Abstract
Mucopolysaccharidosis Type II (MRS II), also known as Hunter syndrome, is a rare X-linked genetic disease caused by mutations in the IDS gene encoding iduronate 2-sulfatase (I2S). This is a multisystem disorder with significant variation in symptoms. Here, we document a human induced pluripotent stem cell (iPSC) line generated from dermal fibroblasts of a patient with Hunter syndrome containing a hemizygous mutation of a 1 bp insertion at nucleotide 208 in exon 2 of the IDS gene. The generation of this line will allow development of cell-based models for drug development, as well as the study of disease pathophysiology.
Resource table.
| Unique stem cell line identifier | TRNDi008-A |
| Alternative name(s) of stem cell line | HT525A |
| Institution | National Institutes of Health National Center for Advancing Translational Sciences Bethesda, Maryland, USA |
| Contact information of distributor | Dr. Wei Zheng Wei.Zheng@nih.gov |
| Type of cell line | iPSC |
| Origin | Human |
| Additional origin info | Age: 3-year-old Sex: Male Ethnicity: Caucasian |
| Cell Source | Skin fibroblasts |
| Clonality | Clonal |
| Method of reprogramming | Integration-free Sendai viral vectors |
| Genetic Modification | NO |
| Type of Modification | N/A |
| Associated disease | Mucopolysaccharidosis Type II |
| Gene/locus | Gene: IDS Locus: Xq28 Mutation: c.208insC (p. H70PfsX29) |
| Method of modification | N/A |
| Name of transgene or resistance | N/A |
| Inducible/constitutive system | N/A |
| Date archived/stock date | 2018 |
| Cell line repository/bank | N/A |
| Ethical approval | NIGMS Informed Consent Form was obtained from patient at time of sample submission. Confidentiality Certificate: CC-GM-15-004 |
Resource utility
The human induced pluripotent stem cells (hiPSC) described here is a useful tool that can be used to investigate disease phenotype and pathophysiology. As a potential cell-based disease model, these cells can be employed for drug development for the treatment of patients with MPS II.
Resource details
MPS II is a rare X-linked genetic disease caused by mutations in the IDS gene encoding iduronate 2-sulfatase (I2S). The I2S enzyme is involved in the lysosomal degradation of two kinds of glycosaminoglycans (GAGs): heparan sulfate and dermatan sulfate (Hopwood et al., 1993). Malfunction of I2S leads to progressive accumulation of GAGs in tissues and organs, causing a variety of clinical symptoms in the patients. The typical manifestations of this disease include respiratory obstruction, cardiomyopathies, joint stiffness and hepatosplenomegaly. Some patients also have central nervous system (CNS) involvement, such as progressive neurological decline and cognitive impairment (Wraith et al., 2008; Whiteman and Kimura, 2017).
In this study, a human iPSC line was established from the fibroblasts of a 3-year-old male patient (GM13203, Coriell Institute) carrying a hemizygous mutation of a 1 bp insertion at nucleotide 208 in exon 2 (208insC) of the IDS gene, resulting in a frameshift with a premature stop codon (H70PfsX29). As described previously (Beers et al., 2015), the OCT3/4, KLF4, SOX2 and C-MYC pluripotency transcription factors were employed to transduce the patient fibroblasts into an iPSC line, named TRNDi008-A. The iPS cells exhibited classical embryonic stem cell morphology (Fig. 1A), characterized by immunofluorescence staining and flow cytometry analysis and expressed major pluripotent protein markers of NANOG, SOX2, OCT4, SSEA4 and TRA-1-60 (Fig. 1A, B). Furthermore, G-banded karyotyping at passage 11 (Fig. 1C) confirmed a normal karyotype (46, XY). The mutation (208insC) in the IDS gene was also verified by Sanger sequencing of the PCR product harboring the single nucleotide variation (SNV) (Fig. 1D). Sendai virus vector (SeV) clearance was determined with reverse transcription polymerase chain reaction (RT-PCR) using SeV-specific primers and the vector disappeared by passage 15 (Fig. 1E). Mycoplasma status was confirmed to be negative (Supplementary Fig. S1) and the cell line was authenticated using STR DNA profiling analysis, which demonstrated matching genotypes at all 18 loci examined (information available with the authors). Finally, pluripotency of TRNDi008-A was confirmed by a teratoma formation experiment, which exhibited the ability to differentiate into all three germ layers (ectoderm, neural tube; mesoderm, smooth muscle; endoderm, gut) in vivo (Fig. 1F)(Table 1).
Fig. 1.
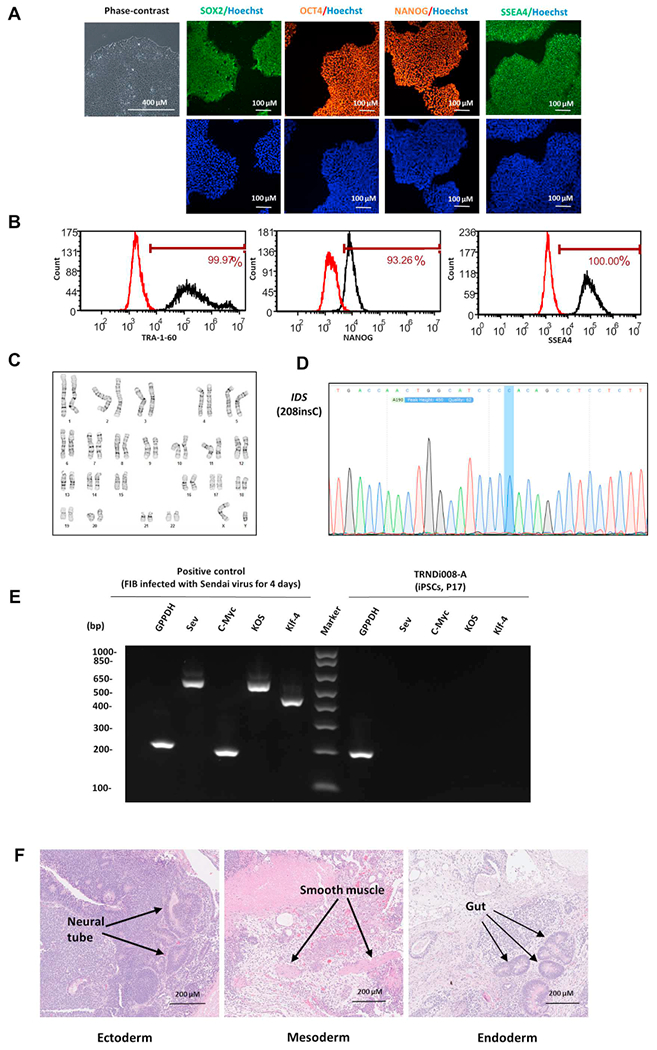
Characterization of TRNDi008-A iPSC line A) From left to right: phase contrast imaging of TRNDi008-A colonies; immunostained TRNDi008-A iPSCs expressing SOX2, OCT4, NANOG and SSEA4. Hoeschst (blue) was used to label the nucleus. B) Pluripotency protein markers (TRA-1-60, NANOG and SSEA4) were assessed by flow cytometry. C) G-banding karyotype analysis confirmed normal karyotype (46, XY). D) Sanger sequencing was used to confirm the mutation in exon 2 of the IDS gene (208insC). E) RT-PCR verification of Sendai virus clearance in TRNDi008-A iPSC line. SeV transduced fibroblasts were used as positive control. F) Histological characterization of teratoma formation, showing normal ectoderm, endoderm and mesoderm differentiation.
Table 1.
Characterization and validation.
| Classification | Test | Result | Data |
|---|---|---|---|
| Morphology | Photography | Normal | Fig. 1 Panel A |
| Phenotype | Immunocytochemistry | SOX2, OCT4, NANOG, SSEA-4 | Fig. 1 Panel A |
| Flow cytometry | TRA-1-60 (99.97%); NANOG (93.26%); SSEA-4 (100%) | Fig. 1 Panel B | |
| Genotype | Karyotype (G-banding) and resolution | 46XY | Fig. 1 Panel C |
| Resolution: 350–400 | |||
| Identity | Microsatellite PCR (mPCR) OR | Not performed | N/A |
| STR analysis | 18 sites tested, all sites matched | Available with the authors | |
| Mutation analysis (IF APPLICABLE) | Sequencing | Hemizygous mutation of IDS | Fig. 1 Panel D |
| Southern Blot OR WGS | N/A | N/A | |
| Microbiology and virology | Mycoplasma | Mycoplasma testing by luminescence. Negative Teratoma with three germlayers formation. Ectoderm (neural tube); Mesoderm (smooth muscle); Endoderm (gut) | Supplementary Fig. S1 |
| Differentiation potential | Teratoma formation | Fig. 1 Panel F | |
| Donor screening (OPTIONAL) | HIV 1 + 2 Hepatitis B, Hepatitis C | N/A | N/A |
| Genotype additional info (OPTIONAL) | Blood group genotyping | N/A | N/A |
| HLA tissue typing | N/A | N/A |
Materials and methods
Cell culture
Patient-derived fibroblasts (GM13203, Coriell Institute) were cultured in DMEM supplemented with 10% fetal bovine serum, 100 units/ml penicillin and 100 μg/ml streptomycin in a humidified incubator with 5% CO2 at 37 °C. Human iPS cells were cultured in StemFlex medium (ThermoFisher) on matrigel (Corning, 354,277)-coated plates at 37 °C in humidified air with 5% CO2 and 5% O2. Cells were passaged with 0.5 mM Ethylenediaminetetraacetic acid (EDTA) upon approaching 80% confluency.
Reprogramming of human skin fibroblasts
As described previously (Li et al., 2019), patient fibroblasts were reprogrammed into iPS cells using non-integrating Sendai virus vector technology (A16517, ThermoFisher).
Genome analysis
Genomic analysis of IDS variants was performed by Applied StemCell (Milpitas, California). Briefly, QuickExtract™ DNA Extraction Solution (Lucigen) was used to extract genomic DNA from TRNDi008-A cells. PCR amplifications (MyTaq™ Red Mix, Bioline) were carried out on a T00 Thermal Cycler (Bio-Rad) using the following program: 95 °C, 2 mins; 35 cycles of [95 °C, 15 s; 60 °C, 15 s; 72 °C], elongation duration varies by amplicon size, 72 °C 5 mins; 4 °C, indefinite. Sanger sequencing analysis was performed for genotyping of the hemizygous mutation of a 1 bp insertion at nucleotide 208 in exon 2 of the IDS gene. The specific primers for gene amplification and sequencing are listed in Table 2.
Table 2.
Reagents details
| Antibodies used for immunocytochemistry/flow-cytometry | |||
|---|---|---|---|
| Antibody | Dilution | Company Cat # and RRID | |
| Pluripotency Markers | Mouse anti-SOX2 | 1:50 | R & D systems, Cat# MAB2018, RRID: AB_358009 |
| Pluripotency Markers | Rabbit anti-NANOG | 1:400 | Cell Signaling Technology, Cat# 4903, RRID: AB_10559205 |
| Pluripotency Markers | Rabbit anti-OCT4 | 1:400 | Thermo Fisher, Cat# A13998, RRID: AB_2534182 |
| Pluripotency Markers | Mouse anti-SSEA4 | 1:1000 | Cell Signaling Technology, Cat# 4755, RRID: AB_1264259 |
| Secondary Antibodies | Donkey anti-Mouse IgG (Alexa Fluor 488) | 1:400 | Thermo Fischer, Cat# A21202, RRID: AB_141607 |
| Secondary Antibodies | Donkey anti-Rabbit IgG (Alexa Fluor 594) | 1:400 | Thermo Fischer, Cat# A21207, RRID: AB_141637 |
| Flow Cytometry Antibodies | Anti-Tra-1-60-DyLight 488 | 1:50 | Thermo Fischer, Cat# MA1–023-D488X, RRID: AB_2536700 |
| Flow Cytometry Antibodies | Anti-Nanog-Alexa Fluor 488 | 1:50 | Millipore, Cat# FCABS352A4, RRID: AB_10807973 |
| Flow Cytometry Antibodies | anti-SSEA-4-Alexa Fluor 488 | 1:50 | Thermo Fischer, Cat# 53-8843-41, RRID: AB_10597752 |
| Flow Cytometry Antibodies | Mouse-IgM-DyLight 488 | 1:50 | Thermo Fischer, Cat# MA1-194-D488, RRID: AB_2536969 |
| Flow Cytometry Antibodies | Rabbit IgG-Alexa Fluor 488 | 1:50 | Cell Signaling Technology, Cat# 4340S, RRID: AB_10694568 |
| Flow Cytometry Antibodies | Mouse IgG3-FITC | 1:50 | Thermo Fischer, Cat# 11-4742-42, RRID: AB_2043894 |
| Primers | ||
|---|---|---|
| Target | Forward/Reverse primer (5′-3′) | |
| Sev specific primers (RT-PCR) | Sev/181 bp | GGA TCA CTA GGT GAT ATC GAG C/ACC AGA CAA GAG TTT AAG AGA TAT GTA TC |
| Sev specific primers (RT-PCR) | KOS/528 bp | ATG CAC CGC TAC GAC GTG AGC GC/ACC TTG ACA ATC CTG ATG TGG |
| Sev specific primers (RT-PCR) | Klf4/410 bp | TTC CTG CAT GCC AGA GGA GCC C/AAT GTA TCG AAG GTG CTC AA |
| Sev specific primers (RT-PCR) | C-Myc/523 bp | TAA CTG ACT AGC AGG CTT GTC G/TCC ACA TAC AGT CCT GGA TGA TGA TG |
| House-Keeping gene (RT-PCR) | GAPDH/197 bp | GGA GCG AGA TCC CTC CAA AAT/GGC TGT TGT CAT ACT TCT CAT GG |
| Targeted mutation analysis (PCR) | IDS (208insC)/357 bp | CGC ACA ATC TGT GCC ATC TG/TGG CTA GGC TGT TAA GGT GC |
Immunocytochemistry
Patient-derived iPSCs were fixed for 15 mins in 4% paraformaldehyde, washed with Dulbecco’s Phosphate-Buffered Saline (DPBS), permeabilized with 0.5% Triton X-100 in DPBS (10 mins), followed by incubation with Image-iT™ FX signal enhancer (ThermoFisher) for 40 mins at ambient temperature in a humidified environment. Primary antibodies (SOX2, OCT4, NANOG and SSEA4) were diluted in the Image-iT™ FX signal enhancer blocking buffer and then incubated with the cells overnight at 4 °C. Following a DPBS wash, corresponding secondary antibodies conjugated with Alexa Fluor 488 or Alex Fluor 594 were added and incubated for 1 h at ambient temperature (Table 2). Cells were stained with Hoechst 33342 (15 mins), washed, and imaged using an INCell Analyzer 2200 (GE Healthcare) and a 20× objective lens with Texas Red, FITC and DAPI filter sets.
Flow cytometry analysis
Human iPSCs were dissociated using TrypLE solution (Thermo Fisher) then fixed and permeabilized for intracellular staining as described previously [5]. The fluorophore conjugated antibodies used in this protocol are listed in Table 2. Samples were analyzed by an AccuriC6 Flow Cytometry system (BD Biosciences).
G-banded karyotyping
Karyotype analysis was performed by WiCell Research Institute (Madison, WI). Twenty randomly selected metaphases were selected and analyzed using G-banding method.
Short tandem repeat (STR) analysis
STR analysis of the patient-derived fibroblasts and patient iPS cells was conducted by the Johns Hopkins University Genetic Resources Facility using a PowerPlex 18D Kit (Promega). The PCR products were electrophoresed on an ABI Prism® 3730x1 Genetic Analyzer. GeneMapper® v 4.0 software (Applied Biosystems) was used to analyze data.
Mycoplasma detection
Mycoplasma status was assessed using the MycoAlert kit (Lonza) per manufacturer instructions. A ratiometric reading of < 0.9 indicates a mycoplasma negative sample.
Testing for Sendai reprogramming vector clearance
Human fibroblasts (GM05659, Coriell) were transfected with Sendai virus for 4 days and used as the positive control. Total RNA was extracted using RNeasy Plus Mini Kit (Qiagen) and 1 μg of RNA was reverse transcribed into cDNA with Superscript™ III First-Strand Synthesis SuperMix kit. PCR was performed using Platinum II Hot-Start PCR Master Mix (ThermoFisher) and the amplifications were carried out as previously described (Li et al., 2019) using the primers listed in Table 2. Products were then loaded to the E-Gel® 1.2% with SYBR Safe™ gel and imaged using a G: Box Chemi-XX6 gel doc system (Syngene).
Teratoma formation assay
TRNDi008-A cells were dissociated from 6-well plates using 0.5 mM EDTA in DPBS. A suspension of 1 × 107 cells in 400 μl medium supplied with 25 mM HEPES (pH 7.4) was kept on ice, and then added with a 50% volume (200 μl) of cold Matrigel (Corning, 354277). The mixture was injected subcutaneously into NSG mice (JAX No. 005557) at 150 μl per injection site. After 6–8 weeks, visible tumors were removed and fixed in 10% Neutral Buffer Formalin. The fixed tumors were embedded in paraffin and stained with hematoxylin and eosin.
Supplementary Material
Acknowledgement
We would like to thank Dr. Zu-xi Yu of the pathology Core of National Heart, Lung and Blood Institute, National Institutes of Health for sectioning and staining the teratoma. We also would like to thank the Research Services Section at National Center for Advancing Translational Sciences for coordinating the STR DNA analysis and mycoplasma testing service. This work was supported by the Intramural Research Program of National Center for Advancing Translational Sciences, National Institutes of Health.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.scr.2019.101451.
References
- Beers J, Linask KL, Chen JA, Siniscalchi LI, Lin Y, Zheng W, Rao M, Chen G, 2015. A cost-effective and efficient reprogramming platform for large-scale production of integration-free human induced pluripotent stem cells in chemically defined culture. Sci. Rep 5, 11319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hopwood JJ, Bunge S, Morris CP, Wilson PJ, Steglich C, Beck M, Schwinger E, Gal A, 1993. Molecular basis of mucopolysaccharidosis type II: mutations in the iduronate-2-sulphatase gene. Hum. Mutat 2, 435–442. [DOI] [PubMed] [Google Scholar]
- Li R, Pradhan M, Xu M, Baskfield A, Farkhondeh A, Cheng YS, Beers J, Zou J, Liu C, Might M, Rodems S, Zheng W, 2019. Generation of an induced pluripotent stem cell line (TRNDi002-B) from a patient carrying compound heterozygous p.Q208X and p.G310G mutations in the NGLY1 gene. Stem Cell Res 34, 101362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whiteman DA, Kimura A, 2017. Development of idursulfase therapy for mucopolysaccharidosis type II (Hunter syndrome): the past, the present and the future. Drug Design Dev. Ther 11, 2467–2480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wraith JE, Scarpa M, Beck M, Bodamer OA, De Meirleir L, Guffon N, Meldgaard Lund A, Malm G, Van der Ploeg AT, Zeman J, 2008. Mucopolysaccharidosis type II (Hunter syndrome): a clinical review and recommendations for treatment in the era of enzyme replacement therapy. Eur. J. Pediatr 167, 267–277. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


