Abstract
Unlike most other cancers, the incidence of melanoma is continuing to increase, particularly in young women, and the disease remains incurable for many due to its aggressive, metastatic nature, and high rate of resistance to conventional, targeted, and immunological agents. Cathepsin proteases are critical for melanoma progression and therapeutic resistance. Intracellular cathepsins cleave/degrade proteins that restrict cancer progression, while extracellular cathepsins directly cleave extracellular matrix and activate pro-invasive proteases in the tumor microenvironment. Cathepsin secretion is dramatically increased in cancer cells; however, the signaling pathways have not been extensively studied. Here, we describe new pathways leading to cathepsin secretion in melanoma cells. We show that Abl and Arg non-receptor tyrosine kinases (Abl/Arg) promote cathepsin B/L secretion by activating transcription factors with key roles in epithelial-mesenchymal transition (EMT), invasion, and therapeutic resistance (Ets1, Sp1, NF-κB/p65). In some melanoma cells, Abl/Arg drive an Ets1/p65->cathepsin pathway via a kinase independent mechanism, whereas, in others, Abl/Arg promote kinase-dependent Sp1/Ets1/p65->cathepsin L and Sp1/p65->cathepsin B pathways. The signaling pathways we identify are functionally and clinically relevant, as Abl/Arg, Sp1, Ets1, and cathepsin mRNAs are correlated in primary melanomas, and Abl/Argdriven invasion and metastasis requires cathepsin secretion. These data are significant as they indicate that drugs targeting Abl kinases, many of which are FDA-approved, might serve as cathepsin secretion inhibitors for melanomas expressing activated Abl kinases. The data also have important translational relevance given the renewed interest in targeting extracellular cathepsins as a therapeutic strategy for aggressive cancers.
Keywords: cathepsin, Abl, Arg, Ets1, Sp1, NF-κB/p65
One Sentence Summary:
Abl and Arg induce cathepsin protein and secretion in melanoma via p65-dependent Ets1/Sp1 pathways, and drive invasion/metastasis in a cathepsin-dependent manner.
INTRODUCTION
Melanoma is a highly aggressive disease, and despite recent advances in the development of targeted and immunotherapies, a significant proportion of patients with metastatic melanoma succumb to the disease (5-year survival rate-18%; https://seer.cancer.gov/data/citation.html.) Cysteine cathepsin proteases (B,C,F,H,L,K,O,S,W,Z) have well-recognized roles during cancer progression, and both intracellular and extracellular cathepsins contribute to metastasis (1). Cathepsins are synthesized as inactive precursors (proforms) in the trans-Golgi network, processed in the acidic late endosome to form single-chain intermediates, and subsequently targeted to lysosomes where they are cleaved into double chain, mature, active forms (1). Cathepsins function within lysosomes to degrade old organelles (autophagy) as well as intracellular and membrane-bound proteins (2). In cancer cells, lysosomal cathepsins promote invasion and metastasis presumably by cleaving intracellular proteins such as Bcl-2, Bcl-xL, and the metastasis suppressor, NM23-H1 (1, 3–6).
In addition to lysosomal functions, cathepsins also have important roles in the nucleus, plasma membrane, and extracellular environment (2). In cancer cells, cathepsin protein and mRNA is often dramatically increased, and excess procathepsins are exocytosed resulting in increased localization at the plasma membrane, in endocytic vesicles, and extracellularly (1, 3–6). Indeed, cysteine cathepsin secretion is induced >200-fold in the media from cancer cells; extracellular cathepsins comprise >40% of total secreted proteins, and secreted cysteine cathepsins are observed in the serum from cancer patients (6). Secreted procathepsins are cleaved/activated in the acidic/hypoxic tumor microenvironment, and promote invasion and metastasis (intravasation, extravasation) by cleaving/activating matrix metalloproteinases (MMPs) and urokinase-type plasminogen activator (uPA), inactivating Tissue Inhibitors of Metalloproteinases (TIMPs), cleaving the extracellular domain of E-cadherin, and directly cleaving/degrading extracellular matrix proteins (such as laminin, collagen, fibronectin, etc.) (1, 3–6). Moreover, secreted cathepsins also are involved in regulation of cell-cell adhesion, angiogenesis, and drive therapeutic resistance (1, 5).
In melanoma, increased cathepsin expression promotes conversion of non-metastatic melanomas into highly aggressive, metastastic melanomas (6). Rab7, which promotes endosome maturation and activation of intracellular cathepsins, is a critical regulator of a phenotypic switch, as induction of Rab7 promotes proliferation early in melanoma development, whereas decreased Rab7 later in the disease promotes cathepsin secretion, which drives invasion and metastasis (7). Indeed, cathepsin B secretion has been negatively associated with survival in melanoma patients (8, 9). Thus, due to their dysregulation during cancer progression and association with therapeutic resistance, cysteine cathepsins are attractive targets. However, unfortunately, few cell-permeable inhibitors targeting cysteine cathepsins have reached the clinic due to issues with toxicity and lack of efficacy, which has been attributed to lack of compound specificity, and the indispensible roles of intracellular cathepsins in other tissues (particularly in the cardiovascular system) (1, 3–6). Moreover, long-term inhibition of single cathepsins could result in compensatory increased expression of other cysteine cathepsins (6). Thus, other strategies designed to inhibit cathepsin expression and/or activity could potentially be more successful as anti-cancer therapies than directly targeting cathepsins.
Abl family non-receptor tyrosine kinases (Abl, Arg), encoded by Abl1 and Abl2 genes, respectively, are best known for their involvement in human leukemia (10). However, evidence accumulated over the past decade indicates that they have critical roles in the development and progression of solid tumors as well (10, 11). Previously, we demonstrated that Abl and Arg are activated in breast cancer and melanoma cell lines and primary tumors, and once activated promote proliferation, survival in response to nutrient deprivation, invasion and metastasis via distinct pathways (4, 12–14). Here, we demonstrate that cathepsin B/L secretion mediated by activated Abl/Arg plays a major role in driving Abl/Arg-dependent melanoma invasion and metastasis. Moreover, we identify the mechanism by showing that Abl/Arg induce cathepsin mRNA expression, promoter activity, and subsequent secretion by activating transcription factors with known roles in epithelial-mesenchymal transition (EMT), invasion, and therapeutic resistance (Ets1, Sp1, NF-kB/p65) (15–25), and drive distinct p65-dependent pathways to induce cathepsin expression/secretion. These data not only uncover new signaling pathways mediated by Abl kinases, which likely is applicable to other cancers, but also support a new strategy for inhibiting secretion of cathepsins, which play critical roles in metastatic progression.
RESULTS
Abl/Arg mRNAs are increased in melanoma cell lines and primary melanomas.
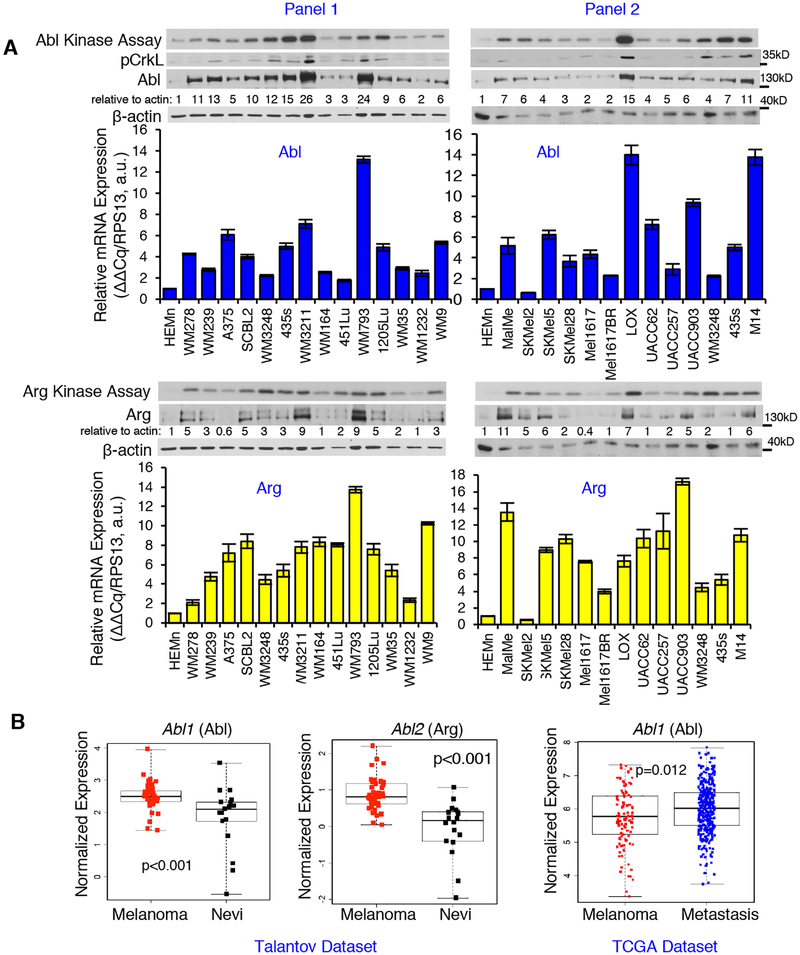
We showed that Abl/Arg were highly expressed in a small panel of melanoma cell lines, and are activated in a subset (12). Moreover, this trend also occurred in a large panel melanoma cell lines (25 lines) when compared to primary human melanocytes (26). Abl/Arg were overexpressed in most cell lines, and a subset (40–60%) also had high kinase activities (assessed directly by in vitro kinase assay and indirectly by examining phosphorylation of Abl/Arg substrates, Crk/CrkL, on Abl/Arg phosphorylation sites; Fig. 1A) (26). Since Abl/Arg proteins were increased in nearly all cell lines examined, we tested whether increased Abl/Arg protein was the result of increased mRNAs. Abl (Abl1) and Arg (Abl2) mRNAs were increased in most but not all cell lines (Fig. 1A), and for the most part, mRNA and protein were correlated (Supplementary Fig. S1A). Abl and Arg mRNAs also were increased in primary melanomas as compared to benign nevi, and Abl mRNA was further increased in metastases from patients (Fig. 1B). Although Abl/Arg activity was not increased in all cell lines containing high amounts of Abl/Arg protein, we observed an association between Abl/Arg mRNA and kinase activities in some lines (Supplementary Fig. S1B; Abl-Panel 2; Arg-Panel 1). Thus, in some cell lines increased mRNA expression is sufficient to activate Abl/Arg, whereas in others, it is likely necessary but not sufficient for activation. Cell lines with high Abl/Arg activities (435s, WM3248, UACC-903, LOX-IVMI) were used for subsequent studies.
Fig. 1. Abl and Arg mRNAs are increased in melanoma cell lines and primary melanomas.
(A) Abl/Arg kinase activities (kinase assay) and protein abundance were assessed in serum-starved melanoma cell lines or primary melanocytes (HEMn). mRNAs were quantitated with qPCR. Mean±SEM, n=2, normalized to RPS13. Western protein blots and kinase assays are part of Supplementary Materials for a recent manuscript (26). The data are reshown since they are critical for comparison to mRNA data. (B) Oncomine (Talantov dataset-left 2 panels) and TCGA data (right panel) were statistically reanalyzed using two-sample t-tests (for normally distributed data; Shapiro Wilk test) or Wilcoxon rank-sum tests. Nevi=benign nevi
Abl and Arg promote cathepsin secretion.
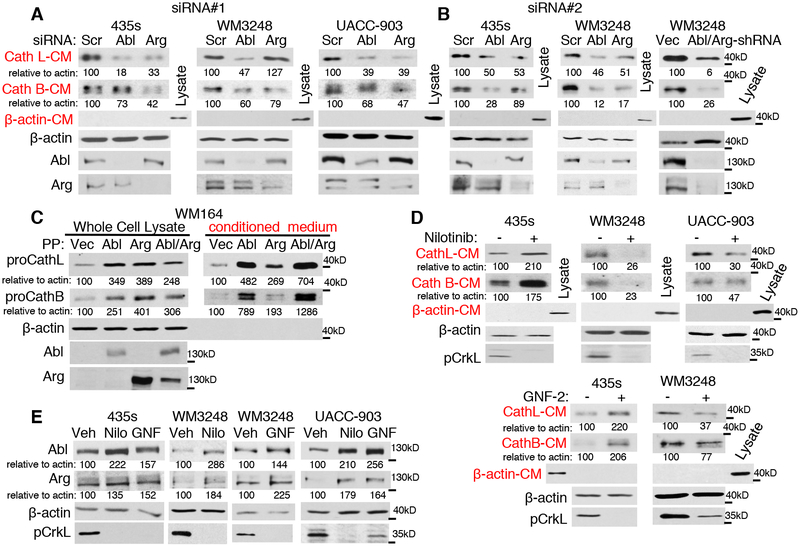
Previously, we showed that Abl/Arg potently increase invasion of breast cancer and melanoma cells, in part, by activating matrix metalloproteinases (MMPs) and inducing degradation of NM23-H1 (4, 12, 13). Since extracellular cathepsins also have important roles in invasion and cancer progression, we tested whether Abl/Arg activation drives cysteine cathepsin secretion. Indeed, silencing Abl or Arg with two independent siRNAs or using an shRNA that silences both Abl and Arg, reduced cysteine cathepsin (B/L) secretion from several melanoma cell lines (435s, WM3248, UACC-903) harboring highly active Abl and Arg (Fig. 2A,B, Supplementary Fig. S2A). Moreover, similar effects were observed in a breast cancer cell line that has activated Abl/Arg (BT-549; Supplementary Fig. S2B)(13, 14) indicating that the effects were not limited to melanoma cells. Gain-of-function studies showed that activation of Abl/Arg also is sufficient to induce cysteine cathepsin secretion, as expression of constitutively active forms (PP)(27) into low-invasive WM164 melanoma cells, which lack activated Abl/Arg (Supplementary Fig. S1A)(26), or into untransformed melan-a melanocytes potently induced cathepsin secretion (Fig. 2C, Supplementary Fig. S2C).
Fig. 2. Abl and Arg induce cathepsin secretion.
Cells were transfected with two independent siRNAs (A,B) an shRNA targeting Abl and Arg (B-right), or treated with Abl/Arg inhibitors, nilotinib (435s-5μM, WM3248–2μM), or GNF-2 (10μM; GNF) for 16h (D,E), serum-starved and conditioned medium (CM) blotted. To adjust for differences in cell number, media was loaded based on cellular protein concentration. Lysate β-actin blots show loading, and β-actin blotting of media demonstrates media purity (lack of lysis). pCrkL blots (recognize Abl/Arg-dependent phosphorylation sites on substrates, Crk and CrkL) demonstrate efficiency of Abl/Arg inhibition by nilotinib/GNF. Nilo=nilotinib (C) Media and lysate from serum-starved WM164 cells, transiently transfected with constitutively active forms of Abl and/or Arg (PP), were blotted.
Abl and Arg contain signaling domains (such as SH2, SH3, polyproline) in addition to a tyrosine kinase domain, and can act using kinase-dependent or kinase-independent mechanisms (11). To test whether Abl/Arg kinase activities are required to induce cathepsin secretion, we treated cells with the Abl/Arg inhibitor, nilotinib (2nd generation ATP-competitive inhibitor). Nilotinib reduced cathepsin secretion in WM3248, UACC-903 and LOX-IVMI melanoma cells containing highly active Abl/Arg but increased cathepsin secretion in 435s and BT-549 (breast) cells (Fig. 2D, Supplementary Fig. S2D,E). This apparent discrepancy in 435s and BT-549 cells was not due to off-target or other on-target effects of nilotinib because GNF-2/GNF-5, highly specific allosteric Abl/Arg inhibitors with no other known targets (28), had similar effects on cathepsin protein/secretion (Fig. 2D, Supplementary Fig. S2E,F). Nilotinib/GNF-2 treatment also caused compensatory increases of Abl and Arg protein (Fig. 2E, Supplementary Fig. S3A). These data suggest that nilotinib/GNF-mediated increase in cathepsin secretion in 435s/BT-549 cells may be due to increased Abl/Arg protein, which are kinase-inactive in the presence of the drugs, but retain their scaffolding functions (11). To test this hypothesis and to examine whether Abl induces cathepsin secretion in a kinase-independent manner in 435s cells, we mocked nilotinib/GNF-induced Abl expression by expressing a kinase-inactive form of Abl (KR). Indeed, Abl-KR expression potently induced secretion of cathepsin B/L (Supplementary Fig. S3B). Thus, Abl/Arg kinase activities are required for cathepsin secretion in WM3248, UACC-903 and LOX-IVMI melanoma lines but their tyrosine kinase activities are not necessary for Abl/Arg-induced cathepsin secretion in 435s cells.
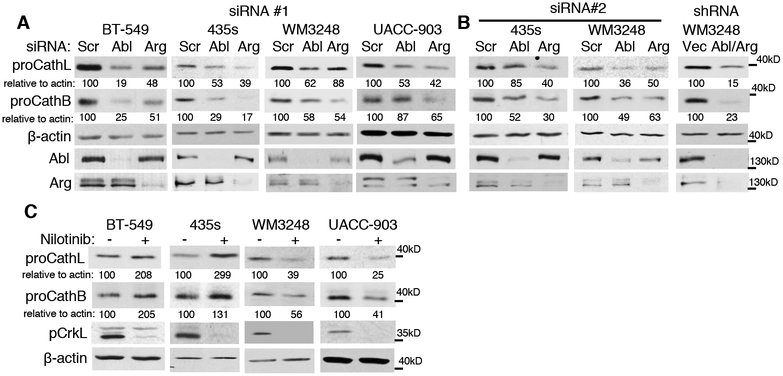
Since excess cathepsin proforms are shunted to the extracellular environment, we hypothesized that Abl/Arg-mediated cathepsin secretion occurs as a result of increased procathepsins. Indeed, silencing Abl and/or Arg with two independent siRNAs or an shRNA targeting both Abl and Arg reduced cathepsin proforms in all cell lines examined (Fig. 3A,B; Supplementary Fig. S3C,D). Introduction of constitutively active Abl (PP) or to a lesser extent Arg, also was sufficient to induce procathepsin B/L in WM164 melanoma cells and in untransformed melan-a melanocytes (Fig. 2C, Supplementary Fig. S2C). Consistent with the effects of Abl/Arg on cathepsin secretion, treatment with nilotinib decreased procathepsins in WM3248 (B and L) and UACC-903 (L) cell lines (similar to Abl/Arg siRNAs), but increased cathepsin proforms in 435s and BT-549 cells (Fig. 3C, Supplementary Fig. S3E), and expression of a kinase-inactive form of Abl induced procathepsin expression in 435s cells (Supplementary Fig. S3B). Moreover, nilotinib reduced all cathepsin forms in WM3248 and UACC-903 cells (proform, intermediate/single chain, mature/double chain) similar to the effect of silencing Abl/Arg (Supplementary Fig. S4A–D). In contrast, in BT-549 and 435s cells, although silencing Abl/Arg also reduced all cathepsin forms, nilotinib treatment reduced active forms (intermediate, mature-double chains), but induced proform accumulation (Supplementary Fig. S4E). These data are consistent with Abl/Arg activating cathepsins (proform->intermediate; intermediate->mature) in a kinase-dependent manner, while inducing cathepsin proforms using a kinase-independent pathway in 435s/BT-549 cells (4). Thus, in most cases, Abl/Arg dependent effects on procathepsins mirror their effects on cathepsin secretion.
Fig. 3. Abl/Arg increase cathepsin proforms.
Cellular lysate from siRNA- or shRNA-transfected (A,B) or nilotinib-treated (C; 16h), serum-starved cells were blotted. pCrkL blots demonstrate efficiency of Abl/Arg inhibition by nilotinib (C).
Abl/Arg induce cathepsin B/L transcription.
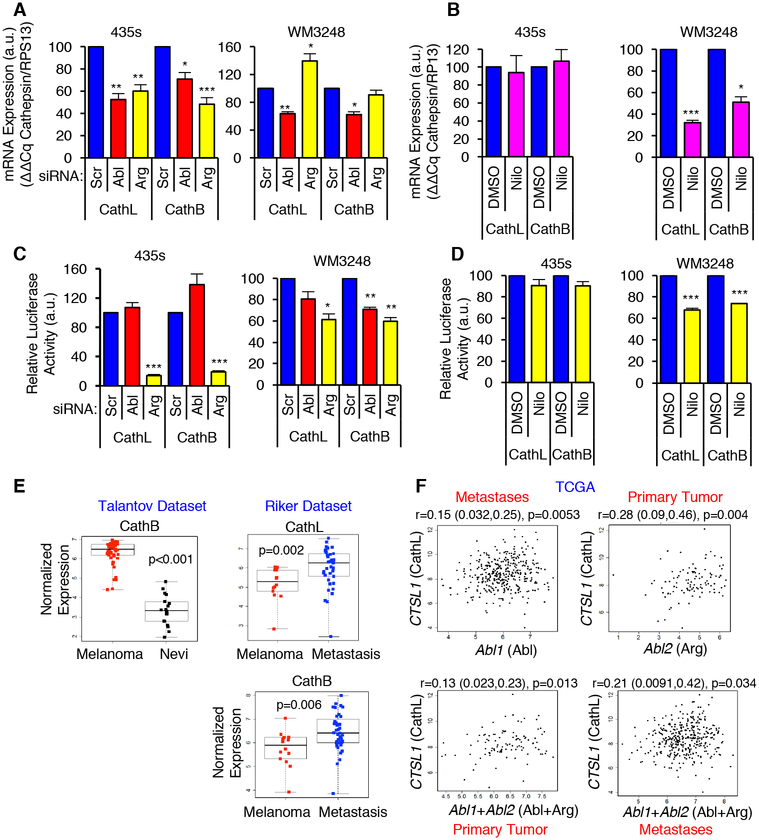
To dissect the mechanism by which Abl/Arg induce kinase-dependent versus kinase-independent effects on procathepsins, we examined the effect of silencing/inhibiting Abl/Arg on cathepsin mRNAs. Knockdown of Abl/Arg reduced cathepsin B/L mRNAs (assessed by qPCR) in cell lines displaying kinase-independent (435s, BT-549) and kinase-dependent (WM3248) procathepsin regulation (Fig. 4A; Supplementary Fig. S5A). In contrast, nilotinib reduced cathepsin mRNAs in WM3248 cells but had no effect in 435s and BT-549 cells (Fig. 4B; Supplementary Fig. S5A). Thus, the kinase-dependent (or independent) effects of Abl/Arg on cathepsin proforms and extracellular cathepsins are likely due to differential regulation of cathepsin mRNAs. Abl/Arg also induced cathepsins K and S mRNAs (Supplementary Fig. S5B), indicating that Abl/Arg regulate multiple pro-metastatic cysteine cathepsins.
Fig. 4. Abl/Arg increase cathepsin mRNAs and transcription.
(A,B) qPCR analyses on RNA from siRNA-transfected or nilotinib-treated cells (see Figs. 2, 3). Mean±SEM, n=3 normalized to RPS13. ***p<0.001, **p<0.01, *p<0.05 using one-sample t-tests and Holm’s adjustment for multiple comparisons. (C,D) Gaussia luciferase activity was measured in the media of siRNA-transfected (C) or nilotinib-treated (D) cells stably expressing cathepsin L or B promoter-luciferase constructs. Mean±SEM, n=3 normalized to total protein in the lysate. ***p<0.001, **p<0.01, *p<0.05 using one-sample t-tests and Holm’s adjustment for multiple comparisons. (E) Oncomine datasets (29, 30) were downloaded and reanalyzed using two-sample t-tests (for normally distributed data; Shapiro-Wilk test) or Wilcoxon ranksum tests. Nevi=benign nevi. (F) Normalized TCGA RNAseq data for primary melanomas and metastases were analyzed with Spearman’s correlation coefficient. Correlation (r), 95% confidence limits (in parentheses), and p-values are shown.
To determine whether changes in cathepsin mRNAs were due to effects of Abl/Arg on cathepsin transcription, we performed luciferase assays using cathepsin B or L promoter constructs. We focused on 435s and WM3248 cell lines since they represent kinase-independent and -dependent mechanisms of Abl/Arg-dependent cathepsin regulation, respectively. In WM3248 cells, silencing Abl or Arg or nilotinib treatment reduced cathepsin B/L promoter activity, which is consistent with Abl/Arg inducing cathepsin transcription in a kinase-dependent manner in this cell line (Fig. 4C,D). In contrast, in 435s cells, silencing Arg reduced cathepsin B/L promoter activity whereas the Abl siRNA and nilotinib had no effect on cathepsin transcription (Fig. 4C,D). Thus, in 435s cells, Arg induces cathepsin transcription in a kinase independent manner, and Abl induction of cathepsin mRNAs does not involve effects on transcription.
To define the translational relevance of Abl/Arg regulation of procathepsins, we examined whether there was an association between Abl/Arg and cathepsin mRNAs in primary melanomas, since increased Abl/Arg mRNA expression often correlated with increased activity (Supplementary Fig. S1). Cathepsin B/L mRNAs were significantly increased in melanomas as compared to benign nevi (Riker, Talantov Oncomine datasets)(29, 30), and cathepsin B mRNA was increased in metastases as compared to primary melanomas (Fig. 4E). Abl (Abl1) and/or Arg (Abl2) mRNAs also correlated with cathepsin B/L mRNAs in Oncomine and TCGA datasets (Fig. 4F, Supplementary Fig. S6).
Abl/Arg impact NF-κB (p65/RelA), Ets1, and Sp1 transcription factors.
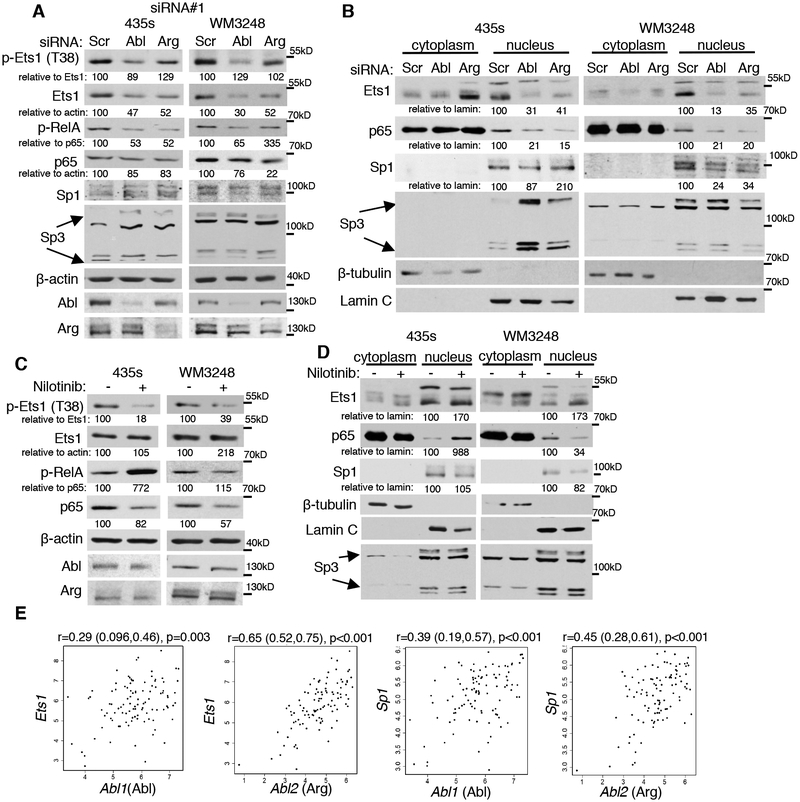
Next, we sought to identify the mechanism by which Abl/Arg induce cathepsin proforms and secretion. Since Abl/Arg effects on cathepsin proforms, for the most part, correlated with effects on secretion, and Abl/Arg induction of cathepsins occurred at the mRNA level, we focused on identifying the mechanism by which Abl/Arg induce cathepsin mRNAs and transcription. Little is known regarding how cysteine cathepsin mRNAs are dramatically induced in human melanoma, although increased cathepsin B mRNA in murine B16 melanoma cells was linked to Sp1, whereas Sp1, Sp3, and NF-Y (nuclear factor-Y) increased cathepsin L in some human melanoma cell lines (31, 32). NF-κB (p65/RelA; denoted p65) and Ets1 have been implicated in cathepsin regulation in other cell types and/or in response to drugs (doxorubicin) or other stimuli (33, 34). The ChIP MAPPER program (UF), indicated that the cathepsin L promoter had putative Sp1 and Ets1 binding sites, whereas cathepsin B had putative NF-κB and Sp1 binding sites. However, the JASPER database suggested the presence of putative Sp1, Ets1, and p65/RelA binding sites in both promoters. Since p65, Sp1/Sp3 and Ets1 have been implicated in inducing cathepsins and have putative binding sites in the cathepsin promoters, we examined whether Abl/Arg impact these transcription factors in melanoma cells. Silencing Abl or Arg reduced Ets1 protein, mRNA, and subsequent phosphorylation and nuclear abundance in WM3248 and 435s cells and Arg induced Ets1 promoter activity in 435s cells (Fig. 5A,B; Supplementary Fig. S7A,C,E and 8A,B). Moreover, in WM3248, silencing Abl or Arg also reduced Sp1, mRNA, and nuclear abundance but had only modest effects on Sp1 promoter activity (Fig. 5B, Supplementary Fig. S7C,F and 8A,B). Knockdown of Abl and/or Arg reduced p65/RelA phosphorylation and nuclear translocation without appreciable effects on p65 protein in 435s cells; however, in WM3248 cells, silencing Abl or Arg reduced p65 abundance in addition to inhibiting its phosphorylation and nuclear abundance (Fig. 5A,B; Supplementary Fig. S7A,C and 8A,B). Thus, in 435s cells, Abl and Arg promote p65 nuclear translocation whereas in WM3248 cells, effects on nuclear p65 may be mediated by changes in protein abundance. For the most part, nilotinib treatment phenocopied Abl/Arg knockdown in WM3248 cells as it reduced Ets1 and Sp1 mRNA, p65 abundance, phosphorylation and nuclear translocation, and also reduced the electrophoretic mobility of nuclear Ets1 (Fig. 5C,D; Supplementary Fig. S7B,D and 8C). In contrast, in 435s cells, nilotinib induced phospho-RelA and nuclear p65 similar to its effect on cathepsins, and had little/less effect on Ets1 mRNA and promoter activity (Fig. 5B,D; Supplementary Fig. S7B,D,E and 8C). These data indicate that Abl/Arg induce p65, Sp1, and Ets1 in a kinase-dependent manner in WM3248 cells, whereas in 435s cells, Abl/Arg effects on Ets1 and p65 likely occur via a kinase-independent mechanism. Consistent with this notion, expression of a kinase-inactive form of Abl (KR) increased nuclear p65 and Ets1 in 435s cells (Supplementary Fig. S8D). Importantly, expression of constitutively active forms of Abl/Arg (PP) (gain-of-function) also was sufficient to induce nuclear p65 and Ets1 and increase the electrophoretic mobility of nuclear Ets1 in untransformed melan-a melanocytes (Supplementary Fig. 8E).
Fig. 5. Abl and Arg induce NF-kB (p65/RelA), Ets1 and Sp1.
Cells transfected with siRNAs (A,B) or treated with nilotinib (C,D) were lysed and blotted (A,C), or fractionated into nuclear and cytoplasmic fractions and blotted (B,D). pCrkL blots demonstrate efficiency of Abl/Arg inhibition by nilotinib. (E) TCGA data was downloaded and correlations assessed as described in Fig. 4.
To determine whether Abl and/or Arg influence the ability of Ets1, Sp1 or p65 to directly bind cathepsin promoters, we performed ChIP assays. Abl/Arg inhibition with nilotinib dramatically reduced the binding of: a) Sp1 to cathepsin B and L promoters; b) Ets1 to the cathepsin L promoter; and c) p65 to the cathepsin B promoter (Supplementary Fig. S9). Thus, Abl/Arg not only impact expression, nuclear translocation, and/or promoter activity of the transcription factors, but they also dramatically promote their binding to cathepsin promoters. These data are critically important since these transcription factors play key roles in melanoma development, progression, and therapeutic resistance (15–25). Finally, we show that Abl/Arg regulation of Ets1 and Sp1 is relevant to human patients as Abl/Arg and Sp1/Ets1 mRNAs were correlated in primary melanomas from Oncomine and TCGA datasets (Fig. 5E, Supplementary Fig. S10A,B).
p65/RelA, Ets1, and Sp1 regulate each other as well as cathepsin abundance in human melanoma cells.
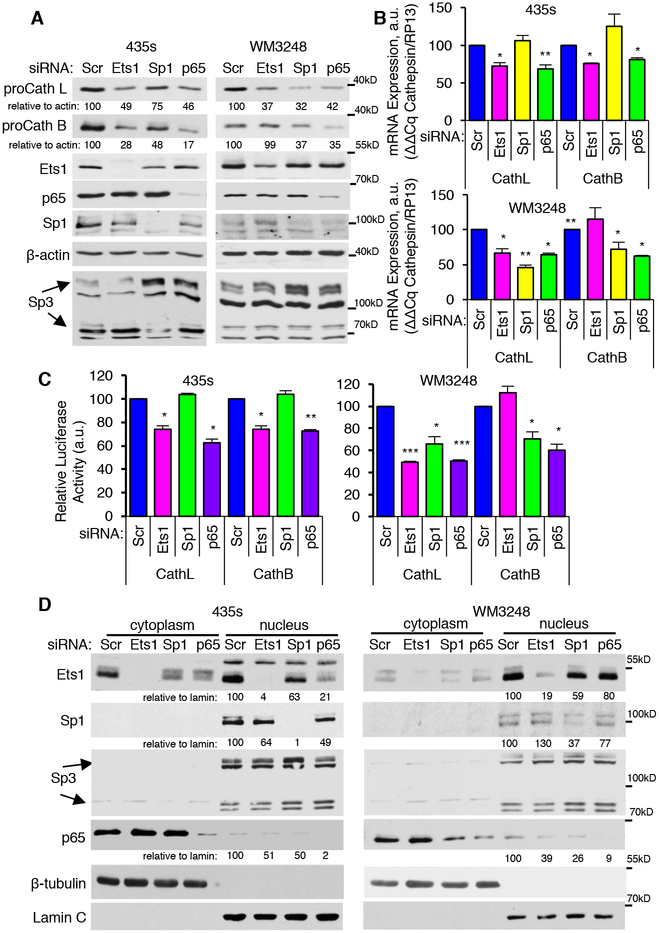
Next, we examined whether p65, Ets1, and Sp1 are required for induction of cathepsins B/L in human melanoma cells. Silencing p65 reduced cathepsin B/L mRNA and promoter activity, proforms, and secreted cathepsins in both cell lines (Fig. 6A–C; Supplementary Fig. S11A–D). In contrast, knockdown of Ets1 reduced cathepsin B/L mRNA and promoter activity as well as intracellular and extracellular cathepsins B and L in 435s cells, but only inhibited cathepsin L mRNA, promoter activity and proforms in WM3248 (Fig. 6A–C; Supplementary Fig. S11A–D). Moreover, silencing Sp1 reduced cathepsin mRNAs, promoter activity and intracellular and extracellular (L) cathepsin abundance in WM3248 cells whereas in 435s cells, silencing Sp1 reduced cathepsin proforms and secretion but had no effect on cathepsin mRNAs (Fig. 6A–C; Supplementary Fig. S11A–D). Thus, p65 induces cathepsins B and L in both lines, whereas effects of Ets1 and Sp1 are cell line and cathepsin-specific. The transcription factors also influenced each others’ nuclear targeting (but not overall abundance) as silencing Ets1 or Sp1 decreased nuclear p65, and silencing p65 inhibited nuclear Ets1 and Sp1 (Fig. 6D, Supplementary Fig. S11E). In summary, Abl/Arg promote cathepsin abundance in a kinase-dependent manner in WM3248 cells via cooperation and bidirectional regulation of Sp1, Ets1, and p65 transcription factors (Supplementary Fig. S12). In contrast, although Sp1 mediates cathepsin expression and secretion in 435s cells, it is not regulated by Abl/Arg in these cells, and Abl/Arg induce procathepsins using a kinase independent mechanism in a manner dependent on Ets1 and p65 (Supplementary Fig. S12).
Fig. 6. NF-kB (p65/RelA), Ets1 and Sp1 transcription factors drive cathepsin expression in melanoma cells.
Transfected cells were subjected to western blot (A), qPCR (B), cathepsin B or L promoter Gaussia luciferase assay (C), or subcellular fractionation/blotting (D). Graphs are Mean±SEM, n=3. ***p<0.001; **p≤0.01; *p<0.05 using one-sample t-tests and Holm’s adjustment for multiple comparisons.
Abl/Arg regulation of p65/RelA drives cathepsin abundance.
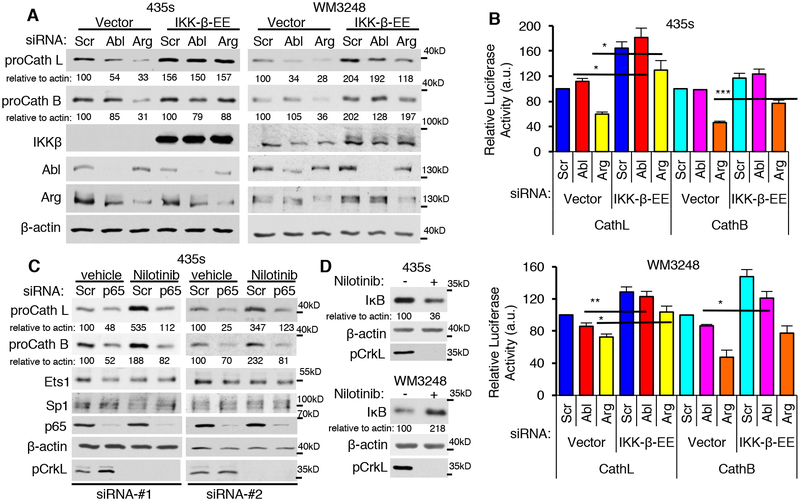
Since Abl/Arg regulate p65, and p65 is required for increased intracellular and extracellular cathepsins in both cell lines, it is likely the convergent point in Abl/Arg regulation of cathepsins. Moreover, nilotinib increases nuclear p65 in 435s cells, but inhibits its nuclear translocation in WM3248, which suggests that Abl/Arg driven kinase-dependent versus kinase-independent regulation of cathepsins is likely mediated by differential effects on p65. To test this hypothesis, we examined whether constitutive activation of the NF-κB pathway rescues effects of silencing Abl/Arg on intracellular and extracellular cathepsins. Indeed, expression of a constitutively active form of IKK-β (inhibitor of nuclear factor kappa-B kinase; S177E/S181E; denoted EE), which phosphorylates and induces degradation of IκB thereby promoting nuclear translocation of p50/p65 dimers, prevented Abl/Arg siRNA-mediated reduction in intracellular (proforms) and extracellular cathepsins (CM) and promoter activity (Fig. 7A,B; Supplementary Fig. S13A–C). Moreover, silencing p65 prevented the nilotinib-mediated increase in proforms and secreted cathepsins in 435s cells (Fig. 7C; Supplementary Fig. S13D,E). Thus, p65 mediates Abl/Arg induction of cathepsins in a kinase-independent manner in 435s cells and in a kinase-dependent manner in WM3248. Moreover, Abl/Arg regulation of p65 likely occurs at the level of IKK-β or upstream of IKK-β rather than by affecting cytoplasmic/nuclear shuttling of p65, as the latter mechanism would not be predicted to be rescued by exogenous IKK-β expression. Indeed, nilotinib treatment stabilized IκB in WM3248 but not in 435s cells (Fig. 7D, Supplementary Fig. S13F). Finally, expression of IKK-β-EE also partially rescued reduction in nuclear Ets1 induced by silencing Arg (Supplementary Fig. S13G,H), indicating that Arg upregulates Ets1 in part by increasing IKK-β induction of nuclear p65.
Fig. 7. Abl/Arg regulate cathepsin proform abundance via activation of NF-kB (p65/RelA).
(A,B) Cells transiently transfected with vector or constitutively active IKKβ-EE were transfected with the indicated siRNAs (72h; #1), serum-starved, and lysates blotted (A), or Gaussia luciferase secretion measured in the media from siRNA-transfected cells expressing cathepsin L or B promoter-luciferase constructs (B). Graphs are Mean±SEM, n=3. ***p<0.001; **p≤0.01; *p<0.05 using two-sample t-tests and Holm’s adjustment for multiple comparisons. (C) Cells transfected with scrambled or two independent p65 siRNAs, were treated with vehicle or nilotinib (16h), and lysates blotted. (D) Nilotinib treated cell lysates were blotted. pCrkL blots demonstrate efficiency of Abl/Arg inhibition by nilotinib.
Abl/Arg drive invasion and metastasis in a cathepsin-dependent manner.
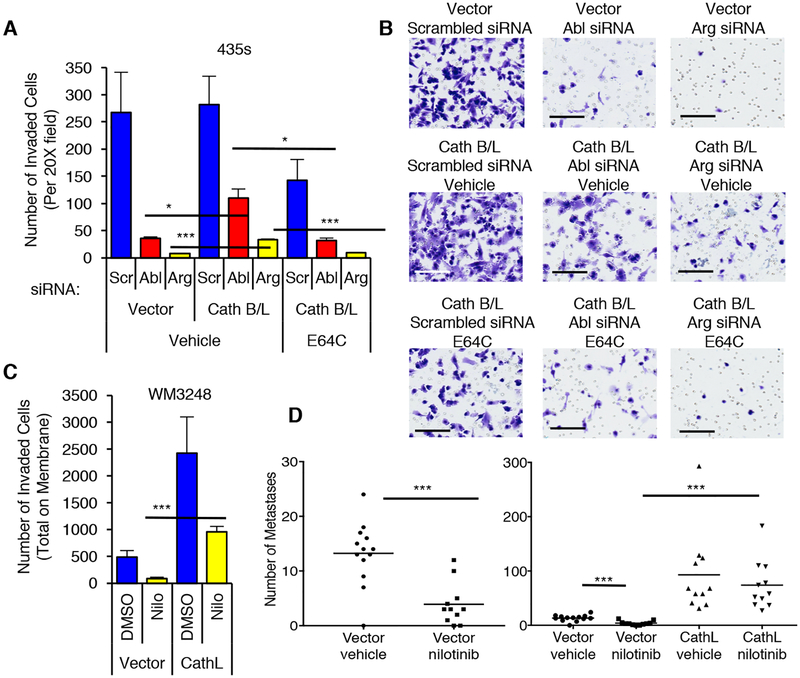
To examine the functional consequence of Abl/Arg induction of cathepsin secretion, we assessed the contribution of extracellular cathepsins B/L to Abl/Arg-driven matrigel invasion. Constitutive expression of cathepsins B/L partially rescued Abl/Arg siRNA-mediated reduction in invasion in 435s cells, indicating that cathepsins play a major role in Abl/Arg–driven invasion (Fig. 8A,B, Supplementary Fig. S14A). Cathepsin B/L expression was more efficient at rescuing Abl-siRNA-mediated inhibition, and only modestly rescued effects of the Arg siRNA, likely due to a reduced ability of cathepsins B/L to rescue Arg siRNA-mediated inhibition of cathepsins B/L (Fig. 8A,B, Supplementary Fig. S14A). In contrast, exogenous cathepsin L expression completely rescued nilotinib-mediated inhibition of endogenous cathepsin L and invasion in WM3248 cells (Fig. 8C, Supplementary Fig. S14B,C). Moreover, rescue of Abl/Arg siRNA or nilotinib-mediated inhibition of invasion was prevented by incubation with E64C, a cysteine cathepsin inhibitor that is impermeable to the cell membrane (Fig. 8A,B; Supplementary Fig. S14C) (35), which indicates that extracellular cathepsins mediate the rescue. As we previously reported, treatment of mice with low doses of nilotinib (doses that show efficacy in a murine leukemia model)(36), dramatically prevent WM3248 lung colonization, in vivo, and here, we demonstrate that exogenous expression of cathepsin L completely rescued the inhibition (Fig. 8D, Supplementary Fig. S15A). Thus, Abl/Arg also promote extravasation in a cathepsin-dependent manner. In addition to increasing lung colonization, exogenous cathepsin L also induced metastasis to another site (likely popliteal lymph node) which was observed in the absence or presence of nilotinib with 100% penetrance (Supplementary Fig. S15B,C).
Fig. 8. Abl/Arg drive invasion and metastasis by regulating cathepsin secretion.
(A,B) 435s cells, stably expressing vectors or cathepsin B and L, were transfected with siRNAs (#1), serum-starved, and utilized in a matrigel invasion assay (1% FBS chemoattractant) in the absence or presence of the cysteine cathepsin inhibitor, E64C (50μM; 24h). An aliquot of cells were lysed and blotted (Supplementary Fig. S14A). (B) Representative fields from A. Size bars=100μm. (C) WM3248 cells stably expressing vector or cathepsin L were serum-starved, treated with nilotinib (2μM; 16h), and utilized in a matrigel invasion assay (IGF-1: 10nM; 36h; bottom). An aliquot of cells was lysed and blotted (Supplementary Fig. S14B), and representative fields are shown in Supplementary Fig. S14C. (A,C) Graphs are Mean±SEM, n=3. ***p<0.001; **p≤0.01; *p<0.05 using two sample t-tests and Holm’s adjustment for multiple comparisons. (D) Quantitation of GFP-positive lung nodules from mice injected i.v. with WM3248 cells stably expressing vector or cathepsin L, treated with vehicle or nilotinib for 33 days. Vector-vehicle=n=13 mice; vector-nilotinib, n=11 mice; CathL-vehicle, n=12 mice; CathL-nilotinib, n=11 mice. ***p<0.001, using Wilcoxon rank sum test and Holm’s adjustment for multiple comparisons. (Left) Vector groups. (Right) All treatment groups.
DISCUSSION
Increased cathepsin abundance and subsequent secretion are critically important for invasion and metastasis of melanoma cells although the signaling pathways underlying this process have not been extensively studied. Here, we identify new signaling pathways by demonstrating that activated Abl and Arg are responsible for inducing cathepsin mRNA, protein, and secretion and do so by influencing the nuclear abundance of NF-κB/p65, Ets1, and Sp1. Although the data shown are focused on cathepsins B/L, Abl/Arg also regulate other cysteine cathepsins that are linked to melanoma dissemination, such as cathepsins K and S (Supplementary Fig. S5B) (37–41), which indicates that Abl/Arg may regulate melanoma metastasis by inducing secretion of a variety of pro-invasive cysteine cathepsins perhaps using similar pathways. Our findings also extend beyond Abl/Arg induction of cathepsins, as Ets1, Sp1 and p65 regulate numerous targets that drive processes crucial for cancer progression such as proliferation, invasion, epithelial-mesenchymal-transition (EMT), regulation of stem cell traits, angiogenesis, metabolic reprogramming, and drug resistance (including to immunotherapy), and their overabundance is associated with a poor prognosis (15–25). Despite their crucial roles in cancer progression, transcription factors are notoriously hard to target. Moreover, few drugs targeting cathepsins have reached the clinic, and those that have done so have many adverse effects. Our data demonstrate that targeting Abl/Arg is a new way not only to prevent cathepsin secretion, but also to reduce the abundance and activity of Ets1, Sp1, and NF-κB/p65, and thus, the data have strong translational relevance.
In addition to showing that Abl/Arg affect the abundance, nuclear translocation, and transcriptional activity of the above transcription factors, we also demonstrate that Abl/Arg dramatically impact the binding of Ets, Sp1, and p65 to cathepsin promoters. We observed no binding of p65 to the cathepsin L promoter, which is consistent with the results of ChIP MAPPER indicating the lack of a p65 binding site. However, since p65 is a binding partner of Sp1 and Ets1, and the transcription factors can cooperate to induce transcription by binding to one or more consensus binding sites (42), p65 may modulate Ets1/Sp1 DNA binding/transcriptional activity without directly binding to its own consensus sequence. Indeed, our finding that p65 increases the nuclear abundance of Ets1/Sp1 are consistent with this hypothesis (Supplementary Fig. S12).
Abl/Arg inhibitors had opposite effects as Abl/Arg siRNAs on cathepsin abundance/secretion in some cell lines (435s, BT-549-breast cancer) whereas nilotinib mimicked RNAi in all other melanoma cell lines examined (WM3248, UACC-903, LOX-IVMI). This apparent discrepancy in 435s and BT-549 cells was not due to off-target or other on-target effects of nilotinib because GNF-2/GNF-5, highly specific allosteric inhibitors with no other known targets, had similar effects as nilotinib on cathepsin abundance/secretion. Thus, we hypothesized that Abl/Arg regulate cathepsin abundance/secretion in a kinase-independent manner in 435s and BT-549 cells. Consistent with this hypothesis, nilotinib/GNF induced compensatory increases of Abl and Arg protein, which are kinase-inactive in the presence of the drugs, and expression of a kinase-inactive form of Abl phenocopied nilotinib/GNF effects. Kinase-inactive forms of Abl are known to retain scaffold/adapter-like signaling functions (11), and although reports describing Abl’s kinase-independent function have previously been described (43), such a role has not previously been documented in cancer cells.
Understanding cell-type specific signaling differences has critical translational relevance as induction of cathepsin secretion by Abl/Arg inhibitors (which occurs in 435s cells) would be predicted to potentially limit their effectiveness as anti-metastatic agents. However, nilotinib effectively reduced late stages of metastasis and inhibited matrigel invasion of 435s cells (4, 12), which indicates that nilotinibmediated inhibition of other pro-invasive pathways is sufficient to at least partially override the opposing effects on cathepsin secretion in these cells. Indeed, Abl/Arg also promote 435s invasion by inducing STAT3-dependent induction of MMP-1 and by promoting degradation of the NM23-H1 metastasis suppressor (4, 12). The existence of these additional pathways may also explain why expression of cathepsins B/L only partially rescued reduction in 435s invasion induced by silencing Abl whereas cathepsin L expression was sufficient to completely rescue nilotinib-mediated reduction in invasion in WM3248 cells. Alternatively, WM3248 cells may have an increased dependence on secreted cathepsins. Kinase-dependent and -independent induction of cathepsin abundance/secretion mediated by Abl/Arg occurs by IKK-β/IκB regulation of NF-κB/p65. Thus, future experiments are aimed at understanding why regulation of IκB stability by Abl/Arg is kinase-independent in some lines and kinase-dependent in others by examining whether Abl/Arg differentially regulate IKK-β abundance, activity and/or interaction with modulators such as IKK-γ-NEMO (44).
In most cases, silencing Abl/Arg reduced cathepsin mRNAs as well as transcriptional activity; however, in some instances mRNA and transcriptional activity were not concordant. For example, silencing Abl reduced cathepsin B/L and Ets1 mRNAs and proforms but had no effect on cathepsin or Ets1 promoter activity in 435s cells. These data suggest that, unlike Arg, which promotes Ets1 and cathepsin transcription in 435s cells, Abl may increase Ets1 and cathepsin mRNA stability. Indeed, Ets1 is regulated by miRNAs, and Abl/Arg induce cathepsin expression via Ets1 in this line (19). Interestingly, in WM3248 cells, Abl also induces Ets1/Sp1 mRNA independent of effects on Ets1/Sp1. Finally, in WM3248 cells, silencing Arg reduced cathepsin promoter activity and proforms but did not reduce cathepsin mRNAs, and cathepsin L mRNA was even increased. These seemingly puzzling data could be due to Arg knockdown increasing expression of an alternatively spliced cathepsin transcript that cannot be distinguished from the full-length mRNA using the primers utilized. Indeed, five cathepsin B and L transcripts have been identified (see www.genecards.org). Moreover, effects of silencing/inhibiting Abl/Arg on Ets1/Sp1 mRNA in WM3248 cells was more dramatic than effects on Ets1/Sp1 promoter activity, indicating that Abl/Arg may also regulate Ets1/Sp1 RNA stability in this line. Silencing/inhibiting Abl/Arg or introduction of constitutively active Abl/Arg (PP) often had more dramatic effects on extracellular as opposed to intracellular cathepsins (Figs. 2,3). These data indicate that, in addition to increasing cathepsin abundance, Abl/Arg also might influence the secretion process itself. We previously showed that Abl and Arg promote vesicular trafficking (endosome maturation), which drives lysosome maturation and intracellular cathepsin activation (4). Vesicular trafficking also is known to be critical for the secretion process. Cathepsin L secretion by melanoma cells requires functional Rab4a, which regulates “fast” transport of vesicles to the plasma membrane, and inhibition of Rab4a reduces melanoma tumorigenicity (45). Moreover, Rab5 to Rab7 switch regulates endosome maturation (46), and Rab7 abundance also has been implicated in lineage-specific phenotypic switching, as silencing Rab7 inhibits melanoma proliferation but increases invasion and extravasation by inducing cathepsin secretion (7). Recently, we found that Abl/Arg induces a switch in epithelial-mesenchymal transcription factor expression, which also drives lineage-specific phenotype switching, invasion, metastasis, and intra-tumor heterogeneity (26, 47, 48). Thus, it is attractive to speculate that Abl/Arg serve as common nodes in these two pathways, which regulate a critical switch between differentiated and invasive states. If so, these findings would have strong translational relevance since intra-tumor heterogeneity has recently been implicated as a key feature driving therapeutic resistance, and targeting heterogeneity has been identified as an important future treatment strategy (48).
MATERIALS AND METHODS
Reagents.
Drugs.
Nilotinib was provided by Novartis, and GNF-2, GNF-5, and E64C were from Selleck. Antibodies were obtained commercially. R&D Systems: cathepsin L (AF952), human cathepsin B (AF953), mouse cathepsin B (AF965); Sigma Aldrich: β-actin, Arg (5C6; western blot), Flag (M2); Santa Cruz Biotechnology: Abl (K12; kinase assay), Abl (8E9; western), Ets1 (N-276), Sp1 (1C6), Sp3 (D-20), IKK-β (H-4), p65/RelA (C-20), and HRP-conjugated secondary antibodies. Cell Signaling: pCrk/CrkL (Y221/Y207), pRelA/p65 (93H1), IκBα (L35A5); BD Biosciences: Abl (8E9; western); Bioworlds: pEts1 (BS4316); ThermoFisher Scientific: β-tubulin; and Millipore: lamin A/C (clone 14), Sp1 (07–645). The Arg antibody used for kinase assays was previously described (49). ChIP antibodies were obtained from Active Motif: Sp1 (39058), Ets1 (39580), p65 (40916).
Plasmids and Cell Lines.
Melanoma lines (mycoplasma tested-8/16; Lonza MycoAlert) were previously described (26).
DNA constructs and stable cell lines.
Constitutively active IKK-β (S177E/S181E; in pCMV2) was provided by Dr. Anjana Rao (La Jolla Institute, La Jolla, CA) via Addgene. The construct was transiently transfected into cells using Lipofectamine 2000, by transfecting cells at 90% confluency, replating the next day, followed by transfection with siRNAs (see below).
Human cathepsin L (11591-PG02), cathepsin B (13951-PG02), Sp1 (3907-PG02), and Ets1 (36631-PG02) Gaussia Luciferase promoter constructs were obtained from GeneCopoeia. The cathepsin L construct contains 1028bp upstream of the transcriptional start site (TSS) and 230bp downstream, whereas the cathepsin B promoter contains 1110bp upstream of the TSS and 222bp downstream. Cell lines stably expressing the promoter constructs, linked to Gaussia Luciferase, were established by transfecting cells (Lipofectamine 2000), selecting with puromycin (435s-1μg/ml; WM3248–2μg/ml), and pooling clones.
psiSTRIKE™-hygro vector containing an shRNA targeting Abl AND Arg (GGGAAATTGCTACCTATGG) was obtained from Dr. Cao (Beijing, China)(50). WM3248 cells were transfected with psiSTRIKE™-hygro or pSTRIKE-Abl/Arg-shRNA using Lipofectamine 2000, selected (hygromycin; 200U/ml), clones expanded, and positive clones (identified by western blot) pooled. Kinase inactive Abl (K290R) cloned into the EcoR1 site of (pSRa)(51, 52) as well as constitutively active Abl (Migr1-Abl-PP) and Arg (PK1-Arg-PP)(4, 51) were previously described.
Cathepsin L and cathepsin B expression constructs were from Addgene (pcDNA3.1), deposited by Dr. Hyeryun Choe (Scripps Clinic, La Jolla, CA). Cathepsin L and B were liberated by cutting with PME1/XhoI, blunting and cathepsin L was cloned into the blunted EcoR1 site of Migr1 (from Warren Pear, University of Pennsylvania)(49) whereas cathepsin L was cloned into the blunted EcoR1 site of MSCV-dsRed2 (obtained from Dr. Yi-Ling Lin; University of California, Los Angeles). MSCR-dsRed2 was created by cloning DsRed2 from pIRES2-DsRed2 (Clontech) into MSCV-hygro by cutting pIRES DsRed2 with Not1, blunting and cutting with BglII and cloning into MSCV-hygro cut with HindIII, blunted, and cut with BglII. 435s cells stably expressing cathepsin B and L were obtained by transfecting Migr1-cathepsin L (1.6μg) and MSCV-DsRed-cathepsin B (1.6μg) constructs or vectors together with PK1 (0.8μg; contains puromycin gene) in a 6-well using Lipofectamine 2000, selecting with puromycin (1μg/ml), pooling positive clones, and sorting GFP/DsRed-positive cells three times. WM3248-cathepsin L cells were obtained by transfecting with Migr1-cathepsin L (3.3μg) together with PK1 (0.67μg), selecting with puromycin (2.5μg/ml), pooling clones, and sorting GFP-positive cells once or twice.
siRNAs and shRNAs.
Transient Silencing.
Cells were transfected with the following siRNAs using Lipofectamine 2000. Abl #1,2 (1336, s886-select), respectively; 20nM), Arg #1,2 (1478, s872-select; 20nM), Ets1 #1,2 (HSS103402-stealth, HS40612-stealth; 10nM), Sp1-#1,2 (VHS49865-stealth, 242578; 10nM), p65 #1,2 (141324; s11915-select; 10nM), scrambled control #1 (control for non-silencer select), silencer select scrambled #1 (for select), or stealth control (12935200). siRNAs were transfected on two consecutive days to increase silencing efficiency. For experiments in which conditioned media was obtained, cells were replated prior to serum-starvation (16h) to obtain equal cell numbers for subsequent media collection.
qPCR.
Primers were blasted with Primer Blast (NCBI website) to rule out homology to other genes. DNase-treated cDNA (50ng; iScript; Bio-Rad) was amplified using SYBER green and gene-specific primers (500nM; 40 cycles, 62°C annealing temperature). Results were analyzed with CFX Manager (Bio-Rad), normalizing to RPS13 or RP2 reference genes (4). Primer sequences are in Supplementary Table S1.
ChIP.
ChIP assays were performed using SimpleChIP Plus Sonication Chromatin IP Kit (Cell Signaling) according to manufacturer’s instructions. Briefly, WM3248 cells (107) treated with DMSO or nilotinib (2μM) were fixed with formaldehyde to crosslink DNA-proteins, chromatin sheared using Microson Ultrasonic Cell Disruptor XL (Misonix) (16 cycles of sonication: 15” each, 2’ rest; amplitude=10, Power=15 watts), chromatin (10μg) incubated with antibodies (Sp1–7μl, Ets1–10ul, p65–10μl, Histone H3-positive control-10μl, IgG-negative control-2μl), and IPs bound to Protein G magnetic beads (30μl). The protein-DNA cross-link was reversed, DNA purified, and enrichment of DNA sequences detected using qPCR and primers listed in Table S2. Data were normalized and analyzed using Fold Enrichment analysis (53).
Kinase Assays and Western Blot Analysis.
Immunoblotting of cell lysates in RIPA buffer (54) was performed using antibody manufacturers’ protocols. Bands were quantified with ImageJ64 (freeware). Kinase assays were previously described (26, 54).
Preparation of Conditioned Media.
Media from serum-starved cells was concentrated (AmiconUltracel-10; ThermoFisher), volumes equalized with basal media, and loaded based on protein concentrations from respective cellular lysates. Actin blots demonstrate media purity.
Luciferase Assays.
Cell lines stably expressing cathepsin B/L Gaussia luciferase promoter constructs were transfected with vector/IKK-b expression plasmids (12h), replated, transfected with siRNAs (56h), washed, and luciferase activity measured in the media 16h later (ThermoFisher kit; 20μl media/50μl luciferase). Values were normalized to cellular protein concentration (BCA, Biorad). For Ets1 and Sp1 promoter luciferase assays, stable cell lines expressing the promoter constructs were transfected with siRNAs or treated with nilotinib (16h) and luciferase activity measured as above.
Subcellular Fractionation.
Cytoplasmic/nuclear lysates were prepared with NE-PER (ThermoFisher) (55).
Matrigel Invasion.
Assays were performed as described (4, 12), using FBS (1%; 24h; 435s) or IGF-1 (10nM; 36h; WM3248) as chemoattractant.
Experimental Metastasis Assays.
WM3248 cells expressing vector (GFP bicistronic vector) or cathepsin L were injected i.v. into female nude mice (Envigo, 2×106 cells/100μl), and mice treated with vehicle (0.5% hydroxymethylcelluose/0.05% Tween-80) or nilotinib (33mg/kg; oral gavage; b.i.d) (4). Mice were euthanized (33 days), lungs perfused (1X PBS; intracardiac injection), formalin-fixed, and metastases quantitated/photographed (Olympus MCX-10; 63X objective). Lymph node tumors were paraffin embedded, stained (H&E), and photographed on an Aperio Scanscope (20X; LEICA). Studies were approved by the University of Kentucky IACUC Committee. Power Analysis. Data simulations (replicated 1000 times; based on preliminary data (4)) and the Wilcoxon rank sum test were used to compare nilotinib groups. Statistical power was calculated as the proportion of replications that achieve statistical significance. A sample of 14 mice/group provided 80% power to detect differences between the groups (5% significance).
Statistical Analyses:
Two sample (comparisons between treatment groups) or one-sample t-tests (comparisons against normalized controls) using the Holm’s method for multiple comparisons were performed. Microarray and clinical data were downloaded from the Oncomine database (28, 29). RNASeq data were downloaded from Genomic Data Commons (GDC) for TCGA Skin Cutaneous Melanoma data (Access date Sep. 2017) and normalized to Transcripts Per Kilobase Million (TPM). Gene expression was compared using two-sample t-tests (for normally distributed data; Shapiro-Wilk test) or Wilcoxon rank-sum tests were utilized. Spearman’s correlation coefficients were used to quantify correlations.
Supplementary Material
Supplementary Fig. S1. Abl and Arg are overexpressed and activated in a large panel of melanoma cell lines.
Supplementary Fig. S2. Effects of silencing or inhibiting Abl/Arg on intracellular and extracellular cathepsins.
Supplementary Fig. S3. Abl/Arg drive cathepsin abundance.
Supplementary Fig. S4. Effects of silencing or inhibiting Abl/Arg on pro-, intermediate and mature (double-chain) cathepsin forms.
Supplementary Fig. S5. Abl and Arg promote cathepsin mRNA expression.
Supplementary Fig. S6. Abl/Arg and cathepsin mRNAs are correlated in primary melanomas.
Supplementary Fig. S7. Abl/Arg alter Ets1 and Sp1 proforms, mRNA and promoter activities.
Supplementary Fig. S8. Abl/Arg alter nuclear localization of p65, Ets1, and Sp1.
Supplementary Fig. S9. Abl/Arg impact the DNA binding capacity of Ets1 and Sp1 on cathepsin promoters.
Supplementary Fig. S10. Abl/Arg and Sp1/Ets1 mRNAs are correlated in primary melanomas.
Supplementary Fig. S11. Ets1, Sp1 and p65 contribute to cathepsin abundance.
Supplementary Fig. S12. Model for Abl/Arg regulation of cathepsin expression and secretion.
Supplementary Fig. S13. Abl/Arg regulate cathepsin expression by activating NF-κB (p65).
Supplementary Fig. S14. Abl/Arg promote invasion in WM3248 cells by inducing cathepsin L secretion.
Supplementary Fig. S15. Abl/Arg promote lung colonization in a cathepsin-L-dependent manner.
Table S1. qPCR primer sequences
Table S2. ChIP primer sequences.
ACKNOWLEDGEMENTS:
University of Kentucky Markey Cancer Center Shared Resources supported this work: Flow Cytometry, Biospecimen Procurement and Translational Pathology, Biostatistics/Bioinformatics (P30CA177558). We thank Dr. Ren Xu for reading the manuscript, and Dr. Li-Ying Lin for the MSCV-dsRed2 construct.
Funding: This work was supported by NIH grants R01CA116784 and R01CA166499 to R.P, and by a Markey Cancer Foundation Women Strong Award to R.P.
Footnotes
Competing Interests: This work was supported by NIH/NCI grants (R01CA116784 and R01CA166499) to R.P. All other authors declare no potential conflicts of interest.
Data and Materials Availability: Nilotinib was provided by Novartis (Switzerland) via an MTA, and melanoma cell lines, provided by Dr. M. Herlyn (Wistar Institute, Philadelphia, PA), were obtained via an MTA (see Materials and Methods).
REFERENCES:
- 1.Olson OC, Joyce JA, Cysteine cathepsin proteases: regulators of cancer progression and therapeutic response. Nat Rev Cancer 15, 712–729 (2015). [DOI] [PubMed] [Google Scholar]
- 2.Turk V, Stoka V, Vasiljeva O, Renko M, Sun T, Turk B, Turk D, Cysteine cathepsins: from structure, function and regulation to new frontiers. Biochim Biophys Acta 1824, 68–88 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Aggarwal N, Sloane BF, Cathepsin B: multiple roles in cancer. Proteomics Clin Appl 8, 427–437 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Fiore LS, Ganguly S, Sledziona J, Cibull ML, Wang C, Richards DL, Neltner JM, Beach C, McCorkle JR, Kaetzel DM, Plattner R, c-Abl and Arg induce cathepsin-mediated lysosomal degradation of the NM23-H1 metastasis suppressor in invasive cancer. Oncogene 33, 4508–4520 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ruan H, Hao S, Young P, Zhang H, Targeting Cathepsin B for Cancer Therapies. Horiz Cancer Res 56, 23–40 (2015). [PMC free article] [PubMed] [Google Scholar]
- 6.Sudhan DR, Siemann DW, Cathepsin L targeting in cancer treatment. Pharmacol Ther 155, 105–116 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Alonso-Curbelo D, Riveiro-Falkenbach E, Perez-Guijarro E, Cifdaloz M, Karras P, Osterloh L, Megias D, Canon E, Calvo TG, Olmeda D, Gomez-Lopez G, Grana O, Sanchez-Arevalo Lobo VJ, Pisano DG, Wang HW, Ortiz-Romero P, Tormo D, Hoek K, Rodriguez-Peralto JL, Joyce JA, Soengas MS, RAB7 controls melanoma progression by exploiting a lineage-specific wiring of the endolysosomal pathway. Cancer Cell 26, 61–76 (2014). [DOI] [PubMed] [Google Scholar]
- 8.Kos J, Stabuc B, Schweiger A, Krasovec M, Cimerman N, Kopitar-Jerala N, Vrhovec I, Cathepsins BH, and L and their inhibitors stefin A and cystatin C in sera of melanoma patients. Clin Cancer Res 3, 1815–1822 (1997). [PubMed] [Google Scholar]
- 9.Zhang H, Fu T, McGettigan S, Kumar S, Liu S, Speicher D, Schuchter L, Xu X, IL-8 and cathepsin B as melanoma serum biomarkers. Int J Mol Sci 12, 1505–1518 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Greuber EK, Smith-Pearson P, Wang J, Pendergast AM, Role of ABL family kinases in cancer: from leukaemia to solid tumours. Nat Rev Cancer 13, 559–571 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ganguly SS, Plattner R, Activation of Abl family kinases in solid tumors. Genes Cancer 3, 414–425 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ganguly SS, Fiore LS, Sims JT, Friend JW, Srinivasan D, Thacker MA, Cibull ML, Wang C, Novak M, Kaetzel DM, Plattner R, c-Abl and Arg are activated in human primary melanomas, promote melanoma cell invasion via distinct pathways, and drive metastatic progression. Oncogene 31, 1804–1816 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Srinivasan D, Plattner R, Activation of Abl tyrosine kinases promotes invasion of aggressive breast cancer cells. Cancer Res 66, 5648–5655 (2006). [DOI] [PubMed] [Google Scholar]
- 14.Srinivasan D, Sims JT, Plattner R, Aggressive breast cancer cells are dependent on activated Abl kinases for proliferation, anchorage-independent growth and survival. Oncogene 27, 1095–1105 (2008). [DOI] [PubMed] [Google Scholar]
- 15.Hong IK, Byun HJ, Lee J, Jin YJ, Wang SJ, Jeoung DI, Kim YM, Lee H, The tetraspanin CD81 protein increases melanoma cell motility by up-regulating metalloproteinase MT1-MMP expression through the pro-oncogenic Akt-dependent Sp1 activation signaling pathways. J Biol Chem 289, 15691–15704 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sachrajda I, Ratajewski M, Mithramycin A suppresses expression of the human melanoma-associated gene ABCB8. Mol Genet Genomics 285, 57–65 (2011). [DOI] [PubMed] [Google Scholar]
- 17.Kubic JD, Little EC, Lui JW, Iizuka T, Lang D, PAX3 and ETS1 synergistically activate MET expression in melanoma cells. Oncogene 34, 4964–4974 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Spangler B, Kappelmann M, Schittek B, Meierjohann S, Vardimon L, Bosserhoff AK, Kuphal S, ETS-1/RhoC signaling regulates the transcription factor c-Jun in melanoma. Int J Cancer 130, 2801–2811 (2012). [DOI] [PubMed] [Google Scholar]
- 19.Mattia G, Errico MC, Felicetti F, Petrini M, Bottero L, Tomasello L, Romania P, Boe A, Segnalini P, Di Virgilio A, Colombo MP, Care A, Constitutive activation of the ETS-1-miR-222 circuitry in metastatic melanoma. Pigment Cell Melanoma Res 24, 953–965 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ashkenazi S, Ortenberg R, Besser M, Schachter J, Markel G, SOX9 indirectly regulates CEACAM1 expression and immune resistance in melanoma cells. Oncotarget, (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Vizcaino C, Mansilla S, Portugal J, Sp1 transcription factor: A long-standing target in cancer chemotherapy. Pharmacol Ther 152, 111–124 (2015). [DOI] [PubMed] [Google Scholar]
- 22.Yang J, Splittgerber R, Yull FE, Kantrow S, Ayers GD, Karin M, Richmond A, Conditional ablation of Ikkb inhibits melanoma tumor development in mice. J Clin Invest 120, 2563–2574 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Murtas D, Piras F, Minerba L, Ugalde J, Piga M, Maxia C, Perra MT, Sirigu P, Nuclear factor-kappaB expression is predictive of overall survival in patients with cutaneous melanoma. Oncol Lett 1, 633–639 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wani AA, Jafarnejad SM, Zhou J, Li G, Integrin-linked kinase regulates melanoma angiogenesis by activating NF-kappaB/interleukin-6 signaling pathway. Oncogene 30, 2778–2788 (2011). [DOI] [PubMed] [Google Scholar]
- 25.Dittmer J, The role of the transcription factor Ets1 in carcinoma. Semin Cancer Biol 35, 20–38 (2015). [DOI] [PubMed] [Google Scholar]
- 26.Jain A, Tripathi R, Turpin CP, Wang C, Plattner R, Abl kinase regulation by BRAF/ERK and cooperation with Akt in melanoma. Oncogene 36, 4585–4596 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Barila D, Superti-Furga G, An intramolecular SH3-domain interaction regulates c-Abl activity. Nature Genet 18, 280–282 (1998). [DOI] [PubMed] [Google Scholar]
- 28.Wang J, Pendergast AM, The Emerging Role of ABL Kinases in Solid Tumors. Trends Cancer 1, 110–123 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Riker AI, Enkemann SA, Fodstad O, Liu S, Ren S, Morris C, Xi Y, Howell P, Metge B, Samant RS, Shevde LA, Li W, Eschrich S, Daud A, Ju J, Matta J, The gene expression profiles of primary and metastatic melanoma yields a transition point of tumor progression and metastasis. BMC Med Genomics 1, 13 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Talantov D, Mazumder A, Yu JX, Briggs T, Jiang Y, Backus J, Atkins D, Wang Y, Novel genes associated with malignant melanoma but not benign melanocytic lesions. Clin Cancer Res 11, 7234–7242 (2005). [DOI] [PubMed] [Google Scholar]
- 31.Sitabkhan Y, Frankfater A, Differences in the expression of cathepsin B in B16 melanoma metastatic variants depend on transcription factor Sp1. DNA Cell Biol 26, 673–682 (2007). [DOI] [PubMed] [Google Scholar]
- 32.Jean D, Guillaume N, Frade R, Characterization of human cathepsin L promoter and identification of binding sites for NF-Y, Sp1 and Sp3 that are essential for its activity. Biochem J 361, 173–184 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yan S, Berquin IM, Troen BR, Sloane BF, Transcription of human cathepsin B is mediated by Sp1 and Ets family factors in glioma. DNA Cell Biol 19, 79–91 (2000). [DOI] [PubMed] [Google Scholar]
- 34.Bien S, Ritter CA, Gratz M, Sperker B, Sonnemann J, Beck JF, Kroemer HK, Nuclear factor-kappaB mediates up-regulation of cathepsin B by doxorubicin in tumor cells. Mol Pharmacol 65, 1092–1102 (2004). [DOI] [PubMed] [Google Scholar]
- 35.Tanida I, Waguri S, Measurement of autophagy in cells and tissues. Methods Mol Biol 648, 193–214 (2010). [DOI] [PubMed] [Google Scholar]
- 36.Manley PW, Zimmermann J, in Polypharmacology in Drug Discovery. (John Wiley & Sons, Inc., 2012), pp. 409–421. [Google Scholar]
- 37.Petricevic SJ, Pavlovic A, Capkun V, Becic K, Durdov MG, Cathepsin K expression in melanoma is associated with metastases. Histol Histopathol, 11833 (2016). [DOI] [PubMed] [Google Scholar]
- 38.Small DM, Burden RE, Jaworski J, Hegarty SM, Spence S, Burrows JF, McFarlane C, Kissenpfennig A, McCarthy HO, Johnston JA, Walker B, Scott CJ, Cathepsin S from both tumor and tumor-associated cells promote cancer growth and neovascularization. Int J Cancer 133, 2102–2112 (2013). [DOI] [PubMed] [Google Scholar]
- 39.Tsai JY, Lee MJ, Chang MD, Wang HC, Lin CC, Huang H, Effects of novel human cathepsin S inhibitors on cell migration in human cancer cells. J Enzyme Inhib Med Chem 29, 538–546 (2014). [DOI] [PubMed] [Google Scholar]
- 40.Rao Q, Wang Y, Xia QY, Shi SS, Shen Q, Tu P, Shi QL, Zhou XJ, Wu B, Cathepsin K in the immunohistochemical diagnosis of melanocytic lesions. Int J Clin Exp Pathol 7, 1132–1139 (2014). [PMC free article] [PubMed] [Google Scholar]
- 41.Rumpler G, Becker B, Hafner C, McClelland M, Stolz W, Landthaler M, Schmitt R, Bosserhoff A, Vogt T, Identification of differentially expressed genes in models of melanoma progression by cDNA array analysis: SPARC, MIF and a novel cathepsin protease characterize aggressive phenotypes. Exp Dermatol 12, 761–771 (2003). [DOI] [PubMed] [Google Scholar]
- 42.Krehan A, Ansuini H, Bocher O, Grein S, Wirkner U, Pyerin W, Transcription factors ets1, NF-kappa B, and Sp1 are major determinants of the promoter activity of the human protein kinase CK2alpha gene. J Biol Chem 275, 18327–18336 (2000). [DOI] [PubMed] [Google Scholar]
- 43.Gonfloni S, Defying c-Abl signaling circuits through small allosteric compounds. Front Genet 5, 392 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hoesel B, Schmid JA, The complexity of NF-kappaB signaling in inflammation and cancer. Mol Cancer 12, 86 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Granger E, McNee G, Allan V, Woodman P, The role of the cytoskeleton and molecular motors in endosomal dynamics. Semin Cell Dev Biol 31, 20–29 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Huotari J, Helenius A, Endosome maturation. EMBO J 30, 3481–3500 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Caramel J, Papadogeorgakis E, Hill L, Browne GJ, Richard G, Wierinckx A, Saldanha G, Osborne J, Hutchinson P, Tse G, Lachuer J, Puisieux A, Pringle JH, Ansieau S, Tulchinsky E, A switch in the expression of embryonic EMT-inducers drives the development of malignant melanoma. Cancer Cell 24, 466–480 (2013). [DOI] [PubMed] [Google Scholar]
- 48.Roesch A, Paschen A, Landsberg J, Helfrich I, Becker JC, Schadendorf D, Phenotypic tumour cell plasticity as a resistance mechanism and therapeutic target in melanoma. Eur J Cancer 59, 109–112 (2016). [DOI] [PubMed] [Google Scholar]
- 49.Plattner R, Kadlec L, DeMali KA, Kazlauskas A, Pendergast AM, c-Abl is activated by growth factors and Src family kinases and has a role in the cellular response to PDGF. Genes Dev 13, 2400–2411 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Li X, Ma Q, Wang J, Liu X, Yang Y, Zhao H, Wang Y, Jin Y, Zeng J, Li J, Song L, Li P, Qian X, Cao C, c-Abl and Arg tyrosine kinases regulate lysosomal degradation of the oncoprotein Galectin-3. Cell Death Differ 17, 1277–1287 (2010). [DOI] [PubMed] [Google Scholar]
- 51.Plattner R, Koleske AJ, Kazlauskas A, Pendergast AM, Bidirectional Signaling Links the Abelson Kinases to the Platelet-Derived Growth Factor Receptor. Mol Cell Biol 24, 2573–2583 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Echarri A, Pendergast AM, Activated c-Abl is degraded by the ubiquitin-dependent proteasome pathway. Curr Biol 11, 1759–1765 (2001). [DOI] [PubMed] [Google Scholar]
- 53.Haring M, Offermann S, Danker T, Horst I, Peterhansel C, Stam M, Chromatin immunoprecipitation: optimization, quantitative analysis and data normalization. Plant Methods 3, 11 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Mitra S, Beach C, Feng GS, Plattner R, SHP-2 is a novel target of Abl kinases during cell proliferation. J Cell Sci 121, 3335–3346 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Sims JT, Ganguly SS, Bennett H, Friend WJ, J. T, Plattner R, Imatinib reverses doxorubicin resistance by affecting activation of STAT3-dependent NF-κB and HSP27/p38/AKT pathways and by inhibiting ABCB1. PLoS One 8, e55509 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Fig. S1. Abl and Arg are overexpressed and activated in a large panel of melanoma cell lines.
Supplementary Fig. S2. Effects of silencing or inhibiting Abl/Arg on intracellular and extracellular cathepsins.
Supplementary Fig. S3. Abl/Arg drive cathepsin abundance.
Supplementary Fig. S4. Effects of silencing or inhibiting Abl/Arg on pro-, intermediate and mature (double-chain) cathepsin forms.
Supplementary Fig. S5. Abl and Arg promote cathepsin mRNA expression.
Supplementary Fig. S6. Abl/Arg and cathepsin mRNAs are correlated in primary melanomas.
Supplementary Fig. S7. Abl/Arg alter Ets1 and Sp1 proforms, mRNA and promoter activities.
Supplementary Fig. S8. Abl/Arg alter nuclear localization of p65, Ets1, and Sp1.
Supplementary Fig. S9. Abl/Arg impact the DNA binding capacity of Ets1 and Sp1 on cathepsin promoters.
Supplementary Fig. S10. Abl/Arg and Sp1/Ets1 mRNAs are correlated in primary melanomas.
Supplementary Fig. S11. Ets1, Sp1 and p65 contribute to cathepsin abundance.
Supplementary Fig. S12. Model for Abl/Arg regulation of cathepsin expression and secretion.
Supplementary Fig. S13. Abl/Arg regulate cathepsin expression by activating NF-κB (p65).
Supplementary Fig. S14. Abl/Arg promote invasion in WM3248 cells by inducing cathepsin L secretion.
Supplementary Fig. S15. Abl/Arg promote lung colonization in a cathepsin-L-dependent manner.
Table S1. qPCR primer sequences
Table S2. ChIP primer sequences.