Abstract
MicroRNA-134 is a brain-enriched small noncoding RNA that has been implicated in diverse neuronal functions, including regulating network excitability. Increased expression of microRNA-134 has been reported in several experimental epilepsy models and in resected brain tissue from temporal lobe epilepsy patients. Rodent studies have demonstrated that reducing microRNA-134 expression in the brain using antisense oligonucleotides can increase seizure thresholds and attenuate status epilepticus. Critically, inhibition of microRNA-134 after status epilepticus can potently reduce the occurrence of spontaneous recurrent seizures. Altered plasma levels of microRNA-134 have been reported in epilepsy patients, suggesting microRNA-134 may have diagnostic value as a biomarker. This review summarises findings on the cellular functions of microRNA-134, as well as the preclinical evidence supporting anti-seizure and disease-modifying effects of targeting microRNA-134 in epilepsy. Finally, we draw attention to unanswered questions and some of the challenges and opportunities involved in preclinical development of a microRNA-based oligonucleotide treatment for epilepsy.
Keywords: Epileptogenesis, Noncoding RNA, Hippocampal sclerosis, Biomarker, RNA therapy
1. Introduction
Epilepsy is a disease of network dysfunction leading to brain hyperexcitability that manifests clinically as seizures. Epilepsy affects over 50 million people worldwide but does not respond adequately to available treatments in around 30% of cases [1]. Physiological brain function requires expression of a multitude of genes that establish the excitable properties of neuronal networks, giving rise to finely balanced excitatory and inhibitory neurotransmission. Protein levels within cells are controlled post-transcriptionally by small noncoding RNAs called microRNAs (miRs) which negatively regulate gene expression [2,3]. This is achieved through Watson-Crick base pairing between the 5′-end of the miR and 3’-UTR of a mRNA, an event mediated in part by Argonaute (AGO) proteins. Because it requires only a 7–8 nucleotide match for a miR to affect its target, individual miRs are predicted to regulate levels of multiple (perhaps hundreds) of mRNAs [4,5]. Overall, the function of miRs appears to be to control cellular protein “noise”, by fine-tuning and buffering protein levels in cells [6].
MiR biogenesis is essential for brain development and function. Brain tissue is particularly enriched in miRs, expressing several uniquely [[7], [8], [9]]. A key role for miRs is shaping the structure and function of synapses [10]. Several miRs have been identified that localise to the synapse where they locally control levels of proteins involved in post-synaptic structures [11]. Some of the biogenesis machinery for miR maturation is also co-located at post-synaptic sites where it is responsive to local changes in calcium signalling down-stream of NMDA receptor activation [12]. Since dendritic spine shape and volume influence synaptic strength [13], certain miRs are critical determinants of brain excitability.
MicroRNA-134 (miR-134) was one of the first miRs to be shown to be enriched in synapses. Originally identified as a brain derived neurotrophic factor (BDNF)-regulated miR, miR-134 was found to negatively regulate dendritic spine volume [11], suggesting potential roles in control of excitability within the brain. Later, increased levels of miR-134 were reported in several animal models of epilepsy and in resected brain tissue from patients with temporal lobe epilepsy (TLE) [14]. Since then, several studies have reported that silencing miR-134 using antisense oligonucleotides (antagomirs) reduces seizure severity in multiple models [14,15] and treatment of rodents with miR-134 inhibitors after an episode of status epilepticus (SE) can prevent or reduce the later occurrence of spontaneous recurrent seizures. Other miRs have been identified as therapeutic targets in epilepsy including miR-124 [16], miR-132 [17], miR-203 [18] and miR-219 [19]. However, miR-134 is a leading candidate because it is brain-enriched (thus limiting off-target effects) and targeting it results in generally superior seizure suppression and this is reported in multiple models [14,15]. miR-134 levels are also altered in blood samples from patients with epilepsy, suggesting diagnostic potential [20]. Here, we summarise our understanding of the role of miR-134 in brain function, its expression in various models of epilepsy and the findings to date on the therapeutic potential of blocking miR-134 for the treatment and prevention of seizures.
1.1. Discovery and genetic localisation
miR-134 was originally discovered among a number of brain-specific miRs in a screen of mouse tissues [21]. The miR was expressed at low levels relative to other brain-enriched miRs, such as miR-124 which is the most abundant. MiR-134 was subsequently shown to be coded within an imprinted locus on mouse distal 12 chromosome (human 14q32), where the mir-134 gene is located in a cluster of conserved A-repeats, close to miR-154 [22]. It was later shown that this locus contains a cluster of ~46 potential miR genes [23]. The expression of the whole locus is strongly controlled by an intergenic germline-derived differentially methylated region (IG-DMR), located between the Dlk1 and Gtl2 genes. The deletion of the IG-DMR from the unmethylated maternally inherited chromosome, but not the methylated paternally inherited chromosome, silences the expression of this gene cluster [24]. Clustering of miR genes is common, as their co-expression is required for a coordinated influence on related biological pathways [25]. Accordingly, the expression of the entire cluster is co-regulated by Mef2 [26]. This is an activity-dependent transcription factor and so suggests a role of miR-134 in controlling neuronal response to ongoing network function.
1.2. Cellular localisation
Schratt et al. [11] performed the first characterisation of miR-134 which strongly implicated miR-134 in the modulation of synaptic development. First, expression of miR-134 in the mouse hippocampus was shown to increase during brain development, reaching its maximum level at p13, the age at which synapses begin to mature. This was in contrast with certain other brain-enriched miRs (eg. miR-124) which were expressed at a constant level throughout development, and so began to show a role for miR-134 in synapse maturation [11]. This was consolidated by in situ hybridisation (ISH) experiments, which showed dendritic localisation of miR-134 [11] and its pre-cursor, pre-miR-134 [27]. Pre-miRs are formed in the nucleus [28] and exported to the cytoplasm [29]. Schratt's team found a specific trafficking mechanism which transports pre-miR-134 to dendrites. An RNA-binding protein, DEAH-box helicase DHX36, recognises a sequence within the pre-miR-134 terminal loop and guides it to the synapse [27], where it is subsequently cleaved by the enzyme Dicer [30] to produce mature miR-134. In recent work, Park and colleagues used atomic force microscopy to estimate the copy number of miR-134 at synapses. They found that immature synapses typically had 10–15 molecules, mainly found at the base of the spine [31]. In contrast, numbers of miR-134 molecules were about half that at mature synapses. Thus, production of active miR-134 is regulated by a combination of trafficking and local-activity-dependent mechanisms and the presence of miR-134 at a synapse may be a strong influence on the maturation and functional state of the local dendrite.
While neurons appear to be the main cell type that expresses miR-134, there remains some uncertainty about which subtypes express miR-134. Schratt's work and ISH performed by our group support a broad expression in both excitatory and inhibitory neurons in the rodent brain [11,14]. Other studies have suggested, however, that miR-134 expression may be enriched in certain inhibitory interneuron populations [32]. Resolving this issue is important since modulating levels of miR-134 might have opposing effects if it is expressed predominantly in inhibitory rather than excitatory neurons.
1.3. What is the function of miR-134 at the synapse?
Several methods have been used to modulate miR-134 expression, revealing insights into its function at the synapse. Schratt et al. [11] overexpressed miR-134 in cultured hippocampal neurons using a pcDNA3 vector, leading to a decrease in dendritic spine width and volume. Correspondingly, in the same study, suppression of endogenous miR-134 using a 2’-O-methylated antisense oligonucleotide (ant-134) caused a specific increase in spine volume, without changing spine density or dendritic complexity. These effects were shown to be mediated via miR-134's interaction with Lim-domain-containing protein kinase 1 (Limk1) mRNA (Fig. 1C), which contains a consensus for miR-134 [11]. Limk1 influences spine dynamics through an interaction with ADF/cofilin [33]. Therefore, in basal conditions, miR-134 seems to have a fairly specific role in modulating spine size. However, the precise role of miR-134 appears to be brain state dependent.
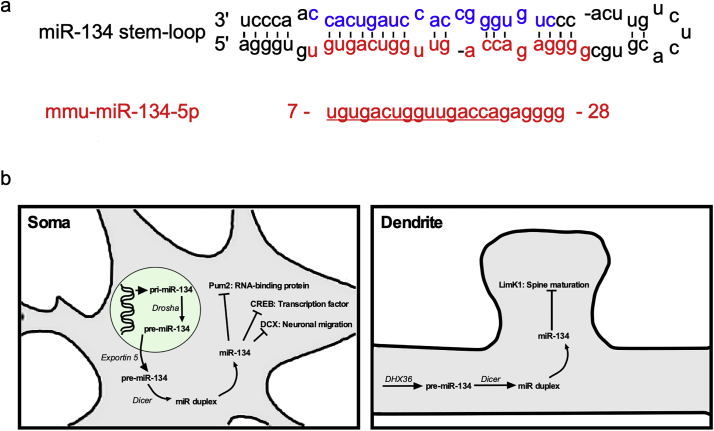
Fig. 1.
Sequence and targets of miR-134. a - Mouse miR-134 stem-loop structure containing both mmu-miR-134-3p (blue text) and mmu-miR-134-5p (red text). b - An overview of the biogenesis and selected mRNA targets of miR-134. Among those most likely to be relevant to epilepsy are CREB [37], DCX [38] and Pum2 [26] within the soma and LimK1 [11,14] in dendritic spines. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
A later study showed that knockdown of miR-134 in cultured hippocampal neurons does not affect dendritic complexity in basal conditions but, critically, can abolish the increase in branching which results from increased neuronal activity [26]. In this context, miR-134 expression is driven by the activity-dependent myocyte enhancing factor 2 (Mef2), which binds upstream of the Gtl2/Dlk1 locus (also referred to as the miR379-410 cluster) and enhances the expression of the entire cluster. The resulting increase in miR-134 promotes activity-dependent dendritogenesis through downregulation of a different target, the RNA-binding protein Pumilio2 (Pum2). This effect is blocked by ant-134. The precise regulation of dendritic outgrowth seems to follow a ‘tuning model’ [34], which requires miR-134 levels to remain within a limited physiologically-relevant range. Indeed, miR-134 overexpression in vivo using a recombinant adeno-associated virus vector also caused a reduction in dendritic outgrowth [35], phenocopying the results seen during miR-134 knockdown in vitro. Further study showed that miR-134 also mediates homeostatic synaptic depression during increased network activity [36]. Using luciferase 3’-UTR constructs, the authors confirmed that this process was mediated by Pum2 during hyperexcitable states [36], and not by Limk1, as in basal conditions [11]. This raises the exciting prospect that miR-134 is able to function differently, through distinct molecular mechanisms, depending on specific brain states.
A number of other targets of miR-134 have been identified which may indirectly affect brain excitability states. Gao and colleagues [37] showed that miR-134 targets cAMP response binding protein (CREB, Fig. 1C), an important regulator of synaptic plasticity. Another target of miR-134 is the neuronal guidance molecule doublecortin (DCX), with miR-134 functioning during brain development to reduce neuronal migration [38] (Fig. 1B).
1.4. miR-134 and epilepsy
Epilepsy is a neurological disease characterised by neuronal network dysfunction that leads to brain hyperexcitability and manifests clinically as the susceptibility to recurrent spontaneous seizures. Epilepsy is also frequently accompanied by significant co-morbidities, including depression and cognitive problems. Epilepsy can be caused by mutations in genes critical to brain development or neuronal function (e.g. ion channels, neurotransmitters) of which over 500 have been discovered, as well as be acquired through damage to the brain from physical or infectious causes [39]. Epileptogenesis is the process which links brain injury and other causative factors to the resulting emergence of epilepsy [[40], [41], [42], [43]]. The pathological underpinnings of epileptogenesis can include select cell loss [44], gliosis [42], neuroinflammation [43], and circuit restructuring [44,45]. The result of epileptogenesis is a network-level disruption which renders brain circuits susceptible to recurrent activation and synchrony of neuronal populations. For a more complete discussion of the mechanisms of epileptogenesis the reader is referred elsewhere [[40], [41], [42], [43]].
Frontline treatments for people with epilepsy are anti-seizure drugs, including channel blockers, receptor modifiers, and other agents which broadly work by dampening network excitability through boosting inhibition or suppressing excitation. Around 30% of patients with epilepsy do not experience seizure freedom with existing treatments [1], leaving an urgent and unmet need for new disease-modifying therapies. Additionally, anti-seizure drugs have limited specificity [46]. This can lead to adverse effects on normal brain function (common side effects include drowsiness/sedation and cognitive difficulties), highlighting the need for novel approaches towards epilepsy management. MiRs represent a highly promising new target in epilepsy therapy, owing to their ability to modify properties of neuronal networks through multiple molecular targets. Since miR-134 shows activity-dependent expression and appears to target network function through several complementary mechanisms in a context-dependent manner, it could represent an ideal target for treatment of seizures. Accordingly, we began to explore the link between miR-134, epileptogenesis and epilepsy.
We began by studying the spatio-temporal relationship between miR-134 levels and pathological brain activity [14]. We used real-time qPCR analysis of brain tissue taken from mice subjected to prolonged seizures induced by microinjection of kainic acid (KA) into the amygdala. This is a well-characterised rodent model of SE which has the benefit of also triggering recurrent spontaneous seizures within a few days, so that changes relating to epilepsy can also be investigated [47]. This revealed an increase in mature miR-134, specifically in the hippocampus ipsilateral to the KA injection. The increase occurred in both the area of damage (CA3 subfield) and the relatively spared CA1 area, suggesting seizures rather than damage are the main driver of this change. Accompanying this change were lower levels of LimK1, as well as CREB, consistent with their expected responses as miR-134 targets. In hippocampal samples from mice that developed epilepsy there was also an increase in miR-134 levels, particularly within the CA1 subfield. The same increase in miR-134 was observed in human specimens of neocortex and hippocampus, from patients undergoing resective surgery for pharmacoresistant TLE [14,15]. This is an important finding because it suggests that miR-134 could have a similar role in human epilepsy and could therefore be exploited clinically. Increased miR-134 levels have since been reported in other seizure models (see Table 1 and Fig. 2C) including pilocarpine [48], pentylenetetrazol (PTZ) [15], and in vitro models [49].
Table 1.
Summary of in vivo results targeting miR-134 in models of epilepsy.
| Model | Treatment time | Delivery mode | Seizure phenotype | Histology | References |
|---|---|---|---|---|---|
| Mouse IAKA | 24 h before SE | i.c.v. | ↓ EEG power in SE | ↓ neuronal death in CA3 | [14] |
| Mouse IAKA | 1 h after SE | i.c.v. | ↓ spontaneous seizures | ↔ neuronal death in CA3 | [14] |
| Mouse PILO | 24 h before SE | i.c.v. | ↓ proportion of mice with SE | N/A | [48] |
| ↑ delay to seizure onset | |||||
| Rat PPS | 24 h before PPS | i.c.v. | SE unchanged | N/A | [15] |
| ↓ spontaneous seizures | |||||
| Rat PPS | Immediately after PPS | i.c.v. | No epilepsy in 6/7 rats | N/A | [15] |
| Mouse PTZ | 24 h before SE | i.c.v. | ↑ delay to seizure onset | N/A | [15] |
| ↓ seizure severity | |||||
| ↓ EEG power | |||||
| Rat i.c.v. KA | Not specified | i.c.v. | ↓ spontaneous seizures | ↓ neuronal death in CA3 | [52] |
| ↓ mossy fibre sprouting | |||||
| High K+ (ex vivo rat) | 2–4 days before slice preparation | i.c.v. | ↑ delay to activity onset | N/A | [15,51] |
Key: IAKA, intraamygdala kainic acid; i.c.v.; intracerebroventricular; KA, kainic acid; PILO, pilocarpine; PPS, perforant pathway stimulation; PTZ, pentylenetetrazol; N/A, not available; SE, status epilepticus.
Fig. 2.
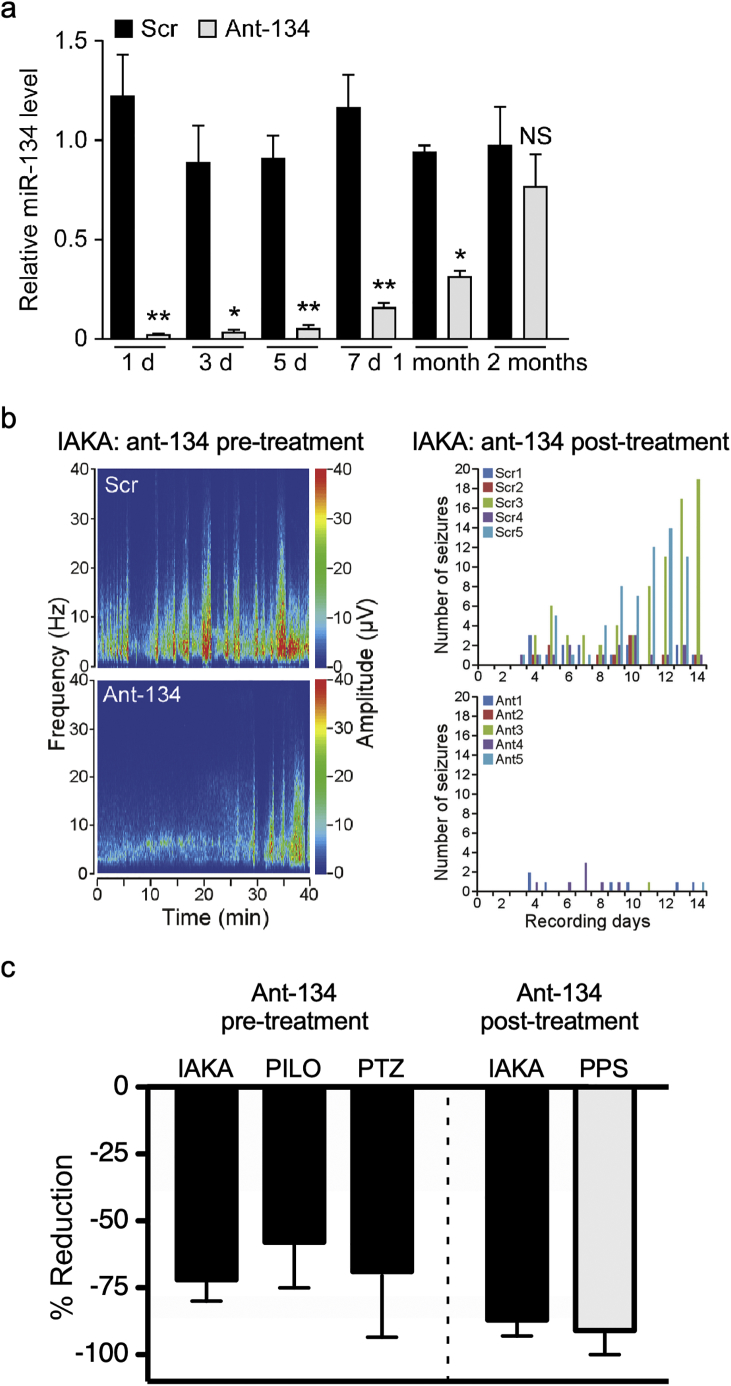
miR-134 knockdown is therapeutic in various in vivo experimental models and species. A - In vivo intracerebroventricular injection of ant-134 in mice mediates a significant knockdown of miR-134 for one month (reproduced with permission from [14]). B - IAKA - intra-amygdala kainic acid (reproduced with permission from [14]): Spectrograms show that status epilepticus is reduced by pre-treatment with ant-134 (left panel). Post-treatment with ant-134 leads to a reduction in spontaneous seizures during the chronic phase of this model (right panel). C - An overview of seizure reduction mediated by ant-134 in different animal models. For pre-treatment studies, we measured the % reduction in total EEG power after KA administration. For post-treatment studies, we measured the % reduction in number of spontaneous seizures in the chronic epilepsy phase of the models. Black bars represent mouse studies and grey bars rat studies. IAKA - intra-amygdala kainic acid; PILO - pilocarpine; PTZ - pentylenetetrazol; PPS - perforant path stimulation.
1.5. Manipulating miR-134 using oligonucleotides: insights into function
We then used ant-134 in the mouse brain and explored whether it affected seizures. The sequence chosen was a 16mer oligonucleotide with a cholesterol tag to enhance cellular uptake. Having found a dose that selectively reduced miR-134 levels without affecting other miRNAs, we measured the timecourse of miR-134 knockdown in mouse hippocampus [14]. Lower miR-134 levels first became detectable 12 h after injection into the ventricle. This route of delivery was required because systemically injected oligonucleotides do not pass through an intact blood-brain barrier [50]. Maximal knockdown (95%) was detected at 24 h and silencing lasted for one month following a single injection (Fig. 2A). This is potentially an important therapeutic advantage of ant-134 since it suggests that a single treatment could be effective for around one month. This is in comparison with anti-epileptic drugs which typically need to be taken at least once per day. When antagomirs were injected 24 h before SE, mice experienced significantly reduced seizure severity in the intraamygdala KA model (Fig. 2B). The hippocampus from these mice was also less damaged. Interestingly, when induction of seizures was delayed for 14 days, a time when miR-134 levels had returned to ~30% of baseline, the mice were no longer refractory to SE. This indicates a need for strong suppression of miR-134 to achieve anti-seizure effects in a model of SE. The in vivo anti-seizure effects of antagomirs targeting miR-134 when given as a pre-treatment have since been reported in other models, including the pilocarpine and PTZ models [15] (Table 1 and Fig. 2C). The same oligonucleotides also suppress epileptiform activity in ex vivo rat brain slices treated with elevated potassium [15,51]. Importantly, a number of independent groups have reported anti-excitability effects of targeting miR-134 in in vitro and in vivo, including work in primary cultures of rat hippocampal neurons exposed to low-magnesium solution [49] and most recently in another in vivo KA rodent model [52].
In a subsequent series of studies, the effects of silencing miR-134 after SE on the development of spontaneous recurrent seizures were explored. SE is an effective method for inducing epileptogenesis and chronic epilepsy, and in the intraamygdala KA model spontaneous seizures begin within 3–5 days, occurring steadily thereafter. Injection of the antagomirs after SE had no effect on the SE itself, consistent with it taking many hours for the miRNA level to be reduced by the antagomirs. Remarkably, when mice injected after SE were tracked for two weeks with continuous EEG monitoring numbers of spontaneous seizures were reduced by over 90% [14]. A second study followed mice out to two months and found seizure rates were still 70% lower (Fig. 2). This was the first demonstration that ant-134 could be therapeutic in epilepsy. An analysis of the mouse brains at the end of studies found evidence of neuroprotection, but the mice still had lesions indicating the disease-modifying action was not solely a function of neuroprotection. This finding has since been extended to rats which underwent perforant pathway stimulation [15]. In that study, silencing miR-134 after SE prevented epilepsy in 6/7 rats whereas all the controls became epileptic. These promising findings suggest that ant-134 can be therapeutic against seizures with differing underlying causes, and so could be effective in a variety of epilepsy syndromes.
1.6. Cognitive effects of targeting miR-134 and safety profile
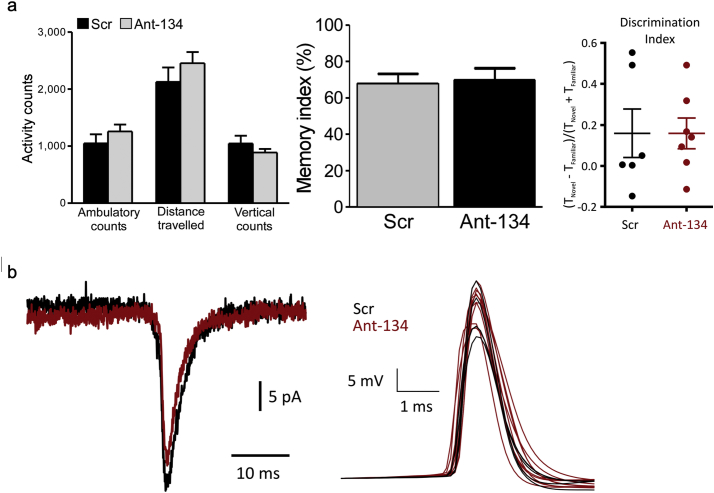
An important aspect in the translation of a new treatment to the clinic is safety and a lack of off-target effects. During early studies we observed that ant-134 causes a small reduction in dendritic spine density in CA3 pyramidal neurons [14]. Further analysis showed that the remaining spines had a greater volume in the ant-134 treated group [48]. Nevertheless, mice injected with antagomirs did not display differences in natural behaviour in an ethogram test [14] (Fig. 3A). We used the novel object location test [53] to probe spatial memory in vivo, since this is thought to be a largely hippocampal dependent task [54]. We observed no difference in object location discrimination between animals injected with ant-134 or vehicle control, in both mice [48] and rats [51], suggesting that ant-134 spares normal hippocampal function (Fig. 3A). Likewise, biophysical properties of pyramidal neurons, along with electrophysiological readouts of glutamatergic neurotransmission, were not changed by ant-134 [51] (Fig. 3B). Further validation of safety will be required, but ant-134 does not appear to have a large impact on normal cognitive function.
Fig. 3.
miR-134 knockdown does not appear to alter normal rodent behaviour or biophysical properties. A - Normal rodent behaviour is unaffected by ant-134 administration. Activity counts in mice (left panel - reproduced with permission from [14]) and novel object location discrimination in mice (middle panel - reproduced with permission from [48]) and rats (right panel - reproduced with permission from [51]) were not changed by ant-134. B - Rat hippocampal pyramidal neuron biophysics are not changed by ant-134. Both miniature excitatory post-synaptic potentials (left panel) and action potentials (right panel) were unchanged (reproduced with permission from [51]).
1.7. Is there an opportunity to co-develop miR-134 as a circulating biomarker?
Several brain-enriched miRNAs including miR-134 have been found to be present in biofluids, including blood, raising the intriguing possibility of their use as diagnostic biomarkers. Overall, miRs are attractive in this regard. In addition to several being brain-specific (and thus their presence in the circulation could reflect damage to the brain or opening of the blood-brain barrier which is known to accompany seizures [55]), they are stable and amenable to rapid detection using techniques such as PCR. While profiling studies have not yet picked out miR-134 as a biomarker of epilepsy, several studies focusing on miR-134 specifically have suggested it may have biomarker potential. We and other groups have detected higher levels of miR-134 in plasma and CSF from patients with epilepsy [20,56]. In contrast, Avansini and coworkers [57] reported lower miR-134 levels in plasma from patients with TLE. Accordingly, more research, perhaps combining samples from different sites, is needed to clarify whether miR-134 is a biomarker of epilepsy. Indeed, other neurological disorders may also cause a spike in miR-134 levels in the circulation meaning it may not be sufficiently specific for epilepsy [58,59]. Finally, we recently showed that injection of a disease-modifying dose of ant-134 into mice could also normalise levels of other circulating miRNA biomarkers [60]. Taken together, these results suggest that a circulating miR-based diagnostic test could be co-developed with a miR-134-targeting therapy for epilepsy.
1.8. Outstanding questions: remaining barriers to clinical translation of ant-134
Targeting miR-134 is an attractive novel approach for seizure control and disease modification. Foremost, it has shown consistent efficacy – seizures are reduced in six of seven tested models (Table 1, Fig. 2). This is as good as and better than the performance of many current AEDs. The apparent anti-epileptogenic actions of ant-134 are highly promising [14,15,52] and it does not appear to have any obvious adverse effects in rodents [48,51]. However, there are still several remaining hurdles before ant-134 can be realistically translated to the epilepsy clinic.
Firstly, in all published studies to date ant-134 was administered either before or immediately after the epileptogenic insult. Whilst this is appropriate for demonstrating anti-seizure and anti-epileptogenic effects, it is not realistic in the clinical setting, where patients usually present with established epilepsy. The more relevant challenge will be to administer ant-134 in rodent models after epilepsy has developed. Although ant-134 has not been tested in such a model, a study recently reported antagomirs targeting miR-135a reduced spontaneous seizures in epileptic rodents [61].
Another obstacle to clinical translation is how to deliver ant-134 to the brain in patients with epilepsy. Rodent studies to date have all used direct administration of ant-134 to the brain via stereotaxic intracerebroventricular injection - a highly invasive surgical procedure. This would not be impossible for use in humans with pharmacorefractory epilepsy - such patients may have to undergo surgery anyway - but a less invasive route of delivery would be highly preferable. Perhaps, if the blood brain barrier is open as a result of recent seizure activity or briefly opened (for example using ultrasound or a hyper-osmotic solution) then a systemic injection could reach the brain. We recently showed this was possible in principal, delivering antagomirs systemically after SE in mice and demonstrating they still have potent, long-lasting anti-seizure effects [62]. Another potential delivery method is via intrathecal injection.
There may be other methods for silencing miR-134. A gene therapy-based approach could use a sponge construct with multiple binding sites for miR-134 to reduce its levels. If delivered via a viral vector it could reach the target tissue and provide lasting, cell-specific miR-134 suppression [63]. Another approach might be to use target site blockers [64]. These are antisense oligonucleotides that block the site within the 3’UTR where a miR binds. This preserves the main targets of the miR while protecting one site. This could reduce off-target effects, particularly if a miR's main effects are through a single target (in the present context, a target site blocker against the miR-134 site in the LimK1 mRNA).
The majority of in vivo studies reported to date have tested antagomirs in adult rodents. Would ant-134 suppress seizures in the developing brain? This could increase the therapeutic use of the molecule since it could be used in paediatric SE or refractory epilepsy. However, the safety of ant-134 in the developing brain requires further validation. This is because miR-134 may play a role in synaptic development and maturation via LimK1 [11] and in neuronal migration via DCX [38].
Could ant-134 be effective in genetic epilepsies? Such conditions are highly pharmacorefractory and often very severe [65], indicative of a strong need for new treatment strategies. There is evidence that other antisense oligonucleotide-based therapies can be therapeutic in genetic epilepsies [66]. Does ant-134 have any effects against the co-morbidities that accompany epilepsy? Cognitive impairments are a feature of several models of acquired epilepsy and it would be important to explore whether treatment with ant-134 has protective effects on these in addition to or independent of seizure control.
Finally, ant-134 has not been tested in human-derived tissue. Whilst the efficacy of ant-134 has been demonstrated in multiple species, there remains a possibility that it could be ineffective in human epilepsy. First, it has yet to be demonstrated that antagomirs can be taken up into human neurons, or that they can effectively target human miRs. Another concern is whether the mRNA pathways targeted by miRs are conserved in humans. One of the most relevant examples is that the 3’UTR of LimK1 mRNA, a key miR-134 target in the context of epilepsy [14], is not well-conserved between rodents and humans [11]. However, ant-134 does also seem to mediate anti-epileptic effects through other targets and so it remains to be seen whether it can be therapeutic in human tissue. These questions can be answered using two types of human-derived tissue. First, induced pluripotent stem cells (iPSCs) can be obtained from patients with epilepsy. These cells can be maintained in culture and reprogrammed to derive the desired cell type, which can then be used to probe the uptake of ant-134 into human cells and its ability to knockdown miR-134 and restore miR-134 targets in single neurons. However, questions remain about the possibility to re-capitulate realistic epileptic networks using human iPSCs. This may be overcome in the future with the use of three-dimensional human neuronal cultures [67,68], which form more (patho)physiologically realistic networks. It would be possible to create such cultures using iPSC-derived neurons donated by patients [69] with genetic epilepsies to realistically mimic a range of epilepsy-related impairments such as neuronal migration (eg RELN mutation [70,71]), transcriptional alterations (eg PURA [72]), or channelopathies (eg SCN1A [73]). Further, there are some doubts about the maturity of iPSC-derived neurons [74]. This limitation is overcome by the second approach: the use of surgically resected tissue from patients with pharmacoresistant epilepsy. This tissue can be used to model epileptiform activity in human brain [75] and is thus the most clinically relevant challenge remaining for ant-134.
2. Conclusion
miRs represent a completely new class of therapeutic target for epilepsy. A leading candidate in this regard is miR-134 for which we now have extensive preclinical data. The barriers to translation are significant. Ant-134 represents an enormously promising novel disease-modifying therapy for epilepsy. It has shown consistent efficacy in multiple disease models with anti-seizure effects that outlast the presence of ant-134 in the brain. Ongoing work will be required to confirm efficacy in humans with established epilepsy and to establish the optimum delivery method for ant-134.
2.1. Search strategy and selection criteria
Sources for this review were identified by searches of PubMed using the search terms ‘miR-134’ or ‘microRNA-134’ and ‘epilepsy’. Further sources were identified from references within these articles. Only articles published in English were included.
Funding sources
The authors are grateful to our funders who have supported research on microRNAs including funding from the Health Research Board Ireland (HRA-POR-2013-325), Science Foundation Ireland (13/IA/1891, 11/TIDA/B1988), fellowships from the Irish Research Council and the European Union Seventh Framework Programme (FP7/2007-2013) under grant agreement n° 602130 (EpimiRNA). This publication has also emanated from research supported in part by a research grant from Service Foundation Ireland (SFI) under Grant Number 16/RC/3948 and co-funded under the European Regional Development Fund and by FutureNeuro industry partners. The funders had no role in the design of the review, decision to publish, or preparation of the manuscript.
Authors' contributions
All authors contributed to literature search, manuscript draft and writing. GM made figures. DH and CRR contributed equally to this manuscript. All authors have approved the final version of this manuscript.
Declaration of Competing Interest
Authors report funding for microRNA-134 and related research supported in part by a research grant from Service Foundation Ireland (SFI) under Grant Number 16/RC/3948 and co-funded under the European Regional Development Fund and by FutureNeuro industry partners. DCH also reports funding from the Health Research Board Ireland (HRA-POR-2013-325), Science Foundation Ireland (13/IA/1891/B1988, 11/TIDA/B1988), and the European Union Seventh Framework Programme (FP7/2007-2013) under grant agreement n° 602,130 (EpimiRNA). CRR reports a fellowship from the Irish Research Council. In addition, Dr. Henshall has a patent awarded on the use of a microRNA-134 inhibitor for the treatment of epilepsy. US patent No. US 9,803,200 B2 “Inhibition of microRNA-134 for the treatment of seizure-related disorders and neurologic injuries”.
Acknowledgements
We thank Eva Jimenez-Mateos for support with extracting data to generate Fig. 2 and current and former colleagues who worked on miR-134 and related projects.
References
- 1.Brodie M.J., Barry S.J.E., Bamagous G.A., Norrie J.D., Kwan P. Patterns of treatment response in newly diagnosed epilepsy. Neurology. 2012;78:1548–1554. doi: 10.1212/WNL.0b013e3182563b19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bartel D.P. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–297. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 3.Bartel D.P. MicroRNAs: target recognition and regulatory functions. Cell. 2009;136:215–233. doi: 10.1016/j.cell.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lim L.P., Lau N.C., Garrett-Engele P., Grimson A., Schelter J.M., Castle J. Microarray analysis shows that some microRNAs downregulate large numbers of target mRNAs. Nature. 2005;433:769–773. doi: 10.1038/nature03315. [DOI] [PubMed] [Google Scholar]
- 5.Selbach M., Schwanhäusser B., Thierfelder N., Fang Z., Khanin R., Rajewsky N. Widespread changes in protein synthesis induced by microRNAs. Nature. 2008;455:58–63. doi: 10.1038/nature07228. [DOI] [PubMed] [Google Scholar]
- 6.Ebert M.S., Sharp P.A. Roles for MicroRNAs in conferring robustness to biological processes. Cell. 2012;149:515–524. doi: 10.1016/j.cell.2012.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hébert S.S., Papadopoulou A.S., Smith P., Galas M.C., Planel E., Silahtaroglu A.N. Genetic ablation of dicer in adult forebrain neurons results in abnormal tau hyperphosphorylation and neurodegeneration. Hum Mol Genet. 2010;19:3959–3969. doi: 10.1093/hmg/ddq311. [DOI] [PubMed] [Google Scholar]
- 8.Miska E.A., Alvarez-Saavedra E., Townsend M., Yoshii A., Sestan N., Rakic P. Microarray analysis of microRNA expression in the developing mammalian brain. Genome Biol. 2004;5:R68. doi: 10.1186/gb-2004-5-9-r68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Nowakowski T.J., Rani N., Golkaram M., Zhou H.R., Alvarado B., Huch K. Regulation of cell-type-specific transcriptomes by microRNA networks during human brain development. Nat Neurosci. 2018;21:1784–1792. doi: 10.1038/s41593-018-0265-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Biever A., Donlin-Asp P.G., Schuman E.M. Local translation in neuronal processes. Curr Opin Neurobiol. 2019;57:141–148. doi: 10.1016/j.conb.2019.02.008. [DOI] [PubMed] [Google Scholar]
- 11.Schratt G.M., Tuebing F., Nigh E.A., Kane C.G., Sabatini M.E., Kiebler M. A brain-specific microRNA regulates dendritic spine development. Nature. 2006;439:283–289. doi: 10.1038/nature04367. [DOI] [PubMed] [Google Scholar]
- 12.Sambandan S., Akbalik G., Kochen L., Rinne J., Kahlstatt J., Glock C. Activity-dependent spatially localized miRNA maturation in neuronal dendrites. Science. 2017;355:634–637. doi: 10.1126/science.aaf8995. [DOI] [PubMed] [Google Scholar]
- 13.Hering H., Sheng M. Dentritic spines: structure, dynamics and regulation. Nat Rev Neurosci. 2001;2:880–888. doi: 10.1038/35104061. [DOI] [PubMed] [Google Scholar]
- 14.Jimenez-Mateos E.M., Engel T., Merino-Serrais P., McKiernan R.C., Tanaka K., Mouri G. Silencing microRNA-134 produces neuroprotective and prolonged seizure-suppressive effects. Nat Med. 2012;18:1087–1094. doi: 10.1038/nm.2834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Reschke C.R., Silva L.F.A., Norwood B.A., Senthilkumar K., Morris G., Sanz-Rodriguez A. Potent anti-seizure effects of locked nucleic acid antagomirs targeting miR-134 in multiple mouse and rat models of epilepsy. Mol Ther - Nucleic Acids. 2017;6:45–56. doi: 10.1016/j.omtn.2016.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wang W., Wang X., Chen L., Zhang Y., Xu Z., Liu J. The microRNA miR-124 suppresses seizure activity and regulates CREB1 activity. Expert Rev Mol Med. 2016;18:1–16. doi: 10.1017/erm.2016.3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jimenez-Mateos E.M., Bray I., Sanz-Rodriguez A., Engel T., McKiernan R.C., Mouri G. MiRNA expression profile after status epilepticus and hippocampal neuroprotection by targeting miR-132. Am J Pathol. 2011;179:2519–2532. doi: 10.1016/j.ajpath.2011.07.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lee S.-T., Jeon D., Chu K., Jung K.-H., Moon J., Sunwoo J. Inhibition of miR-203 reduces spontaneous recurrent seizures in mice. Mol Neurobiol. 2017;54:3300–3308. doi: 10.1007/s12035-016-9901-7. [DOI] [PubMed] [Google Scholar]
- 19.Zheng H., Tang R., Yao Y., Ji Z., Cao Y., Liu Z. MiR-219 protects against seizure in the kainic acid model of epilepsy. Mol Neurobiol. 2016;53:1–7. doi: 10.1007/s12035-014-8981-5. [DOI] [PubMed] [Google Scholar]
- 20.McArdle H., Jimenez-Mateos E.M., Raoof R., Carthy E., Boyle D., ElNaggar H. “TORNADO” – theranostic one-step RNA detector; microfluidic disc for the direct detection of microRNA-134 in plasma and cerebrospinal fluid. Sci Rep. 2017;7 doi: 10.1038/s41598-017-01947-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lagos-Quintana M., Rauhut R., Yalcin A., Meyer J., Lendeckel W., Tuschl T. Identification of tissue-specific MicroRNAs from mouse. Curr Biol. 2002;12:735–739. doi: 10.1016/s0960-9822(02)00809-6. [DOI] [PubMed] [Google Scholar]
- 22.Seitz H., Youngson N., Lin S.P., Dalbert S., Paulsen M., Bachellerie J.P. Imprinted microRNA genes transcribed antisense to a reciprocally imprinted retrotransposon-like gene. Nat Genet. 2003;34:261–262. doi: 10.1038/ng1171. [DOI] [PubMed] [Google Scholar]
- 23.Seitz H., Royo H., Bortolin M.-L., Lin S.-P., Ferguson-Smith A.C., Cavaillé J. A large imprinted microRNA gene cluster at the mouse Dlk1-Gtl2 domain. Genome Res. 2004;14:1741–1748. doi: 10.1101/gr.2743304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lin S.P., Youngson N., Takada S., Seitz H., Reik W., Paulsen M. Asymmetric regulation of imprinting on the maternal and paternal chromosomes at the Dlk1-Gtl2 imprinted cluster on mouse chromosome 12. Nat Genet. 2003;35:97–102. doi: 10.1038/ng1233. [DOI] [PubMed] [Google Scholar]
- 25.He L., Thomson J.M., Hemann M.T., Hernando-Monge E., Mu D., Goodson S. A microRNA polycistron as a potential human oncogene. Nature. 2005;435:828–833. doi: 10.1038/nature03552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Fiore R., Khudayberdiev S., Christensen M., Siegel G., Flavell S.W., Kim T.K. Mef2-mediated transcription of the miR379-410 cluster regulates activity-dependent dendritogenesis by fine-tuning Pumilio2 protein levels. EMBO J. 2009;28:697–710. doi: 10.1038/emboj.2009.10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bicker S., Khudayberdiev S., Weiß K., Zocher K., Baumeister S., Schratt G. The DEAH-box helicase DHX36 mediates dendritic localization of the neuronal precursor-microRNA-134. Genes Dev. 2013;27:991–996. doi: 10.1101/gad.211243.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lee Y., Ahn C., Han J., Choi H., Kim J., Yim J. The nuclear RNase III Drosha initiates microRNA processing. Nature. 2003;425:415–419. doi: 10.1038/nature01957. [DOI] [PubMed] [Google Scholar]
- 29.Yi R, Qin Y, Macara IG, Cullen BR. Exportin-5 mediates the nuclear export of pre-microRNAs and short hairpin RNAs Exportin-5 mediates the nuclear export of pre-microRNAs and short hairpin RNAs 2003:3011–6. doi: 10.1101/gad.1158803. [DOI] [PMC free article] [PubMed]
- 30.Denli A.M., Tops B.B.J., Plasterk R.H.A., Ketting R.F., Hannon G.J. Processing of primary microRNAs by the microprocessor complex. Nature. 2004;432:231–235. doi: 10.1038/nature03049. [DOI] [PubMed] [Google Scholar]
- 31.Park I., Kim H.J., Kim Y., Hwang H.S., Kasai H., Kim J.-H. Nanoscale imaging reveals miRNA-mediated control of functional states of dendritic spines. Proc Natl Acad Sci U S A. 2019:1–26. doi: 10.1073/pnas.1819374116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chai S., Cambronne X.A., Eichhorn S.W., Goodman R.H. MicroRNA-134 activity in somatostatin interneurons regulates H-Ras localization by repressing the palmitoylation enzyme, DHHC9. Proc Natl Acad Sci. 2013;110:17898–17903. doi: 10.1073/pnas.1317528110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bamburg J.R. Proteins of the ADF/cofilin family: essential regulators of actin dynamics. Annu Rev Cell Dev Biol. 1999;15:185–230. doi: 10.1146/annurev.cellbio.15.1.185. [DOI] [PubMed] [Google Scholar]
- 34.Hobert O. miRNAs play a tune. Cell. 2007;131:22–24. doi: 10.1016/j.cell.2007.09.031. [DOI] [PubMed] [Google Scholar]
- 35.Christensen M., Larsen L.A., Kauppinen S., Schratt G. Recombinant Adeno-associated virus-mediated microRNA delivery into the postnatal mouse brain reveals a role for miR-134 in Dendritogenesis in vivo. Front Neural Circuits. 2010;3 doi: 10.3389/neuro.04.016.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Fiore R., Rajman M., Schwale C., Bicker S., Antoniou A., Bruehl C. MiR-134-dependent regulation of Pumilio-2 is necessary for homeostatic synaptic depression. EMBO J. 2014;33:2231–2246. doi: 10.15252/embj.201487921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Gao J., Wang W.Y., Mao Y.W., Gräff J., Guan J.S., Pan L. A novel pathway regulates memory and plasticity via SIRT1 and miR-134. Nature. 2010;466:1105–1109. doi: 10.1038/nature09271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Gaughwin P., Ciesla M., Yang H., Lim B., Brundin P. Stage-specific modulation of cortical neuronal development by Mmu-miR-134. Cereb Cortex. 2011;21:1857–1869. doi: 10.1093/cercor/bhq262. [DOI] [PubMed] [Google Scholar]
- 39.Devinsky O., Vezzani A., O'Brien T.J., Jette N., Scheffer I.E., de Curtis M. Epilepsy. Nat Rev Dis Prim. 2018;4 doi: 10.1038/nrdp.2018.24. [DOI] [PubMed] [Google Scholar]
- 40.Goldberg E.M., Coulter D.A. Mechanisms of epileptogenesis: a convergence on neural circuit dysfunction. Nat Rev Neurosci. 2013;14:337–349. doi: 10.1038/nrn3482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Pitkänen A., Lukasiuk K. Mechanisms of epileptogenesis and potential treatment targets. Lancet Neurol. 2011;10:173–186. doi: 10.1016/S1474-4422(10)70310-0. [DOI] [PubMed] [Google Scholar]
- 42.Devinsky O., Vezzani A., Najjar S., De Lanerolle N.C., Rogawski M.A. Glia and epilepsy: excitability and inflammation. Trends Neurosci. 2013;36:174–184. doi: 10.1016/j.tins.2012.11.008. [DOI] [PubMed] [Google Scholar]
- 43.Vezzani A., French J., Bartfai T., Baram T.Z. The role of inflammation in epilepsy. Nat Rev Neurol. 2011;7:31–40. doi: 10.1038/nrneurol.2010.178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Mello L.E.A.M., Cavalheiro E.A., Tan A.M., Kupfer W.R., Pretorius J.K., Babb T.L. Circuit mechanisms of seizures in the pilocarpine model of chronic epilepsy: cell loss and mossy fiber sprouting. Epilepsia. 1993;34:985–995. doi: 10.1111/j.1528-1157.1993.tb02123.x. [DOI] [PubMed] [Google Scholar]
- 45.Sutula T., Xiao-Xian H., Cavazos J., Scott G. Synaptic reorganization in the hippocampus induced by abnormal functional activity. Science (80-) 1988;239:1147–1150. doi: 10.1126/science.2449733. [DOI] [PubMed] [Google Scholar]
- 46.Sirven J.I., Noe K., Hoerth M., Drazkowski J. Antiepileptic drugs 2012: recent advances and trends. Mayo Clin Proc. 2012;87:879–889. doi: 10.1016/j.mayocp.2012.05.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Mouri G., Jimenez-Mateos E., Engel T., Dunleavy M., Hatazaki S., Paucard A. Unilateral hippocampal CA3-predominant damage and short latency epileptogenesis after intra-amygdala microinjection of kainic acid in mice. Brain Res. 2008;1213:140–151. doi: 10.1016/j.brainres.2008.03.061. [DOI] [PubMed] [Google Scholar]
- 48.Jimenez-Mateos E.M., Engel T., Merino-Serrais P., Fernaud-Espinosa I., Rodriguez-Alvarez N., Reynolds J. Antagomirs targeting microRNA-134 increase hippocampal pyramidal neuron spine volume in vivo and protect against pilocarpine-induced status epilepticus. Brain Struct Funct. 2015;220:2387–2399. doi: 10.1007/s00429-014-0798-5. [DOI] [PubMed] [Google Scholar]
- 49.Wang X.M., Jia R.H., Wei D., Cui W.Y., Jiang W. MiR-134 blockade prevents status epilepticus like-activity and is neuroprotective in cultured hippocampal neurons. Neurosci Lett. 2014;572 doi: 10.1016/j.neulet.2014.04.049. [DOI] [PubMed] [Google Scholar]
- 50.Straarup E.M., Fisker N., Hedtjärn M., Lindholm M.W., Rosenbohm C., Aarup V. Short locked nucleic acid antisense oligonucleotides potently reduce apolipoprotein B mRNA and serum cholesterol in mice and non-human primates. Nucleic Acids Res. 2010;38:7100–7111. doi: 10.1093/nar/gkq457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Morris G., Brennan G.P., Reschke C.R., Henshall D.C., Schorge S. Spared CA1 pyramidal neuron function and hippocampal performance following antisense knockdown of microRNA-134. Epilepsia. 2018:1–9. doi: 10.1111/epi.14475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Gao X., Guo M., Meng D., Sun F., Guan L., Cui Y. Silencing MicroRNA-134 alleviates hippocampal damage and occurrence of spontaneous seizures after intraventricular kainic acid-induced status epilepticus in rats. Front Cell Neurosci. 2019;13:1–11. doi: 10.3389/fncel.2019.00145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Hattiangady B., Mishra V., Kodali M., Shuai B., Rao X., Shetty A.K. Object location and object recognition memory impairments, motivation deficits and depression in a model of gulf war illness. Front Behav Neurosci. 2014;8:1–10. doi: 10.3389/fnbeh.2014.00078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.O'Keefe J., The Nadel L. Oxford University Press; Oxford: 1978. Hippocampus as a cognitive map. [Google Scholar]
- 55.Rüber T., David B., Lüchters G., Nass R.D., Friedman A., Surges R. Evidence for peri-ictal blood-brain barrier dysfunction in patients with epilepsy. Brain. 2018;141:2952–2965. doi: 10.1093/brain/awy242. [DOI] [PubMed] [Google Scholar]
- 56.Wang X., Luo Y., Liu S., Tan L., Wang S., Man R. MicroRNA-134 plasma levels before and after treatment with valproic acid for epilepsy patients. Oncotarget. 2017;8:72748–72754. doi: 10.18632/oncotarget.20292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Avansini S.H., De Sousa Lima B.P., Secolin R., Santos M.L., Coan A.C., Vieira A.S. MicroRNA hsa-miR-134 is a circulating biomarker for mesial temporal lobe epilepsy. PLoS One. 2017;12 doi: 10.1371/journal.pone.0173060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Zhou J., Chen L., Chen B., Huang S., Zeng C., Wu H. Increased serum exosomal miR-134 expression in the acute ischemic stroke patients. BMC Neurol. 2018;18 doi: 10.1186/s12883-018-1196-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Rong H., Liu T.B., Yang K.J., Yang H.C., Wu D.H., Liao C.P. MicroRNA-134 plasma levels before and after treatment for bipolar mania. J Psychiatr Res. 2011;45:92–95. doi: 10.1016/j.jpsychires.2010.04.028. [DOI] [PubMed] [Google Scholar]
- 60.Raoof R., Bauer S., El Naggar H., Connolly N.M.C., Brennan G.P., Brindley E. Dual-center, dual-platform microRNA profiling identifies potential plasma biomarkers of adult temporal lobe epilepsy. EBioMedicine. 2018;38:127–141. doi: 10.1016/j.ebiom.2018.10.068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Vangoor V.R., Reschke C.R., Senthilkumar K., van de Haar L.L., de Wit M., Giuliani G. Antagonizing increased miR-135a levels at the chronic stage of experimental TLE reduces spontaneous recurrent seizures. J Neurosci. 2019:3014–3018. doi: 10.1523/JNEUROSCI.3014-18.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Reschke C.R., Jimenez-Mateos E., Rosso M., Vangoor V., Sanz-Rodriguez A., Batool A. 12th ECE, Czech Repub. 2016. Systemic delivery of antagomir-134 produces anti-epileptogenic effects; pp. 6–225. [Google Scholar]
- 63.Brown B.D., Naldini L. Exploiting and antagonizing microRNA regulation for therapeutic and experimental applications. Nat Rev Genet. 2009;10:578–585. doi: 10.1038/nrg2628. [DOI] [PubMed] [Google Scholar]
- 64.Sonneville F., Ruffin M., Coraux C., Rousselet N., Le Rouzic P., Blouquit-Laye S. MicroRNA-9 downregulates the ANO1 chloride channel and contributes to cystic fibrosis lung pathology. Nat Commun. 2017;8 doi: 10.1038/s41467-017-00813-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Guerrini R., Aicardi J. Epileptic encephalopathies with myoclonic seizures in infants and children (severe myoclonic epilepsy and myoclonic-astatic epilepsy) J Clin Neurophysiol. 2003;20:449–461. doi: 10.1097/00004691-200311000-00007. [DOI] [PubMed] [Google Scholar]
- 66.Petrou S., Li M., Jancovsk N., Paymaan J., Burbano L., Nemiroff A. Antisense oligonucleotide therapy for SCN2A gain-of-function epilepsies. Am Epilepsy Soc Annu Meet. 2018;2018 [Abst 1.466] [Google Scholar]
- 67.D'Aiuto L., Naciri J., Radio N., Tekur S., Clayton D., Apodaca G. Generation of three-dimensional human neuronal cultures: application to modeling CNS viral infections. Stem Cell Res Ther. 2018;9:1–9. doi: 10.1186/s13287-018-0881-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Velasco S., Kedaigle A.J., Simmons S.K., Nash A., Rocha M., Quadrato G. Individual brain organoids reproducibly form cell diversity of the human cerebral cortex. Nature. 2019 doi: 10.1038/s41586-019-1289-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Parent J.M., Anderson S.A. Reprogramming patient-derived cells to study the epilepsies. Nat Neurosci. 2015;18:360–366. doi: 10.1038/nn.3944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Hong S.E., Shugart Y.Y., Huang D.T., Al Shahwan S., Grant P.E., Hourihane J.O. Autosomal recessive lissencephaly with cerebellar hypoplasia is associated with human RELN mutations. Nat Genet. 2000;26:93–96. doi: 10.1038/79246. [DOI] [PubMed] [Google Scholar]
- 71.Michelucci R., Pulitano P., Di Bonaventura C., Binelli S., Luisi C., Pasini E. The clinical phenotype of autosomal dominant lateral temporal lobe epilepsy related to reelin mutations. Epilepsy Behav. 2017;68:103–107. doi: 10.1016/j.yebeh.2016.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Lalani S.R., Zhang J., Schaaf C.P., Brown C.W., Magoulas P., Tsai A.C.H. Mutations in PURA cause profound neonatal hypotonia, seizures, and encephalopathy in 5q31.3 microdeletion syndrome. Am J Hum Genet. 2014;95:579–583. doi: 10.1016/j.ajhg.2014.09.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Wolff M., Cassé-Perrot C., Dravet C. Severe myoclonic epilepsy of infants (Dravet syndrome): natural history and neuropsychological findings. Epilepsia. 2006;47:45–48. doi: 10.1111/j.1528-1167.2006.00688.x. [DOI] [PubMed] [Google Scholar]
- 74.Engle S.J., Blaha L., Kleiman R.J. Best practices for translational disease modeling using human iPSC-derived neurons. Neuron. 2018;100:783–797. doi: 10.1016/j.neuron.2018.10.033. [DOI] [PubMed] [Google Scholar]
- 75.Cohen I., Navarro V., Clemenceau S., Baulac M., Miles R. On the origin of Interictal activity in human temporal lobe epilepsy in vitro. Science (80-) 2002;298:1418–1421. doi: 10.1126/science.1076510. [DOI] [PubMed] [Google Scholar]