Abstract
Type III interferon (IFN-λ) is important for innate immune protection at mucosal surfaces and has therapeutic benefit against influenza A virus (IAV) infection. However, the mechanisms by which IFN-λ programs adaptive immune protection against IAV are undefined. Here we found that IFN-λ signaling in dendritic cell (DC) populations was critical for the development of protective IAV-specific CD8+ T cell responses. Mice lacking the IFN-λ receptor (Ifnlr1–/–) had blunted CD8+ T cell responses relative to wildtype and exhibited reduced survival after heterosubtypic IAV re-challenge. Analysis of DCs revealed IFN-λ signaling directed the migration and function of CD103+ DCs for development of optimal anti-viral CD8+ T cell responses, and bioinformatic analyses identified IFN-λ regulation of a DC IL-10 immunoregulatory network. Thus, IFN-λ serves a critical role in bridging innate and adaptive immunity from lung mucosa to lymph nodes to program DCs to direct effective T cell immunity against IAV.
Introduction
Type III interferons (IFN-λ) are members of the interferon (IFN) family and critical mediators of antiviral defenses1, 2, 3. IFN-λ family members have functions similar to type I IFN, and are structurally related to the IL-10 cytokine family, utilizing the IL-10 receptor as a part of their heterodimeric receptor with IFN-λR1 chain4. Although IFN-λ has overlapping functions with type I IFNs, several studies have revealed a unique, non-redundant role for IFN-λ in mediating protection against disease and pathology in the liver and at mucosal and barrier surfaces5, 6, 7, 8, 9, 10.
Influenza A virus (IAV) is a respiratory pathogen that causes seasonal epidemics and also presents the potential for major pandemic outbreaks and global disease through continual emergence and re-emergence from avian reservoirs. The high incidence of IAV infection, coupled with low vaccine effectiveness during the 2017–2018 season, highlights the necessity to gain a better understanding of the processes that regulate the immune responses against infection to guide efforts aimed at developing effective therapeutic and vaccine adjuvants to protect against infection11.
As an innate immune and immune-regulatory cytokine, IFN-λ has the capacity to direct protective immunity against IAV infection. When administered intransally following IAV challenge, IFN-λ restricts IAV infection without inducing excessive, tissue damaging inflammation typically associated with type I IFN administration12, 13, 14, 15. The protective and non-injurious effect of IFN-λ are attributed in part to the selective expression of IFN-λR1 compared to the broad expression of the type I IFN receptor (IFNAR), wherein IFN-λR1 is selectively expressed on epithelial cells, neutrophils, plasmacytoid dendritic cells and potentially natural killer (NK) cells that contribute to IAV immunity12, 13, 16, 17, 18, 19, 20.
Although IFN-λ directs innate immune defenses that can restrict IAV infection, the contribution of IFN-λ signaling to adaptive immunity is less clear21, 22, 23. IFN-λ has been shown to enhance TH1 T cell responses in the lung, suggesting it has a role in regulating immune polarization during IAV infection21, 22. Despite these observations, the direct function of IFN-λ on programming and maintenance of adaptive immune responses during IAV infection remains largely unknown. Here, we utilized murine infection models to investigate the role of IFN-λ (of which IFN-λ2 and IFN-λ3 are conserved between mice and humans) in adaptive immunity against IAV. We demonstrate that IFN-λ signaling is critical for the development of virus-specific CD8+ T cells and generation of protective immunity that is able to provide cross-protection against heterosubtypic IAV re-challenge. We found that the blunted CD8+ T cell response in IFN-λ-deficient (Ifnlr1–/–) mice is due to a defect in the activation, migration, and function of dendritic cells (DCs) linked with aberrant expression of a DC-derived IL-10 immunoregulatory gene network. IFN-λ specifically programs migratory, antigen presenting CD103+ DCs to mediate CD8+ T cell activation for broad protection against IAV infection, and for heterosubtypic virus challenge that underscores contemporary IAV outbreaks in humans. Thus, IFNλ, through the recruitment and function of CD103+ DCs and regulation of IL-10 production, is an essential component of effective immunity against IAV infection and for cross-protection against heterosubtypic IAV emergence.
Results
IFN-λ signaling is required to generate protective memory T cell responses
To assess the role of IFN-λ signaling downstream of IFN-λR1 in controlling protection against secondary IAV infection we examined the response to heterosubtypic IAV challenge and determined the ability of Ifnlr1–/– mice to mount a protective homosubtypic and heterosubtypic (T cell-dependent) memory response against IAV infection. We interrogated both homo- and hetero-subtypic re-challenge as neutralizing antibody responses have been demonstrated to effectively prevent homosubtypic IAV infection, but heterosubtypic IAV infection is a common occurrence in humans and other susceptible species during pandemic and seasonal outbreaks caused by exposure to related heterosubtypic IAV strains. It has been established in mouse models of heterosubtypic IAV re-challenge, as well as during human infection, that neutralizing antibody responses are not effective at preventing heterosubtypic IAV infection and disease24, 25, 26. In contrast, CD8+ T cells are critical mediators of protection against heterosubtypic IAV infection while antibodies are dispensible24, 27, 28, 29. As in previous heterosubtypic IAV challenge studies of mice, we mock-infected animals, challenged them with a low dose of murine-adapted IAV A/PR/8/34 (IAV-PR8) that leads to clinical respiratory illness and weight loss with complete survival and recovery30, 31, or challenged with an equivalent infectious dose of recombinant H3N2 IAV containing the HA and NA of A/HK/1/68 and the remaining segments from IAV-PR8 (IAV-X31). The dose of IAV-PR8 utilized consistently lead to a minor, yet statistically significant increase in virus titers, and caused similar weight loss and respiratory illness, but no lethality in wildtype and Ifnlr1–/– mice as described by others13, 15, 32 (Supplementary Fig. 1). On day 45 following initial infection, all mice were challenged with a completely lethal dose of 2000 PFU IAV-PR8 (Fig. 1a), thus modeling IAV cross-exposure as occurs during seasonal epidemics. Both wildtype and Ifnlr1–/– mice succumbed to secondary infection following mock primary infection while prior exposure to IAV-PR8 provided immune memory that protected against disease (Figure 1b–d). Antibodies likely contribute to this protection against the homologous 2000 PFU challenge as we observed similar levels of IAV-specific IgM, IgG, IgG1, and IgG2a antibodies in bronchiole alveolar lavage fluid of wildtype and Ifnlr1–/– mice, and a significant increase in IgA in Ifnlr1–/–, on day 35 following sublethal IAV-PR8 infection (Supplementary Figure 2). Prior infection with IAV-X31 resulted in weight loss and illness but no change in overall susceptibility to IAV-PR8 heterosubtypic infection in wildtype mice (Figure 1b–d). Importantly, Ifnlr1–/– mice previously infected with IAV-X31 were increasingly susceptible to IAV-PR8 heterosubtypic infection and exhibited a significant reduction in survival concomitant with accelerated and prolonged weight loss and disease symptoms, with a significant increase in illness score in Ifnlr–/– mice on days 2–5 post heterosubtypic IAV challenge compared to wildtype (Figure 1b–d). These results indicate that IFN-λ signaling is critical in generating T cell-mediated cross-protective immunity against heterologous IAV infection.
Figure 1. IFN-λ signaling is critical for protection against heterologous IAV challenge.
a. wildtype (WT) (solid lines) and Ifnlr1–/– (dashed lines) mice were mock treated (green lines) or infected intranasally (i.n.) with 40 PFU PR8 (blue lines) or X31 (a mouse adapted H3N2 strain containing HA and NA from A/HK/3/68 and all other gene segments from PR8, red lines). 45 days post infection (p.i.), mice were challenged i.n. with high dose (2000 PFU) PR8. Following high dose challenge, weight (a), illness (b), and survival (c) were monitored for 14 days. Dashed line in b indicates presence of respiratory illness. For a-c data from two pooled experiments shown. Error bars show mean ± SEM. For Mock-Mock n=2 mice/group, Mock-PR8 n=8 (WT) or 9 (Ifnlr1–/–) mice/group, PR8-PR8 n=4 (WT) or 9 (Ifnlr1–/–) mice/group, X31-PR8 n=21 (Ifnlr1–/–) or 22 (WT) mice/group. Significance comparing WT X31-PR8 to WT PR8-PR8 and Ifnlr1–/– X31-PR8 to WT X31-PR8 was determined using log-rank test. n.s. indicates p=0.3720, * indicates p<0.01
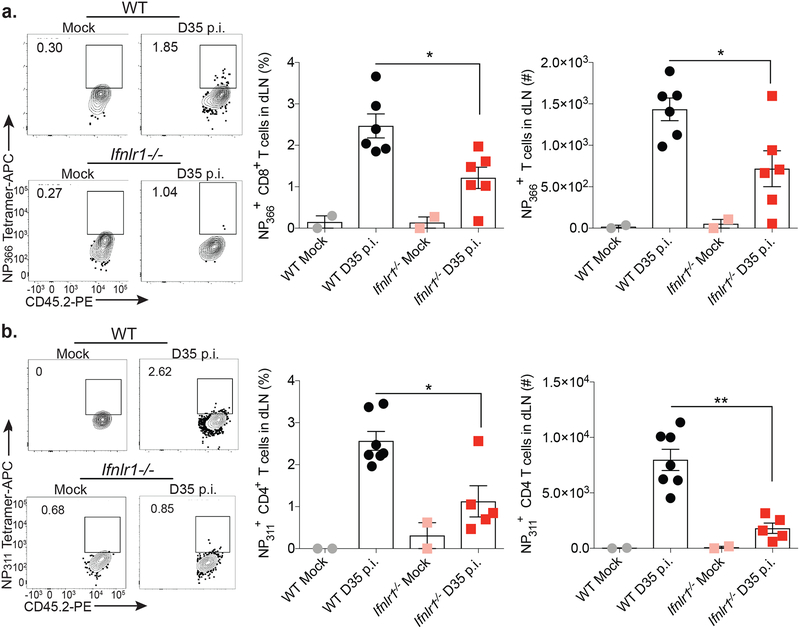
To determine whether populations of IAV-specific T cells were functionally altered in Ifnlr1–/– mice compared to wildtype at a memory time point (day 35 post infection) following IAV infection, we challenged wildtype and Ifnlr1–/– mice with IAV-PR8 and assessed T cell responses in lung-draining mediastinal lymph nodes (dLN) on day 35 post infection. We observed a significant reduction in the frequency and numbers of IAV-specific CD4+ and CD8+ T cells in dLN of Ifnlr1–/– mice compared to wildtype on day 35 post infection (Figure 2a and 2b). These results suggest that insufficient memory T cells likely account for the enhanced susceptibility of Ifnlr1–/– mice upon heterosubtypic virus secondary challenge.
Figure 2. IFN-λ signaling is critical for memory T cell responses.
WT and Ifnlr1–/– mice were mock- or IAV-infected i.n. with PBS or 40 PFU PR8. On day 35 p.i., lung-draining lymph nodes (dLN) were harvested and IAV NP366-specific CD8+ T cell (a) and NP311-specific CD4+ T cell (b) frequency (% of CD8+ or CD4+ T cells) and numbers were determined by flow cytometry. Data from two, pooled experiments shown. For (a) 4 (Mock) and 12 (WT and Ifnlr1–/– D35 p.i.) mice/group with half as many data points shown as each point shows pooled dLN from two mice. For (b) 4 (Mock), 14 (WT D35 p.i.), or 10 (Ifnlr1–/– D35 p.i.) mice/group with half as many data points shown as each point shows pooled dLN from two mice. For (a) and (b) error bars represent mean ± SEM. Significance was determined using one-way ANOVA followed by Tukey’s multiple comparisons test between WT D35 and Ifnlr1–/– D35. * indicates p<0.05, ** indicates p<0.001
IFN-λ signaling is essential for the development of the IAV-specific CD8+ T cell response
To determine the role of IFN-λ signaling in programming the effector T cell response against IAV infection, we measured the IAV-specific CD8+ T cell response in the lungs of wildtype and Ifnlr1–/– mice following IAV infection. On day 9 post infection, the frequency and numbers of CD8+ T cells specific to the IAV immunodominant NP366-epitope were significantly reduced in Ifnlr1–/– animals compared to wildtype (Figure 3a). Additionally, the lung CD8+ T cells displayed reduced amounts of IFN-γ and TNF production in Ifnlr1–/– compared to wildtype mice during IAV infection (Figure 3b–d). A similar, significant reduction in the PA224-specific response of Ifnlr1–/– mice infected with IAV was also observed (data not shown). In contrast, no difference in the frequency or numbers of IAV-specific CD4+ T cells or their cytokine production following stimulation with NP311 IAV peptide ex vivo was found between wildtype and Ifnlr1–/– mice on day 9 post infection (Supplementary Figure 3). Together, these findings identify a requirement for IFN-λ signaling in the generation of an optimal IAV-specific CD8+ T cell response.
Figure 3. IFN-λ signaling is required for viral control and generation of the CD8+ T cell response during IAV infection.
a-d. WT and Ifnlr1–/– mice were mock- or IAV-infected i.n. with PBS or 40 PFU PR8 expressing a 2W epitope, respectively. a. Lungs were harvested on day 9 p.i. and frequency (% of CD8+ T cells) and numbers IAV-specific CD8+ T cells were determined by flow cytometry. b-d. On day 9 p.i., whole lung homogenates were stimulated with media, IAV (NP366) peptide, or PMA + ionomycin for 6 hrs in the presence of brefeldin A. Following stimulation, IFN-γ (b) and TNF (c) single- or co-production (d) was determined by flow cytometry. For a-b, two pooled independent experiments are shown. For (a) n=2 (Ifnlr1–/– Mock), 3 (WT Mock), 5 (WT-IAV), or 7 (Ifnlr1–/–-IAV) mice/group. For (b) n=6 (WT-IAV), or 8 (Ifnlr1–/–-IAV) mice/group. For a-d error bars represent mean ± SEM. Significance was determined using one-way ANOVA followed by Tukey’s multiple comparisons test between WT-IAV and Ifnlr1–/–-IAV.
IFN-λ signaling in DCs is important for function and priming of IAV-specific CD8+ T cells
Previous studies have shown that T cells lack IFN-λ receptor expression and do not respond directly to IFN-λ23. However, given the defect in the IAV-specific CD8+ T cell response in Ifnlr1–/– mice, we opted to assess whether IFN-λ signaling, intrinsic or extrinsic to CD8+ T cells, was responsible for the aberrant CD8+ T cell response in the Ifnlr1–/– mice. Stimulation of lung homogenates with phorbol 12-myristate 13-acetate (PMA) and ionomycin yielded equivalent amounts of IFN-γ and TNF produced by wildtype and Ifnlr1–/– CD8+ T cells (Figure 3b–d), suggesting that CD8+ T cells are fully capable of producing these cytokines in the absence of Ifnlr1 and IFN-λ signaling. Further, we purified CD8+ T cells from the spleens of wildtype and Ifnlr1–/– mice and stimulated them with plate bound α-CD3/CD28. Both wildtype and Ifnlr1–/– CD8+ T cells underwent robust cell division and expressed the activation markers CD44 and CD69 when stimulated ex vivo (Figure 4a). Thus, the defect in the activation of the IAV-specific CD8+ T cell response in Ifnlr1–/– is not T cell-intrinsic.
Figure 4. DCs, but not CD8+ T cells, lacking Ifnlr1 have intrinsic defects.
a. WT and Ifnlr1–/– splenic CD8+ T cells were CFSE-labeled, stimulated for 72 hrs with α-CD3/CD28, and cell division and CD44 and CD69 upregulation were determined. Data representative of two independent experiments. b. 106 CFSE-labeled, P14 transgenic CD8+ T cells were transferred to WT and Ifnlr1–/– mice and infected i.n. with 40 PFU PR8-GP33. dLN were harvested 84 hrs p.i., to determine division of P14 cells. Two pooled independent experiments shown where each data point=dLN pooled from two mice. n=6 (Ifnlr1–/–) or 8 (WT) mice/group. c-e. WT and Ifnlr1–/– BMDCs were infected with PR8 (MOI=0.5) for 22 hrs. c. Ifnlr1, Ifnar1, and Il10rb message was determined by qRT-PCR. Two pooled independent experiments shown. n= 3 (Mock) or 4 (PR8). d. CD40 expression was determined by flow cytometry. Three pooled independent experiments shown. n=3 (WT) or 4 (Ifnlr1–/–) b-d. Bars show mean ± SEM. e. IAV NP message was determined by qRT-PCR. Two pooled independent experiments shown. Bars show mean ± SEM. n=2 (Mock), 4 (IAV) f. WT and Ifnlr1–/– BMDCs were incubated with 10 ug/ml DQ-Ova for 30 min, 1 hr, or 2 hr at 37° C or 2 hr on ice, and uptake of DQ-Ova was measured using flow cytometry. One representative of three independent experiments shown where n=3. Mean ± SD shown. For b, d, and f statistical significance determined using an unpaired two-sided t-test comparing WT and Ifnlr1–/–. For c significance determined using one-way ANOVA and Tukey’s multiple comparisons test. * indicates p<0.05, ** indicates p<0.001, *** indicates p<0001
To verify that the defect in the IAV-specific CD8+ T cell response was due to a T cell-extrinsic effect that impacts T cell activation and functional differentiation, we transferred wildtype, CFSE-labeled, GP33-specific CD8+ T cells expressing a disparate CD45 molecule into wildtype and Ifnlr1–/– mice, and infected the mice with IAV-PR8 containing a GP33 epitope. After 3.5 days in Ifnlr1–/– hosts, the average number of divisions activated GP33-specific CD8+ T cells had undergone was significantly reduced in dLN compared to GP33-specific CD8+ T cells transferred to WT mice (Figure 4b). Together, these results demonstrate that the defect in the CD8+ T cell response against IAV occurs early at the time of antigen priming and is caused by a T cell-extrinsic process in Ifnlr1–/– mice.
IFN-λ signaling in DCs is required for their function and migration to dLN following IAV infection
To determine if a defect in DC function could be responsible for the aberrant CD8+ T cell response in Ifnlr1–/– mice, we evaluated the responses of wildtype and Ifnlr1–/– bone marrow-derived DCs (BMDCs) to several stimuli. We found that BMDCs expressed Ifnlr1 mRNA with increasing levels upon IAV infection in contrast to their levels of Ifnar1 expression, which was similar in both wildtype and Ifnlr1–/– BMDCs irrespective of infection (Figure 4c). Stimulation of BMDCs with 500 ng IFN-λ3 or 10 ng IFNα2 lead to similar phosphorylation of the transcription factor STAT1, which is downstream of IFN-λR1 (Supplementary Figure 4a), and responded to IFN-λ in a dose-dependent manner (Supplementary Figure 4b), indicating that BMDCs respond directly to IFN-λ stimulation. Further, these BMDCs expressed interferon stimulated genes (ISGs) 12 hrs following treatment with IFN-λ3 (Supplemental Figure 4c). However, we found that Ifnlr1–/– BMDCs were unable to increase CD40 surface protein expression following IAV infection (Figure 4d). These changes in activation upon infection were not due to changes in viral burden in BMDCs, as wildtype and Ifnlr1–/– BMDCs expressed similar levels of IAV NP at this time following infection (Figure 4e). Further, Ifnlr1–/– BMDCs ability to uptake and process antigen as measured by DQ ovalbumin (DQ-Ova) fluorescence was reduced compared to wildtype BMDCs (Figure 4f). Thus, IFN-λ signaling is intact in BMDCs, and is essential for DC maturation and function following IAV infection.
To determine if DC responses were defective in vivo in Ifnlr1–/– mice, we assessed the ability of DCs to migrate from the lungs to dLN. 5(6)-Carboxyfluorescein N-hydroxysuccinimidyl ester (CFSE) was administered intranasally to the lungs of wildtype and Ifnlr1–/– mice to label resident cells33. Migration of antigen presenting cells (APCs) from the lungs to dLN following IAV-PR8 infection was monitored by measuring the accumulation of CFSE+ CD11c+ cells in the lung dLN (Figures 5a–5c). Although pulmonary APCs accumulated in dLN of WT mice 24 hrs following IAV infection, the migration of APCs to the lung dLN in Ifnlr1–/– mice was significantly reduced (Figures 5b & 5c). We further assessed the lung to dLN migration of specific DC subsets including inflammatory DCs (iDC), bulk conventional DCs (cDCs), CD4+ DCs, cross-presenting CD8α+ DCs and CD103+ DCs, noting that the latter have been shown to take up antigen in the lung and migrate to dLN to effectively cross-present antigen for activation of CD8+ T cells34, 35. There was no difference in iDC migration between wildtype and Ifnlr1–/– mice but we did observe a significant reduction in accumulation of cDCs in Ifnlr1–/– compared to WT (Figure 5e). We found a significant reduction in migratory CD103+ DCs and cross-presenting CD8α+ DCs in dLN of Ifnlr1–/– mice following IAV infection (Figure 5f & 5g). These findings show that IFN-λ signaling is required for DC function and specific DC subset migration to the dLN following IAV infection.
Figure 5. IFN-λ signaling is essential for APC migration and accumulation of CD103+ DC and CD8α+ DC subsets in dLN during IAV infection.
a-c. WT and Ifnlr1–/– mice were administered CFSE i.n. and infected with 40 PFU PR8 i.n.. 24 hrs post infection, lungs and dLN were harvested. a. CFSE labeling of the lung as determined by flow cytometry. b. CFSE+/CD11c+ cells (upper right quadrant) in dLN determined by flow cytometry. c. Quantification of the frequency of CFSE+/CD11c+ cells in dLN. a-c. Data from two pooled independent experiments shown. n=4 mice/group d-g. WT and Ifnlr1–/– mice were infected with 40 PFU PR8 i.n., and 72 hrs post infection dLN were harvested. The frequency and total number of the following DC populations in dLN was determined by flow cytometry utilizing the gating strategy in d. e. iDCs (CD3−/CD19−/F480−/CD11b+/CD11c+/MHCII+), cDCs (CD3−/CD19−/F480−/CD11b−/CD11c+/MHCII+), f. CD4+ DCs (CD3−/CD19−/F480−/CD11b+/CD11c+/MHCII+/CD8α−/CD4+), CD8α+ DCs (CD3−/CD19−/F480−/CD11b+/CD11c+/MHCII+/CD4−/CD8α+), g. CD103+ DCs (CD3−/CD19−/F480−/CD11b+/CD11c+/MHCII+/CD103+). Data pooled from two independent experiments. n=3 (Mock), 9 (WT IAV), or 7 (Ifnlr1–/– IAV) mice/group. h. WT mice were infected with 40 PFU PR8 i.n.. 72 hrs post infection dLN were harvested and the expression of IFNAR on iDCs, CD8α+ DCs, and CD103+ DCs was determined by flow cytometry. Two pooled independent experiments shown. n=5 mice/group. a-h. Bars show mean ± SEM. Significance determined using one-way ANOVA followed by Tukey’s multiple comparisons test. i. iDCs and CD103+ DCs were sorted from naive lungs of WT mice, RNA was isolated and Ifnar1 and Ifnlr1 message was determined. Data from two individual mice shown. N.D.=not detected. Bars show mean ± SD. * indicates p<0.05, ** indicates p<0.01, *** indicates p<0.0001
To determine if CD103+ DC and CD8α+ DC subsets were functionally and differentially dependent on IFN-λ or type I IFN signaling, we assessed the protein and mRNA level of type I IFN receptor expression of each DC subset. In particular, CD103+ DCs have been described to be resistant to type I IFN signaling36, and therefore postulated that CD103+ DC and CD8α+ DC subsets might be dependent on IFN-λ signaling for their maturation, migration, and antigen presentation functions. Compared to iDCs that are known to respond to type I IFN, IFNAR1 protein levels were reduced on the surface of CD8α+ DCs and CD103+ DCs found in dLN 3 days following IAV-PR8 infection (Figure 5h). We note that a reliable antibody or an immunoassay to detect IFN-λR1 on the surface of cells is not currently available; therefore we utilized RT-qPCR analysis to measure Ifnlr1 gene expression. We found that both sorted iDCs and CD103+ DCs from the lungs of naive mice express Ifnar1 mRNA, whereas Ifnlr1 mRNA expression was detected only in CD103+ DCs (Figure 5i). These results were similar to relative Ifnar1 and Ifnlr1 mRNA expression from lung CD103+ DCs previously defined by microarray analysis and available in the Immunological Genome Project database37. In summary, these data further indicate that IFN-λ signaling induces a central program that directs the migration of lung DCs to accumulate in dLN during acute IAV infection. Thus, IFN-λ might impart a gene expression program in DCs to regulate their function and downstream adaptive immune responses during IAV infection.
IFN-λ signaling is responsible for regulation of an IL-10 immunoregulatory network in DCs
To define the DC gene expression profile associated with IFNλ signaling and response we conducted RNA sequencing analysis of mRNA from cDCs (CD3–/CD19−/MHC-II+CD11b–CD11c+) sorted from dLN of wildtype and Ifnlr1–/– mice on day 4 following IAV infection. This time point corresponded with DC accumulation from the lung to the dLN to prime the CD8+ T cell response. This examination of DCs allowed for identifying differentially expressed (DE) genes within the interface of innate and adaptive immunity that could impart function to program DC actions through response to IFN-λ (Figure 6a). A Venn diagram of significantly induced (upregulated) or suppressed (down-regulated) genes showed overlapping and distinct genes amongst DCs from IAV-infected wildtype and Ifnlr1–/– mice compared to mock-infected controls of each genotype (Figure 6b). This indicates there is a global transcriptional program in Ifnlr1–/– cDCs that is distinct from wildtype cDCs in dLN, demonstrating IFN-λ signaling contributes to the functional programming of the DC responsible for presenting antigen to activate IAV-specific CD8+ T cell responses. A scatter plot showing all genes undergoing significant changes in expression for each genotype compared to mock infection (listed in Supplementary Table 1) indicated that a large number of genes, including a number of interferon stimulated genes (ISGs), were induced to equivalent levels in wildtype and Ifnlr1–/– DC upon IAV infection in vivo (Figure 6c). This similar upregulation of at least a subset of ISGs suggested that it was not simply a defect in general and canonical interferon signaling that was driving the defect observed in Ifnlr1–/– animals. Notably, the upper left quadrant of this scatter plot represented a divergent response of genes that were downregulated in wildtype DCs during IAV infection but upregulated in Ifnlr1–/– DCs. The genes in this quadrant that were highly upregulated in Ifnlr1–/–, but downregulated in wildtype included Loc101055769, Rgs9bp, Tmem246, Wdr31, and Il10. Analysis of genes involved in DC maturation also identified striking differences in transcriptional profiles in Ifnlr1–/– compared to wildtype (Figure 6d). For example, Il10, which was also identified as a gene whose expression was highly divergent between wildtype and Ifnlr1–/–, was of particular interest as the cytokine actions of IL-10 have been recently described to reduce CD8+ T cell antigen sensitivity and function in the context of chronic infection38. Further analyses revealed a non-canonical cell-mediated immune network of genes with IL-10 as a central regulatory hub in Ifnlr1–/– cDCs from dLN (Figure 6e). Many genes in this network were not induced to statistical significance in wildtype cDCs in vivo as indicated by white nodes (Figure 6e). To determine whether the change in Il10 mRNA translated into a difference at the protein level, we infected wildtype and Ifnlr1–/– BMDCs with IAV and assessed IL-10 concentration in culture supernatants. Although BMDCs from wildtype mice did not secrete detectable amounts of IL-10, IL-10 production and/or secretion was detected in Ifnlr1–/– BMDCs following IAV infection (Figure 6f), validating the transcriptional upregulation of Il10 to program an IL-10 response network in dLN cDCs from Ifnlr1–/– mice. Overall, these data indicate that during IAV infection the cDCs in dLN undergo gene induction patterns that are dependent on IFN-λ signaling, and that IFN-λ signaling is essential for the negative regulation of IL-10 and its immunoregulatory network that links with aberrant CD8+ T cell priming and responses during IAV infection.
Figure 6. IFN-λ signaling is essential for APC migration to dLN during IAV infection.
a. WT and Ifnlr1–/– mice were infected with 40 PFU PR8 or administered PBS i.n., and dLN were harvested 96 hrs later. DCs were enriched using MACS separation, and cDCs were sorted using gating strategy shown in Figure 5. RNA was isolated, and mRNA was prepared to perform mRNA sequencing. Following data cleanup, significant changes in gene expression for each genotype were determined compared to their respective mock-infected samples using limma package in R (threshold: log2 fold change ≥ 1.5 and FDR ≤ 0.05) and plotted as shown. b. Venn diagram of genes up- or down-regulated in cDC during IAV infection compared to mock-infected cDCs for each genotype. c. Scatter plots showing genes significantly changed in expression during infection for either genotype compared to their mock. Named genes in dark blue indicated ISGs. Il10 is highlighted in red. d. DC maturation genes significantly up- or down-regulated compared to mock for either genotype were plotted in a heat map. Red=upregulated genes, blue=downregulated genes. e. A non-canonical cell-mediated immune network was identified in Ifnlr1–/– samples and plotted. The same network is shown for WT cDCs. Red=upregulated genes, blue=downregulated genes, white=not significant. a-e. n=3 samples from dLN of two mice pooled for each condition. f. WT and Ifnlr1–/– BMDCs were infected with IAV at an MOI of 0.5 for 24hrs. Supernatants were collected and IL-10 levels were determined by Luminex. n=2 biological replicates from individual mice of one experiment representative of two independent experiments. Bars show mean ± SD.
IFN-λ signaling in CD11c+ cells is required for the generation of pulmonary IAV-specific CD8+ T cells
Our observations support a pivotal role for IFN-λ signaling in DCs via regulation of a unique transcriptional profile and gene actions that direct DC function for the proper priming of CD8+ T cells against IAV infection. To determine if cDC-specific IFN-λ signaling (including CD8α+ DCs and CD103+ DCs) was required to impart the IAV-specific CD8+ T cell response, we examined the response to IAV infection in conditional knockout mice where IFN-λR1 expression in CD11c+ cells is absent (Ifnlr1fl/flItgax-cre)39. Upon IAV-PR8 infection there was no difference in weight loss between Ifnlr1fl/fl wildtype controls and Ifnlr1fl/flItgax-cre, similar to full Ifnlr1–/– (Figure 7a). Loss of IFN-λR1 specifically in CD11c+ cells phenocopied global Ifnlr1–/– mice, as we found a significant reduction in the IAV-specific CD8+ T cell response relative to wildtype controls (Figure 7b). There was no defect in the CD4+ T cell response between Ifnlr1fl/fl wildtype controls and Ifnlr1fl/flItgax-cre mice, similar to wildtype and Ifnlr1–/– mice (Figure 7c). In addition to DCs, alveolar macrophages, splenic macrophage subsets, NK cells, and to a much lesser extent a few other immune cell subsets also express CD11c40. To address the potential impact of the deletion of Ifnlr1 from macrophages that also occurs within the Ifnlr1fl/flItgax-cre, mice, we utilized Ifnlr1fl/flLyz2-cre mice specifically lacking Ifnlr1 in macrophages (and neutrophils), and evaluated the CD8+ T cell response to IAV infection. Ifnlr1fl/flLyz2-cre mice had CD4+ and CD8+ T cell responses similar to Ifnlr1fl/fl wildtype controls (Supplementary Figure 5). Thus, IFN-λ signaling in DCs but not macrophages serves to direct their actions to program CD8+ T cells against IAV infection. Taken together, our study reveals an important requirement for IFN-λ signaling in programming DCs for activation, maturation, and migration from lung to dLN during IAV infection to prime an optimal CD8+ T cell response that is required for effective immune memory and protection against heterosubtypic IAV infection (Supplementary Figure 6).
Figure 7. IFN-λ signaling in CD11c+ cells is required for generation of the CD8+ T cell response during IAV infection.
Ifnlr1fl/fl and Ifnlr1fl/flItgax-cre mice were administered 40 PFU PR8 containing a 2W epitope i.n. and weight loss (a) was monitored. Symbols show mean and error bars represent SEM. Lungs were harvested on day 9 post infection and frequency (% of CD8+ or CD4+ T cells) and numbers of PA224-specific CD8+ T cells (b) and 2W1S-specific CD4+ T cells (c) were assessed by flow cytometry. For (b) and (c) two pooled, independent experiments shown. n=9 (Ifnlr1fl/flItgax-cre) or 10 (Ifnlr1fl/fl) mice/group. Bars show mean ± SEM. Significance was determined using unpaired two-sided t-test. * indicates p <0.05, ** indicates p<0.01
Discussion
Herein we investigated the role of IFN-λ signaling in programming adaptive immune responses, in particular we examined DC function and CD8+ T cell-mediated immunity that is key for clearance of IAV41. Our results highlight the critical function of IFN-λ signaling in DCs at the innate/adaptive immune interface required for activation, function, and maintenance of IAV-specific CD8+ T cell responses critical for heterosubtypic IAV protection. Our studies reveal that IFN-λ signaling in DCs is essential for optimal antigen uptake and processing, upregulation of costimulatory molecules, regulation of IL-10 production by DCs, DC migration to dLN, and CD8+ T cell activation. Intriguingly, we found that IFN-λ actions are specific to DC subsets (CD8α+ DCs and CD103+ DCs) important for migration to the dLN and regulation of CD8+ T cells in vivo during IAV infection. However, multiple DC subsets are capable of priming IAV-specific CD4+ T cell responses42, 43, which likely explains why we observed similar effector CD4+ T cell response in the lungs of wildtype and Ifnlr1–/– mice in the lungs during IAV infection. This outcome suggests a unique requirement of IFN-λ signaling for the optimal migration and function of CD103+ DCs to program effective CD8+ T cell-mediated protective immunity. Thus our results uncover a unique and non-redundant role for IFNλ signaling in adaptive immunity.
Coupled with publicly available microarray data, our results reveal that CD103+ DCs express low level of IFNAR compared to other DC subtypes and are resistant to type I IFN36, 37. Conversely, lung CD103+ DCs express high Ifnlr1 levels relative to other DC subsets37. Unfortunately, specific and sensitive antibodies required to measure the surface expression of IFN-λR1 are currently unavailable, but our results indicate DCs are able to respond directly to IFN-λ3 (an IFN-λ subtype conserved in both mice and humans). We hypothesize CD103+ DCs rely upon IFN-λ signaling for optimal activation due to higher levels of IFN-λ receptor expression and their specific defect in cell migration to dLN during IAV infection in vivo in Ifnlr1–/– compared to other DC subsets. This specific responsiveness of CD103+ DC subsets to IFN-λ signaling represents a novel potential target for development of IFNλ-based adjuvants to IAV vaccination.
We identified IL-10 and an Il10 immunoregulatory gene network containing Rel and Il27 that was specifically in cDCs in Ifnlr1–/–, distinct from canonical IFN signaling, and linked with the defective CD8+ T cell response against IAV infection in vivo. Rel/NF-kB signaling in DC has been shown to lead to upregulation of IL-27 and subsequently IL-10, which performs an immunoregulatory function in shaping the CD4+ T cell repertoire44. Additionally, IL-10 has recently been shown to direct immunosuppressive actions to reduce CD8+ T cell activation and function in the context of chronic viral infection38, and IL-10 production from DCs was linked to T cell suppression in response to anti-tumor vaccination45. In the context of IAV infection, we propose a model in which IFN-λ signaling regulates and restricts IL-10 production in antigen cross-presenting CD103+ DCs to facilitate optimal activation of CD8+ T cell responses that are critical for clearance of infection.
The aberrant/immunosuppressive function of CD103+ DCs and reduced CD8+ T cell responses in Ifnlr1–/– mice during IAV infection ultimately culminated in enhanced mortality during heterosubtypic IAV re-challenge. This heterosubtypic IAV challenge model is dependent upon CD8+ T cell responses for protection and mimics the mode of protection that can occur in the human population during the emergence of novel, recombined IAV strains, such as the pandemic H1N1 strain in 200926, 46. In addition to this natural frequency of occurrence in the human population, the emerging threat of avian influenza IAV highlight the necessity for developing a comprehensive understanding of host factors that favor development of protective immunity. Given the importance of CD103+ DCs in delivery of IAV antigen for cross-priming of CD8+ T cell responses30, 34, 35, and the requirement for IFN-λ to impart DCs function, our results suggest IFN-λ could have application as a immune adjuvant to IAV vaccination. Future studies will focus on revealing the kinetics and requirements for development of CD8+ and CD4+ T cell memory responses in Ifnlr1–/– and in the context of activation during vaccination in conjunction with IFN-λ. Although several recent studies have highlighted the potential for IFNλ to impart superior protective immunity over type I IFN against IAV infection12, 13, 14, our data reveals that IFN-λ signaling is essential in programming DCs function during IAV infection. In particular, our findings support a model in which IFN-λ signaling functions to limit IL-10 immunosuppressive actions, thus facilitating DCs function for enhanced CD8+ T cell protection against heterosubtypic IAV challenge. Overall, our study highlights the important role of IFN-λ signaling to program an innate-adaptive immune interface in which an IFN-λ-DC-CD8+ T cell axis elicits protective adaptive immunity against IAV infection and exposure to reemergent, heterosubtypic IAV.
Materials/Methods
Specific details are listed below and in the Life Sciences Reporting Summary.
Mice
WT C57BL/6/J and Ifnar1–/– mice were purchased from The Jackson Laboratory or bred in house from breeders obtained from The Jackson Laboratory. Ifnlr1–/– mice were kindly provided by Dr. Michael Diamond and maintained as a colony in the Gale Laboratory47. The entire Ifnlr coding sequence is ablated in these mice48. Ifnlr1fl/fl, Lyz2-cre, and Itgax-cre mice were provided by Dr. Herbert Virgin and maintained in the Gale Laboratory39. Briefly, these conditional-ready Ifnlr1fl/fl mice contain two loxP sites around Exon 2 of Ifnlr1. Upon expression of Cre-recombinase, the excision of this floxed site leads to a change in the amino acid sequence and a premature stop at amino acid 4239. Male and female mice were between the ages of 8 and 10 wks at the onset of experiments. All experiments were performed according to approved IACUC protocols at the University of Washington.
Viruses
All viruses were propagated in embryonated chicken eggs as previously described49. X31, PR8, PR8-GP33, PR8–2W. PR8 viruses expressing GP33 (lymphocytic choreomeningitis virus) or 2W1S (from Salmonella typhimurium) epitopes were engineered into a site within the stalk of NA known to be tolerant to insertions50 and generated as previously described51. The PR8–2W virus was utilized in experiments to evaluate T cell responses as CD4+ T cells specific for the 2W1S epitope in C57Bl/6/J mice undergo a large expansion relative to other CD4+ T cell specificities52, allowing for easier analysis of CD4+ T cell responses specifically activated by IAV infection.
Murine Infections, Tissue Harvest, and Processing
For all primary infection studies, mice were challenged with 40 PFU PR8, PR8-GP33, or PR8–2W as noted in each figure legend. This dose of 40 PFU represents a commonly utilized infection dose that in our model leads to weight loss and development of clinical respiratory illness, but where all animals go on to recover from infection30, 31. For heterologous re-challenge studies, Mice were mock-infected or challenged with 40 PFU PR8 or X31. 45 days following initial infection, animals were challenged with 2000 PFU PR8. Mice were anesthetized using ketamine/xylazine and administered virus diluted in 50ul PBS or 50ul PBS along for mock infections via intranasal administration49.
Virus titers
At designated times after infection, mice were euthanized, perfused with PBS, and lungs were harvested in 1mL PBS + 0.5% BSA in Precellys tubes. Tissues were homogenized using a Precellys homogenizer. Samples were then centrifuged and supernatants were aliquoted and snap frozen. Pulmonary virus titers were determined on frozen lung aliquots by plaque assay as previously described49.
Preparation of samples for Flow Cytometry and Acquisition of Samples
At the designated times after infection, mice were euthanized, and tissues were perfused with PBS by cardiac injection. Lungs and/or dLN were harvested and single cell suspensions were created by enzymatic digestion with type IV collagenase (Fisher/Worthington) and DNase I (Sigma) and GentleMacs (Miltemyi) processing.
For flow cytometry staining, single cell homogenates were blocked in 2% rat serum for 10 min at room temperature, incubated with specified antibodies for 30 min on ice, fixed with BD FACS Lysis buffer for 10 min at room temperature, resuspended in PBS, and run on the LSRII (BD). Samples were analyzed using FlowJo software (BD).
The following anti-mouse antibodies conjugated to fluorochromes were utilized for flow cytometry: (company, clone) α-CD3ε (eBioscience/Invitrogen, 145–2C11), α-CD4 (Biolegend, GK1.5), α-CD8α (Biolegend, 53–6.7), α-CD11b (eBioscience/Invitrogen, M1/70), α-CD11c (eBioscience/Invitrogen, N418), α-CD19 (eBioscience/Invitrogen, 1D3), α-CD40 (Biolegend, HM40–3), α-CD44 (BD, IM7), α-CD45.1 (eBioscience/Invitrogen, A20), α-CD45.2 (Biolegend, 104), α-CD69 (Biolegend, H1.2F3), α-CD103 (Biolegend, 2E7), α-F4/80 (eBioscience/Invitrogen, BM8), α-IFNAR1 (Biolegend, MAR1–5A3), α-IFNγ (eBioscience/Invitrogen, XMG1.2), α-MHCII (eBioscience/Invitrogen, M5/114.15.2), α-TNFα (eBioscience/Invitrogen, MP6-XT22).
Analysis of T cell responses
WT, Ifnlr1–/–, Ifnlr1fl/fl, Ifnlr1fl/flItgax-cre, or Ifnlr1fl/flLyz2-cre mice were infected i.n. with 40 PFU PR8 or PR8 containing the 2W1S epitope or administered an equivalent volume of PBS. Lung single cell homegnates from day 9 post infection were stained with NP366 MHCI tetramer, the PA224 MHCI tetramer, the NP311 MHCII tetramer, the 2W1S MHCII tetramer, or the control CLIP MHC II tetramer (NIH Tetramer Core) to enumerate IAV-specific CD4+ and CD8+ T cells from lung homogenates harvested on day 9 and 35 (WT and Ifnlr1–/– only) post infection. For MHCII tetramer staining, CD4+ T cell frequencies shown in graphs are for NP311 or 2W1S minus the CLIP negative control. For WT and Ifnlr1–/– infection experiments harvested on day 9, a portion of the cells were stimulated with NP366, PA224, NP311 peptides (Genemed Synthesis, Inc), PMA + Ionomycin (Sigma), or incubated with media only for 6 hours in the presence of brefeldin A (Sigma). Following incubation, cytokine production was determined by staining for flow cytometry.
in vivo T cell activation and proliferation
For in vivo T cell activation and proliferation experiments, 1×106 CFSE-labeled P14 T cells specific for the GP33 epitope on the CD45.1 background were adoptively transferred i.v. to WT or Ifnlr1–/– mice. Approximately 24 hours later, mice were infected i.n. with 40 PFU PR8 containing the GP33 epitope or administered an equivalent volume of PBS. Lung-draining lymph nodes were harvest 92 hrs post infection. Single cell suspensions of tissues were created and surface staining for flow cytometry was performed.
BMDC Generation, virus infection, sample harvest and DQ-OVA
BMDCs were generated from WT and Ifnlr1–/– bone marrow as previously described53. Briefly, bone marrow cells from WT and Ifnlr1–/– mice were isolated and cultured for 7 days in complete RPMI-1640 (Gibco) containing 20 ng/mLGM-CSF (Peprotech) and 20 ng/mL IL-4 (Peprotech). BMDCs were infected at an MOI 0.5 with PR8 in PBS containing Ca/Mg and 0.5% BSA for 1hr at 37° rocking. Following infection, BMDCs were incubated in complete RPMI until harvest. For qRT-PCR analysis, samples were harvested in RLT buffer, and RNA was isolated using an RNeasy Extraction Kit (Qiagen). cDNA was sythensized (BioRad iScript Kit), and relative gene expression was determined by qRT-PCR on a VIIA7 (Life Technologies) using the TaqMan Fast Advanced Master Mix (ThermoFisher) and primers specific for murine Ifnlr1 (Thermofisher Mm00558035_m1), murine Ifnar1 (Thermofisher Mm00439544_m1), murine Il10rb (ThermoFisher Mm00434157_m1), murine Gapdh (ThermoFisher Mm99999915_g1), and IAV NP (IDT). For IL-10 cytokine analysis supernatants were harvested and subsequent IL-10 levels were determined by multiplex immunoassay (Affymetrix Procartaplex).
For western blot analysis, BMDCs were stimulated with 500ng/mL recombinant murine IFN-λ3 (R&D) or 10ng/mL recombinant murine IFNα2 (R&D) for 1 hr, or 0.1, 1, 10, or 100ng/mL recombinant murine IFN-λ3 (R&D) for 30 min. Following stimulation, cell pellets were collected in RIPA buffer (ThermoFisher) with 1x HALT Protease/Phosphatase inhibitor added (ThermoFisher). Samples were run on an SDS-Page gel, transferred to PVDF, and probed for phospho-STAT1 at site Y701 (Cell Signaling, 58D6), total STAT1 (Cell Signaling, #9172), and Actin (Cell Signaling, D6A8). For phospho- and total STAT1 α-rabbit HRP (Jackson ImmunoResearch) and SuperSignal West Femto Maximum Sensitivity Substrate (ThermoFisher) were used and chemiluminescent signal detected on X-Ray film. For Actin, α-rabbit AF680 (Jackson ImmunoResearch) or α-rabbit HRP (Jackson ImmunoResearch) was used to detect fluorescent signal on a LI-Cor Odyssey or chemiluminescent signal using a ChemiDoc XRS+ (BioRad).
To determine ISG gene induction by BMDCs following stimulation, WT BMDCs were stimulated with 500ng/mL recombinant murine IFN-λ3 (R&D) for 12 hrs. Following stimulation samples were harvested in RLT buffer, and RNA was isolated using an RNeasy Extraction Kit (Qiagen). cDNA was sythensized (BioRad iScript Kit), and relative gene expression was determined by qRT-PCR on a VIIA7 (Life Technologies) using the SYBR Green Master Mix (ThermoFisher) and the following murine-specific primers (IDT): mChmp2a Fwd (5’-AGACGCCAGAGGAACTACTTC-3’), mChmp2a Rev (5’-ACCAGGTCTTTTGCCATGATTC-3’), mIfi44 Fwd (5’-GTAACACAGCAATGCCTCTTGT-3’), mIfi44 Rev (5’-AACTGACTGCTCGCAATAATGT-3’), mIfit1 Fwd (5’-CTGAGATGTCACTTCACATGGAA-3’), mIfit1 Rev (5’-GTGCATCCCCAATGGGTTCT-3’), mIsg15 Fwd (5’-GGTGTCCGTGACTAACTCCAT-3’), mIsg15 Rev (5’-TGGAAAGGGTAAGACCGTCCT-3’), mOas3 Fwd (5’-TCTGGGGTCGCTAAACATCAC-3’), and mOas3 Rev(5’-GGCAATCCTTATCACTCTTGGTC-3’).
For antigen uptake/processing assay, BMDCs were incubated with 10 μg/mL DQ-OVA (ThermoFisher) in PBS for 30 min, 1 hr, or 2 hr at 37° or 2 hr on ice as a control as previously described54. For flow cytometric analysis, samples were harvest at designated times post infection or DQ-OVA administration and stained as described above.
Lung CFSE labeling for DC migration
Mice were anesthetized using ketamine/xylazine and administered 8 mM CFSE (ThermoFisher) intranasally as previously described55. Shortly after CFSE administration mice were infected with PR8. 24hrs post infection, lungs and dLN were harvested and prepared for analysis by flow cytometry as described above.
Isolation of iDC and CD103+ DCs for gene expression analysis
iDCs and CD103+ DCs were sorted from naive lungs of WT mice using a FACS Aria with the gating strategy shown in Figure 5. RNA was harvested from these cells and cDNA was amplified utilizing the Cells-to-ct kit (ThermoFisher) according to manufacturers instructions. Ifnar1 and Ifnlr message was determined by RT-qPCR as described above.
mRNA sequencing
On day 4 following IAV infection, dLN were isolated from WT and Ifnlr1–/– mice. Single cell suspensions were prepared as described above. DC were enriched using a StemCell DC enrichment kit separation (StemCell), and cDC were sorted into RLT buffer using a FACS Aria with the gating strategy shown in Figure 5. RNA was isolated (Qiagen RNeasy Micro Kit) and quality of RNA was assessed using an RNA 6000 Pico Kit run on a Bioanalyzer (Agilent). cDNA was amplified (Takara SMART-Seq v4 Ultra Low Input RNA Kit), purified (Zymo DNA Clean and Concentrator), and analyzed (Qubit). Samples were tagmented and amplified (Nextera XT DNA Library Prep Kit). The samples were then cleaned using AMPure XP beads and the quality of final samples was determined using the Bioanalyzer DNA High Sensitivity Kit (Agilent) before sequencing on an Illumina MySeq. All kits for sample preparation were used according to manufacturers protocols.
RNAseq Data Processing and Analysis
Raw RNAseq data (Fastq files) were demultiplexed and checked for quality using FastQC (0.11.3). Next rRNA was digitally removed using Bowtie2 (2.3.4). Sufficient host reads (~twenty million) were then mapped to the Mouse genome (MM10) using STAR (2.5.3a) and converted into gene counts with ht-seq (0.6.1). Both the genome sequence (fasta) and gene transfer files (gtf) for mouse were obtained using Illumina’s igenomes website (https://support.illumina.com/sequencing/sequencing_software/igenome.html). Gene counts were then loaded in the R statistical programming language (3.4.3) and filtered by a row sum of fifty or more across all samples. Exploratory analysis and statistics were also run using R and bioconductor (3.4). The gene count matrix was normalized using voom through the limma bioconductor package (3.34.8). Statistical analysis (including differential expression) was performed using the limma package56. A Venn diagram was created using the Venn counts function through the limma bioconductor package in R. Scatterplot was generated in Spotfire (7.11.1) using log2 fold changes values where genes were deemed statistically significant in at least one of the two comparisons (WT vs. Mock and Ifnlr1–/– vs. Mock) and displayed. To produce co-expression heat maps, we performed co-expression analysis on genes statistically significant in the DE analysis (threshold: log2 fold change ≥ | 1.5 | and false discovery rate (FDR) ≤ 0.05) in at least one comparison (WT and Ifnlr1–/–) against their mock control. The union of these DE genes were loaded into R. Correlations (ward.2 clustering and euclidean distance) were run on the union of log2FC using the WGCNA (1.61) and gplots (3.0.1) bioconductor packages in R56, 57. To produce the Il10 gene network, we first identified a cell mediated immunological network containing Il10 using Ingenuity Pathway Analysis (IPA). The network was derived from known connections in the Ingenuity knowledge base, which include published gene-to-gene correlations, knockout studies, and protein-protein interactions. The network was overlayed using log2 fold change expression values from WT and Ifnlr1–/– and then exported into cytoscape (3.2.1) for visualization. This data set is available on GEO under accession GSE124399, and the markdown of our analysis of this data is available at https://hemann.galelab.org/.
Statistics
Statistics were calculated using GraphPad Prism 7.0. Specific statistical analyses used are described in legend for each figure and include log-rank test, one-way analysis of variance (ANOVA) with Tukey’s multiple comparisons test, and unpaired two-sided t-test.
Data Availability Statement
The data that support the findings of this study are available from the corresponding author upon reasonable request. The RNAseq data set is available on GEO under accession GSE124399, and the markdown of our analysis of this data is available at https://hemann.galelab.org/.
Supplementary Material
Acknowledgements
We would like to thank the Cell Analysis Facility Flow Cytometry in the Department of Immunology at the University of Washington and Elise Smith for technical assistance.
Funding
EAH is supported by American Heart Association award 17POST33660907. This work was also supported by National Institutes of Health grants T32AI7509 (EAH), AI132962 (RAL), AI108765 (RS), AI104002 (MG), AI118916 (MG), AI083019 (MG), and AI127463 (MG).
Footnotes
Competing interests
The authors declare no competing interests.
References
- 1.Kotenko SV, Gallagher G, Baurin VV, Lewis-Antes A, Shen M, Shah NK, et al. IFN-lambdas mediate antiviral protection through a distinct class II cytokine receptor complex. Nature immunology 2003, 4(1): 69–77. [DOI] [PubMed] [Google Scholar]
- 2.Sheppard P, Kindsvogel W, Xu W, Henderson K, Schlutsmeyer S, Whitmore TE, et al. IL-28, IL-29 and their class II cytokine receptor IL-28R. Nature immunology 2003, 4(1): 63–68. [DOI] [PubMed] [Google Scholar]
- 3.Prokunina-Olsson L, Muchmore B, Tang W, Pfeiffer RM, Park H, Dickensheets H, et al. A variant upstream of IFNL3 (IL28B) creating a new interferon gene IFNL4 is associated with impaired clearance of hepatitis C virus. Nature genetics 2013, 45(2): 164–171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gad HH, Dellgren C, Hamming OJ, Vends S, Paludan SR, Hartmann R. Interferon-lambda is functionally an interferon but structurally related to the interleukin-10 family. The Journal of biological chemistry 2009, 284(31): 20869–20875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Donnelly RP, Kotenko SV. Interferon-lambda: a new addition to an old family. Journal of interferon & cytokine research : the official journal of the International Society for Interferon and Cytokine Research 2010, 30(8): 555–564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hemann EA, Gale M Jr, Savan R. Interferon Lambda Genetics and Biology in Regulation of Viral Control. Frontiers in immunology 2017, 8: 1707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lazear HM, Nice TJ, Diamond MS. Interferon-lambda: Immune Functions at Barrier Surfaces and Beyond. Immunity 2015, 43(1): 15–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lee S, Baldridge MT. Interferon-Lambda: A Potent Regulator of Intestinal Viral Infections. Frontiers in immunology 2017, 8: 749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Levy DE, Marie IJ, Durbin JE. Induction and function of type I and III interferon in response to viral infection. Current opinion in virology 2011, 1(6): 476–486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Odendall C, Kagan JC. The unique regulation and functions of type III interferons in antiviral immunity. Current opinion in virology 2015, 12: 47–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Flannery B, Chung JR, Belongia EA, McLean HQ, Galglani M, Murthy K, et al. Interim Estimates of 2017–18 Seasonal Influenza Vaccine Effectiveness — United States, February 2018. MMWR Morb Mortal Wkly Rep 2018, 67: 180–185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Davidson S, McCabe TM, Crotta S, Gad HH, Hessel EM, Beinke S, et al. IFNlambda is a potent anti-influenza therapeutic without the inflammatory side effects of IFNalpha treatment. EMBO molecular medicine 2016, 8(9): 1099–1112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Galani IE, Triantafyllia V, Eleminiadou EE, Koltsida O, Stavropoulos A, Manioudaki M, et al. Interferon-lambda Mediates Non-redundant Front-Line Antiviral Protection against Influenza Virus Infection without Compromising Host Fitness. Immunity 2017, 46(5): 875–890 e876. [DOI] [PubMed] [Google Scholar]
- 14.Kim S, Kim MJ, Kim CH, Kang JW, Shin HK, Kim DY, et al. The Superiority of IFN-lambda as a Therapeutic Candidate to Control Acute Influenza Viral Lung Infection. American journal of respiratory cell and molecular biology 2017, 56(2): 202–212. [DOI] [PubMed] [Google Scholar]
- 15.Klinkhammer J, Schnepf D, Ye L, Schwaderlapp M, Gad HH, Hartmann R, et al. IFN-lambda prevents influenza virus spread from the upper airways to the lungs and limits virus transmission. eLife 2018, 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Broggi A, Tan Y, Granucci F, Zanoni I. IFN-lambda suppresses intestinal inflammation by non-translational regulation of neutrophil function. Nature immunology 2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Crotta S, Davidson S, Mahlakoiv T, Desmet CJ, Buckwalter MR, Albert ML, et al. Type I and type III interferons drive redundant amplification loops to induce a transcriptional signature in influenza-infected airway epithelia. PLoS pathogens 2013, 9(11): e1003773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mordstein M, Neugebauer E, Ditt V, Jessen B, Rieger T, Falcone V, et al. Lambda interferon renders epithelial cells of the respiratory and gastrointestinal tracts resistant to viral infections. Journal of virology 2010, 84(11): 5670–5677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wang Y, Li T, Chen Y, Wei H, Sun R, Tian Z. Involvement of NK Cells in IL-28B-Mediated Immunity against Influenza Virus Infection. Journal of immunology 2017, 199(3): 1012–1020. [DOI] [PubMed] [Google Scholar]
- 20.Yin Z, Dai J, Deng J, Sheikh F, Natalia M, Shih T, et al. Type III IFNs are produced by and stimulate human plasmacytoid dendritic cells. Journal of immunology 2012, 189(6): 2735–2745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Egli A, Santer DM, O’Shea D, Barakat K, Syedbasha M, Vollmer M, et al. IL-28B is a key regulator of B- and T-cell vaccine responses against influenza. PLoS pathogens 2014, 10(12): e1004556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Koltsida O, Hausding M, Stavropoulos A, Koch S, Tzelepis G, Ubel C, et al. IL-28A (IFN-lambda2) modulates lung DC function to promote Th1 immune skewing and suppress allergic airway disease. EMBO molecular medicine 2011, 3(6): 348–361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Misumi I, Whitmire JK. IFN-lambda exerts opposing effects on T cell responses depending on the chronicity of the virus infection. Journal of immunology 2014, 192(8): 3596–3606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Benton KA, Misplon JA, Lo CY, Brutkiewicz RR, Prasad SA, Epstein SL. Heterosubtypic immunity to influenza A virus in mice lacking IgA, all Ig, NKT cells, or gamma delta T cells. Journal of immunology 2001, 166(12): 7437–7445. [DOI] [PubMed] [Google Scholar]
- 25.Effros RB, Doherty PC, Gerhard W, Bennink J. Generation of both cross-reactive and virus-specific T-cell populations after immunization with serologically distinct influenza A viruses. The Journal of experimental medicine 1977, 145(3): 557–568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sridhar S, Begom S, Bermingham A, Hoschler K, Adamson W, Carman W, et al. Cellular immune correlates of protection against symptomatic pandemic influenza. Nature medicine 2013, 19(10): 1305–1312. [DOI] [PubMed] [Google Scholar]
- 27.Liang S, Mozdzanowska K, Palladino G, Gerhard W. Heterosubtypic immunity to influenza type A virus in mice. Effector mechanisms and their longevity. Journal of immunology 1994, 152(4): 1653–1661. [PubMed] [Google Scholar]
- 28.Bennink J, Effros RB, Doherty PC. Influenzal pneumonia: early appearance of cross-reactive T cells in lungs of mice primed with heterologous type A viruses. Immunology 1978, 35(3): 503–509. [PMC free article] [PubMed] [Google Scholar]
- 29.Effros RB, Bennink J, Doherty PC. Characteristics of secondary cytotoxic T-cell responses in mice infected with influenza A viruses. Cellular immunology 1978, 36(2): 345–353. [DOI] [PubMed] [Google Scholar]
- 30.Langlois RA, Varble A, Chua MA, Garcia-Sastre A, tenOever BR. Hematopoietic-specific targeting of influenza A virus reveals replication requirements for induction of antiviral immune responses. Proceedings of the National Academy of Sciences of the United States of America 2012, 109(30): 12117–12122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Waring BM, Sjaastad LE, Fiege JK, Fay EJ, Reyes I, Moriarity B, et al. MicroRNA-Based Attenuation of Influenza Virus across Susceptible Hosts. Journal of virology 2018, 92(2). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mordstein M, Kochs G, Dumoutier L, Renauld JC, Paludan SR, Klucher K, et al. Interferon-lambda contributes to innate immunity of mice against influenza A virus but not against hepatotropic viruses. PLoS pathogens 2008, 4(9): e1000151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.McGill J, Van Rooijen N, Legge KL. Protective influenza-specific CD8 T cell responses require interactions with dendritic cells in the lungs. The Journal of experimental medicine 2008, 205(7): 1635–1646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ho AW, Prabhu N, Betts RJ, Ge MQ, Dai X, Hutchinson PE, et al. Lung CD103+ dendritic cells efficiently transport influenza virus to the lymph node and load viral antigen onto MHC class I for presentation to CD8 T cells. Journal of immunology 2011, 187(11): 6011–6021. [DOI] [PubMed] [Google Scholar]
- 35.Kim TS, Braciale TJ. Respiratory dendritic cell subsets differ in their capacity to support the induction of virus-specific cytotoxic CD8+ T cell responses. PloS one 2009, 4(1): e4204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Moltedo B, Li W, Yount JS, Moran TM. Unique type I interferon responses determine the functional fate of migratory lung dendritic cells during influenza virus infection. PLoS pathogens 2011, 7(11): e1002345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Heng TS, Painter MW, Immunological Genome Project C. The Immunological Genome Project: networks of gene expression in immune cells. Nature immunology 2008, 9(10): 1091–1094. [DOI] [PubMed] [Google Scholar]
- 38.Smith LK, Boukhaled GM, Condotta SA, Mazouz S, Guthmiller JJ, Vijay R, et al. Interleukin-10 Directly Inhibits CD8(+) T Cell Function by Enhancing N-Glycan Branching to Decrease Antigen Sensitivity. Immunity 2018, 48(2): 299–312 e295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Baldridge MT, Lee S, Brown JJ, McAllister N, Urbanek K, Dermody TS, et al. Expression of Ifnlr1 on Intestinal Epithelial Cells Is Critical to the Antiviral Effects of Interferon Lambda against Norovirus and Reovirus. Journal of virology 2017, 91(7). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Abram CL, Roberge GL, Hu Y, Lowell CA. Comparative analysis of the efficiency and specificity of myeloid-Cre deleting strains using ROSA-EYFP reporter mice. Journal of immunological methods 2014, 408: 89–100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Topham DJ, Tripp RA, Doherty PC. CD8+ T cells clear influenza virus by perforin or Fas-dependent processes. Journal of immunology 1997, 159(11): 5197–5200. [PubMed] [Google Scholar]
- 42.Krishnaswamy JK, Gowthaman U, Zhang B, Mattsson J, Szeponik L, Liu D, et al. Migratory CD11b(+) conventional dendritic cells induce T follicular helper cell-dependent antibody responses. Science immunology 2017, 2(18). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Mount AM, Smith CM, Kupresanin F, Stoermer K, Heath WR, Belz GT. Multiple dendritic cell populations activate CD4+ T cells after viral stimulation. PloS one 2008, 3(2): e1691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Aparicio-Siegmund S, Garbers C. The biology of interleukin-27 reveals unique pro- and anti-inflammatory functions in immunity. Cytokine & growth factor reviews 2015, 26(5): 579–586. [DOI] [PubMed] [Google Scholar]
- 45.Llopiz D, Ruiz M, Infante S, Villanueva L, Silva L, Hervas-Stubbs S, et al. IL-10 expression defines an immunosuppressive dendritic cell population induced by antitumor therapeutic vaccination. Oncotarget 2017, 8(2): 2659–2671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Guo H, Santiago F, Lambert K, Takimoto T, Topham DJ. T cell-mediated protection against lethal 2009 pandemic H1N1 influenza virus infection in a mouse model. Journal of virology 2011, 85(1): 448–455. [DOI] [PMC free article] [PubMed] [Google Scholar]
Methods-only References
- 47.Lazear HM, Daniels BP, Pinto AK, Huang AC, Vick SC, Doyle SE, et al. Interferon-lambda restricts West Nile virus neuroinvasion by tightening the blood-brain barrier. Science translational medicine 2015, 7(284): 284ra259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ank N, Iversen MB, Bartholdy C, Staeheli P, Hartmann R, Jensen UB, et al. An important role for type III interferon (IFN-lambda/IL-28) in TLR-induced antiviral activity. Journal of immunology 2008, 180(4): 2474–2485. [DOI] [PubMed] [Google Scholar]
- 49.Cottey R, Rowe CA, Bender BS. Influenza virus. Current protocols in immunology 2001, Chapter 19: Unit 19 11. [DOI] [PubMed] [Google Scholar]
- 50.Heaton NS, Sachs D, Chen CJ, Hai R, Palese P. Genome-wide mutagenesis of influenza virus reveals unique plasticity of the hemagglutinin and NS1 proteins. Proceedings of the National Academy of Sciences of the United States of America 2013, 110(50): 20248–20253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Schmidt ME, Knudson CJ, Hartwig SM, Pewe LL, Meyerholz DK, Langlois RA, et al. Memory CD8 T cells mediate severe immunopathology following respiratory syncytial virus infection. PLoS pathogens 2018, 14(1): e1006810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Moon JJ, Chu HH, Pepper M, McSorley SJ, Jameson SC, Kedl RM, et al. Naive CD4(+) T cell frequency varies for different epitopes and predicts repertoire diversity and response magnitude. Immunity 2007, 27(2): 203–213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Suthar MS, Ramos HJ, Brassil MM, Netland J, Chappell CP, Blahnik G, et al. The RIG-I-like receptor LGP2 controls CD8(+) T cell survival and fitness. Immunity 2012, 37(2): 235–248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Daro E, Pulendran B, Brasel K, Teepe M, Pettit D, Lynch DH, et al. Polyethylene glycol-modified GM-CSF expands CD11b(high)CD11c(high) but notCD11b(low)CD11c(high) murine dendritic cells in vivo: a comparative analysis with Flt3 ligand. Journal of immunology 2000, 165(1): 49–58. [DOI] [PubMed] [Google Scholar]
- 55.McGill J, Legge KL. Cutting edge: contribution of lung-resident T cell proliferation to the overall magnitude of the antigen-specific CD8 T cell response in the lungs following murine influenza virus infection. Journal of immunology 2009, 183(7): 4177–4181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Smyth GK. Linear models and empirical bayes methods for assessing differential expression in microarray experiments. Statistical applications in genetics and molecular biology 2004, 3: Article3. [DOI] [PubMed] [Google Scholar]
- 57.Loraine AE, Blakley IC, Jagadeesan S, Harper J, Miller G, Firon N. Analysis and visualization of RNA-Seq expression data using RStudio, Bioconductor, and Integrated Genome Browser. Methods in molecular biology 2015, 1284: 481–501. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data that support the findings of this study are available from the corresponding author upon reasonable request. The RNAseq data set is available on GEO under accession GSE124399, and the markdown of our analysis of this data is available at https://hemann.galelab.org/.