Abstract
Context
Visceral fat (VF), more than fat elsewhere in the body [mostly subcutaneous fat (SF)], promotes systemic inflammation and related disease. The mechanisms of preferentially visceral accumulation of body fat are largely unknown.
Objective
To identify genetic loci and mechanistic pathways of preferential accumulation of VF and associated low-grade systemic inflammation.
Design
Genome-wide association study (GWAS).
Setting and Participants
Population-based cohort of 1586 adolescents (aged 12 to 19 years) and adults (aged 36 to 65 years).
Main Outcome Measures
Abdominal VF and SF were measured with MRI, total body fat (TBF) was assessed with bioimpedance, and low-grade systemic inflammation was examined by serum C-reactive protein (CRP) measurement.
Results
This GWAS of preferential accumulation of VF identified a significant locus on chromosome 6 at rs803522 (P = 1.1 × 10−9 or 4.3 × 10−10 for VF adjusted for SF or TBF, respectively). The major allele was associated with more VF; the association was similar in adolescents and adults. The allele was also associated with higher CRP level, but this association was stronger in adults than adolescents (P for interaction = 4.5 × 10−3). In adults, VF was a significant mediator (P = 1.9× 10−4) in the association between the locus and CRP, explaining 30% of the mediation. The locus was near ATG5, encoding an autophagy molecule reported to modulate adipocyte size and macrophage polarization.
Conclusion
A genetic locus near ATG5 regulates preferential accumulation of VF (vs SF) in youth and adulthood and contributes to the development of systemic inflammation in adulthood.
We identified a genetic locus of preferential visceral accumulation of body fat and related low-grade systemic inflammation.
Excess body fat (i.e., overweight and obesity) is a major health problem in the developed world. It has become highly prevalent (>60% of adults and >35% of children are obese or overweight), and it increases risk for multiple disorders, including the cardiovascular disease, a major cause of mortality. As a result, life expectancy in the developed world may decline for the first time since the Great Depression (1).
Health consequences of overweight and obesity are determined not only by excess body fat but also by the site of body-fat accumulation. Thus, fat deposited around internal organs in the abdominal cavity [visceral fat (VF)] rather than fat stored elsewhere in the body [mostly subcutaneous fat (SF)] is linked to disease (2–6). Much of the disease risk in this context is related to low-grade systemic inflammation that develops as a result of proinflammatory molecules secreted from expanding adipose tissue; VF compared with SF has a more proinflammatory secretory profile (2–4). The differential biology of VF and SF is supported by separate embryonic origins of the two types of fat tissue (7).
Compared with women, men have a greater propensity for visceral accumulation of body fat (8). Adult body-fat distribution occurs during adolescence. Large interindividual differences exist in the capacity for visceral (vs subcutaneous) accumulation of body fat. Mechanisms behind these differences are largely unknown. Genome-wide association studies (GWASs) may provide valuable insights (9, 10). We performed a GWAS of preferentially visceral accumulation of body fat in a population-based cohort of 1586 adolescents and middle-aged adults from the Saguenay Youth Study (SYS) (11), in whom we measured VF with MRI.
Methods
Cohort
The study sample included 989 adolescents (aged 12 to 19 years; 52% female) and 600 adults (aged 36 to 65 years; 53% female) of French-Canadian ancestry recruited in the SYS (11). The SYS is a population-based study of cardiometabolic and brain health (11). Characteristics of the participants are presented in Table 1. Written consent (from parents) and assent (from adolescents) were obtained in accordance with the research ethics committees of the Chicoutimi Hospital (Chicoutimi, Quebec, Canada) and the Hospital for Sick Children (Toronto, Ontario, Canada). Recruitment, selection, and exclusion criteria have been described previously (11). The 989 adolescents and 600 middle-aged adults studied here all had a complete set of MRI, bioelectrical impedance, and genotype data. For secondary serum C-reactive protein (CRP) analyses, 33 participants with CRP level >10 mg/L were excluded to avoid possible confounding by acute-phase inflammatory response, as suggested previously (12).
Table 1.
Characteristics of Participants
| Median (Minimum, Maximum), % | ||||
|---|---|---|---|---|
| Adolescents | Adults | |||
| Girls (n = 515) | Boys (n = 474) | Women (n = 317) | Men (n = 283) | |
| Age, y | 14.9 (12.0, 19.2) | 14.8 (12.0, 19.0) | 48.3 (36.4, 59.2) | 50.9 (38.9, 65.4) |
| Height, cm | 159.7 (134.5, 178.0) | 169.0 (140.5, 189.0) | 161.3 (146.5, 178.0) | 173.0 (156.0, 195.0) |
| Body weight, kg | 54.0 (28.2, 108.1) | 59.5 (28.8,153.5) | 69.6 (42.8, 137.1) | 84.4 (53.9,139.2) |
| VF, cm3 | 16.3 (4.4, 102) | 13.9 (1.7, 171) | 53.5 (3.1, 313) | 88.7 (8.1, 386) |
| SF, cm3 | 116 (18.9, 636) | 61.7 (15.4, 622) | 292 (51, 863) | 233 (53.6, 723) |
| TBF, kg | 13.1 (1.74, 56.4) | 7.92 (1.29, 59.9) | 24.4 (4.5, 76.8) | 20.4 (2.0, 49.0) |
| TBF, % body weight | 24 (5,49) | 14 (4, 40) | 34 (11, 56) | 24 (3, 39) |
| CRP, mg/L | 0.47 (0.15, 6.04) | 0.30 (0.15, 4.12) | 1.45 (0.19, 10.0) | 1.20 (0.19, 9.20) |
Measurements
VF and SF
VF and SF were measured with MRI from T1-weighted images of the abdomen acquired on a Gyroscan NT 1.0-T scanner (Philips Healthcare, Best, Netherlands) in adolescents and an Avanto 1.5-T scanner (Siemens, Erlangen, Germany) in adults. A 10-mm–thick axial slice at the level of the umbilicus was selected to quantify VF and SF, in cubic centimeters, with a semiautomatic method described previously (11).
Total body fat
Total body fat (TBF) was assessed using multifrequency bioimpedance analysis (in adolescents, Xitron Technologies, San Diego, CA; in adults, Tanita BC-418, Arlington Heights, IL) (13). Participants were asked to refrain from caffeine, alcohol, and vigorous activity 24 hours before the measurement. The measurement was made after a 20-minute stabilization period, during which the participants were resting in a supine position.
Serum CRP levels
Using a high-sensitivity assay (Beckman Coulter) with detection limits of 0.08 to 80 mg/L, concentrations of CRP were measured in sera from morning blood samples collected after overnight fasting.
Genotypes
The SYS adolescents and parents were genotyped in two waves. First, samples from the initial group of recruited adolescents (n = 592) were genotyped with the Illumina Human610-Quad BeadChip [n = 582,892 single nucleotide polymorphisms (SNPs)] at the Centre National de Génotypage (Paris, France). Second, samples from the remaining adolescents and all parents were genotyped with the HumanOmniExpress BeadChip (Illumina; n = 729,295 SNPs) at the Genome Analysis Centre of Helmholtz Zentrum Mümchen (Munich, Germany). For both chips, SNPs with a call rate of <95%, minor allele frequency of <0.01, or not in Hardy-Weinberg equilibrium (P < 1.0 × 10−6) were excluded. After this quality control, 542,345 SNPs on the first chip and 644,283 SNPs on the second chip were available for the additional analyses.
To equate the set of SNPs genotyped on each chip and increase the SNP density, we performed genotype imputations, which have been widely used in GWASs to boost statistical power and fine mapping of association signals (14). Haplotype phasing was performed with SHAPEIT, version 2.790 (15), using the subset of 313,653 post–quality control SNPs that were present on both genotyping platforms and the 1000 Genomes European reference panel (phase 1, release 3). Imputation was conducted on the phased data with IMPUTE2 (16). To improve the accuracy of phasing, imputation was performed on the whole cohort of adolescents and parents. Markers with low imputation quality (information score <0.5) or minor allele frequency <0.01 were removed. After this quality control of imputation, a total of 8,511,049 SNPs were analyzed.
Female menopause
Adult women answered questions related to reproductive health from the Ontario Health Study (www.ontariohealthstudy.ca) incorporated in to a medical questionnaire. Adult women were classified as postmenopausal (n = 113), if they self-reported menopause, had missed 12 or more periods within the last year, or were 55 years of age or older.
Statistical analyses
Primary analyses
We carried out a GWAS of preferential accumulation of VF [i.e., VF adjusted for either SF (VFSF) or TBF (VFTBF)]. For this purpose, we used a computationally efficient, mixed model–based approach implemented in ProbABEL (17). The tested mixed models were also adjusted for age, age group (parent vs adolescent), sex and height as fixed effects, and family relatedness as a random effect. All adiposity measures (i.e., VF, SF, and TBF) were transformed using log base 10.
Secondary analyses
We performed association tests of the top locus using the linear mixed-effects regression implemented in the lmekin function from the coxme R package that allowed additional adjusting for common family environment as a random effect (18). These tests examined associations of the locus with preferential accumulation of VF and other fat, and with low-grade systemic inflammation. Preferential accumulation of VF was assessed as VFSF and VFTBF, and preferential accumulation of other fat than VF was examined as SFVF and TBFVF. In addition, we tested VF, SF, and TBF without adjusting for any other fat depot. Low-grade systemic inflammation was assessed by measuring CRP level (12). CRP values were transformed using the rank-based inverse normal transformation (19). Similar to VF analysis, this analysis was performed while adjusting for SF or TBF. We also examined whether VF was a mediator in the association between the locus and CRP. We estimated the proportion of effect of VF on CRP, explained by the mediation, using the medflex package in R (20). We assessed the significance of the mediation effect using a permutation test with 105 replications.
Finally, we investigated the regulatory genomic elements of the identified locus using Encyclopedia of DNA Elements database (21). We also examined mRNA expression levels of the genes closest to the identified locus (upstream and downstream), using the Genotype-Tissue Expression (GTEx) project data (22), and protein-protein networks for the genes, using STRING, version10, which uses previously published experimental and text data (23).
Results
GWAS of preferential accumulation of VF and systemic inflammation
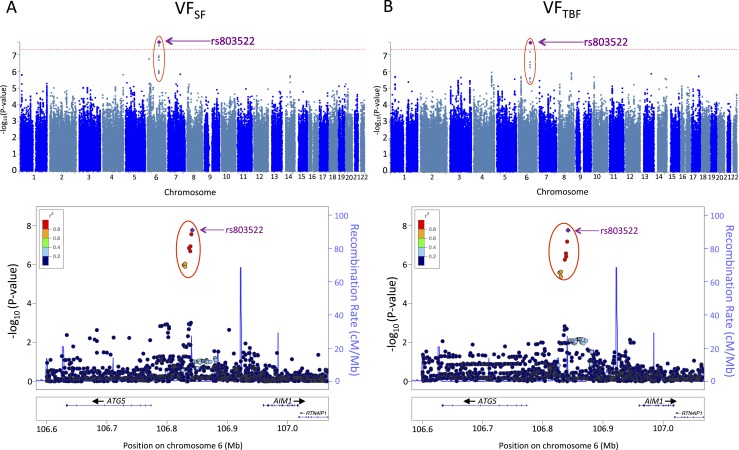
The whole cohort of the studied 1586 participants is characterized in Table 1 and Fig. 1. GWAS of VFSF carried out in this cohort identified one genome-wide significant locus on chromosome 6 (rs803522; P = 1.1 × 10−9; Table 2). This single locus explained 2.2% of the variance of VFSF. Remaining suggestive associations with P < 1 × 10−5 are reported in Supplemental Table 1 an online repository (24). The association of the top locus (i.e., rs803522) was similar for VFTBF (P = 4.3 × 10−10; Table 2). The major allele of the locus was associated with higher VFSF and VFTBF (Table 2). In addition, the major allele was associated with lower SFVF (P = 5.7 × 10−8), as well as lower TBFVF (P = 1.7 × 10−8). Furthermore, the allele was not associated with any major differences in nonadjusted VF (P = 0.02), TBF (P = 0.10), or SF (P = 0.24; Table 2). These results suggest that the locus regulates mainly the site of fat accumulation (visceral vs subcutaneous) and not the quantity of the accumulation. Based on the direction of effect, the major allele stimulates visceral (vs subcutaneous) accumulation of body fat.
Figure 1.
Manhattan and regional association plots from our GWAS of (A) VFSF and (B) VFTBF. Upper panels: The red dotted line indicates the genome-wide significance level of 5.0 × 10−8. The most significant locus is highlighted with red circle. The top SNP rs803522 is indicated by the purple diamond. The major allele frequency of this SNP is 0.87 in our cohort. Lower panels: Each point indicates an SNP tested in the GWAS within the shown genomic region. The top SNP rs803522 is indicated by the purple diamond. The right-side axis represents the estimated recombination rates from the HapMap Project samples (ftp://ftp.ncbi.nlm.nih.gov/hapmap/recombination/2008-03_rel22_B36/). The red-to-blue colors indicate the degree of linkage disequilibrium (LD) between each SNP and rs803522. The LD was based on the pairwise squared allelic correlation, r2, estimated using SYS parent data. The plots were created using LocusZoom (http://locuszoom.org/).
Table 2.
Associations Between rs803522 and VF, SF, and TBF With or Without Adjustment for Other Fat Depots
| Phenotype | Estimatea | SE | P | No. |
|---|---|---|---|---|
| Primary analyses | ||||
| VFSF | 0.152 | 0.025 | 1.1 × 10−9 | 1577 |
| VFTBF | 0.170 | 0.027 | 4.3 × 10−10 | 1538 |
| Secondary analyses | ||||
| SFVF | −0.128 | 0.024 | 5.7 × 10−8 | 1577 |
| TBFVF | −0.121 | 0.021 | 1.7 × 10−8 | 1538 |
| VF | 0.102 | 0.042 | 1.5 × 10−2 | 1589 |
| SF | −0.047 | 0.039 | 2.4 × 10−1 | 1577 |
| TBF | −0.054 | 0.032 | 9.8 × 10−2 | 1619 |
Estimate indicates the change in log10(VF, SF, or TBF) with each additional copy of the major allele of rs803522.
Additional subset analyses demonstrated that the effect of the major allele on VFSF and VFTBF was highly similar and nominally significant in all four sex- and age group–defined subsets (i.e., male and female adolescents and male and female adults; Table 3). Furthermore, the effect of the major allele on VFSF remained similar after additional adjusting for menopause [unadjusted: 0.18 (0.06) vs adjusted: 0.18 (0.07) log10 (cm3)]. These results indicate that the locus may operate already in adolescents and in both sexes. The consistency of effect across the four subsets and with two modalities of measurement of VF-independent body fat (i.e., SF by MRI and TBF by bioimpedance) support the robustness of the identified locus.
Table 3.
Associations Between rs803522 With Preferentially Visceral Accumulation of Body Fat in Male and Female Adolescents and Adults
| Female Adolescents | Male Adolescents | Female Adults | Male Adults | |||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Estimatea | SE | P | No. | Estimatea | SE | P | No. | Estimatea | SE | P | No. | Estimatea | SE | P | No. | |
| VFSF | 0.112 | 0.035 | 1.3 × 10−3 | 515 | 0.156 | 0.037 | 2.8 × 10−5 | 473 | 0.177 | 0.061 | 3.7 × 10−3 | 308 | 0.167 | 0.067 | 1.3 × 10−2 | 281 |
| VFTBF | 0.166 | 0.039 | 2.5 × 10−5 | 498 | 0.163 | 0.049 | 8.9 × 10−4 | 447 | 0.163 | 0.056 | 3.6 × 10−3 | 313 | 0.134 | 0.060 | 2.5 × 10−2 | 280 |
Estimate indicates the change in log10(VF, SF, or TBF) with an additional copy of the major allele of rs803522.
It is known that VF, more so than SF, promotes the development of low-grade systemic inflammation (2–4). Therefore, we tested whether our top locus of VFSF (i.e., rs803522) was also associated with serum CRP, a commonly used marker of systemic inflammation (12). These analyses showed that, indeed, the major allele associated with more VF was also associated with higher levels of serum CRP (VFSFP = 2.5 × 10−3; Table 4). They also showed that this association was stronger in adults than in adolescents (age group by genotype interaction P = 4.5 × 10−3; Table 4). Finally, in adults, VF was a significant mediator in the association between the locus and CRP; it explained 30% of the mediation (P = 1.9 × 10−4). Similar results were obtained when data were adjusted for TBF (Table 4).
Table 4.
Associations Between rs803522 and CRP Adjusted for SF and TBF
| Adolescents | Adults | All | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Estimatea | SE | P | No. | Estimate | SE | P | No. | Estimate | SE | P | P b | P c | No. | |
| SF | 0.036 | 0.066 | 0.58 | 847 | 0.134 | 0.037 | 3.1 × 10−4 | 578 | 0.103 | 0.048 | 0.03 | 6.9 × 10−3 | 2.5 × 10−3 | 308 |
| TBF | 0.059 | 0.069 | 0.39 | 806 | 0.241 | 0.079 | 2.2 × 10−3 | 641 | 0.099 | 0.047 | 0.04 | 8.8 × 10−2 | 2.5 × 10−2 | 313 |
Estimate indicates the change in inverse-normal-transformed CRP level with each additional copy of the major allele of rs80352.
P value for the age group by rs803522 interaction.
P value for the overall effect of rs803522 [i.e., joint test of the main and interaction (with age group) effects of rs803522].
Identified locus: genomic context, gene expression, and protein-protein interactions
The identified locus (rs803522) was located between the autophagy-related gene 5 (ATG5) and the crystallin β-γ domain containing 1 gene (CRYBG1), being positioned upstream of both genes (Fig. 1). The Encyclopedia of DNA Elements database (21) revealed that the region of high linkage disequilibrium (r2 > 0.60) contained binding sites of the transcription factors STAT3, MYC, and FOS (24), suggesting the locus may play a role in regulating gene expression.
Publicly available data (22) indicate ATG5 and CRYBG1 are expressed in both VF and SF (24) and in several alternative transcript forms. Also based on the GTEx project data (22), rs803522 is not associated with mRNA expression of either gene, but these initial explorations could not take into account alternative transcripts or consider possible effects of age, sex, and adiposity.
Finally, we analyzed protein-protein networks, using previously published experimental and text data [STRING, version10 (23)], which showed that (1) the most significant biological process of the ATG5-related protein network is macroautophagy [false discovery rate–corrected P = 1.4 × 10−39 (24)], and (2) the most significant biological process of the CRYBG1-related protein network is regulation of fat-cell differentiation [false discovery rate–corrected P = 8 × 10−4 (24)].
Discussion
In the current study, we identified a locus of preferential accumulation of visceral (vs subcutaneous) body fat that promotes the development of low-grade systemic inflammation. The association of the locus with adiposity was similar in adolescents and adults, but the association of the locus with related systemic inflammation was stronger in adults than in adolescents.
Genetic basis of visceral (vs subcutaneous) accumulation of body fat has not been studied extensively. This is mainly because VF is quantifiable accurately only with imaging techniques, such MRI or CT (25). One previous GWAS of 10,557 middle-aged and older individuals identified a locus of VF and SF at LYPLAL1 (rs11118316; P = 3.1 × 10−9) (9). In our study of adolescents and middle-aged adults, this locus was not associated with VFSF, VFTBF, VF, SF, or TBF (P = 0.37, 0.54, 0.87, 0.72, and 0.95, respectively). Interestingly, despite the preferential accumulation of body fat viscerally in males vs females, rs11118316 and the locus identified in the current study (rs803522) did not vary by sex (9).
The locus we identified was located near ATG5, which modulates adipocyte size and macrophage polarization (26–28). ATG5 encodes a key molecule of autophagy, which is a cellular process of the autophagosome-lysosome degradation of cytoplasm that is involved in the maintenance of cellular energy balance and homeostasis; it removes damaged or senescent organelles to maintain basal energy balance and mobilizes intracellular nutrients (including lipids) to meet energy requirements in the case of nutrient deficiency (29–33). ATG5 is involved in the early stages of autophagosome formation (34).
Regarding a role of ATG5 in regulating accumulation of body fat, it has been shown in mice that a global overexpression of Atg5 decreases body adiposity (and prolongs lifespan) (26), and adipocyte-specific deletion of Atg5 increases body adiposity through its influence on adipocytes’ capacity to store vs burn intracellular lipids (27). The latter phenotype is also seen with adipocyte-specific deletion of Atg7, which is a molecular partner of ATG5 (35). Furthermore, in humans (as well as in mice), ATG5 expression in adipose tissue is higher in obese vs lean individuals (36–38) and decreases in obese and increases in lean individuals in response to caloric restriction (or bariatric surgery) (38). Moreover, a carefully conducted study of ∼400 individuals with paired samples of VF and SF showed that ATG5 expression is higher in VF vs SF, and that this difference is more pronounced in obese vs lean individuals, and among obese individuals, in viscerally vs subcutaneously obese individuals (36). The GTEx data (22) explored in the current study do not to confirm the VF vs SF differential expression of ATG5 (24), but closer examination that would consider overall body adiposity and relative quantities of VF and SF was not possible.
Regarding a role of ATG5 in modulating adiposity-related systemic inflammation, it has been shown that ATG5 influences macrophage polarization (39), with macrophage-specific Atg5 deletion promoting a proinflammatory phenotype of this cell (28). This may be relevant to the results of the current study, because macrophage infiltration and polarization toward a proinflammatory phenotype are key to the involvement of adipose tissue in obesity-related systemic inflammation (2–4). VF shows a greater infiltration by macrophages than SF (2–4).
Based on the aforementioned previous research, we envision that the major allele of the locus identified in the current could downregulate ATG5 and thus enhance both visceral adiposity and systemic inflammation. Furthermore, the other gene located close to the locus identified in the current study was CRYBG1, which encodes an actin cytoskeleton-binding protein, studied previously as a tumor suppressor gene in cancer (40). To our knowledge, CRYBG1 has not been investigated for a role in regulating adiposity; however, our exploratory analysis of protein-protein interactions identified regulation of fat-cell differentiation as the most important biological process of the CRYBG1-related protein network.
Finally, the locus reported here was identified in a population isolate of French-Canadian origin (11). It was not significant in a previous GWAS carried out in a cohort of broader European ancestry (9). Additional research is required to test if the locus is specific to the French-Canadian population isolate.
In summary, we identified a genetic locus of preferentially visceral accumulation of body fat and related systemic inflammation. The effect of the locus on VF is already seen in adolescence, whereas the adverse health consequences of VF accumulation are only observed in adulthood.
Acknowledgments
We thank the following individuals for their contributions in data acquisition and management in the Saguenay Youth Study (SYS): Manon Bernard (database architect, The Hospital for Sick Children), Hélène Simard and her team of research assistants (CÉGEP de Jonquière), Jacynthe Tremblay and her team of research nurses (Chicoutimi Hospital), and Diane Brisson and her team of research nurses (ECOGENE-21). We thank all participants who took part in the Saguenay Youth Study.
Financial Support: The Saguenay Youth Study has been funded by the Canadian Institutes of Health Research (T.P., Z.P.), the National Institutes of Health (Z.P.), Heart and Stroke Foundation of Canada (Z.P.), and the Canadian Foundation for Innovation (Z.P.). C.S. is funded by the SickKids Research Institute.
Disclosure Summary: The authors have nothing to disclose.
Glossary
Abbreviations:
- ATG5
autophagy-related gene 5
- CRP
C-reactive protein
- CRYBG1
crystallin β-γ domain containing 1 gene
- GWAS
genome-wide association study
- SF
subcutaneous fat
- SNP
single nucleotide polymorphism
- SYS
Saguenay Youth Study
- TBF
total body fat
- VF
visceral fat
- VFSF
visceral fat adjusted for subcutaneous fat
- VFTBF
visceral fat adjusted for total body fat
References
- 1. Olshansky SJ, Passaro DJ, Hershow RC, Layden J, Carnes BA, Brody J, Hayflick L, Butler RN, Allison DB, Ludwig DS. A potential decline in life expectancy in the United States in the 21st century. N Engl J Med. 2005;352(11):1138–1145. [DOI] [PubMed] [Google Scholar]
- 2. Pou KM, Massaro JM, Hoffmann U, Vasan RS, Maurovich-Horvat P, Larson MG, Keaney JF Jr, Meigs JB, Lipinska I, Kathiresan S, Murabito JM, O’Donnell CJ, Benjamin EJ, Fox CS. Visceral and subcutaneous adipose tissue volumes are cross-sectionally related to markers of inflammation and oxidative stress: the Framingham Heart Study. Circulation. 2007;116(11):1234–1241. [DOI] [PubMed] [Google Scholar]
- 3. Fontana L, Eagon JC, Trujillo ME, Scherer PE, Klein S. Visceral fat adipokine secretion is associated with systemic inflammation in obese humans. Diabetes. 2007;56(4):1010–1013. [DOI] [PubMed] [Google Scholar]
- 4. Harman-Boehm I, Blüher M, Redel H, Sion-Vardy N, Ovadia S, Avinoach E, Shai I, Klöting N, Stumvoll M, Bashan N, Rudich A. Macrophage infiltration into omental versus subcutaneous fat across different populations: effect of regional adiposity and the comorbidities of obesity. J Clin Endocrinol Metab. 2007;92(6):2240–2247. [DOI] [PubMed] [Google Scholar]
- 5. Tchkonia T, Thomou T, Zhu Y, Karagiannides I, Pothoulakis C, Jensen MD, Kirkland JL. Mechanisms and metabolic implications of regional differences among fat depots. Cell Metab. 2013;17(5):644–656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Kaess BM, Pedley A, Massaro JM, Murabito J, Hoffmann U, Fox CS. The ratio of visceral to subcutaneous fat, a metric of body fat distribution, is a unique correlate of cardiometabolic risk. Diabetologia. 2012;55(10):2622–2630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Chau YY, Bandiera R, Serrels A, Martínez-Estrada OM, Qing W, Lee M, Slight J, Thornburn A, Berry R, McHaffie S, Stimson RH, Walker BR, Chapuli RM, Schedl A, Hastie N. Visceral and subcutaneous fat have different origins and evidence supports a mesothelial source. Nat Cell Biol. 2014;16(4):367–375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Pausova Z. Visceral fat and hypertension: sex differences. In: Watson RR, ed. Nutrition in the Prevention and Treatment of Abdominal Obesity. Cambridge, MA: Academic Press Elsevier; 2014:99–112. [Google Scholar]
- 9. Fox CS, Liu Y, White CC, Feitosa M, Smith AV, Heard-Costa N, Lohman K, Johnson AD, Foster MC, Greenawalt DM, Griffin P, Ding J, Newman AB, Tylavsky F, Miljkovic I, Kritchevsky SB, Launer L, Garcia M, Eiriksdottir G, Carr JJ, Gudnason V, Harris TB, Cupples LA, Borecki IB, Harris TB, Cupples LA, Borecki IB; GIANT Consortium; MAGIC Consortium; GLGC Consortium. Genome-wide association for abdominal subcutaneous and visceral adipose reveals a novel locus for visceral fat in women. PLoS Genet. 2012;8(5):e1002695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Shungin D, Winkler TW, Croteau-Chonka DC, Ferreira T, Locke AE, Mägi R, Strawbridge RJ, Pers TH, Fischer K, Justice AE, Workalemahu T, Wu JMW, Buchkovich ML, Heard-Costa NL, Roman TS, Drong AW, Song C, Gustafsson S, Day FR, Esko T, Fall T, Kutalik Z, Luan J, Randall JC, Scherag A, Vedantam S, Wood AR, Chen J, Fehrmann R, Karjalainen J, Kahali B, Liu CT, Schmidt EM, Absher D, Amin N, Anderson D, Beekman M, Bragg-Gresham JL, Buyske S, Demirkan A, Ehret GB, Feitosa MF, Goel A, Jackson AU, Johnson T, Kleber ME, Kristiansson K, Mangino M, Leach IM, Medina-Gomez C, Palmer CD, Pasko D, Pechlivanis S, Peters MJ, Prokopenko I, Stančáková A, Sung YJ, Tanaka T, Teumer A, Van Vliet-Ostaptchouk JV, Yengo L, Zhang W, Albrecht E, Ärnlöv J, Arscott GM, Bandinelli S, Barrett A, Bellis C, Bennett AJ, Berne C, Blüher M, Böhringer S, Bonnet F, Böttcher Y, Bruinenberg M, Carba DB, Caspersen IH, Clarke R, Daw EW, Deelen J, Deelman E, Delgado G, Doney AS, Eklund N, Erdos MR, Estrada K, Eury E, Friedrich N, Garcia ME, Giedraitis V, Gigante B, Go AS, Golay A, Grallert H, Grammer TB, Gräßler J, Grewal J, Groves CJ, Haller T, Hallmans G, Hartman CA, Hassinen M, Hayward C, Heikkilä K, Herzig KH, Helmer Q, Hillege HL, Holmen O, Hunt SC, Isaacs A, Ittermann T, James AL, Johansson I, Juliusdottir T, Kalafati IP, Kinnunen L, Koenig W, Kooner IK, Kratzer W, Lamina C, Leander K, Lee NR, Lichtner P, Lind L, Lindström J, Lobbens S, Lorentzon M, Mach F, Magnusson PK, Mahajan A, McArdle WL, Menni C, Merger S, Mihailov E, Milani L, Mills R, Moayyeri A, Monda KL, Mooijaart SP, Mühleisen TW, Mulas A, Müller G, Müller-Nurasyid M, Nagaraja R, Nalls MA, Narisu N, Glorioso N, Nolte IM, Olden M, Rayner NW, Renstrom F, Ried JS, Robertson NR, Rose LM, Sanna S, Scharnagl H, Scholtens S, Sennblad B, Seufferlein T, Sitlani CM, Smith AV, Stirrups K, Stringham HM, Sundström J, Swertz MA, Swift AJ, Syvänen AC, Tayo BO, Thorand B, Thorleifsson G, Tomaschitz A, Troffa C, van Oort FV, Verweij N, Vonk JM, Waite LL, Wennauer R, Wilsgaard T, Wojczynski MK, Wong A, Zhang Q, Zhao JH, Brennan EP, Choi M, Eriksson P, Folkersen L, Franco-Cereceda A, Gharavi AG, Hedman ÅK, Hivert MF, Huang J, Kanoni S, Karpe F, Keildson S, Kiryluk K, Liang L, Lifton RP, Ma B, McKnight AJ, McPherson R, Metspalu A, Min JL, Moffatt MF, Montgomery GW, Murabito JM, Nicholson G, Nyholt DR, Olsson C, Perry JR, Reinmaa E, Salem RM, Sandholm N, Schadt EE, Scott RA, Stolk L, Vallejo EE, Westra HJ, Zondervan KT, Amouyel P, Arveiler D, Bakker SJ, Beilby J, Bergman RN, Blangero J, Brown MJ, Burnier M, Campbell H, Chakravarti A, Chines PS, Claudi-Boehm S, Collins FS, Crawford DC, Danesh J, de Faire U, de Geus EJ, Dörr M, Erbel R, Eriksson JG, Farrall M, Ferrannini E, Ferrières J, Forouhi NG, Forrester T, Franco OH, Gansevoort RT, Gieger C, Gudnason V, Haiman CA, Harris TB, Hattersley AT, Heliövaara M, Hicks AA, Hingorani AD, Hoffmann W, Hofman A, Homuth G, Humphries SE, Hyppönen E, Illig T, Jarvelin MR, Johansen B, Jousilahti P, Jula AM, Kaprio J, Kee F, Keinanen-Kiukaanniemi SM, Kooner JS, Kooperberg C, Kovacs P, Kraja AT, Kumari M, Kuulasmaa K, Kuusisto J, Lakka TA, Langenberg C, Le Marchand L, Lehtimäki T, Lyssenko V, Männistö S, Marette A, Matise TC, McKenzie CA, McKnight B, Musk AW, Möhlenkamp S, Morris AD, Nelis M, Ohlsson C, Oldehinkel AJ, Ong KK, Palmer LJ, Penninx BW, Peters A, Pramstaller PP, Raitakari OT, Rankinen T, Rao DC, Rice TK, Ridker PM, Ritchie MD, Rudan I, Salomaa V, Samani NJ, Saramies J, Sarzynski MA, Schwarz PE, Shuldiner AR, Staessen JA, Steinthorsdottir V, Stolk RP, Strauch K, Tönjes A, Tremblay A, Tremoli E, Vohl MC, Völker U, Vollenweider P, Wilson JF, Witteman JC, Adair LS, Bochud M, Boehm BO, Bornstein SR, Bouchard C, Cauchi S, Caulfield MJ, Chambers JC, Chasman DI, Cooper RS, Dedoussis G, Ferrucci L, Froguel P, Grabe HJ, Hamsten A, Hui J, Hveem K, Jöckel KH, Kivimaki M, Kuh D, Laakso M, Liu Y, März W, Munroe PB, Njølstad I, Oostra BA, Palmer CN, Pedersen NL, Perola M, Pérusse L, Peters U, Power C, Quertermous T, Rauramaa R, Rivadeneira F, Saaristo TE, Saleheen D, Sinisalo J, Slagboom PE, Snieder H, Spector TD, Stefansson K, Stumvoll M, Tuomilehto J, Uitterlinden AG, Uusitupa M, van der Harst P, Veronesi G, Walker M, Wareham NJ, Watkins H, Wichmann HE, Abecasis GR, Assimes TL, Berndt SI, Boehnke M, Borecki IB, Deloukas P, Franke L, Frayling TM, Groop LC, Hunter DJ, Kaplan RC, O’Connell JR, Qi L, Schlessinger D, Strachan DP, Thorsteinsdottir U, van Duijn CM, Willer CJ, Visscher PM, Yang J, Hirschhorn JN, Zillikens MC, McCarthy MI, Speliotes EK, North KE, Fox CS, Barroso I, Franks PW, Ingelsson E, Heid IM, Loos RJ, Cupples LA, Morris AP, Lindgren CM, Mohlke KL; ADIPOGen Consortium; CARDIOGRAMplusC4D Consortium; CKDGen Consortium; GEFOS Consortium; GENIE Consortium; GLGCICBP International Endogene Consortium; LifeLines Cohort Study; MAGIC Investigators; MuTHER Consortium; PAGE Consortium; ReproGen Consortium. New genetic loci link adipose and insulin biology to body fat distribution. Nature. 2015;518(7538):187–196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Pausova Z, Paus T, Abrahamowicz M, Bernard M, Gaudet D, Leonard G, Peron M, Pike GB, Richer L, Séguin JR, Veillette S. Cohort profile: the Saguenay Youth Study (SYS). Int J Epidemiol. 2017;46(2):e19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Ridker PM. C-reactive protein. A simple test to help predict risk of heart attack and stroke. Circulation. 2003;108(12):e81–e85. [DOI] [PubMed] [Google Scholar]
- 13. Cornier MA, Després JP, Davis N, Grossniklaus DA, Klein S, Lamarche B, Lopez-Jimenez F, Rao G, St-Onge MP, Towfighi A, Poirier P; American Heart Association; Obesity Committee of the Council on Nutrition, Physical Activity and Metabolism; Council on Arteriosclerosis, Thrombosis and Vascular Biology; Council on Cardiovascular Disease in the Young; Council on Cardiovascular Radiology and Intervention; Council on Cardiovascular Nursing; Council on Epidemiology and Prevention; Council on the Kidney in Cardiovascular Disease; and Stroke Council. Assessing adiposity: a scientific statement from the American Heart Association. Circulation. 2011;124(18):1996–2019. [DOI] [PubMed] [Google Scholar]
- 14. Marchini J, Howie B. Genotype imputation for genome-wide association studies. Nat Rev Genet. 2010;11(7):499–511. [DOI] [PubMed] [Google Scholar]
- 15. Delaneau O, Zagury JF, Marchini J. Improved whole-chromosome phasing for disease and population genetic studies. Nat Methods. 2013;10(1):5–6. [DOI] [PubMed] [Google Scholar]
- 16. Howie BN, Donnelly P, Marchini J. A flexible and accurate genotype imputation method for the next generation of genome-wide association studies. PLoS Genet. 2009;5(6):e1000529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Aulchenko YS, Struchalin MV, van Duijn CM. ProbABEL package for genome-wide association analysis of imputed data. BMC Bioinformatics. 2010;11(1):134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Therneau T. The lmekin function. Available at: https://cran.r-project.org/web/packages/coxme/vignettes/lmekin.pdf. Accessed 12 February 2019.
- 19. Beasley TM, Erickson S, Allison DB. Rank-based inverse normal transformations are increasingly used, but are they merited? Behav Genet. 2009;39(5):580–595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Steen J, Loeys T, Moerkerke B, Vansteelandt S. medflex: An R package for flexible mediation analysis using natural effect models. J Stat Softw. 2017;76(11):1–46. [Google Scholar]
- 21. ENCODE Project Consortium. An integrated encyclopedia of DNA elements in the human genome. Nature. 2012;489(7414):57–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Lonsdale J, Thomas J, Salvatore M, Phillips R, Lo E, Shad S, Hasz R, Walters G, Garcia F, Young N, Foster B, Moser M, Karasik E, Gillard B, Ramsey K, Sullivan S, Bridge J, Magazine H, Syron J, Fleming J, Siminoff L, Traino H, Mosavel M, Barker L, Jewell S, Rohrer D, Maxim D, Filkins D, Harbach P, Cortadillo E, Berghuis B, Turner L, Hudson E, Feenstra K, Sobin L, Robb J, Branton P, Korzeniewski G, Shive C, Tabor D, Qi L, Groch K, Nampally S, Buia S, Zimmerman A, Smith A, Burges R, Robinson K, Valentino K, Bradbury D, Cosentino M, Diaz-Mayoral N, Kennedy M, Engel T, Williams P, Erickson K, Ardlie K, Winckler W, Getz G, DeLuca D, MacArthur D, Kellis M, Thomson A, Young T, Gelfand E, Donovan M, Grant G, Mash D, Marcus Y, Basile M, Liu J, Zhu J, Tu Z, Cox NJ, Nicolae DL, Gamazon ER, Kyung H, Konkashbaev A, Pritchard J, Stevens M, Flutre T, Wen X, Dermitzakis T, Lappalainen T, Guigo R, Monlong J, Sammeth M, Koller D, Battle A, Mostafavi S, McCarthy M, Rivas M, Maller J, Rusyn I, Nobel A, Wright F, Shabalin A, Feolo M, Sharopova N, Sturcke A, Paschal J, Anderson JM, Wilder EL, Derr LK, Green ED, Struewing JP, Temple G, Volpi S, Boyer JT, Thomson EJ, Guyer MS, Ng C, Abdallah A, Colantuoni D, Insel TR, Koester SE, Little AR, Bender PK, Lehner T, Yao Y, Compton CC, Vaught JB, Sawyer S, Lockhart NC, Demchok J, Moore HF; GTEx Consortium The Genotype-Tissue Expression (GTEx) project. Nat Genet. 2013;45(6):580–585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Szklarczyk D, Franceschini A, Wyder S, Forslund K, Heller D, Huerta-Cepas J, Simonovic M, Roth A, Santos A, Tsafou KP, Kuhn M, Bork P, Jensen LJ, von Mering C. STRING v10: protein-protein interaction networks, integrated over the tree of life. Nucleic Acids Res. 2015;43(D1):D447–D452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Shin J, Syme C, Wang D, Richer L, Pike GB, Gaudet D, Paus T, Pausova Z Data from: Novel genetic locus of visceral fat and systemic inflammation. figshare 2019. Deposited 11 March 2019. 10.6084/m9.figshare.7827923.v3. [DOI] [PMC free article] [PubMed]
- 25. Hu HH, Nayak KS, Goran MI. Assessment of abdominal adipose tissue and organ fat content by magnetic resonance imaging. Obes Rev. 2011;12(5):e504–e515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Pyo JO, Yoo SM, Ahn HH, Nah J, Hong SH, Kam TI, Jung S, Jung YK. Overexpression of Atg5 in mice activates autophagy and extends lifespan. Nat Commun. 2013;4(1):2300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Altshuler-Keylin S, Shinoda K, Hasegawa Y, Ikeda K, Hong H, Kang Q, Yang Y, Perera RM, Debnath J, Kajimura S. Beige adipocyte maintenance is regulated by autophagy-induced mitochondrial clearance. Cell Metab. 2016;24(3):402–419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Razani B, Feng C, Coleman T, Emanuel R, Wen H, Hwang S, Ting JP, Virgin HW, Kastan MB, Semenkovich CF. Autophagy links inflammasomes to atherosclerotic progression. Cell Metab. 2012;15(4):534–544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Kim KH, Lee MS. Autophagy--a key player in cellular and body metabolism. Nat Rev Endocrinol. 2014;10(6):322–337. [DOI] [PubMed] [Google Scholar]
- 30. Moulis M, Vindis C. Autophagy in metabolic age-related human diseases. Cells. 2018;7(10):149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Fernández AF, Sebti S, Wei Y, Zou Z, Shi M, McMillan KL, He C, Ting T, Liu Y, Chiang WC, Marciano DK, Schiattarella GG, Bhagat G, Moe OW, Hu MC, Levine B. Disruption of the beclin 1-BCL2 autophagy regulatory complex promotes longevity in mice [published correction appears in Nature. 2018;561(7723):E30] Nature. 2018;558(7708):136–140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Nakamura S, Yoshimori T. Autophagy and longevity. Mol Cells. 2018;41(1):65–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Singh R, Kaushik S, Wang Y, Xiang Y, Novak I, Komatsu M, Tanaka K, Cuervo AM, Czaja MJ. Autophagy regulates lipid metabolism. Nature. 2009;458(7242):1131–1135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Füllgrabe J, Klionsky DJ, Joseph B. The return of the nucleus: transcriptional and epigenetic control of autophagy. Nat Rev Mol Cell Biol. 2014;15(1):65–74. [DOI] [PubMed] [Google Scholar]
- 35. Singh R, Xiang Y, Wang Y, Baikati K, Cuervo AM, Luu YK, Tang Y, Pessin JE, Schwartz GJ, Czaja MJ. Autophagy regulates adipose mass and differentiation in mice. J Clin Invest. 2009;119(11):3329–3339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Kovsan J, Blüher M, Tarnovscki T, Klöting N, Kirshtein B, Madar L, Shai I, Golan R, Harman-Boehm I, Schön MR, Greenberg AS, Elazar Z, Bashan N, Rudich A. Altered autophagy in human adipose tissues in obesity. J Clin Endocrinol Metab. 2011;96(2):E268–E277. [DOI] [PubMed] [Google Scholar]
- 37. Kosacka J, Kern M, Klöting N, Paeschke S, Rudich A, Haim Y, Gericke M, Serke H, Stumvoll M, Bechmann I, Nowicki M, Blüher M. Autophagy in adipose tissue of patients with obesity and type 2 diabetes. Mol Cell Endocrinol. 2015;409:21–32. [DOI] [PubMed] [Google Scholar]
- 38. Nuñez CE, Rodrigues VS, Gomes FS, Moura RF, Victorio SC, Bombassaro B, Chaim EA, Pareja JC, Geloneze B, Velloso LA, Araujo EP. Defective regulation of adipose tissue autophagy in obesity. Int J Obes. 2013;37(11):1473–1480. [DOI] [PubMed] [Google Scholar]
- 39. Ye X, Zhou XJ, Zhang H. Exploring the role of autophagy-related gene 5 (ATG5) yields important insights into autophagy in autoimmune/autoinflammatory diseases. Front Immunol. 2018;9:2334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Haffner MC, Esopi DM, Chaux A, Gürel M, Ghosh S, Vaghasia AM, Tsai H, Kim K, Castagna N, Lam H, Hicks J, Wyhs N, Biswal Shinohara D, Hurley PJ, Simons BW, Schaeffer EM, Lotan TL, Isaacs WB, Netto GJ, De Marzo AM, Nelson WG, An SS, Yegnasubramanian S. AIM1 is an actin-binding protein that suppresses cell migration and micrometastatic dissemination. Nat Commun. 2017;8(1):142. [DOI] [PMC free article] [PubMed] [Google Scholar]