., Zeller K. I., Dang C. V., Mendell J. T., c-Myc-regulated microRNAs modulate E2F1 expression. Nature 435, 839–843 (2005). [DOI] [PubMed] [Google Scholar]
- 59.Tay Y., Rinn J., Pandolfi P. P., The multilayered complexity of ceRNA crosstalk and competition. Nature 505, 344–352 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Cesana M., et al. , A long noncoding RNA controls muscle differentiation by functioning as a competing endogenous RNA. Cell 147, 358–369 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Yang F., Zhang H., Mei Y., Wu M., Reciprocal regulation of HIF-1α and lincRNA-p21 modulates the Warburg effect. Mol. Cell 53, 88–100 (2014). [DOI] [PubMed] [Google Scholar]
- 62.Yoon J. H., et al. , LincRNA-p21 suppresses target mRNA translation. Mol. Cell 47, 648–655 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Huarte M., et al. , A large intergenic noncoding RNA induced by p53 mediates global gene repression in the p53 response. Cell 142, 409–419 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Rhodes G. H., Valbracht J. R., Nguyen M. D., Vaughan J. H., The p542 gene encodes an autoantigen that cross-reacts with EBNA-1 of the Epstein Barr virus and which may be a heterogeneous nuclear ribonucleoprotein. J. Autoimmun. 10, 447–454 (1997). [DOI] [PubMed] [Google Scholar]
- 65.Rossi A., et al. , Identification and dynamic changes of RNAs isolated from RALY-containing ribonucleoprotein complexes. Nucleic Acids Res. 45, 6775–6792 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Sallam T., et al. , Feedback modulation of cholesterol metabolism by the lipid-responsive non-coding RNA LeXis. Nature 534, 124–128 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.McKeown M. R., Bradner J. E., Therapeutic strategies to inhibit MYC. Cold Spring Harb. Perspect. Med. 4, a014266 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Chen H., Liu H., Qing G., Targeting oncogenic Myc as a strategy for cancer treatment. Signal Transduct. Target. Ther. 3, 5 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Posternak V., Cole M. D., Strategically targeting MYC in cancer. F1000 Res. 5, 408 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Yang Y., et al. , TRMP, a p53-inducible long noncoding RNA, regulates G1/S cell cycle progression by modulating IRES-dependent p27 translation. Cell Death Dis. 9, 886 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Livak K. J., Schmittgen T. D., Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods 25, 402–408 (2001). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The RNA sequencing data have been deposited in the National Center for Biotechnology Information Sequence Read Archive with accession codes SRP171977 and SRP171802.