Abstract
Background
Root morphology is known to be affected by light quality, quantity and direction. Light signal is perceived at the shoot, translocated to roots through vasculature and further modulates the root development. Photoreceptors are differentially expressed in both shoot and root cells. The light irradiation to the root affects shoot morphology as well as whole plant development. The current work aims to understand the white light intensity dependent changes in root patterning and correlate that with the global gene expression profile.
Results
Different fluence of white light (WL) regulate overall root development via modulating the expression of a specific set of genes. Phytochrome A deficient Arabidopsis thaliana (phyA-211) showed shorter primary root compared to phytochrome B deficient (phyB-9) and wild type (WT) seedlings at a lower light intensity. However, at higher intensity, both mutants showed shorter primary root in comparison to WT. The lateral root number was observed to be lowest in phyA-211 at intensities of 38 and 75 μmol m − 2 s − 1. The number of adventitious roots was significantly lower in phyA-211 as compared to WT and phyB-9 under all light intensities tested. With the root phenotypic data, microarray was performed for four different intensities of WL light in WT. Here, we identified ~ 5243 differentially expressed genes (DEGs) under all light intensities. Gene ontology-based analysis indicated that different intensities of WL predominantly affect a subset of genes having catalytic activity and localized to the cytoplasm and membrane. Furthermore, when root is irradiated with different intensities of WL, several key genes involved in hormone, light signaling and clock-regulated pathways are differentially expressed.
Conclusion
Using genome wide microarray-based approach, we have identified candidate genes in Arabidopsis root that responded to the changes in light intensities. Alteration in expression of genes such as PIF4, COL9, EPR1, CIP1, ARF18, ARR6, SAUR9, TOC1 etc. which are involved in light, hormone and clock pathway was validated by qRT-PCR. This indicates their potential role in light intensity mediated root development.
Electronic supplementary material
The online version of this article (10.1186/s12864-019-5933-5) contains supplementary material, which is available to authorized users.
Keywords: Root, Light signaling, Intensity, Gene expression, Auxin, Hormone
Background
Light is an essential parameter for the optimal growth and survival of plants. The quality, quantity, direction and duration of light are important factors required for various aspects of plant development [1]. Root development comprises of different aspects such as primary root elongation, lateral root elongation, lateral root branching, root geotropism, root hair formation etc. Root patterning beneath the soil plays a crucial role in penetration, anchorage and gravitropism leading to absorption of water and nutrient. To perceive light, plants have evolved with many canonical photoreceptors such as phytochromes (PHYs), cryptochromes (CRYs), phototropins (PHOTs) and UVB-resistance 8 (UVR8) [2, 3]. Light regulates the patterning of shoot as well as root system [4]. It has been shown to regulate all the aforesaid aspects of root development at different stages of plant life-cycle [5–8]. Light signal from shoot can translocate through phloem to root and alters light-mediated responses [9]. Although, roots grown beneath the soil generally don’t experience direct light, still few parts of the root are exposed to some amount of light seeping through the cracks, pores of the soil and affects the overall root development. Light percolates through the soil and reaches the root which leads to the production of reactive oxygen species (ROS) and promotes root growth when present in an optimal level. Ha et.al., 2018 has shown the correlation of photoreceptors with ROS-mediated root growth. The shoot localized PHYs have been reported to mediate this response through ROS accumulation in roots. A shoot-localized abscisic acid (ABA) signaling component is also shown to be involved in PHYB-mediated primary root elongation. It has been shown that when roots are exposed to light, shoot PHYs induce ABA biosynthesis and signaling mediator which further promote primary root growth. PHY controls the translocation of ABA signals from shoot to root which increase the expression of ABA INSENSITIVE 5 (ABI5) gene, which activates PEROXIDASE 1 (PER1) in the root. ABI5 encodes a basic leucine-rich zipper transcription factor while PER1 encodes a peroxidase that detoxifies ROS. When root is exposed to light, activation of PER1 leads to detoxification of ROS and maintains its level, which promotes root growth. This showed that PHYB promotes primary root growth through regulating ABI5 and PER1 activity and ROS accumulation [10]. ELONGATED HYPOCOTYL 5 (HY5), one of the major transcription factors, downstream to PHYs has been shown to be a mobile signal. It has been documented that, HY5 is activated in shoot, translocate to roots and regulates the root architecture [11]. Intrinsic component such as phytohormones are also involved in regulating root patterning. Auxin is one of the important phytohormones that plays a major role in root development. HY5 has also been shown to be a mediator of PHY and auxin signaling. Along with phytohormones, nutrients such as sugar also plays an important role in root development. The cross-talk of sugar and phytohormone such as auxin has been shown to modulate the root growth and development [12]. MEDIATOR (MED) complex is one of the most important candidate that couples sugar and auxin signalling pathways in root development. In support of this, MED12 and MED13 genes have been shown to promote primary root length, root hair number and root hair length by enhancing the cell elongation, cell division and auxin response. Addition of sucrose compliments the root defect in med12 and med13 mutants. Thus, MED12 and MED13 are the important candidate genes which link auxin signaling and nutritional status of the root [13]. On the contrary, med18 mutant has been reported to show shorter primary root, lesser number of lateral roots with longer and denser root hairs. The alteration of root architecture in med18 mutant is because of altered auxin response and its distribution in primary root. Although in natural condition, roots grow under relative darkness still there is always some communication and signal translocation between shoot to root, historically known as light piping. In med18 mutant, it has been shown that shoot perceives the light and causes the death of root meristem cells. However, cell death at root meristem occurred irrespective of direct light exposure to the roots, which suggested a long-distance communication between shoot and root is plausible. This indicated that root growth is affected in similar fashion irrespective of its light irradiation [14]. There are very few available reports which document the light and hormone cross-talk in root development [15–18]. The light quality and quantity both affect the root patterning [19]. Although, the effect of light intensity on regulation of root development has not been well investigated at molecular level and needs a systematic study.
Nicotiana tabacum plants grown under variable intensity of WL for different durations, showed altered root growth, leaf biomass, sugar content and chlorophyll level. It has been reported that, plants constantly grown either for 14 or 18 days under 60 μmol m− 2 s− 1 (condition A) light, they have lesser fresh weight of root and shoot as compared to plants grown under 300 μmol m− 2 s− 1 (condition B) light. However, plants grown for first 14 days under 60 μmol m− 2 s− 1 light and then subsequent 4 days under 300 μmol m− 2 s− 1 light (condition C), after a total of 18 days of light treatment, the fresh weight of root and shoot were observed to be intermediate of that of continuous 18 days under 60 μmol m− 2 s− 1 and 300 μmol m− 2 s− 1 light intensity. This suggested an additive effect of light fluence and duration. Further, glucose, fructose and sucrose levels were observed to be highest in plants grown under condition C compared to condition A and B. Then, it was concluded that the promotion of root growth under higher light intensity is due to enhanced carbohydrate transport from shoot to root and independent of sugar content [20]. In tomato, Solanum lycopersicum cyclophilins (SlCyp1) are peptidyl-prolyl cis/trans isomerases which play an important role in plant development. It is transported from shoot to root as a phloem mobile signal. The trafficking of SlCyp1 is enhanced with increasing light intensity, leading to profound root growth. [21]. It has been reported that, in gymnosperms such as Pinus sylvestris L. (Scots pine), plants grown in the presence of different intensities of red (R) and far-red (FR) light (1, 10, 25 and 100 μmol m− 2 s− 1) showed variation in root growth. Irradiation of R light had no significant effect on root length whereas low intensity of FR light (1 and 10 μmol m− 2 s− 1) has resulted shorter root in comparison to the seedlings grown under complete darkness. However, under highest intensity of FR light, root length was significantly higher as compared to other light conditions. Therefore, it can be concluded that equal intensity of monochromatic light didn’t show similar type of effect on plant development [22]. In a recent report, Arabidopsis thaliana seedlings grown in two different experimental setups, in one the roots were directly exposed to WL while in another, root was covered and the shoot was exposed to light (D-root system). In this report, it has been shown that the spatial expression of the photoreceptor genes such as UVR8, CRY1, CRY2, PHOT1, PHOT2, PHYA and PHYB varied in different segments of roots under the above mentioned experimental setups [23]. Although light intensity has been shown to influence the root development, how it correlates with the gene expression, leading to root patterning needs to be examined in detail. In the present work whole seedlings were grown under 38, 75, 112 and 150 μmol m− 2 s− 1 intensities of WL with 16 h light and 8 h dark. The root patterning was analysed in WT and PHY mutants. Using microarray-based approach, we analysed the variation in the global transcription profile of the root tissue of WT seedlings. Further, we evaluated the expression pattern of various genes that play important role in light signaling, hormone signaling and clock-regulated pathways.
Results
Phytochrome mutants showed variation in root patterning under different intensities of white light
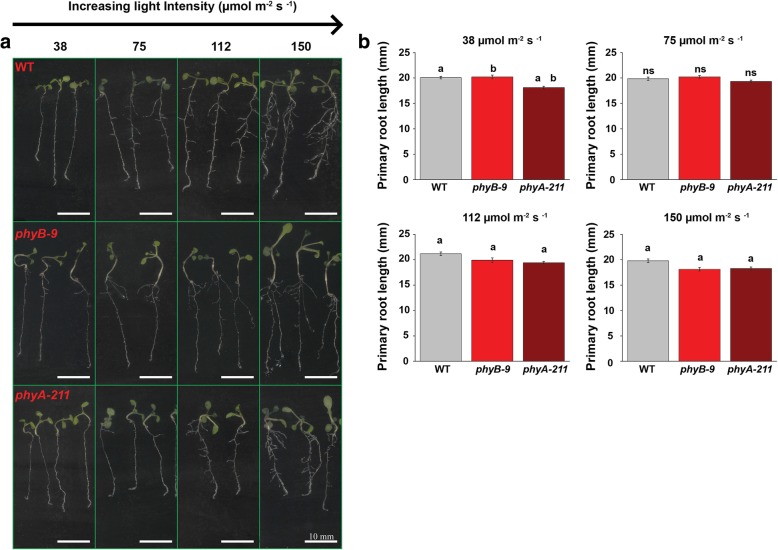
Arabidopsis seedlings, WT, phyB-9 and phyA-211 showed variable root growth responses under different light intensities of 38, 75, 112 and 150 μmol m − 2 s − 1 WL (Fig. 1a). Primary root length in 6-days old seedlings was found to be slightly shorter in case of phyA-211 as compared to phyB-9 and WT under 38 μmol m − 2 s − 1 light intensity. Under 150 and 112 μmol m − 2 s − 1 light, the primary root length was observed to be marginally shorter in both the PHY mutants as compared to WT (Fig. 1b). It was also observed that lateral and adventitious root growth varied in PHY mutants under different light intensity in comparison to WT (Fig. 2a). The zoomed image for qualitative details of lateral root growth has been shown in Additional file 1: Figure S1. When light intensity increased from 38 to 150 μmol m − 2 s − 1, the number of lateral roots approximately increased by two times in case of WT, phyB-9 and three times in phyA-211. However, under 112 μmol m − 2 s − 1 light, phyB-9 has significantly lesser number of lateral root and about 16.7 and 30.3% reduction were observed in comparison to 75 and 150 μmol m − 2 s − 1 light intensity respectively. The lateral root number in phyA-211 decreased by 12.5 and 35.7% as compared to WT under 38 μmol m − 2 s − 1 and 75 μmol m − 2 s − 1 light respectively (Fig. 2b). Interestingly, under 112 μmol m − 2 s − 1 light, the lateral root number in case of phyB-9 was reduced by 39.5 and 24.5% in comparison to WT and phyA-211 respectively. At higher intensity of 150 μmol m − 2 s − 1, no significant difference was found in lateral root number as all the genotypes showed ~ 14–16 rootlets (Fig. 2b).
Fig. 1.
Light intensity dependent root growth varies in phytochrome mutants. a Phenotypic differences in root architecture. b Primary root length (mm) for WT, phyB-9 and phyA-211 under four different WL intensities of 38, 75, 112 and 150 μmol m − 2 s − 1. The analysis was done in 6-days old seedlings. Error bars represent SE. The means were compared using Tukey test with p ≤ 0.05. Same letters denote statistical significance. There were five technical replicates, each replicate consists of 20 seedlings. Scale bar = 10 mm
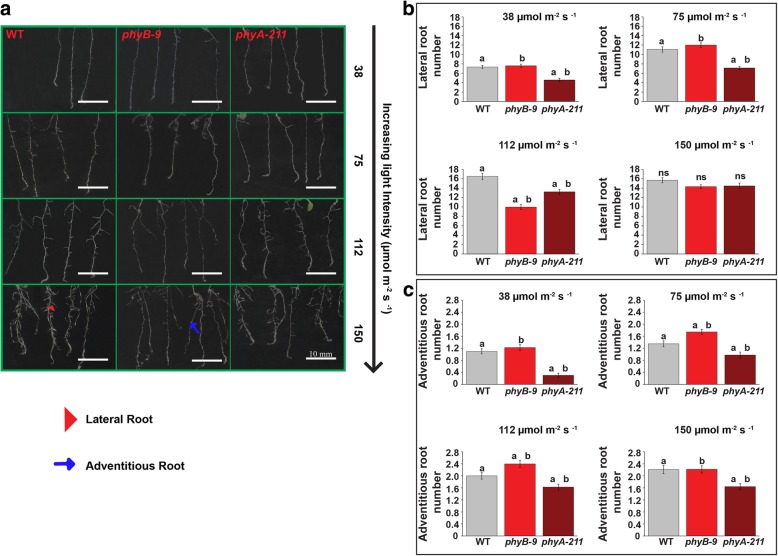
Fig. 2.
Phytochrome mutants show variability in light intensity dependent lateral and adventitious root growth. a Lateral and adventitious root architectural differences. The lateral and adventitious root have been indicated by arrowheads and arrows respectively. b Phytochrome mutants show variation in lateral root number. c Adventitious root number varies among the genotypes under all four WL intensity. The analysis has been done in 6-days old seedlings. Error bars represent SE. The means were compared using Tukey test with p ≤ 0.05. Same letters denote statistical significance There were three technical replicates, each replicate consists of 20 seedlings. Scale bar is 10 mm
The number of adventitious roots was shown to increase by increasing light intensity till 75 μmol m − 2 s − 1 and apparently saturates in between 112 μmol m − 2 s − 1 and 150 μmol m − 2 s − 1. It has been observed that the adventitious root number was least in case of phyA-211 as compared to WT and phyB-9 under all light intensities (Fig. 2c). The adventitious root number in case of phyA-211 was least under 38 μmol m − 2 s − 1 as compared to other light intensities. The microscope images of lateral root growth for qualitative visualization in 6-days old seedling have been shown in Additional file 2: Figure S2.
Identification of differentially expressed genes under four different intensities of white light in WT root
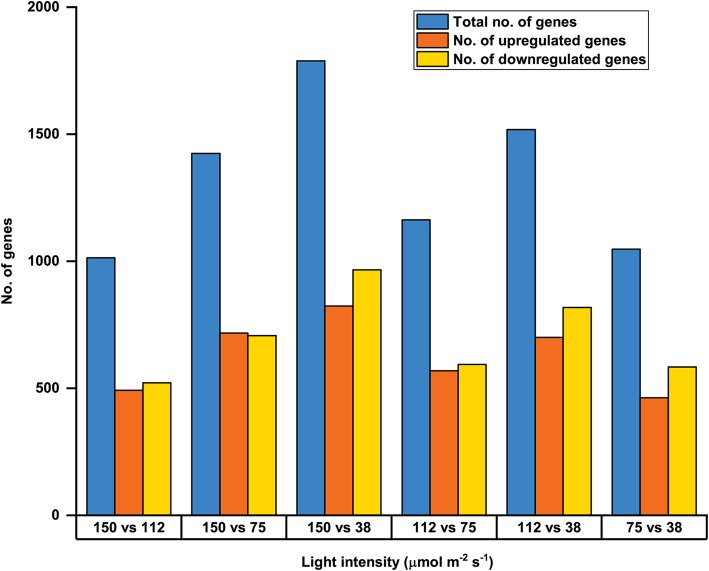
In all the genotypes root architectural differences were observed to follow a pattern when grown under different intensities of WL. With this information, microarray was carried out with root samples of 5-days old WT seedlings grown under four different WL intensities (mentioned earlier). Differential expression analysis was performed to identify the differentially expressed genes (DEGs) from microarray data. The DEGs having minimum fold change (FC) of ~ 1.2 and False Discovery Rate (FDR) < 0.05 were selected for further analysis. The DEGs under comparative light conditions such as 150 vs 112, 150 vs 75, 150 vs 38, 112 vs 75, 112 vs 38 and 75 vs 38 μmol m − 2 s − 1 intensities of WL were taken in consideration for the study. Largest number of DEGs were found between 150 and 38 μmol m − 2 s − 1 while least number was observed in between 150 and 112 μmol m − 2 s − 1 light. The details of the DEGs upregulated and downregulated have been summarized in Table 1. The graphical representation of upregulated and downregulated genes for different comparative light intensities has been shown in Fig. 3.
Table 1.
Number of Differentially Expressed Genes identified under various white light intensities
| Light intensity (μmol m − 2 s − 1) | 150 vs 112 | 150 vs 75 | 150 vs 38 | 112 vs 75 | 112 vs 38 | 75 vs 38 |
|---|---|---|---|---|---|---|
| Total no. of genes | 1014 | 1424 | 1789 | 1163 | 1518 | 1047 |
| No. of genes upregulated | 492 | 717 | 823 | 569 | 700 | 463 |
| No. of genes downregulated | 522 | 707 | 966 | 594 | 818 | 584 |
Fig. 3.
DEGs found under different comparative light intensities from microarray. Total number of DEGs, number of upregulated and downregulated DEGs under comparative light intensity of 150 vs 112, 150 vs 75, 150 vs 38, 112 vs 75, 112 vs 38 and 75 vs 38 μmol m − 2 s − 1 obtained from 5-days old WT root microarray analysis
To identify overlapping and unique genes, R program analysis was performed and presented in the Venn diagrams between 150 vs 112, 150 vs 75 and 150 vs 38 μmol m − 2 s − 1 light (Fig. 4a), 150 vs 112, 112 vs 75 and 112 vs 38 μmol m − 2 s − 1 light (Fig. 4b), 150 vs 75, 112 vs 75 and 75 vs 38 μmol m − 2 s − 1 light (Fig. 4c) and 150 vs 38, 112 vs 38 and 75 vs 38 μmol m − 2 s − 1 light (Fig. 4d) conditions. With respect to 150 μmol m − 2 s − 1 intensity, under 112, 75 and 38 μmol m − 2 s − 1, 68 common DEGs were identified. When light conditions of 150 vs 112, 112 vs 75 and 75 vs 38 μmol m − 2 s − 1 were compared, the overlapping number of DEGs was 96. When all light intensities were investigated with respect to 75 μmol m − 2 s − 1 light, 65 common DEGs were found. The total number of common DEGs was 136, when 150 vs 38, 112 vs 38, 75 vs 38 μmol m − 2 s − 1 light intensity were taken into consideration. CYTOCHROME P450 81F2 (CYP81F2), At2g39445, At5g22555 and At2g44130 genes have been found to be common and altered under all six comparative light conditions. CYP81F2 is a membrane-localized protein, known to play a role in indole glucosinolate biosynthesis and offers resistance to fungus named Plectosphaerella cucumerina [24]. At2g39445 encodes for phosphatidylinositol n-acetylglucosaminyltransferase, At5g22555 is a putative transmembrane protein while At2g44130 encodes a F-box protein KISS ME DEADLY 3 (KMD3). KMD3 has been reported to be induced by Meloidogyne incognita (root-knot nematode) and makes the plant susceptible towards this nematode [25]. The genes which were unique and common in two, three, four or five comparative light conditions have been summarized in supplementary excel files (Additional files 3 and 4).
Fig. 4.
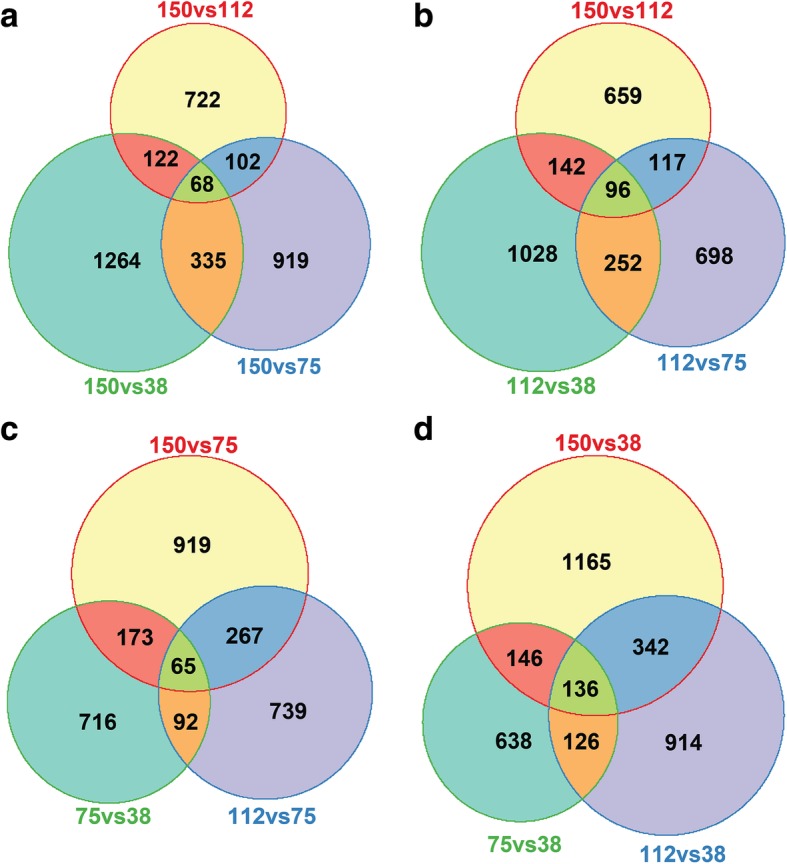
Venn diagram representation of overlapping and differential DEGs under variable white light intensity. a DEGs for 150 vs 112, 150 vs 75 and 150 vs 38 μmol m − 2 s − 1. b DEGs for 150 vs 112, 112 vs 75 and 112 vs 38 μmol m − 2 s − 1. c DEGs for 150 vs 75, 112 vs 75 and 75 vs 38 μmol m − 2 s − 1. d DEGs for 150 vs 38, 112 vs 38 and 75 vs 38 μmol m − 2 s − 1 light intensity from microarray analysis of WT root
Gene ontology enrichment analysis of over-represented differentially expressed genes
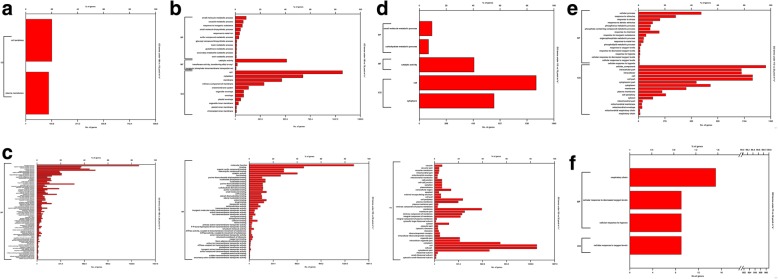
Gene ontology (GO) enrichment analysis was performed to understand how light intensity affects different biological phenomena and processes in roots. GO analysis is based on the gene products and the related functions at molecular and cellular level with available literature databases. The analysis type used was PANTHER Over-representation Test and annotation version was GO Ontology Database [26, 27]. The categorization has been done on the basis of Fisher’s exact with FDR multiple test correction type. GO pathway analysis classifies DEGs into three categories/domains named Biological Process (BP), Molecular Function (MF) and Cellular Component (CC). The categorization is based on the gene or gene product functions and site of their functions. The BP category consists of the outcome of gene function and the major pathways involved. The MF domain represents the function of gene products at molecular level or activities of the gene at molecular level. The CC category explains about the site of gene functioning. Each category consists of various specific and broad terms based on the reported information and available database. The categorization of DEGs has been performed under 150 vs 112 μmol m − 2 s − 1 (Fig. 5a), 150 vs 75 μmol m − 2 s − 1 (Fig. 5b), 150 vs 38 μmol m − 2 s − 1 (Fig. 5c), 112 vs 75 μmol m − 2 s − 1 (Fig. 5d), 112 vs 38 μmol m − 2 s − 1 (Fig. 5e) and 75 vs 38 μmol m − 2 s − 1 (Fig. 5f) light intensities. The composite figure describing detailed GO analysis of DEGs has been presented in Additional file 5: Figure S3.
Fig. 5.
Classification of DEGs under different categories and terms of GO annotation analysis. a DEGs under 150 vs 112 μmol m − 2 s − 1 b DEGs under 150 vs 75 μmol m − 2 s − 1 c DEGs under 150 vs 38 μmol m − 2 s − 1 d DEGs under 112 vs 75 μmol m − 2 s − 1 e DEGs under 112 vs 38 μmol m − 2 s − 1 f DEGs under 75 vs 38 μmol m − 2 s− 1 light intensity from microarray analysis for the functionality of DEGs in 5-days old WT root
All the GO categories and terms which were highly enriched or over-represented under different light conditions have been summarized in Table 2. In case of 150 vs 112 μmol m − 2 s − 1 light condition, significant DEGs were found only under CC category, plasma membrane and cell periphery were the most affected GO terms. When the DEGs were analysed for 150 vs 75 μmol m − 2 s − 1 light, the enriched DEGs were categorized under CC and MF domains. Under this light condition, the most enriched terms were cell, cytoplasm and membrane under CC and catalytic activity was the most enriched term under MF category. The largest number of DEGs were found under 150 vs 38 μmol m − 2 s − 1 light condition and were classified under BP, CC and MF categories. The highly enriched terms under BP category were biological process, cellular process, metabolic process, cellular metabolic process and organic substance metabolic process. In this comparative light conditions, cell, cytoplasm, membrane, integral component of membrane, intrinsic component of membrane and cell periphery were the most enriched GO terms of CC category. In MF category, highly enriched terms were ion binding, heterocyclic compound binding, organic cyclic compound binding, molecular function and catalytic activity.
Table 2.
Summary of highly affected GO categories and terms under different intensities of white light
| Light intensity (μmol m − 2 s − 1) | Category | Terms | GO Accession |
|---|---|---|---|
| 150 vs 112 | CC | Plasma membrane | GO:0005886 |
| Cell periphery | GO:0071944 | ||
| 150 vs 75 | CC | Cell | GO:0005623 |
| Cytoplasm | GO:0005737 | ||
| Membrane | GO:0016020 | ||
| MF | Catalytic activity | GO:0003824 | |
| 150 vs 38 | BP | Cellular process | GO:0009987 |
| Metabolic process | GO:0008152 | ||
| Cellular metabolic process | GO:0044237 | ||
| Organic substance metabolic process | GO:0071704 | ||
| Biological process | GO:0008150 | ||
| CC | Cell | GO:0005623 | |
| Cytoplasm | GO:0005737 | ||
| Membrane | GO:0016020 | ||
| Integral component of membrane | GO:0016021 | ||
| Intrinsic component of membrane | GO:0031224 | ||
| Cell periphery | GO:0071944 | ||
| MF | Ion binding | GO:0043167 | |
| Heterocyclic compound binding | GO:1901363 | ||
| Organic cyclic compound binding | GO:0097159 | ||
| Molecular function | GO:0003674 | ||
| Catalytic activity | GO:0003824 | ||
| 112 vs 75 | CC | Cytoplasm | GO:0005737 |
| Cell | GO:0005623 | ||
| MF | Catalytic activity | GO:0003824 | |
| 112 vs 38 | BP | Cellular process | GO:0009987 |
| CC | Membrane | GO:0016020 | |
| Cytoplasm | GO:0005737 | ||
| Cell | GO:0005623 | ||
| Intracellular | GO:0005622 |
The DEGs under 112 vs 75 μmol m − 2 s − 1 condition were significantly categorized under BP, CC and MF, however the enriched terms were found under CC and MF categories only. Cell and cytoplasm terms were highly enriched under CC category and catalytic activity was the most affected term in MF category. In case of 112 vs 38 μmol m − 2 s − 1 light, the DEGs were grouped in BP and CC categories. In BP category, the most enriched term was cellular process and under CC category, cell, cytoplasm, membrane and intracellular were the most enriched terms. The DEGs under 75 vs 38 μmol m − 2 s − 1 light condition were classified under CC and BP categories and in CC category respiratory chain was the only over-represented term. The DEGs under this comparative light condition were found to be less enriched.
Kyoto encyclopedia of genes and genomes colour pathway analysis of DEGs to investigate the gene functionality
Kyoto Encyclopedia of Genes and Genomes (KEGG) analysis represents the gene function and their utilities in biological, cellular and molecular process [28]. This analysis is based on the present database consisting of various genome sequencing, bioinformatic information etc. The DEGs obtained from microarray were further subjected to KEGG colour pathway analysis. The total number of DEGs under 150 vs 112 μmol m − 2 s − 1 light condition was 1014 and only 184 DEGs were identified with KEGG analysis. Under 150 vs 75 μmol m − 2 s − 1 light, 1424 DEGs were obtained from microarray data. In this light condition, the number of DEGs identified through KEGG colour pathway was 290. In case of 150 vs 38 μmol m − 2 s − 1, 420 genes were identified by KEGG out of 1789 DEGs obtained from microarray analysis. Under 112 vs 75 μmol m − 2 s − 1 light, 1163 DEGs were found from microarray, out of these, 249 genes were identified through KEGG analysis. In case of 112 vs 38 μmol m − 2 s − 1 light, out of 1518 DEGs only 314 genes were identified by KEGG analysis. Under 75 vs 38 μmol m − 2 s − 1 light, 1047 DEGs were detected from microarray and KEGG analysis identified only 172 genes.
The detailed KEGG analysis of DEGs has been presented in Table 3. This analysis showed that the metabolic pathway was the major ones affected with largest number of genes under all comparative light intensities.
Table 3.
Categorization of DEGs on the basis of KEGG analysis
| Light condition (75 μmol m − 2 s − 1) | Total no. of genes | No. of genes identified by KEGG | Major pathways involved | No. of genes in each pathway |
|---|---|---|---|---|
| 150 vs 112 | 1014 | 184 | Metabolic pathway | 71 |
| Secondary metabolite biosynthesis | 40 | |||
| Plant hormone signal transduction | 19 | |||
| Protein processing in ER | 15 | |||
| MAPK signaling | 11 | |||
| Carbon metabolism | 11 | |||
| 150 vs 75 | 1424 | 290 | Metabolic pathway | 143 |
| Secondary metabolite synthesis | 82 | |||
| Carbon metabolism | 24 | |||
| Amino acid biosynthesis | 19 | |||
| Glycolysis | 15 | |||
| Plant-pathogen interaction | 13 | |||
| Glutathione metabolism | 13 | |||
| RNA transport | 13 | |||
| Starch and sucrose metabolism | 13 | |||
| Oxidative phosphorylation | 12 | |||
| Purine metabolism | 12 | |||
| Amino sugar and nucleotide sugar metabolism | 10 | |||
| Plant hormone signal transduction | 10 | |||
| 150 vs 38 | 1789 | 420 | Metabolic pathway | 79 |
| Secondary metabolite biosynthesis | 114 | |||
| Ribosome | 51 | |||
| Carbon metabolism | 27 | |||
| Amino acid biosynthesis | 25 | |||
| Oxidative phosphorylation | 21 | |||
| Phenylpropanoid biosynthesis pathway | 20 | |||
| RNA transport | 17 | |||
| Starch and sucrose metabolism | 17 | |||
| Spliceosome | 17 | |||
| Plant-pathogen interaction | 17 | |||
| Glutathione metabolism | 16 | |||
| Plant hormone signal transduction | 15 | |||
| Endocytosis | 13 | |||
| Glycine, serine and threonine metabolism | 13 | |||
| Glycolysis | 13 | |||
| Purine metabolism | 12 | |||
| Amino sugar and nucleotide sugar metabolism | 12 | |||
| Cysteine and methionine metabolism | 11 | |||
| 2-oxocarboxylic acid metabolism | 10 | |||
| Protein processing in ER | 10 | |||
| Ubiquitin mediated proteolysis | 10 | |||
| Photosynthesis | 10 | |||
| 112 vs 75 | 1163 | 249 | Metabolic pathway | 122 |
| Biosynthesis of secondary metabolites | 63 | |||
| Carbon metabolism | 18 | |||
| Starch and sucrose metabolism | 15 | |||
| Plant hormone signal transduction | 14 | |||
| Amino acid biosynthesis | 12 | |||
| Protein processing in the ER | 11 | |||
| Phenylpropanoid biosynthesis pathway | 10 | |||
| 112 vs 38 | 1518 | 314 | Metabolic pathway | 131 |
| Secondary metabolite biosynthesis | 59 | |||
| Ribosome | 30 | |||
| Plant hormone signal transduction | 27 | |||
| Carbon metabolism | 22 | |||
| Oxidative phosphorylation | 18 | |||
| Protein processing in ER | 13 | |||
| MAPK signaling | 13 | |||
| Spliceosome | 12 | |||
| Purine metabolism | 12 | |||
| Glyoxylate and dicarboxylate metabolism | 11 | |||
| Amino acid biosynthesis pathway | 10 | |||
| 75 vs 38 | 1047 | 172 | Metabolic pathway | 64 |
| Biosynthesis of secondary metabolite | 37 | |||
| Plant hormone signaling | 10 | |||
| Plant-pathogen interaction | 10 |
Validation of microarray result by qRT-PCR for DEGs involved in hormonal, light signaling and clock regulated pathways
We further validated the microarray result by qRT-PCR of few selected genes. Selection of genes for qRT-PCR validation was based on their i) expression level data obtained from microarray analysis and ii) significance and potential role in root development as per the KEGG pathway analysis. Therefore, the genes invoved in hormone signaling, light signaling and clock-regulated pathways were chosen for qRT-PCR validation. The root development is affected by various factors such as light, phytohormones, circadian clock etc. Photoreceptors such as PHYA and PHYB play major role in root positive phototropism [7]. PHYs have also been shown to be involved in root growth as well as its gravitropic response [5]. The phytohormones also regulate the lateral root initiation in a dose dependent manner [29]. They have also been shown to control root hair initiation and its growth [30]. High intensity of light and exogenous auxin stimulate adventitious rooting in Eucalyptus [31]. On the other hand, circadian clock genes have also been shown to regulate lateral root emergence [32].
Genes such as AUXIN RESPONSE FACTOR 2 (ARF2), AUXIN RESPONSE FACTOR 4 (ARF4) and AUXIN RESPONSE FACTOR 18 (ARF18), LIKE AUX 2 (LAX2), SMALL AUXIN UPREGULATED RNA 9 (SAUR9), SMALL AUXIN UPREGULATED RNA 26 (SAUR26) and IAA7 were selected to evaluate the effect of WL intensity on auxin homeostasis, while, TYPE A RESPONSE REGULATOR 6 (ARR6), KISS ME DEADLY 1(KMD1) and COP1 INTERACTING PROTEIN 1 (CIP1) were analysed to understand the impact of WL intensity on cytokinin and ABA signaling, respectively. Genes involved in light signaling such as PHYTOCHROME RAPIDLY REGULATED 2 (PAR2), HY5, PHYTOCHROME INTERACTING FACTOR 4 (PIF4), EARLY PHYTOCHROME RESPONSE 1 (EPR1), CONSTANS LIKE 3 (COL3), CONSTANS LIKE 9 (COL9), COP9 SIGNALOSOME COMPLEX SUBUNIT 6A (CSN6A) and COP9 SIGNALOSOME COMPLEX SUBUNIT 6B (CSN6B) were selected to investigate the correlation of WL intensities with expression level of downstream light signaling components. Three clock-associated genes such as CIRCADIAN CLOCK-ASSOCIATED 1 (CCA1), TIMING OF CAB EXPRESSION 1 (TOC1) and PSEUDO RESPONSE REGULATOR 9 (PRR9) were also chosen to understand the response of light quantity on circadian clock regulation. The possible functions of all the above-mentioned genes and respective primers have been compiled in Table 4 and supplementary file (Additional file 6) respectively. The qRT-PCR was performed with the same RNA samples used for microarray analysis of WT. The gene expression of all the light signaling, hormone signaling and clock-regulated genes mentioned earlier were analysed for 150 vs 112, 150 vs 75, 150 vs 38, 112 vs 75, 112 vs 38 and 75 vs 38 μmol m − 2 s − 1 comparative light intensity (Figs. 6, 7, 8, 9, 10 and 11). The FC of gene expression data has been summarized in Table 5.
Table 4.
List of gene functions
| Gene names | Gene function |
|---|---|
| PAR2 | Negative regulator of shade avoidance syndrome responses and transcriptional repressor of SAUR15 and SAUR68 [33]. |
| ARR6 | Type-A response regulator, acts as negative regulator of cytokinin signaling [34]. |
| HY5 | Transcription factor playing downstream to photoreceptor signaling, promotes photomorphogenesis. It is also involved in root greening and gravitropism [15, 35]. |
| ARF18 | Auxin response factor acting as transcriptional repressor for auxin responsive genes [36]. |
| ARF2 | Auxin response factor acting as transcriptional repressor for auxin responsive genes [37]. |
| ARF4 | Auxin response factor acting as transcriptional repressor for auxin responsive genes [38]. |
| KMD1 | Type-A response regulator which targets type-B response regulator and acts as negative regulator of cytokinin signaling [39]. |
| PIF4 | Transcription factor playing negatively in phytochrome signaling pathway [40]. |
| IAA7 | Auxin inducible gene which negatively regulates auxin signaling [41]. |
| EPR1 | Transcriptional factor playing role in PHYA-mediated cotyledon opening and it is regulated by circadian clock [42]. |
| COL3 | Zn finger protein which is a positive regulator of red light signaling and photomorphogenesis. Also regulates root and shoot development [43]. |
| SAUR9 | Early auxin responsive gene induced on auxin application [44]. |
| SAUR26 | Early auxin responsive gene induced on auxin application [45]. |
| LAX2 | Auxin influx carrier involved in leaf venation [46]. |
| COL9 | CONSTANS like protein, acts as negative regulator of flowering [46]. |
| CIP1 | COP1 interaction protein, plays positive role in ABA response [47]. |
| CSN6A | Component of COP 9 signalosome complex regulating ubiquitin conjugation pathway and plays role in repression of photomorphogenesis in dark [48]. |
| CSN6B | Component of COP 9 signalosome complex regulating ubiquitin conjugation pathway and plays role in repression of photomorphogenesis in dark [48]. |
| CCA1 | Morning loop gene which functions along with LHY1 and it represses TOC1 [49]. |
| TOC1 | Evening complex gene, regulates plant fitness by controlling the clock output through repressing morning and evening loop of circadian clock [50]. |
| PRR9 | Temperature sensitive component of circadian clock, interacting with TOC1 [51]. |
Fig. 6.
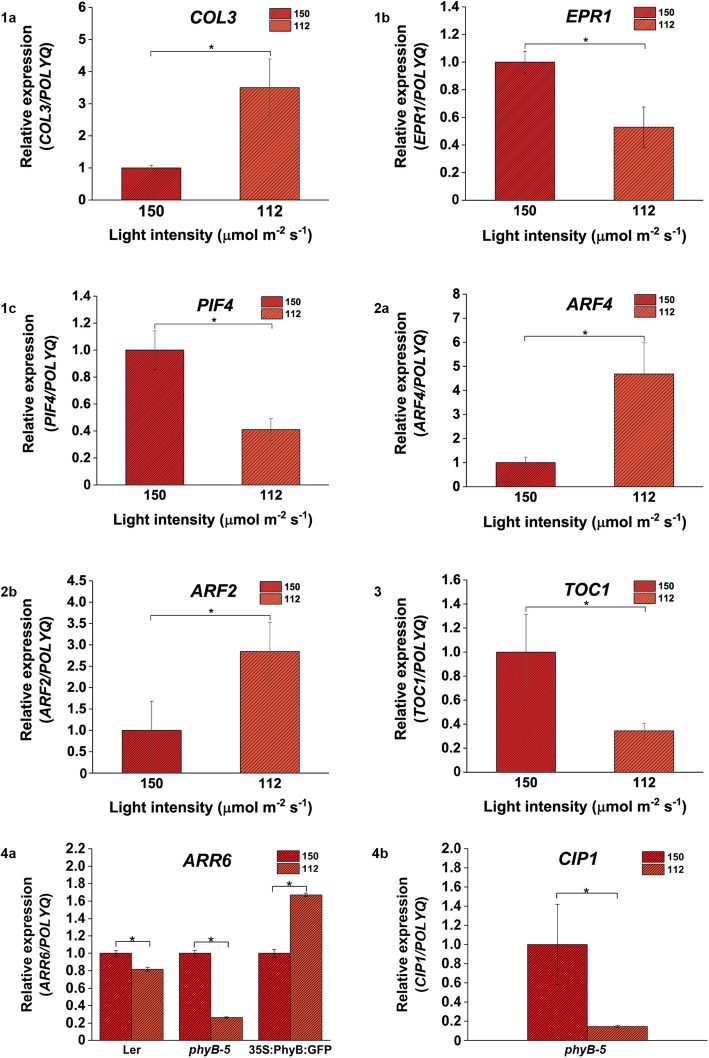
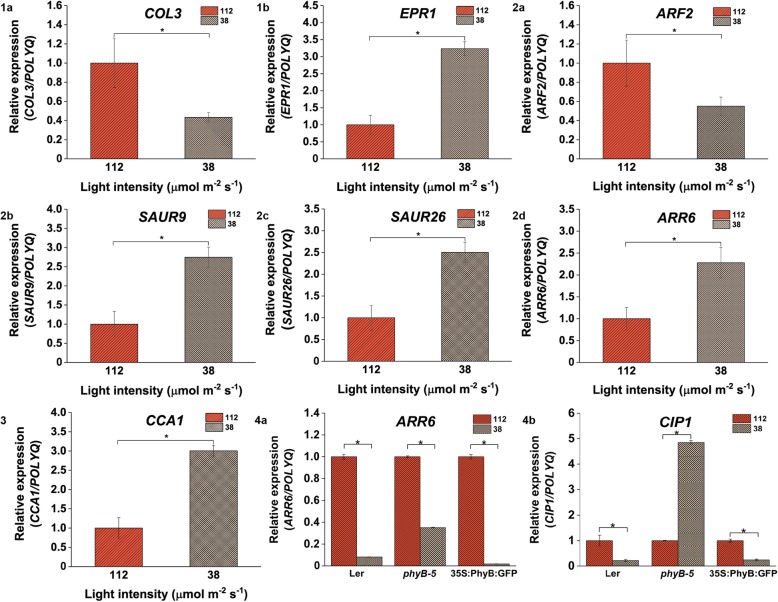
Genes involved in light, hormone and clock-regulated pathways are differentially expressed under 150 vs 112 μmol m − 2 s − 1 light intensity. Light signaling genes (1a) COL3, (1b) EPR1 and (6.2c) PIF4 showed differential regulation; (2a) ARF4 and (2b) ARF2 genes involved in auxin signaling were differentially expressed; (3) TOC1 gene showed variability in its expression in case of WT; (4a) ARR6 and (4b) CIP1 genes were differentially expressed in Ler, 35S::PhyBGFP, phyB-5 and phyB-5 respectively under 150 vs 112 μmol m − 2 s − 1 light intensity. qRT-PCR was performed with root samples of 5-days old seedlings. Analysis has been described in the method section
Fig. 7.
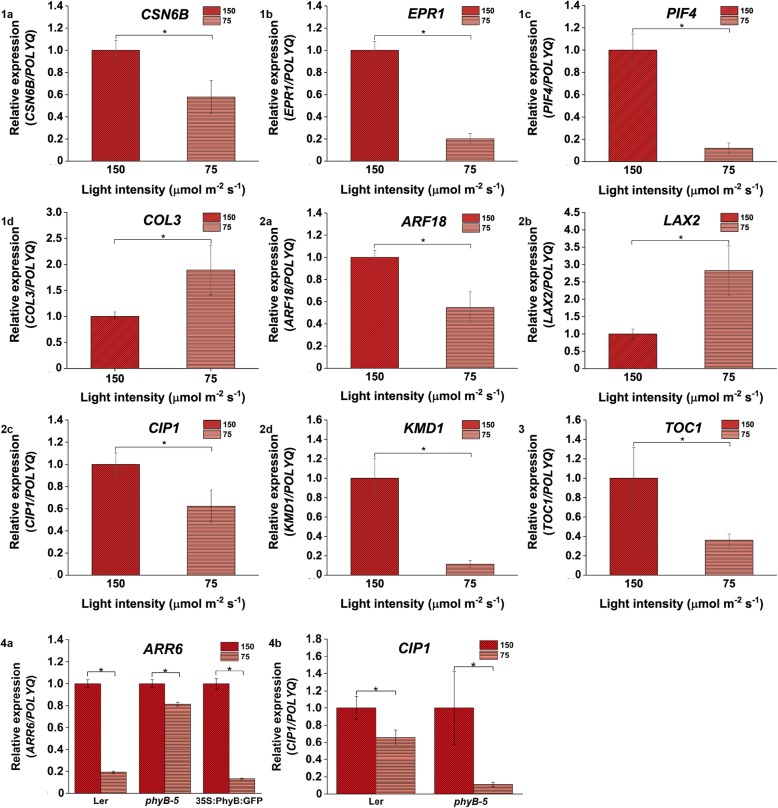
Relative expression of genes involved in light, hormone and clock-regulated pathways is influenced under 150 vs 75 μmol m − 2 s − 1 light intensity. Differential expression of (1a) CSN6B, (1b) EPR1, (1c) PIF4 and (1d)COL3 genes; Variation in gene expression of (2a) ARF18, (2b) LAX2, (2c) CIP1 and (2d) KMD1 ; (3) TOC1 gene regulation in WT; (4a) ARR6 showed variation in its regulation in all genotypes but (4b) CIP1 expression varied only in case of Ler and phyB-5 under 150 vs 75 μmol m − 2 s − 1 light intensity. qRT-PCR was performed with root samples of 5-days old seedlings. Analysis has been described in the method section
Fig. 8.
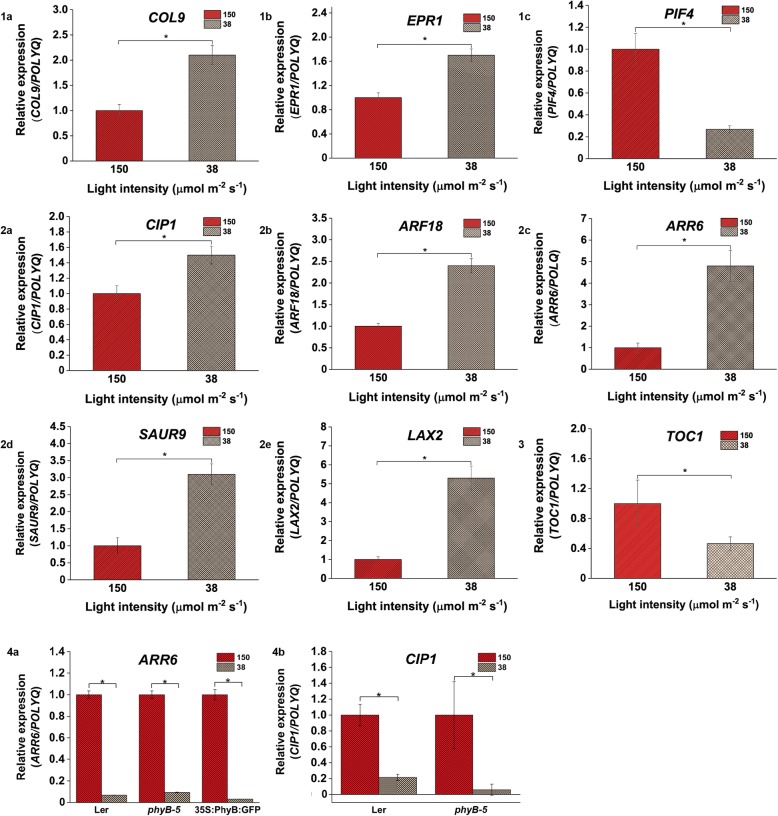
Light intensity of 150 vs 38 μmol m − 2 s − 1 affects the expression profile of genes involved in light, hormone and clock-regulated pathways. Variation in expression of (1a) COL9, (1b) EPR1 and (1c) PIF4 candidate genes; (2a) CIP1, (2b) ARF18, (8.2c) ARR6, (2d) SAUR9 and (2e) LAX2 genes showed variable expression; (3) Regulation in TOC1 gene expression in WT root; (4a) ARR6 expression varied in all genotypes but (4b) CIP1 expression varied only in case of Ler and phyB-5 under 150 vs 38 μmol m − 2 s − 1 light intensity. qRT-PCR was performed with root samples of 5-days old seedlings. Analysis has been described in the method section
Fig. 9.
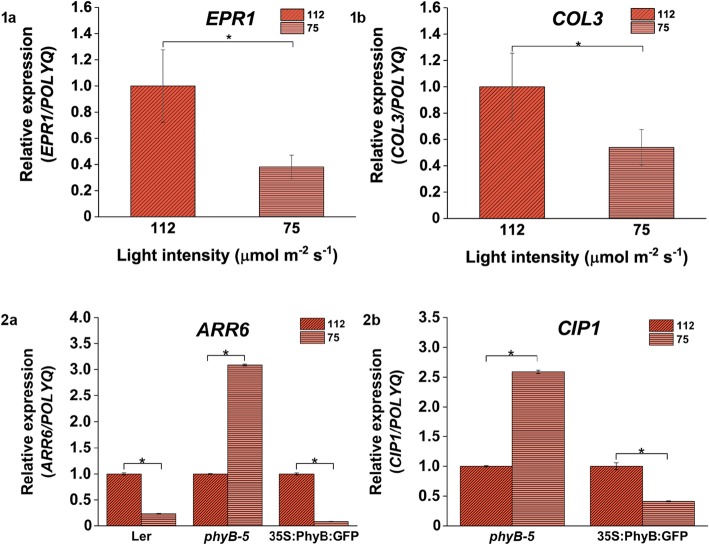
Relative expression of genes involved in light signaling pathways under 112 vs 75 μmol m − 2 s − 1 light. Gene expression profiling of (1a) EPR1 and (1b) COL3 varied in WT root; Expression of (2a) ARR6 varied in all seed lines and (2b) CIP1 was differentially expressed in 35S::PhyBGFP and phyB-5 only under 112 vs 75 μmol m − 2 s − 1 light intensity. The qRT-PCR was performed with root samples of 5-days old seedlings. Analysis has been described in the method section
Fig. 10.
Expression profiling of genes involved in light, hormone and clock-regulated pathways is influenced under 112 vs 38 μmol m − 2 s − 1 light intensity. (1a) COL3 and (1b) EPR1 genes were differentially expressed; Variation in expression profile of (2a) ARF2, (2b) SAUR9, (2c) SAUR26 and (2d) ARR6 genes; (3) CCA1 gene expression varied in WT; Expression profiling of (4a) ARR6 and (4b) CIP1 genes varied in all seed lines under 112 vs 38 μmol m − 2 s − 1 light intensity. qRT-PCR was performed with root samples of 5-days old seedlings. Analysis has been described in the method section
Fig. 11.
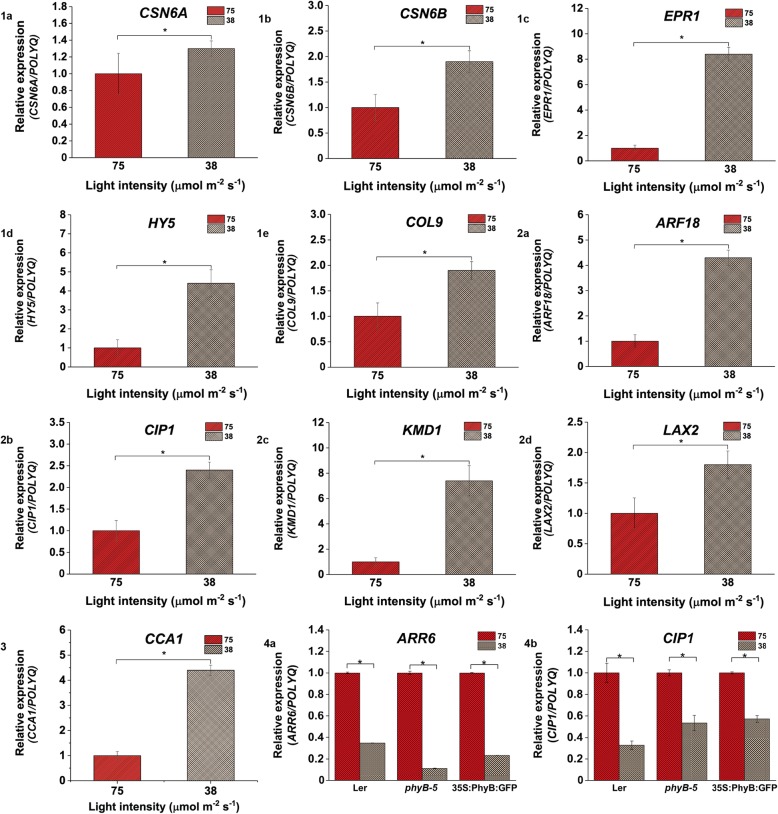
Change in relative expression of genes involved in light, hormone and clock-regulated pathways under 75 vs 38 μmol m − 2 s − 1 light intensity. Gene expression profile of (1a) CSN6A, (1b) CSN6B, (1c) EPR1, (1d) HY5 and (1e) COL9 varied; (2a) ARF18, (2b) CIP1, (2c) KMD1 and (2d) LAX2 genes showed variable expression; (3) CCA1 gene showed variable expression; Expression profiling of (4a) ARR6 and (4b) CIP1 genes differ in all seed lines under 75 vs 38 μmol m − 2 s − 1 light intensity. qRT-PCR was performed with root samples of 5-days old seedlings. Analysis has been described in the method section
Table 5.
Relative expression of selected genes analysed by qRT-PCR
| Gene names | Light Intensity (μmol m − 2 s − 1) | Fold change |
|---|---|---|
| ARR6 | 150 vs 38 | 4.7 |
| 112 vs 38 | 2.2 | |
| HY5 | 75 vs 38 | 4.4 |
| ARF18 | 150 vs 75 | −1.8 |
| 150 vs 38 | 2.4 | |
| 75 vs 38 | 4.3 | |
| ARF2 | 150 vs 112 | 2.8 |
| 112 vs 38 | − 1.8 | |
| ARF4 | 150 vs 112 | 4.6 |
| KMD1 | 150 vs 75 | −8.7 |
| 75 vs 38 | 7.4 | |
| PIF4 | 150 vs 112 | −2.4 |
| 150 vs 75 | −8.3 | |
| 150 vs 38 | −3.7 | |
| EPR1 | 150 vs 112 | −1.8 |
| 150 vs 75 | −4.9 | |
| 150 vs 38 | 1.7 | |
| 112 vs 75 | − 2.6 | |
| 112 vs 38 | 3.2 | |
| 75 vs 38 | 8.4 | |
| COL3 | 150 vs 112 | 3.5 |
| 150 vs 75 | 1.8 | |
| 112 vs 75 | −1.8 | |
| 112 vs 38 | − 2.3 | |
| SAUR9 | 150 vs 38 | 3.1 |
| 112 vs 38 | 2.7 | |
| SAUR26 | 112 vs 38 | 2.5 |
| LAX2 | 150 vs 75 | 2.8 |
| 150 vs 38 | 5.3 | |
| 75 vs 38 | 1.8 | |
| COL9 | 150 vs 38 | 2.1 |
| 75 vs 38 | 1.9 | |
| CIP1 | 150 vs 75 | −1.6 |
| 150 vs 38 | 1.5 | |
| 75 vs 38 | 2.4 | |
| CSN6A | 75 vs 38 | 1.3 |
| CSN6B | 150 vs 75 | −1.7 |
| 75 vs 38 | 1.9 | |
| CCA1 | 112 vs 38 | 3 |
| 75 vs 38 | 4.3 | |
| TOC1 | 150 vs 112 | −2.9 |
| 150 vs 75 | −2.7 | |
| 150 vs 38 | −2.1 |
qRT-PCR analysis showed that in comparison to 150 μmol m − 2 s − 1 light intensity, under 112 μmol m − 2 s − 1, a positive regulator of red light signaling, COL3 was significantly upregulated (Fig. 6.1a) whereas other transcription factors involved in PHY signaling such as EPR1 and PIF4 were downregulated (Fig. 6.1b and .1c). Auxin responsive factors such as ARF2 and ARF4 were upregulated under the mentioned light intensity (Fig. 6.2a and .2b). The clock-regulated genes such as TOC1 was downregulated under 112 μmol m − 2 s − 1 light intensity compared to 150 μmol m − 2 s − 1 light intensity (Fig. 6.3). Under 75 μmol m − 2 s − 1 compared to 150 μmol m − 2 s − 1 intensity of light, the expression level of light signaling genes such as CSN6B, EPR1, and PIF4 genes was reduced (Fig. 7.1a, .1b and .1c) however, COL3 expression was enhanced (Fig. 7.1d). Auxin influx carrier, LAX2 was upregulated (Fig. 7.2b) whereas ARF18, CIP1 and KMD1 were downregulated (Fig. 7.2a, .2c and .2d) under this comparative light condition. TOC1 was downregulated under 75 μmol m − 2 s − 1 in comparison to 150 μmol m − 2 s − 1 intensity (Fig. 7.3). Under 38 μmol m − 2 s − 1 light in comparison to 150 μmol m − 2 s − 1, the expression of light signaling genes like COL9 and EPR1 was upregulated (Fig. 8.1a and .1b) whereas PIF4 expression was downregulated (Fig. 8.1c). On the other hand, genes involved in hormone signaling such as CIP1, ARF18, ARR6, SAUR9, and LAX2 were upregulated (Fig. 8.2a, .2b, .2c, .2d and .2e). TOC1 was downregulated under 38 μmol m − 2 s − 1 light as compared to 150 μmol m − 2 s − 1 (Fig. 8.3). Overall, it was observed that TOC1 is downregulated in all lower light intensities compared to highest intensities.
In comparison to 112 μmol m − 2 s − 1, under 75 μmol m − 2 s − 1 light condition, the expression of EPR1 and COL3 was downregulated (Fig. 9a and b). There was no significant change observed in the expression pattern of hormonal signaling and clock-regulated genes under this comparative light condition. With respect to 112 μmol m − 2 s − 1 under 38 μmol m − 2 s − 1 light intensity, COL3 expression was reduced whereas EPR1 gene was enhanced (Fig. 10.1a and .1b). When hormone signaling pathway genes were focused, it was observed that the expression of SAUR9, SAUR26 and ARR6 was upregulated (Fig. 10.2b, .2c and .2d) whereas ARF2 expression was downregulated (Fig. 10.2a). The expression of clock regulated gene such as CCA1 was upregulated under this comparative light condition (Fig. 10.3).
In comparison to 75 μmol m − 2 s − 1 under low light intensity of 38 μmol m − 2 s − 1, the expression of CSN6A, CSN6B, EPR1, HY5 and COL9 genes was higher (Fig. 11.1a, .1b, .1c, .1d and .1e) and hormone pathway genes such as ARF18, CIP1, KMD1 and LAX2 were also upregulated (Fig. 11.2a, .2b, .2c and .2d). CCA1 expression was also shown to be upregulated under this low light condition (Fig. 11.3). The genes which were induced or repressed under low light intensity in comparison to high intensity of light have been summarized in Table 6.
Table 6.
Genes induced and repressed in roots under lower light versus higher light intensity
| Light intensities (μmol m − 2 s − 1) | Genes Upregulated | Genes downregulated |
|---|---|---|
| 150 vs 38 | COL9, EPR1, CIP1, ARF18, ARR6, SAUR9 and LAX2 | PIF4 and TOC1 |
| 112 vs 38 | EPR1, SAUR9, SAUR26, ARR6 and CCA1 | COL3 and ARF2 |
| 75 vs 38 | CSN6A, CSN6B, EPR1, HY5, COL9, ARF18, CIP1, KMD1, LAX2 and CCA1 |
Further to validate the transcriptomics data, we performed the qRT-PCR for ARR6 and CIP1 genes in Landsberg erecta (Ler), constitutively overexpressor (35S::PhyB:GFP) and mutant (phyB-5) lines of PHYB in Ler background. qRT-PCR was performed with root samples of 5-days old seedlings grown under all four different light intensities. It has been observed that the expression of ARR6 and CIP1 was downregulated in case of Ler and phyB-5 under 112 μmol m − 2 s − 1 in comparison to 150 μmol m − 2 s − 1 light (Fig. 6.4a and .4b) whereas ARR6 has been shown to be significantly upregulated in case of 35S::PhyBGFP (Fig. 6.4b). When the expression of these two genes were analysed in presence of 75 μmol m − 2 s − 1 in comparison to 150 μmol m − 2 s−1 light intensity, ARR6 was downregulated in all three genotypes (Fig. 7.4a) and CIP1 showed downregulation only in case of phyB-5 genotype (Fig. 7.4b). Similarly, under low light of 38 μmol m − 2 s − 1 intensity in comparison to 150 μmol m − 2 s − 1, both these genes were downregulated. However, CIP1 did show a significant downregulation only in case of Ler and phyB-5 (Fig. 8.4a and .4b).
When the relative expression pattern of ARR6 and CIP1 was analysed under 75 μmol m − 2 s − 1 in comparison to 112 μmol m − 2 s−1light intensity, ARR6 was found to be downregulated in Ler and 35S::PhyBGFP whereas it was upregulated in case of phyB-5 (Fig. 9.2a). On the other hand, CIP1 was upregulated in phyB-5 but downregulated in case of 35S::PhyBGFP (Fig. 9.2b). Under 38 μmol m − 2 s − 1 as compared to 112 μmol m − 2 s−1 light intensity, ARR6 expression was downregulated in all genotypes tested (Fig. 10.4a). CIP1 was downregulated in case of Ler and 35S::PhyBGFP whereas upregulated in phyB-5 (Fig. 10.4b). Both the genes were downregulated under 38 μmol m − 2 s − 1 in comparison to 75 μmol m − 2 s − 1 light intensity in all three genotypes (Fig. 11.4a and .4b). These data suggested that ARR6 and CIP1 are differentially regulated by PHYB under variable WL intensities.
Discussion
Although earlier studies have investigated the effect of light intensity as well as light quality on root development, yet the molecular players that fine tune the output response are poorly understood [1, 17]. Light signaling mediated through phytohormones modulate plant development [52]. Under shade, where the R:FR ratio is very low, the light is primarily sensed by PHYs and their downstream factors [53]. This leads to change in plant phenotype such as elongated hypocotyl, longer petiole, small leaves, apical hook formation, etc. On the other hand, phytohormones such as gibberellin (GA), auxin (IAA), ethylene (ET) and brassinosteroid (BR) have been shown to regulate plant development under relative dark and shade conditions. GA promotes hypocotyl elongation and suppresses other photomorphogenic features, these phenotypic differences appear through inactivation of HY5 and other light signaling transcription factors such as PIFs. Light controls GA synthesis by downregulating GA biosynthetic enzymes and stimulating GA-inactivation enzymes [54]. It has also been reported that shade (low R:FR) induces hypocotyl growth via enhanced TRYPTOPHAN AMINOTRANSFERASE OF ARABIDOPSIS 1 (TAA1)-dependent auxin biosynthesis [53]. PIF5 promotes ethylene-mediated apical hook formation, one of the characteristics of shade avoidance syndrome in dark [55]. Along with light, circadian clock also regulates phytohormone signaling and synthesis. Ethylene production is regulated by circadian clock genes such as TOC1 and CCA1, similarly ACC SYNTHASE 8 (ACS8) is also controlled by light and circadian clock [56]. Auxin signaling and responses are also regulated by circadian clock [57]. It has been reported that cytokinin t-zeatin treatment induces expression of TOC1, GIGANTEA (GI) in morning and CCA1 in the evening. Active NAA or ABA treatment has been shown to downregulate CCA1 expression in morning [58]. TOC1 and other circadian clock-associated genes have been reported to control the expression of genes involved in GA biosynthesis [59]. Clock regulates the transcript level of PIF4 and PIF5 genes [60]. In the present work, we have shown that different WL intensities (150, 112, 75 and 38 μmol m − 2 s − 1) show variable effects on the root development in case of WT and PHY mutants. Light quantity influenced the root architecture as well as the expression profile of the genes playing role in light signaling, phytohormone related and clock-regulated pathways. It has been reported that, although only aerial shoot portion perceive light, some amount of light gets translocated down through light piping and influences root patterning [9]. In the current work, we explored the candidate genes involved in light intensity-based root patterning over direct exposure of seedlings to light.
Primary, lateral and adventitious roots are affected by different intensities of white light
Different intensities of light as well as their quality have been shown to affect root development [20–22, 17]. PHYs sense the quality as well quantity of light and have been shown to influence phototropism, gravitropism and elongation of root [5]. Under shade condition, where the ratio of R:FR is reduced, the hypocotyl elongates, however, the effect of shade on root development is poorly understood [61]. We have shown here that under different light intensity, the root patterning changes significantly. Primary root elongation was more influenced in case of PHY mutants. Higher light intensity (150 μmol m − 2 s − 1) induced larger number of lateral as well as adventitious roots. PHYB predominantly affected the adventitious root growth whereas PHYA has been shown to be involved in primary root growth based on variable light intensity. Both PHYA and PHYB are involved in sensing different intensities of WL that affect the root architecture accordingly. The analysis of root wave, root coiling, root hair density etc., need to be carried out in detail in PHY mutants under different WL intensity. These aspects should also be addressed under different light intensities of monochromatic light to understand the correlation of different quality and quantity of light on root architecture.
Light intensity influences root development and also affects the gene expression profile in root
When light signal translocates through the shoot to the root or seedling directly exposed to light, the gene expression profile of root changes differently. It has been shown that the spatial expression profile of photoreceptors changes throughout the root when shoot is directly exposed to light [23]. With microarray analysis, we found about 5243 DEGs under four different light intensities (150, 112, 75 and 38 μmol m − 2 s − 1) and highest and lowest number of DEGs were detected under 150 vs 38 μmol m − 2 s − 1 light condition and 150 vs 112 μmol m − 2 s − 1 light intensity respectively. The change of light intensity from high to low, severely affected the gene expression pattern of root whereas the slight change of 150 to 112 μmol m − 2 s − 1 light didn’t have significant effect on gene expression profile. Interestingly, CYP81F2 (Cytochrome P450 in indole glucosinolate biosynthesis), At2g39445 (phosphatidylinositol n-acetylglucosaminyltransferase), At5g22555 (transmembrane protein) and At2g44130 (F-box protein) genes were altered in all comparative light intensity. The functions of these candidate genes should be studied in detail as this can correlate the light intensity-based root development with glucosinolate biosynthesis, nucleotide sugar and glycolipid signaling and SCF ubiquitin-mediated pathways.
GO enrichment and KEGG colour pathway analysis of DEGs highlight the potential role of specific pathways involved in light intensity-based root development
GO analysis suggested about functionality of the genes altered under variable light intensities, their specific locations and their involvement in the major biological, cellular and molecular processes. The change in light intensity affected the root development possibly by influencing the catalytic activities of the genes present in root. Different intensities of light also altered the expression of gene predominantly localized in cytoplasm, cell periphery and membrane part of the cell. KEGG analysis has shown that various pathways such as metabolic, secondary metabolite biosynthesis, ribosome-mediated, carbon metabolism, plant hormone signaling, protein processing in ER, MAPK signaling, amino acid biosynthesis, starch and sucrose metabolic, oxidative phosphorylation, purine metabolic, amino and nucleotide sugar metabolic, etc. were altered in roots on irradiation of different WL intensities.
Light intensity affects the expression pattern of genes involved in the light signaling, hormone related and clock-regulated pathways in roots
Earlier reports showed that light affects root development through photoreceptors. PHYA promotes root elongation under R, FR, B while PHYB enhances root length under R light only [61, 18]. Lateral root growth is promoted by PHYA, PHYB and PHYE whereas the high light sensor PHYD inhibits this phenomenon [4]. Root hair formation is also promoted by light where PHYA and PHYB have been shown to stimulate root hair initiation [62]. Therefore, few genes known to play major role in light signaling were analysed to investigate that how they are regulated by different light intensities. Remarkably, the expression of PHYA and PHYB was unaltered under all tested light intensities. This could be possibly because of post translational modification of PHYs under these light intensities, which may further lead to their instability and degradation in roots. It also indicates that the expression level of PHYs may change under other intensities of light not investigated in the present work. Furthermore, light dependent mRNA stability and subsequent splicing events adding to another layer of complex regulation may be possible. Then, expression profile of other light signaling genes were analyzed and it was observed that the expression of PIF4 was maximum at higher intensity of 150 μmol m − 2 s − 1 light whereas the expression of HY5, CSN6A, CSN6B, EPR1 and COL9 was highest under 38 μmol m − 2 s − 1 light. The expression of positive regulator of red light signaling and suppressor of flowering, COL3 was observed to be highest under 112 μmol m − 2 s − 1 light. These genes are most probable candidate for sensing the light quality along with quantity to affect the root architecture. Expression of PHYs didn’t change, however, their downstream genes were found to be altered with different light intensities irradiated on root. Light affects the hormone synthesis, signaling and transport and known to be involved in root development [63]. Hence, few genes involved in auxin, cytokinin and ABA pathways were analysed in this current work. The expression level of auxin responsive factors such as ARF2 and ARF4 was observed to be maximum under 112 μmol m − 2 s − 1 light. Auxin influx carrier; LAX2, negative regulator of cytokinin signaling; ARR6 and a positive regulator of ABA signaling; CIP1 have shown maximum expression under low light intensity of 38 μmol m − 2 s − 1. Specific light intensity may alter certain metabolic processes that might account for the observed output gene regulation. Early auxin-responsive genes such as SAUR9 and SAUR26 were upregulated with decreasing light intensity and their expression was maximum at 38 μmol m − 2 s −1 light. This differential gene expression profiling showed that various hormonal pathways are also influenced by different light quantity that further lead to the differences in root architecture. Circadian clock has a major role in plant development and it has been reported to affect root growth [64, 32]. Clock genes such as CCA1, PRR9 and TOC1 were analyzed in this current work and it was observed that, the expression of CCA1 and TOC1 was maximum under 38 μmol m − 2 s − 1 and 150 μmol m − 2 s − 1 light intensity respectively. They are associated with evening and morning complex and make sense to peak at low light intensities normally represented as dawn and dusk. However, their upregulation at high light intensity may be a result of light stress response. Strikingly, expression of PRR9 was not significantly changed under any light condition, suggesting that PRR9 oscillation is robust and not influenced by the concerned component. It shows that both light intensity and clock-regulated pathways interact to regulate the root architecture. A detailed study of these genes needs to be done for their involvement in intensity dependent root growth. Difference in light intensity alters various light signaling, hormone associated and clock-regulated genes localized in root leading to change in its phenotype. Most of the genes analysed in by qRT-PCR had shown maximum expression at 38 μmol m − 2 s − 1 light. Very few genes analysed in this study such as PIF4 and TOC1 showed highest expression at higher light intensity of about 150 μmol m − 2 s − 1 light. From the transcriptome validation data, it was observed that expression profile of few of the genes such as ARR6 and CIP1 are light intensity as well as PHYB dependent. In Ler, ARR6 and CIP1 showed antagonistic regulation with respect to WT under few light intensities. As under 38 μmol m − 2 s − 1, ARR6 was downregulated in case of Ler when compared with 150 and 112 μmol m − 2 s − 1 whereas it has shown upregulation in WT. Similarly, CIP1 showed downregulation under 38 μmol m − 2 s − 1 in comparison to 150 and 75 μmol m − 2 s − 1 light in Ler on the other hand, it was upregulated in case WT. These variable gene expression in different genetic lines suggest that the light intensity mediated gene expression in roots may be partly dependent on ecotypes. Similar reports are available, which have shown that timing to flower varies in different ecotypes. Under variable light intensities, the net amount of Pr and Pfr varies and are most likely regulated in spatio-temporal manner. The amount of endogenous level of active and inactive forms of photoreceptor at a given temperature can perhaps add an extra layer of complexity to generate a transcriptomics pattern as a read-out.
The investigation of genes involved in other phytohormone signaling, synthesis and transport under variable light intensities could be carried out. Few of the light signaling genes such as PIF5, FAR-RED ELONGATED HYPOCOTYL 1 (FHY1), FHY1-LIKE (FHL), CONSTITUTIVE PHOTOMORPHOGENESIS 1 (COP1) etc. could also be analysed with respect to light intensity based root growth. The expression profiling of the candidate genes playing role in shade avoidance syndrome like HY5, LONG HYPOCOTYL IN FAR-RED LIGHT 1 (HFR1), Arabidopsis thaliana HOMEOBOX PROTEIN 4 (ATHB4) etc. could also be studied under changing light intensity [53]. The expression of these genes could be analysed in the shoot as well as root under the four WL intensities mentioned or other suitable intensities. These studies could be extended by performing different sets of experiments with variable intensity of R, FR, B light and then analyzing the expression of genes involved in light, phytohormone signaling, clock regulation as well as other related pathways.
Conclusion
Our current work showed the effect of different WL intensity on the root phenotype of WT as well as phyA and phyB mutants. The primary root length was shorter in case of phyA-211 under all four variable light intensities, however under higher intensities of 112 and 150 μmol m − 2 s − 1 both the PHY mutants had shown shorter primary root in comparison to WT. phyB-9 had shown more adventitious roots in comparison to phyA-211 and WT under same light intensity. Under 38 and 75 μmol m − 2 s − 1 light intensity, phyA-211 had lesser number of lateral roots whereas under 112 μmol m − 2 s − 1 light, phyB-9 showed lesser number of lateral roots in comparison to phyA-211 and WT. Both adventitious as well as lateral root number were similar in all genotypes under 150 μmol m − 2 s − 1. Higher intensity of WL such as 150 μmol m − 2 s − 1 and lower intensity of light such as 38 μmol m − 2 s − 1 had the most significant effects on the gene expression profile of root as the number of DEGs was found to be highest in case of 150 vs 38 μmol m − 2 s − 1 light. KEGG colour analysis pathway has shown that most of the genes found from microarray belonged to metabolic, secondary metabolite synthesis, carbon metabolic and plant hormone signaling pathway. Genes such as PIF4, EPR1, COL3, CIP1, TOC1 etc. were shown to be differentially expressed under almost all WL intensities. These genes may act as possible candidates in correlation of light intensity dependent root development. This study showed that different quantity of light affects the transcript abundance of light, hormone and clock pathway genes in the root tissues, indicating their potential role in light intensity mediated root development. Transcriptomics profiling using laser capture microdissection at single cell/tissue level could be valuable and would add in depth understanding of the regulatory network.
Methods
Plant material and growth conditions
Columbia Wild type (WT), phytochrome mutants such as phyB-9 and phyA-211 seed lines of Arabidopsis thaliana were used for root architectural studies. For microarray analysis, WT root samples were used. The initial qRT-PCR validation was performed with WT and for further transcriptome validation, Landsberg erecta (Ler) wild type, overexpressor line, 35S::PhyBGFP (Landsberg erecta) and mutant line, phyB-5 (Landsberg erecta) of PHYB were used. These seed lines were a kind gift from Prof. Eberhard Schäfer, Albert Ludwigs University of Freiburg, Germany [65, 66]. The seeds were surface sterilized with 70 and 100% ethanol sequentially and dried under sterile bench. The sterilized seeds were sown on half-strength Murashige and Skoog salt (Himedia, Cat. No. PT021) supplemented with 1% sucrose (w/v) (MP biomedical, Cat. No. 194018) and 0.8% agar (w/v) (Himedia, Cat. No. PCT0901) in square plates and then kept for 72 h of cold stratification at 4 °C in dark. The plates were kept vertically at 90 °and grown under four different WL intensities (Long day, 16 h light/8 h dark) in the range of 150, 112, 75 to 38 μmol m − 2 s − 1 light in Percival light cabinets (Model No. CU36L6) at 22 °C with relative humidity of ~ 70%. Root phenotyping and RNA extraction were done with 6-days old and 5-days old seedlings respectively. Sample harvesting was approximately done at ZT = 12. The germination induction resulted in uniform germination among all genetic lines tested and confirmed by light microscopy, after 2 days of transferring in light. The root growth comparison was performed in 6-days old seedlings.
RNA extraction and microarray analysis
RNA was extracted from the root tissues of 5-days old WT seedlings grown under different light intensities (38, 75, 112 and 150 μmol m − 2 s − 1) with TRIzol reagent (Thermofisher Scientific, Cat. No.15596026), following the manufacturer’s protocol. RNA samples were subjected to DNaseI treatment with TURBO DNA free kit (Thermofisher Scientific, Cat. No. AM1907). For microarray analysis, the RNA quality was analysed with the Bioanalyzer (Agilent Bioanalyzer 2100 system). cDNA was prepared from RNA samples with RevertAid H Minus First Strand cDNA Synthesis Kit (Thermofisher Scientific, Cat. No. K1631). Microarray analysis was performed following Affymetrix GeneChip® WT PLUS Reagent Kit Manual Target Preparation for GeneChip® Whole Transcript (WT) Expression Arrays. Microarray experiment was done at ILS laboratory, Gurgaon, Haryana, India. Two independent biological replicates for each condition were used for microarray experiment in addition to this, three technical replicates were utilized. DEGs obtained from microarray were analysed with One-Way Between-Subject ANOVA (unpaired) test. Gene Level Differential Expression Analysis was performed and genes having minimum FC of 1.2 with FDR < 0.05 were selected for further analysis. The DEGs were categorized in six comparative light intensities such 150 vs 112 μmol m − 2 s − 1 light, 150 vs 75 μmol m − 2 s − 1 light, 150 vs 38 μmol m − 2 s − 1 light, 112 vs 75 μmol m − 2 s − 1 light, 112 vs 38 μmol m − 2 s − 1 light and 75 vs 38 μmol m − 2 s − 1 light.
Venn diagram, gene enrichment (GO) pathway analysis and KEGG analysis
Venn diagrams were prepared for overlapping genes considering a combination of three comparative light conditions at a time. Gene lists from the microarray data were sorted and used for plotting Venn diagram using a custom script for R [67] using packages gdata [68], Vennerable [69], gplots [70], RBGL [71] and graph [72]. The script is available online on GitHub https://github.com/debadutta-patra/Venn-Diagram. GO pathway analysis was performed using Panther Over-representation test and the annotation version is GO Ontology database. The FISHER test was used and the categorization of DEGs under different GO terms was done considering FDR < 0.05. The KEGG search and colour pathway analysis was performed considering only a small group of DEGs.
qRT-PCR analysis for DEGs
For quantitative qRT-PCR analysis, an aliquot of the cDNA samples was prepared from the same RNA used for microarray of WT. For validation of transcriptome data, qRT-PCR was performed with Ler, 35S::PhyBGFP and phyB-5 root samples. The qRT-PCR was performed using Biorad Evagreen kit (Bio-Rad SSoFast EvaGreen Supermix, Cat. No. 172–5203) and Bio-Rad C1000 Touch™ thermal cycler was used for qRT-PCR (CFX384™ Real Time System). The total volume of the reaction was 10 μl with 25 ng cDNA for WT, 50 ng cDNA for Ler, 35S::PhyBGFP and phyB-5 and each primer of 5 μM concentration was used. The software used for data analysis was Bio-Rad CFX Manager 3.81, the gene expression was analysed with FC of 1.2 and p-value < 0.05. The transcript level was normalized with a pair of primer specific for POLYUBIQ gene. qRT-PCR was done at least in triplicate and the data shown represent the mean + SE.
The forward and reverse primers are listed in the supplementary data (Additional file 6).
Measurement and statistical analysis
The plates showing root phenotype were scanned at 600 dpi using HP Scanjet G4010 Flatbed Scanner. The lateral root growth pictures were captured with Nikon stereo zoom microscope (Nikon SMZ745T). The lateral root and adventitious root number were quantified under the stereo zoom microscope. The scanned images were used for measuring primary root length using ImageJ [73]. Mean comparison was done by one-way ANOVA with Tukey test for multiple mean comparison and with a probability threshold of 0.05. All the statistical analysis and graphs plotting were done using OriginPro (OriginLab, Northampton, MA).
Additional files
Figure S1. Detailed zoomed pictures of lateral root growth in 6-day old WT, phyB-9 and phyA-211 seedlings grown under WL intensities of 38, 75, 112 and 150 μmol m − 2 s − 1. Scale bar = 5 mm. Arrow head represents lateral roots. (PNG 1250 kb)
Figure S2. Microscope images of lateral root growth for qualitative visualization in 6-day old WT, phyB-9 and phyA-211 seedlings grown under WL intensities of 38, 75, 112 and 150 μmol m − 2 s − 1. Scale bar = 5 mm. (PNG 4917 kb)
List of DEGs Common in two, three, four or five comparative light conditions. (xlsx 1970 kb)
List of Unique DEGs in specific comparative light conditions. (XLSX 40kb)
Figure S3. Composite figure of GO analysis of DEGs under (a) 150 vs 112 μmol m − 2 s − 1 (b) DEGs under 150 vs 75 μmol m − 2 s − 1 (c) DEGs under 150 vs 38 μmol m − 2 s − 1 (d) DEGs under 112 vs 75 μmol m − 2 s − 1 (e) DEGs under 112 vs 38 μmol m − 2 s − 1 (f) DEGs under 75 vs 38 μmol m − 2 s − 1 light intensity. (PNG 40 kb)
List of primers used for qRT-PCR. (DOCX 14 kb)
Acknowledgments
We are thankful to Prof. Eberhard Schäfer, Albert Ludwigs University of Freiburg, Germany for the seed lines. The authors would like to thank Durga Prasad Biswal, NISER for few of the experimental strategies. The authors would also like to thank Dr. Chandan Goswami, NISER for his critical comments to improve the manuscript. We would also like to acknowledge the CIF facility, NIPGR.
Abbreviations
- ACS8
ACC synthase 8
- ARF18
Auxin response factor 18
- ARF2
Auxin response factor 2
- ARF4
Auxin response factor 4
- ARR6
Type A response regulator 6
- ATHB4
Arabidopsis thaliana homeobox protein 4
- BP
Biological Processes
- CC
Cellular component
- CCA1
Circadian clock-associated 1
- CIP1
COP1 interacting protein1
- COL3
Constans like 3
- COL9
Constans like 9
- COP1
Constitutive photomorphogenesis 1
- CRY1
Cryptochrome 1
- CRY2
Cryptochrome 2
- CSN6A
COP 9 signalosome complex subunit 6 A
- CSN6B
COP 9 signalosome complex subunit 6 B
- CYP81F2
Cytochrome P450 81F2
- DEGs
Differentially expressed genes
- EPR1
Early phytochrome response 1
- FC
Fold change
- FDR
False Discovery Rate
- FHL
FHY1-like
- FHY1
Far-red elongated hypocotyl 1
- GO
Gene ontology
- HFR1
Long hypocotyl in far-red light 1
- HY5
Elongated hypocotyl 5
- KEGG
Kyoto Encyclopedia of Genes and Genomes
- KMD1
Kiss me deadly 1
- LAX2
Like aux 2
- LHY1
Late elongated hypocotyl 1
- MF
Molecular Function
- PAR2
Phytochrome rapidly regulated 2
- PHOT1
Phototropin 1
- PHOT2
Phototropin 2
- PHYA
Phytochrome A
- PHYB
Phytochrome B
- PIF4
Phytochrome interacting factor 4
- PRR9
Pseudo response regulator 9
- RIN
RNA Integrity No
- SAUR26
Small auxin upregulated RNA 26
- SAUR9
Small auxin upregulated RNA 9
- SlCyp1
Solanum lycopersicum cyclophilin
- TAA1
Tryptophan aminotransferase of arabidopsis1
- TOC1
Timing of cab expression 1
- UVR8
UVB-Resistance 8
- WL
White light
Authors’ contributions
KCSP, AKS and SK have planned the experiments. SK, SY, DP and SS have performed all the experiments. The data analysis was done by SK, SY, DP and SS. The manuscript writing has been done by SK and picture editing was done by SK and DP. SY and DP have equally contributed in this work. The authors have reviewed the manuscript, contributed to its improvement and approved it for publication.
Funding
This work was funded by the Department of biotechnology (DBT), Ministry of Science and Technology, India to KCSP (BT/PBA/MF2014). AKS acknowledges NIPGR core grant. SK, DP, SY and SS were supported by Department of Atomic Energy (DAE), INSPIRE, UGC and CSIR respectively for fellowship. The funders were not involved in the planning of this research work, data analysis or manuscript writing.
Availability of data and materials
The datasets which support the conclusion of this article have been included in the article and additional files have been provided separately.
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Footnotes
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Sandeep Yadav and Debadutta Patra contributed equally to this work.
Contributor Information
Sony Kumari, Email: sony.kumari@niser.ac.in.
Sandeep Yadav, Email: sandeep_yadav@nipgr.ac.in.
Debadutta Patra, Email: debadutta.p@niser.ac.in.
Sharmila Singh, Email: sharmil86singh45@gmail.com.
Ananda K. Sarkar, Email: aksarkar@nipgr.ac.in
Kishore C. S. Panigrahi, Phone: +91-674-2494140/4226, Email: panigrahi@niser.ac.in
References
- 1.Kelly JWG, Landhäusser SM, Chow PS. The impact of light quality and quantity on root-to-shoot ratio and root carbon reserves in aspen seedling stock. New For. 2015;46(4):527–545. doi: 10.1007/s11056-015-9473-9. [DOI] [Google Scholar]
- 2.Fankhauser C, Staiger D. Photoreceptors in Arabidopsis thaliana: light perception, signal transduction and entrainment of the endogenous clock. Planta. 2002;216(1):1–16. doi: 10.1007/s00425-002-0831-4. [DOI] [PubMed] [Google Scholar]
- 3.Briggs WR. Photoreceptors in plant Photomorphogenesis to date. Five Phytochromes, two Cryptochromes, one Phototropin, and one Superchrome. Plant Physiol. 2001;125(1):85–88. doi: 10.1104/pp.125.1.85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Salisbury FJ, Hall A, Grierson CS, Halliday KJ. Phytochrome coordinates Arabidopsis shoot and root development. Plant J. 2007;50(3):429–438. doi: 10.1111/j.1365-313X.2007.03059.x. [DOI] [PubMed] [Google Scholar]
- 5.Correll MJ, Kiss JZ. The roles of phytochromes in elongation and gravitropism of roots. Plant Cell Physiol. 2005;46(2):317–323. doi: 10.1093/pcp/pci038. [DOI] [PubMed] [Google Scholar]
- 6.Burbach C, Markus K, Zhang Y, Schlicht M, Baluška F. Photophobic behavior of maize roots. Plant Signal Behav. 2012;7(7):874–878. doi: 10.4161/psb.21012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kiss JZ, Mullen JL, Correll MJ, Hangarter RP. Phytochromes a and B mediate red-light-induced positive phototropism in roots. Plant Physiol. 2003;131(3):1411–1417. doi: 10.1104/pp.013847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Canamero RC, Bakrim N, Bouly JP, Garay A, Dudkin EE, Habricot Y. Cryptochrome photoreceptors cry1 and cry2 antagonistically regulate primary root elongation in Arabidopsis thaliana. Planta. 2006;224(5):995–1003. doi: 10.1007/s00425-006-0280-6. [DOI] [PubMed] [Google Scholar]
- 9.Lee H-J, Ha J-H, Kim S-G, Choi H-K, Kim ZH, Han Y-J. Stem-piped light activates phytochrome B to trigger light responses in Arabidopsis thaliana roots. Sci Signal. 2016;9(452):106. doi: 10.1126/scisignal.aaf6530. [DOI] [PubMed] [Google Scholar]
- 10.Ha JH, Kim JH, Kim SG, Sim HJ, Lee G, Halitschke R. Shoot phytochrome B modulates reactive oxygen species homeostasis in roots via abscisic acid signaling in Arabidopsis. Plant J. 2018;94(5):790–798. doi: 10.1111/tpj.13902. [DOI] [PubMed] [Google Scholar]
- 11.Chen X, Yao Q, Gao X, Jiang C, Harberd NP, Fu X. Shoot-to-root Mobile transcription factor HY5 coordinates plant carbon and nitrogen acquisition. Curr Biol. 2016;26(5):640–646. doi: 10.1016/j.cub.2015.12.066. [DOI] [PubMed] [Google Scholar]
- 12.Singh M, Gupta A, Laxmi A. Glucose control of root growth direction in Arabidopsis thaliana. J Exp Bot. 2014;65(12):2981–2993. doi: 10.1093/jxb/eru146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Raya-González J, López-Bucio JS, Prado-Rodríguez JC, Ruiz-Herrera LF, Guevara-García ÁA, López-Bucio J. The MEDIATOR genes MED12 and MED13 control Arabidopsis root system configuration influencing sugar and auxin responses. Plant Mol Biol. 2017;95(1–2):141–156. doi: 10.1007/s11103-017-0647-z. [DOI] [PubMed] [Google Scholar]
- 14.Raya-González J, Oropeza-Aburto A, López-Bucio JS, Guevara-García AÁ, de Veylder L, López-Bucio J. MEDIATOR 18 influences Arabidopsis root architecture, represses auxin signaling and is a critical factor for cell viability in root meristems. Plant J. 2018;96(5):895–909. doi: 10.1111/tpj.14114. [DOI] [PubMed] [Google Scholar]
- 15.Kobayashi K, Baba S, Obayashi T, Sato M, Toyooka K, Keranen M. Regulation of root greening by light and Auxin/Cytokinin signaling in Arabidopsis. Plant Cell. 2012;24(3):1081–1095. doi: 10.1105/tpc.111.092254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Más P, Alabadí D, Yanovsky MJ, Oyama T, Kay SA. Dual role of TOC1 in the control of circadian and photomorphogenic responses in Arabidopsis. Plant Cell. 2003;15(1):223–236. doi: 10.1105/tpc.006734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Meng L, Song W, Liu S, Dong J, Zhang Y, Wang C. Light quality regulates lateral root development in tobacco seedlings by shifting Auxin distributions. J Plant Growth Regul. 2015;34(3):574–583. doi: 10.1007/s00344-015-9491-z. [DOI] [Google Scholar]
- 18.Pierik R, De Wit M. Shade avoidance: Phytochrome signalling and other aboveground neighbour detection cues. J Exp Bot. 2014;65(11):2815–2824. doi: 10.1093/jxb/ert389. [DOI] [PubMed] [Google Scholar]
- 19.Ragonezi C, Castro MR, Klimaszewska K, Lima M, Zavattieri MA. Influence of light quality and intensity on adventitious root formation in microshoots of Pinus pinea L. Acta Hortic. 2010;865:287–292. doi: 10.17660/ActaHortic.2010.865.38. [DOI] [Google Scholar]
- 20.Nagel KA, Schurr U, Walter A. Dynamics of root growth stimulation in Nicotiana tabacum in increasing light intensity. Plant Cell Environ. 2006;29(10):1936–1945. doi: 10.1111/j.1365-3040.2006.01569.x. [DOI] [PubMed] [Google Scholar]
- 21.Spiegelman Z, Ham BK, Zhang Z, Toal TW, Brady SM, Zheng Y. A tomato phloem-mobile protein regulates the shoot-to-root ratio by mediating the auxin response in distant organs. Plant J. 2015;83(5):853–863. doi: 10.1111/tpj.12932. [DOI] [PubMed] [Google Scholar]
- 22.Razzak A, Ranade SS, Strand Å, García-Gil MR. Differential response of scots pine seedlings to variable intensity and ratio of red and far-red light. Plant Cell Environ. 2017;40(8):1332–1340. doi: 10.1111/pce.12921. [DOI] [PubMed] [Google Scholar]
- 23.Qu Y, Liu S, Bao W, Xue X, Ma Z, Yokawa K. Expression of root genes in Arabidopsis seedlings grown by standard and improved growing methods. Int J Mol Sci. 2017;18(5):951. doi: 10.3390/ijms18050951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bednarek P, Piślewska-Bednarek M, Svatoš A, Schneider B, Doubský J, Mansurova M. A glucosinolate metabolism pathway in living plant cells mediates broad-spectrum antifungal defense. Science. 2009;323(5910):101–106. doi: 10.1126/science.1163732. [DOI] [PubMed] [Google Scholar]
- 25.Curtis RHC. Pankaj, powers SJ, Napier J, Matthes MC. The Arabidopsis F-box/Kelch-repeat protein At2g44130 is Upregulated in Giant cells and promotes nematode susceptibility. Mol Plant-Microbe Interact. 2013;26(1):36–43. doi: 10.1094/MPMI-05-12-0135-FI. [DOI] [PubMed] [Google Scholar]
- 26.Mi H, Huang X, Muruganujan A, Tang H, Mills C, Kang D. PANTHER version 11: expanded annotation data from gene ontology and Reactome pathways, and data analysis tool enhancements. Nucleic Acids Res. 2017;45(D1):D183–D189. doi: 10.1093/nar/gkw1138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Harris MA, Clark J, Ireland A, Lomax J, Ashburner M, Foulger R. The gene ontology (GO) database and informatics resource. Nucleic Acids Res. 2004;32:D258–D261. doi: 10.1093/nar/gkh066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Salomé PA, McClung CR, Salome PA, McClung CR. PSEUDO-RESPONSE REGULATOR 7 and 9 are partially redundant genes essential for the temperature responsiveness of the Arabidopsis circadian clock. Plant Cell. 2005;17(3):791–803. doi: 10.1105/tpc.104.029504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Moriwaki T, Miyazawa Y, Kobayashi A, Uchida M, Watanabe C, Fujii N. Hormonal regulation of lateral root development in Arabidopsis modulated by MIZ1 and requirement of GNOM activity for MIZ1 function. Plant Physiol. 2011;157(3):1209–1220. doi: 10.1104/pp.111.186270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhang S, Huang L, Yan A, Liu Y, Liu B, Yu C. Multiple phytohormones promote root hair elongation by regulating a similar set of genes in the root epidermis in Arabidopsis. J Exp Bot. 2016;67(22):6363–6372. doi: 10.1093/jxb/erw400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Fogaça CM, Fett-Neto AG. Role of auxin and its modulators in the adventitious rooting of Eucalyptus species differing in recalcitrance. Plant Growth Regul. 2005;45(1):1–10. doi: 10.1007/s10725-004-6547-7. [DOI] [Google Scholar]
- 32.Voß U, Wilson MH, Kenobi K, Gould PD, Robertson FC, Peer WA. The circadian clock rephases during lateral root organ initiation in Arabidopsis thaliana. Nat Commun. 2015;6:7641. doi: 10.1038/ncomms8641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gentleman R, Whalen E, Huber W, Flacon S. graph: graph: A package to handle graph data structures. 2018. [Google Scholar]
- 34.Roig-Villanova I, Bou-Torrent J, Galstyan A, Carretero-Paulet L, Portolés S, Rodríguez-Concepción M. Interaction of shade avoidance and auxin responses: a role for two novel atypical bHLH proteins. EMBO J. 2007;26(22):4756–4767. doi: 10.1038/sj.emboj.7601890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kim HJ, Chiang Y-H, Kieber JJ, Schaller GE. SCFKMD controls cytokinin signaling by regulating the degradation of type-B response regulators. Proc Natl Acad Sci. 2013;110(24):10028–10033. doi: 10.1073/pnas.1300403110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.To JPC. Haberer G, Ferreira FJ, Deruère J, Mason MG, Schaller GE. Type-a Arabidopsis response regulators are partially redundant negative regulators of cytokinin signaling. Plant Cell. 2004;16(3):658–671. doi: 10.1105/tpc.018978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tokitaka Oyama YS. And KO. The Arabidopsis HY5gene encodes a bZIP protein that regulates stimulus-induced development of root and hypocotyl. Genes Dev. 1997;11(22):2983–2995. doi: 10.1101/gad.11.22.2983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Liu J, Hua W, Hu Z, Yang H, Zhang L, Li R. Natural variation in ARF18 gene simultaneously affects seed weight and silique length in polyploid rapeseed. Proc Natl Acad Sci. 2015;112(37):E5123–E5132. doi: 10.1073/pnas.1502160112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Rasband W. ImageJ . U S Natl institutes heal Bethesda, Maryland, USA. 2017. [Google Scholar]
- 40.Williams L, Carles CC, Osmont KS, Fletcher JC. A database analysis method identifies an endogenous trans-acting short-interfering RNA that targets the Arabidopsis ARF2, ARF3, and ARF4 genes. Proc Natl Acad Sci. 2005;102(27):9703–9708. doi: 10.1073/pnas.0504029102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Piya S, Shrestha SK, Binder B, Stewart CN, Hewezi T. Protein-protein interaction and gene co-expression maps of ARFs and aux/IAAs in Arabidopsis. Front Plant Sci. 2014;5:744. doi: 10.3389/fpls.2014.00744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Pham VN, Kathare PK, Huq E. Phytochromes and Phytochrome interacting factors. Plant Physiol. 2017;176(2):1025–1038. doi: 10.1104/pp.17.01384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lavy M, Estelle M. Mechanisms of auxin signaling. Development. 2016;143(18):3226–3229. doi: 10.1242/dev.131870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kuno N, Møller SG, Shinomura T, Xu X, Chua NH, Furuya M. The novel MYB protein EARLY-PHYTOCHROME-RESPONSIVE1 is a component of a slave circadian oscillator in Arabidopsis. Plant Cell. 2003;15(10):2476–2488. doi: 10.1105/tpc.014217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Datta S, Hettiarachchi GH, Deng XW, Holm M. Arabidopsis CONSTANS-LIKE3 is a positive regulator of red light signaling and root growth. Plant Cell. 2006;18(1):70–84. doi: 10.1105/tpc.105.038182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Chen Y, Hao X, Cao J. Small auxin upregulated RNA (SAUR) gene family in maize: identification, evolution, and its phylogenetic comparison with Arabidopsis, rice, and sorghum. J Integr Plant Biol. 2014;56(2):133–150. doi: 10.1111/jipb.12127. [DOI] [PubMed] [Google Scholar]
- 47.Moreno-Piovano GS, Moreno JE, Cabello JV, Arce AL, Otegui ME, Chan RL. A role for LAX2 in regulating xylem development and lateral-vein symmetry in the leaf. Ann Bot. 2017;120(4):577–590. doi: 10.1093/aob/mcx091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Cheng XF, Wang ZY. Overexpression of COL9, a CONSTANS-LIKE gene, delays flowering by reducing expression of CO and FT in Arabidopsis thaliana. Plant J. 2005;43(5):758–768. doi: 10.1111/j.1365-313X.2005.02491.x. [DOI] [PubMed] [Google Scholar]
- 49.Ren C, Zhu X, Zhang P, Gong Q. Arabidopsis COP1-interacting protein 1 is a positive regulator of ABA response. Biochem Biophys Res Commun. 2016;477(4):847–853. doi: 10.1016/j.bbrc.2016.06.147. [DOI] [PubMed] [Google Scholar]
- 50.Peng Z, Serino G, Deng XW. Molecular characterization of subunit 6 of the COP9 signalosome and its role in multifaceted developmental processes in Arabidopsis. Plant Cell. 2001;13(11):2393–2407. doi: 10.1105/tpc.010248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Alabadí D, Oyama T, Yanovsky MJ, Harmon FG, Más P, Kay SA. Reciprocal regulation between TOC1 and LHY / CCA1 within the Arabidopsis circadian clock. Science. 2001;293(5531):880–883. doi: 10.1126/science.1061320. [DOI] [PubMed] [Google Scholar]
- 52.Zdarska M, Dobisová T, Gelová Z, Pernisová M, Dabravolski S, Hejátko J. Illuminating light, cytokinin, and ethylene signalling crosstalk in plant development. J Exp Bot. 2015;66(16):4913–4931. doi: 10.1093/jxb/erv261. [DOI] [PubMed] [Google Scholar]
- 53.Casal JJ. Shade Avoidance. Arabidopsis Book. 2012;10:e0157. doi: 10.1199/tab.0157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Alabadí D, Gallego-Bartolomé J, Orlando L, García-Cárcel L, Rubio V, Martínez C. Gibberellins modulate light signaling pathways to prevent Arabidopsis seedling de-etiolation in darkness. Plant J. 2008;53(2):324–335. doi: 10.1111/j.1365-313X.2007.03346.x. [DOI] [PubMed] [Google Scholar]
- 55.Khanna R, Shen Y, Marion CM, Tsuchisaka A, Theologis A, Schafer E. The basic helix-loop-helix transcription factor PIF5 acts on ethylene biosynthesis and Phytochrome signaling by distinct mechanisms. Plant Cell. 2007;19(12):3915–3929. doi: 10.1105/tpc.107.051508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Thain SC, Vandenbussche F, Laarhoven LJJ, Dowson-Day MJ, Wang Z-Y, Tobin EM. Circadian rhythms of ethylene emission in Arabidopsis. Plant Physiol. 2004;136(3):3751–3761. doi: 10.1104/pp.104.042523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Covington MF, Harmer SL. The circadian clock regulates auxin signaling and responses in Arabidopsis. PLoS Biol. 2007;5(8):1773–1784. doi: 10.1371/journal.pbio.0050222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Hanano S, Domagalska MA, Nagy F, Davis SJ. Multiple phytohormones influence distinct parameters of the plant circadian clock. Genes Cells. 2006;11(12):1381–1392. doi: 10.1111/j.1365-2443.2006.01026.x. [DOI] [PubMed] [Google Scholar]
- 59.Blázquez MA, Trénor M, Weigel D. Independent control of gibberellin biosynthesis and flowering time by the circadian clock in Arabidopsis. Plant Physiol. 2002;130(4):1770–1775. doi: 10.1104/pp.007625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Nozue K, Covington MF, Duek PD, Lorrain S, Fankhauser C, Harmer SL. Rhythmic growth explained by coincidence between internal and external cues. Nature. 2007;448(7151):358–361. doi: 10.1038/nature05946. [DOI] [PubMed] [Google Scholar]
- 61.Correll Melanie J., Coveney Katrina M., Raines Steven V., Mullen Jack L., Hangarter Roger P., Kiss John Z. Phytochromes play a role in phototropism and gravitropism in Arabidopsis roots. Advances in Space Research. 2003;31(10):2203–2210. doi: 10.1016/S0273-1177(03)00245-X. [DOI] [PubMed] [Google Scholar]
- 62.Costigan SE, Warnasooriya SN, Humphries BA, Montgomery BL. Root-localized Phytochrome Chromophore synthesis is required for Photoregulation of root elongation and impacts root sensitivity to Jasmonic acid. Plant Physiol. 2011;157(3):1138–1150. doi: 10.1104/pp.111.184689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.De Simone S, Oka Y, Inoue Y. Effect of light on root hair formation in Arabidopsis thaliana Phytochrome-deficient mutants. J Plant Res. 2000;113(1):63–69. doi: 10.1007/PL00013917. [DOI] [Google Scholar]
- 64.Alabadí D, Blázquez MA. Molecular interactions between light and hormone signaling to control plant growth. Plant Mol Biol. 2009;69(4):409–417. doi: 10.1007/s11103-008-9400-y. [DOI] [PubMed] [Google Scholar]
- 65.Liu J, Rowe J, Lindsey K. Hormonal crosstalk for root development: a combined experimental and modeling perspective. Front Plant Sci. 2014;5:1–8. doi: 10.3389/fpls.2014.00116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Farré EM. The regulation of plant growth by the circadian clock. Plant Biol. 2012;14(3):401–410. doi: 10.1111/j.1438-8677.2011.00548.x. [DOI] [PubMed] [Google Scholar]
- 67.Medzihradszky M, Bindics J, Adam E, Viczian A, Klement E, Lorrain S. Phosphorylation of Phytochrome B inhibits light-induced signaling via accelerated dark reversion in Arabidopsis. Plant Cell. 2013;25(2):535–544. doi: 10.1105/tpc.112.106898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Rausenberger J, Tscheuschler A, Nordmeier W, Wüst F, Timmer J, Schäfer E. Photoconversion and nuclear trafficking cycles determine phytochrome A’s response profile to far-red light. Cell. 2011;146(5):813–825. doi: 10.1016/j.cell.2011.07.023. [DOI] [PubMed] [Google Scholar]
- 69.R Development Core Team R . R: A Language and Environment for Statistical Computing . Vol. 1, R Foundation for Statistical Computing. 2011. [Google Scholar]
- 70.Warnes GR, Bolker B, Bonebakker L, Gentleman R, Liaw WHA, Lumley T. gdata: Various R Programming Tools for Data Manipulation. 2017. [Google Scholar]
- 71.Swinton J. Vennerable: Venn and Euler area-proportional diagrams. 2016. [Google Scholar]
- 72.Warnes GR, Bolker B, Bonebakker L, Gentleman R, Liaw WHA, Lumley T. Package “gplots”: Various R programming tools for plotting data. R Packag version 2170. 2016. pp. 1–68. [Google Scholar]
- 73.Carey V, Long L, Gentleman R. RBGL: An interface to the BOOST graph library. 2018. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1. Detailed zoomed pictures of lateral root growth in 6-day old WT, phyB-9 and phyA-211 seedlings grown under WL intensities of 38, 75, 112 and 150 μmol m − 2 s − 1. Scale bar = 5 mm. Arrow head represents lateral roots. (PNG 1250 kb)
Figure S2. Microscope images of lateral root growth for qualitative visualization in 6-day old WT, phyB-9 and phyA-211 seedlings grown under WL intensities of 38, 75, 112 and 150 μmol m − 2 s − 1. Scale bar = 5 mm. (PNG 4917 kb)
List of DEGs Common in two, three, four or five comparative light conditions. (xlsx 1970 kb)
List of Unique DEGs in specific comparative light conditions. (XLSX 40kb)
Figure S3. Composite figure of GO analysis of DEGs under (a) 150 vs 112 μmol m − 2 s − 1 (b) DEGs under 150 vs 75 μmol m − 2 s − 1 (c) DEGs under 150 vs 38 μmol m − 2 s − 1 (d) DEGs under 112 vs 75 μmol m − 2 s − 1 (e) DEGs under 112 vs 38 μmol m − 2 s − 1 (f) DEGs under 75 vs 38 μmol m − 2 s − 1 light intensity. (PNG 40 kb)
List of primers used for qRT-PCR. (DOCX 14 kb)
Data Availability Statement
The datasets which support the conclusion of this article have been included in the article and additional files have been provided separately.