Abstract
Background: A recent study has revealed that miR-106b-5p might promote hepatocellular carcinoma (HCC) stemness maintenance and metastasis by targeting PTEN via PI3K/Akt pathway based on HCC cell lines and animal models. Its clinical relevance remains unknown.
Purpose: Herein, we aimed to evaluate associations of miR-106b-5p dysregulation with various clinicopathological features of HCC patients and investigate its functions during HCC progression.
Patients and methods: At first, miR-106b-5p expression in 130 pairs of HCC and adjacent normal liver tissues was detected by quantitative PCR. Chi-square test was then performed to determine clinical significance. Further investigations on its functions were performed by miRNA target prediction and validation, as well as cellular experiments.
Results: miR-106b-5p levels in HCC tissues were significantly higher than those in the adjacent normal liver tissues (P<0.001). High miR-106b-5p expression was significantly associated with advanced tumor stage (P=0.02) and high tumor grade (P=0.03). In addition, Friend of GATA 2 (FOG2) was identified as a direct target of miR-106b-5p in HCC cells. Moreover, the clinical relevance to HCC progression of the combined high miR-106b-5p and low FOG2 expression was more significant than high miR-106b-5p alone. Functionally, enforced expression of miR-106b-5p reduced FOG2 expression and promoted the proliferation and invasion of HCC cells. Furthermore, co-transfection of FOG2 restored the oncogenic roles of miR-106b-5p over-expression.
Conclusion: Our data offer the convincing evidence that miR-106b-5p upregulation may promote the aggressive progression of HCC. miR-106b-5p overexpression may promote HCC cell proliferation and invasion by suppressing FOG2, implying its potentials as a promising therapeutic target for HCC patients.
Keywords: hepatocellular carcinoma, microRNA-106b-5p, friend of GATA 2, tumor progression, prognosis
Introduction
Hepatocellular carcinoma (HCC) represents one of the most common malignant cancer and a major cause of cancer-related mortality, accounting for nearly 700,000 deaths worldwide annually.1 Despite the advanced development in early diagnosis and targeted therapeutics, most HCC patients display unfavorable prognoses because of the high incidence of intrahepatic metastasis and recurrence following curative therapeutic intervention.2 Growing number of studies have identified more and more biomarkers or therapeutic targets with high sensitivity and specificity for HCC diagnosis and therapy; however, none has justified by clinical practice.3 Therefore, it is of great clinical significance to better understand the cellular and molecular mechanisms underlying HCC development and progression, which may be necessary to screen novel diagnostic markers and identify potential therapeutic targets.
miRNAs, which are a class of short non-coding RNAs with 17–26 nucleotides in length, function as a kind of gene expression repressors by base-pairing to the 3‘-untranslated region of the corresponding target mRNAs at a post-transcriptional level.4 Accumulating studies have revealed that miRNAs may be involved in various biological processes, such as embryonic development, cell proliferation, differentiation, apoptosis, cell death, cell migration and metabolism.5,6 Pathologically, the abnormal expression of miRNAs has also been indicated to play crucial roles in the progression of various types of human cancers by regulating the specific genes.7 miRNAs may function as either onco-miRs or tumor suppressors.8 miR-106-5p, transcribed from the miR-106b~25 cluster, is located on chromosome 7 and has been indicated to be involved and overexpressed in multiple tumor types.9 Especially, Shi et al10 found that the upregulation of miR-106b-5p expression in HCC tissues and cell lines compared to that in non-tumor tissues and hepatocytes, respectively. They also confirmed that miR-106b-5p overexpression exhibited a promoting role in cancer stem cell properties, cell migration and activation of phosphatidylinositol-3 kinase (PI3K)/Akt signaling in vitro, as well as in lung metastasis in vivo, implying the important role of miR-106b-5p during HCC progression. However, its clinical relevance remains unknown.
Thus, in the current study, we aimed to evaluate the associations of miR-106b-5p dysregulation with various clinicopathological features of HCC patients, and further investigate its oncogenic functions and the underlying molecular mechanisms.
Materials and methods
Patients and tissue samples
The current research was approved by the Research Ethics Committee of 302nd Hospital of PLA, Beijing, China (No. 2016187E). All patients have signed the informed consent.
This retrospective study enrolled a total of 130 patients with primary HCC who underwent a curative liver resection at the 302nd Hospital of PLA. This clinical cohort was the same as our previous studies.11,12 The detailed information of our clinical cohort is provided in Supplementary material. The clinicopathological features of 130 patients are summarized in Table 1.
Table 1.
Association of miR-106b-5p overexpression with clinicopathological features of 130 hepatocellular carcinoma patients
| Clinicopathological Features | Case | miR-106b-5p-high (n, %) | P |
|---|---|---|---|
| Age (years) | |||
| ≤50 | 72 | 36 (50.00) | NS |
| >50 | 58 | 30 (51.72) | |
| Gender | |||
| Male | 96 | 47 (48.96) | NS |
| Female | 34 | 19 (55.88) | |
| Serum AFP | |||
| Positive | 72 | 43 (59.72) | NS |
| Negative | 58 | 25 (43.10) | |
| Tumor stage | |||
| T1 | 23 | 1 (4.35) | 0.02 |
| T2 | 40 | 12 (30.00) | |
| T3 | 52 | 38 (73.08) | |
| T4 | 15 | 15 (100.00) | |
| Tumor grade | |||
| G1 | 31 | 6 (19.35) | 0.03 |
| G2 | 76 | 40 (52.63) | |
| G3 | 23 | 20 (86.96) | |
| Growth pattern | |||
| Trabecular | 101 | 52 (51.49) | NS |
| Nontrabecular | 29 | 14 (48.28) | |
| Cirrhosis | |||
| Yes | 86 | 42 (48.84) | NS |
| No | 44 | 24 (54.55) | |
Note: “NS” refers to the differences among groups have no statistical significance.
Cell culture and transfection
Normal liver cell line (L-02), HCC cell lines HepG2 and MHCC-97H, purchased from the Cell Type Culture Collection of the Chinese Academy of Sciences (Shanghai, China) were used to detect the expression levels of miR-106b-5p and its target gene-Friend of GATA 2 (FOG2), as well as to investigate the functions of miR-106b-5p and FOG2 in HCC. Both cell lines were cultured in high glucose DMEM with 10% FBS at 37°C with 5% CO2.
miR-106b-5p mimics (106b-mimics), mimics control (NC-mimics), FOG2 overexpression plasmid (pcDNA3.1-FOG2) and empty pcDNA3.1 plasmid (pcDNA3.1-NC) were synthesized by Genepharma, Shanghai, China. Cells were seeded into 6-well plates at 60–70% confluence 24 hrs prior to cell transfection of oligonucleotides or plasmids, which was conducted using Lipofectamine 2000 (Invitrogen; Thermo Fisher Scientific, Inc., Waltham, MA, USA) according to the manufacturer’s protocol. The transfection effects were confirmed by Quantitative-PCR described in the following section.
Dual luciferase reporter gene assay
Wild-type FOG2 3‘-UTR and mutant type FOG2 3‘-UTR were inserted into PmirGLO plasmid. pmirGLO-FOG2-wt/pmirGLO-FOG2-mut and miR-106b-5p mimics/NC-mimics were co-transfected into HepG2 or MHCC-97H cells. At 24 hrs after transfection, dual-Luciferase reporter assay system (Promega, Madison, USA) was applied to detect the luciferase activity. All experiments were performed in triplicate.
Cell Counting Kit-8 (CCK-8) assay
Cells in different groups were seeded into 96-well plates with a density of 3,000 cells/well, and then incubated for 0, 24, 48 or 96 h at 37°C. CCK-8 solution (10 μL, Dojindo Molecular Technologies, Inc., Kumamoto, Japan) was added into individual wells at indicated time points and incubated for another 2 h at 37 °C. The absorbance was determined at 450 nm wavelength using an EnSpireTM 2300 Multilabel Reader (PerkinElmer, Inc., Waltham, MA, USA) according to the manufacturer’s protocol. All experiments were performed in triplicate.
Transwell cell invasion assay
Transwell cell invasion assay was performed to evaluate cell invasive ability using 24-well transwell boyden chambers (8.0 μm pore size; Costar, Cambridge, MA) according to the manufacturer’s protocol. Briefly, 5×104 cells in different groups were suspended in FBS-free DMEM and plated onto the upper chambers. The lower chambers were filled with 500 μL of DMEM containing 20% FBS and served as a chemoattractant. After another incubation at 37°C for 2 hrs, the non-invaded cells remained in the upper chambers and scraped off gently using a cotton swab. The invaded cells were fixed with 100% methanol, stained with 0.5% crystal violet solution (Beyotime Institute of Biotechnology, Shanghai, China), washed with PBS, dried in air and counted the cell number under the light microscope (Nikon, Chiyoda-Ku, Japan). All experiments were performed in triplicate.
Quantitative PCR assay
Total RNA was extracted from HCC tissues and cells in different groups using Total RNA mini kit (Qiagen, Valencia, CA, USA) based on the manufacturer’s protocol. Nanodrop spectrophotometry was used to examine the concentration and purity of total RNA. Then, RNA was reverse-transcribed into cDNA using High-Capacity cDNA Reverse Transcription kit (Applied Biosystems, Foster City, CA, USA). Quantitative PCR was performed using a quantitative RT-PCR Detection Kit (GeneCopoeia, Rockville, MD, USA) based on the manufacturer’s instructions. U6 and GAPDH were used as internal controls for miR-106b-5p and FOG2 mRNA expression detection, respectively. The sequence of primers for miRNAs and mRNAs were listed as follows:
miR-106b-5p, 5‘-TGC GGC AAC ACC AGT CGA TGG-3‘ (sense) and 5‘-CCA
GTGCAGGGTCCGAGGT-3‘ (antisense).
U6, 5‘- GCT TCG GCA GCA CAT ATA CTA AAA T −3‘ (sense) and 5‘- CGC TTC ACG AAT TTG CGT GTC AT −3‘ (antisense).
FOG2, 5‘-GAG CTG CGA AGA CGT GGA GT-3‘ (sense) and 5‘-CCA GGC TGT CCT GGT TTG TC-3‘(antisense).
GAPDH, 5‘-CTC CTC CAC CTT TGA CGC T-3‘ (sense) and 5‘-GGG TCT CTC TCT TCC TCT TGT G-3‘(antisense).
All experiments were performed in triplicate.
Statistical analysis
The statistical analyses in the current study were performed using the software of SPSS version 13.0 for Windows (SPSS Inc, IL, USA). The Student’s t-test was used for comparisons between groups. The chi-squared test was used to show differences in categorical variables. Spearman’s correlation analysis was used to assess the correlation between miR-106b-5p and FOG2 mRNA level in HCC tissues. Differences were considered statistically significant when P was less than 0.05.
Results
Overexpression of miR-106b-5p in HCC tissue associates with aggressive clinicopathological features of patients
The expression levels of miR-106b-5p in 130 self-pairs of HCC and adjacent nonneoplastic liver tissues were detected by quantitative PCR assay. As shown in Figure 1, the expression levels of miR-106b-5p in HCC tissues were significantly higher than those in the adjacent nonneoplastic liver tissues (mean ± SD: 2.89±0.80 vs 1.23±0.22, P<0.001, Figure 1A), which was consistent with the data based on HCC cell lines and normal liver cell line (HepG2 vs L-02, mean ± SD: 3.04±0.47 vs 1.06±0.31, P<0.001; MHCC-97H vs L-02, mean ± SD: 3.33±0.44 vs 1.06±0.31, P<0.001; Figure 1B).
Figure 1.
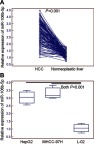
Expression levels of miR-106b-5p in HCC tissues and cells detected by quantitative PCR assay. (A) Expression levels of miR-106b-5p in HCC tissues were significantly higher than those in the adjacent nonneoplastic liver tissues (mean ± S.D.: 2.89±0.80 vs 1.23±0.22, P<0.001). (B) Expression levels of miR-106b-5p in HCC cell lines were significantly higher than those in normal liver cell line (HepG2 vs L-02, mean ± S.D.: 3.04±0.47 vs 1.06±0.31, P<0.001; MHCC-97H vs L-02, mean ± S.D.: 3.33±0.44 vs 1.06±0.31, P<0.001).
To evaluate the associations of miR-106b-5p expression with various clinicopathological features of HCC patients, all 130 patients were divided into miR-106b-5p-low (n=64, 49.23%) and miR-106b-5p-high (n=66, 50.77%) groups using the median value of its expression level as a cutoff point. As shown in Table 1, high miR-106b-5p expression was more frequently occurred in HCC patients with advanced tumor stage (P=0.02) and high tumor grade (P=0.03).
FOG2 was an direct target of miR-106b-5p
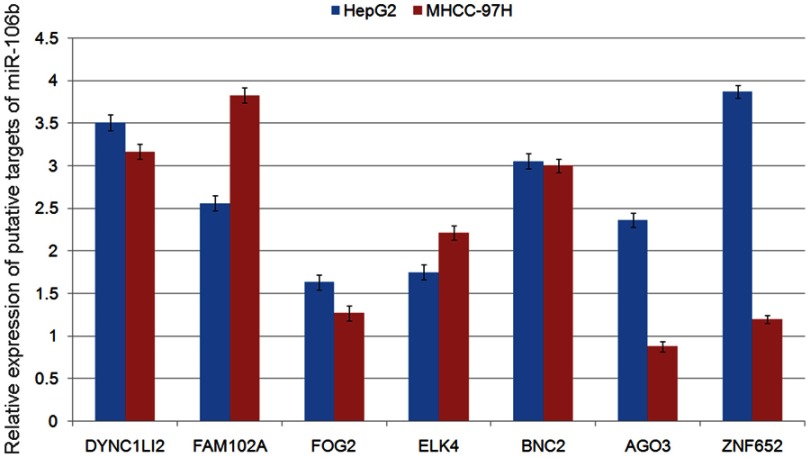
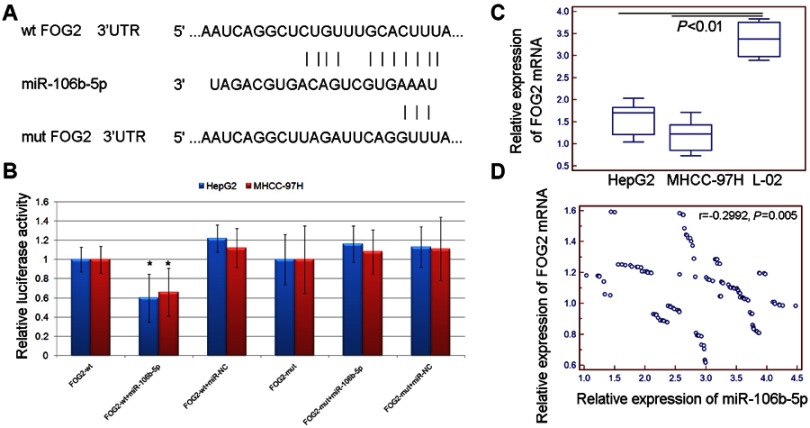
To investigate the underlying mechanisms of miR-106b-5p, its putative target genes were collected from TargetScan (Release 5.2, http://www.targetscan.org/vert_50/). A total of 7 genes (DYNC1LI2, FAM102A, FOG2, ELK4, BNC2, AGO3, ZNF652) with more than three conserved sites were predicted as the putative targets of miR-106b-5p. As shown in Figure S1, the expression levels of FOG2 mRNA were lowest in both HepG2 and MHCC-97H cells. In addition, the involvement of FOG2 in HCC remains unknown. Therefore, we chose FOG2 for the following experiments. Figure 2A shows that FOG2 3’UTR contained a miR-106b-5p-binding site. The dual luciferase reporter gene assay demonstrated that the luciferase activities of HepG2 and MHCC-97H cells cotransfected with the FOG2-WT-3‘-UTR reporter vector and miR-106b-5p mimics were both significantly lower than those in control groups (both P<0.05, Figure 2B). However, there were no obvious changes in luciferase activity when the FOG2-MUT-3‘-UTR and miR-106b-5p mimics were cotransfected (Figure 2B).
Figure S1.
Relative expression levels of putative targets of miR-106b-5p.
Figure 2.
FOG2 was a direct target of miR-106b-5p. (A) FOG2 3’UTR contains a miR-106b-5p binding site. (B) The luciferase activities of HepG2 and MHCC-97H cells cotransfected with the FOG2-WT/MUT-3‘-UTR reporter vector and miR-106b-5p/NC mimics (* P<0.05, FOG2-WT-3‘-UTR+miR-106b-5p mimics vs FOG2-MUT-3‘-UTR+miR-106b-5p). (C) Expression levels of FOG2 mRNA in HCC cells (all P<0.01). (D) The Spearman correlation analysis showed that the expression levels of miR-106b-5p in HCC tissues were negatively correlated with those of FOG2 mRNA (r=−0.2992, P=0.005).
In addition, quantitative PCR assay indicated that the expression levels of FOG2 mRNA in HCC tissues and cells were all lower than those in the adjacent normal liver tissues and normal liver cells (all P<0.01, Figure 2C). The Spearman correlation analysis showed that the expression levels of miR-106b-5p in HCC tissues were negatively correlated with those of FOG2 mRNA (r=−0.2992, P=0.005, Figure 2D). These findings suggest that FOG2 was a direct target of miR-106b-5p in HCC cells.
The median value of the expression levels of FOG2 in HCC tissues was used as a cutoff point. All 130 patients were divided into FOG2-low (n=66, 50.77%) and FOG2-high (n=64, 49.23%) groups. Table 2 shows the groups with the combined expression of miR-106b-5p and FOG2: miR-106b-5p-low/FOG2-low (n=26, 20.00%), miR-106b-5p-low/FOG2-high (n=38, 29.23%), miR-106b-5p-high/FOG2-low (n=40, 30.77%) and miR-106b-5p-high/FOG2-high (n=26, 20.00%). Statistically, HCC patients with advanced tumor stage and tumor grade had higher frequency of miR-106b-5p-high/FOG2-low than those with low tumor stage and tumor grade (both P<0.01, Table 3).
Table 2.
Expression of miR-106b-5p and FOG2 proteins in 130 hepatocellular carcinoma patients
| FOG2 expression | P | |||
|---|---|---|---|---|
| High (n=64) | Low (n=66) | |||
| miR-106b-5p expression | High (n=66) | 26 | 40 | 0.01 |
| Low (n=64) | 38 | 26 | ||
Table 3.
Association of miR-106b-5p/FOG2 dysregulation with clinicopathological features of 130 hepatocellular carcinoma patients
| Clinicopathological Features | Case | miR-106b-5p-high/FOG2-low (n, %) | P |
|---|---|---|---|
| Age (years) | |||
| ≤50 | 72 | 26 (36.11) | NS |
| >50 | 58 | 14 (24.14) | |
| Gender | |||
| Male | 96 | 30 (31.25) | NS |
| Female | 34 | 10 (29.41) | |
| Serum AFP | |||
| Positive | 72 | 27 (37.50) | NS |
| Negative | 58 | 13 (22.41) | |
| Tumor stage | |||
| T1 | 23 | 1 (4.35) | <0.01 |
| T2 | 40 | 7 (23.33) | |
| T3 | 52 | 20 (38.46) | |
| T4 | 15 | 12 (80.00) | |
| Tumor grade | |||
| G1 | 31 | 4 (12.90) | <0.01 |
| G2 | 76 | 18 (23.68) | |
| G3 | 23 | 18 (78.26) | |
| Growth pattern | |||
| Trabecular | 101 | 32 (31.68) | NS |
| Nontrabecular | 29 | 8 (27.59) | |
| Cirrhosis | |||
| Yes | 86 | 28 (32.55) | NS |
| No | 44 | 12 (27.27) | |
Note: “NS” refers to the differences among groups have no statistical significance.
miR-106b-5p promoted the proliferation and invasion of HCC cells via reducing FOG2 expression
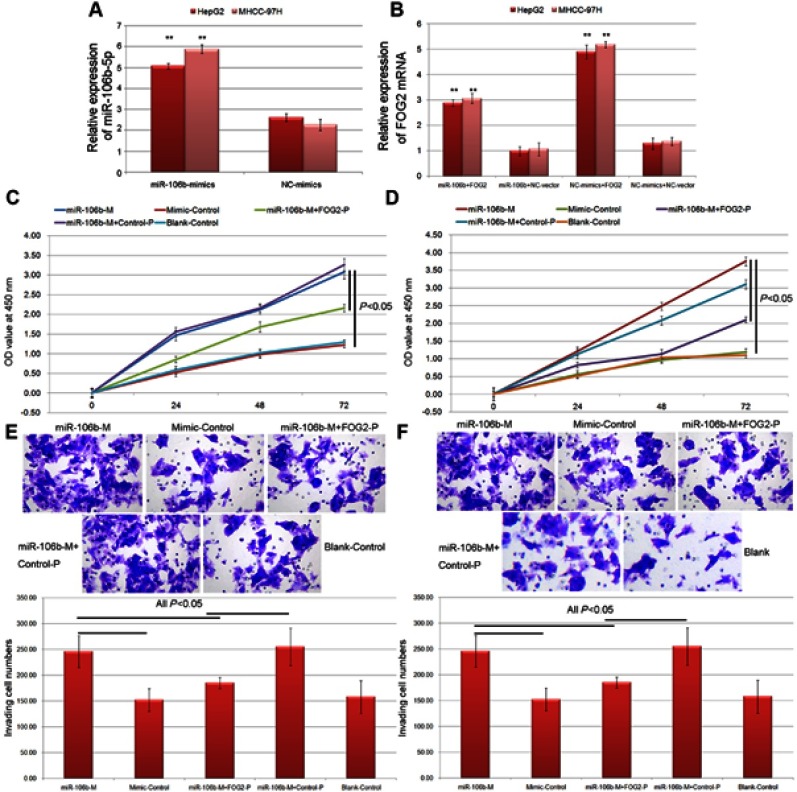
To determine the effects of miR-106b-5p-FOG2 axis on cell proliferation and invasion of HCC cells, the CCK-8 and the transwell assays were performed based on both HepG2 and MHCC-97H cells transfected with miR-106b-5p-mimics or cotransfected with miR-106b-5p-mimics and FOG2 expression vector. As shown in Figure 3A, the expression levels of miR-106b-5p and FOG2 mRNA in HCC cells transfected with miR-106b-5p-mimics were, respectively, higher and lower than those in HCC cells transfected with NC-mimics (both P<0.01). Following the cotransfection with miR-106b-5p-mimics and FOG2 expression vector, the expression levels of FOG2 mRNA in HCC cells were significantly higher than those cotransfected with miR-106b-5p-mimics and NC-expression vector (P<0.05, Figure 3B).
Figure 3.
miR-106b-5p promoted the proliferation and invasion of HCC cells via reducing FOG2 expression. (A) Expression levels of miR-106b-5p in HCC cells with different transfection (** P<0.01, miR-106b-5p mimics vs NC mimics). (B) Expression levels of FOG2 mRNA in HCC cells with different transfection (** P<0.01, miR-106b-5p mimics+FOG2 vs miR-106b-5p mimics+NC, or NC mimics+FOG2 vs NC mimics+NC). (C and D) The cell proliferation of HepG2 and MHCC-97H cells with different transfection. (E and F) The cell invasion abilities of HepG2 and MHCC-97H cells detected by Transwell assay.
Functionally, the results of CCK-8 assay indicated that the enforced expression of miR-106b-5p significantly inhibited the proliferation of both HepG2 and MHCC-97H cells, while the cotransfection of miR-106b-5p mimics and FOG2 expression vector could reduce the inhibitory effects of miR-106b-5p on HCC cell proliferation (all P<0.05, Figure 3C and D). As for Transwell assay shown in Figure 3E and F, the numbers of both HepG2 and MHCC-97H cells passing through the membrane were dramatically higher in miR-106b-5p mimics-transfection group than those in control group (both P<0.05, Figure 3E and F). Moreover, the overexpression of FOG2 impaired the promoting role of miR-106b-5p in HCC cell invasion of cotransfection group compared with miR-106b-5p mimics-transfection group (both P<0.05, Figure 3E and F).
Discussion
Due to its increased incidence and unsatisfactory curative effects, to elucidate the underlying molecular mechanisms of HCC development and progression as well as to develop novel therapeutic targets may be of great clinical significance. In the current study, we demonstrated that miR-106b-5p expression levels in HCC tissues were significantly higher than those in the adjacent normal liver tissues. High miR-106b-5p expression was significantly associated with advanced tumor stage and high tumor grade. In addition, we identified FOG2 as a direct target of miR-106b-5p in HCC cell lines. Functionally, the enforced expression of miR-106b-5p reduced FOG2 expression and promoted the proliferation and invasion of HCC cells. Furthermore, co-transfection of FOG2 restored the oncogenic roles of miR-106b-5p over-expression. These findings suggest that miR-106b-5p may play oncogenic roles in HCC, at least partly, via directly targeting FOG2.
miR-106b-5p belongs to the miR-106b seed family and functions as an oncogenic miRNA in multiple cancer types. Yang et al13 observed that the downregulation of miR-106b in Y79 retinoblastoma cells decreased cell viability and migration, as well as induced cell apoptosis via regulating its target gene Runx3. Liu et al14 found that miR-106b-5p was distinctly upregulated in glioma tumor samples and cell lines compared with normal brain tissues, and its expression level correlated with the pathological grading. miR-106b-5p overexpression in glioma cells dramatically promoted cell proliferation, although inhibited cell apoptosis in vitro and in vivo. Lu et al15 reported that miR-106b-5p promoted renal cell carcinoma aggressiveness and stem-cell-like phenotype by activating Wnt/β-catenin signaling. Wei et al16 found a relatively higher miR-106b-5p expression in non-small cell lung cancer specimens and cell lines. In vitro, miR-106b-5p overexpression promoted proliferation and inhibited apoptosis. In vivo, miR-106b-5p promoted xenograft tumor formation. Xu et al17 reported that miR-106b promoted the proliferation and invasion of laryngeal carcinoma cells by directly targeting RUNX3. Recent studies have revealed that miRNAs are involved in chemoresistance, leading to the failure of chemotherapy. Yu et al18 indicated that miR-106b-5p regulated cisplatin chemosensitivity by targeting polycystic kidney disease-2 in non-small-cell lung cancer. Consistently, we observed the overexpression of miR-106b-5p in both HCC cells and tissues compared to normal controls, implying that it might play a role in the development of HCC. Statistically, miR-106b-5p upregulation was dramatically associated with the aggressive clinicopathological features of patients with HCC, suggesting its involvement in the progression of HCC. The significant clinical relevance of miR-106b-5p as mentioned above prompted us to further investigate its underlying molecular mechanisms.
Each miRNA can have multiple target genes, and several miRNA can regulate the same gene.19 On this basis, we identified the target genes of miR-106b-5p by combining miRNA target prediction, dual luciferase reporter gene assay, correlation analysis based on miRNA and target mRNA expression in HCC tissues, clinical significance evaluation of combined miRNA and target mRNA expression, as well as functional experiments. As a result, FOG2 was identified as a direct target of miR-106b-5p. FOG2 is broadly expressed in various adult tissues including the brain, heart, liver, lung, testes and skeletal muscle, and is a co-regulator of transcriptional factor GATA family.20 Functionally, FOG2 has been indicated to be involved in the regulation of multiple physiological processes, such as coronary vascular development, heart morphology, insulin signaling regulation and lipid metabolism.21 Recent studies have revealed that FOG2 may be a novel inhibitor of PI3K/Akt signaling.22–24 Guo et al25 found that the increased miR-200 expression might contribute to early lung tumorigenesis and could activate AKT in lung adenocarcinoma cells through a FOG2-independent mechanism involving IRS-1. Ma et al26 indicated that the expression of FOG2, as a target of miR-200s, was downregulated in human hepatic stellate cells with the stable expression of miR-200c. Moreover, miR-200c accelerated hepatic stellate cell-induced liver fibrosis via targeting the FOG2/PI3K Pathway. In this study, we elucidated the clinical significance of miR-106b-5p-FOG2 combined expression after verifying the regulatory effects of miR-106b-5p on FOG2. Our data showed the downregulation of FOG2 mRNA in HCC tissues and cells, which was negatively correlated with that of miR-106b-5p. Importantly, the combination of high miR-106-5p and low FOG2 expression was more significantly associated with the advanced tumor stage and high tumor grade than high miR-106b-5p alone, suggesting the crucial roles of miR-106b-5p-FOG2 axis during the development and progression of HCC. Moreover, we also observed that the overexpression of FOG2 might attenuate the oncogenic roles of miR-106b-5p.
In conclusion, our data offer the convincing evidence that miR-106b-5p upregulation may promote the aggressive tumor progression of HCC. miR-106b-5p overexpression may promote the proliferation and invasion of HCC cells by suppressing FOG2, implying its potentials as a promising therapeutic target for HCC patients.
Disclosure
The authors clarified that there were no conflicts of interest in the current study.
Supplementary material
References
- 1.Quaglia A. Hepatocellular carcinoma: a review of diagnostic challenges for the pathologist. J Hepatocell Carcinoma. 2018;5:99–108. doi: 10.2147/JHC.S159808 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Choi WT, Ramachandran R, Kakar S. Immunohistochemical approach for the diagnosis of a liver mass on small biopsy specimens. Hum Pathol. 2017;63:1–13. doi: 10.1016/j.humpath.2016.12.025 [DOI] [PubMed] [Google Scholar]
- 3.Llovet JM, Montal R, Sia D, Finn RS. Molecular therapies and precision medicine for hepatocellular carcinoma. Nat Rev Clin Oncol. 2018;15:599–616. doi: 10.1038/s41571-018-0073-4 [DOI] [PubMed] [Google Scholar]
- 4.Mens MMJ, Ghanbari M. Cell cycle regulation of stem cells by MicroRNAs. Stem Cell Rev. 2018;14:309–322. doi: 10.1007/s12015-018-9808-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wu J, Ding J, Yang J, Guo X, Zheng Y. MicroRNA roles in the nuclear factor kappa B signaling pathway in cancer. Front Immunol. 2018;9:546. doi: 10.3389/fimmu.2018.00546 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Liu H, Lei C, He Q, Pan Z, Xiao D, Tao Y. Nuclear functions of mammalian MicroRNAs in gene regulation, immunity and cancer. Mol Cancer. 2018;17:64. doi: 10.1186/s12943-018-0765-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Curtale G. MiRNAs at the crossroads between innate immunity and cancer: focus on macrophages. Cells. 2018;7:pii: E12. doi: 10.3390/cells7020012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Han B, Chao J, Yao H. Circular RNA and its mechanisms in disease: from the bench to the clinic. Pharmacol Ther. 2018;187:31–44. doi: 10.1016/j.pharmthera.2018.01.010 [DOI] [PubMed] [Google Scholar]
- 9.Ivanovska I, Ball AS, Diaz RL, et al. MicroRNAs in the miR-106b family regulate p21/CDKN1A and promote cell cycle progression. Mol Cell Biol. 2008;28:2167–2174. doi: 10.1128/MCB.01977-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Shi DM, Bian XY, Qin CD, Wu WZ. miR-106b-5p promotes stem cell-like properties of hepatocellular carcinoma cells by targeting PTEN via PI3K/Akt pathway. Onco Targets Ther. 2018;11:571–585. doi: 10.2147/OTT.S152611 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhang Y, Guo X, Li Z, et al. A systematic investigation based on microRNA-mediated gene regulatory network reveals that dysregulation of microRNA-19a/Cyclin D1 axis confers an oncogenic potential and a worse prognosis in human hepatocellular carcinoma. RNA Biol. 2015;12:643–657. doi: 10.1080/15476286.2015.1022702 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhang Y, Guo X, Xiong L, et al. Comprehensive analysis of microRNA-regulated protein interaction network reveals the tumor suppressive role of microRNA-149 in human hepatocellular carcinoma via targeting AKT-mTOR pathway. Mol Cancer. 2014;13:253. doi: 10.1186/1476-4598-13-253 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yang G, Fu Y, Zhang L, Lu X, Li Q. miR106b regulates retinoblastoma Y79 cells through Runx3. Oncol Rep. 2017;38:3039–3043. doi: 10.3892/or.2017.5931 [DOI] [PubMed] [Google Scholar]
- 14.Liu F, Gong J, Huang W, et al. MicroRNA-106b-5p boosts glioma tumorigensis by targeting multiple tumor suppressor genes. Oncogene. 2014;33:4813–4822. doi: 10.1038/onc.2013.428 [DOI] [PubMed] [Google Scholar]
- 15.Lu J, Wei JH, Feng ZH, et al. miR-106b-5p promotes renal cell carcinoma aggressiveness and stem-cell-like phenotype by activating Wnt/β-catenin signalling. Oncotarget. 2017;8:21461–21471. doi: 10.18632/oncotarget.15591 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wei K, Pan C, Yao G, et al. MiR-106b-5p promotes proliferation and inhibits apoptosis by regulating BTG3 in non-small cell lung cancer. Cell Physiol Biochem. 2017;44:1545–1558. doi: 10.1159/000485650 [DOI] [PubMed] [Google Scholar]
- 17.Xu Y, Wang K, Gao W, et al. MicroRNA-106b regulates the tumor suppressor RUNX3 in laryngeal carcinoma cells. FEBS Lett. 2013;587:3166–3174. doi: 10.1016/j.febslet.2013.05.069 [DOI] [PubMed] [Google Scholar]
- 18.Yu S, Qin X, Chen T, Zhou L, Xu X, Feng J. MicroRNA-106b-5p regulates cisplatin chemosensitivity by targeting polycystic kidney disease-2 in non-small-cell lung cancer. Anticancer Drugs. 2017;28:852–860. doi: 10.1097/CAD.0000000000000524 [DOI] [PubMed] [Google Scholar]
- 19.Van Roosbroeck K, Calin GA. Cancer hallmarks and MicroRNAs: the therapeutic connection. Adv Cancer Res. 2017;135:119–149. doi: 10.1016/bs.acr.2017.06.002 [DOI] [PubMed] [Google Scholar]
- 20.Becker LE, Takwi AA, Lu Z, Li Y. The role of miR-200a in mammalian epithelial cell transformation. Carcinogenesis. 2015;36:2–12. doi: 10.1093/carcin/bgu202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mei S, Xin J, Liu Y, et al. MicroRNA-200c promotes suppressive potential of myeloid-derived suppressor cells by modulating PTEN and FOG2 expression. PLoS One. 2015;10:e0135867. doi: 10.1371/journal.pone.0135867 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Xiao Y, Wang J, Chen Y, et al. Up-regulation of miR-200b in biliary atresia patients accelerates proliferation and migration of hepatic stallate cells by activating PI3K/Akt signaling. Cell Signal. 2014;26:925–932. doi: 10.1016/j.cellsig.2014.01.003 [DOI] [PubMed] [Google Scholar]
- 23.Park JT, Kato M, Yuan H, et al. FOG2 protein down-regulation by transforming growth factor-β1-induced microRNA-200b/c leads to Akt kinase activation and glomerular mesangial hypertrophy related to diabetic nephropathy. J Biol Chem. 2013;288:22469–22480. doi: 10.1074/jbc.M113.453043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Dou L, Zhao T, Wang L, et al. miR-200s contribute to interleukin-6 (IL-6)-induced insulin resistance in hepatocytes. J Biol Chem. 2013;288:22596–22606. doi: 10.1074/jbc.M112.423145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Guo L, Wang J, Yang P, Lu Q, Zhang T, Yang Y. MicroRNA-200 promotes lung cancer cell growth through FOG2-independent AKT activation. IUBMB Life. 2015;67:720–725. doi: 10.1002/iub.1412 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ma T, Cai X, Wang Z, et al. miR-200c accelerates hepatic stellate cell-induced liver fibrosis via targeting the FOG2/PI3K pathway. Biomed Res Int. 2017;2017:2670658. doi: 10.1155/2017/2670658 [DOI] [PMC free article] [PubMed] [Google Scholar]