Abstract
The influenza A virus (IAV) genome is incorporated into newly produced virions through a tightly orchestrated process that is one of the best studied examples of genome packaging by a segmented virus. Despite the remarkable selectivity and efficiency of this process, it is clear that the vast majority of IAV virions are unable to express the full set of essential viral gene products and are thus incapable of productive replication in the absence of complementation. Here, we attempt to reconcile the widespread production of these semi-infectious particles (SIPs) with the high efficiency and selectivity of IAV genome packaging. We also cover what is known and what remains unknown about the consequences of SIP production for the replication and evolution of viral populations.
Introduction
Influenza A virus (IAV) is a respiratory pathogen that remains a serious threat to public health. Seasonal epidemics of IAV continue to cause significant morbidity and mortality worldwide, causing hundreds of thousands of deaths annually, while the threat of pandemic strains entering the population and causing millions of excess deaths continually looms [1]. IAV populations exhibit high levels of diversity both in terms of nucleotide sequence (genetic diversity) and in the specific viral gene products expressed by individual particles (genomic diversity) [2]. While the consequences of genetic diversity have been explored to some extent in a number of other studies, the role that genomic diversity plays in IAV biology is just beginning to be appreciated [2,3].
Like many viruses, IAV has long been known to have a high particle to PFU ratio [4]. Only a small fraction of IAV particles (~1–30% depending on the strain examined) are capable of initiating a productive replication cycle, with the remaining bulk of the population consisting of defective interfering particles (DIPs) and semi-infectious particles (SIPs) [2,5–8,9••]. While DIPs are defined by a specific molecular signature (large internal deletions in one or more gene segments), SIPs lack such a signature and are instead defined by a functional phenotype (failure to express one or more gene products) [2]. Here, we attempt to reconcile the existence of SIPs with the highly selective packaging mechanism of IAV, and highlight outstanding questions regarding the role of SIPs in IAV biology.
IAV genome packaging
The organization and packaging scheme of the IAV genome is central to the biology of SIPs, and to understanding broader patterns of viral genomic diversity. The IAV genome is divided into eight negative sense, single-stranded RNA segments that each encode one or more gene products, and expression from all eight gene segments is required to initiate a productive infection [10]. Each individual segment is generally maintained as a viral nucleoprotein complex (vRNP), in which the viral genomic RNA is bound by multiple copies of NP and associates with a single copy of the viral RNA-dependent RNA polymerase complex [10].
Genome segmentation imposes the challenge of packaging the full set of IAV gene segments required for the next round of replication. While many specific details of the influenza packaging mechanism remain a mystery, a multitude of studies using a variety of approaches have clearly demonstrated the existence of a highly selective and efficient genome packaging mechanism [11••]. Structural analyses using electron microscopy revealed that vRNPs are arranged in a highly ordered structure within the virion. Cross sections of budding particles demonstrated that the 8 vRNPs are organized into a ‘7+1’ array, with 7 vRNPs surrounding 1 central vRNP [12•,13,14]. The formation of this array likely depends upon specific physical interactions between the viral genomic RNAs that occur prior to and during the packaging process, likely sometime between nuclear export and viral budding [15–18]. Interestingly, the formation of the 7+1 array appears to be crucial for virion assembly, as recombinant viruses lacking a single gene segment recruit host ribosomal RNAs to complete the array [19•]. Further, influenza C and D viruses, which only have 7 genome segments in total, also form a 7+1 array [20]. These EM studies were complemented by the work of Chou et al., which used single molecule fluorescent in situ hybridization (smFISH) to demonstrate that most IAV particles contain a single copy of each of the 8 distinct vRNAs [21••].
The IAV packaging process depends upon segment-specific packaging signals located at both the 5′ and 3′ ends of each vRNA [11••,22]. Specifically, the promoter sequences and non-coding regions together serve as incorporation signals that ensure the incorporation of that segment, while the terminal coding regions serve as bundling signals that ensure the incorporation of the full set of 8 distinct segments [23]. Additional sequence elements outside of the canonical packaging signals also appear to be important for achieving maximum packaging efficiency [17,24]. Mutations in packaging signals can decrease the incorporation frequency of the mutated segment, as well as other segments due to the essential role of intersegment interactions during packaging [11••].
Mechanisms of SIP production
Given the vast multitude of data demonstrating the efficiency and specificity of the IAV packaging process, why do so few virions actually encode the full set of essential viral genes? Below, we review the most likely mechanisms behind the generation of SIPs and their predominance in IAV populations. Please note that these mechanisms are not mutually exclusive.
Imperfect genome packaging
Despite being incredibly selective and efficient, the IAV genome packaging process is not perfect. EM-based examination of a collection of IAV and IBV strains revealed that up to 20% of virions contained fewer than eight RNA segments, suggesting a greater degree of flexibility in packaging fidelity than was previously thought [25•]. This number is likely an underestimation, given the observation from the same group that non-viral RNAs can be packaged into the 7 +1 array within virions in some cases [19•]. In addition, we found that mutations within NP that emerge during host adaptation can selectively modulate the packaging efficiency of individual segments, likely through changes in the overall intracellular abundance of the genomic RNAs [26•].
This flexibility in IAV packaging fidelity does not fully account for the ten-or-more-fold higher proportion of non-infectious to fully infectious particles, however. It is likely that many SIPs arise from the failure to complete the early steps in the viral life cycle that are necessary for successful gene segment expression.
Failure of gene segment transport
IAV vRNPs must successfully enter the nucleus to initiate gene expression. This entails transit across the cytosol from the point of fusion to the nuclear pore, as well as successful translocation through the nuclear pore complex. The success rates of these individual steps, and the specific factors that may influence them, remain poorly defined. Numerous studies have indicated that cytosolic trafficking of IAV vRNPs and the nuclear import of model cargo molecules are far from perfect [27–29]. Further, interferon inducible antiviral effectors such as Mx1 and PLSCR1 may actively interfere with vRNP import [30,31]. In the only study to date to specifically examine the co-trafficking of individual IAV vRNPs, Chou et al. used smFISH to quantify the degree of co-localization between vRNPs during early infection [32•]. By comparing segment co-localization within the cytosol following inhibition of membrane fusion or nuclear export, they concluded that the vRNPs enter the nucleus as a complex and that dissociation of individual vRNPs prior to nuclear entry was minimal. This suggests that degradation or loss of vRNPs prior to nuclear entry is a minor contributor to the SIP phenotype. By contrast, Heldt et al. used an experimentally parameterized computational model to predict that 38% of infections were non-productive as a result of vRNA degradation [[32•]]. More work is clearly needed to reconcile these results and to determine whether the success rates of these steps vary under different conditions, in different cell types, and for different viral genotypes.
Failure of gene segment expression
Incomplete viral gene expression patterns may also arise through the failure to initiate vRNA transcription and replication in the nucleus or produce a functional gene product. Russell et al. used single cell mRNA sequencing to estimate that ~50–65% of IAV infected cells fail to express one or more viral mRNAs as under low MOI conditions [33]. The high mutation rate of influenza viruses (predicted to be as high as 1.8 × 10−4, or ~2–3 mutations per replicated genome), is likely to contribute to viral gene expression failure [34]. For instance, an incoming vRNA may carry a mutation in a critical cis-acting regulatory sequence that prevents replication and transcription of the gene segment or mRNA. Additionally, non-synonymous mutations in coding sequences may result in the production of truncated or misfolded proteins that will not be detected by antibody staining.
Measurements of IAV mutant frequencies are generally much lower than the above rate estimate, around 1.5 × 10−5 to 2 × 10−6 mutations per nucleotide per infectious cycle, not high enough to explain observed SIP frequencies [35–39]. Further, if random mutations were the main mechanism underlying the SIP phenotype, the gene expression frequencies would be roughly inversely proportional to segment length. Instead, what is observed is that gene expression frequencies are nearly equivalent across segments of different length, or vary in ways that do not track with segment length [9••,26•].
Altogether, the small degree of flexibility in IAV packaging fidelity likely accounts for a portion of the SIP population, with the remaining population most likely generated through a combination of failures in gene segment trafficking or expression. These post-entry failure mechanisms may be probabilistic rather than deterministic in many cases. Thus, a perfectly intact virion containing eight gene segments of wild type sequence can end up being a SIP due to stochastic events following entry. Many open questions remain, including why different IAV strains vary so widely in SIP production, and whether the observed frequencies of fully infectious particles are really the best the virus can do [26•].
Potential biological roles of SIPs
Are SIPs merely defective particles generated as an unavoidable byproduct of infection, or do they play a more complex role during IAV infection? SIPs are produced during infection of mice, guinea pigs, and possibly humans [9••,25•,26•]. Several studies have shown that both laboratory and clinical IAV strains vary significantly in the numbers and specific gene expression patterns of SIPs produced during infection [9••,25•,26•]. This raises the possibility that differences in SIP production between strains may influence other viral phenotypes. Below, we cover what is currently known about how SIPs can modulate the behavior of viral populations.
Co-infection and reassortment
Because SIPs require complementation to replicate, viral populations with more SIPs will be more dependent upon co-infection to propagate. Since co-infection results in reassortment, we hypothesized that SIP production might influence reassortment rates by imposing a requirement for co-infection on the majority of the population [9••]. Fon-ville et al. directly tested this hypothesis by using UV irradiation to artificially increase the SIP content within a viral population [40•]. They found that as the proportion of SIPs within a population increased, the proportion of productively infected cells that were co-infected also increased, as did the overall reassortment rate [40•]. These studies suggest that IAV strains with distinct SIP production phenotypes may differ in their reassortment potential. If this is the case, SIP production could be seen as acting as a type of recombination modifier for IAV [41,42].
Gene dosage effects
Variation in functional gene content between individual virions can result in the delivery of differential copy numbers of individual viral genes to multiple-infected cells, altering viral gene dose ratios. Modulating gene dosage is a well-established mechanism for influencing gene expression across diverse biological systems [43]. The importance of gene dosing in promoting optimal viral replication and transmission has been demonstrated in multiple virus systems. For instance, in multipartite plant viruses, there appears to be optimal ratio of the individual gene segments that is associated with maximal viral replication [44]. For vaccinia virus, selection under conditions in which the viral K3L protein was singularly responsible for antagonizing the antiviral effects of PKR resulted in an expansion in copy number of the K3L gene, leading to an increase in relative K3L expression [45].
We demonstrated that modulation of relative NA gene segment abundance at the population level (mirroring changes in gene segment packaging rates observed for a mutant that increased SIP production) was sufficient to significantly affect the relative expression level of NA in infected cells [26•]. More work is needed to determine whether population-level changes in functional gene segment abundance play a significant role in the regulation of viral gene expression.
Defining the individual
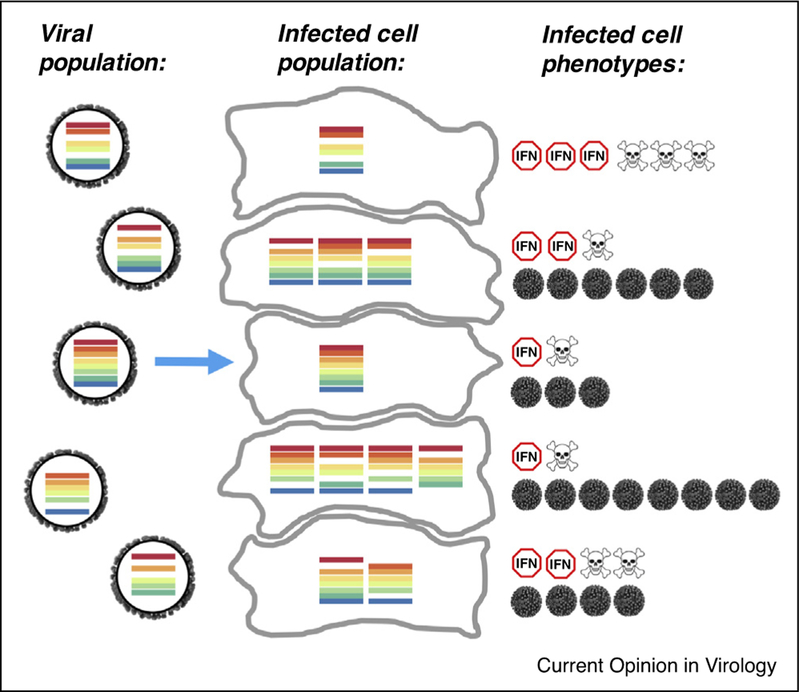
While SIPs cannot propagate on their own, they can contribute to replication and the genetic makeup of the next generation when complemented through co-infection. This means that the traditional focus on fully infectious virions may be misleading. We propose that infected cells rather than virions represent the relevant individuals during IAV infection. Heterogeneity in the functional gene content delivered by individual virions, combined with variation in co-infection frequencies over time and space, mean that the viral genomic content of individual infected cells can vary significantly. This variation in genomic content between infected cells is likely to result in substantial heterogeneity in virus production and the infection response between individual infected cells (Figure 1). As a result, the phenotypes of the infected cell population, at the single cell level as well as collectively, will be determined in part by the relative production and gene expression patterns of SIPs.
Figure 1:
Heterogeneity in viral genomic content of individual infected cells. Variation in both the genomic content of infecting virions and the effective multiplicity of infection result in heterogeneity in the copy numbers of individual viral gene segments (depicted as colored bars) within individual infected cells. Differences in these infected cell genotypes may result in distinct single cell phenotypes in virus production (indicated by the number of grey virions), host transcriptional responses to infection (indicated by the number of interferon (IFN) symbols), and cell fate (indicated by the number of skulls).
Conclusions and open questions
It is now clear that the remarkable selectivity and efficiency of influenza virus genome packaging does not translate into a scenario where all virions carry a complete functional set of viral genes, as was long believed. SIP production is clearly a fundamental feature of IAV biology, and an enormous amount of work is still needed to define both the mechanistic basis of SIP production and the effects of SIPs on the transmission, evolution, and pathogenic potential of IAV populations.
A number of critical open questions remain. First, what are the viral genetic determinants that give rise to strain difference in SIP production and what can they tell us about the specific mechanisms involved? Second, how do differences in SIP production influence emergent viral phenotypes like transmissibility and pathogenicity? Third, can SIP production be beneficial to the virus, and if so, how? Fourth, does selection act upon SIP production or other features of viral genomic diversity like DIP production? Addressing these questions will help shed light on how the unique genomic organization of IAV populations shapes their infectious and evolutionary potential.
Finally, the production of large numbers of SIPs by IAV evokes the unique genome organization of multipartite viruses, where each genome segment is independently encapsulated and viral replication can only occur through complementation [46]. While IAV clearly does not qualify as a multipartite virus, given that it does actually co-package the full genome at some efficiency, key questions concerning the viability of a multipartite lifestyle also apply to the costs and benefits of SIP production [47]. Comparative analysis of influenza viruses with multipartite viruses will likely provide deeper insight into the origins and consequences of different genome organization strategies.
References and recommended reading
Papers of particular interest, published within the period of review, have been highlighted as
• of special interest
•• of outstanding interest
- 1.Taubenberger JK, Kash JC: Influenza virus evolution, host adaptation, and pandemic formation. Cell Host Microbe 2010, 7:440–451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Brooke CB: Population diversity and collective interactions during influenza virus infection. J Virol 2017, 91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Xue KS, Moncla LH, Bedford T, Bloom JD: Within-host evolution of human influenza virus. Trends Microbiol 2018. 10.1016/j.tim.2018.02.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Donald HB, Isaacs A: Counts of influenza virus particles. J Gen Microbiol 1954, 10:457–464. [DOI] [PubMed] [Google Scholar]
- 5.Brooke CB: Biological activities of “noninfectious” influenza A virus particles. Future Virol 2014, 9:41–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Nayak DP, Chambers TM, Akkina RK: Defective-interfering (DI) RNAs of influenza viruses: origin, structure, expression, and interference. Curr Top Microbiol Immunol 1985, 114:103–151. [DOI] [PubMed] [Google Scholar]
- 7.Dou D, Hernández-Neuta I, Wang H, Östbye H, Qian X, Thiele S, Resa-Infante P, Kouassi NM, Sender V, Hentrich K et al. : Analysis of IAV replication and co-infection dynamics by a versatile rna viral genome labeling method. Cell Rep 2017, 20:251–263. [DOI] [PubMed] [Google Scholar]
- 8.Heldt FS, Kupke SY, Dorl S, Reichl U, Frensing T: Single-cell analysis and stochastic modelling unveil large cell-to-cell variability in influenza A virus infection. Nat Commun 2015, 6:8938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Brooke CB, Ince WL, Wrammert J, Ahmed R, Wilson PC, Bennink JR, Yewdell JW: Most influenza A virions fail to express at least one essential viral protein. J Virol 2013, 87:3155–3162. [DOI] [PMC free article] [PubMed] [Google Scholar]; •• This study was the first to describe the nature and production of semi-infectious particles by different IAV strains.
- 10.Eisfeld AJ, Neumann G, Kawaoka Y: At the centre: influenza A virus ribonucleoproteins. Nat Rev Microbiol 2015, 13:28–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hutchinson EC, von Kirchbach JC, Gog JR, Digard P: Genome packaging in influenza A virus. J Gen Virol 2010, 91:313–328. [DOI] [PubMed] [Google Scholar]; •• Great review on influenza genome packaging.
- 12.Noda T, Sagara H, Yen A, Takada A, Kida H, Cheng RH, Kawaoka Y: Architecture of ribonucleoprotein complexes in influenza A virus particles. Nature 2006, 439:490–492. [DOI] [PubMed] [Google Scholar]; • Original paper revealing the now classic 7+1 architecture of RNPs within the influenza virion.
- 13.Noda T, Sugita Y, Aoyama K, Hirase A, Kawakami E, Miyazawa A, Sagara H, Kawaoka Y: Three-dimensional analysis of ribonucleoprotein complexes in influenza A virus. Nat Commun 2012, 3:639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sugita Y, Sagara H, Noda T, Kawaoka Y: Configuration of viral ribonucleoprotein complexes within the influenza A virion. J Virol 2013, 87:12879–12884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fournier E, Moules V, Essere B, Paillart J-C, Sirbat J-D, Isel C, Cavalier A, Rolland J-P, Thomas D, Lina B et al. : A supramolecular assembly formed by influenza A virus genomic RNA segments. Nucleic Acids Res 2012, 40:2197–2209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fournier E, Moules V, Essere B, Paillart J-C, Sirbat J-D, Cavalier A, Rolland J-P, Thomas D, Lina B, Isel C et al. : Interaction network linking the human H3N2 influenza A virus genomic RNA segments. Vaccine 2012, 30:7359–7367. [DOI] [PubMed] [Google Scholar]
- 17.Gavazzi C, Yver M, Isel C, Smyth RP, Rosa-Calatrava M, Lina B, Moules V, Marquet R: A functional sequence-specific interaction between influenza A virus genomic RNA segments. Proc Natl Acad Sci 2013, 110:16604–16609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gavazzi C, Isel C, Fournier E, Moules V, Cavalier A, Thomas D, Lina B, Marquet R: An in vitro network of intermolecular interactions between viral RNA segments of an avian H5N2 influenza A virus: comparison with a human H3N2 virus. Nucleic Acids Res 2013, 41:1241–1254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Noda T, Murakami S, Nakatsu S, Imai H, Muramoto Y, Shindo K, Sagara H, Kawaoka Y: Importance of the 1+7 configuration of ribonucleoprotein complexes for influenza A virus genome packaging. Nat Commun 2018, 9:54. [DOI] [PMC free article] [PubMed] [Google Scholar]; • This study revealed that host RNAs can be incorporated into the 7+1 array observed within influenza virions.
- 20.Nakatsu S, Murakami S, Shindo K, Horimoto T, Sagara H, Noda T, Kawaoka Y: Influenza C and D viruses package eight organized ribonucleoprotein complexes. J Virol 2018, 92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chou Y, Vafabakhsh R, Doǧanay S, Gao Q, Ha T, Palese P: One influenza virus particle packages eight unique viral RNAs as shown by FISH analysis. Proc Natl Acad Sci 2012, 109:9101–9106. [DOI] [PMC free article] [PubMed] [Google Scholar]; •• Used single-molecule FISH to determine the viral RNA composition of single virions, and directly showed that most influenza A virions contain one copy of each RNA segment.
- 22.Fujii Y, Goto H, Watanabe T, Yoshida T, Kawaoka Y: Selective incorporation of influenza virus RNA segments into virions. Proc Natl Acad Sci 2003, 100:2002–2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Goto H, Muramoto Y, Noda T, Kawaoka Y: The genome-packaging signal of the influenza a virus genome comprises a genome incorporation signal and a genome-bundling signal. J Virol 2013, 87:11316–11322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Williams GD, Townsend D, Wylie KM, Kim PJ, Amarasinghe GK, Kutluay SB, Boon ACM: Nucleotide resolution mapping of influenza A virus nucleoprotein–RNA interactions reveals RNA features required for replication. Nat Commun 2018, 9:465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Nakatsu S, Sagara H, Sakai-Tagawa Y, Sugaya N, Noda T, Kawaoka Y: Complete and incomplete genome packaging of influenza A and B viruses. mBio 2016, 7 e01248–16. [DOI] [PMC free article] [PubMed] [Google Scholar]; • EM analysis of different IAV and IBV strains that revealed more flexibility in influenza genome packaging fidelity than was previously thought.
- 26.Brooke CB, Ince WL, Wei J, Bennink JR, Yewdell JW: Influenza A virus nucleoprotein selectively decreases neuraminidase gene-segment packaging while enhancing viral fitness and transmissibility. Proc Natl Acad Sci 2014, 111:16854–16859. [DOI] [PMC free article] [PubMed] [Google Scholar]; • Revealed that specific mutations can modulate semi-infectious particle production through selective changes in genome packaging efficiency, and surprisingly showed that viruses with decreased genome packaging efficiency could actually be more fit and transmissiblein vivo.
- 27.Babcock HP, Chen C, Zhuang X: Using single-particle tracking to study nuclear trafficking of viral genes. Biophys J 2004, 87:2749–2758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lowe AR, Siegel JJ, Kalab P, Siu M, Weis K, Liphardt JT: Selectivity mechanism of the nuclear pore complex characterized by single cargo tracking. Nature 2010, 467:600–603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Schelker M, Mair CM, Jolmes F, Welke R-W, Klipp E, Herrmann A, Flöttmann M, Sieben C: Viral RNA degradation and diffusion act as a bottleneck for the influenza A virus infection efficiency. PLOS Comput Biol 2016, 12:e1005075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Götz V, Magar L, Dornfeld D, Giese S, Pohlmann A, Höper D, Kong B-W, Jans DA, Beer M, Haller O et al. : Influenza A viruses escape from MxA restriction at the expense of efficient nuclear vRNP import. Sci Rep 2016, 6:23138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Luo W, Zhang J, Liang L, Wang G, Li Q, Zhu P, Zhou Y, Li J, Zhao Y, Sun N et al. : Phospholipid scramblase 1 interacts with influenza A virus NP, impairing its nuclear import and thereby suppressing virus replication. PLoS Pathog 2018, 14:e1006851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chou Y, Heaton NS, Gao Q, Palese P, Singer R, Lionnet T: Colocalization of different influenza viral RNA segments in the cytoplasm before viral budding as shown by single-molecule sensitivity FISH analysis. PLOS Pathog 2013, 9 e1003358. [DOI] [PMC free article] [PubMed] [Google Scholar]; • Used single molecule FISH to track the movements of individual genome segments between entry and nuclear import. Suggested that the success rate of pre-nuclear transport of segments is high.
- 33.Russell AB, Trapnell C, Bloom JD: Extreme heterogeneity of influenza virus infection in single cells. eLife 2018, 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Pauly MD, Procario MC, Lauring AS: A novel twelve class fluctuation test reveals higher than expected mutation rates for influenza A viruses. eLife 2017, 6:e26437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Parvin JD, Moscona A, Pan WT, Leider JM, Palese P: Measurement of the mutation rates of animal viruses: influenza A virus and poliovirus type 1. J Virol 1986, 59:377–383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Suárez-López P, Ortín J: An estimation of the nucleotide substitution rate at defined positions in the influenza virus haemagglutinin gene. J Gen Virol 1994, 75(Pt 2):389–393. [DOI] [PubMed] [Google Scholar]
- 37.Bloom JD: An experimentally determined evolutionary model dramatically improves phylogenetic fit. Mol Biol Evol 2014, 31:1956–1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Nobusawa E, Sato K: Comparison of the mutation rates of human influenza A and B viruses. J Virol 2006, 80:3675–3678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Suárez P, Valcárcel J, Ortín J: Heterogeneity of the mutation rates of influenza A viruses: isolation of mutator mutants. J Virol 1992, 66:2491–2494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Fonville JM, Marshall N, Tao H, Steel J, Lowen AC: Influenza virus reassortment is enhanced by semi-infectious particles but can be suppressed by defective interfering particles. PLOS Pathog 2015, 11:e1005204. [DOI] [PMC free article] [PubMed] [Google Scholar]; • This study suggests that increased production of semi-infectious particles may result in increased frequency of reassortment.
- 41.Charlesworth B: Recombination modification in a flucturating environment. Genetics 1976, 83:181–195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Becks L, Agrawal AF: The evolution of sex is favoured during adaptation to new environments. PLoS Biol 2012, 10:e1001317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Birchler JA, Veitia RA: Gene balance hypothesis: connecting issues of dosage sensitivity across biological disciplines. Proc Natl Acad Sci U S A 2012, 109:14746–14753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sicard A, Yvon M, Timchenko T, Gronenborn B, Michalakis Y, Gutierrez S, Blanc S: Gene copy number is differentially regulated in a multipartite virus. Nat Commun 2013, 4:2248. [DOI] [PubMed] [Google Scholar]
- 45.Elde NC, Child SJ, Eickbush MT, Kitzman JO, Rogers KS, Shendure J, Geballe AP, Malik HS: Poxviruses deploy genomic accordions to adapt rapidly against host antiviral defenses. Cell 2012, 150:831–841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Fulton RW: Biological significance of multicomponent viruses. Annu Rev Phytopathol 1980, 18:131–146. [Google Scholar]
- 47.Sicard A, Michalakis Y, Gutiérrez S, Blanc S: The strange lifestyle of multipartite viruses. PLOS Pathog 2016, 12:e1005819. [DOI] [PMC free article] [PubMed] [Google Scholar]